Xiao et al. demonstrate that formation of super-enhancers at Il9 locus is critical for robust IL-9 expression and Th9 cell induction, and assembly of Il9 super-enhancers is driven by OX40-mediated chromatin acetylation.
Abstract
Th9 cells are prominently featured in allergic lung inflammation, but the mechanism that regulates IL-9 induction in T helper cells remains poorly defined. Here we demonstrate that formation of super-enhancers (SEs) is critical in robust induction of IL-9 and that assembly of the Il9 SEs in Th cells requires OX40-triggered chromatin acetylation. Mechanistically, we found that OX40 costimulation induces RelB expression, which recruits the histone acetyltransferase p300 to the Il9 locus to catalyze H3K27 acetylation. This allows binding of the SE factor Brd4 to organize assembly of the SE complex, which in turn drives robust IL-9 expression and Th9 cell induction. Thus, Th9 cells are strongly induced upon OX40 stimulation, and disruption of SEs abolished Th9 cell induction in vitro and inhibited Th9 cell–mediated allergic airway inflammation in vivo. Together, our data suggest that formation of SEs is essential in IL-9 expression and Th9 cell induction. These findings may have important clinical implications.
Introduction
CD4+ Th cells have the capacity to differentiate into diverse Th cell subsets after activation (e.g., Th1, Th2, Th9, Th17, Tfh, iTreg cells), and by acquiring different cytokine profiles, they define the nature as well as the outcomes of immune responses (Li et al., 2014). Th9 cells are characterized by the production of IL-9, a pleiotropic cytokine with diverse effects (Schmitt et al., 2014). Functionally, Th9 cells provide both beneficial and deleterious effects, depending on the models and the context of their induction. Specifically, Th9 cells mediate protection against parasitic infections and show strong anticancer immunity. However, they also induce allergic inflammation, asthma, and autoimmune diseases (Kaplan, 2013). Thus, development of strategies to therapeutically modulate Th9 cells is an important and clinically relevant issue.
However, we remain poorly informed about how Th9 cells are induced and maintained, especially under in vivo conditions (Li et al., 2017). The Il9 locus by itself has been well characterized (Perumal and Kaplan, 2011). The coding region of the Il9 locus contains five exons, with an additional three conserved noncoding sequences (CNS0–2). CNS0 is located ∼6 kb upstream of the Il9 transcription start site (TSS; −6 kb), whereas CNS2 is ∼5.4 kb downstream of the TSS (+5.4 kb). CNS1 denotes the promoter region that contains binding sites for multiple transcription factors (Kaplan, 2017). Surprisingly, little is known about how IL-9 is controlled in Th9 cells or under Th9 cell–inducing conditions (Li et al., 2017). As compared with other Th cell subsets, a “lineage-specific” or a “lineage-defining” transcription factor for Th9 cells has not been identified thus far, despite tremendous efforts devoted to this area. Instead, a myriad of transcription factors are shown to facilitate Th9 induction under various conditions, and such factors include PU.1, IRF4, STAT5, STAT6, NFAT, GATA1, GATA3, Smads, Etv5, and Notch, as well as NF-κB, BATF, and AP-1 (Zhao et al., 2013). Of note, none of those transcription factors are Th9 cell–specific (Tan and Gery, 2012), making a detailed study of Th9 cells a challenging task. It remains contentious whether Th9 cells are a distinct Th cell subset or simply intermediaries of other Th cell subsets. In most models, Th9 cells are best induced by TGF-β and IL-4 in vitro (Schmitt et al., 1994), which individually promotes Foxp3+ Treg cells and Th2 cells, respectively. It is puzzling that TGF-β and IL-4 often convert only a very small fraction of naive CD4+ T cells into Th9 cells, and in some settings, they coexpress other cytokines such as IL-10 and IL-21, cytokines that are associated with Th2 and Th17 cells (Dardalhon et al., 2008; Nowak et al., 2009). Additionally, Th9 cells, once induced, are metastable; they tend to lose IL-9 expression within days (Tan and Gery, 2012), although in some studies, adoptively transferred Th9 cells show prolonged effects in vivo (Lu et al., 2012). Together, these data provide certain evidence that Th9 cells may be controlled by very different mechanisms. Indeed, we and others recently reported that, besides the cytokines TGF-β and IL-4, costimulatory molecules in the TNFR superfamily (OX40, GITR, and TL1A) are powerful inducers of Th9 cells (Meylan and Siegel, 2017). OX40 in particular shows exceptional potency in supporting the induction of Th9 cells, and together with the polarizing cytokines, OX40 drives the generation of massive number of Th9 cells (Xiao et al., 2012a). As a member of the TNFR superfamily, OX40 traditionally signals through the NF-κB pathway (Sun, 2017). But how the NF-κB family members, which are highly induced in all activated T cells, shows proclivity to Th9 cells, especially in the context of other Th9 cell–related transcription factors, remains completely unknown.
In the present study, we took a different approach and examined genome-wide the intragenic and intergenic regulatory elements involved in the induction of Th9 cells by OX40 costimulation, and identified super-enhancer (SE) elements at the Il9 locus that are critically required for robust Th9 cell induction. These SEs are induced by OX40, showing an exceptionally high degree of open chromatin configuration and densely populated with transcription cofactors. We also found that formation of Il9 SEs depends on RelB and the histone acetyltransferase p300 and that Il9 SEs are critical in robust Th9 cell induction in vitro and Th9 cell–mediated allergic airway inflammation in vivo.
Results
Induction of SEs upon OX40 signaling
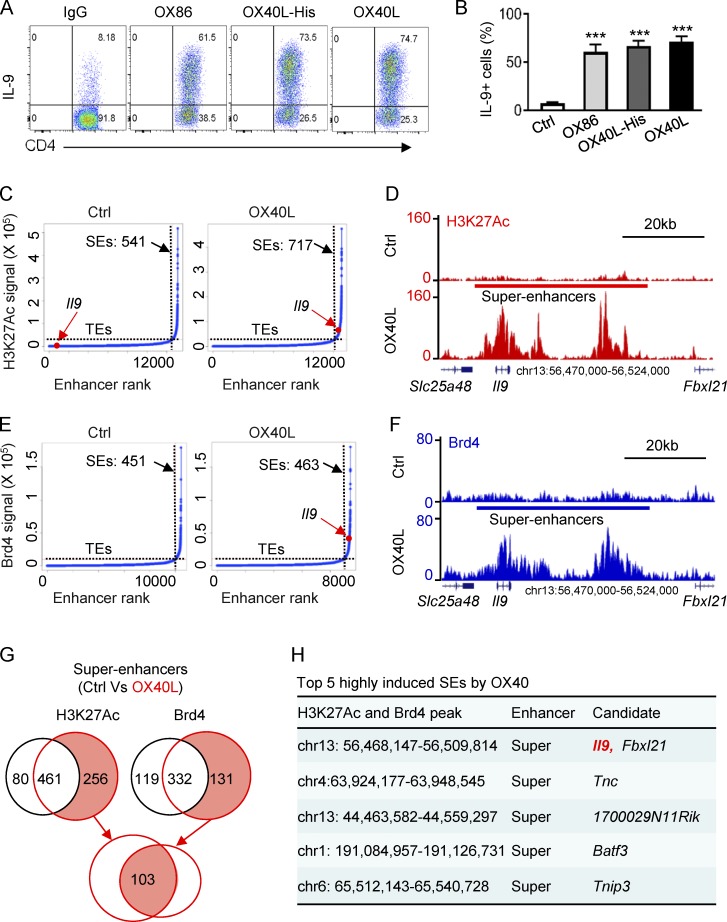
As previously reported (Xiao et al., 2012a), we FACS-sorted naive CD4+ T cells (CD62L+CD44−Foxp3−) from Foxp3gfp reporter mice and activated them with anti-CD3 plus syngeneic APCs under Th9 cell–culturing conditions (TGF-β+IL-4). In these cultures we used an agonistic anti-OX40 mAb (OX86), OX40L transgenic APCs, or OX40L-His fusion protein (followed by anti-His cross-linking) to trigger OX40 signaling. Induction of Th9 cells was examined 3 d later by flow cytometry. As shown in Fig. 1 A, in the presence of OX40 costimulation, up to 70% of the CD4+ T cells became Th9 cells, which contrasted sharply to the result without OX40 stimulation, in which only ∼8% of the CD4+ T cells were converted to Th9 cells. This robust Th9 induction was highly consistent regardless of using OX86, OX40L-His, or OX40Ltg APCs to engage the OX40 receptor (Fig. 1 B), thus demonstrating a remarkable potency of OX40 in the induction of Th9 cells.
Figure 1.
Identification of SEs in Th9 cells induced by OX40 stimulation. (A) The FACS plots depict Th9 cells induced by TGF-β and IL-4 with or without OX40 stimulation. Naive CD4+ T cells were activated by anti-CD3/APCs, and OX40 stimulation was provided by an agonist anti-OX40 mAb (OX86), OX40 ligand-His recombinant protein (OX40L-His), or OX40 ligand transgenic APC (OX40L). An isotype control IgG was used as control. Representative plots from one of five independent experiments. (B) Summary of Th9 cells induced as described in A from five independent experiments. (C) Distribution of SEs based on H3K27Ac tag signals and enhancer ranks in control Th9 (Ctrl) and OX40-induced Th9 cells (OX40L). The number denotes the SEs induced in Th9 cells with or without OX40 costimulation. The red dot shows the positions of Il9 TE and Il9 SE. (D) ChIP-seq profiles showing levels of genome-wide H3K27 acetylation in control Th9 cells (Ctrl) and OX40-induced Th9 cells (OX40L). The position of SE and the Il9 locus are highlighted. (E) Distribution of SE in control Th9 (Ctrl) and OX40-induced Th9 cells (OX40L) based Brd4 tag signals and enhancer ranks. The number denotes the SEs identified in Th9 cells, and the red dot shows the position of Il9 SE. (F) Brd4 ChIP-seq profiles showing enrichment of Brd4 in Th9 cells induced with or without OX40 costimulation. The position of SE and Il9 locus is shown. (G) The Venn diagrams show the number of unique and overlapping SEs induced by OX40 based on the H3K27Ac and Brd4 ChIP-seq signals. (H) The top five SEs induced by OX40 ranked by fold changes of both H3K27Ac and Brd4 ChIP-seq signals. Graphs in B depict mean ± SEM of five experiments, each with triplicate cultures. Statistical differences between groups were determined by the Student’s t test. ***, P < 0.001.
As a Th9 cell–specific transcription factor has not been identified (Li et al., 2017), we focused on the intragenic and intergenic regulatory elements in the Il9 locus, and by using H3K27Ac as an indicator of active enhancers (Kitagawa et al., 2017), we performed genome-wide chromatin immunoprecipitation sequencing (ChIP-seq) assay in Th9 cells induced with or without OX40 signaling. Following standard protocols (Hnisz et al., 2013), we defined SEs as stretches of highly acetylated chromatin segments within 12.5 kb in size and used the ROSE algorithm to stitch the H3K27Ac peaks together. We then ranked such enhancers by H3K27Ac tag intensity and plotted the tag intensity (y-axis) against the enhancer ranks (x-axis), which gave rise to the resulting curve shown in Fig. 1 C. We then used the tangent of the curve to distinguish SEs from typical enhancers (TEs; Hnisz et al., 2013). In this setting, we identified 717 SEs in Th9 cells induced by OX40 costimulation (OX40-induced Th9 cells) and 541 SEs in those without OX40 stimulation (control Th9 cells). Interestingly, as compared with control Th9 cells, OX40 stimulation resulted in the induction of prominent Il9 SE in Th9 cells (Fig. 1 C). A genome-wide profile of H3K27Ac ChIP-seq signals revealed striking differences in the levels, as well as the extent, of H3K27 acetylation at the Il9 locus in Th9 cells induced with or without OX40 stimulation (Fig. 1 D). Specifically, as compared with control Th9 cells, OX40-induced Th9 cells showed exceptionally high levels of H3K27 acetylation, which spanned a large genomic segment (∼33 kb region). This segment included the Il9 coding region and was flanked by the Slc25a48 and Fbxl21 loci. As shown in Fig. 1 D, we observed three striking Il9 SE clusters within this segment in relation to the Il9 TSS.
