Capucha et al. demonstrate that mucosal Langerhans cell (LC) differentiation from pre–dendritic cells and monocytes involves consecutive BMP7 and TGF-β1 signaling in separate anatomical locations. Moreover, mucosal microbiota regulates the development of LCs that in turn shape microbial and immunological homeostasis.
Abstract
Mucosal Langerhans cells (LCs) originate from pre–dendritic cells and monocytes. However, the mechanisms involved in their in situ development remain unclear. Here, we demonstrate that the differentiation of murine mucosal LCs is a two-step process. In the lamina propria, signaling via BMP7-ALK3 promotes translocation of LC precursors to the epithelium. Within the epithelium, TGF-β1 finalizes LC differentiation, and ALK5 is crucial to this process. Moreover, the local microbiota has a major impact on the development of mucosal LCs, whereas LCs in turn maintain mucosal homeostasis and prevent tissue destruction. These results reveal the differential and sequential role of TGF-β1 and BMP7 in LC differentiation and highlight the intimate interplay of LCs with the microbiota.
Introduction
Langerhans cells (LCs) represent a special type of APC embedded in stratified squamous epithelia of skin and mucosa. Skin and mucosal LCs share similar transcriptomic signatures and immunological functions, and both express langerin (CD207) as well as high levels of epithelial cell adhesion molecule (EpCAM [CD326]; Nagao et al., 2009; Arizon et al., 2012; Capucha et al., 2015). Despite these similarities, skin and mucosal LCs greatly differ in their origins and developmental pathways. Skin LCs originate from embryonic precursors that seed the epidermis prenatally and expand rapidly after birth while differentiating into a radioresistant and self-renewing population (Merad et al., 2002; Chorro et al., 2009; Hoeffel et al., 2012). Mucosal LCs, on the other hand, originate from bone marrow (BM) precursors (pre–dendritic cells [pre-DCs] and monocytes), which gradually differentiate in the epithelium after birth and are continuously replenished from the circulation (Capucha et al., 2015). In vivo and in vitro data have established that skin LCs require TGF-β1 for their development. For instance, skin LCs are absent in mice lacking TGF-β1, Id2, or Runx3, the last two being transcription factors controlled by TGF-β1 (Borkowski et al., 1996; Hacker et al., 2003; Fainaru et al., 2004). Furthermore, ablation of TGF-β receptor I (ALK5) in CD11c-expressing cells impairs both postnatal differentiation and maintenance of immature LCs in the skin (Kel et al., 2010). A reduction in skin LCs was also observed after ablation of TGF-β receptor II or TGF-β1 in langerin-expressing cells, indicating that autocrine signaling via TGF-β1 is required for the maintenance of fully differentiated LCs (Kaplan et al., 2007; Bobr et al., 2010). It was also shown that differentiation of LCs from monocytes by TGF-β1 involves repression of Kruppel-like factor 4 (Jurkin et al., 2017). Nevertheless, recent studies have questioned the role of TGF-β1 in LC development. First, deletion of the canonical TGF-β1–Smad signaling pathway had no effect on cutaneous LC homeostasis (Xu et al., 2012; Li et al., 2016) Second, bone morphogenetic protein 7 (BMP7), a member of the TGF-β superfamily, induces potent differentiation of LC-like cells from human CD34+ hematopoietic progenitor cells by activating the BMP type I receptor (ALK3; Yasmin et al., 2013). Moreover, the ability of TGF-β1 to generate human LC-like cells is mediated by ALK3, whereas simultaneous activation of ALK5 abrogated their differentiation. Although these findings illustrate the controversy regarding the contribution of TGF-β1 and BMP7 to LC differentiation in the skin, the mechanisms mediating mucosal LC development are largely unknown.
Besides molecular instructions encoded by the host genome, LC differentiation might be also shaped by environmental factors. Epithelial tissues are in close contact with commensal microbiota, which is known to modulate mucosal immunity and steady-state hematopoiesis (Ouchi et al., 2011; Naik et al., 2012, 2015; Khosravi et al., 2014). We recently reported that the microbiota induces expression of growth arrest protein 6 (GAS6) in the oral epithelium after birth, expression that was crucial for maintaining mucosal homeostasis (Nassar et al., 2017). GAS6 is a potent ligand of AXL, a tyrosine kinase receptor acting downstream of TGF-β1 that regulates epidermal LC development (Bauer et al., 2012). Because mucosal LCs developed gradually in the oral epithelium after birth, we hypothesized that oral symbiotic bacteria, which are required for postnatal maturation of the epithelium, will also regulate the differentiation of oral mucosal LCs. In this study, we demonstrate that sequential BMP7 and TGF-β1 signaling regulated by the local microbiota controls the development of mucosal LCs.
