TRAF6 is essential for RANK-mediated NF-κB activation and is involved in the development of several types of cells. Kanaya et al. demonstrate that RANK–TRAF6-mediated NF-κB is essential for the development of M cells and FAE.
Abstract
M cells are located in the follicle-associated epithelium (FAE) that covers Peyer’s patches (PPs) and are responsible for the uptake of intestinal antigens. The differentiation of M cells is initiated by receptor activator of NF-κB. However, the intracellular pathways involved in M cell differentiation are still elusive. In this study, we demonstrate that the NF-κB pathway activated by RANK is essential for M cell differentiation using in vitro organoid culture. Overexpression of NF-κB transcription factors enhances the expression of M cell–associated molecules but is not sufficient to complete M cell differentiation. Furthermore, we evaluated the requirement for tumor necrosis factor receptor–associated factor 6 (TRAF6). Conditional deletion of TRAF6 in the intestinal epithelium causes a complete loss of M cells in PPs, resulting in impaired antigen uptake into PPs. In addition, the expression of FAE-associated genes is almost silenced in TRAF6-deficient mice. This study thus demonstrates the crucial role of TRAF6-mediated NF-κB signaling in the development of M cells and FAE.
Introduction
The mucosal surface of the intestinal tract is exposed to variety of foreign antigens, including harmful pathogens for host animals. To avoid the infectious risks posed by these pathogens, the mucosal tissue uses multiple layers of barrier mechanisms. For instance, the tightly integrated intestinal epithelial cell (IEC) monolayer physically blocks the invasion of macromolecules, including bacteria—the mucus layer generated by goblet cells physicochemically impairs the attachment of pathogens to the epithelial cell surface—and antimicrobial proteins mainly produced by Paneth cells sterilize the mucosal surface (Gallo and Hooper, 2012; Zhang et al., 2015). Recently, it has also been reported that epithelial fucosylation contributes to the protection against intestinal pathogens (Goto et al., 2014). These epithelial barriers are indispensable for the maintenance of intestinal homeostasis.
IECs also contribute to mucosal immune responses. The follicle-associated epithelium (FAE) is composed of specialized IECs that cover the luminal side of the lymphoid follicles of gut-associated lymphoid tissue (GALT; Neutra et al., 2001), such as Peyer’s patches (PPs), colonic patches, cecal patches, and isolated lymphoid follicles distributed throughout the intestine. A principal role of the FAE is the uptake and transport of luminal antigens into GALT, and this task is thought to be single-handedly accomplished by microfold or membranous cells (M cells) located in the FAE (Kraehenbuhl and Neutra, 2003). M cells possess a high phagocytic and transcytotic capacity, which is responsible for the rapid transport of bacterial antigens to antigen-presenting cells in GALT (Neutra et al., 2001). We previously demonstrated that this M cell–mediated antigen transport largely contributes the antigen-specific immune responses, such as the activation of T cells and the production of IgA from plasma cells (Hase et al., 2009; Kanaya et al., 2012; Rios et al., 2015).
Despite the important role of M cells in mucosal immune responses, the mechanisms for the development of M cells are not well characterized because of their rarity in the intestine (Kanaya and Ohno, 2014). M cells are a subset of IECs derived from Lgr5-positive epithelial intestinal stem cells (ISCs) located at the bottom of crypts (de Lau et al., 2012). M cells are restricted to FAE that is closely associated with GALT stromal cells and immune cells, implying that these cells influence M cell differentiation from ISCs. Indeed, receptor activator of NF-κB (RANK) ligand (RANKL) produced by stromal cells underneath the FAE was shown to critically regulate M cell differentiation (Knoop et al., 2009). Notably, exogenous RANKL administration elicits ectopic M cell differentiation in villous epithelium (VE) normally devoid of M cells (Knoop et al., 2009). Taking advantage of this phenomenon, we screened the profile of RANKL-responsive genes in IECs to identify a transcription factor essential for M cell differentiation, the Ets-family transcription factor Spi-B (Kanaya et al., 2012). Our study also revealed that Spi-B is indispensable for the expression of some M cell–associated molecules but not sufficient for the expression of other M cell–specific molecules, suggesting that additional factors are required for M cell differentiation (de Lau et al., 2012; Kanaya et al., 2012; Sato et al., 2013).
The ligation of RANK is known to activate both canonical and noncanonical NF-κB signaling pathways (Akiyama et al., 2008). Canonical NF-κB signaling is transient and rapid and is involved in inflammatory responses, whereas noncanonical NF-κB signaling is slow and persistent and contributes to cellular differentiation. The RANKL–RANK-mediated noncanonical NF-κB pathway activates the NF-κB transcription factor RelB, which is required for the development of medullary thymic epithelial cells (mTECs; Akiyama et al., 2008) and osteoclasts (Vaira et al., 2008). Likewise, it has recently been reported that RelB is essential for the initiation of RANKL-induced ectopic M cell differentiation (Kimura et al., 2015). However, the molecular basis of RANKL–RANK-mediated M cell differentiation has not been established.
Here, we evaluated the significance of RANK-induced NF-κB in M cell differentiation using an in vitro M cell differentiation system established using cultured intestinal organoids. Inactivation of NF-κB by the deletion of RelB and RelA abolishes lymphoid tissues, including GALT (Weih and Caamaño, 2003), resulting in a lack of FAE and M cells; as a result, there is a limitation in in vivo analyses to determine the significance of the NF-κB pathway in the development of M cells. In organoids, M cell differentiation is activated by the ligation of RANK on the epithelium with exogenous RANKL, which enables us to evaluate the significance of epithelial-intrinsic activation of NF-κB in vitro independently of any defects in GALT architecture in the mice from which the organoids are derived. Consequently, we demonstrate here that M cell differentiation requires the activation of both canonical and noncanonical NF-κB. We also examine the role of TNF receptor (TNFR)–associated factor 6 (TRAF6) in M cell differentiation. A conditional deletion of TRAF6 in IECs causes a complete loss of M cell–lineage commitment, resulting in impairments in antigen uptake into PPs and antigen-specific IgA responses. We further show that TRAF6 is also critical for the expression of FAE-associated molecules, which provides novel insights into the molecular basis for the development of both M cells and FAE enterocytes.
Results
Epithelial-intrinsic noncanonical NF-κB is essential to evoke M cell differentiation
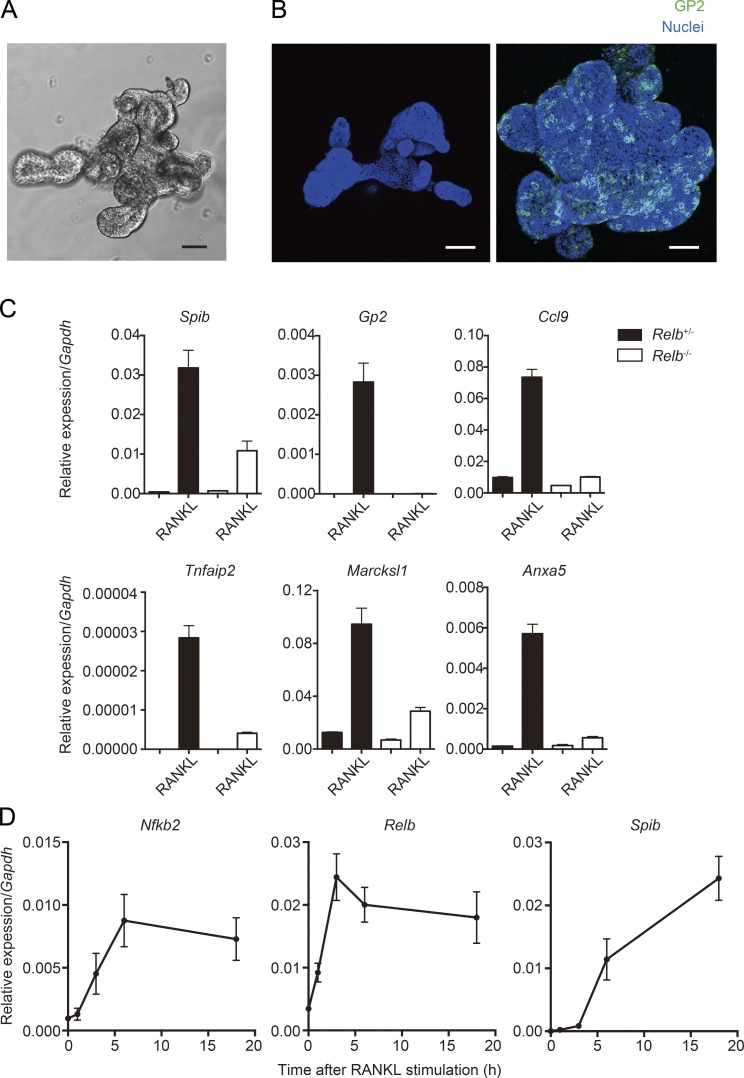
We and others identified Spi-B as a crucial transcription factor for M cell differentiation downstream of RANKL–RANK signaling (de Lau et al., 2012; Kanaya et al., 2012; Sato et al., 2013). In addition, our recent study has indicated that noncanonical NF-κB transcription factor RelB is essential for RANKL-induced ectopic M cell differentiation (Kimura et al., 2015). To further establish the molecular basis of M cell differentiation, we took advantage of an in vitro M cell differentiation model. When intestinal crypts harboring ISCs are cultured under defined conditions, they grow and form an organoid structure (Fig. 1 A; Sato et al., 2009). ISCs are maintained in these organoids, and M cells can be induced in organoids upon addition of RANKL (Fig. 1 B; de Lau et al., 2012; Rios et al., 2015). This in vitro M cell differentiation is a suitable model for determining the epithelial-intrinsic factors required for M cell differentiation because organoids can exclude the involvement of factors derived from non-IECs such as stromal and immune cells.
Figure 1.
Epithelial-intrinsic noncanonical NF-κB is essential to evoke M cell differentiation in organoids. (A) Small intestinal crypts were isolated from WT mice and embedded into Matrigel. After 5-d culture with the optimal crypt growth medium, crypts grew and formed a branching organoid structure. Bar, 50 µm. (B) Organoids were stimulated with RANKL for 3 d and fixed with 4% paraformaldehyde. Organoids were then immunostained with anti–GP2 antibody. Green shows GP2, and blue shows nuclei. Bars, 100 µm. (C) Organoids generated from Relb+/−and Relb−/− (top) mice were stimulated with RANKL for 3 d. The expression of M cell markers was examined by quantitative PCR analysis (normalized with Gapdh, n = 3, mean ± SEM). (D) Organoids from WT mice were stimulated with RANKL for indicated time course (1, 3, 6, and 18 h), and the expressions of Spib, Relb, and Nfkb2 were analyzed by quantitative PCR (normalized with Gapdh, n = 3, mean ± SD). Data are representative of two independent experiments.
The RelB-deficient (Relb−/−) mouse is well known to completely lack activation of the noncanonical NF-κB pathway (Sun, 2012). Examining M cell formation in Relb−/− mice is one experimental approach to assess the importance of the noncanonical NF-κB pathway in M cell differentiation. However, these mice completely lack PPs and, therefore, FAE because of the requirement of the noncanonical NF-κB in lymphoid tissue generation (Miyawaki et al., 1994; Yilmaz et al., 2003), precluding the examination of M cell differentiation in vivo using these mice. To overcome this problem, we generated organoids from the small intestine of Relb−/− mice and stimulated with RANKL for 3 d to evaluate their potential to differentiate into M cells. The expression of M cell marker genes was prominently induced in the organoids from Relb+/− mice as positive controls. On the other hand, Gp2 was not induced by RANKL in the organoids from Relb−/− mice, and induction of other M cell markers, including Spib, was drastically decreased compared with the controls (Fig. 1 C).
