Lee et al. show that established obesity alters the composition and long-term fitness of the hematopoietic stem cell (HSC) compartment, in part through a Gfi1-dependent HSC regulatory program that is activated by the chronic oxidative stress associated with this condition.
Abstract
Obesity is a chronic organismal stress that disrupts multiple systemic and tissue-specific functions. In this study, we describe the impact of obesity on the activity of the hematopoietic stem cell (HSC) compartment. We show that obesity alters the composition of the HSC compartment and its activity in response to hematopoietic stress. The impact of obesity on HSC function is progressively acquired but persists after weight loss or transplantation into a normal environment. Mechanistically, we establish that the oxidative stress induced by obesity dysregulates the expression of the transcription factor Gfi1 and that increased Gfi1 expression is required for the abnormal HSC function induced by obesity. These results demonstrate that obesity produces durable changes in HSC function and phenotype and that elevation of Gfi1 expression in response to the oxidative environment is a key driver of the altered HSC properties observed in obesity. Altogether, these data provide phenotypic and mechanistic insight into durable hematopoietic dysregulations resulting from obesity.
Introduction
Obesity is a major epidemic in the first world (Flegal et al., 2012; Ogden et al., 2014) and is considered to be a leading cause for proinflammatory metabolic syndrome, cancer development, and increased mortality (Goodwin and Stambolic, 2015; Arnold et al., 2016; Grundy, 2016). Among its numerous physiological consequences, obesity affects bone marrow (BM) homeostasis. A high-fat diet (HFD) qualitatively and quantitatively modifies the composition of the adipocyte tissue in the BM while disrupting the ability of mesenchymal progenitors to generate osteoblastic cells (Krings et al., 2012; Styner et al., 2014; Chen et al., 2016). In addition to its local effects, obesity is associated with profound systemic dysregulations. Adipose tissue acts as an active endocrine organ that secretes a plethora of bioactive substances (Iyengar et al., 2015). As a consequence, obesity contributes to adipokine and hormone imbalance. In parallel, obesity triggers the infiltration of activated immune cells into the adipose tissue, leading to a chronic inflammatory phenotype. Altogether, obesity can be considered as a chronic and complex pathological state associated with systemic and BM-specific stresses.
Previous studies have demonstrated the effect of diet and obesity on the hematopoietic system (Claycombe et al., 2008; Trottier et al., 2012; Adler et al., 2014b; Mihaylova et al., 2014). Conditions associated with metabolic dysregulations such as adipose tissue accumulation, hyperglycemia, and hypercholesterolemia have been linked to hematopoietic disruption and particularly to myeloid skewing (Nagareddy et al., 2013, 2014; Adler et al., 2014b; Tie et al., 2014). Recent research studying the direct effect of obesity on the hematopoietic stem and progenitor cells (HSPCs) specifically focused on the signals induced by the obese inflammatory state (Singer et al., 2014, 2015; van den Berg et al., 2016). Other dysregulations in HSPC compartments were associated with disruptions in the BM microenvironment. Research identified the expansion of the BM adipocytes as a key limiting factor of the hematopoietic activity upon transplantation (Naveiras et al., 2009). Similarly, diabetes has been shown to affect the mobilization capacity of hematopoietic stem cells (HSCs) by altering chemokine expression in the BM niche (Ferraro et al., 2011). Finally, a study has linked diet-induced modification of the microbiota to alteration of the BM endosteal niche and hematopoietic dysregulation (Luo et al., 2015). Although they describe specific effects of obesity on the hematopoietic system, these studies do not address its long-term effect on the fitness of the HSC compartment, the HSC-specific regulatory mechanisms that are disrupted in this condition, or whether these effects can persist upon weight loss.
The HSC compartment is highly heterogeneous, being composed of multiple cell subsets with variable levels of quiescence, self-renewal capability, and potential for differentiation (Wilson et al., 2008; Challen et al., 2010; Benz et al., 2012; Yamamoto et al., 2013). Contribution of these various HSC subsets to steady-state and emergency hematopoiesis is still a matter of debate (Sun et al., 2014; Busch et al., 2015; Sawai et al., 2016). However, maintenance of a healthy HSC pool is essential to sustaining a normal long-term hematopoiesis. Pathophysiological conditions such as aging, which are associated with a restriction of the diversity of the HSC compartment and the accumulation of myeloid-biased HSCs, correlate with hematopoietic disruptions and an increased susceptibility to hematological malignancies (Akunuru and Geiger, 2016). Although obesity has also been associated with hematological pathologies (Bhaskaran et al., 2014), its impact on the global fitness of the HSC compartment remains poorly understood. In this study, we show that obesity alters the cellular architecture of the HSC compartment and modifies its functional properties in response to acute hematopoietic stresses. We show that obesity has a progressive effect on the HSC compartment but also that some of the acquired properties in these conditions can persist upon weight loss or exposure to a normal environment. Molecularly, we establish that the transcription factor Gfi1 is a key regulator of the HSC fate in obesity by controlling its quiescence status and contributing to its aberrant stress response. Finally, we establish the role of the chronic oxidative stress in regulating Gfi1 expression in HSCs. Altogether, this work identifies key cellular and molecular mechanisms by which obesity, viewed as a chronic stress, affects the HSC compartment and could promote durable disruption of its fitness.
Results
Obesity modifies the composition of the HSC compartment
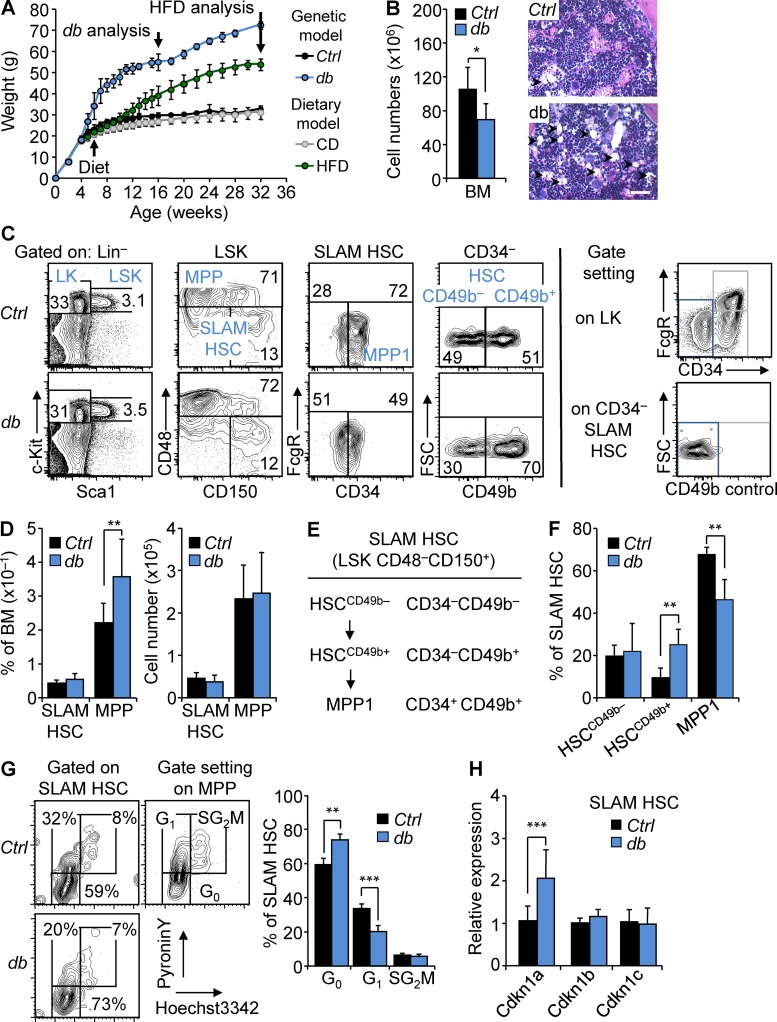
Obesity is a complex and multicausal condition. To fully recapitulate its phenotype, we conducted our studies in two different mouse models of obesity (Lutz and Woods, 2012). First, we used a genetic obese mouse model (hereafter denoted as db mice) carrying an inactivating mutation in the leptin receptor (Lepr) gene (Chua et al., 1996). Leptin is an adipokine secreted by adipocytes, which is part of a negative feedback loop that modulates fat tissue by regulating appetite and energy expenditure. Inactivation of the leptin signaling in the central nervous system induces hyperphagia, leading to rapid and homogeneous weight gain (Fig. 1 A). We analyzed db mice at 16 wk of age to allow for the interaction of adult HSCs with the environment of established obesity. To validate our results, we used a dietary mouse model in which WT mice are fed with HFD (Khandekar et al., 2011). Mice in this model take up to 5 mo to achieve significant weight gain (Fig. 1 A). As such, we performed our analyses on 8-mo-old HFD-fed WT mice with the potential caveat of adding age as a confounding parameter (Young et al., 2016). At these time points, obese db and HFD-fed mice showed reduced BM cellularity associated with the accumulation of adipocytes in the BM (Figs. 1 B and S1 A). Consistent with previous research on the db model, we observed that obesity correlated with myeloid–lymphoid imbalance in steady-state hematopoiesis (Nagareddy et al., 2014), with (A) significant reduction of the B cell progenitor populations (Claycombe et al., 2008; Adler et al., 2014a) and (B) skewing of the myeloid-committed progenitor compartment (lineage–/c-Kit+/Sca-1–; LK) toward the granulocyte–macrophage progenitors (GMPs) at the expense of the megakaryocyte–erythroid progenitors (unpublished data; Nagareddy et al., 2014). Of note, these results appear milder than previously reported in the HFD model (unpublished data; Luo et al., 2015), suggesting that environmental factors might modulate the immune phenotype of this model.
Figure 1.
HSCs maintain a highly quiescent state in obesity. (A) Kinetics of weight gain in genetic (db) and dietary (HFD) mouse models of obesity. db, n = 5; HFD, n = 8. (B) BM cellularity of 4-mo-old db mice compared with Ctrl littermates. n = 13/group. Images show H&E staining of the BM section from Ctrl and db mice (n = 3). Arrowheads indicate BM adipocytes. Bars, 100 µm. (C) FACS plots of HSPC populations in the BM of 4-mo-old Ctrl and db mice (n = 6). Right panels show strategies used to position CD34– and CD49b– gates. (D) Mean percentages (left) and absolute numbers (right) ± SD of HSPC populations in the BM of 4-mo-old Ctrl and db mice. n = 13/group. (E and F) Phenotypic definition and mean percentages ± SD of HSC subsets in the BM of 4-mo-old Ctrl and db mice. n = 6/group. (G) Representative FACS plots (left) and mean percentages ± SD (right) of Ctrl and db SLAM HSC distribution in cell cycle phases. n = 4/group. Two independent experiments. (H) qRT-PCR analyses for Cdkn1a, Cdkn1b, and Cdkn1c gene expression in SLAM HSCs isolated from 4-mo-old db mice. Results are expressed as fold change ± SD relative to Ctrl SLAM HSCs. n = 12 pools of 100 cells. Student’s t test; *, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤ 0.0005. Two independent experiments.