Results
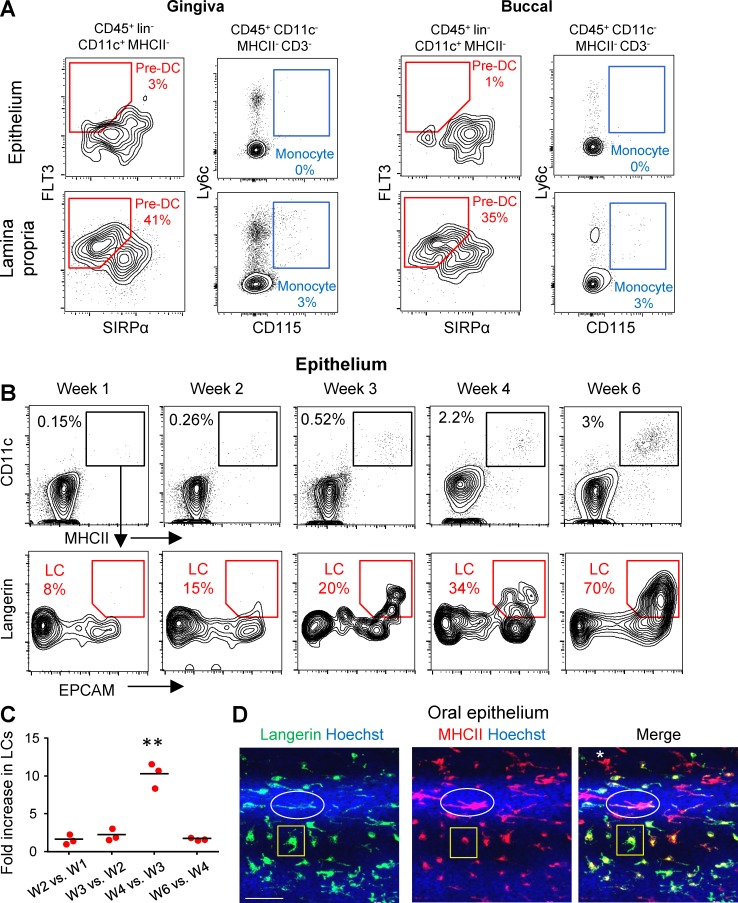
LC precursors enter the murine mucosal epithelium as MHCII+CD11c+ cells and then sequentially express EpCAM and langerin
To dissect the mechanism of mucosal LC differentiation, we first characterized the location of LC precursors in the mucosa. Epithelial and lamina propria layers were separated from the gingiva and buccal mucosa and then processed and stained with antibodies to identify pre-DCs (CD45+linnegCD11cintMHCIInegFlt3+Sirpαint) and monocyte (CD45+CD11cnegMHCIInegCD3negLy6C+CD115+) precursors. As demonstrated in Fig. 1 A, LC precursors were clearly detected in the lamina propria but could not be identified in the epithelium. We then took advantage of the gradual differentiation of oral LCs postnatally to characterize the acquisition kinetics of surface LC markers. As early as 1 wk after birth, a small population of MHCII+CD11c+ cells was observed in the mucosal epithelium (Fig. 1 B). The frequency of these cells increased slowly in the following weeks, as well as the level of CD11c expression. Examination of EpCAM and langerin expression revealed that a fraction of the MHCII+CD11c+ population starts to express EpCAM followed by langerin. We next quantified the relative increment in LC seeding during the early weeks of life (Fig. 1 C). A steady increase in LC differentiation was observed throughout the 2 wk after birth (approximately one- to twofold); however, between the third and fourth week, a massively increased differentiation rate of >10-fold was detected, which returned to steady levels in the following weeks. In a direct correlation with this differentiation kinetics, we observed the highest frequency of pre-DCs 2 wk after birth (Fig. S1 A). With regard to the lamina propria, a relatively large population of MHCII+CD11c+ cells was found 1 wk after birth, and its size increased in a fashion similar to that observed in the epithelium (Fig. S1, B and C). Nevertheless, in contrast to the epithelium, these cells did not express EpCAM or langerin.