Noncanonical NF-κB directly targets Spib expression
We further corroborated the causal relationship between noncanonical NF-κB and Spi-B. Organoids generated from WT mice were stimulated with RANKL for relatively short durations of time, and expression of Spib and noncanonical NF-κB–associated genes, Relb and Nfkb2, was examined. Stimulation for 3 h with RANKL was sufficient to induce the expression of Relb and Nfkb2, whereas Spib required 6 h to be induced (Fig. 1 D). Taken together, these observations confirmed that epithelial-intrinsic noncanonical NF-κB is required for M cell differentiation downstream of RANK and upstream of Spi-B.
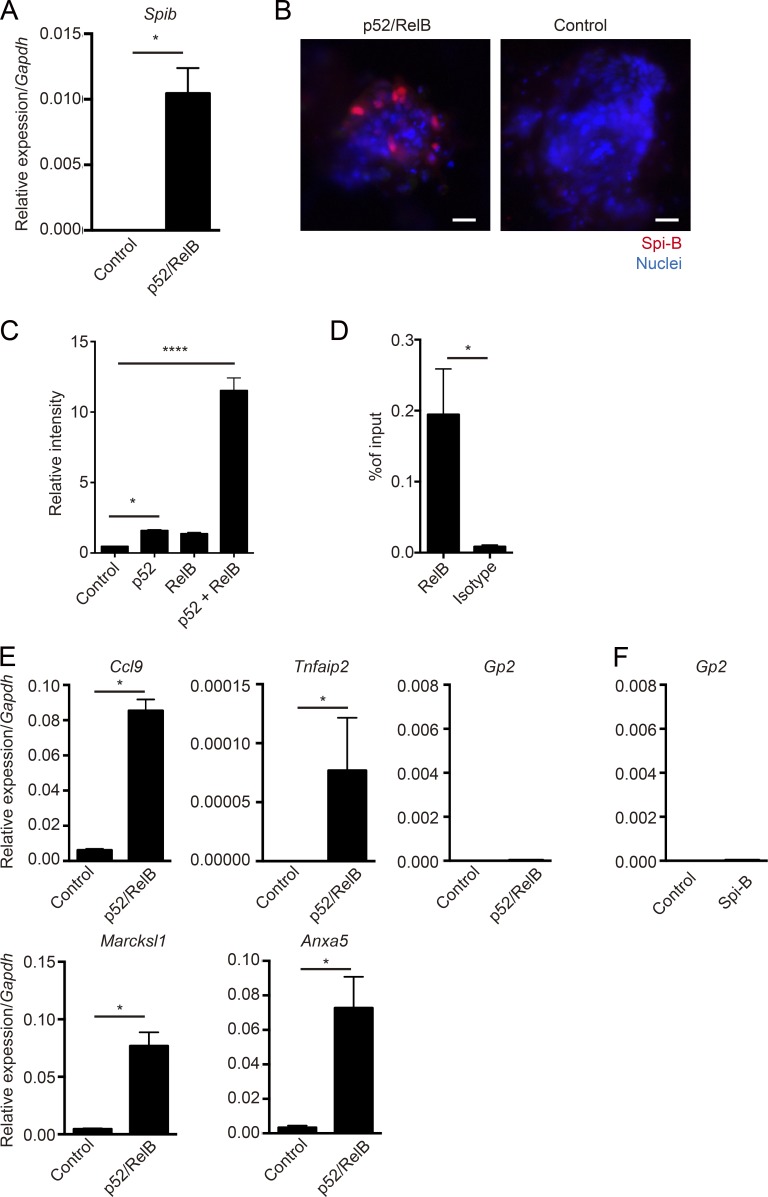
We next assessed whether noncanonical NF-κB is directly involved in induction of Spib expression. Organoids were dissociated and treated with separate lentiviruses coding p52 and RelB, resulting in a significant increase in Spib transcripts and protein in organoids (Fig. 2, A and B). We further determined whether noncanonical NF-κB targeted the Spib promoter using a luciferase assay. Overexpression of p52-RelB prominently enhanced Spib promoter activity in a mouse IEC line (Fig. 2 C). In addition, chromatin immunoprecipitation (ChIP) in RANKL-stimulated organoids revealed the specific binding of RelB to the Spib promoter region (Fig. 2 D). These data suggest that the noncanonical NF-κB transcription factor RelB directly binds to the Spib promoter to induce Spib expression.
Figure 2.
Noncanonical NF-κB directly targets Spib but is not sufficient to induce Gp2 expression. (A) Organoids from WT mice were dissociated and exposed to lentivirus encoding p52 and RelB for 3 d, and the expression of Spib was analyzed by quantitative PCR (normalized with Gapdh, mean ± SD). *, P < 0.05 (Mann-Whitney U test, n = 4). (B) Organoids exposed to lentivirus encoding p52 and RelB were stained with anti–Spi-B antibody. Red shows Spi-B, and blue shows nuclei. Data are representative of three independent experiments. Bars, 20 µm. (C) Spib promoter containing pGL3-Basic reporter vector and pcDNA3-expression vector containing p52, RelB, or a combination of p52 and RelB was overexpressed in mICcl2 cells. After overnight culture, cell lysates were harvested, and the activity of luciferase was examined. Results show the fold-change against the control group (without expression vector; n = 4, mean ± SD; *, P < 0.05; ****, P < 0.0001, one-way ANOVA with Bonferroni’s post-test). (D) Organoids were stimulated for 6 h with RANKL, and cross-linked chromatin was immunoprecipitated with anti–RelB antibody. Immunoprecipitated Spib promoter region was quantified by quantitative PCR (n = 4, mean ± SEM; *, P < 0.05, Mann-Whitney U test). (E and F) Organoids from WT mice were dissociated and exposed to lentivirus encoding p52 and RelB (E) or Spi-B (F) for 3 d, and the expression of M cell markers were analyzed by quantitative PCR (normalized with Gapdh, n = 4, mean ± SD; *, P < 0.05, Mann-Whitney U test). Data are representative of four independent experiments.
Noncanonical NF-κB is not sufficient for M cell differentiation
On learning that the noncanonical NF-κB pathway is essential for M cell differentiation, we next asked whether this pathway is sufficient for M cell differentiation. To this end, we examined the expression of M cell–associated genes upon overexpression of p52-RelB. The significant increase in Spib transcripts under these conditions suggested that Spi-B–dependent M cell markers could also be induced. Indeed, Ccl9 and Tnfaip2 were significantly up-regulated by the transduction of p52-RelB (Fig. 2 E). We also examined the expression of Marcksl1 and Anxa5, which are expressed in the M cell lineage independently of Spi-B (Kanaya et al., 2012; Sato et al., 2013), and found that both genes were up-regulated by p52-RelB (Fig. 2 E). On the other hand, the expression of Gp2, whose expression also requires the presence of Spi-B, was not induced by the overexpression of p52-RelB despite the induction of Spi-B expression (Fig. 2 E). Indeed, overexpression of Spi-B itself in organoids did not induce Gp2 expression (Fig. 2 F). This is consistent with a previous study, which suggests the insufficiency of Spi-B to induce Gp2 (de Lau et al., 2012). These results suggest that noncanonical NF-κB is not alone sufficient for full M cell differentiation, especially as assessed by expression of Gp2, although it is required for early steps, including Spib induction.
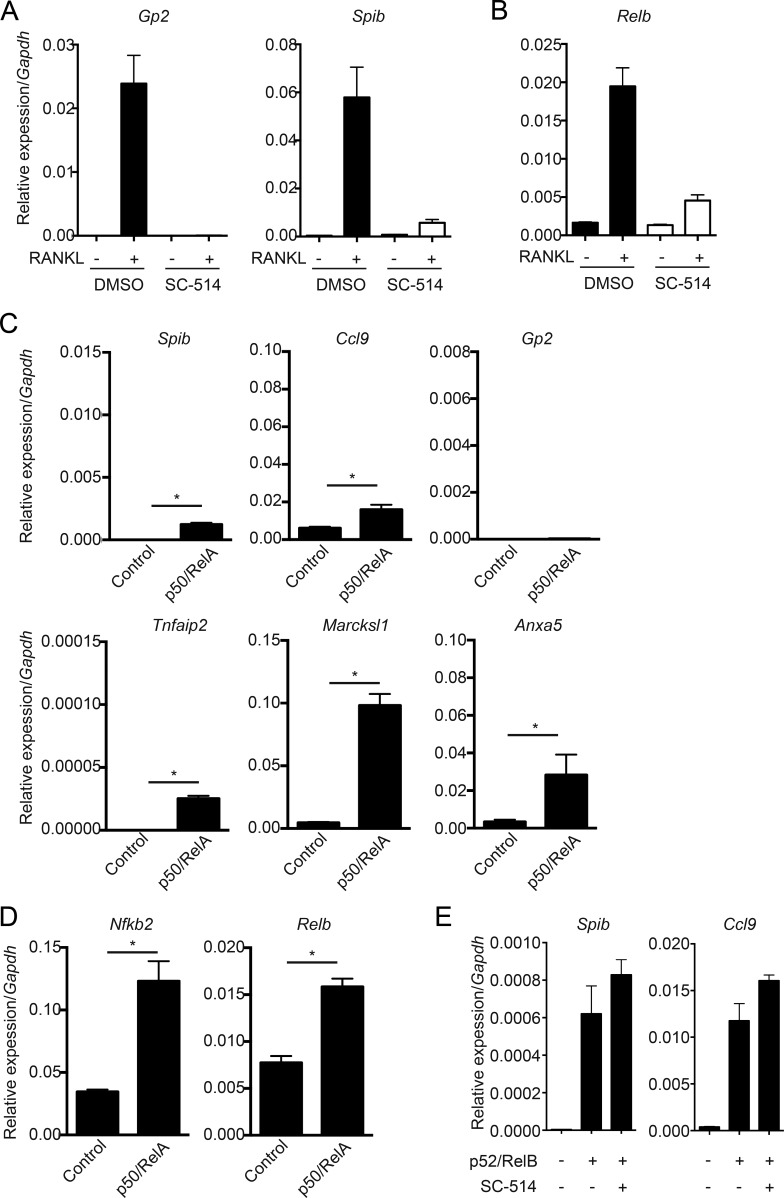
Canonical NF-κB is also required for RANKL–RANK-induced M cell differentiation
RANKL–RANK is known to activate canonical as well as noncanonical NF-κB pathways for the development of mTECs (Akiyama et al., 2008). We therefore examined the importance of canonical NF-κB in M cell differentiation. To verify this, SC-514, a specific inhibitor of IκB kinase-β (Kishore et al., 2003), was used. Organoids generated from WT mice were stimulated with RANKL in the presence or absence of SC-514. SC-514 silenced the expression of Gp2 and Spib, indicating that canonical NF-κB is also required for M cell differentiation (Fig. 3 A). We also performed overexpression of p50-RelA to assess the contribution of canonical NF-κB in the expression of M cell markers. The overexpression of p50-RelA significantly increased the expression of M cell markers, with the exception of Gp2; however, the efficiency was much lower than that induced when p52-RelB was transduced (Fig. 3 C). In contrast to the other M cell markers examined, Marcksl1 was strongly induced by p50-RelA, suggesting that canonical NF-κB largely contributes to the expression of Marcksl1 but less to that of other M cell markers. We next examined a direct linkage between canonical and noncanonical NF-κB, because a previous study demonstrated that canonical NF-κB directly targets Relb transcription (Bren et al., 2001). The administration of SC-514 drastically suppressed RANKL-induced Relb induction (Fig. 3 B). Consistently, the overexpression of p50-RelA significantly up-regulated the transcription of Relb and Nfkb2 (Fig. 3 D). Furthermore, SC-514 could not suppress the induction of Spib and Ccl9 in the organoids transduced with p52-RelB (Fig. 3 E), suggesting that the noncanonical NF-κB pathway can induce M cell markers independently of the canonical pathway. Collectively, these results demonstrated that canonical NF-κB plays a crucial role in M cell differentiation upstream of noncanonical NF-κB but independently enhances the expression of Marcksl1.