We also performed ChIP-seq analysis using Brd4 as a tag. Brd4 is a BET protein that binds acetylated H3K27, which typically initiates assembly of SE complexes (Pott and Lieb, 2015). Using the same approach (Hnisz et al., 2013), we plotted the Brd4 tag intensity against the enhancer ranks obtained from Th9 cells induced with or without OX40 costimulation, and the resulting curve showed that in OX40-induced Th9 cells (but not in control Th9 cells), Il9 SEs were also strongly induced (Fig. 1 E). As shown in Fig. 1 F, Brd4 ChIP-seq signals revealed that in the same Il9 chromatin segment (aligned with H3K27 ChIP-seq sequence), an extremely high level of Brd4 was enriched in OX40-induced Th9 cells, showing a similar pattern of active chromatin structure as revealed by the H3K27Ac ChIP-seq (Fig. 1 D). Of note, Brd4 enrichment at this region was not observed in control Th9 cells. The Venn graph shown in Fig. 1 G summarizes the number of SEs specifically induced by OX40 costimulation under Th9 cell conditions, calculated solely based on H3K27Ac and Brd4 intensity, and among the 103 SEs ranked by fold changes of both H3K27Ac and Brd4 signals, the top five highly induced SEs by OX40 are shown in Fig. 1 H. A complete list of SE tags was deposited in GEO under the accession no. GSE102940. Together, these data demonstrate a striking role for OX40 in the induction of Il9 SE in Th9 cells.
SEs are functionally involved in robust Th9 cell induction
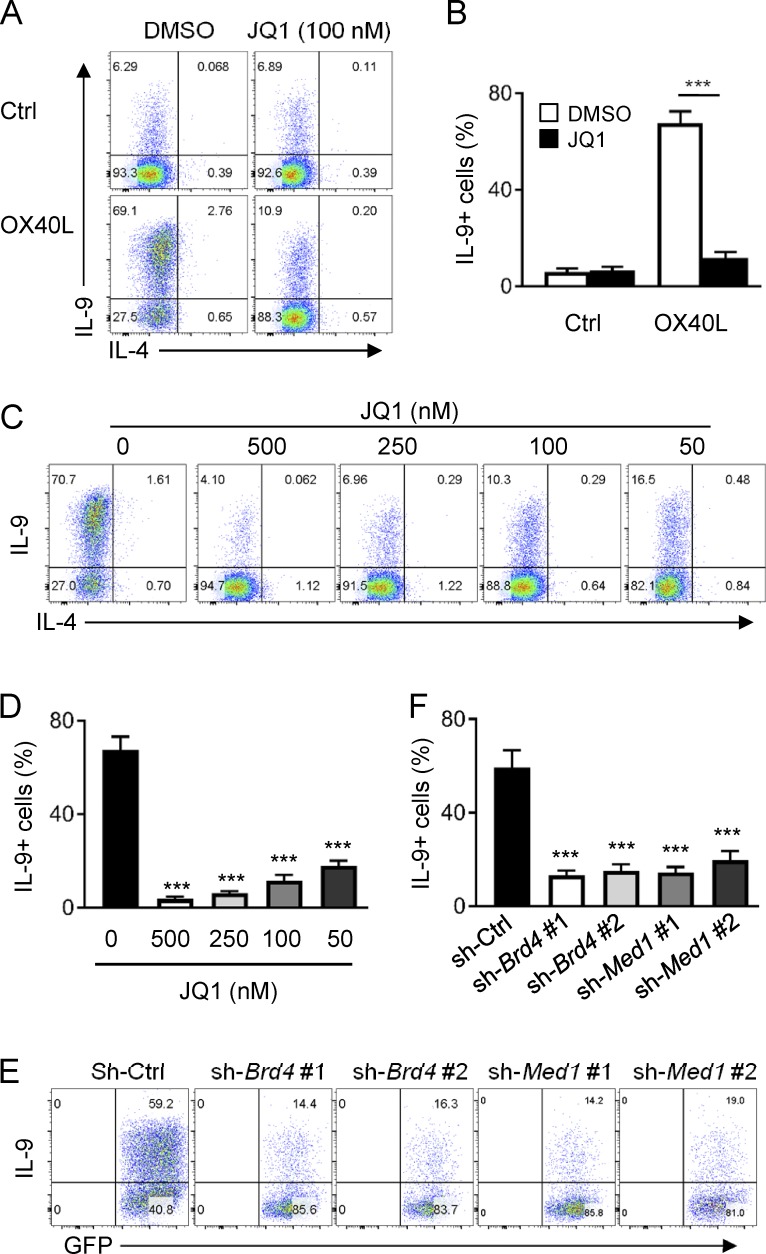
To determine the role of SEs in OX40-mediated Th9 cell induction, we focused on Brd4, as Brd4 is a “hot spot” in formation of SEs (Pott and Lieb, 2015), and took several approaches to address this issue. We first used a pharmacological inhibitor of Brd4 called JQ1 and examined whether JQ1 would affect Th9 cell induction by OX40. JQ1 binds with extremely high affinity to the hydrophobic pocket of Brd4, which prevents Brd4 from binding to the acetylated H3K27 sites, and therefore subsequent assembly of SEs is prevented (Lovén et al., 2013). To this end, we activated naive CD4+ T cells under Th9 cell conditions (TGF-β+IL-4) with or without OX40 costimulation, and in these cultures we added JQ1 or DMSO as a vehicle control, and then examined induction of Th9 cells 3 d later. As shown in Fig. 2, in Th9 cells induced without OX40 costimulation, JQ1 (100 nM) did not show significant effects; ∼7% of Th9 cells were induced from naive CD4+ T cells regardless of JQ1 addition (Fig. 2, A and B). OX40 costimulation resulted in robust Th9 cell induction (∼70% of Th9 cells), and strikingly, this induction was strongly inhibited by JQ1 (∼11% of Th9 cells). This inhibitory effect was not a result of cell death, as CD4+ T cells showed comparable viability regardless of JQ1 addition (not depicted). A careful titration of JQ1 (from 500 to 50 nM) showed that JQ1 had exceptional potency in suppressing OX40-mediated induction of Th9 cells (Fig. 2, C and D).
Figure 2.
Role of the BET inhibitor JQ1 in OX40-mediated induction of Th9 cells. (A) A representative FACS plot showing induction of Th9 cells by OX40 in the presence or absence of JQ1. DMSO was included as a vehicle control. (B) The bar graphs are summaries of Th9 cells induced by OX40 with or without JQ1 from five individual experiments performed as in A. (C) A representative FACS plot showing induction of Th9 cells by TGF-β and IL-4 plus OX40 stimulation where graded doses of JQ1 were added. (D) A summary of Th9 cells induced by OX40 as in C from five individual experiments. (E) FACS analysis of Th9 cells induced by OX40 stimulation from activated CD4+ T cells transduced with Brd4 and Med1 specific shRNAs. A empty vector (sh-Ctrl) was used as a control. Plots shown were gated on GFP+ cells. (F) The bar graphs show summary of Th9 cells induced as described in E from five independent experiments. Graphs in B, D, and F depict mean ± SEM of five experiments with triplicate cultures. Statistical differences between groups were determined by the Student’s t test. ***, P < 0.001.
Next, we designed a set of Brd4-specific shRNA and used the gene knockdown approach to examine the specific role of Brd4 in Th9 cell induction by OX40. We overexpressed control shRNA or Brd4-specific shRNA in activated CD4+ T cells, and then examined the induction of Th9 cells in the presence of OX40 stimulation. We first optimized the knockdown efficiency of this shRNA approach (not depicted). Indeed, knockdown Brd4 in activated CD4+ T cells strongly inhibited the induction of Th9 cells by OX40 (∼15%), whereas in control shRNA-transduced cells, Th9 cells were induced in large numbers (∼60%), further demonstrating the role of Brd4 in OX40-mediated induction of Th9 cells (Fig. 2, E and F). Last, we used the same shRNA approach to specifically knock down Med1, another key molecule in SE formation (Lovén et al., 2013), and examined the induction of Th9 cells by OX40. Similar to that observed in Brd4 knockdown cells, knockdown of Med1 also inhibited the induction of Th9 cells by OX40, showing a similar potency to Brd4 knockdown in suppressing Th9 cells (Fig. 2, E and F). Collectively, these data suggest that formation of SEs is functionally involved in Th9 induction by OX40.
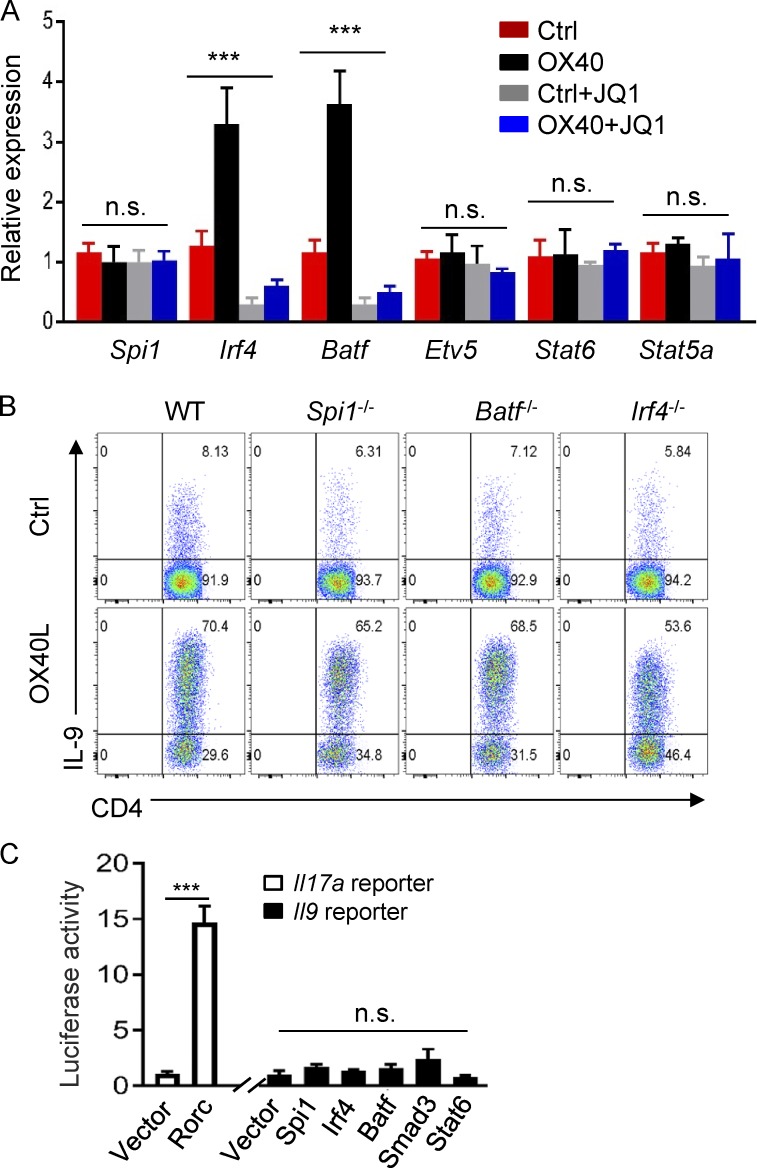
We also examined the role of JQ1 in expression of Th9 cell–associated transcription factors in OX40-stimulated T cells, testing whether inhibition of Th9 cell induction by JQ1 is also a result of suppression of key Th9 cell–associated transcription factors. As shown in Fig. 3 A, OX40 strongly up-regulated Irf4 and Batf expression in activated CD4+ T cells, whereas expression of Spi1 (PU.1), Etv5, Stat6, and Stat5a did not show marked changes with or without OX40 stimulation. Interestingly, addition of JQ1 into the cultures completely abolished the expression of Irf4 and Batf3 in OX40-stimulated T cells. We then FACS-sorted naive CD4+ T cells from WT B6, Spi1−/−, Batf−/−, and Irf4−/− mice, activated them in vitro under Th9 conditions with or without OX40 stimulation, and assessed Th9 cell induction side by side 3 d later. We observed that deficiency of Spi1 (PU.1), Batf, or Irf4 did not affect the induction of Th9 cells by OX40, and as compared with WT B6 CD4+ T cells, similar levels of Th9 cells (∼60–70%) were induced from Spi1-, Batf-, and Irf4-deficient CD4+ T cells in the presence of OX40 stimulation (Fig. 3 B). We also constructed a luciferase reporter assay in which the Il9 core promoter region (without SE segments) was genetically linked to the luciferase gene and used this assay to test the role of individual transcription factors in driving IL-9 gene expression. As a positive control, we included an Il17a gene reporter in our assays. As shown in Fig. 3 C, introduction of Rorc into the reporter assay induced robust Il17a activities. In contrast, introduction of Spi1, Irf4, Batf, Smad3, or Stat6 individually failed to provoke marked Il9 reporter activities. Thus, in addition to transcription factors, formation of SE complex is also critical in robust Il9 gene transcription.