Figure 1.
Mucosal LC precursors enter the epithelium as CD11c+MHCII+ cells and acquire LC phenotype in the epithelium. (A) Epithelial and lamina propria layers were separated from gingival and buccal tissues of 6-wk-old B6 mice (n = 4), representative flow cytometry plots present the frequencies of live pre-DCs (PI−CD45+Lin−CD11c+FLT3+SIRPαint) and monocytes (PI−CD45+CD11c−MHCII−CD3−Ly6C+CD115+) in each layer. Data are representative of three independent experiments. (B) Gingival epithelial layers were prepared from B6 mice at the noted weeks after birth for flow cytometry analysis. After gating on live CD45+ cells, the frequencies of CD11c+MHCII+ cells at each week, as well as the capacity of this population to express EpCAM and langerin, are presented. (C) Graph presents the fold change in total LC numbers between the successive weeks examined as the mean (n = 3). One representative out of three independent experiments is shown. (D) Gingival epithelial layer of 6-wk-old B6 mice stained for MHCII (red), langerin (green), and Hoechst (blue) for nuclear visualization. Shown are two LCs subsets with distinct cellular expression patterns are indicated (ellipse and rectangle), as well cells expressing MHC only (asterisk). Representative confocal microscopy images are presented representing one out of three independent experiments. Bar, 50 µm. **, P < 0.001.
Next, we monitored the capacity of mucosal LCs to proliferate in the epithelium by analyzing cell cycle phases using flow cytometry (Fig. S1 C) and by immunofluorescence staining against langerin and Ki67 (Fig. S1 D). The vast majority of LCs in the oral epithelium were in G1 phase and negative to Ki67, suggesting that after differentiation, oral LCs are hardly proliferating. Next, we used confocal laser scanning microscopy on whole-mount gingival epithelial tissues to visualize expression of MHCII and langerin. As depicted in Fig. 1 D, two phenotypically distinct LC subsets were identified: the first subset expresses MHCII all over the cell body, with langerin expressed on the cell surface. In the second subset, expression of MHCII was mainly perinuclear, with langerin expressed mostly in the cell body. These expression patterns are in agreement with the reported dual origin of BM-derived epidermal LCs (Seré et al., 2012). An additional type of APC expressing MHCII, but not langerin, was observed in the epithelium, representing LC precursors and/or macrophages (Capucha et al., 2015). Altogether, these results suggest that pre-DCs and monocytes acquire a MHCII+CD11c+ phenotype in the lamina propria. Within the epithelium, they express EpCAM and later on langerin as they develop into LCs. This differentiation process is abruptly accelerated around the third week of life, most likely because of elevated entry of precursors rather than proliferation of LCs.
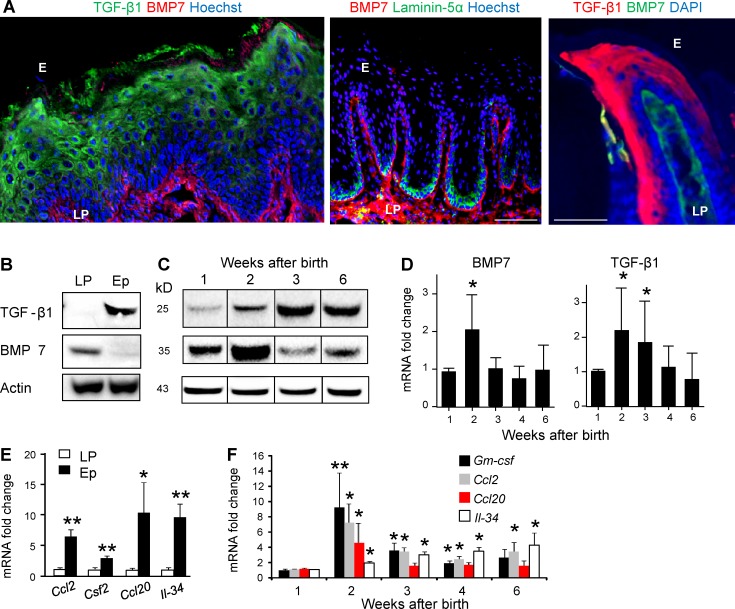
TGF-β1 and BMP7 are differentially expressed in the murine oral mucosa