Figure 3.
Blocking canonical NF-κB impairs RANKL-induced M cell differentiation in organoids. (A) Organoids from WT mice were stimulated with RANKL for 3 d in the absence or presence of 125 µM SC-514. Expression of Gp2 and Spib was analyzed by quantitative PCR (normalized with Gapdh, n = 3, mean ± SD). (B) Relb expression was analyzed in almost the same experiments as A with a slight difference (culture time was 18 h). (C and D) Organoids from WT mice were dissociated and exposed to lentivirus encoding p50 and RelA for 3 d, and the expression of M cell markers (C) and noncanonical NF-κB genes was analyzed by quantitative PCR (normalized with Gapdh, n = 4, mean ± SD; *, P < 0.05, Mann-Whitney U test). (E) Organoids from WT mice were dissociated and exposed to lentivirus encoding p52 and RelB in the presence of SC-514 for 3 d, and the expression of Spib and Ccl9 was analyzed by quantitative PCR. Data are representative of two independent experiments.
TRAF6 is essential for M cell lineage commitment
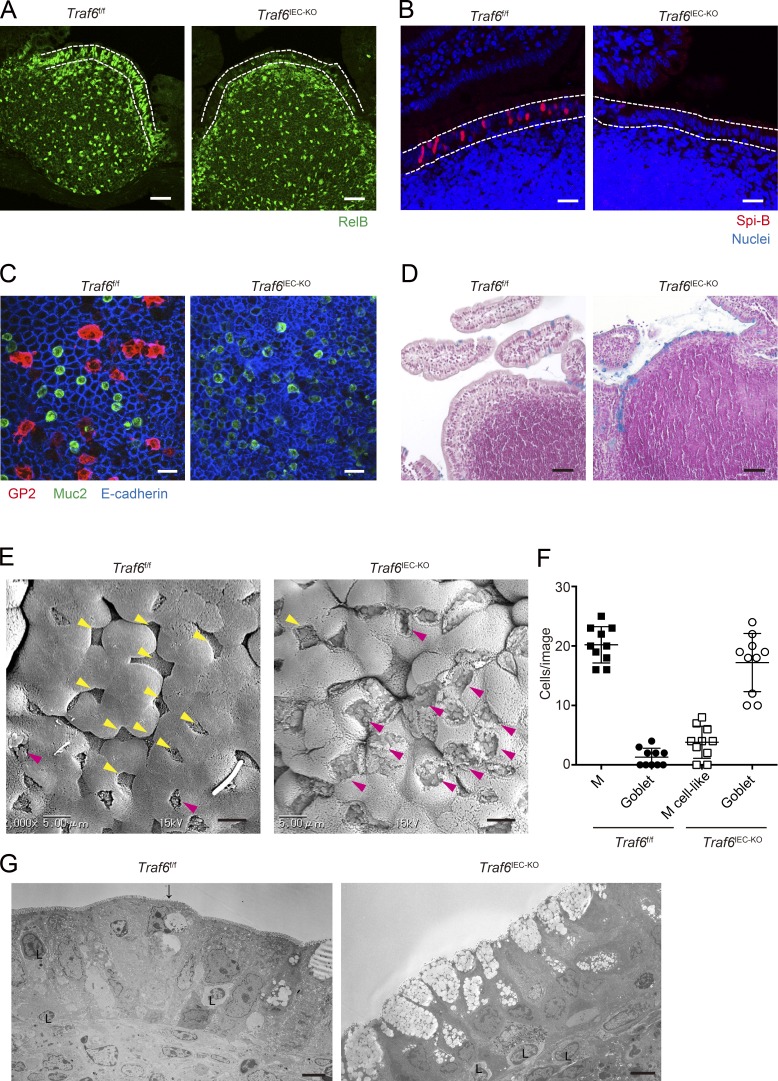
To further evaluate the importance of canonical NF-κB in M cell differentiation in vivo, we focused on a regulatory molecule of RANK-mediated canonical NF-κB activation. TRAFs are recruited to the cytoplasmic domain of TNFRs upon ligand stimulation. Among them, TRAF6 is known to play an essential role in the RANK-mediated canonical NF-κB pathway (Walsh and Choi, 2014). In addition, a previous study demonstrated that TRAF6 has a critical role in the expression of RelB in mTECs (Akiyama et al., 2005). This strongly suggests that TRAF6 has a pivotal role in M cell differentiation upstream of canonical and noncanonical NF-κB. To examine the requirement of TRAF6 in M cell differentiation, we used TRAF6-deficient mice. Because systemic TRAF6 deficiency causes early mortality (17–19 d) in mice (Akiyama et al., 2005), we used mice harboring LoxP-flanked Traf6 alleles (Traf6f/f) and crossed these mice with Villin-Cre transgenic mice to obtain mice lacking TRAF6 specifically in IECs (Traf6IEC-KO mice; Fig. S1 A). In the presence of TRAF6, RelB is predominantly observed in the nuclei of PP FAE (Fig. 4 A), consistent with a previous study (Yilmaz et al., 2003). In contrast, nuclear RelB was drastically decreased in PP FAE of Traf6IEC-KO mice (Fig. 4 A), indicating that the activation of noncanonical NF-κB is severely impaired in IECs in the absence of TRAF6. Consistent with the severe reduction of nuclear RelB, expression of the M cell markers GP2, Spi-B, Marcksl1, and Annexin A5 was silenced in PP FAE of Traf6IEC-KO mice (Fig. 4, B and C; and Fig. S1 B). M cells develop not only in PP FAE but also throughout the GALT, including colonic patches. We therefore evaluated whether TRAF6 deficiency also affects M cell development in colonic patches. GP2+ M cells were identified in colonic patches of Traf6f/f mice, but not in those of Traf6IEC-KO mice (Fig. S1 C), indicating that TRAF6 is essential for M cell differentiation throughout the GALT, including the large intestine.
Figure 4.
TRAF6 deficiency in IECs causes a loss of M cells in PP FAE. (A) PPs of Traf6f/f and Traf6IEC-KO mice were immunostained with anti-RelB antibody (green). Dotted lines indicate the FAE. Bars, 40 µm. (B) PPs of Traf6f/f and Traf6IEC-KO mice were immunostained with anti–Spi-B antibody. Red shows Spi-B, and blue shows nuclei. Dotted lines indicate the FAE. Bars, 20 µm. (C) Isolated FAE was immunostained with anti–GP2 (red), anti–Muc2 (green), and anti–E-cadherin (blue) antibodies. Bars, 50 µm. (D) Sections of PPs from Traf6f/f and Traf6IEC-KO mice were stained with alcian blue to visualize mucus-secreting cells. Bars, 50 µm. (E) PP FAE was examined by scanning electron microscopy. Bars, 5 µm. Yellow arrowheads indicate M cells in Traf6f/f or M cell–like cells in Traf6IEC-KO mice with sparse microvilli compared with surrounding enterocytes. Magenta arrowheads indicate goblet cells harboring large granules. (F) The number of M cells, M cell–like cells and goblet cells were quantified. (G) PP FAE was examined by TEM. The arrow indicates an M cell. L, lymphocyte. Bars, 5 µm. Data are representative of two independent experiments.
We next evaluated the loss of M cells in PP FAE of Traf6IEC-KO mice using scanning electron microscopy. Cells featuring sparse and irregular microvilli typical of normal M cells were only rarely detected in Traf6IEC-KO mice (Fig. 4 E, yellow arrowheads), indicating that morphologically mature M cells are mostly absent in Traf6IEC-KO mice. We could identify cells morphologically similar to M cells in terms of their surface structure (M cell–like cells) in Traf6IEC-KO mice (Fig. 4 E, yellow arrowheads); however, they do not seem to be authentic M cells because they lack the expression of M cell markers in immunofluorescence staining (Fig. 4, B and C; and Fig. S1 B). We also noticed that the number of goblet cells harboring large mucus granules was drastically increased in PP FAE of Traf6IEC-KO mice (Fig. 4, E and F, magenta arrowheads). To precisely evaluate the increase in goblet cells in Traf6IEC-KO mice, we imaged the PP FAE of Traf6IEC-KO mice using alcian blue staining and transmission electron microscopy (TEM) and confirmed that goblet cells are markedly increased in PP FAE of Traf6IEC-KO mice (Fig. 4, D and G). However, the hyperplasia of goblet cells was not observed in the FAE of SpibIEC-KO mice, which completely lack mature M cells (Fig. S2). These suggest that TRAF6 deficiency not only causes the loss of M cells but also affects the differentiation of other enterocyte subsets in the FAE.
RANK–TRAF6 is responsible for the expression of FAE-associated molecules
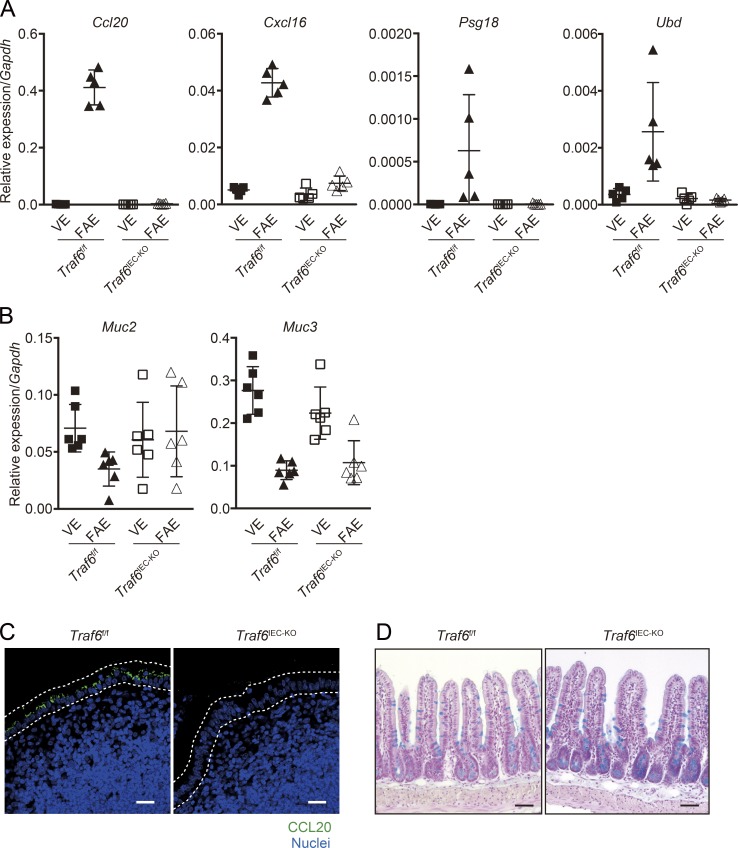
Based on our observation that goblet cells increased in the FAE of Traf6IEC-KO mice, we examined the effect of TRAF6 deficiency on the properties of the FAE. It is known that the features of FAE enterocytes are distinct from those of VE enterocytes. For instance, FAE enterocytes constitutively express some chemokines, such as CCL20 and CXCL16, at substantially higher levels than VE enterocytes (Rumbo et al., 2004; Hase et al., 2006). In addition, we have previously reported additional genes preferentially expressed in the FAE, but not in villi (Hase et al., 2005; Kawano et al., 2007). We found that all FAE-associated genes examined (i.e., Ccl20, Cxcl16, and Psg18 [pregnancy-specific glycoprotein 18; Kawano et al., 2007] and Ubd [ubiquitin D; Hase et al., 2005]) were almost totally silenced in Traf6IEC-KO mice (Fig. 5 A). We also observed the complete loss of CCL20 protein in FAE of Traf6IEC-KO mice (Fig. 5 C). These results suggest that the RANK–TRAF6-mediated NF-κB signaling is required for the expression of FAE-associated molecules. To further verify the altered properties of FAE in the absence of TRAF6, we examined the expression of mucin genes, which are normally suppressed in FAE compared with VE (Jinnohara et al., 2017). The expression of Muc3 was suppressed in the FAE of Traf6IEC-KO mice as well as in that of Traf6f/f mice. This suggests that the suppression of Muc3 is independent of RANK–TRAF6–mediated NF-κB signaling. On the other hand, Muc2 exhibited a slight increase in the FAE of Traf6IEC-KO mice (Fig. 5 B), which coincided with the observation that goblet cells were increased in the FAE of Traf6IEC-KO mice (Fig. 4, D–G). This raised the possibility that TRAF6 might be involved in the developmental regulation of goblet cells; however, the number of goblet cells in the VE appeared comparable between Traf6f/f and Traf6IEC-KO mice (Fig. 5 D). It is therefore likely that the increase in the number of goblet cells in the FAE of Traf6IEC-KO mice is due to secondary effects caused by perturbations in FAE differentiation.