Figure 3.
Role of JQ1 in expression of Th9 cell–associated transcription factors and their roles in Th9 induction. (A) Quantitative RT-PCR data showing expression of Th9 cell–associated transcription factors in control Th9 cells and OX40-induced Th9 cells with or without JQ1 at 2 d after T cell activation. Data were normalized to GAPDH expression. (B) Induction of Th9 cells from WT B6, Spi1−/−, Batf−/−, and Irf4−/− CD4+ T cells with or without OX40 stimulation. The FACS plots depict the percentage of IL-9+ cells among total CD4+ T cells 3 d after Th9 cell induction. (C) Luciferase reporter assay showing Il9 promoter activities induced by Th9 cell–associated transcription factors in HEK293T cells. The Il17a reporter, which is driven by Rorc, was used as a positive control. Graphs in A and C depict mean ± SEM of three independent experiments, each with triplicate cultures. Data in B are representative of five independent experiments. Statistical differences between groups were determined by one-way ANOVA or between two groups by the Student’s t test. ***, P < 0.001.
Genetic and functional mapping of Il9 SE clusters
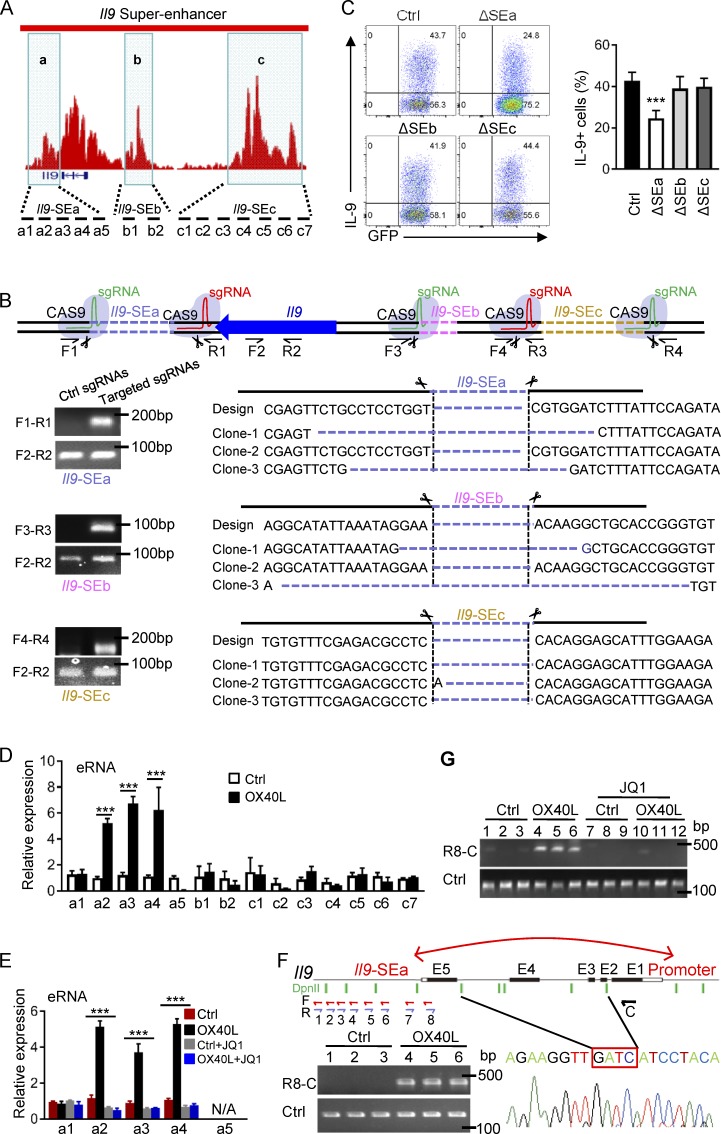
Based on H3K27Ac ChIP-seq signals, we segregated the SE elements (excluding the Il9 coding and promoter regions) into three SE clusters, namely the Il9 SEa, SEb, and SEc (Fig. 4 A). The Il9 SEa is ∼3.0 kb downstream of Il9 TSS, spanning the ∼3.5-kb chromatin segment, and the SEb is ∼5 kb upstream of the TSS, covering a ∼4.0-kb segment. Interestingly, SEc spans a ∼8-kb chromatin segment, located ∼18 kb upstream of Il9 TSS. To probe the roles of SE clusters in Th9 cell induction, we designed sequence-specific guide RNA (sgRNA) targeting individual Il9 SE regions and used the CRISPR/Cas9 approach to selectively delete such regions (Fig. 4 B). We introduced pairs of sgRNA, flanking the designated SE regions, into Cas9 transgenic CD4+ T cells via retroviral-mediated gene transfer (Sander and Joung, 2014). The retroviral vectors carry either a Gfp or a Rfp reporter gene to specifically mark transduced T cells. CD4+ T cells that were successfully transduced by both sgRNA, which are marked by expression of both GFP and RFP tags (Fig. S1 A), were selected for further analyses. Clearly, fractions of activated CD4+ T cells expressed either GFP or RFP, but only in GFP and RFP double positive CD4+ T cells can Cas9 mediate deletion of target segments (Fig. 4 B). Using pairs of specific primer sets that across individual SE clusters (F1R1 pair for SEa, F3R3 for SEb, and F4R3 for SEc) where deletion of a desired SE cluster is marked by amplification of the corresponding PCR product, we confirmed that the Cas9/sgRNA approach could result in deletion of individual SE clusters (Fig. 4 B). In these experiments, we used primers that span the Il9 coding region (F2R2), which consistently amplified the target sequence regardless of sgRNA expression, as controls (Fig. 4 B). We also examined the deletion efficiency using primers against the SEa–c sequences in sgRNA transduced CD4+ T cells and then normalized against the WT Il9 coding sequence, and showed that the deletion efficiency was around 50–60% in sgRNA transduced CD4+ T cells (Fig. S1 B).
Figure 4.
Functional mapping of Il9 SE clusters. (A) Schematic representation of Il9 SE clusters based on H3K27Ac ChIP-seq signals, namely, Il9 SEa, SEb, and SEc, is shown in shaded areas. The location of PCR primer sets designed to cover all three SE clusters is shown at the bottom. (B) Top: shows the locations of sgRNAs designed to target individual SE clusters where CRISPR/Cas9-mediated deletion of SEa, SEb, and SEc occurred. The primer sets used to validate the deletion is indicated. F, forward primer; R, reverse primer. Bottom: PCR-based products and DNA sequencing data showing specific deletion of target SE sequences in activated CD4+ T cells. (C) Induction of Th9 cells by OX40 upon CRISPR/Cas9-mediated deletion of individual SE clusters showing in representative FACS plots and summary of three independent experiments (mean and SD of n = 5). Cas9 transgenic CD4+ T cells were transduced with sgRNA vectors marked by GFP and RFP, and T cells positive for both GFP and RFP were selectively gated for analysis. (D) Expression of eRNA from individual SE clusters in control Th9 cells and OX40-stimulated Th9 cells was quantified by quantitative real-time PCR using position-specific primer sets as indicated in A. Data shown are mean ± SEM of three independent experiments with triplicate cultures. (E) Real-time qPCR data showing eRNA expression in control Th9 cells and OX40-induced Th9 cells with or without addition of JQ1 inhibitor. Data shown are mean ± SEM of three independent experiments. (F) 3C assays showing formation of chromatin loop between Il9 SEa and Il9 promoter regions. DNA from control Th9 cells (Ctrl) and OX40-indcued Th9 cells (OX40L) was digested by DpnII and ligated with T4 DNA ligase. Primer sets covering all potential ligation events were used to detect ligation products between Il9 promoter and Il9 SEa. R8 and C primer sets produced a PCR amplicon corresponding to a specific ligation event, which is confirmed by DNA sequencing. Data from one of three experiments are shown. (G) 3C assays measuring chromatin loop formations between Il9 promoter and Il9 SEa regions in control Th9 cells (Ctrl) and OX40-induced Th9 cells (OX40L) with or without the JQ1 inhibitor. The image shows representative data from one of three independent experiments. ***, P < 0.001.
Next, we transduced Cas9 transgenic CD4+ T cells with pairs of SE-specific sgRNA as described in Fig. 4 B, after a brief stimulation with anti-CD3/CD28 beads (for 1 d), and further cultured them under Th9 cell conditions (TGF-β+IL-4) in the presence of OX40 stimulation for another 2 d. Induction of Th9 cells was examined by gating onto GFP and RFP double-positive cells (Fig. S1 A). As shown in Fig. 4 C, deletion of Il9 SEa (ΔSEa) markedly reduced the induction of Th9 cells (by ∼50%), whereas deletion of either SEb (ΔSEb) or SEc (ΔSEc) did not show significant effects in suppressing Th9 cell induction, suggesting that the Il9 SEa clusters may play a more important role in Th9 cell induction by OX40. We also examined the expression of enhancer RNA (eRNA) by Th9 cells induced with or without OX40 stimulation across all three SE clusters. In contrast to mRNA, eRNA lacks the poly A tail and can only be reverse transcribed to cDNA with random primers (Witte et al., 2015; Fig. S1 C). We observed that eRNA was highly expressed in OX40-induced Th9 cells, but not in control Th9 cells, and such eRNA expression was confined to SEa clusters (Fig. 4 D). Interestingly, analysis of the role of JQ1 in eRNA expression in Th9 cells showed that JQ1 was very effective in blocking eRNA expression in Th9 cells induced by OX40 costimulation (Fig. 4 E), which is highly consistent with the effects of JQ1 in suppressing OX40-mediated Th9 cell induction (Fig. 2, A–D).
A key function of SEs is their ability to form chromatin loops to engage the promoter region of target genes (Jin et al., 2013). To examine whether the Il9 SEa cluster could loop around to engage the Il9 promoter region, we performed the chromatin conformation capture assay (3C assay), which specifically detects formation of chromatin loop (Grob and Grossniklaus, 2017). We polarized naive CD4+ T cells to Th9 cells in the presence of OX40 costimulation for 2 d, extracted the nuclear DNA, digested with DpnII, and followed by ligation with T4 DNA ligase. As shown in Fig. 4 F, sequence-specific primers covering the entire SEa and the promoter region were used for PCR amplification. In this setting, only the C (forward) and R8 (reverse) primer set, which was across the entire 3kb Il9 coding region, produced a specific PCR amplicon, indicating the formation of chromatin loop between the Il9 promoter and the Il9 SEa region (Fig. 4 F and Fig. S1 D). This chromatin loop formation was observed only in OX40-induced Th9 cells, and the loop formation was readily abolished by JQ1 treatment (Fig. 4 G). These data demonstrate that OX40 signaling induces prominent Il9 SE under Th9 cell conditions and identify the SEa cluster as functionally important in Th9 cell induction.