We then analyzed the BM immature lineage–/c-Kit+/Sca-1+ (LSK) compartment, subfractionated based on the expression of the signaling lymphocyte activation molecules (SLAM), CD150 and CD48 (Fig. 1 C; Kiel et al., 2005). In db mice, we noticed a slight expansion of the LSK compartment caused by the increase of the short-term LSK CD48+ CD150+/− multipotent progenitors (MPPs; Fig. 1 D). This increase was found in all myeloid and lymphoid-biased MPP subpopulations (known as MPP2/3 and MPP4, respectively), suggesting that obesity did not affect the differentiation pathways within the MPP compartments (not depicted; Pietras et al., 2015). In contrast with MPPs, the frequency of the LSK CD48−CD150+ SLAM HSCs was not significantly affected in obese mice. Overall, the absolute number of SLAM HSCs and MPPs remained unchanged in db mice despite the overall loss of BM cellularity (Fig. 1 D). Interestingly, detailed analyses based on the CD34 and CD49b markers revealed changes in the cellular subsets that compose the db SLAM HSC compartment (Fig. 1, E and F). First, we observed in db mice the expansion of the CD34–LSK CD48−CD150+ compartment that contains the most quiescent and self-renewing HSC population as opposed to the more cycling and short-term CD34+LSK CD48−CD150+ compartment (known as MPP1; Fig. 1 C; Wilson et al., 2008). We then used the CD49b marker, which was previously shown to define intermediate-term HSC potential within the CD34–HSC compartment (Wagers and Weissman, 2006; Wilson et al., 2008; Benveniste et al., 2010; Qian et al., 2016). In obesity, we found the specific expansion of the intermediary CD49b+CD34–HSC (HSCCD49b+) population, suggesting that a dysregulation of the differentiation process occurs at the transition between the quiescent HSC (HSCCD49b-) and the active MPP1 compartments (Fig. 1, C and F). Importantly, the relevance of these HSC subsets defined by the CD34 and CD49b markers was highlighted by gene expression analyses. Notably, phenotypically defined HSCCD49b– expressed high levels of the transcription factors Mecom and Ndn, which are involved in the regulation of HSC quiescence and self-renewal (Fig. S2 A; Asai et al., 2011, 2012; Kataoka et al., 2011). Conversely, MPP1 cells expressed high levels of Myc, a key regulator of HSC differentiation (Fig. S2 A; Wilson et al., 2004). HSCCD49b+ showed an intermediary phenotype for these molecular features compatible with its position in the SLAM HSC compartment. Overall, the expression levels of Mecom, Ndn, and Myc in the HSC subsets were consistent with previous studies comparing SLAM HSC subpopulations (Fig. S2 B; Cabezas-Wallscheid et al., 2014, 2017) but were not significantly affected in obesity (Fig. S2 A). In contrast, obesity was associated with an increased quiescence of the overall SLAM HSC compartment as indicated by intracellular Hoechst/pyronin Y staining (Fig. 1 G) and an increase in the percentage of side population LSK cells with high dye efflux capacity (not depicted; Goodell et al., 1996; Arai et al., 2004). These changes in quiescence status were consistent with the modification in the cellular structure of the SLAM HSC compartment in obesity and particularly with the expansion of the CD34– fraction. In addition, in vitro tracking of cell division kinetics suggests that obesity could also affect the intrinsic properties of these subsets. As expected, these assays showed different division kinetics between HSC subsets (Fig. S2 C, left). They also uncovered that obesity promotes a slight delay (∼4 h) in the onset of the first division of the MPP1 cells, compatible with an increased quiescence of this subset (Fig. S2 C, right). Finally, increased quiescence in the whole SLAM HSC compartment was correlated with a specific up-regulation of Cdkn1a, a cyclin-dependent kinase inhibitor previously shown to regulate HSC stress response (Fig. 1 H; Cheng et al., 2000; van Os et al., 2007).
The BM LSK compartment in the dietary mouse model recapitulated some of the phenotypic characteristics found in the db model. Notably, we observed a significant expansion of the MPP compartment in HFD-fed mice (Fig. S1, B and C). However, consistent with their age (8 mo), SLAM HSCs showed increased frequency and higher CD150 expression in both chow diet (CD)- and HFD-fed mice (Rossi et al., 2005; Beerman et al., 2010). We also observed an expansion of the CD34– SLAM HSC population in both conditions, a phenotype precluding the analysis of the HSC subsets defined by the CD49b marker (Fig. S1 D; Beerman et al., 2010). Altogether, our results recapitulate previous studies that describe the effects of obesity on hematopoietic lymphoid- and myeloid-committed progenitors (Adler et al., 2014a; Nagareddy et al., 2014). These analyses indicate that obesity does not dramatically change the overall size of the SLAM HSC compartment. However, in the genetic model, obesity is associated with changes in the cellular structure and the functional properties of this compartment that together lead to an overall increased quiescence.
Obesity promotes an aberrant HSC response to stress
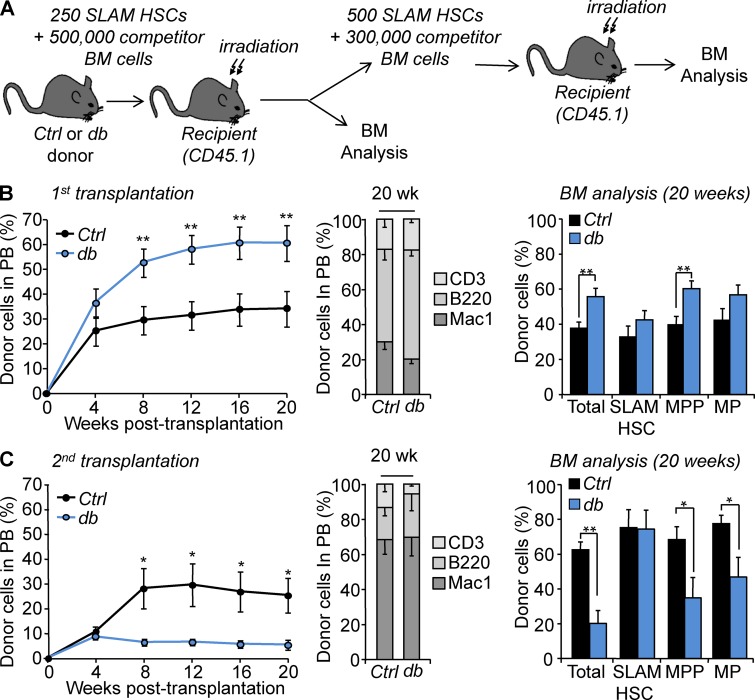
To explore the functional consequences of these changes in vivo, we performed competitive transplantation assays using purified SLAM HSCs isolated from Ctrl or db mice (Fig. 2 A). Analysis of the peripheral blood (PB) showed a dramatically increased chimerism in recipient mice transplanted with db SLAM HSCs (Fig. 2 B, left). Engraftment was stable over a 5-mo period with myeloid and lymphoid cell production indistinguishable from the controls (Fig. 2 B, middle). BM analyses at 5 mo after transplantation did not reveal any major differences between groups, notwithstanding an increase of chimerism in the db MPP compartment that mimicked the phenotype observed in primary obese mice (Fig. 2 B, right). Donor-derived db SLAM HSCs used for secondary competitive transplantation showed sustained but dramatically reduced reconstitution ability (Fig. 2 C, left). Analyses at 5 mo after transplantation showed reduced chimerism in all BM hematopoietic compartments with the exception of the SLAM HSC compartment, which displayed a high degree of chimerism similar to controls (Fig. 2 C, right). At this time point, Ctrl and db SLAM HSCs displayed similar high CD150 levels (not depicted) and similar skewing toward the production of myeloid cells (Ctrl, 67.3 ± 8.2%, versus db, 68.4 ± 10.4%; Fig. 2 C, middle; Beerman et al., 2010). Similar but milder results were obtained with the 8-mo-old dietary mouse model (Fig. S3 A). Similar to the db condition, SLAM HSCs isolated from HFD-fed mice displayed a hyperactive phenotype in primary transplantations and enhanced MPP production (Fig. S3 B). In contrast, no differences were observed in secondary transplantations as CD and HFD SLAM HSCs showed similar low engraftment (Fig. S3 C). Altogether, these results demonstrate that obesity affects the ability of HSCs to respond to the hematopoietic stresses associated with transplantation. These results show that obesity drives an aberrant biphasic HSC stress response that includes enhanced reconstitution ability in primary recipients and functional exhaustion after serial transplantation. Collectively, these results demonstrate that obesity triggers cell-intrinsic mechanisms that affect the long-term fitness of the HSC compartment.
Figure 2.
Functional impact of obesity on HSCs. (A) Experimental scheme for serial transplantation assay. (B and C) Hematopoietic reconstitution in primary and secondary recipients. The left graph indicates PB chimerism over time. The middle graph shows myeloid and lymphoid PB chimerism 20 wk after transplantation. The right graph shows the percentage of donor-derived total BM cells, SLAM HSCs, MPPs, and myeloid progenitors (MPs) 20 wk after transplantation. Results are expressed as means ± SEM. n = 8 and 4. Student’s t test; *, P ≤ 0.05; **, P ≤ 0.01. Four and two independent experiments, respectively.
Obesity is associated with up-regulation of Gfi1 expression in HSCs
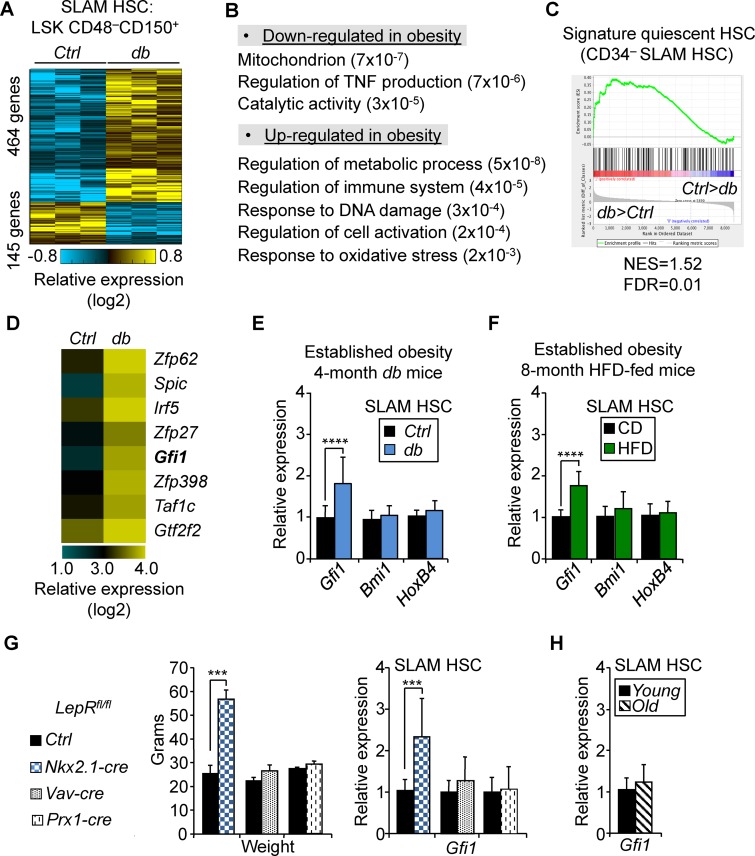
We next investigated the molecular mechanisms that could contribute to the abnormal function of the HSC compartment in obesity. We performed genomewide gene expression analysis on 20,000 SLAM HSCs isolated from 4-mo-old Ctrl and db mice (Fig. 3 A and Table S3). Gene set enrichment analyses (GSEAs) indicated a specific down-regulation in the obesity of genes associated with metabolic processes (e.g., mitochondrion and generation of metabolite precursor and energy), a set of functions potentially related to their increased quiescent status (Fig. 3 B). In parallel, we observed the up-regulation of genes linked with several stress-response pathways (e.g., regulation of immune system process, response to DNA damage stimulus, regulation of cell activation, and response to oxidative stress), suggesting that HSCs were sensitive to some of the environmental stresses associated with obesity. Consistent with their cell surface phenotype (Fig. 1 C), db SLAM HSCs were also significantly enriched for the gene signature associated with CD34– SLAM HSCs (Fig. 3 C; Qian et al., 2016). We then focused our analysis on hematopoietic-relevant transcription factors (Rossi et al., 2012) whose expression was affected in obesity. Among the transcriptional factors up-regulated in obesity (Fig. 3 D), we noted Gfi1, a transcriptional repressor known to regulate HSC quiescence and self-renewal (Hock et al., 2004; Zeng et al., 2004). Gfi1 has been previously described as being highly expressed in the most quiescent CD34– SLAM HSC subset (Cabezas-Wallscheid et al., 2014), a compartment that we found expanded in obesity (Fig. 1 F). We confirmed by quantitative RT-PCR (qRT–PCR) analyses the higher level of Gfi1 expression in SLAM HSCs isolated from the db and dietary mouse models (Fig. 3, E and F). Increased Gfi1 expression in SLAM HSCs was also found in 4-mo-old obese mice with neuron-specific Lepr deletion (denoted Nkx2.1-cre::Leprfl/fl; Ring and Zeltser, 2010) but not in SLAM HSCs isolated from 4-mo-old nonobese mice carrying hematopoietic or BM stromal cell–specific Lepr deletions (denoted Vav1-cre::Leprfl/fl and Prx1-cre::Leprfl/fl, respectively; Fig. 3 G; Ding and Morrison, 2013; Zhou et al., 2014). As such, these results further show that Gfi1 up-regulation in HSCs is correlated with obesity and not linked to the disruption of the leptin-signaling pathway in the hematopoietic cells or the BM microenvironment. In addition, Gfi1 up-regulation was not observed in 2-yr-old control mice (Fig. 3 H), confirming that this feature is not part of an aging gene signature (Rossi et al., 2005; Chambers et al., 2007).