Figure 5.
TRAF6 deficiency abolishes the expression of FAE-associated genes. (A) The expression of FAE-associated genes (Ccl20, Cxcl16, Psg18, and Ubd) in Traf6f/f and Traf6IEC-KO mice was analyzed (normalized with Gapdh, mean ± SD; n = 5). Solid symbols indicate the data of Traf6f/f mice, whereas open ones indicate those of Traf6IEC-KO mice. (B) In the same sample sets as A, the expression of mucus-associated genes was examined by quantitative PCR. (C) PPs of Traf6f/f and Traf6IEC-KO mice were immunostained with anti–CCL20 antibody (green). Nuclei were counterstained with DAPI (blue). Dotted lines indicate the FAE. Bars, 20 µm. (D) Sections of the small intestine from Traf6f/f and Traf6IEC-KO mice were stained with alcian blue to visualize mucus-secreting cells. Bars, 50 µm. Data are representative of two independent experiments.
We also evaluated the significance of RANK in the expression of FAE-associated genes using organoids. Administration of RANKL enhanced the expression of all of FAE-associated genes examined in organoids (Fig. S3), which supports our finding that RANK–TRAF6 is important for the expression of FAE-associated molecules. On the other hand, a previous study indicates that LTβR signaling contributes to the development of the FAE (Rumbo et al., 2004). We therefore assessed the contribution of LTβR in the expression of FAE-associated genes in organoids. Indeed, LTα1β2, the ligand of LTβR, also increased the expression of FAE-associated genes in organoids, although the efficiency was much lower than RANKL (Fig. S3). Together, these results suggest that RANK–TRAF6 is predominantly responsible for the expression of FAE-associated molecules.
The uptake of particulate antigens is impaired in Traf6IEC-KO mice
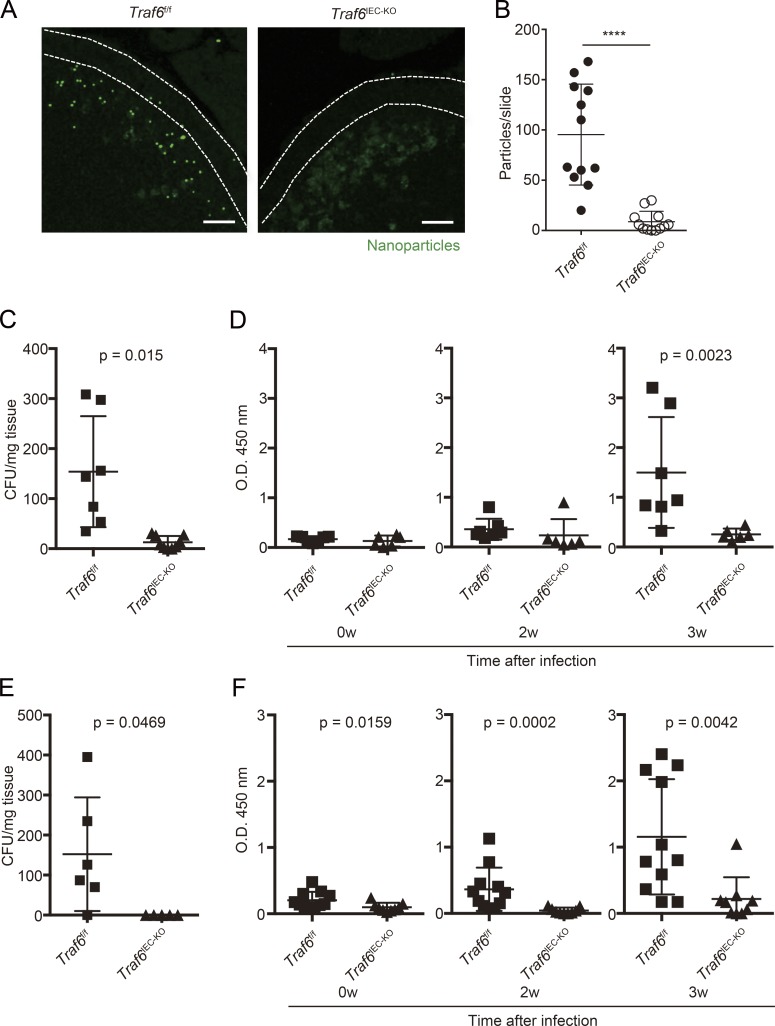
The loss of M cells in Traf6IEC-KO mice raises the possibility that uptake of nanoparticles and bacteria into PPs is impaired in Traf6IEC-KO mice, because M cells are critical for the uptake of such luminal particulate antigens (Hase et al., 2009; Kanaya et al., 2012; Rios et al., 2015). We therefore examined the capacity for particulate antigen transport, which is a principal function of M cells, in Traf6IEC-KO mice. Fluorescent nanoparticles were orally administered, and the number of particles incorporated into PPs was quantified. Consistent with the lack of M cell marker expression and morphologically defined M cells in Traf6IEC-KO mice, fluorescent nanoparticles were much less efficiently incorporated into PPs in Traf6IEC-KO mice than Traf6f/f mice (Fig. 6, A and B). We next evaluated the uptake of orally administered Salmonella enterica serovar Typhimurium and Yersinia enterocolitica into PPs. As expected, minimal uptake of S. Typhimurium and Y. enterocolitica was detected in PPs of Traf6IEC-KO mice compared with control Traf6f/f mice (Fig. 6, C and E). This reduced uptake of pathogenic bacteria implies that the M cell–mediated IgA response might also be impaired in Traf6IEC-KO mice. To evaluate this, we administered an attenuated S. Typhimurium strain lacking the aroA gene or Y. enterocolitica to Traf6f/f and Traf6IEC-KO mice and examined the amount of fecal IgA. Consistent with the significant decrease of S. Typhimurium or Y. enterocolitica uptake into PPs, the amount of S. Typhimurium– or Y. enterocolitica–specific IgA in feces was significantly decreased in Traf6IEC-KO mice compared with their Traf6f/f counterparts (Fig. 6, D and F). Together, these findings demonstrated that the loss of M cells caused by TRAF6 deficiency impaired M cell–associated mucosal immune responses.
Figure 6.
The uptake of nanoparticles, S. Typhimurium, and Y. enterocolitica into PPs and S. Typhimurium– or Y. enterocolitica–specific IgA responses are impaired in Traf6IEC-KO mice. (A) Traf6f/f and Traf6IEC-KO mice were orally gavaged with fluorescent nanoparticles. 4 h after administration, PPs were collected. Cryosections of PPs from Traf6f/f and Traf6IEC-KO mice were examined using a fluorescent microscope. Dotted lines indicate the FAE. (B) The number of nanoparticles incorporated into PPs was quantified using MetaMorph software (mean ± SD, n = 12; ****, P < 0.0001; Mann-Whitney U test). (C) Traf6f/f and Traf6IEC-KO mice were intragastrically gavaged with S. Typhimurium (χ3306), and 24 h after infection, PPs were collected. PPs were then homogenized and plated on LB plates to calculate the amount of S. Typhimurium taken up into PPs (mean ± SD, n = 7, Mann-Whitney U test). (D) Traf6f/f (n = 8) and Traf6IEC-KO (n = 7) mice were intragastrically gavaged with ΔaroA S. Typhimurium (UF20). After 2 and 3 wk, feces were collected, and S. Typhimurium–specific IgA was analyzed by ELISA (Mann-Whitney U test). (E) Traf6f/f and Traf6IEC-KO mice were intragastrically gavaged with Y. enterocolitica, and 24 h after infection, PPs were collected. PPs were then homogenized and plated on Yersinia selection plates to calculate the number of Y. enterocolitica taken up into PPs (mean ± SD, n = 6, Mann-Whitney U test). (F) Traf6f/f (n = 11) and Traf6IEC-KO (n = 9) mice were intragastrically gavaged with Y. enterocolitica. After 2 and 3 wk, feces were collected, and Y. enterocolitica–specific IgA was analyzed by ELISA (Mann-Whitney U test). Data are representative of two independent experiments.
TRAF6 deficiency selectively blocks RANK-mediated canonical NF-κB activation in IECs
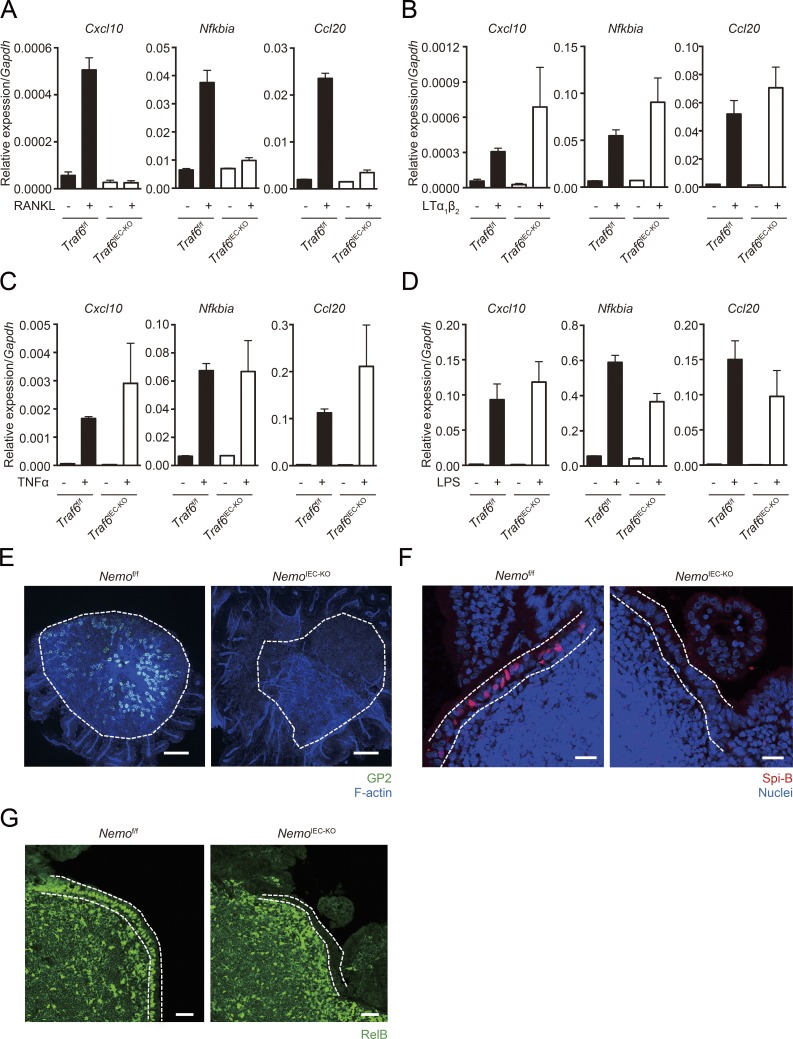
Because canonical NF-κB pathway plays important roles in IEC functions to maintain intestinal homeostasis (Nenci et al., 2007; Zaph et al., 2007), we examined the potential of TRAF6-deficient IECs to activate canonical NF-κB in response to multiple stimuli. As expected, the inflammatory genes (Cxcl10, Nfkbia, and Ccl20) targeted by canonical NF-κB failed to respond to RANKL stimulation in the organoids generated from Traf6IEC-KO mice (Fig. 7 A). By contrast, the ligations of TNFR1, lymphotoxin β receptor (LTβR), and toll-like receptor 4 (TLR4) were almost intact, even in TRAF6-deficient IECs (Fig. 7, B–D). These data suggest that TRAF6 deficiency selectively blocks RANKL-mediated canonical NF-κB activation in IECs.