The NF-κB family member RelB mediates formation of Il9 SEs
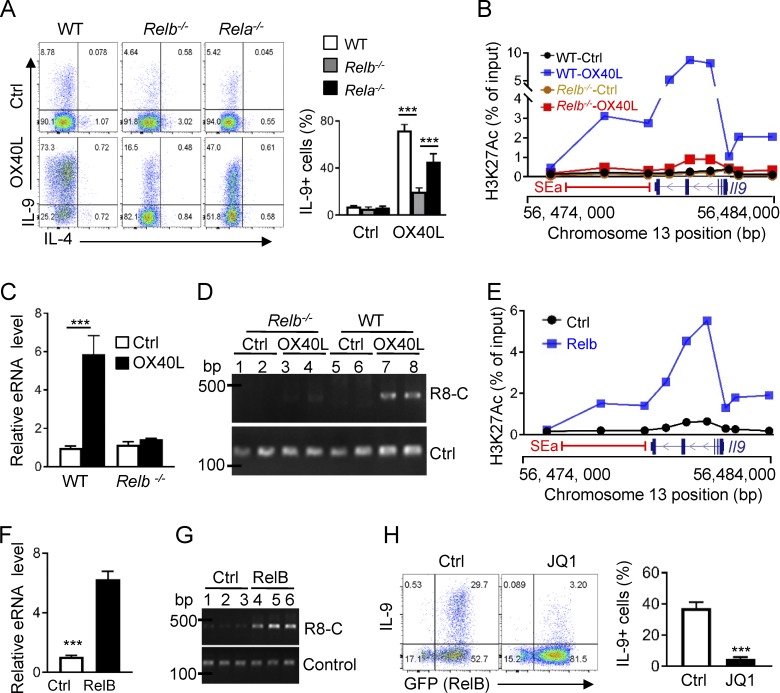
As a TNFR superfamily member, OX40 typically signals through the NF-κB pathway (Croft, 2010). To determine the mechanisms by which OX40 induces Il9 SE and subsequently Th9 cells, we activated naive CD4+ T cells from WT B6, Rela−/−, and Relb−/− mice under Th9 conditions, and examined the role of OX40 in Th9 cell induction by flow cytometry. As compared with the WT B6 controls, deficiency of either RelA or RelB did not affect Th9 cell induction from naive CD4+ T cells in the absence of OX40 stimulation (Fig. 5 A). However, in the presence of OX40 stimulation, deficiency of RelB strongly inhibited the induction of Th9 cells (∼16%) as compared with WT B6 controls (∼75%). Interestingly, in RelA-deficient CD4+ T cells, a substantial number of Th9 cells could still be induced by OX40 stimulation (∼50%). ChIP assay showed that in Relb−/− CD4+ T cells, OX40 stimulation failed to trigger H3K27 acetylation, as well as binding of Brd4 to the Il9 SEa and the coding region under Th9 cell–inducing conditions, which was in striking contrast to WT CD4+ T cells, in which OX40 costimulation produced robust and extensive H3K27 acetylation and strong Brd4 binding to such regions under Th9 cell–culturing conditions (Fig. 5 B and Fig. S2 A). Furthermore, when Relb−/− CD4+ T cells were cultured under Th9 cell conditions, we failed to detect eRNA expression (Fig. 5 C) or formation of chromatin loop (Fig. 5 D) regardless of OX40 stimulation, whereas in WT CD4+ T cells cultured under identical Th9 conditions, OX40 stimulation induced robust eRNA expression and prominent chromatin loop formation (Fig. 5, C and D).
Figure 5.
Critical role of RelB in SE formation and Th9 cell induction. (A) Representative FACS plots on Th9 cell induction and quantification of Th9 cells from WT B6, Relb−/−, and Rela−/− CD4+ T cells under Th9 cell–inducing conditions with or without OX40 signal. (B) H3K27Ac ChIP assay showing levels of H3K27Ac at the SEa and Il9 coding regions from WT B6 and Relb−/− Th9 cells induced with or without OX40 stimulation. (C) Real-time qPCR assay showing Il9 SEa (a4) eRNA expression by WT B6 and Relb−/− Th9 cells induced with or without OX40 stimulation. (D) Data shown are 3C assays measuring DNA loops between Il9 promoter and Il9 SEa regions in WT B6 and Relb−/− Th9 cells induced with (OX40L) or without OX40 stimulation (Ctrl). Primer sets amplifying the DNA fragment without DpnII sites were used as controls. (E) H3K27Ac ChIP assay showing levels of H3K27 acetylation at the Il9 locus and the SEa region in CD4+ T cells transduced with control vector or with RelB expressing vector and cultured under Th9 cell conditions. (F) Expression of eRNA from Il9 SEa (a4) by CD4+ T cells transduced by control vector and RelB expressing vector and cultured under Th9 cell conditions. (G) 3C assays based on R8 and C primer sets and qPCR measuring DNA loop formations between the Il9 promoter and Il9 SEa regions in CD4+ T cells transduced with Ctrl- or RelB-expressing vector. (H) A representative FACS plot and quantification of Th9 cells induced under Th9 cell conditions from CD4+ T cells transduced with Ctrl- or RelB-expressing vector and treated with JQ1. Graphs are representative images from at least three independent experiments and shown as mean ± SEM of three experiments, each with triplicate cultures. ***, P < 0.001.
In another set of experiments, we overexpressed RelB in CD4+ T cells using retroviral-mediated gene transfer and examined induction of SEs, as well as Th9 cells by TGF-β and IL-4 (without OX40 stimulation). As compared with control vector transduced cells, overexpression of RelB in activated CD4+ T cells resulted in prominent H3K27 acetylation at the Il9 locus, as well as robust expression of SEa eRNA and formation of chromatin loop (Fig. 5, E–G), suggesting the formation of SEs. Indeed, forced expression of RelB in activated CD4+ T cells resulted in a substantial number of Th9 cells when cultured in TGF-β and IL-4 alone without OX40 costimulation (∼40%), which was strongly inhibited by JQ1 (Fig. 5 H and Fig. S2 B). Collectively, data from such “loss of function” and “gain of function” studies suggest that RelB plays a critical role in SE formation and Th9 cell induction by OX40 stimulation.
SE formation requires the histone acetyltransferase p300
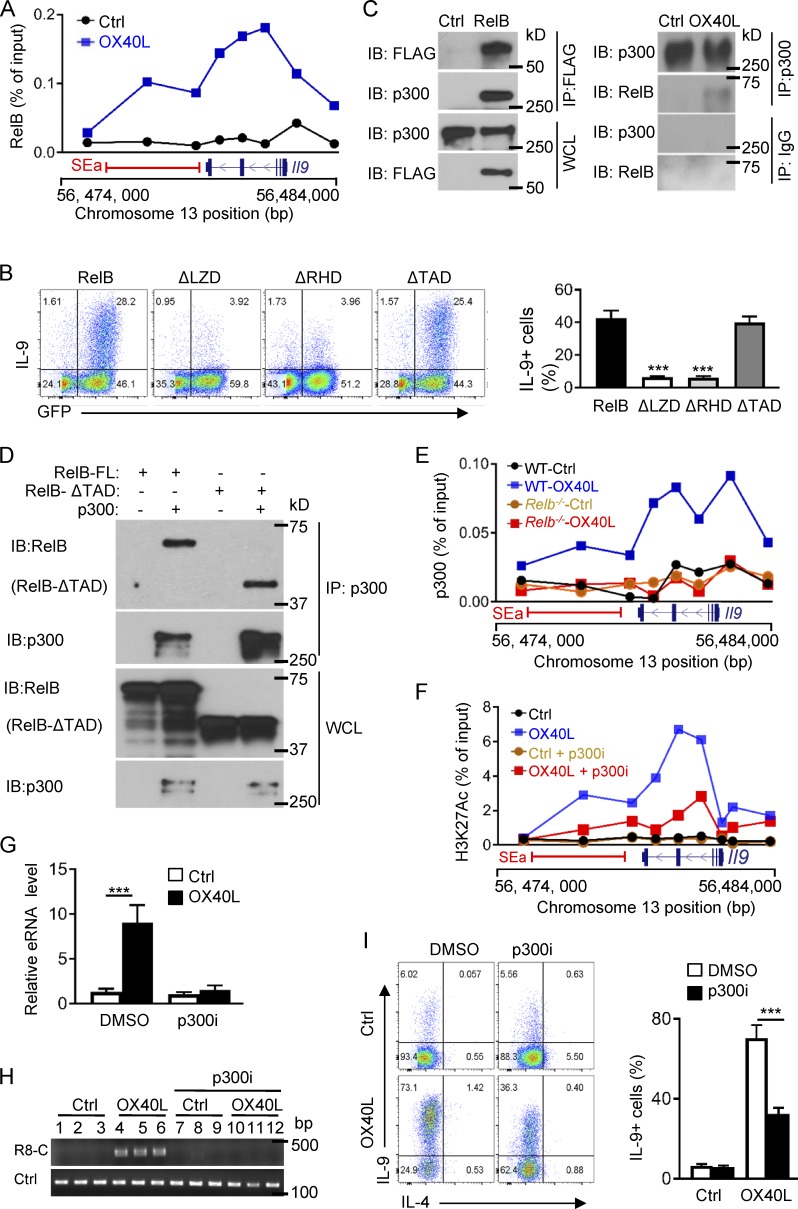
RelB is best known as a transcription factor, and upon dimerizing with p52, it binds κB sites in the genome and drives target gene expression (Vallabhapurapu and Karin, 2009). Sequence analysis showed that the Il9 locus, including the SEa segment, contains multiple κB sites. Indeed, ChIP assay revealed that RelB was highly enriched at the Il9 SEa and the Il9 coding region in OX40-induced Th9 cells, and this enrichment was not observed in control Th9 cells (Fig. 6 A). Structurally, RelB contains a leucine zipper domain (LZD), a Rel homology domain (RHD), and a transactivation domain (TAD), which are involved in protein dimerization, DNA binding, and transcriptional activities, respectively (Vallabhapurapu and Karin, 2009). We made a series of RelB mutants, with deletions of specific domains, and overexpressed such RelB mutants in activated CD4+ T cells, so that their roles in Th9 cell induction could be studied under Th9 cell–inducing conditions. As shown in Fig. 6 B, overexpression of full-length RelB led to robust induction of Th9 cells by TGF-β and IL-4, which was abolished upon deletion of the LZD or RHD. Interestingly, deletion of TAD did not affect the induction of Th9 cells by TGF-β and IL-4, suggesting that unlike its transcriptional activities, RelB likely triggers a very different mechanism in promoting robust Th9 cell induction.
Figure 6.
Formation of SEs requires the histone acetyltransferase p300. (A) ChIP assay showing RelB enrichment at the Il9 coding region and the SEa region in Th9 cells induced with (OX40L) and without OX40 stimulation (Ctrl). (B) Wt B6 CD4+ T cells transduced with retroviral vectors expressing full-length RelB or RelB mutants, with specific deletion of the LZD (ΔLZD), RHD (ΔRHD), or TAD (ΔTAD), and then cultured under Th9 cell conditions for 3 d. The FACS plots depict a representative experiment in Th9 cell induction. The bar graphs on the right side show quantification of Th9 cells from three independent experiments. (C) Coimmunoprecipitation analysis showing RelB and p300 interactions. FLAG-tagged RelB was introduced into CD4+ T cells and cultured under Th9 cell conditions. Left: RelB was immunoprecipitated with anti-FLAG mAb, followed by immunoblotting for p300 using anti-p300 mAb. Right: P300 was immunoprecipitated from control Th9 cells and OX40-induced Th9 cells, followed by immunoblotting for RelB. WCL was included as a positive control. (D) Full-length RelB or RelB with TAD deletion (RelB-ΔTAD) was introduced into 293 T cells, along with p300, and binding of RelB to p300 was determined by Co-IP experiments. The blots show binding of p300 to full length and TAD-deleted RelB. WCL was included as a control. (E) ChIP assay showing binding p300 to the Il9 coding region and the SEa region in WT B6 and Relb−/− Th9 cells induced with or without OX40 stimulation. (F) H3K27Ac ChIP data showing levels of H3K27 acetylation at Il9 locus and the SEa region in control Th9 cells and OX40-induced Th9 cells with or without addition of the p300 inhibitor SGC-CBP30 (p300i). (G) Expression of eRNA from Il9 SEa (a4) by control Th9 cells and OX40-induced Th9 cells treated with the p300 inhibitor or DMSO vehicle control. (H) 3C assay measuring DNA loop formations based PCR products amplified by R8 and C primer sets in control Th9 cells and OX40-induced Th9 cells with or without the p300 inhibitor. (I) A representative FACS plot showing induction of Th9 cells by OX40 in the presence of the p300 inhibitor or DMSO as a control. The bar graphs on the right show summary of Th9 cells with or without the p300i inhibitor. All graphs are representative images from at least three independent experiments, and bar graphs shown are mean ± SEM of three independent experiments with triplicate cultures. ***, P < 0.001.