Figure 3.
Obesity is associated with up-regulation of Gfi1 expression in HSCs. (A) Differential gene signature in SLAM HSCs isolated from in 4-mo-old Ctrl and db mice. (B) GSEA for dysregulated genes in db SLAM HSCs. Numbers in parentheses indicate adjusted p-values. (C) Enrichment of quiescent CD34– SLAM HSC gene signature (Qian et al., 2016). FDR, false discovery rate; NES, normalized enrichment score. (D) Heat map showing examples of differentially expressed transcription factors in Ctrl and db SLAM HSCs (ordered by db/Ctrl expression ratio). (E and F) qRT-PCR analyses for Gfi1, Bmi1, and HoxB4 gene expression in SLAM HSCs isolated from 4-mo-old db mice (E) or from 8-mo-old HFD-fed mice (F). Results are expressed as fold change ± SD relative to their respective controls. (G) Body weight (left) and qRT-PCR analyses for Gfi1 expression in SLAM HSCs (right) of Nkx2.1-cre::Leprfl/fl, Vav1-cre::Leprfl/fl, and Prx1-cre::Leprfl/fl mice. Body weights are expressed as means ± SD (n = 3–5). qRT-PCR results are expressed as fold change ± SD relative to their respective controls. Student’s t test; ***, P ≤ 0.005; ****, P ≤ 0.0005. (H) qRT-PCR analyses for Gfi1 gene expression in old SLAM HSCs (20–24 mo). Results are expressed as fold change ± SD relative to young controls. n = 18 pools of 100 cells. Three independent experiments.
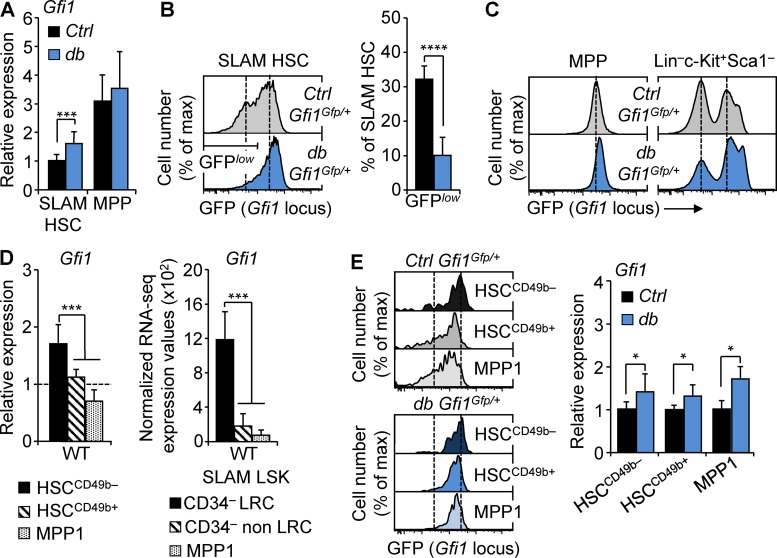
Further analyses in the db model showed that Gfi1 up-regulation in obesity was specific to SLAM HSCs and did not occur in the downstream MPP compartment (Fig. 4 A). To expand this result, we used a knock-in Gfi1 reporter mouse carrying a GFP cassette inserted in the endogenous Gfi1 locus, allowing for the monitoring of the activity of this locus at the single cell level (Yücel et al., 2004). We crossed these mice with Ctrl or db mice to generate two models (hereafter denoted Ctrl::Gfi1Gfp/+ and db::Gfi1Gfp/+) monitoring the impact of obesity on the Gfi1 locus. In the Ctrl::Gfi1Gfp/+ model, SLAM HSCs showed a heterogeneous low-to-intermediary level of GFP expression positioned between the nonexpressing megakaryocyte–erythroid progenitors and the high-expressing GMP populations found in the LSK progenitors (Fig. 4, B and C). db::Gfi1Gfp/+ mice showed similar weight gain to the parental db mice (not depicted). Consistent with our gene expression analyses, we found that obesity in this model promotes an increase of GFP expression in SLAM HSCs but not in MPPs (Fig. 4, B and C). A similar phenotype was found in Ctrl::Gfi1Gfp/+ mice fed with HFD for 8 mo, further confirming the link between obesity and the activity of the Gfi1 locus in HSCs (Fig. S4, A and B). Consistent with previous research (Cabezas-Wallscheid et al., 2017), we found that Gfi1 was highly expressed in the most quiescent HSC subset (Fig. 4 D). However, Gfi1 up-regulation in obesity was not simply a result of change in SLAM HSC composition, as both GFP detection in reporter mice and gene expression analyses showed that obesity significantly affects Gfi1 expression in all HSC subsets (Fig. 4 E). Altogether, these analyses indicate a correlation between obesity and the up-regulation of Gfi1 in HSCs in three distinct mouse models. They identify two cooccurring effects of obesity: one affecting the architecture of the SLAM HSC compartment (with a relative increase of HSCCD49+/− at the expense of MPP1 cells) and the other affecting the level of Gfi1 expression in all subsets composing this compartment. As such, our results indicate that Gfi1 up-regulation is a cell-intrinsic hallmark of the HSCs in obesity and suggest that this molecular feature might contribute to the long-term effect of obesity on the HSC compartment.
Figure 4.
Gfi1 up-regulation in obesity affects all HSC subsets. (A) qRT-PCR analyses for Gfi1 gene expression in SLAM HSCs and MPPs isolated from 4-mo-old Ctrl and db mice. Results are expressed as fold change ± SD relative to Ctrl SLAM HSCs. n = 12 pools of 100 cells. (B) Representative FACS plots showing GFP fluorescence in SLAM HSCs of 4-mo-old Ctrl::Gfi1Gfp/+ and db::Gfi1Gfp/+ mice. Graph indicates mean percentages ± SD of GFPlow SLAM HSCs. n = 4/group. (C) Representative FACS plots showing GFP fluorescence in MPPs (left) and in LK progenitors (right) of Ctrl::Gfi1Gfp/+ and db::Gfi1Gfp/+ mice. n = 4/group. (D) qRT-PCR analyses for Gfi1 gene expression in HSC subsets isolated from WT mice. Results are expressed as fold change ± SD relative to total SLAM HSCs. n = 6 pools of 100 cells. The right panel shows mean RNA-seq expression for Gfi1 transcripts in dormant long-term label-retaining cell (LRC) HSCs, active non-LRC HSCs, and MPP1 cells (n = 3; Cabezas-Wallscheid et al., 2017). (E) Representative FACS plots showing GFP fluorescence in HSC subsets of 4-mo-old Ctrl::Gfi1Gfp/+ and db::Gfi1Gfp/+ mice. n = 3/group. The right panel shows qRT-PCR analyses for Gfi1 gene expression in HSC subsets isolated from db mice. Results are expressed as fold change ± SD relative to their respective controls. n = 6 pools of 100 cells. Student’s t test (A, B, and E) or one-way ANOVA with Tukey’s post hoc test (D); *, P ≤ 0.05; ***, P ≤ 0.0005; ****, P ≤ 0.00005. Two independent experiments.
Obesity has a progressive but long-lasting impact on the HSC compartment
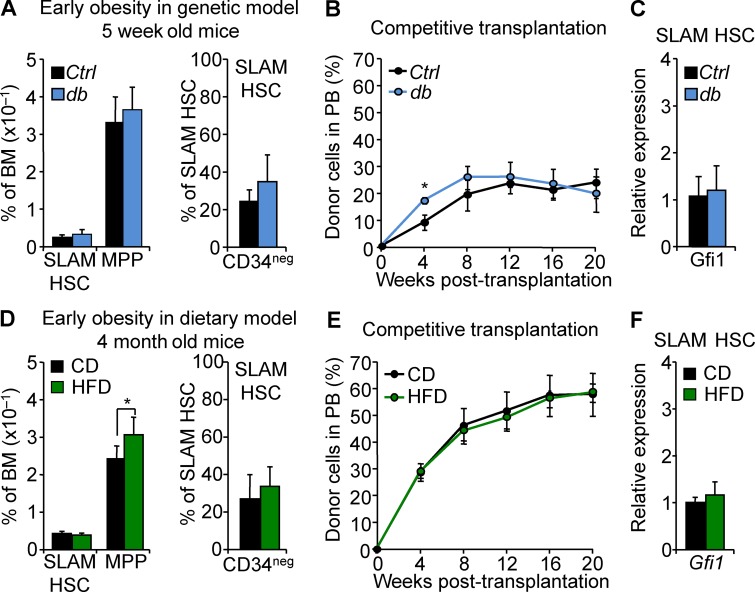
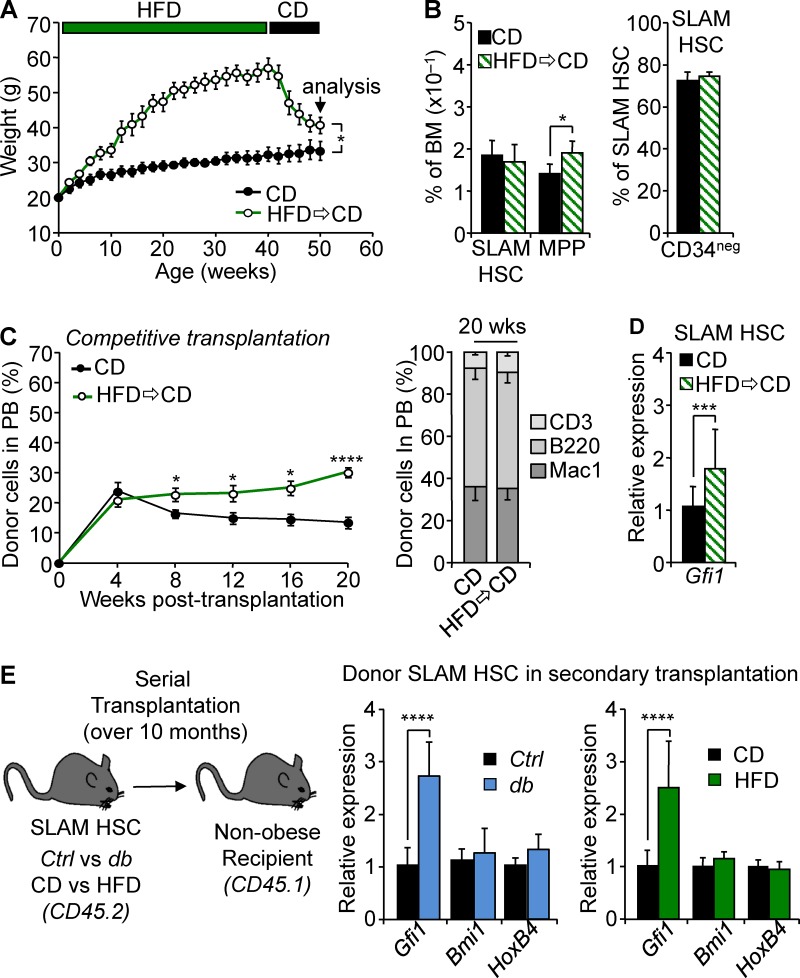
Our results showed that prolonged exposure to obesity affects the HSC compartment and that this effect could be molecularly associated with Gfi1 up-regulation. To reinforce this link, we next sought to establish the conditions of acquisition for these functional and molecular characteristics. We first examined the HSC compartment at obesity onset in 5-wk-old db mice or in 4-mo-old HFD-fed mice (Fig. 1 A). In both models, we found that the HSPC compartment isolated in the early stage of obesity showed minimal phenotypic disruption (Fig. 5, A and D). This was associated with a normal SLAM HSC function in competitive transplantation assays (Fig. 5, B and E). Finally, at obesity onset, we found that SLAM HSCs display levels of Gfi1 expression indistinguishable from those of controls (Fig. 5, C and F). Next, we sought to determine whether the effect of obesity was reversible by analyzing the consequences of dietary reversion on HSCs. WT mice were fed with HFD for 10 mo to induce obesity before being switched to CD. This diet reversion dramatically impacted the overall mouse weight, leading toward weight normalization (Fig. 6 A). Although mouse weight dramatically changed in these conditions, hematological parameters did not reverse even 10 wk after reintroduction of regular chow. As expected for their age (50 wk; Beerman et al., 2010), SLAM HSCs in these mice showed increased frequency and low CD34 expression (Fig. 6 B). Despite this phenotype, we still observed an expansion of the MPP compartment, one of the hallmarks of the LSK compartment in obesity. In addition, SLAM HSCs isolated from the mice after diet reversion maintained a hyperactive phenotype in competitive transplantation assays (Fig. 6 C). Interestingly, this abnormal function was once again associated with up-regulation of Gfi1 expression (Fig. 6 D), suggesting that a high Gfi1 level is a stable cell-intrinsic property of these HSCs. To reinforce this latter result, we analyzed the SLAM HSCs isolated from db and HFD conditions and transplanted into nonobese congenic recipient mice. Strikingly, we found that up-regulation of Gfi1 expression associated with obesity in primary db and HFD-fed mice was maintained in HSCs even after these cells were serially transplanted into a nonobese environment for an extended period of time (Fig. 6 E). Altogether, these results indicate that obesity promotes a progressive change in the HSC compartment during the establishment of obesity but also that this cell-intrinsic effect is long lasting and can persist upon weight loss. These data show that up-regulation of Gfi1 expression in the HSC compartment, once acquired, is a stable feature that correlated with their aberrant activity upon transplantation.