Because mucosal LCs acquire their final phenotype within the epithelium, we next examined the expression of molecules known to be involved in LC differentiation such as TGF-β1 and BMP7. As depicted in Fig. 2 (A and B) and S1E, whereas TGF-β1 was expressed in the mucosal epithelium at the suprabasal layers, BMP7 expression was restricted to the lamina propria adjacent to the basal membrane. Staining with laminin-5 to visually separate the epithelium from the lamina propria clearly demonstrates the distinct anatomical localization of TGF-β1 and BMP7. Quantifying protein and mRNA levels of these molecules postnatally revealed that a relatively low level of TGF-β1 protein is present in the tissue in the first week of life, with an increased expression on the second and third weeks to reach steady-state protein levels (Fig. 2, C and D). Levels of BMP7 protein, on the other hand, were relatively high 1 wk after birth, and RNA and protein levels increased considerably during the second week of life but returned to steady-state levels by week 3. We next characterized expression of GM-CSF, CCL20, CCL2, and IL-34, which are also involved in LC development (Romani et al., 2010; Clausen and Stoitzner, 2015). These cytokines and chemokines were expressed at higher levels in the epithelium relative to the lamina propria (Fig. 2 E). In particular, expression of GM-CSF, CCL20 and CCL2 was sharply up-regulated during the second week of life, after which it gradually declined to steady-state levels (Fig. 2 F). IL-34, on the other hand, did not follow this pattern and instead increased constantly after birth throughout the examined period of time. These data thus suggest that the mucosal epithelium delivers the developmental instructions to attract circulating LC precursors and promote their differentiation in situ. Furthermore, the association of BMP7 with the basal membrane suggests that similar to other systems, it might play a role in enabling the transition of LC precursors to the mucosal epithelium (Zeisberg et al., 2005; Hieronymus et al., 2015).
Figure 2.
Expression profiles of cytokines and chemokines involved in LC differentiation in the oral mucosa. (A) Buccal mucosa cross sections of 6-wk-old B6 mice were stained for TGF-β1 (green), BMP7 (red), and Hoechst (blue) or alternatively with laminin-5α (green), BMP7 (red), and Hoechst (blue) for immunofluorescence analysis. Gingival cross sections (right) were stained for TGF-β1 (red), BMP7 (green), and DAPI (blue). Representative confocal microscopy images from one out of four independent experiments. E, epithelium; LP, lamina propria. (B) Western blot analysis showing the presence of TGF-β1, BMP7, and Actin in the epithelium (E) and lamina propria (LP) of gingival tissue of 6-wk-old B6 mice (n = 3). (C) TGF-β1 and BMP7 proteins in whole gingival tissues excised from B6 mice (n = 3) at the noted weeks after birth were analyzed by Western blot. Representative results of one out of three independent experiments are shown. (D) Relative expression levels of Tgfb1 and Bmp7 in whole gingival tissues isolated at the indicated time points were analyzed by quantitative RT-PCR. (E and F) Expression of Ccl2, Csf2, Ccl20, and Il-34 in gingival Ep or LP was quantified by quantitative RT-PCR in 6-wk-old B6 mice (E), as well as their expression kinetics postnatally (F). RNA expression data were pooled from three independent experiments and are presented as the mean ± SEM (n = 6–12 mice). Bars, 50 µm. *, P < 0.05; **, P < 0.001.