As RelB itself lacks chromatin-modifying activities, RelB must partner with chromatin modifiers to mediate SE formation. We tagged RelB with a FLAG tag and overexpressed the FLAG-RelB in activated CD4+ T cells using retroviral mediated gene transfer. Using FLAG-tagged RelB as a bait, we immunoprecipitated RelB from OX40-induced Th9 cells and then performed Co-IP experiments to search for RelB binding proteins. As shown in Fig. 6 C, we found that the histone acetyltransferase p300 was strongly associated with RelB. In a reciprocal experiment, we cultured CD4+ T cells under Th9 cell conditions with or without OX40 stimulation, and immunoprecipitated p300 first using anti-p300 mAb. Subsequent immunoblotting experiments identified RelB in the p300 immunoprecipiates (Fig. 6 C), suggesting that RelB and p300 can physically interact with each other in Th9 cells. Indeed, overexpression of RelB and p300 in 293T cells resulted in prominent association between RelB and p300 (Fig. S3, A and B). Interestingly, binding of p300 to RelB does not rely on the TAD domain, as deletion of the TAD domain did not affect RelB from binding to p300, as shown by RelB/p300 Co-IP experiments using the RelB-ΔTAD mutant (Fig. 6 D). Furthermore, ChIP assay revealed that in OX40-induced Th9 cells, p300 was highly enriched at the Il9 locus and the SEa region, and this enrichment was absent in Relb−/− CD4+ T cells regardless of OX40 stimulation (Fig. 6 E), suggesting that p300 recruitment is dependent on RelB expression.
To further ascertain the role of p300 in Il9 SE formation and Th9 cell induction, we investigated the effect of a pharmacological inhibitor of p300 (SGC-CBP30) in OX40-mdiated Th9 cell induction. We activated naive CD4+ T cells under Th9 cell–inducing conditions together with OX40 stimulation, and in these cultures we added the p300 inhibitor, or DMSO as a vehicle control. At different time points, we examined Th9 cell induction, as well as H3K27 acetylation, eRNA expression, and chromatin loop formation at the Il9 locus. We observed that addition of the p300 inhibitor resulted in a marked reduction of H3K27 acetylation at the Il9 SEa and the coding region in the presence of OX40 stimulation, as shown by H3K27Ac ChIP assay (Fig. 6 F). Similarly, induction of eRNA expression and chromatin loop formation by OX40 was also strongly inhibited by the p300 inhibitor (Fig. 6, G and H). Flow cytometry analysis showed that inhibition of p300 substantially reduced the induction of Th9 cells by OX40 stimulation (Fig. 6 I). Furthermore, studies using shRNA-mediated knockdown of p300 showed similar results in suppression of Th9 cell induction (Fig. S3, C and D), demonstrating a critical role for p300 in OX40-mediated Il9 SE formation and Th9 cell induction.
The BET protein inhibitor JQ1 suppresses allergic airway inflammation in vivo
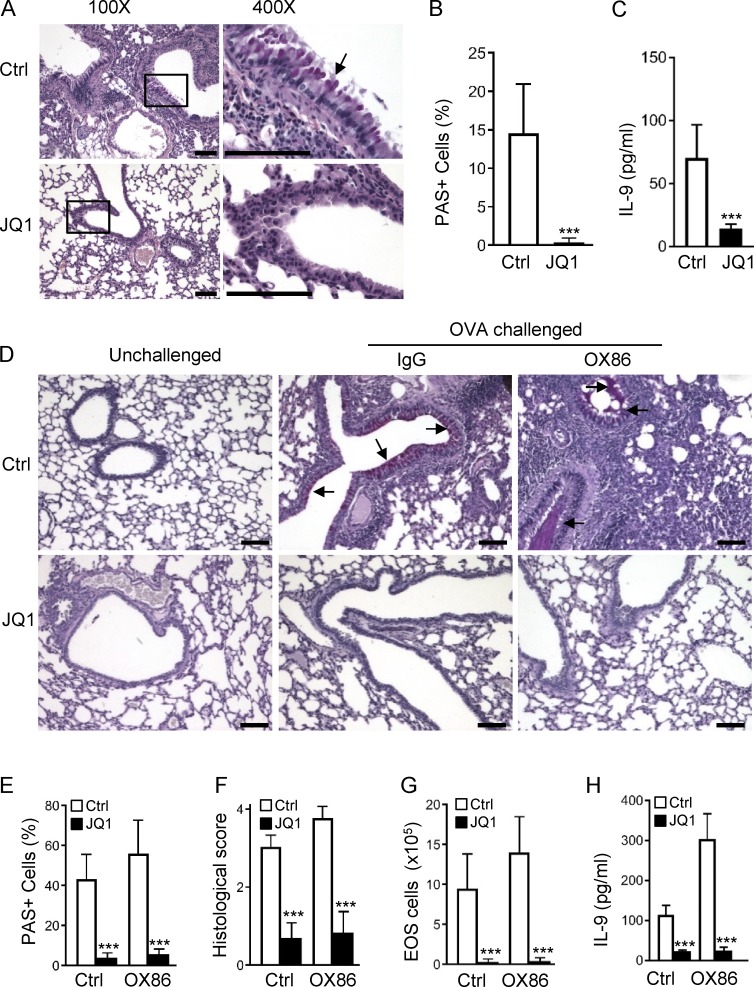
Th9 cells are prominently featured in allergic airway inflammation, especially in the proliferation of mucin-producing cells in the airway epithelium (Townsend et al., 2000). We then examined roles of the BET inhibitor JQ1 in allergic lung inflammation using several animal models. First, we treated OX40Ltg mice, which spontaneously develop severe airway inflammation (Xiao et al., 2012b), with JQ1 (50 mg/kg, i.p. for 14 d), and examined histological changes in the lungs. As shown in Fig. 7 A, treatment with JQ1 strongly inhibited hyperplasia of mucin-producing cells in the airways, as well as inflammatory cell infiltration in the lungs, and as compared with vehicle control–treated mice, the improvement in lung histology after JQ1 treatment was very striking. Moreover, quantitative assessments showed that proliferation of the mucin-producing cells (PAS+) was completely inhibited by JQ1 (Fig. 7 B), which was associated with a marked reduction of IL-9 in the bronchioalveolar lavage (BAL) fluid, as assessed by ELISA (Fig. 7 C).
Figure 7.
Disruption of SEs by the BET inhibitor JQ1 suppresses allergic airway inflammation in vivo. (A) Representative lung histology sections showing tissue inflammation and hyperplasia of airway epithelia in OX40Ltg mice treated with JQ1 (50 mg/kg) or vehicle control DMSO for 2 wk. The square represents areas that are shown under high power (400×). The arrow shows mucin-producing cells in the airway epithelia stained purple-red using PAS analysis. Bars, 100 µm. (B) Frequency of PAS+ mucin-producing cells in the airway epithelia in control or JQ1-treated OX40Ltg mice. (C) ELISA analysis of IL-9 in the BAL in OX40Ltg mice treated with or without JQ1. (D) Lung tissue sections from WT B6 mice immunized with OVA in alum, followed by challenges with aerosolized OVA with or without JQ1 treatment. Groups of mice were given OX86 or an isotype control IgG at the time of OVA challenge. Arrows denote hyperplasia of mucin-producing cells in airway epithelia. Bars, 100 µm. (E) Quantitation of PAS+ mucin-producing cells in control and JQ1-treated mice. (F) Histological scores based on cell infiltration, epithelial hyperplasia, and tissue damage. (G and H) The number of eosinophils (EOS; G) and IL-9 levels (H) in the BAL in mice treated with JQ1. Mice treated with the vehicle control DMSO were included as controls. Data in A and D are representative of two independent experiments, and data in B, C, and E–H are pooled from two independent experiments with five to seven mice per group in each experiment (mean and SD of n = 10–14). ***, P < 0.001.
In a different setting, we immunized WT B6 mice with OVA in alum adjuvant, and challenged them with aerosolized OVA 3 wk later for five consecutive days. In some experiments, the immunized mice were given two doses of OX86 at the time of OVA challenge (Fig. S4 A). Groups of mice were also treated with JQ1 starting 2 d before OVA challenge for 7 d. All mice were sacrificed on day 26 for analyses. As shown in Fig. 7 D, challenge of OVA-sensitized mice induced extensive inflammatory cell infiltration in the lungs, as well as strong hyperplasia of mucin-producing cells in the airway epithelium (Fig. 7 D and Fig. S4 B). OX86 treatment further enhanced interstitial lung inflammation, along with enhanced proliferation of mucin-producing cells in the airways (often with mucin plugs in the small airways; Fig. 7, D–F). These changes were associated with a marked increase in IL-9 production in the BAL (Fig. 7 H), features that are consistent with Th9 cell induction (Xiao et al., 2012a). Strikingly, JQ1 treatment produced dramatic improvements in the lungs, as assessed by tissue histology and disease scores, as well as eosinophilia and IL-9 production (Figs. 7, D–H). Of note, proliferation of the mucin-producing cells is driven by Th9 cells in vivo, and JQ1 treatment showed remarkable potency in suppressing proliferation of such mucin-producing cells, especially in the OX86-treated mice.
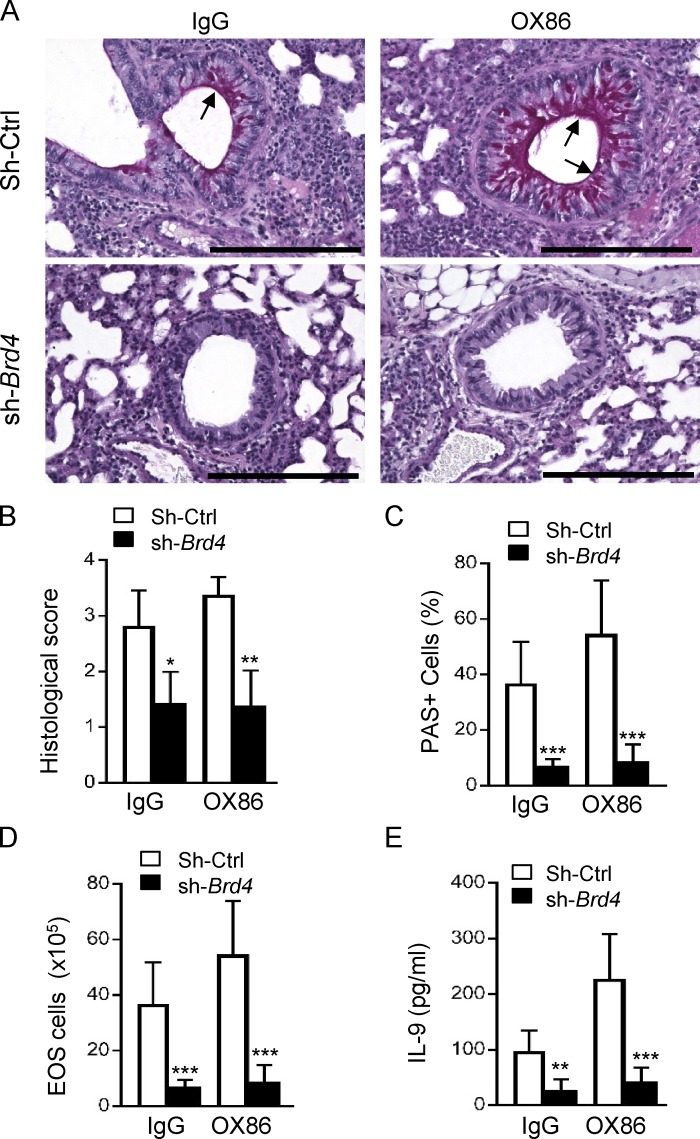
To specifically address the role of Th9 cells in vivo, we adoptively transferred OT-II cells into syngeneic Rag-1–deficient mice (2 × 106 cells/host), and groups of mice also received OT-II cells where Brd4 was selectively knocked down using Brd4-specific shRNA (or shRNA controls). The host mice were challenged with aerosolized OVA (5%) daily for 6 d and sacrificed on day 7 for analysis. As shown in Fig. 8, challenge of the Rag-1−/− mice with OVA that were transferred with OT-II cells induced proliferation of the mucin-producing cells in the airways (PAS+ cells), which was dramatically expanded after OX68 treatment, and knockdown of Brd4 in OT-II cells prevented the expansion of PAS+ cells in the airway epithelium (Fig. 8 C), as well as infiltration of eosinophils and production of IL-9 in the BAL fluid (Fig. 8, D and E).