Figure 5.
Progressive effect of obesity on the HSC compartment. (A) Mean percentages ± SD of SLAM HSC/MPPs (left) and CD34– SLAM HSCs (right) in the BM of 5-wk-old Ctrl and db mice. n = 3–4/group. (B) PB chimerism in competitive reconstitution assays with SLAM HSCs isolated from Ctrl and db 5-wk-old mice. Results are expressed as means ± SEM. n = 4/group. (C) qRT-PCR analyses for Gfi1 in SLAM HSCs isolated from 5-wk-old db mice. Results are expressed as fold change ± SD relative to Ctrl SLAM HSCs. n = 12 pools of 100 cells. (D) Mean percentages ± SD of SLAM HSC/MPPs (left) and CD34– SLAM HSCs (right) in the BM of 4-mo-old mice fed with HFD. n = 8/group. Student’s t test; *, P ≤ 0.05. (E) PB chimerism in competitive reconstitution assays with SLAM HSCs isolated from 4-mo-old mice fed with HFD. Results are expressed as means ± SEM. n = 4/group. (F) qRT-PCR analyses for Gfi1 in SLAM HSCs isolated from 4-mo-old mice fed with HFD. Results are expressed as fold change ± SD relative to CD SLAM HSCs. n = 12 pools of 100 cells. Two independent experiments.
Figure 6.
Long-lasting effect of obesity on the HSC compartment. (A) Kinetics of weight variation after diet change. Data are represented as means ± SD. n = 4/group. (B) Mean percentages ± SD of SLAM HSC/MPPs (left) and CD34– SLAM HSCs (right) in the BM of mice constantly fed with CD or experiencing diet change. n = 4/group. (C) PB chimerism in competitive reconstitution assays with SLAM HSCs isolated from mice constantly fed with CD or experiencing diet change. The right graph shows myeloid and lymphoid PB chimerism 20 wk after transplantation. Results are expressed as means ± SEM. n = 11–12/group. (D) qRT-PCR analyses for Gfi1 gene expression in SLAM HSCs isolated from mice experiencing diet change. Results are expressed as fold change ± SD relative to controls. n = 16 pools of 100 cells. (E) qRT-PCR analyses for Gfi1, Bmi1, and HoxB4 genes in db (left) and HFD SLAM HSCs (right) after serial transplantations. Results are expressed as fold change ± SD relative to their respective controls. n = 12 pools of 100 cells. Student’s t test; *, P ≤ 0.05; ***, P ≤ 0.005; ****, P ≤ 0.0005. Two independent experiments.
Gfi1 up-regulation mediates HSC-functional dysregulations in obesity
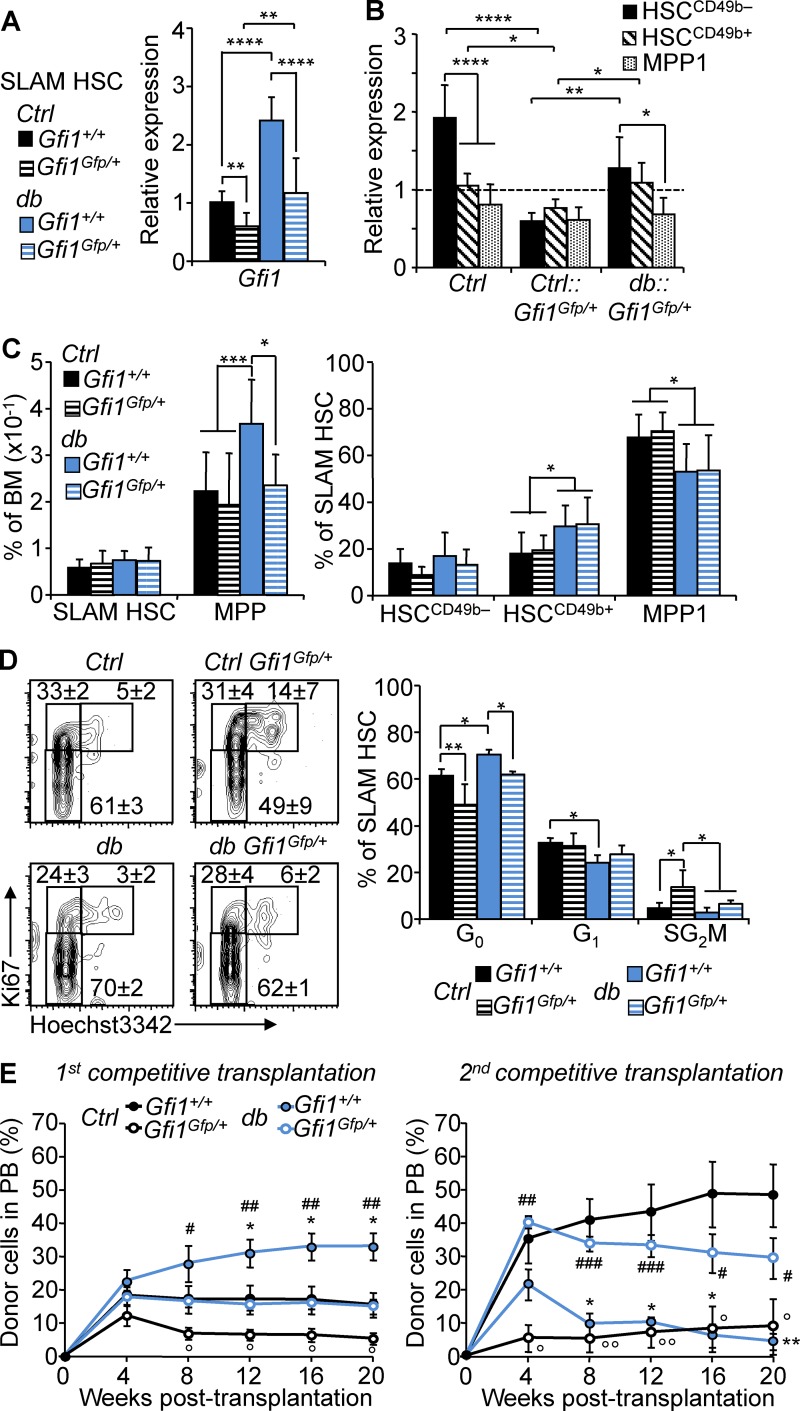
To determine whether Gfi1 up-regulation directly affects the function of HSCs in obesity, we used a loss-of-function approach with the aforementioned Ctrl::Gfi1Gfp/+ and db::Gfi1Gfp/+ reporter mice in which GFP insertion leads to the disruption of one Gfi1 allele (Yücel et al., 2004). In nonobese conditions, we found that Gfi1 haploinsufficiency led to reduced Gfi1 expression in SLAM HSCs, a result consistent with published studies in other cellular contexts (Fig. 7, A and B; Yücel et al., 2004; Fraszczak et al., 2016; Hönes et al., 2016). Haploinsufficiency in the obese condition blunted the Gfi1 up-regulation and led to a normalization of Gfi1 expression in the overall SLAM HSC compartment and in the subsets that compose this compartment. Modulation of Gfi1 expression in obesity normalized the size of the MPP compartment but failed to affect the obesity-associated phenotype in SLAM HSCs (Fig. 7 C). In contrast, we observed that Gfi1 haploinsufficiency affects SLAM HSC cell cycle status by reducing quiescence both in the Ctrl and db background (Fig. 7 D). Particularly, Gfi1 haploinsufficiency led to a normalization of the SLAM HSC cell cycle status in obesity. Alteration of Gfi1 gene dosage also correlated with the normalization of the expression of the cell cycle regulator Cdkn1a (p21) but not Cdkn1c (p57; Fig. S5). Using competitive transplantation assays (as shown in Fig. 2 A), we then assessed the impact of Gfi1 haploinsufficiency on the HSC function (Fig. 7 E). Ctrl::Gfi1Gfp/+ SLAM HSCs displayed a significantly reduced reconstitution ability compared with Ctrl SLAM HSCs in both primary and secondary transplantation. In obesity, Gfi1 haploinsufficiency also affected the SLAM HSC activity, leading to the normalization of their reconstitution capability. Notably, we found that Gfi1 haploinsufficiency reversed the hyperactive phenotype of the db SLAM HSCs in primary transplantation as well as the associated functional exhaustion detected in secondary transplantation. Altogether, these results show that Gfi1 haploinsufficiency does not affect the phenotypic characteristics of the db SLAM HSC compartment but normalizes its function (as measured by cell cycle status and reconstitution capability). Therefore, these results indicate that a tight control of Gfi1 expression is critical to HSC function, particularly during stress, and show that Gfi1 is one of the key determinants of HSC function in obesity.
Figure 7.
Gfi1 controls HSC fate in obesity. (A) qRT-PCR analyses for Gfi1 gene expression in SLAM HSCs isolated from 4-mo-old db, Ctrl::Gfi1Gfp/+, and db::Gfi1Gfp+ mice. Results are expressed as fold change ± SD relative to total Ctrl SLAM HSCs. n = 6–12 pools of 100 cells. Student’s t test; **, P ≤ 0.01; ****, P ≤ 0.0005. Three independent experiments. (B) qRT-PCR analyses for Gfi1 gene expression in HSC subsets isolated from 4-mo-old Ctrl, Ctrl::Gfi1Gfp/+, and db::Gfi1Gfp/+ mice. Results are expressed as fold change ± SD relative to total Ctrl SLAM HSCs. n = 6–12 pools of 100 cells. Two-way ANOVA with Tukey’s post hoc test; *, P ≤ 0.05; **, P ≤ 0.005; ****, P ≤ 0.0001. Two independent experiments. (C) Mean percentages ± SD of SLAM HSC/MPPs (left) and HSC subsets (right) in the BM of 4-mo-old Ctrl, db, Ctrl::Gfi1Gfp/+, and db::Gfi1Gfp+ mice. n = 8–18. (D) Representative FACS plots (left) and mean percentages ± SD (right) showing SLAM HSC distribution in cell cycle phases. n = 4–6 mice/group. One-way ANOVA with Tukey’s post hoc test; *, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤ 0.001. Two independent experiments. (E) PB chimerism in primary (left) and secondary (right) competitive reconstitution assays using SLAM HSCs isolated from Gfi1Gfp/+ and db compound mice. Results are expressed as means ± SEM. n = 4–8/group. Student’s t test; Ctrl vs. db: *, P ≤ 0.05; **, P ≤ 0.005; Ctrl vs. Ctrl::Gfi1Gfp/+: °, P ≤ 0.05; °°, P ≤ 0.05; db vs. db::Gfi1Gfp/+: #, P ≤ 0.05; ##, P ≤ 0.005; ###, P ≤ 0.0005. Two independent experiments.