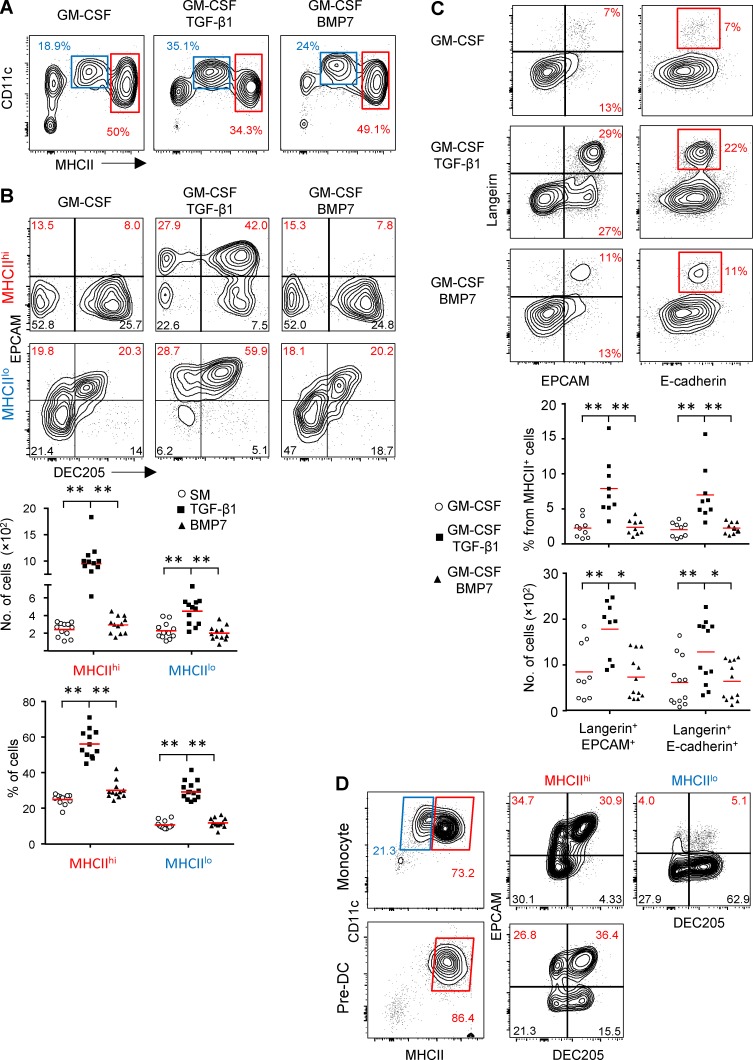
TGF-β1, but not BMP7, induces LC differentiation from murine BM cells in vitro
To further examine the role of BMP7 and TGF-β1 in LC development, we used in vitro differentiation cultures using murine BM cells that contain pre-DCs and monocytes, the established precursors of mucosal LCs (Capucha et al., 2015). BM cells were cultured for 5 d in serum-free media supplemented with SCF, FLT3L and TNF-α (supplemented medium [SM]) containing GM-CSF in the presence or absence of TGF-β1 or BMP7. As demonstrated in Fig. 3 A, two CD11c+MHCII+ populations expressing MHCIIlo or MHCIIhi levels were generated upon exposure to GM-CSF, GM-CSF + TGF-β1, or GM-CSF + BMP7. Whereas the numbers of MHCIIhi cells were similar among the various differentiation cultures, the number of MHCIIlo population was significantly larger in GM-CSF + TGF-β1 culture (Fig. S2 A). Because we observed earlier that EpCAM is expressed in vivo before langerin (Fig. 1 B), we next quantified the generation of LCs by gating on EpCAM+DEC205+ cells among the CD11c+MHCII+ population because these were previously proven to represent LC-like cells (Chopin et al., 2013). Within the MHCIIhi population, GM-CSF + TGF-β1 was capable of inducing large amounts of EpCAM+DEC205+ cells, whereas GM-CSF alone or GM-CSF + BMP7 failed to do so (Fig. 3 B). Moderate differentiation of EpCAM+DEC205+ cells was detected in MHCIIlo cells after exposure to GM-CSF or GM-CSF + BMP7, but the addition of TGF-β1 to the GM-CSF cultures resulted in higher expression of EpCAM as well as larger numbers of EpCAM+DEC205+ cells. Similar to the situation in vivo, LC-like cells induced by TGF-β1 first expressed EpCAM and subsequently langerin (Fig. 3 C). The incapability of BMP7 to facilitate differentiation of LC-like cells was also evident in cultures containing serum (Fig. S2 B). Of note, all cytokines in the SM were required for efficient LC differentiation. Using different gating strategies based on the coexpression of langerin and E-cadherin or, alternatively, langerin and EpCAM, we confirmed the superior capacity of TGF-β1 to induce langerin+E-cadherin+ or langerin+EpCAM+ cells among the total CD11c+MHCII+ population (Fig. 3 C).
Figure 3.
TGF-β1, but not BMP7, induces LC-like cell differentiation from murine BM precursors. (A–D) BM cells were incubated for 5 d in serum-free SM in the presence of GM-CSF with or without TGF-β1 or BMP7. The cultures were analyzed by flow cytometry, and representative plots and graphs present the percentages of CD11c+MHCIIlo and CD11c+MHCIIhi population in each culture (A), the frequencies and absolute numbers of LC-like cells based on expression of CD205 and EpCAM (B) or, alternatively, expression of langerin and EpCAM or E-cadherin+ cells (C). Representative flow cytometry plots and graph of one out of six independent experiments are shown as mean. Each experiment included at least three independent BM preparations. (D) Pre-DCs and monocytes were sorted from BM cells and incubated for 5 d with serum containing media in the presence of GM-CSF and TGF-β1 (n = 4). Representative flow cytometry plots demonstrate the differentiation of MHCIIlo and MHCIIhi populations and further the generation LC-like cells based on the expression of CD11c, MHCII, CD205, and EpCAM. Representative data of one out of three independent experiments are shown. *, P < 0.05; **, P < 0.001.