Figure 7.
TRAF6 deficiency selectively blocks RANK-mediated canonical NF-κB. (A–C) Organoids generated from Traf6f/f and Traf6IEC-KO mice were stimulated with RANKL, LTα1β2 or TNF-α for 90 min. The organoids were harvested and the expression of inflammatory genes were analyzed by quantitative PCR (normalized with Gapdh, n = 3, mean ± SEM). (D) LPS was intraperitoneally injected to Traf6f/f and Traf6IEC-KO mice. After 90 min, mice were sacrificed, and IECs were isolated with EDTA treatment. The expressions of inflammatory genes were analyzed by quantitative PCR (normalized with Gapdh, n = 3, mean ± SEM). Solid bars indicate the expression profiles of Traf6f/f mice, and open ones indicate those of Traf6IEC-KO mice. (E) Isolated FAE from Nemof/f and NemoIEC-KO mice were immunostained with anti-GP2 antibody. Green color shows GP2, and blue shows F-actin. Dotted lines indicate the FAE. Bars, 100 µm. (F) PPs of Nemof/f and NemoIEC-KO mice were immunostained with anti–Spi-B antibody. Red shows Spi-B, and blue shows nuclei. Dotted lines indicate the FAE. Bars, 20 µm. (G) PPs of Nemof/f and NemoIEC-KO mice were immunostained with anti–RelB antibody (green). Dotted lines indicate the FAE. Bars, 40 µm. Data are representative of two independent experiments.
The crucial role of TRAF6 in canonical NF-κB has been well examined; on the other hand, it has been reported that TRAF6 has potential to directly activate noncanonical NF-κB (Xiao et al., 2012). This raises the possibility that the lack of M cells in Traf6IEC-KO mice is not directly caused by the loss of TRAF6-mediated canonical NF-κB activation but instead results from an inability to activate noncanonical NF-κB. To assess this possibility, we analyzed M cell differentiation using mice lacking NF-κB essential modulator (NEMO), a molecule critical for canonical NF-κB activation that is not involved in noncanonical NF-κB activation. We crossed Nemo floxed mice with Villin-Cre transgenic mice to obtain mice lacking NEMO specifically in IECs (NemoIEC-KO mice). We used these mice and examined the expression of M cell markers in PP FAE. The expression of GP2 and Spi-B was absent in NemoIEC-KO mice as well as in Traf6IEC-KO mice (Fig. 7, E and F), indicating that canonical NF-κB signaling is important for M cell differentiation. Additionally, we found that nuclear RelB was nearly absent from the FAE of NemoIEC-KO mice, indicating that canonical NF-κB plays a crucial role in the activation of noncanonical NF-κB in vivo.
TRAF6 is essential for M cell differentiation in human organoids
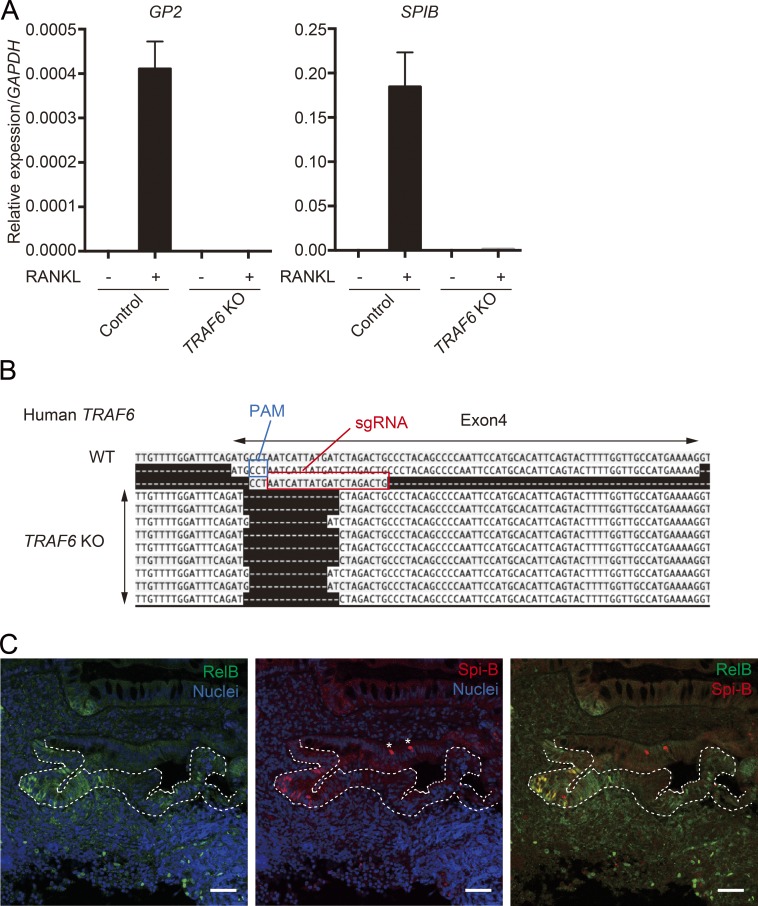
We also evaluated the requirements for TRAF6 in human M cell differentiation using human small intestinal organoids. RANKL stimulation could induce M cell differentiation in human intestinal organoids (Fig. 8 A; Rouch et al., 2016). We therefore generated TRAF6-deficient human organoids using a CRISPR/Cas9 system (Fig. 8 B) and examined the expression of M cell markers upon RANKL stimulation. As expected, the expression of GP2 and SPIB was silenced in TRAF6-deficient human organoids (Fig. 8 A), indicating that TRAF6 is essential for human M cell differentiation. Additionally, we found nuclear RelB-positive cells in the FAE of human PPs, and their distribution colocalized with Spi-B–positive cells (Fig. 8 C). Together, these data suggest that the RANK–TRAF6–NF-κB pathway is essential for M cell differentiation in humans as well as in mice.
Figure 8.
Requirement for NF-κB in M cell differentiation is conserved between mice and humans. (A) Normal and TRAF6-KO human organoids were stimulated with RANKL for 3 d, and the expression of GP2 and SPIB was analyzed by quantitative PCR (normalized with GAPDH, n = 4, mean ± SEM). (B) The introduction of deletion mutations in the TRAF6 gene was confirmed by transit-amplifying cloning of PCR products spanning the targeted region. PAM, protospacer adjacent motif. (C) Human PP biopsy samples were stained with antibodies to RelB and Spi-B. Green shows RelB, red shows Spi-B, and blue shows nuclei. Dotted lines indicate the FAE. Bars, 40 µm. Data are representative two independent experiments.
Discussion
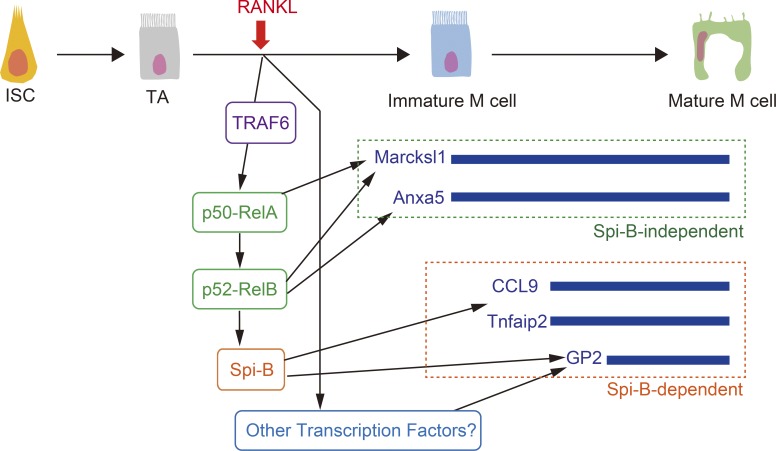
In this study, we have demonstrated that epithelial-intrinsic NF-κB plays a crucial role in M cell differentiation by taking advantage of the intestinal organoids culture system. Our previous work has demonstrated the significance of noncanonical NF-κB in M cell differentiation using RANKL-induced ectopic in vivo M cell differentiation (Kimura et al., 2015). This in vivo M cell differentiation assay is useful for analyzing mice completely lacking PPs such as Relb−/− mice; however, it could not exclude the possibility that the inactivation of noncanonical NF-κB in non-IECs might affect M cell differentiation. In contrast, in vitro M cell differentiation in organoids demonstrates the significance of epithelial-intrinsic factors for M cell differentiation, because organoids are predominantly composed of IECs. In addition, M cells induced in organoids mimic the features of in vivo FAE M cells, especially in their expression of the full spectrum of FAE M cell markers and transcription factors (Haber et al., 2017), suggesting that in vitro–induced M cells in organoids are a powerful tool for understanding the M cell differentiation pathway. We generated organoids from Relb−/− mice and successfully demonstrated a requirement for IEC-intrinsic noncanonical NF-κB in M cell differentiation. The in vitro organoids system also permitted the use of lentivirus-mediated gene transduction for overexpression of transcription factors implicated in M cell differentiation. We found that expression of Spi-B in enterocytes is not sufficient for terminal differentiation of M cells. Furthermore, we found that overexpression of neither p52-RelB nor p50-RelA is sufficient to complete M cell differentiation, as assessed by expression of Gp2, indicating that an additional factor activated by RANKL–RANK might be required to complete M cell differentiation (Fig. 9).
Figure 9.
Updated model of M cell differentiation. RANKL activates canonical NF-κB through TRAF6. Canonical NF-κB transcription factor (p50-RelA) is essential for the activity of noncanonical NF-κB transcription factor (p52-RelB), and largely contributes the expression of Marcksl1, which is one of Spi-B–independent M cell markers. p52-RelB directly binds Spib promoter and is responsible for Spi-B expression. On the other hand, both canonical and noncanonical NF-κB is insufficient to induce GP2. Unknown transcription factors activated by RANKL are required for the development of mature M cells. TA, transit amplifying.
We have also identified that TRAF6, the upstream transducer of canonical NF-κB activation by RANKL, is critical for M cell differentiation using Traf6IEC-KO mice. TRAF6 also plays an essential role in the development of mTECs (Akiyama et al., 2005) and osteoclasts (Kobayashi et al., 2001). Notably, a previous study demonstrated that Spi-B is also induced during mTEC development and that noncanonical NF-κB transcription factors could bind Spib promoter in mTECs (Akiyama et al., 2014). The shared dependence of M cell and mTEC differentiation on RANKL-induced Spib expression suggests that there may also be other shared mechanisms regulating the differentiation of these two specialized types of epithelial cells. In this regard, MAPK and AP-1 have been implicated to be involved, alongside NF-κB, downstream of TRAF6 (Kobayashi et al., 2003; Oeckinghaus et al., 2011); therefore, these pathways could cooperate with the NF-κB–Spi-B axis to lead to terminal differentiation of M cells with GP2 expression.