Oxidative stress is a driver for HSC dysregulation in obesity
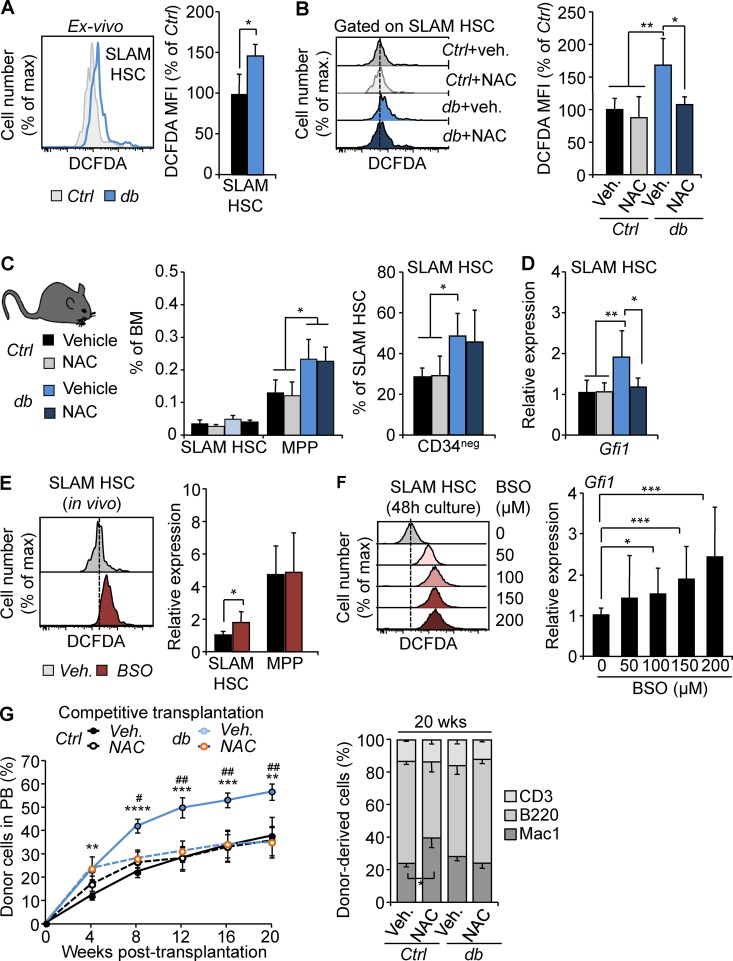
Although Gfi1 haploinsufficiency normalizes the db HSC functions, we found that it also negatively impacts the Ctrl HSCs. To rule out a potential nonspecific effect of Gfi1 haploinsufficiency, we investigated the upstream mechanisms of Gfi1 regulation in obesity. We reasoned that the initial activation of the Gfi1 locus could be driven by cellular stresses associated with obesity. We found that SLAM HSCs isolated from 4-mo-old db mice displayed a significantly increased level of reactive oxygen species (ROS; Fig. 8 A). As expected, the treatment of db mice with the antioxidant N-acetyl cysteine (NAC; 60 mM in drinking water administered ad libitum for 4 wk) during the establishment of the obese phenotype (3–4 mo of age) led to a normalization of the ROS level (Fig. 8 B). NAC treatment during this limited period did not affect the expansion of the db MPP compartment or the phenotype of the db SLAM HSCs (Fig. 8 C). In contrast, removal of this oxidative stress led to the normalization of Gfi1 expression in db SLAM HSCs (Fig. 8 D), suggesting that Gfi1 expression could be regulated by oxidative stress in HSCs. Consistent with this idea, we found that in vivo induction of ROS in WT mice using buthionine sulfoximine (BSO) treatment (Smith et al., 1989) led to the up-regulation of Gfi1 expression in SLAM HSCs but not in the downstream MPP compartment (Fig. 8 E). A similar effect was found when purified SLAM HSCs were treated in vitro with BSO (Li et al., 2003; Ito et al., 2006), indicating a direct and dose-dependent effect of ROS on Gfi1 expression in HSCs (Fig. 8 F). Finally, we found that normalization of Gfi1 expression in obesity after NAC treatment fully rescues the HSC hyperactive activity upon transplantation (Fig. 8 G). Importantly, NAC treatment did not disrupt Gfi1 expression in Ctrl HSCs and did not affect their function upon transplantation. Altogether, these results fully mimic those obtained with Gfi1 haploinsufficient mice and therefore independently confirm the link between Gfi1 up-regulation and HSCs’ aberrant function in obesity. These results indicate that oxidative stress is a key contributor to the initial dysregulation of the Gfi1 locus during the establishment of obesity and identify Gfi1 as an oxidative stress-response gene in HSCs.
Figure 8.
Oxidative stress is a key driver of HSC dysregulation in obesity. (A) Representative FACS plots (left) and mean fluorescence intensity (MFI) ± SD (right) showing ROS level detected by 2’,7’–dichlorofluorescin diacetate (DCFDA) staining in SLAM HSCs isolated from 4-mo-old Ctrl and db mice. n = 3/group. Student’s t test; *, P ≤ 0.05. Two independent experiments. (B) Representative FACS plots (left) and mean fluorescence intensity ± SD (right) showing ROS level in SLAM HSCs isolated from NAC-treated Ctrl and db mice. n = 4–7/group. (C) Mean percentages ± SD of SLAM HSC/MPPs (left) and CD34– SLAM HSCs (right) in the BM isolated from NAC-treated Ctrl and db mice. n = 5/group. (D) qRT-PCR analyses for Gfi1 gene expression in SLAM HSCs isolated from NAC-treated Ctrl and db mice. Results are expressed as fold change ± SD relative to vehicle-treated Ctrl SLAM HSCs. n = 6–12 pools of 100 cells. One-way ANOVA with Tukey's post hoc test; *, P ≤ 0.05; **, P ≤ 0.005. Two independent experiments. (E) Representative FACS plots (left) and qRT-PCR analyses (right) showing ROS level and Gfi1 gene expression in SLAM HSCs isolated from BSO-treated WT mice. Results are expressed as fold change ± SD relative to vehicle-treated mice. n = 6–12 pools of 100 cells. Student’s t test; *, P ≤ 0.05. Two independent experiments. (F) Representative FACS plots (left) and qRT-PCR analyses (right) showing ROS level and Gfi1 gene expression in SLAM HSCs cultured for 48 h in presence of BSO. Results are expressed as fold change ± SD relative to vehicle-treated mice. n = 5–9 pools of 100 cells. One-way ANOVA with Dunnett’s post hoc test; *, P ≤ 0.05; ***, P = 0.0005. Three independent experiments. (G) PB chimerism in competitive reconstitution assays using SLAM HSCs isolated from vehicle- or NAC-treated Ctrl and db mice. The right graph shows myeloid and lymphoid PB chimerism 20 wk after transplantation. Results are expressed as means ± SEM. n = 7/group. Student’s t test; Ctrl + Veh. vs. db + Veh.: **, P ≤ 0.005; ***, P ≤ 0.0005; ****, P ≤ 0.00005; db + Veh. vs. db + NAC: #, P ≤ 0.01; ##, P ≤ 0.005. Two independent experiments.
Discussion
Using independent mouse models, our results demonstrate that obesity has a durable effect on the HSC compartment. Although HSCs in obesity only display limited phenotypic and functional perturbations at steady-state, they show an exacerbated proliferative response to hematopoietic stresses along with intermediate-term hematopoietic reconstitution capability. Mechanistically, we found that this aberrant HSC activity is linked to the acquisition of specific molecular features, particularly the up-regulation of Gfi1 expression. We show that these molecular and functional abnormal HSC features are progressively acquired during the establishment of obesity but remain stable even when the stresses associated with obesity are relieved after weight loss or transplantation into a normal recipient. Finally, we demonstrate that Gfi1 is one of the key regulators of HSC fate in obesity in response to the abnormal oxidative environment associated with this condition. In sum, obesity produces sustained changes in HSC function and phenotype that are mirrored at the molecular level by durable elevation in Gfi1 expression, which is genetically linked to the changes in HSC function but not phenotype.
Although obesity and aging share many common immunological, metabolic, and hormonal characteristics (Adler et al., 2014b; Akunuru and Geiger, 2016), our results draw a sharp contrast in the impact of these two conditions on the HSC compartment. Although aging is associated with the accumulation of a myeloid-biased CD150high HSC population with a low capacity for hematopoietic reconstitution (Rossi et al., 2005; Beerman et al., 2010), none of these characteristics were found in obesity. In contrast, our results show that obesity promotes changes in the cellular architecture of the HSC compartment and lead to an exacerbated proliferative response upon transplantation. Consistent with this idea, the HSC gene expression profile associated with obesity is different from the gene signatures previously associated with aging (Rossi et al., 2005; Chambers et al., 2007). Notably, we identify the up-regulation of Gfi1 as a specific HSC characteristic in obesity that it is not found in aging. As such, our results indicate that the effect of obesity on the HSC compartment is distinct from premature aging. Similarly, our data indicate that the effects of obesity on the HSC compartment do not mimic the effects of acute and chronic inflammatory conditions, which are associated with HSC cell cycle activation and rapid functional exhaustion (Essers et al., 2009; Baldridge et al., 2010; Esplin et al., 2011). In this study, we show that HSCs in obesity acquired an immature CD34neg phenotype and remain quiescent. Although inflammatory signals present in obesity have been shown to impact the downstream GMP compartment (Nagareddy et al., 2014), these results suggest that HSCs could be refractory to the low-grade inflammation associated with obesity. We can speculate that this property is caused by a higher threshold of activation or the presence of cell-intrinsic protective mechanisms activated in obesity (see below). In any case, our results indicate that the effect of obesity on the HSC compartment cannot be solely reduced to its systemic inflammatory components. In contrast, they establish obesity as a singular chronic pathophysiological state. The conditions that are actually responsible for the dysregulation of the HSC compartment in obesity remain to be fully established. Of note, the phenotype, the quiescence status, and the reconstitution pattern described for HSCs in obesity are reminiscent of the characteristics found in HSCs isolated from BM adipocyte-rich regions (Naveiras et al., 2009). This suggests that changes in the adipose tissues in the BM could be among the major drivers of the HSC phenotype in obesity (Ambrosi et al., 2017).