A recent study suggested that upon stimulation with GM-CSF, BM precursors differentiate into MHCIIlo cells from monocytes and into MHCIIhi cells from both pre-DCs and monocytes (Helft et al., 2015). To examine whether LC-like cells generated in vitro from MHCIIlo and MHCIIhi populations originate from monocytes and/or pre-DCs, we purified monocytes and pre-DCs from BM cells by flow cytometry and cultured them with GM-CSF + TGF-β1. As depicted in Fig. 3 D, purified monocytes were able to give rise to both MHCIIlo and MHCIIhi cells that further differentiated into EpCAM+DEC205+ cells. Purified pre-DCs differentiated only into MHChi cells that were also capable to generate EpCAM+DEC205+ cells. Another study had reported that BMP7 is a potent inducer of LC-like cells from enriched CD34+ human umbilical cord blood cells (Yasmin et al., 2013). To examine whether BMP7 can induce LC differentiation from even more primitive murine precursors, we established a differentiation culture from fetal liver cells. Fetal livers were collected on embryonic day 13.5 (E13.5) or E14.5 and treated with SM containing GM-CSF ± TGF-β1 or BMP7 as described in Fig. 3 A (Fig. S2 C). In all culture conditions, the majority of the differentiated CD11c+MHCII+ cells were MHCIIlo, probably because the fetal liver contains many embryonic monocytes (Hoeffel et al., 2012). However, similar to adult BM cells, the addition of TGF-β1 resulted in higher frequencies of EpCAM+CD205+ cells as compared with GM-CSF ± BMP7. Collectively, the superior capacity of TGF-β1 to differentiate monocytes and pre-DCs into LC-like cells in vitro, and its presence in the mucosal epithelium, suggest that TGF-β1 is essential for mucosal LC differentiation.
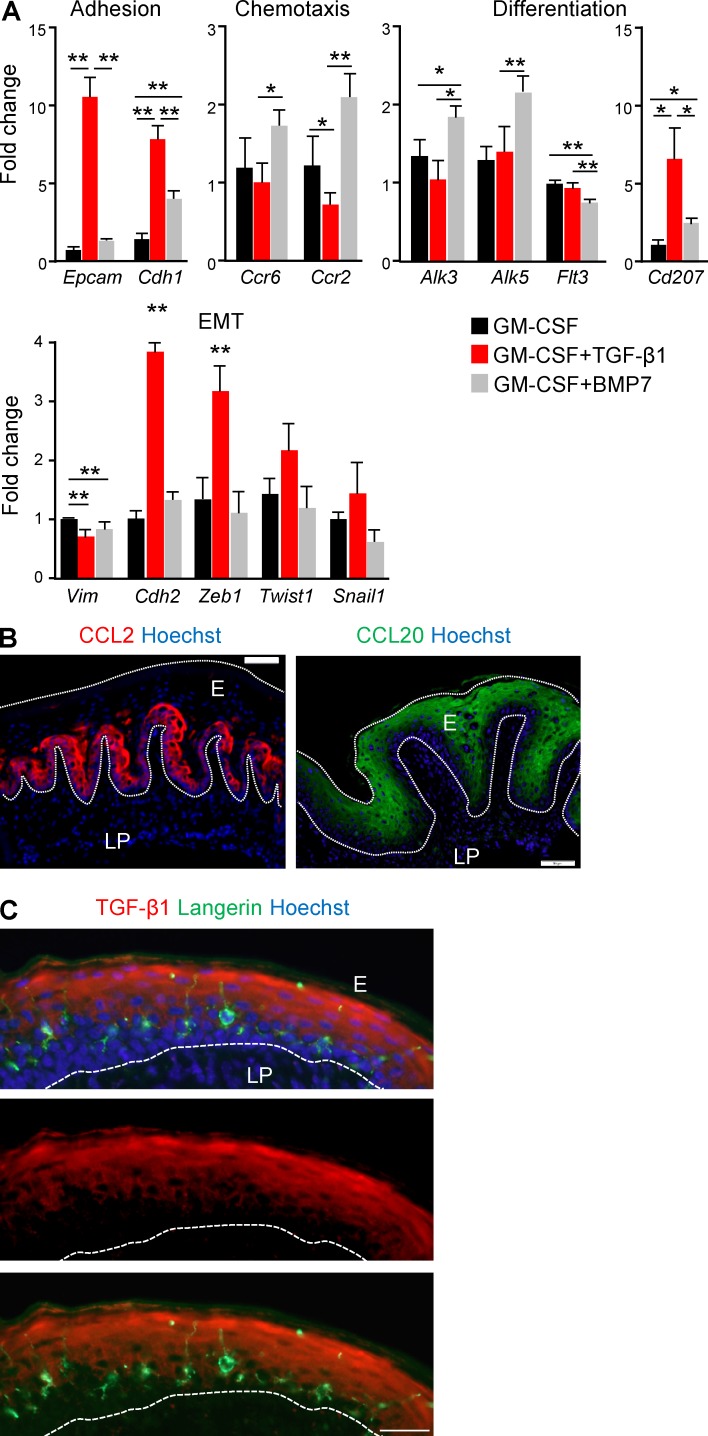
BMP7 up-regulates the expression of genes required for the transition of LC precursors into the epithelium
Although BMP7 was not capable of inducing differentiation of LC-like cells from BM precursors in vitro, its anatomical association with the basal membrane as well as its reported role in mesenchymal-to-epithelial transition (MET) programming (Zeisberg et al., 2005) led us to hypothesize that BMP7 might promotes the transition of LC precursors to the epithelium. Thus, we examined whether BMP7 has an impact on expression of genes associated with epithelial transition. First, quantitative RT-PCR analysis revealed that the addition of BMP7 to the GM-CSF cultures up-regulated expression of E-cadherin, a known MET marker