Figure 8.
Role of Brd4 knockdown in allergic airway inflammation in an adoptive transfer model. (A) Lung histology showing inflammatory cell infiltration and proliferation of mucin producing cells in the airway epithelia in Rag-1−/− hosts adoptively transferred with WT OT-II cells and Brd4 knockdown OT-II cells. Arrows show airway epithelia and mucin-producing cells. The picture is representative of two experiments with five mice per group in each experiment. Bars, 100 µm. (B) The bar graph shows the lung histological score in control IgG and OX86-treated mice. (C) Quantitation of PAS+ mucin-producing cells in control IgG and OX86-treated mice. (D and E) The eosinophil numbers (EOS; D) and IL-9 levels (E) in the BAL of mice treated with control IgG and OX86. Data in A are representative of two independent experiments, and data in B–E are pooled from two independent experiments with five to six mice per group in each experiment (mean and SD of n = 10–12). *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Discussion
In the present study, we demonstrated a novel and potentially important mechanism in regulation of Th cell generation, i.e., formation of SEs as a key event in IL-9 expression, and subsequently in Th9 cell induction, and characterized in greater detail the structure and function relationships between Il9 SEs, IL-9 expression, and Th9 cell induction. We also demonstrated a potent role for OX40 in mediating SE formation, especially in driving chromatin remodeling required for SE assembly, and thus markedly expanded its roles as a T cell costimulatory molecule. We provided strong evidence that besides acting as a key transcription factor, RelB also drives extensive chromatin modifications that are mechanistically different from its transcriptional activities. Furthermore, we showed that the BET protein inhibitor JQ1, which acts primarily by disrupting SEs (Lovén et al., 2013), exhibits remarkable potency in suppressing allergic lung inflammation in vivo, highlighting the potential importance of BET protein inhibitors in the clinic.
A number of studies have shown that SEs are uniquely important in defining cell identities, as well as in lineage specification of stem cells (Hnisz et al., 2013; Fang et al., 2015; Kitagawa et al., 2017), and their involvement in IL-9 expression and Th9 cell induction is rather unexpected. Traditionally, induction of Th cell subsets depends on the expression of “lineage-defining” transcription factors, which are often induced by specific cytokines (Dong, 2008). However, Th9 cells appear to be an exception, as all the known transcription factors identified thus far are not Th9 cell–specific. Instead, a plethora of transcription factors can facilitate Th9 cell induction to various degrees (Kaplan, 2017). Moreover, besides the polarizing cytokines (TGF-β, IL-4), some TNFR superfamily members, especially OX40, exhibit remarkable potency in promoting Th9 cell induction (Xiao et al., 2012a). Because of that, uncertainty remains as to whether Th9 cells are truly a distinct Th cell subset. In this regard, the demonstration of SEs in IL-9 expression and Th9 cell induction is a significant step forward. In our model, induction of Il9 SEs by OX40 is remarkably robust, and targeted interference of SEs with either the BET inhibitor JQ1, or selective knockdown of Brd4 or Med1, proteins that are key for SE assembly (Vahedi et al., 2015), all abolished Th9 cell induction under conditions of OX40 stimulation. Of note, this shRNA-mediated knockdown of Brd4 or Med1 did not affect signaling of the polarizing cytokines, nor affected the OX40 signaling (unpublished data), but rather disrupted formation of SEs, as clearly revealed by the eRNA expression and chromatin loop formation assays. One potential limitation of this approach is that in vivo targeting of Brd4 or Med1, which are global regulators of SEs, may also affect other cytokines or other cell types. Nonetheless, our data suggest a model in which establishment of SEs is a prerequisite and likely precedes the transcription factors in robust induction of Th9 cells. Our data also suggest that without early chromatin remodeling and subsequent assembly of SEs, robust IL-9 expression may not be achieved, even under optimal Th9 cell polarizing conditions. This by no means negates the importance of transcription factors in Th9 cell differentiation, but rather provides additional mechanisms to explain the unique dynamics of Th9 cells. In fact, the multiplicity of transcription factors identified in Th9 cell induction suggests that some of those transcription factors may have very different roles in Th9 cell induction, beyond their transcription activities, and may likely participate in SE formation. This is consistent with the notion that SEs provide a key platform for assembly of transcription factors and transcription cofactors in which robust gene expression can be best achieved (Siersbæk et al., 2014). This is also supported by the strong enrichment of Brd4 and Med1 at the intragenic Il9 locus, as well as at the intergenic enhancer elements after OX40 costimulation. Clearly, the requirement of SEs for Th9 cell induction separates Th9 cells from other Th cell subsets and may help explain the broad number of transcription factors involved in Th9 cell induction; it may also explain that in certain models, Th9 cell–associated transcription factors alone show limited efficacy in Th9 cell induction (Li et al., 2017).
Our data emphasize the importance of intergenic regulatory elements, especially the Il9 SEa cluster, in robust Th9 cell induction. The Il9 coding region contains five exons, spanning a ∼3-kb genomic segment. Besides the promoter region (CNS1), there are two additional CNS regions identified in the mouse (Perumal and Kaplan, 2011). Interestingly, the SEa region extends from the fifth exon to include the CNS2 region, covering a ∼3.5-kb segment, and this region is especially important in robust Th9 cell induction. We provide evidence that the Il9 SEa can loop around to engage the Il9 promoter, and deletion of this region resulted in a significant reduction in Th9 cell induction by OX40 costimulation. Thus, our data provide an explanation that in two lines of Il9 reporter mice, insertion of reporter genes around the Il9 ATG codon may disrupt the interaction between SEa and Il9 promoter, explaining the poor efficacy in reporting Il9 expression in vivo (Wilhelm et al., 2011; Ulrich et al., 2017). Other genetic strategies that disrupt the integrity of SEa to generate Il9 reporter mice may also fail to faithfully report Il9 expression in vivo. Sequence alignment showed that the SEb clusters correspond to the CNS0 region, which shows features of SEs by ChIP-seq analysis, but fail to transcribe eRNA. Also, Cas9/sgRNA-mediated deletion of SEb had little impact on Th9 cell induction by OX40, thus questioning the exact role of this enhancer segment in our model. Similar questions remain for the SEc segment, which shows the most dramatic chromatin modifications. Nonetheless, the discrepancy between Brd4 knockdown and SEa deletion in Th9 cell induction by OX40 also suggests certain limitations of the Cas9/sgRNA approach used in our studies. Considering the challenges in coordinating two sets of sgRNA in binding to designated sites in primary T cells simultaneously, and the deletion efficacy of the CRISPR/Cas9 system in our studies (∼60%), the possibility that the CRISPR/Cas9 approach may not robust enough to allow accurate report of the roles of SE clusters in Th9 cells remains. However, the SEa produced high levels of eRNA, whereas SEb and SEc clusters did not, and therefore, the function of SEb and SEc as SEs remains to be clarified. Whether they are involved in other aspects of Th9 cells, such as function or stability, remains to be defined in future studies.
Our finding that in Th9 cells, extensive chromatin modifications at the Il9 locus are driven by RelB and p300, is highly interesting. Clearly, the dual role of RelB as a critical transcription factor and a key chromatin modifier makes RelB an interesting “hot spot” in chromatin remodeling and gene regulation (Brown et al., 2014). The Il9 locus and the intergenic regulatory regions are especially rich in κB sites, and we showed that RelB can productively recruit p300 to such sites to drive profound chromatin acetylation, as targeting p300, either with pharmacological inhibitors or shRNA-mediated knockdown, strongly inhibited H3K27 acetylation and, consequently, preventing SE formation as well as Th9 cell induction. This effect does not rely on its transactivation domain, which is very different from its transcription activities. In a different model, we showed that RelB is also capable of recruiting histone methyltransferases G9a and Setdb1 to mediate gene repression (Xiao et al., 2016), further supporting the role of RelB as a critical chromatin modifier. However, what drives the divergence of RelB from its classic role as a transcription factor to a chromatin modifier remains unknown, and whether other TNFR members that also activate RelB induce SE formation in promoting Th9 cells requires further investigation.
In summary, our study demonstrates that SEs are critically involved in Th9 cell induction, and formation of Il9 SEs is driven by OX40 signaling, primarily through RelB/p300-mediated chromatin acetylation. Conceptually, it is highly interesting that robust IL-9 expression and Th9 cell induction can be controlled at the level of SEs, in addition to the induction of transcription factors. This may place the Th9 cells as a unique cell type among a growing family of Th cell subsets. Our data also suggest that targeting SEs using BET protein inhibitors is highly efficacious in suppressing allergic lung inflammations, in which Th9 cells are prominently involved (Xiao et al., 2012a), highlighting the therapeutic potentials of such inhibitors in the clinic.
Materials and methods
Animals
Rela flox/flox, H11LSL-Cas9, and Cd4cre mice were obtained from The Jackson Laboratory. Relb−/−, OX40L-Tg, and Foxp3gfp reporter mice have been previously described (Xiao et al., 2012a). Some strains were crossed with Foxp3gfp mice to genetically mark Foxp3+ T regulatory cells for cell sorting purposes. All animals were maintained in a specific pathogen-free facility at Houston Methodist Research Institute in Houston, TX. Animal use and care were approved by the Houston Methodist Animal Care Committee in accordance with institutional animal care and use guidelines.
Polarization of naive CD4+ T cells in vitro
Naive CD4+ T cells (CD62L+CD44−Foxp3−) were FACS-sorted from foxp3gfp reporter mice and stimulated with mAb to CD3e (2 µg/ml, 2C11; eBioscience) plus equal numbers of syngeneic APCs in 96-well tissue-culture plates (Sigma-Aldrich) at 5 × 104 cells per well. APCs were prepared by depletion of T cells from total spleen cells with PE–anti-CD3 (2C11; Miltenyi Biotec) and anti-PE microbeads (Miltenyi Biotec), followed by a brief treatment with mitomycin C (50 µg/ml; Sigma-Aldrich) before each experiment. For induction of Th9 cells in vitro, cells were activated in the presence of human TGF-β1 (3 ng/ml) and mouse IL-4 (10 ng/ml). All recombinant cytokines were from R&D Systems. To stimulate OX40 signaling on activated T cells, OX40L transgenic APCs prepared from OX40L transgenic mice (OX40Ltg) were used. As an alternative method, OX40 agonist antibody OX86 (5 µg/ml, BioXcell) or recombinant mouse OX40L-His protein (100 ng/ml, along with 10 µg/ml of anti-His mAb; both from R&D Systems) was also used to activate OX40 signaling.
In some cases, cells were plated as above in the presence of Brd4 inhibitor (JQ1) or p300 inhibitor (SGC-CBP30; both from Sigma-Aldrich) at the concentrations as indicated. CD4+ T cells cultured for 2–4 d under Th9 cell–polarizing conditions were collected and assessed by intracellular IL-9 staining, ChIP, immunoblot, and quantitative real-time RT-PCR analyses.