Our work describes a novel ROS–Gfi1 pathway linking oxidative stress response and mechanisms controlling HSC fate in obesity. Although obesity is associated with an increased level of intracellular ROS in HSCs, we found that the functional consequences of this phenomenon are mild in obesity compared with previous research, which describes ROS inducing a rapid HSC exhaustion through induction of the p38 mitogen-activated protein kinase (Ito et al., 2006). This could suggest that the level of intracellular ROS present in obesity may not be sufficient to fully activate this pathway. Alternatively, it could also indicate that the progressive acquisition of the oxidative stress during the establishment of obesity leads to specific adaptive response mechanisms in HSCs. In this study, we show that the oxidative stress associated with obesity directly impacts on Gfi1 expression in HSCs but not in downstream MPPs. Gfi1 expression is tightly controlled in HSCs and is a key regulator of HSC quiescence as Gfi1-knockout HSCs fail to remain quiescent and undergo rapid functional exhaustion (Hock et al., 2004; Zeng et al., 2004). We found that Gfi1 haploinsufficiency also reduces HSC quiescence and limits their hematopoietic reconstitution ability upon transplantation. Conversely, we found that a nominally modest two- to threefold up-regulation of Gfi1 suffices to affect HSC function. As such, Gfi1 acts as a rheostat able to fine-tune the properties of the HSC compartment. As others, we found that Gfi1 expression level correlates with Cdkn1a (p21) expression in HSCs (Hock et al., 2004; Zeng et al., 2004). However, it remains unclear whether p21 expression is directly linked to Gfi1 activity and actually contributes to the HSC properties (Liu et al., 2009). In obesity, we show that Gfi1 plays a dual role contributing to reinforce HSC quiescence at steady state while also affecting their response to stress assayed in serial transplantations. Importantly, modulation of Gfi1 expression through gene dosage or antioxidant treatment is effective in restoring normal HSC functional properties. These results demonstrate that the ROS–Gfi1 axis controls the fitness of the overall HSC compartment in obesity. Consistent with this point, Gfi1 modulation during the establishment of the obese phenotype in the haploinsufficient model also prevents the expansion of the downstream MPP compartments. In contrast, Gfi1 modulation fails to reverse the HSC phenotypic features associated with obesity and, notably, the change in the proportion of the different HSC subsets. This disconnect between HSC phenotype and function suggests that other factors are controlling the transition between these different subsets but that Gfi1 up-regulation is dominant in modulating the overall activity of these subsets. Further studies will be needed to determine whether the ROS–Gfi1 axis has a differential impact on each of the HSC subsets. Altogether, our work highlights the unique importance of Gfi1 in HSC biology and the need to decipher its downstream regulatory programs in this compartment. It also indicates that deciphering the molecular pathway linking oxidative stress response to Gfi1 expression could be key to understanding the dysregulation of HSC functions in chronic stress conditions.
Finally, our work establishes that some of the alterations in the HSC compartment were long lasting after weight loss or transplantation into a normal environment. Thus, the status of the HSCs during established obesity dramatically differs from the reversible HSC activation observed upon acute BM injury or granulocyte colony-stimulating factor (G-CSF) treatment (Wilson et al., 2008). Molecularly, these changes in the HSC compartment were associated with and supported by the activation of the Gfi1 locus. Although progressively acquired and initially dependent on obesity-induced ROS level during obesity establishment, Gfi1 up-regulation persists even as HSCs are transferred into a normal environment, suggesting that obesity also promotes stable alterations in the Gfi1 locus. These results are reminiscent of the chromatin accessibility and epigenetic changes induced by HFD in liver tissue, and they suggest that epigenetic alterations could also contribute to the change in function of the HSC compartment in obesity (Leung et al., 2014, 2016). Altogether, our results indicate that obesity is associated with an alteration of the regulatory programs in the different HSC pools along with ROS accumulation and dysregulation of the stress response. Interestingly, the same alterations have been linked to age-related hematological pathologies (Akunuru and Geiger, 2016). In the clinic, obesity is associated with multiple hematological disorders ranging from immunodeficiency (Falagas and Kompoti, 2006; Huttunen and Syrjanen, 2013) to increased susceptibility to hematological malignancies (Pan et al., 2004; Reeves et al., 2007; Parr et al., 2010; Bhaskaran et al., 2014). Future studies will need to address the relevance of the HSC alterations on the fitness of the hematopoietic system in obese patients and their potential contribution to the development of hematopoietic pathologies.
Materials and methods
Mice
WT, Leprdb/db(db), LepRfl/fl, Vav-cre, Nkx2-1-cre, Prx1-cre, and Gfi1Gfp/+ mice (all in C57BL/6 background) were purchased from The Jackson Laboratory and housed at the Association for Assessment and Accreditation of Laboratory Animal Care International–accredited animal facility of the Cincinnati Children’s Hospital Medical Center. Nonmutant littermates were used as controls for each experiment. For the dietary model of obesity, WT C57BL/6 mice were maintained in control diet with 13 Kcal% fat (5010; Lab Diets) or switched to an HFD with 60 Kcal% fat (D12492; Research Diet, Inc.) at 5 wk of age for a duration of 8 mo. 8–12-wk-old C57BL/6 congenic mice (Ptprca Pepcb/BoyJ) were used as recipients for cell transplantation experiments. All animal experiments were approved by the Cincinnati Children’s Institutional Animal Care and Use Committee.
Flow cytometry
Cell surface staining and HSC isolation procedures were performed as described previously (Reynaud et al., 2011). In brief, BM cells were obtained by crushing long bones and hips, treating with ACK (150 mM NH4Cl and 10 mM KHCO3) to lyse red blood cells, and purifying the cells by Ficoll separation (Histopaque-1119; Sigma-Aldrich). Unfractionated or autoMACS (Miltenyi Biotec)-enriched c-kit+ cells were stained with unconjugated rat lineage-specific antibodies (Lin, Ter-119, Mac1, Gr-1, B220, CD5, CD3, CD4, CD8, and CD127), followed by staining with goat anti–rat PE-Cy5 secondary antibody. Cells were then stained with c-kit–APC-eFluor780, Sca1-PB, CD48–Alexa Fluor 647, CD150-PE, CD34-FITC, CD49b-biotin/streptavidin-PE-Cy7, and FcγR-PerCP-eFluor710 antibodies. Blood- and BM-differentiated cells were stained with Ter119-PE-Cy5, CD3-APC, B220-APC-eFluor780, Mac1-PE-Cy7, and Gr-1-PB antibodies. B progenitors were stained with B220-BV421 and IgM-APC-eFluor780 antibodies. CD45.1–Alexa Fluor 700 and CD45.1-FITC antibodies were included for blood chimerism analysis and isolation of donor-derived HSCs after transplantation. All antibodies used in this study can be found in Table S1. Stained cells were resuspended in Hank’s buffered salt solution with 2% heat-inactivated FBS and 1 µg/ml propidium iodide for dead cell exclusion. For cell cycle status, surface-stained cells (Lin-PeCy5, c-kit–APC-eFluor780, Sca1-PE-Cy7, CD48-PercP-cy5.5, and CD150-APC were incubated at 37°C for 30 min with Hoechst 33342 (5 µg/ml; Molecular Probes) in the presence of verapamil (50 µg/ml; Sigma-Aldrich) and pyronin Y (0.05 µg/ml; Sigma-Aldrich). For Hoechst 33342/Ki67 staining, surface-stained cells were fixed and permeabilized using a Cytofix/Cytoperm kit (BD) before staining with a PE-conjugated anti-Ki67 antibody (BioLegend) and Hoechst 33342 (5 µg/ml). For measuring intracellular ROS levels, surface-stained cells were incubated with 1 µM 2,7-dichlorofluorescin diacetate (Sigma-Aldrich) for 15 min in a 37°C water bath before flow cytometry analysis. Cell sorting and analysis were performed on a FACS ARIAII and a FACS LSRII or LSRFortessa (BD). Data analysis was performed using FlowJo software (BD).
In vivo assays
For competitive transplantation assays, 8–12-wk-old CD45.1 C57BL/6 congenic recipient mice were lethally irradiated (11 Gy delivered in split doses 3 h apart) and retroorbitally injected with 250 or 500 purified SLAM HSCs (CD45.2+) together with 500,000 or 300,000 BoyJ whole BM cells (CD45.1+) as competitors. Transplanted mice were kept on antibiotic-containing water (neomycin sulfate/polymyxin B sulfate; Sigma-Aldrich) for 4 wk and analyzed for donor-derived chimerism by monthly bleeding. Peripheral blood was collected by retroorbital bleeding, treated with ACK, and stained with specific antibodies for flow cytometry analysis. For ROS inhibition, db and littermate control mice were given 10 mg/ml of NAC (Sigma-Aldrich) in drinking water that was changed daily for 4 wk. For in vivo glutathione depletion and induction of intracellular ROS, mice were intraperitoneally injected for 5 d with the glutathione synthesis inhibitor BSO (800 mg/kg daily; Sigma-Aldrich) or vehicle (PBS). Blood and BM phenotypes were analyzed, and HSCs were purified for molecular and functional analysis at the end of treatment.
In vitro assays
Purified SLAM HSCs were cultured in StemPro-34 media (Invitrogen) complemented by glutamine and cytokines (stem cell factor, 20 ng/ml; thrombopoietin, 20 ng/ml). For division-tracking experiments, cells were sorted directly into 60-well terasaki plates (one cell per well) and visually monitored at the indicated time points to assess the kinetics of the first division. For in vitro glutathione depletion and induction of intracellular ROS, purified SLAM HSCs were cultured in 96-well plates (5,000–10,000 per well) in the presence of indicated concentration of BSO (50–200 µM). After 48 h in culture, cells were analyzed for ROS level and gene expression.
RNA sequencing
20,000 SLAM HSCs were isolated from db and littermate control mice by FACS sorting directly into TRIzol (Invitrogen). After chloroform extraction, total RNA was purified using a RNeasy microKit column system (QIAGEN). Three independent replicates were isolated in parallel to ensure reproducibility and allow statistical analysis. RNA quality was controlled using a Bioanalyzer (Agilent Technologies) before being processed for retrotranscription, linear amplification, and cDNA library generation. The whole transcriptome was amplified using the SMARTer Ultra Low RNA kit for Illumina Sequencing (Takara Bio Inc.). cDNA libraries were prepared using Nextera XT DNA Sample preparation reagents. Fragmented and tagged libraries were pooled and examined using a high-sensitivity DNA chip (Agilent Technologies). Libraries were sequenced on a HiSeq 2500 platform (Illumina) using the pair-end 75-bp sequencing strategy.
Bioinformatics analyses
RNA-Seq data were aligned and quantified (transcript per million estimates) using the software BowTie2 and RNA-seq by expectation maximization with the mm10 UCSC transcriptome reference. All differentially expressed genes were identified based on an empirical Bayes-moderated t test p-value <0.05. For all heatmaps, log2-transformed transcripts per million values were normalized to the median expression of each gene across all compared cells/samples. GSEAs were performed using AltAnalyze (GO-Elite algorithm) using gene sets from the embedded Gene Ontology, ImmGen, and Pathway Commons database (Zambon et al., 2012). Comparative analyses with previously published signatures were performed with differentially expressed genes using GSEA v2.0 software (Subramanian et al., 2005). The GEO accession number for RNA sequencing data was GSE90725.
qRT-PCR array
qRT-PCR arrays were performed in multiplex assays as previously described (Reynaud et al., 2011). For each experiment, pools of 100 cells were directly sorted into 5 µl of resuspension buffer from Cellsdirect One-Step qRT-PCR kits (Invitrogen). Samples were used for gene-specific based reverse transcription (SuperScript III; Thermo Fisher Scientific) and preamplified for 18 cycles with Platinum Taq DNA polymerase in presence of multiplex primer mix (50 nM each). Resulting cDNA products were diluted fivefold in TE buffer (10 mM Tris and 0.1 mM EDTA, pH 8.0) and analyzed on a 7900 HT Fast Real-Time PCR System (Applied Biosystems) using gene-specific primers and SYBR green reagents (Applied Biosystems). Each measurement was performed in duplicate with a β-actin (Actb) housekeeping gene used for normalization. Primers used for preamplification and quantitative measurement were intron-spanning and validated on serial dilution of total mouse cDNA to ensure linear amplification and PCR specificity. Sequences for qRT-PCR primers can be found in Table S2. Results were validated by classical qRT-PCR using total RNA isolated from ∼0.5–2 × 104 cells. RNA was treated with DNase I and reverse-transcribed using SuperScript III kit and random hexamers (Invitrogen). cDNA equivalent of 200 cells per reaction were used for quantitative PCR assays with SYBR green reagents.
Statistics
All results are expressed as means with error bars reflecting SD or SEM as indicated. n represents the number of animals per experiment. Differences between two groups were assessed using unpaired two-tailed Student’s t tests. Data involving more than two groups were assessed by one-way or two-way ANOVAs with Tukey’s or Dunnett’s post hoc tests.
Online supplemental material
Fig. S1 shows hematopoietic disruption associated with established obesity in the dietary (HFD) mouse model. Fig. S2 documents gene expression and kinetics of division of different HSC subsets. Fig. S3 shows the validation of the HSC functional dysregulation in dietary mouse model. Fig. S4 shows the activity of the Gfi1 locus in Ctrl::Gfi1Gfp/+ knock-in mice fed with HFD for 8 mo. Fig. S5 shows the effect of Gfi1 gene dosage on the expression of Cdkn1a and Cdkn1c. Tables S1 and S2 list the antibodies and the primer sequences used for flow cytometry and qRT-PCR analyses, respectively. Table S3 provides source data for the heatmap of gene expression and GSEAs generated from 4 mo Ctrl and db HSCs.