Our use of intestinal organoids in these studies provided an opportunity to expand our findings to human enterocytes. Gene targeting using a CRISPR/Cas9 system enabled us to generate TRAF6-KO human organoids, and evaluate the role of TRAF6 in human M cell differentiation. Our study demonstrates the conserved requirement of TRAF6 for M cell differentiation in mice and humans.
Intriguingly, we found that goblet cells increased in the FAE of Traf6IEC-KO mice, and M cells seemed to be replaced by goblet cells in the lack of M cell lineage commitment. Goblet cells, along with Paneth and enteroendocrine cells, are secretory lineage cells, implying that M cells might also belong to the secretory lineage; alternatively, the commitment to an M cell lineage might be closely related to that of goblet cells. It is well known that Notch signaling regulates secretory or nonsecretory cell fate decision in IECs, although little is known about the contribution of this lineage commitment in M cell differentiation. Mice harboring a spontaneous mutation in Notch ligand Delta-3 (pudgy mutant strain) exhibited an increase in the number of cells with abnormal apical membrane morphology, which are speculated to be the intermediates between M cells and their precursors (Mach et al., 2005). Independently of this, Hsieh and Lo (2012) demonstrated the requirements of Notch signaling in M cell differentiation using mice lacking Notch1 or its ligand, Jagged1. The conditional deletion of Notch1 increased the number of M cells as well as that of goblet cells, and conversely, Jagged1 deletion decreased their numbers. These studies indicate that Notch signaling and lateral inhibition affect M cell differentiation and support our hypothesis that M cells might belong to the secretory lineage. However, the increase in goblet cells was not observed in the FAE of SpibIEC-KO mice. We have previously demonstrated that Spi-B is essential for full M cell differentiation but dispensable for the expression of Annexin A5 and Marcksl1 (Kanaya et al., 2012), suggesting that M cell lineage commitment can be initiated in the absence of Spi-B. The lack of goblet cell hyperplasia in the FAE of SpibIEC-KO mice suggests that once IECs have committed to the M cell lineage, they cannot convert to goblet cells, even in the absence of further maturation into M cells. In addition, we did not observe hyperplasia of goblet cells in the FAE of RankIEC-KO mice (unpublished data; Rios et al., 2015), which is contradictory to our finding in Traf6IEC-KO mice. However, TRAF6 is known as a signal transducer for IL-1R and MyD88-dependent TLRs (Kawagoe et al., 2008). Thus, the observed goblet cell hyperplasia may not occur when only the RANKL–RANK signaling pathway is interrupted but may occur because of the disruption of other signaling pathways that converge further downstream of IEC TRAF6.
In the absence of TRAF6, the nuclear RelB remained (Fig. 4 A). In contrast, nuclear RelB was completely abolished in NemoIEC-KO mice (Fig. 7 G). These observations suggest that a certain molecule, which does not require TRAF6, might be involved in the residual activation of noncanonical NF-κB in the FAE of Traf6IEC-KO mice. We speculate that LTβR signaling is responsible for the residual activation of noncanonical NF-κB, because a previous study has reported the possibility that LTβR activates noncanonical NF-κB independently of TRAF6 (Qin et al., 2007). In addition, the present study demonstrated that the administration of LTα1β2 increased the expression of the genes associated with noncanonical NF-κB, FAE, and M cells, but its efficiency was much lower than that in organoids stimulated with RANKL (Fig. S3). Together, these data suggest that the residual noncanonical NF-κB activated by LTβR in the absence of TRAF6 is not sufficient for the development of FAE and M cells.
Collectively, we demonstrated that IEC-intrinsic NF-κB activation evoked by the RANK–TRAF6–NF-κB axis is essential for the development of M cells and FAE and thus makes a significant contribution to the role of IEC-intrinsic NF-κB in maintaining intestinal homeostasis.
Material and methods
Animals
BALB/c and C57BL/6J mice were purchased from CLEA. Traf6f/f (Kobayashi et al., 2003), Spibf/f (KOMP Repository), and Nemof/f mice (Schmidt-Supprian et al., 2000) were crossed with Villin-Cre transgenic (Jackson) mice to obtain Traf6IEC-KO, SpibIEC-KO, and NemoIEC-KO mice, respectively. Relb−/− mice were purchased from The Jackson Laboratory. Mice were weaned at 4 wk old, and the genotypes were determined by PCR assays. To avoid the development of changes in the microbiota composition of mice with different genotypes, Traf6f/f and Traf6IEC-KO mice were cohoused. For the infectious experiments, 8–12-wk-old Traf6f/f and Traf6IEC-KO mice were used at the same time, which included littermates and nonlittermates. All mice were maintained under specific pathogen-free conditions at RIKEN Center for Integrative Medical Sciences Animal Facility or conventional facility at Yokohama City University. All animal experiments were approved by the Institutional Animal Care and Use Committees of RIKEN Yokohama Branch and Yokohama City University.
ISC culture (organoids)
Proximal small intestine was incubated with 2 mM EDTA/PBS for 30 min on ice. Then, intestinal crypts were isolated by vigorous pipetting and embedded in Matrigel (Corning). Crypts were cultured in a previously described optimal medium (Sato et al., 2009) consisting of Advanced DMEM/F12 (Life Technologies) containing 10 mM Hepes, Glutamax (Invitrogen), 10 ng/ml epidermal growth factor (Peprotech), R-spondin1–conditioned medium (Ootani et al., 2009), 100 ng/ml Noggin (Peprotech), N2 and B27 supplement (Invitrogen), and 1 mM N-acetylcysteine (Sigma). Organoids were passaged every 3–4 d by vigorous pipetting with a Pasteur pipette.
Reagents for stimulation of organoids
For the stimulation of organoids with recombinant proteins or other reagents, we used organoids capable of stable growth after passage. To induce M cell differentiation, the glutathione S-transferase–mouse RANKL fusion protein GST-RANKL (Knoop et al., 2009) was added into organoids at a concentration of 500 ng/ml. To evoke the activation of canonical NF-κB, 100 ng/ml mouse TNF-α (Peprotech) and 1 µg/ml human LTα1β2 (R&D) were added into organoid cultures. To block the activity of IκB kinase-β, 125 µM SC-514 (Calbiochem) was used (Akiyama et al., 2008).
Isolation of epithelial cells
For the preparation of VE and FAE, small pieces of the ileum and PPs were dissected from mice (Hase et al., 2005). Tissues were incubated with 30 mM EDTA/HBSS for 15 min on ice, and epithelial cells were peeled off from lamina propria or PPs. To isolate the FAE regions, surrounding VE tissue was carefully removed by stereomicroscope-assisted dissection.
Histological analyses
Intestinal tissues were dissected, washed with cold PBS, and fixed with periodate lysine paraformaldehyde solution for 4–5 h on ice. Then, tissues were dehydrated with a graded series of ethanol and embedded in paraffin. 5-μm-thick sections were prepared, and these sections were used for immunohistochemistry and alcian blue staining.
Antibodies
Rat anti–mouse GP2 antibody was generated in our laboratory (2F11-3). Rabbit anti–Muc2 (H-300), rabbit anti–TRAF6 (H-274), and goat anti–Annexin V (R-20) antibodies were purchased from Santa Cruz Biotechnology. Rabbit anti–RelB monoclonal antibody (D7D7W) was purchased from Cell Signaling Technology. Rabbit anti–Marcksl1 antibody was purchased from Proteintech. Goat anti–E-cadherin, goat anti–CCL20, and sheep anti-Spi-B antibodies were obtained from R&D Systems. Donkey Dylight549 anti–rat IgG antibody and Dylight488 anti–rabbit IgG were purchased from Jackson ImmunoResearch. Donkey Alexa Fluor 555 and 633 anti–goat IgG were purchased from Life Technologies.
Immunohistochemistry
Paraffin sections were rehydrated and washed with PBS. Then, tissues were incubated with 1% BSA/PBS supplemented with 5% normal serum donkey serum to quench the nonspecific binding of antibodies. Antigen retrieval with citrate buffer, pH 7.0 (121°C for 5 min), was required for staining with anti–RelB and anti–Marcksl1 antibodies. Primary antibodies were incubated at 4°C overnight. For whole-mount immunostaining, isolated FAE and organoids were fixed with 4% paraformaldehyde for 1 h on ice and then incubated with primary antibodies. The binding of primary antibodies was followed by fluorescent-dye–conjugated secondary antibodies. Nuclei were stained by DAPI. The sections were examined using a confocal microscope (SP5; Leica) or a light and fluorescent microscope (AF6000; Leica).
Scanning electron microscopy
PPs were fixed in 2.5% glutaraldehyde/0.1 M phosphate buffer, pH 7.4, for 2 h and dehydrated with a graded ethanol series. Then, tissues were substituted with t-butyl alcohol. After being freeze dried, tissues were metal-coated using a magnetron sputter (MSP-1S; Vacuum Device), and examined by scanning electron microscopy (VE-7800; Keyence).
TEM
PPs were fixed in 2.5% glutaraldehyde/0.1 M phosphate buffer, pH 7.4, for 2 h. After rinsing in phosphate buffer, tissues were post-fixed in 1% OsO4 for 90 min, dehydrated through a graded series of ethanol, and embedded in epon according to a conventional procedure. Ultrathin sections were prepared and stained with aqueous solutions of uranyl acetate and lead citrate for observation under an electron microscope (H-7100; Hitachi).
Expression analysis
Total RNA from each sample was extracted using the RNeasy mini kit (Qiagen) and reverse transcribed with ReverTra Ace (Toyobo). Gene expression was analyzed by real-time quantitative PCR with SYBR Premix Ex Taq (Takara) and specific primers (Table S1). The expression of target genes was assessed by a comparative cycling threshold method using expression of Gapdh or GAPDH. Primer sequences are summarized in Table S1.
Oral gavage of fluorescent nanoparticles
Traf6f/f and Traf6IEC-KO mice were fasted for 3 h, and 0.2-µm-diameter fluorescent nanoparticles (ThermoFisher) were orally gavaged. After 4 h, mice were sacrificed, and distal PPs were collected. PPs tissues were fixed with 3.7% formalin/PBS for 2 h. Fixed tissues were incubated overnight with 30% sucrose/PBS and finally embedded in OCTcompound (Sakura Fintech). Cryosections were observed by fluorescent microscope (AF6000; Leica), and the number of nanoparticles taken up into PPs was quantified by MetaMorph software (Molecular Devices).
Immunoprecipitation and Western blot analysis
Isolated epithelial cells and splenocytes from Traf6f/f and Traf6IEC-KO mice were lysed with lysis buffer. Cell lysates were incubated with rabbit anti-TRAF6 antibody, and TRAF6 proteins were immunoprecipitated with protein G magnetic beads (Millipore). Purified proteins were loaded on SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane. Then, membrane was incubated with anti-TRAF6 antibody, and followed with HRP-conjugated secondary antibody (TrueBlot; Rockland). Signal development was performed by chemiluminescent kit (Millipore).