enabling adhesion to epithelial cells (Fig. 4 A). TGF-β1 up-regulated E-cadherin expression even further and was also able to induce EpCAM expression, in agreement with our flow cytometry results (Fig. 3 B). Second, BMP7 also increased the expression of CCR2 and CCR6, two chemokine receptors that are critically involved in epithelial differentiation of mucosal LCs (Capucha et al., 2015). As depicted in Fig. 4 B, the ligands of these chemokine receptors (CCL2 and CCL20, respectively) are differentially expressed in the oral epithelium; CCL2 is expressed in the basal layers, whereas CCL20 is located suprabasally. Intriguingly, TGF-β1 had no impact on CCR6 expression but significantly reduced the expression of CCR2 in comparison to GM-CSF–treated cultures. This capacity of TGF-β1 is in line with the localization of LCs in the epithelium (Fig. 4 C), because after reaching the epithelial zone in which TGF-β1, but not CCL2, is expressed, signaling via CCR2 is likely to be dispensable. Next, we measured genes involved in LC differentiation and discovered that BMP7, but not TGF-β1, was able to up-regulate the expression of ALK3 and ALK5, the receptors of BMP7 (ALK3) and TGF-β1 (ALK5 and ALK3). This suggests that BMP7 facilitates the ability of LC precursors to receive signals via both receptors upon their arrival to the epithelium. BMP7 also induced low langerin expression but attenuated FLT3 expression, probably representing a reduced hematopoietic potential of LC precursors. We then determined the expression of genes associated with the opposite programming (i.e., epithelial-to-mesenchymal transition [EMT]). Both BMP7 and TGF-β1 reduced vimentin expression in line with the general MET that LC-like cells undergo. However, TGF-β1 significantly up-regulated expression of Zeb1 (but not Snail1 or Twist1), a key EMT transcriptional regulator involved in LC maturation (Konradi et al., 2014; Fig. 4 C). Moreover, up-regulation of N-cadherin (Cdh2) was observed upon exposure to TGF-β1, but not BMP7. Such an induction of EMT-associated genes in GM-CSF + TGF-β1 cultures could be attributed to the presence of TNF-α in our SM because together, TNF-α and TGF-β1 facilitate EMT in various systems (Takahashi et al., 2010; Asiedu et al., 2011). Collectively, these results strongly suggest that BMP7 is involved in modulating the expression of genes required for the transition of LC precursors to the mucosal epithelium, in line with its known capacity to induce MET.
Figure 4.