Supplementary Material
Acknowledgments
We acknowledge the assistance of the Cincinnati Children’s Hospital Medical Center Research Flow Cytometry and the DNA Sequencing Cores.
This work was supported by a Cincinnati Children’s Hospital Medical Center Arnold W. Strauss Postdoctoral Fellowship Award to J.-M. Lee, a Concern Foundation research award, a National Institutes of Health grant (R56 RHL131861A) and Department of Defense Peer Reviewed Cancer Research Program award (DOD#W81XWH-15-1-0344) to D. Reynaud, and a National Institutes of Health grant (R01HL122661) to H.L. Grimes.
The authors declare no competing financial interests.
Author contributions: J.-M. Lee, V. Govindarajah, and B. Goddard performed and analyzed all the experiments. A. Hinge and D.E. Muench provided assistance for cell cycle and flow cytometry analyses. B. Aronow provided assistance for the initial bioinformatics analyses. N. Salomonis performed all bioinformatics analyses and contributed to the writing of the associated sections. N. Salomonis, H.L. Grimes, J.A. Cancelas, M.D. Filippi, and B. Aronow provided critical insights and review of the manuscript. D. Reynaud conceived and supervised the project. J.-M. Lee and D. Reynaud wrote the manuscript.
References
- Adler B.J., Green D.E., Pagnotti G.M., Chan M.E., and Rubin C.T.. 2014a High fat diet rapidly suppresses B lymphopoiesis by disrupting the supportive capacity of the bone marrow niche. PLoS One. 9:e90639 10.1371/journal.pone.0090639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler B.J., Kaushansky K., and Rubin C.T.. 2014b Obesity-driven disruption of haematopoiesis and the bone marrow niche. Nat. Rev. Endocrinol. 10:737–748. 10.1038/nrendo.2014.169 [DOI] [PubMed] [Google Scholar]
- Akunuru S., and Geiger H.. 2016. Aging, Clonality, and Rejuvenation of Hematopoietic Stem Cells. Trends Mol. Med. 22:701–712. 10.1016/j.molmed.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosi T.H., Scialdone A., Graja A., Gohlke S., Jank A.M., Bocian C., Woelk L., Fan H., Logan D.W., Schürmann A., et al. 2017. Adipocyte Accumulation in the Bone Marrow during Obesity and Aging Impairs Stem Cell-Based Hematopoietic and Bone Regeneration. Cell Stem Cell. 20:771–784. 10.1016/j.stem.2017.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai F., Hirao A., Ohmura M., Sato H., Matsuoka S., Takubo K., Ito K., Koh G.Y., and Suda T.. 2004. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 118:149–161. 10.1016/j.cell.2004.07.004 [DOI] [PubMed] [Google Scholar]
- Arnold M., Leitzmann M., Freisling H., Bray F., Romieu I., Renehan A., and Soerjomataram I.. 2016. Obesity and cancer: An update of the global impact. Cancer Epidemiol. 41:8–15. 10.1016/j.canep.2016.01.003 [DOI] [PubMed] [Google Scholar]
- Asai T., Liu Y., Bae N., and Nimer S.D.. 2011. The p53 tumor suppressor protein regulates hematopoietic stem cell fate. J. Cell. Physiol. 226:2215–2221. 10.1002/jcp.22561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Liu Y., Di Giandomenico S., Bae N., Ndiaye-Lobry D., Deblasio A., Menendez S., Antipin Y., Reva B., Wevrick R., and Nimer S.D.. 2012. Necdin, a p53 target gene, regulates the quiescence and response to genotoxic stress of hematopoietic stem/progenitor cells. Blood. 120:1601–1612. 10.1182/blood-2011-11-393983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge M.T., King K.Y., Boles N.C., Weksberg D.C., and Goodell M.A.. 2010. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature. 465:793–797. 10.1038/nature09135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman I., Bhattacharya D., Zandi S., Sigvardsson M., Weissman I.L., Bryder D., and Rossi D.J.. 2010. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc. Natl. Acad. Sci. USA. 107:5465–5470. 10.1073/pnas.1000834107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste P., Frelin C., Janmohamed S., Barbara M., Herrington R., Hyam D., and Iscove N.N.. 2010. Intermediate-term hematopoietic stem cells with extended but time-limited reconstitution potential. Cell Stem Cell. 6:48–58. 10.1016/j.stem.2009.11.014 [DOI] [PubMed] [Google Scholar]
- Benz C., Copley M.R., Kent D.G., Wohrer S., Cortes A., Aghaeepour N., Ma E., Mader H., Rowe K., Day C., et al. 2012. Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell. 10:273–283. 10.1016/j.stem.2012.02.007 [DOI] [PubMed] [Google Scholar]
- Bhaskaran K., Douglas I., Forbes H., dos-Santos-Silva I., Leon D.A., and Smeeth L.. 2014. Body-mass index and risk of 22 specific cancers: a population-based cohort study of 5·24 million UK adults. Lancet. 384:755–765. 10.1016/S0140-6736(14)60892-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K., Klapproth K., Barile M., Flossdorf M., Holland-Letz T., Schlenner S.M., Reth M., Höfer T., and Rodewald H.R.. 2015. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 518:542–546. 10.1038/nature14242 [DOI] [PubMed] [Google Scholar]
- Cabezas-Wallscheid N., Klimmeck D., Hansson J., Lipka D.B., Reyes A., Wang Q., Weichenhan D., Lier A., von Paleske L., Renders S., et al. 2014. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell. 15:507–522. 10.1016/j.stem.2014.07.005 [DOI] [PubMed] [Google Scholar]
- Cabezas-Wallscheid N., Buettner F., Sommerkamp P., Klimmeck D., Ladel L., Thalheimer F.B., Pastor-Flores D., Roma L.P., Renders S., Zeisberger P., et al. 2017. Vitamin A-Retinoic Acid Signaling Regulates Hematopoietic Stem Cell Dormancy. Cell. 169:807–823. 10.1016/j.cell.2017.04.018 [DOI] [PubMed] [Google Scholar]
- Challen G.A., Boles N.C., Chambers S.M., and Goodell M.A.. 2010. Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-beta1. Cell Stem Cell. 6:265–278. 10.1016/j.stem.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S.M., Shaw C.A., Gatza C., Fisk C.J., Donehower L.A., and Goodell M.A.. 2007. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 5:e201 10.1371/journal.pbio.0050201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Shou P., Zheng C., Jiang M., Cao G., Yang Q., Cao J., Xie N., Velletri T., Zhang X., et al. 2016. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ. 23:1128–1139. 10.1038/cdd.2015.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T., Rodrigues N., Shen H., Yang Y., Dombkowski D., Sykes M., and Scadden D.T.. 2000. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 287:1804–1808. 10.1126/science.287.5459.1804 [DOI] [PubMed] [Google Scholar]
- Chua S.C. Jr., Chung W.K., Wu-Peng X.S., Zhang Y., Liu S.M., Tartaglia L., and Leibel R.L.. 1996. Phenotypes of mouse diabetes and rat fatty due to mutations in the OB (leptin) receptor. Science. 271:994–996. 10.1126/science.271.5251.994 [DOI] [PubMed] [Google Scholar]
- Claycombe K., King L.E., and Fraker P.J.. 2008. A role for leptin in sustaining lymphopoiesis and myelopoiesis. Proc. Natl. Acad. Sci. USA. 105:2017–2021. 10.1073/pnas.0712053105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., and Morrison S.J.. 2013. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 495:231–235. 10.1038/nature11885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esplin B.L., Shimazu T., Welner R.S., Garrett K.P., Nie L., Zhang Q., Humphrey M.B., Yang Q., Borghesi L.A., and Kincade P.W.. 2011. Chronic exposure to a TLR ligand injures hematopoietic stem cells. J. Immunol. 186:5367–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers M.A., Offner S., Blanco-Bose W.E., Waibler Z., Kalinke U., Duchosal M.A., and Trumpp A.. 2009. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 458:904–908. 10.1038/nature07815 [DOI] [PubMed] [Google Scholar]
- Falagas M.E., and Kompoti M.. 2006. Obesity and infection. Lancet Infect. Dis. 6:438–446. 10.1016/S1473-3099(06)70523-0 [DOI] [PubMed] [Google Scholar]
- Ferraro F., Lymperi S., Méndez-Ferrer S., Saez B., Spencer J.A., Yeap B.Y., Masselli E., Graiani G., Prezioso L., Rizzini E.L., et al. 2011. Diabetes impairs hematopoietic stem cell mobilization by altering niche function. Sci. Transl. Med. 3:104ra101 10.1126/scitranslmed.3002191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegal K.M., Carroll M.D., Kit B.K., and Ogden C.L.. 2012. Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999-2010. JAMA. 307:491–497. 10.1001/jama.2012.39 [DOI] [PubMed] [Google Scholar]
- Fraszczak J., Helness A., Chen R., Vadnais C., Robert F., Khandanpour C., and Möröy T.. 2016. Threshold Levels of Gfi1 Maintain E2A Activity for B Cell Commitment via Repression of Id1. PLoS One. 11:e0160344 10.1371/journal.pone.0160344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodell M.A., Brose K., Paradis G., Conner A.S., and Mulligan R.C.. 1996. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J. Exp. Med. 183:1797–1806. 10.1084/jem.183.4.1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin P.J., and Stambolic V.. 2015. Impact of the obesity epidemic on cancer. Annu. Rev. Med. 66:281–296. 10.1146/annurev-med-051613-012328 [DOI] [PubMed] [Google Scholar]
- Grundy S.M. 2016. Metabolic syndrome update. Trends Cardiovasc. Med. 26:364–373. 10.1016/j.tcm.2015.10.004 [DOI] [PubMed] [Google Scholar]
- Hock H., Hamblen M.J., Rooke H.M., Schindler J.W., Saleque S., Fujiwara Y., and Orkin S.H.. 2004. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 431:1002–1007. 10.1038/nature02994 [DOI] [PubMed] [Google Scholar]
- Hönes J.M., Botezatu L., Helness A., Vadnais C., Vassen L., Robert F., Hergenhan S.M., Thivakaran A., Schütte J., Al-Matary Y.S., et al. 2016. GFI1 as a novel prognostic and therapeutic factor for AML/MDS. Leukemia. 30:1237–1245. 10.1038/leu.2016.11 [DOI] [PubMed] [Google Scholar]
- Huttunen R., and Syrjanen J.. 2013. Obesity and the risk and outcome of infection. Int. J. Obes. 37:333–340. 10.1038/ijo.2012.62 [DOI] [PubMed] [Google Scholar]
- Ito K., Hirao A., Arai F., Takubo K., Matsuoka S., Miyamoto K., Ohmura M., Naka K., Hosokawa K., Ikeda Y., and Suda T.. 2006. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat. Med. 12:446–451. 10.1038/nm1388 [DOI] [PubMed] [Google Scholar]
- Iyengar N.M., Hudis C.A., and Dannenberg A.J.. 2015. Obesity and cancer: local and systemic mechanisms. Annu. Rev. Med. 66:297–309. 10.1146/annurev-med-050913-022228 [DOI] [PubMed] [Google Scholar]
- Kataoka K., Sato T., Yoshimi A., Goyama S., Tsuruta T., Kobayashi H., Shimabe M., Arai S., Nakagawa M., Imai Y., et al. 2011. Evi1 is essential for hematopoietic stem cell self-renewal, and its expression marks hematopoietic cells with long-term multilineage repopulating activity. J. Exp. Med. 208:2403–2416. 10.1084/jem.20110447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandekar M.J., Cohen P., and Spiegelman B.M.. 2011. Molecular mechanisms of cancer development in obesity. Nat. Rev. Cancer. 11:886–895. 10.1038/nrc3174 [DOI] [PubMed] [Google Scholar]
- Kiel M.J., Yilmaz O.H., Iwashita T., Yilmaz O.H., Terhorst C., and Morrison S.J.. 2005. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 121:1109–1121. 10.1016/j.cell.2005.05.026 [DOI] [PubMed] [Google Scholar]