ChIP
For ChIP-PCR, mouse organoids were stimulated with RANKL for 6 h. Chromatin was cross-linked with 1% formaldehyde for 15 min at room temperature. The reaction was stopped by the addition of glycine to final concentration of 125 mM and then centrifuged to obtain the organoids pellet. Organoids were suspended with lysis buffer 1 (50 mM Hepes, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, and 0.25% Triton X-100) supplemented with cOmplete protease inhibitor cocktail (Roche) and 0.1 mM PMSF for 10 min on ice with rotation. Nuclei pellet was washed with lysis buffer 2 (10 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, and 0.5 mM EGTA) supplemented with cOmplete protease inhibitor cocktail for 10 min on ice with rotation. Nuclei pellet was resuspended with lysis buffer 3 (10 mM Tris-HCl, pH 8.0, 300 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% sodium deoxycholate, and 0.5% N-laurylsarcosine sodium salt) supplemented with cOmplete protease inhibitor cocktail (Roche) and 0.1 mM PMSF and then sonicated with Bioruptor for 5 min (30-s pulse and 30-s rest) five times. Debris was removed from sonicated chromatin by centrifugation, and Triton X-100 was added to the final concentration of 1%. Obtained chromatin solution was preincubated with Dynabeads M-280 Sheep anti–rabbit IgG (ThermoFisher) for 30 min at 4°C and then incubated with anti–RelB antibody overnight at 4°C with rotation. Dynabeads M-280 Sheep anti–rabbit IgG was added to chromatin solution and incubated for 1 h at 4°C with rotation. To obtain immunoprecipitated chromatin conjugated to magnetic beads, supernatant was removed using a magnetic stand. Beads were washed with low-salt buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.1 b% SDS, and 1% Triton X-100), high-salt buffer (20 mM Tris-HCl, pH 8.0, 400 mM NaCl, 2 mM EDTA, 0.1 b% SDS, and 1% Triton X-100), and RIPA-LiCl buffer (50 mM Hepes, pH 7.5, 500 mM LiCl, 1 mM EDTA, 1% NP-40, and 0.5% sodium deoxycholate). Finally, ChIP DNA was eluted with elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, and 1% SDS) by incubation overnight at 65°C with vigorous shaking. Input DNA and ChIP DNA were treated with RNase A (ThermoFisher) at 37°C for 30 min, followed by incubation with Proteinase K (Roche) at 55°C for 30 min. DNA was purified by phenol/chloroform extraction. ChIP DNA was quantified by real-time quantitative PCR with SYBR Premix Ex Taq and specific primers for the Spib promoter region.
Lentivirus infection
Lentiviral supernatants were generated by transient transfection of RelB, RelA, p50, or p52-containing SIN vector (CSII-CMV-RfA-IRES2-Venus; 17 µg), 10 µg pCAG-HIVgp, and 10 µg pCMV-VSV-G-Rev into Lenti-X 293T (5 × 106/15 ml; Clontech) using Polyethylenimine Max (Polysciences) according to the manufacturer’s protocol. All of these vectors were provided by Dr. Miyoshi (Keio University, Tokyo, Japan) and RIKEN BRC. Transfected host cells were incubated for 24 h, and another 48-h culture after changing the culture medium containing 10 µM forskolin allowed those cells to produce free virions. Viruses were purified after removal of host cells and ultracentrifugation, and viral titers were measured using Lenti-X p24 Rapid Titer kit (Clontech) according to manufacturer’s protocol. Dissociated organoids were infected by mixing with 250 µl virus-including medium followed by centrifugation at 600 g for 1 h at 32°C. Those cells were then embedded in Matrigel and incubated for 3 d.
Luciferase assay
The coding sequences of mouse RelB and p52 (Nfkb2) were amplified from FAE-derived cDNA and subcloned into pcDNA3 vector (Life Technologies). The putative Spib promoter region (approximately −1,930 to 0) was amplified from a bacterial artificial chromosome (BAC) clone (RP23-455C13 purchased from Life Technologies), and subcloned into pGL3Basic vector (Promega). To determine the direct interaction of noncanonical NF-κB and Spib promoter, mICcl2 (kindly provided by Dr. Jean-Pierre Kraehenbuhl; Bens et al., 1996) cells were transfected with the combination of these expression and luciferase reporter vectors by Lipofectamine 2000 (Life Technologies). After the overnight culture, cells were lysed. Then luciferase activity was measured by the dual-luciferase reporter assay system (Promega).
Quantification of S. Typhimurium taken up into PPs
Mice were orally administered with 109 CFU S. Typhimurium (χ3306; Gulig et al., 1998). After 24 h, PPs were dissected and incubated for 30 min at 20°C with gentle shaking in sterile PBS containing 100 µg/ml gentamicin. Pooled PP tissue was weighed and homogenized in sterile PBS. The homogenates were serially diluted in sterile PBS and plated on LB agar plates containing 25 µg/ml nalidixic acid.
Evaluation of S. Typhimurium or Y. enterocolitica–specific IgA in feces
Mice were orally administered ΔaroA S. Typhimurium (UF20, 5 × 109 CFU/mouse; Gulig and Doyle, 1993; the major defect of ΔaroA strains is the requirement for para-amino benzoic acid, which cannot be supplied by the host in sufficient amounts, resulting in a deficiency in replication) or Y. enterocolitica (108 CFU/mouse). After 2 and 3 wk of infection, feces were collected. Feces were suspended in sterile PBS (100 µl to 10 mg of feces), homogenized and centrifuged at 12,000 g for 10 min, and collect the supernatant. The supernatants were analyzed using a mouse IgA ELISA kit (Bethyl Laboratories) to measure the amount of S. Typhimurium–specific IgA. In this study, 96-well plates were coated with lysates prepared from S. Typhimurium or Y. enterocolitica to capture S. Typhimurium– or Y. enterocolitica–specific IgA in supernatant.
Human subjects
Human small intestinal organoids were established using tissue from healthy human small intestine. This procedure was approved by the Committee on Human Subjects in RIKEN and Keio University. Human organoids were cultured in the optimal medium consisting of Advanced DMEM/F12 (Life Technologies) containing 10 mM Hepes, Glutamax (Invitrogen), 10 ng/ml epidermal growth factor (Peprotech), R-spondin1–conditioned medium (Ootani et al., 2009), Wnt3a-conditioning medium (L-Wnt3a cells were a gift from the H. Clevers laboratory, Hubrecht Institute, Utrecht, Netherlands), 100 ng/ml Noggin (Peprotech), B27 supplement (Invitrogen), 1 mM N-acetylcysteine (Sigma), 500 nM A83-01 (Tocris), and 10 µM SB202190 (Sigma). TRAF6 KO human organoids were generated using a CRISPR/cas9 system according to a previous study (Fujii et al., 2015). To induce M cell differentiation, organoids were stimulated with 1 µg/ml sRANKL (Oriental Yeast) for 3 d. Biopsy samples of human PPs were collected from healthy small intestinal tissues and analyzed by immunohistochemistry. This procedure was approved by the Committee on Human Subjects in RIKEN and Chiba University.
Statistical analyses
Student’s t test, Mann-Whitney U test, and one-way analysis of variance followed by the Bonferroni post-hoc test were used. P-values of less than 0.05 were considered statistically significant.
Online supplemental material
Fig. S1 demonstrates that M cells are also absent in the FAE of colonic patches of Traf6IEC-KO mice. Fig. S2 demonstrates that the hyperplasia of goblet cell does not occur in the FAE of SpibIEC-KO mice. Fig. S3 demonstrates that the expression of FAE-associated genes is more dependent on RANKL than LTβR. Table S1 shows the primer sequences used for quantitative PCR analysis.
Supplementary Material
Acknowledgments
We thank the members of laboratory for Intestinal Ecosystem for technical support and experimental assistance. We also thank Dr. Manolis Pasparakis (University of Cologne, Cologne, Germany) for providing Nemo floxed mice, Dr. Calvin Kuo (Stanford University, Stanford, CA) for the R-spondin1–producing cell line, Dr. Hans Clevers for L-Wnt3a cells, Dr. Hidenori Matsui (Kitasato University, Tokyo, Japan) for S. Typhimurium, Dr. Hiroyuki Miyoshi for lentivirus vectors, Dr. Atsushi Miyawaki (RIKEN, Kanagawa, Japan) for Venus cDNA, Dr. Jean-Pierre Kraehenbuhl (HSeT Foundation, Epalinges, Switzerland) for mICcl2 cells, and Dr. Yasutaka Motomura (RIKEN) for the protocol for ChIP experiments.
This work was supported in part by the Japan Society for the Promotion of Science KAKENHI (grant 26460584 to T. Kanaya and grant 24249029 to H. Ohno), Ministry of Education, Culture, Sports, Science and Technology KAKENHI (grant 15H01165 to T. Kanaya and grant 20113003 to H. Ohno), Uehara Memorial Foundation (grant to T. Kanaya), Takeda Science Foundation (grant to T. Kanaya), and Mochida Memorial Foundation for Medical and Pharmaceutical Research (grant to T.K.).
The authors declare no competing financial interests.
Author contributions: T. Kanaya conceived the study, designed experiments, analyzed data, and wrote manuscript. S. Sakakibara performed a large part of experiments. T. Jinnohara and N. Tachibana performed infection experiments. M. Hachisuka generated lentivirus vectors. S. Hidano and T. Kobayashi provided Traf6 flox mice. S. Kimura and T. Iwanaga performed the TEM analysis. T. Nakagawa, T. Katsuno. and N. Kato collected human biopsy samples. T. Akiyama helped interpret the data. T. Sato generated TRAF6 KO organoids. I.R. Williams helped interpret the data and edited the manuscript. H. Ohno directed the research and wrote the manuscript.