Intracellular staining
For cytokine staining, CD4+ T cells activated under various polarizing conditions were restimulated for 4 h with phorbol 12-myristate 13-acetate (50 ng/ml) and ionomycin (550 ng/ml; Sigma-Aldrich) in the presence of GolgiStop (BD PharMingen). Cells were then fixed and permeablized with Foxp3 staining buffer set or Cytofix/Cytoperm solution to preserve the GFP signal, and then stained with fluorochrome conjugated anti–IL-9 (RM9A4), anti–IL-4 (11B11), and anti-CD4 (GK1.5) antibodies (all from Biolegend) according to the manufacturer’s instructions. All samples were acquired using LSRII, and data were analyzed with FlowJo v10 software (Xiao et al., 2016).
Immunoblot and immunoprecipitation analysis
Activated CD4+ T cells under various designed conditions were resuspended in culture medium and harvested by centrifugation. Harvested T cells were washed in cold PBS once and then lysed in RIPA lysis buffer (89901; Fisher Scientific) with proteases and phosphatase inhibitor cocktail for 30 min. After 15,000g centrifugation for 15 min, the supernatants were carefully transferred to new Eppendorf tubes as whole-cell protein extracts. For immunoprecipitation (IP) assay, cells were lysed in IP lysis buffer (87788; Fisher Scientific) with proteases and phosphatase inhibitor cocktail for 30 min. After collecting the lysate by centrifugation (15,000g for 15 min at 4°C), precleaning was performed by adding protein A beads into collected lysate and incubated with constant rotation for 1 h at 4°C, followed by another 15,000g centrifugation for 15 min at 4°C. The supernatants were then transferred into new Eppendorf tubes and ready for immunoprecipitation. The first step involved addition of primary antibodies to the supernatants, followed by incubation with constant rotation overnight at 4°C. The following antibodies against mouse antigens were used: anti-p300 (SC-32244, SC-585), and anti-RelB (SC-48379; all from Santa Cruz Biotechnology); anti-Med1 (A300-793A; Bethyl Laboratories); and anti-FLAG (F1804; Sigma-Aldrich). After adding the protein A agarose beads suspension, the mixture was further incubated with constant rotation for 3 h at 4°C. The precipitates were washed four times by using cold IP lysis buffer. After final washing, the beads were resuspended in sample buffer and boiled for 10 min. The immunoprecipitates or the whole cell lysates (WCLs) were resolved by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were probed with appropriate antibodies. The IgG horseradish peroxidase–conjugated antibodies were used as the secondary antibodies. The results were detected using the ECL-Plus immunoblotting detection system (Xiao et al., 2015).
Real-time quantitative RT-PCR analysis
Total RNA from targeted T cells was extracted with the RNeasy mini kit (74106; Qiagen) according to the manufacturer's instructions. In experiments for eRNA detection, potential DNA contamination in total RNA was removed by in-tube digestion. RNA concentration was determined using a Nano drop apparatus, and a total of 400 ng RNA were used for reversed transcription using a High-Capacity cDNA Reverse Transcription kit (4375575; ABI) with random primers. Reverse transcription was also performed by using oligo dT primers or without reverse transcription in all experiments for eRNA detection. Quantitative real-time PCR was performed by using specific primers (Table S1) and SYBR Green Mix, and analyzed using a Bio-Rad CFX96 real-time PCR system. Data were normalized to the expression of housekeeping GAPDH gene and the relative abundance of transcripts was calculated by the 2-ΔΔCt method.
Luciferase reporter assay
A luciferase reporter assay was performed as previously described (Xiao et al., 2012a). In brief, the Il9 promoter region consisting of ∼1 kb in size upstream of the transcription start site was cloned into pGL3 luciferase reporter plasmid (Promega). HEK293 T cells were transfected with the reporter constructs together with expression plasmids carrying cDNA encoding individual Th9 cell–related transcription factors using the Lipofectamine 2000 method (Invitrogen). The Il-17a reporter was used as a positive control. After 48 h, the luciferase activities of WCLs were analyzed using the dual-luciferase reporter assay system (Promega). Cotransfection with the Renilla-luciferase expression vector pRL-TK was performed in all reporter assays. For all samples, the relative luciferase activity was calculated by dividing the firefly luciferase activity to the Renilla luciferase activity.
Retrovirus-mediated gene expression
The cDNA fragments encoding mouse RelB (full-length or various RelB mutants) were amplified by PCR and further cloned into the pMYs-IRES-EGFP retroviral vector (Cell Biolabs). Retroviral particles were prepared by transfection of these vectors into packaging Plat-E cells according to the manufacturer’s recommendations (Cell Biolabs). For the transduction of T cells, naive CD4+ T cells were activated for 24 h with the Dynabeads of Mouse CD3/CD28 T Cell Expander (11453D; Invitrogen) at first, followed by incubation with freshly prepared retroviral particles by centrifugation for 2 h at 780g and 32°C in the presence of 10 µg/ml polybrene (Sigma-Aldrich). After centrifugation, cells were cultured for 6 h at 32°C, followed by culture of the transduced T cells under various polarizing conditions for additional 3 d in complete 1640 medium at 37°C. The induction of Th9 cells from naive CD4+ T cells was assessed by intracellular staining and flow cytometry, as previously reported (Xiao et al., 2012a).
shRNA-mediated gene knockdown in T cells
The shRNA sequences (Table S1) to targeting genes were designed by using an online tool (https://rnaidesigner.thermofisher.com). Five top-listed shRNA sequences for each gene were selected and synthesized at Integrated DNA Technologies (IDT; Coralville, Iowa). Two complementary oligos were annealed at annealing buffer and ligated into shRNA vector (pSIREN-RetroQ-ZsGreen) from Clontech. After BamHI and EcoRI digestion (both from NEB), positive colonies were further verified by direct sequencing. The knockdown efficiency for each shRNA was measured in NIH3T3 cells at first by immunoblot, and the two most powerful shRNAs were selected for further experiments in T cells. The knockdown efficiency in T cells was further verified by immunoblot analysis using FACS-sorted shRNA transduced T cells before individual experiments (Xiao et al., 2016).
CRISPR/CAS9-mediated gene deletion
For the design of sequence-specific guide RNA (sgRNA), ∼200 bp length DNA sequence at both sides of the targeted DNA fragment was used as a reference sequence. CRISPR crRNA was designed using an online tool (http://crispr.mit.edu/). Based the score and the number of off-targeted sites, one sequence for generation of crRNA in each site was selected. The sgRNA oligos (Table S1) containing the crRNA and the transactivating CRISPR RNA (tracrRNA) sequences were directly synthesized by the IDT company. After annealing, the sgRNA oligos were cloned into pSIREN-RetroQ-ZsGreen or RNAi-Ready pSIREN-RetroQ-ZsRed vector (both from Clontech Laboratories) by using BamHI and EcoRI sites. The sgRNA sequence was confirmed by direct sequencing. A pair of sgRNA targeting both sites along with the Cas9 encoding plasmids were transfected into NIH3T3 cells to examine the cleavage efficiency by PCR-based genotyping. Highly efficient sgRNAs were selected and transfected into Platinum-E cells for generation of retrovirus. Retroviruses were used to infect Cd4creCas9 LSL transgenic CD4+ T cells to induce specific deletion of targeted genomic sequences. Deletion of targeted DNA fragments was further confirmed by PCR-based genotyping as well as DNA sequencing (Sander and Joung, 2014). The efficiency of deletion was determined by real-time PCR using specific primers (Table S1) to the deleted sequences, as well as primers to the Il9 coding sequence in the same reactions, which were further used for normalization.
ChIP-PCR and ChIP-seq
ChIP was performed by using a ChIP-IT PBMC kit (53042; Active Motif). In brief, targeted T cells were harvested and fixed with 1% formaldehyde for 15 min and quenched with 0.125 M glycine. Cells were resuspended in swelling buffer with detergent for 30 min and centrifuged for 10 min at 3,200g at 4°C. Cell pellets were resuspended in 500 µl ChIP buffer for chromatin sonication. Genomic DNA was sonicated to a mean length of 300–500 bp. The input genomic DNA was prepared by treating aliquots of chromatin with RNase and proteinase K, after heating for de–cross-linking, and then precipitated by ethanol. An aliquot of chromatin (25–30 µg) was precleaned with protein A agarose beads. Epigenetically modified or transcription factor–binding genomic DNA was isolated using specific antibodies (4 µg). The following antibodies against mouse antigens were used: anti-Drd4 (A301-985A50; Bethyl Laboratiories); anti-H3K27Ac (39133; Active Motif); and anti-p300 (SC-585) and anti-RelB (SC-226; both from Santa Cruz Biotechnology). DNA and protein complexes were washed, eluted from the beads with SDS buffer, and subjected to RNase and proteinase K treatment. Cross-links were reversed by incubation overnight at 65°C, and ChIP DNA was purified by phenol-chloroform extraction and ethanol precipitation. Quantitative PCR (qPCR) reactions were performed in triplicate to detect specific genomic regions using SYBR Green Supermix (1725274; Bio-Rad) and specific primers (Table S1). The resulting signals were normalized to signals from input DNA using the same primers.
Illumina sequencing libraries were prepared from the ChIP and input DNAs by the standard consecutive enzymatic steps for end-polishing, dA-addition, and adaptor ligation. After a final PCR amplification step, the resulting DNA libraries were quantified and sequenced on Illumina’s NextSeq 500 (75 nt reads, single end). Reads were aligned to the mouse genome (mm10) using the BWA algorithm (default settings; Li and Durbin, 2009). Duplicate reads were removed, and only uniquely mapped reads (mapping quality ≥ 25) were used for further analysis. Alignments were extended in silico at their 3′-ends to a length of 200 bp, which is the mean genomic fragment length in the size-selected library, and assigned to 32-nt bins along the genome. The resulting histograms (genomic “signal maps”) were stored in BigWig files. Peak locations were determined using the MACS algorithm (v1.4.2) with a cutoff of P value of 10−7 (Zhang et al., 2008). Rank Ordering of SEs (ROSE) was performed with default settings (i.e., no promoter exclusion) using MACS peaks as input (Hnisz et al., 2013).
3C assay
The 3C assay was performed according to the published protocol (Grob and Grossniklaus, 2017). In brief, 10 million activated CD4+ T cells were fixed by 2% formaldehyde for 10 min at room temperature. The cross-linking reaction was quenched by adding cold 1-M glycine solution. After centrifugation, the cell pellet was lysed in 5 ml cold IP lysis buffer and was incubated for 10 min on ice. The nuclei were obtained by discarding the supernatant after centrifuge for 5 min at 400g and further digested by DpnII restriction enzyme (R0543L; NEB) overnight at 37°C. The digested DNA was diluted 10-fold and ligated by T4 DNA ligase for 5 h at room temperature. The cross-links were reversed by overnight incubation at 65°C in the presence of 5 mg/ml proteinase K. Ligated DNA was extracted by phenol–chloroform methods. The DNA was dissolved and quantified by Nanodrop. The extracted DNA was further determined by the agarose gel–based method as well as qPCR using primers to amplify no-DpnII site fragment. After that, the 3C sample DNA was adjusted to a similar concentration and further used to detect all possible ligation products by qPCR with all possible primer combinations (Table S1). PCR products were run on 1.5% agarose gels containing 0.75 mg/ml ethidium bromide. Ligation products that were detected by PCR showing the expected size were further sequenced to confirm the specific ligation events.