- Krings A., Rahman S., Huang S., Lu Y., Czernik P.J., and Lecka-Czernik B.. 2012. Bone marrow fat has brown adipose tissue characteristics, which are attenuated with aging and diabetes. Bone. 50:546–552. 10.1016/j.bone.2011.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A., Parks B.W., Du J., Trac C., Setten R., Chen Y., Brown K., Lusis A.J., Natarajan R., and Schones D.E.. 2014. Open chromatin profiling in mice livers reveals unique chromatin variations induced by high fat diet. J. Biol. Chem. 289:23557–23567. 10.1074/jbc.M114.581439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A., Trac C., Du J., Natarajan R., and Schones D.E.. 2016. Persistent Chromatin Modifications Induced by High Fat Diet. J. Biol. Chem. 291:10446–10455. 10.1074/jbc.M115.711028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Ragheb K., Lawler G., Sturgis J., Rajwa B., Melendez J.A., and Robinson J.P.. 2003. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J. Biol. Chem. 278:8516–8525. 10.1074/jbc.M210432200 [DOI] [PubMed] [Google Scholar]
- Liu Y., Elf S.E., Miyata Y., Sashida G., Liu Y., Huang G., Di Giandomenico S., Lee J.M., Deblasio A., Menendez S., et al. 2009. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 4:37–48. 10.1016/j.stem.2008.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Chen G.L., Hannemann N., Ipseiz N., Krönke G., Bäuerle T., Munos L., Wirtz S., Schett G., and Bozec A.. 2015. Microbiota from Obese Mice Regulate Hematopoietic Stem Cell Differentiation by Altering the Bone Niche. Cell Metab. 22:886–894. 10.1016/j.cmet.2015.08.020 [DOI] [PubMed] [Google Scholar]
- Lutz T.A., and Woods S.C.. 2012. Overview of animal models of obesity. Curr. Protocols Pharmacol. 58:61 10.1002/0471141755.ph0561s58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova M.M., Sabatini D.M., and Yilmaz O.H.. 2014. Dietary and metabolic control of stem cell function in physiology and cancer. Cell Stem Cell. 14:292–305. 10.1016/j.stem.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagareddy P.R., Murphy A.J., Stirzaker R.A., Hu Y., Yu S., Miller R.G., Ramkhelawon B., Distel E., Westerterp M., Huang L.S., et al. 2013. Hyperglycemia promotes myelopoiesis and impairs the resolution of atherosclerosis. Cell Metab. 17:695–708. 10.1016/j.cmet.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagareddy P.R., Kraakman M., Masters S.L., Stirzaker R.A., Gorman D.J., Grant R.W., Dragoljevic D., Hong E.S., Abdel-Latif A., Smyth S.S., et al. 2014. Adipose tissue macrophages promote myelopoiesis and monocytosis in obesity. Cell Metab. 19:821–835. 10.1016/j.cmet.2014.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveiras O., Nardi V., Wenzel P.L., Hauschka P.V., Fahey F., and Daley G.Q.. 2009. Bone-marrow adipocytes as negative regulators of the haematopoietic microenvironment. Nature. 460:259–263. 10.1038/nature08099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden C.L., Carroll M.D., Kit B.K., and Flegal K.M.. 2014. Prevalence of childhood and adult obesity in the United States, 2011-2012. JAMA. 311:806–814. 10.1001/jama.2014.732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S.Y., Johnson K.C., Ugnat A.M., Wen S.W., and Mao Y.. Canadian Cancer Registries Epidemiology Research Group . 2004. Association of obesity and cancer risk in Canada. Am. J. Epidemiol. 159:259–268. 10.1093/aje/kwh041 [DOI] [PubMed] [Google Scholar]
- Parr C.L., Batty G.D., Lam T.H., Barzi F., Fang X., Ho S.C., Jee S.H., Ansary-Moghaddam A., Jamrozik K., Ueshima H., et al. Asia-Pacific Cohort Studies Collaboration . 2010. Body-mass index and cancer mortality in the Asia-Pacific Cohort Studies Collaboration: pooled analyses of 424,519 participants. Lancet Oncol. 11:741–752. 10.1016/S1470-2045(10)70141-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras E.M., Reynaud D., Kang Y.A., Carlin D., Calero-Nieto F.J., Leavitt A.D., Stuart J.M., Göttgens B., and Passegué E.. 2015. Functionally Distinct Subsets of Lineage-Biased Multipotent Progenitors Control Blood Production in Normal and Regenerative Conditions. Cell Stem Cell. 17:35–46. 10.1016/j.stem.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian P., He X.C., Paulson A., Li Z., Tao F., Perry J.M., Guo F., Zhao M., Zhi L., Venkatraman A., et al. 2016. The Dlk1-Gtl2 Locus Preserves LT-HSC Function by Inhibiting the PI3K-mTOR Pathway to Restrict Mitochondrial Metabolism. Cell Stem Cell. 18:214–228. 10.1016/j.stem.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves G.K., Pirie K., Beral V., Green J., Spencer E., and Bull D.. Million Women Study Collaboration . 2007. Cancer incidence and mortality in relation to body mass index in the Million Women Study: cohort study. BMJ. 335:1134 10.1136/bmj.39367.495995.AE [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud D., Pietras E., Barry-Holson K., Mir A., Binnewies M., Jeanne M., Sala-Torra O., Radich J.P., and Passegué E.. 2011. IL-6 controls leukemic multipotent progenitor cell fate and contributes to chronic myelogenous leukemia development. Cancer Cell. 20:661–673. 10.1016/j.ccr.2011.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring L.E., and Zeltser L.M.. 2010. Disruption of hypothalamic leptin signaling in mice leads to early-onset obesity, but physiological adaptations in mature animals stabilize adiposity levels. J. Clin. Invest. 120:2931–2941. 10.1172/JCI41985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D.J., Bryder D., Zahn J.M., Ahlenius H., Sonu R., Wagers A.J., and Weissman I.L.. 2005. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl. Acad. Sci. USA. 102:9194–9199. 10.1073/pnas.0503280102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi L., Lin K.K., Boles N.C., Yang L., King K.Y., Jeong M., Mayle A., and Goodell M.A.. 2012. Less is more: unveiling the functional core of hematopoietic stem cells through knockout mice. Cell Stem Cell. 11:302–317. 10.1016/j.stem.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai C.M., Babovic S., Upadhaya S., Knapp D.J.H.F., Lavin Y., Lau C.M., Goloborodko A., Feng J., Fujisaki J., Ding L., et al. 2016. Hematopoietic Stem Cells Are the Major Source of Multilineage Hematopoiesis in Adult Animals. Immunity. 45:597–609. 10.1016/j.immuni.2016.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer K., DelProposto J., Morris D.L., Zamarron B., Mergian T., Maley N., Cho K.W., Geletka L., Subbaiah P., Muir L., et al. 2014. Diet-induced obesity promotes myelopoiesis in hematopoietic stem cells. Mol. Metab. 3:664–675. 10.1016/j.molmet.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer K., Maley N., Mergian T., DelProposto J., Cho K.W., Zamarron B.F., Martinez-Santibanez G., Geletka L., Muir L., Wachowiak P., et al. 2015. Differences in Hematopoietic Stem Cells Contribute to Sexually Dimorphic Inflammatory Responses to High Fat Diet-induced Obesity. J. Biol. Chem. 290:13250–13262. 10.1074/jbc.M114.634568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.C., Liao J.T., Page J.G., Wientjes M.G., and Grieshaber C.K.. 1989. Pharmacokinetics of buthionine sulfoximine (NSC 326231) and its effect on melphalan-induced toxicity in mice. Cancer Res. 49:5385–5391. [PubMed] [Google Scholar]
- Styner M., Thompson W.R., Galior K., Uzer G., Wu X., Kadari S., Case N., Xie Z., Sen B., Romaine A., et al. 2014. Bone marrow fat accumulation accelerated by high fat diet is suppressed by exercise. Bone. 64:39–46. 10.1016/j.bone.2014.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Ramos A., Chapman B., Johnnidis J.B., Le L., Ho Y.J., Klein A., Hofmann O., and Camargo F.D.. 2014. Clonal dynamics of native haematopoiesis. Nature. 514:322–327. 10.1038/nature13824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie G., Messina K.E., Yan J., Messina J.A., and Messina L.M.. 2014. Hypercholesterolemia induces oxidant stress that accelerates the ageing of hematopoietic stem cells. J. Am. Heart Assoc. 3:e000241 10.1161/JAHA.113.000241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier M.D., Naaz A., Li Y., and Fraker P.J.. 2012. Enhancement of hematopoiesis and lymphopoiesis in diet-induced obese mice. Proc. Natl. Acad. Sci. USA. 109:7622–7629. 10.1073/pnas.1205129109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg S.M., Seijkens T.T., Kusters P.J., Beckers L., den Toom M., Smeets E., Levels J., de Winther M.P., and Lutgens E.. 2016. Diet-induced obesity in mice diminishes hematopoietic stem and progenitor cells in the bone marrow. FASEB J. 30:1779–1788. 10.1096/fj.201500175 [DOI] [PubMed] [Google Scholar]
- van Os R., Kamminga L.M., Ausema A., Bystrykh L.V., Draijer D.P., van Pelt K., Dontje B., and de Haan G.. 2007. A Limited role for p21Cip1/Waf1 in maintaining normal hematopoietic stem cell functioning. Stem Cells. 25:836–843. 10.1634/stemcells.2006-0631 [DOI] [PubMed] [Google Scholar]
- Wagers A.J., and Weissman I.L.. 2006. Differential expression of alpha2 integrin separates long-term and short-term reconstituting Lin-/loThy1.1(lo)c-kit+ Sca-1+ hematopoietic stem cells. Stem Cells. 24:1087–1094. 10.1634/stemcells.2005-0396 [DOI] [PubMed] [Google Scholar]
- Wilson A., Murphy M.J., Oskarsson T., Kaloulis K., Bettess M.D., Oser G.M., Pasche A.C., Knabenhans C., Macdonald H.R., and Trumpp A.. 2004. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 18:2747–2763. 10.1101/gad.313104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A., Laurenti E., Oser G., van der Wath R.C., Blanco-Bose W., Jaworski M., Offner S., Dunant C.F., Eshkind L., Bockamp E., et al. 2008. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 135:1118–1129. 10.1016/j.cell.2008.10.048 [DOI] [PubMed] [Google Scholar]
- Yamamoto R., Morita Y., Ooehara J., Hamanaka S., Onodera M., Rudolph K.L., Ema H., and Nakauchi H.. 2013. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 154:1112–1126. 10.1016/j.cell.2013.08.007 [DOI] [PubMed] [Google Scholar]
- Young K., Borikar S., Bell R., Kuffler L., Philip V., and Trowbridge J.J.. 2016. Progressive alterations in multipotent hematopoietic progenitors underlie lymphoid cell loss in aging. J. Exp. Med. 213:2259–2267. 10.1084/jem.20160168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yücel R., Kosan C., Heyd F., and Möröy T.. 2004. Gfi1:green fluorescent protein knock-in mutant reveals differential expression and autoregulation of the growth factor independence 1 (Gfi1) gene during lymphocyte development. J. Biol. Chem. 279:40906–40917. 10.1074/jbc.M400808200 [DOI] [PubMed] [Google Scholar]
- Zambon A.C., Gaj S., Ho I., Hanspers K., Vranizan K., Evelo C.T., Conklin B.R., Pico A.R., and Salomonis N.. 2012. GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics. 28:2209–2210. 10.1093/bioinformatics/bts366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Yücel R., Kosan C., Klein-Hitpass L., and Möröy T.. 2004. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 23:4116–4125. 10.1038/sj.emboj.7600419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.O., Yue R., Murphy M.M., Peyer J.G., and Morrison S.J.. 2014. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 15:154–168. 10.1016/j.stem.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.