References
- Akiyama N., Shinzawa M., Miyauchi M., Yanai H., Tateishi R., Shimo Y., Ohshima D., Matsuo K., Sasaki I., Hoshino K., et al. . 2014. Limitation of immune tolerance-inducing thymic epithelial cell development by Spi-B-mediated negative feedback regulation. J. Exp. Med. 211:2425–2438. 10.1084/jem.20141207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama T., Maeda S., Yamane S., Ogino K., Kasai M., Kajiura F., Matsumoto M., and Inoue J.. 2005. Dependence of self-tolerance on TRAF6-directed development of thymic stroma. Science. 308:248–251. 10.1126/science.1105677 [DOI] [PubMed] [Google Scholar]
- Akiyama T., Shimo Y., Yanai H., Qin J., Ohshima D., Maruyama Y., Asaumi Y., Kitazawa J., Takayanagi H., Penninger J.M., et al. . 2008. The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity. 29:423–437. 10.1016/j.immuni.2008.06.015 [DOI] [PubMed] [Google Scholar]
- Bens M., Bogdanova A., Cluzeaud F., Miquerol L., Kernéis S., Kraehenbuhl J.P., Kahn A., Pringault E., and Vandewalle A.. 1996. Transimmortalized mouse intestinal cells (m-ICc12) that maintain a crypt phenotype. Am. J. Physiol. 270:C1666–C1674. [DOI] [PubMed] [Google Scholar]
- Bren G.D., Solan N.J., Miyoshi H., Pennington K.N., Pobst L.J., and Paya C.V.. 2001. Transcription of the RelB gene is regulated by NF-kappaB. Oncogene. 20:7722–7733. 10.1038/sj.onc.1204868 [DOI] [PubMed] [Google Scholar]
- de Lau W., Kujala P., Schneeberger K., Middendorp S., Li V.S.W., Barker N., Martens A., Hofhuis F., DeKoter R.P., Peters P.J., et al. . 2012. Peyer’s patch M cells derived from Lgr5(+) stem cells require SpiB and are induced by RankL in cultured “miniguts”. Mol. Cell. Biol. 32:3639–3647. 10.1128/MCB.00434-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M., Matano M., Nanki K., and Sato T.. 2015. Efficient genetic engineering of human intestinal organoids using electroporation. Nat. Protoc. 10:1474–1485. 10.1038/nprot.2015.088 [DOI] [PubMed] [Google Scholar]
- Gallo R.L., and Hooper L.V.. 2012. Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 12:503–516. 10.1038/nri3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y., Obata T., Kunisawa J., Sato S., Ivanov I.I., Lamichhane A., Takeyama N., Kamioka M., Sakamoto M., Matsuki T., et al. . 2014. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. 345:1254009 10.1126/science.1254009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulig P.A., and Doyle T.J.. 1993. The Salmonella typhimurium virulence plasmid increases the growth rate of salmonellae in mice. Infect. Immun. 61:504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulig P.A., Doyle T.J., Hughes J.A., and Matsui H.. 1998. Analysis of host cells associated with the Spv-mediated increased intracellular growth rate of Salmonella typhimurium in mice. Infect. Immun. 66:2471–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber A.L., Biton M., Rogel N., Herbst R.H., Shekhar K., Smillie C., Burgin G., Delorey T.M., Howitt M.R., Katz Y., et al. . 2017. A single-cell survey of the small intestinal epithelium. Nature. 551:333–339. 10.1038/nature24489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase K., Ohshima S., Kawano K., Hashimoto N., Matsumoto K., Saito H., and Ohno H.. 2005. Distinct gene expression profiles characterize cellular phenotypes of follicle-associated epithelium and M cells. DNA Res. 12:127–137. 10.1093/dnares/12.2.127 [DOI] [PubMed] [Google Scholar]
- Hase K., Murakami T., Takatsu H., Shimaoka T., Iimura M., Hamura K., Kawano K., Ohshima S., Chihara R., Itoh K., et al. . 2006. The membrane-bound chemokine CXCL16 expressed on follicle-associated epithelium and M cells mediates lympho-epithelial interaction in GALT. J. Immunol. 176:43–51. 10.4049/jimmunol.176.1.43 [DOI] [PubMed] [Google Scholar]
- Hase K., Kawano K., Nochi T., Pontes G.S., Fukuda S., Ebisawa M., Kadokura K., Tobe T., Fujimura Y., Kawano S., et al. . 2009. Uptake through glycoprotein 2 of FimH(+) bacteria by M cells initiates mucosal immune response. Nature. 462:226–230. 10.1038/nature08529 [DOI] [PubMed] [Google Scholar]
- Hsieh E.-H., and Lo D.D.. 2012. Jagged1 and Notch1 help edit M cell patterning in Peyer’s patch follicle epithelium. Dev. Comp. Immunol. 37:306–312. 10.1016/j.dci.2012.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinnohara T., Kanaya T., Hase K., Sakakibara S., Kato T., Tachibana N., Sasaki T., Hashimoto Y., Sato T., Watarai H., Kunisawa J., Shibata N., Williams I.R., Kiyono H., and Ohno H.. 2017. IL-22BP dictates characteristics of Peyer’s patch follicle-associated epithelium for antigen uptake. J. Exp. Med. 214:1607–1618. 10.1084/jem.20160770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya T., and Ohno H.. 2014. The Mechanisms of M-cell Differentiation. Biosci. Microbiota Food Health. 33:91–97. 10.12938/bmfh.33.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya T., Hase K., Takahashi D., Fukuda S., Hoshino K., Sasaki I., Hemmi H., Knoop K.A., Kumar N., Sato M., et al. . 2012. The Ets transcription factor Spi-B is essential for the differentiation of intestinal microfold cells. Nat. Immunol. 13:729–736. 10.1038/ni.2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagoe T., Sato S., Matsushita K., Kato H., Matsui K., Kumagai Y., Saitoh T., Kawai T., Takeuchi O., and Akira S.. 2008. Sequential control of Toll-like receptor-dependent responses by IRAK1 and IRAK2. Nat. Immunol. 9:684–691. 10.1038/ni.1606 [DOI] [PubMed] [Google Scholar]
- Kawano K., Ebisawa M., Hase K., Fukuda S., Hijikata A., Kawano S., Date Y., Tsuneda S., Itoh K., and Ohno H.. 2007. Psg18 is specifically expressed in follicle-associated epithelium. Cell Struct. Funct. 32:115–126. 10.1247/csf.07014 [DOI] [PubMed] [Google Scholar]
- Kimura S., Yamakami-Kimura M., Obata Y., Hase K., Kitamura H., Ohno H., and Iwanaga T.. 2015. Visualization of the entire differentiation process of murine M cells: suppression of their maturation in cecal patches. Mucosal Immunol. 8:650–660. 10.1038/mi.2014.99 [DOI] [PubMed] [Google Scholar]
- Kishore N., Sommers C., Mathialagan S., Guzova J., Yao M., Hauser S., Huynh K., Bonar S., Mielke C., Albee L., et al. . 2003. A selective IKK-2 inhibitor blocks NF-kappa B-dependent gene expression in interleukin-1 beta-stimulated synovial fibroblasts. J. Biol. Chem. 278:32861–32871. 10.1074/jbc.M211439200 [DOI] [PubMed] [Google Scholar]
- Knoop K.A., Kumar N., Butler B.R., Sakthivel S.K., Taylor R.T., Nochi T., Akiba H., Yagita H., Kiyono H., and Williams I.R.. 2009. RANKL is necessary and sufficient to initiate development of antigen-sampling M cells in the intestinal epithelium. J. Immunol. 183:5738–5747. 10.4049/jimmunol.0901563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N., Kadono Y., Naito A., Matsumoto K., Yamamoto T., Tanaka S., and Inoue J.. 2001. Segregation of TRAF6-mediated signaling pathways clarifies its role in osteoclastogenesis. EMBO J. 20:1271–1280. 10.1093/emboj/20.6.1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Walsh P.T., Walsh M.C., Speirs K.M., Chiffoleau E., King C.G., Hancock W.W., Caamano J.H., Hunter C.A., and Scott P.. 2003. TRAF6 Is a Critical Factor for Dendritic Cell Maturation and Development. Immunity. 19:353–363. 10.1016/S1074-7613(03)00230-9 [DOI] [PubMed] [Google Scholar]
- Kraehenbuhl J.-P., and Neutra M.R.. 2003. Epithelial M cells: Differentiation and Function. Annu. Rev. Cell Dev. Biol. 16:301–332. 10.1146/annurev.cellbio.16.1.301 [DOI] [PubMed] [Google Scholar]
- Mach J., Hshieh T., Hsieh D., Grubbs N., and Chervonsky A.. 2005. Development of intestinal M cells. Immunol. Rev. 206:177–189. 10.1111/j.0105-2896.2005.00281.x [DOI] [PubMed] [Google Scholar]
- Miyawaki S., Nakamura Y., Suzuka H., Koba M., Yasumizu R., Ikehara S., and Shibata Y.. 1994. A new mutation, aly, that induces a generalized lack of lymph nodes accompanied by immunodeficiency in mice. Eur. J. Immunol. 24:429–434. 10.1002/eji.1830240224 [DOI] [PubMed] [Google Scholar]
- Nenci A., Becker C., Wullaert A., Gareus R., van Loo G., Danese S., Huth M., Nikolaev A., Neufert C., Madison B., et al. . 2007. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 446:557–561. 10.1038/nature05698 [DOI] [PubMed] [Google Scholar]
- Neutra M.R., Mantis N.J., and Kraehenbuhl J.-P.. 2001. Collaboration of epithelial cells with organized mucosal lymphoid tissues. Nat. Immunol. 2:1004–1009. 10.1038/ni1101-1004 [DOI] [PubMed] [Google Scholar]
- Oeckinghaus A., Hayden M.S., and Ghosh S.. 2011. Crosstalk in NF-κB signaling pathways. Nat. Immunol. 12:695–708. 10.1038/ni.2065 [DOI] [PubMed] [Google Scholar]
- Ootani A., Li X., Sangiorgi E., Ho Q.T., Ueno H., Toda S., Sugihara H., Fujimoto K., Weissman I.L., Capecchi M.R., and Kuo C.J.. 2009. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 15:701–706. 10.1038/nm.1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Konno H., Ohshima D., Yanai H., Motegi H., Shimo Y., Hirota F., Matsumoto M., Takaki S., Inoue J., and Akiyama T.. 2007. Developmental stage-dependent collaboration between the TNF receptor-associated factor 6 and lymphotoxin pathways for B cell follicle organization in secondary lymphoid organs. J. Immunol. 179:6799–6807. 10.4049/jimmunol.179.10.6799 [DOI] [PubMed] [Google Scholar]
- Rios D., Wood M.B., Li J., Chassaing B., Gewirtz A.T., and Williams I.R.. 2015. Antigen sampling by intestinal M cells is the principal pathway initiating mucosal IgA production to commensal enteric bacteria. Mucosal Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouch J.D., Scott A., Lei N.Y., Solorzano-Vargas R.S., Wang J., Hanson E.M., Kobayashi M., Lewis M., Stelzner M.G., Dunn J.C.Y., et al. . 2016. Development of Functional Microfold (M) Cells from Intestinal Stem Cells in Primary Human Enteroids. PLoS One. 11:e0148216 10.1371/journal.pone.0148216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbo M., Sierro F., Debard N., Kraehenbuhl J.-P., and Finke D.. 2004. Lymphotoxin beta receptor signaling induces the chemokine CCL20 in intestinal epithelium. Gastroenterology. 127:213–223. 10.1053/j.gastro.2004.04.018 [DOI] [PubMed] [Google Scholar]
- Sato S., Kaneto S., Shibata N., Takahashi Y., Okura H., Yuki Y., Kunisawa J., and Kiyono H.. 2013. Transcription factor Spi-B-dependent and -independent pathways for the development of Peyer’s patch M cells. Mucosal Immunol. 6:838–846. 10.1038/mi.2012.122 [DOI] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., and Clevers H.. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459:262–265. 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- Schmidt-Supprian M., Bloch W., Courtois G., Addicks K., Israël A., Rajewsky K., and Pasparakis M.. 2000. NEMO/IKK gamma-deficient mice model incontinentia pigmenti. Mol. Cell. 5:981–992. 10.1016/S1097-2765(00)80263-4 [DOI] [PubMed] [Google Scholar]
- Sun S.-C. 2012. The noncanonical NF-κB pathway. Immunol. Rev. 246:125–140. 10.1111/j.1600-065X.2011.01088.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaira S., Johnson T., Hirbe A.C., Alhawagri M., Anwisye I., Sammut B., O’Neal J., Zou W., Weilbaecher K.N., Faccio R., and Novack D.V.. 2008. RelB is the NF-kappaB subunit downstream of NIK responsible for osteoclast differentiation. Proc. Natl. Acad. Sci. USA. 105:3897–3902. 10.1073/pnas.0708576105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh M.C., and Choi Y.. 2014. Biology of the RANKL-RANK-OPG System in Immunity, Bone, and Beyond. Front. Immunol. 5:511 10.3389/fimmu.2014.00511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weih F., and Caamaño J.. 2003. Regulation of secondary lymphoid organ development by the nuclear factor-kappaB signal transduction pathway. Immunol. Rev. 195:91–105. 10.1034/j.1600-065X.2003.00064.x [DOI] [PubMed] [Google Scholar]
- Xiao X., Balasubramanian S., Liu W., Chu X., Wang H., Taparowsky E.J., Fu Y.-X., Choi Y., Walsh M.C., and Li X.C.. 2012. OX40 signaling favors the induction of T(H)9 cells and airway inflammation. Nat. Immunol. 13:981–990. 10.1038/ni.2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz Z.B., Weih D.S., Sivakumar V., and Weih F.. 2003. RelB is required for Peyer’s patch development: differential regulation of p52-RelB by lymphotoxin and TNF. EMBO J. 22:121–130. 10.1093/emboj/cdg004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaph C., Troy A.E., Taylor B.C., Berman-Booty L.D., Guild K.J., Du Y., Yost E.A., Gruber A.D., May M.J., Greten F.R., et al. . 2007. Epithelial-cell-intrinsic IKK-beta expression regulates intestinal immune homeostasis. Nature. 446:552–556. 10.1038/nature05590 [DOI] [PubMed] [Google Scholar]
- Zhang K., Hornef M.W., and Dupont A.. 2015. The intestinal epithelium as guardian of gut barrier integrity. Cell. Microbiol. 17:1561–1569. 10.1111/cmi.12501 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.