Acute allergic lung inflammation model
WT B6 mice at 6–12 wk old were sensitized by 100 µg of OVA (S7951; Sigma-Aldrich) in 100 µl of Imject Alum (77161; Fisher Scientific) i.p. The sensitizing solution was given at day 0 and day 14. At day 20, the sensitized mice were randomly divided into four groups. All mice were then challenged with aerosolized OVA (1% in PBS) for 30 min over a period of five consecutive days (days 21, 22, 23, 24, and 25). The OVA aerosol was generated using a Bennett nebulizer at a flow rate of 10 liters/min. Two of four groups of mice were also treated with two doses (0.5 mg) of agonist anti-OX40 antibody (OX86) at day 21 and day 23. Before challenge with aerosolized OVA, two groups of mice with or without OX86 treatment were also treated with JQ1 given i.p. at 50 mg/kg per mouse. In other experiments, FACS-sorted sh-Brd4 transduced (GFP+) OT-II cells (2 × 106/mouse) were injected via the tail vein into syngeneic hosts deficient in recombination-activating gene 1. sh-Ctrl–transduced OT-II cells were used as controls. The next day, host mice were challenged 30 min daily for 6 d with aerosolized OVA (5%) via the airways. Groups of mice were also treated with OX86 or control IgG at day 2 and day 5 during OVA challenge. One day after the final challenge, all the mice were sacrificed for analyses. Specifically, the lungs were dissected, and the trachea was cannulated with a polyethylene tube for lavage. BAL was collected with 1 ml PBS and centrifuged at 500g to separate cells from liquid. BAL fluid was used to determine IL-9 levels by ELISA (88-8092-88; eBioscience). The total number of BAL cells was determined by trypan blue exclusion, and then differential cell counts were assessed by cytospins and staining with a Hema 3 staining kit (Fisher Scientific), a modified Wright-Giemsa stain. Lung tissue sections were also used for H&E and PAS staining. For quantification of tissue histopathology as well as peribronchial and perivascular cellular infiltration, samples were graded by a semiquantitative scoring system (0–4, where 0 indicates no inflammation). For each slide, five randomly chosen areas were assigned scores. Mucin-producing cells were assessed by the frequency of epithelial cells that stained strongly positive for PAS. Individual slides were examined independently. Pictures were processed using a ZEISS AxioImager Z1 and LEICA DM 6000 B microscope (Xiao et al., 2012a).
Statistical analysis
The Mann-Whitney test was used for analysis of histology scores. A two-tailed Student’s t test or one-way ANOVA was used to generate P value data, as specified in the figure legends. A p-value <0.05 was considered statistically significant.
Data availability
ChIP-seq data for this project have been deposited at NCBI’s Gene Expression Omnibus under the following accession no.: GSE102940.
Online supplemental information
Fig. S1 shows the CRISPR/Cas9 strategy in deletion of Il9 SE in activated CD4+ T cells. Fig. S2 shows the RelB-dependent enrichment of Brd4 at the Il9 SE regions. Fig. S3 demonstrates a critical role of p300 in the formation of Il9 SEs. Fig. S4 includes a schematic of the OVA-induced asthma model and PAS staining of lung sections. Table S1 shows qPCR primers and oligos used in this study.
Supplementary Material
Acknowledgments
We thank Professor Naoto Ishii at Tohoku University (Sendai, Japan) for the generous gift of OX40Ltg mice. We also acknowledge the excellent services from the flow cytometry core and the pathology core at Houston Methodist Hospital in Houston, TX.
This work was supported by grants from the National Institutes of Health (R01AI129906 and R01AI106200).
The authors declare no competing financial interests.
Author contributions: X. Xiao and Y. Fan designed and performed experiments and analyzed the data. J. Li bred mouse colonies used in the study and helped with some experiments. L. Minze provided operational support. Y. Xiao performed ChIP-seq analysis. X. Lou, X. Zhang, P. Lan, X. Shi, and Y. Dou performed some experiments and provided helpful discussions. X.C. Li supervised the studies and wrote the paper.
References
- Brown J.D., Lin C.Y., Duan Q., Griffin G., Federation A., Paranal R.M., Bair S., Newton G., Lichtman A., Kung A., et al. 2014. NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol. Cell. 56:219–231. 10.1016/j.molcel.2014.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft M. 2010. Control of immunity by the TNFR-related molecule OX40 (CD134). Annu. Rev. Immunol. 28:57–78. 10.1146/annurev-immunol-030409-101243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardalhon V., Awasthi A., Kwon H., Galileos G., Gao W., Sobel R.A., Mitsdoerffer M., Strom T.B., Elyaman W., Ho I.C., et al. 2008. IL-4 inhibits TGF-β–induced-Foxp3+ T cells, and together with TGF-β, generates IL-9+ IL-10+ Foxp3−effector T cells. Nat. Immunol. 9:1347–1355. 10.1038/ni.1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C. 2008. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat. Rev. Immunol. 8:337–348. 10.1038/nri2295 [DOI] [PubMed] [Google Scholar]
- Fang Z., Hecklau K., Gross F., Bachmann I., Venzke M., Karl M., Schuchhardt J., Radbruch A., Herzel H., and Baumgrass R.. 2015. Transcription factor co-occupied regions in the murine genome constitute T-helper-cell subtype-specific enhancers. Eur. J. Immunol. 45:3150–3157. 10.1002/eji.201545713 [DOI] [PubMed] [Google Scholar]
- Grob S., and Grossniklaus U.. 2017. Chromatin Conformation Capture-Based Analysis of Nuclear Architecture. Methods Mol. Biol. 1456:15–32. 10.1007/978-1-4899-7708-3_2 [DOI] [PubMed] [Google Scholar]
- Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., and Young R.A.. 2013. Super-enhancers in the control of cell identity and disease. Cell. 155:934–947. 10.1016/j.cell.2013.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.A., Schmitt A.D., Espinoza C.A., and Ren B.. 2013. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 503:290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan M.H. 2013. Th9 cells: differentiation and disease. Immunol. Rev. 252:104–115. 10.1111/imr.12028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan M.H. 2017. The transcription factor network in Th9 cells. Semin. Immunopathol. 39:11–20. 10.1007/s00281-016-0600-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa Y., Ohkura N., Kidani Y., Vandenbon A., Hirota K., Kawakami R., Yasuda K., Motooka D., Nakamura S., Kondo M., et al. 2017. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat. Immunol. 18:173–183. 10.1038/ni.3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chen S., Xiao X., Zhao Y., Ding W., and Li X.C.. 2017. IL-9 and Th9 cells in health and diseases-From tolerance to immunopathology. Cytokine Growth Factor Rev. 37:47–55. 10.1016/j.cytogfr.2017.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Spolski R., Liao W., and Leonard W.J.. 2014. Complex interactions of transcription factors in mediating cytokine biology in T cells. Immunol. Rev. 261:141–156. 10.1111/imr.12199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., and Young R.A.. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 153:320–334. 10.1016/j.cell.2013.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Hong S., Li H., Park J., Hong B., Wang L., Zheng Y., Liu Z., Xu J., He J., et al. 2012. Th9 cells promote antitumor immune responses in vivo. J. Clin. Invest. 122:4160–4171. 10.1172/JCI65459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan F., and Siegel R.M.. 2017. TNF superfamily cytokines in the promotion of Th9 differentiation and immunopathology. Semin. Immunopathol. 39:21–28. 10.1007/s00281-016-0612-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak E.C., Weaver C.T., Turner H., Begum-Haque S., Becher B., Schreiner B., Coyle A.J., Kasper L.H., and Noelle R.J.. 2009. IL-9 as a mediator of Th17-driven inflammatory disease. J. Exp. Med. 206:1653–1660. 10.1084/jem.20090246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumal N.B., and Kaplan M.H.. 2011. Regulating Il9 transcription in T helper cells. Trends Immunol. 32:146–150. 10.1016/j.it.2011.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott S., and Lieb J.D.. 2015. What are super-enhancers? Nat. Genet. 47:8–12. 10.1038/ng.3167 [DOI] [PubMed] [Google Scholar]
- Sander J.D., and Joung J.K.. 2014. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32:347–355. 10.1038/nbt.2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt E., Germann T., Goedert S., Hoehn P., Huels C., Koelsch S., Kühn R., Müller W., Palm N., and Rüde E.. 1994. IL-9 production of naive CD4+ T cells depends on IL-2, is synergistically enhanced by a combination of TGF-beta and IL-4, and is inhibited by IFN-gamma. J. Immunol. 153:3989–3996. [PubMed] [Google Scholar]
- Schmitt E., Klein M., and Bopp T.. 2014. Th9 cells, new players in adaptive immunity. Trends Immunol. 35:61–68. 10.1016/j.it.2013.10.004 [DOI] [PubMed] [Google Scholar]
- Siersbæk R., Rabiee A., Nielsen R., Sidoli S., Traynor S., Loft A., Poulsen L.C., Rogowska-Wrzesinska A., Jensen O.N., and Mandrup S.. 2014. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Reports. 7:1443–1455. 10.1016/j.celrep.2014.04.042 [DOI] [PubMed] [Google Scholar]
- Sun S.C. 2017. The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 17:545–558. 10.1038/nri.2017.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C., and Gery I.. 2012. The unique features of Th9 cells and their products. Crit. Rev. Immunol. 32:1–10. 10.1615/CritRevImmunol.v32.i1.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend J.M., Fallon G.P., Matthews J.D., Smith P., Jolin E.H., and McKenzie N.A.. 2000. IL-9-deficient mice establish fundamental roles for IL-9 in pulmonary mastocytosis and goblet cell hyperplasia but not T cell development. Immunity. 13:573–583. 10.1016/S1074-7613(00)00056-X [DOI] [PubMed] [Google Scholar]
- Ulrich B.J., Verdan F.F., McKenzie A.N., Kaplan M.H., and Olson M.R.. 2017. STAT3 Activation Impairs the Stability of Th9 Cells. J. Immunol. 198:2302–2309. 10.4049/jimmunol.1601624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahedi G., Kanno Y., Furumoto Y., Jiang K., Parker S.C., Erdos M.R., Davis S.R., Roychoudhuri R., Restifo N.P., Gadina M., et al. 2015. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature. 520:558–562. 10.1038/nature14154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S., and Karin M.. 2009. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 27:693–733. 10.1146/annurev.immunol.021908.132641 [DOI] [PubMed] [Google Scholar]
- Wilhelm C., Hirota K., Stieglitz B., Van Snick J., Tolaini M., Lahl K., Sparwasser T., Helmby H., and Stockinger B.. 2011. An IL-9 fate reporter demonstrates the induction of an innate IL-9 response in lung inflammation. Nat. Immunol. 12:1071–1077. 10.1038/ni.2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte S., O’Shea J.J., and Vahedi G.. 2015. Super-enhancers: Asset management in immune cell genomes. Trends Immunol. 36:519–526. 10.1016/j.it.2015.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Balasubramanian S., Liu W., Chu X., Wang H., Taparowsky E.J., Fu Y.X., Choi Y., Walsh M.C., and Li X.C.. 2012a OX40 signaling favors the induction of T(H)9 cells and airway inflammation. Nat. Immunol. 13:981–990. 10.1038/ni.2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Gong W., Demirci G., Liu W., Spoerl S., Chu X., Bishop D.K., Turka L.A., and Li X.C.. 2012b New insights on OX40 in the control of T cell immunity and immune tolerance in vivo. J. Immunol. 188:892–901. 10.4049/jimmunol.1101373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Shi X., Fan Y., Zhang X., Wu M., Lan P., Minze L., Fu Y.X., Ghobrial R.M., Liu W., and Li X.C.. 2015. GITR subverts Foxp3(+) Tregs to boost Th9 immunity through regulation of histone acetylation. Nat. Commun. 6:8266 10.1038/ncomms9266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Shi X., Fan Y., Wu C., Zhang X., Minze L., Liu W., Ghobrial R.M., Lan P., and Li X.C.. 2016. The Costimulatory Receptor OX40 Inhibits Interleukin-17 Expression through Activation of Repressive Chromatin Remodeling Pathways. Immunity. 44:1271–1283. 10.1016/j.immuni.2016.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Xiao X., Ghobrial R.M., and Li X.C.. 2013. IL-9 and Th9 cells: progress and challenges. Int. Immunol. 25:547–551. 10.1093/intimm/dxt039 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq data for this project have been deposited at NCBI’s Gene Expression Omnibus under the following accession no.: GSE102940.