Differential induction of genes involved in LC development by TGF-β1 and BMP7. (A) BM cells were incubated in serum-free SM in the presence of GM-CSF with or without TGF-β1 or BMP7. 5 d later, total RNA was prepared from the cultures and expression of the noted genes was quantified using quantitative RT-PCR. Bar graphs present the fold change in mRNA levels among the various treatments ± SEM (n = 5). Results of one experiments out of two independent experiments are presented. (B) Buccal cross sections prepared from 8-wk-old B6 mice were stained for CCL2 (red) or CCL20 (green) and Hoechst (blue). (C) Gingival cross section stained for TGF-β1 (red), langerin (green), and Hoechst (blue). Representative immunofluorescence image from one out of two-three independent experiments are presented. E, epithelium; LP, lamina propria. Dotted white lines indicate the basal and/or epical ends of the epithelium. Bars, 50 µm. *, P < 0.05; **, P < 0.001.
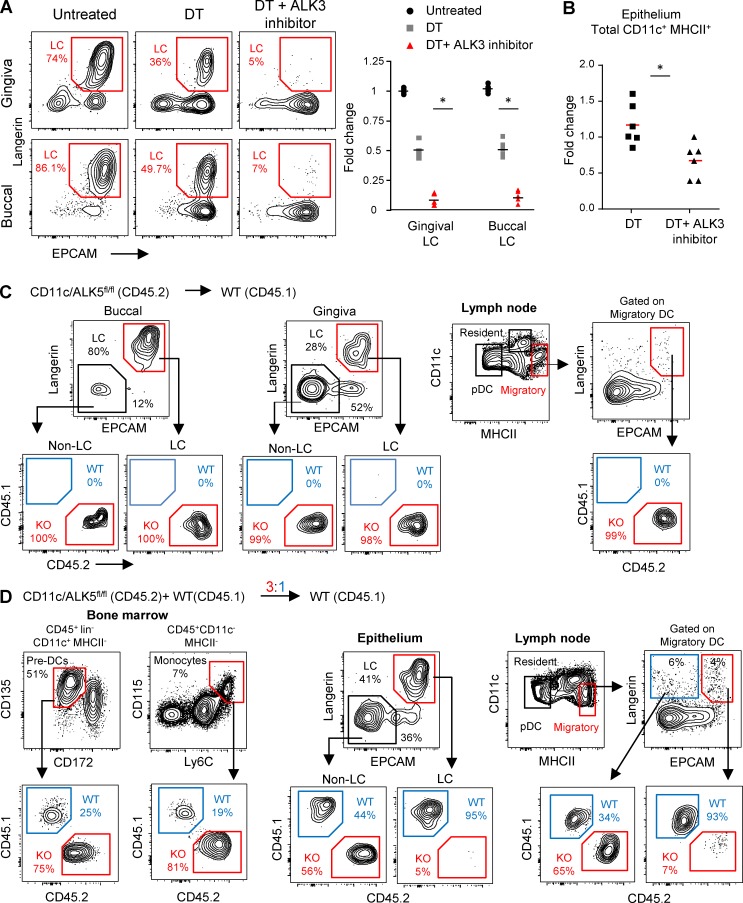
Signaling via both ALK3 and ALK5 is required for differentiation of mucosal LCs in vivo
We next sought to examine the role of ALK3 and ALK5 in the development of mucosal LCs in vivo. First, we depleted langerin-expressing cells from adult langerin/diphtheria toxin receptor (DTR) mice using diphtheria toxin (DT) and monitored the repopulation of LCs in the oral mucosa with or without daily administration of dorsomorphin, a selective inhibitor of ALK2, 3, and 6. As demonstrated in Fig. 5 A, 6 d after depletion, LCs began to rapidly repopulate the epithelium of the gingiva and buccal mucosa, whereas dorsomorphin treatment significantly reduced LC differentiation. Quantifying CD11c+MHCII+ cells in the epithelium revealed that mice treated with DT and dorsomorphin had significantly lower numbers of these cells than mice treated with DT only (Fig. 5 B). This suggests that signaling via ALK3 controls the transition of LC precursors to the epithelium, in line with our hypothesis that BMP7, the ALK3 ligand, mediates this process. We then tested the role of ALK5 using adult CD45.2+ CD11c-cre/ALK5fl/fl mice, in which ALK5 is deleted specifically in cells expressing CD11c (e.g., LCs and DCs; Kel et al., 2010). BM cells purified from these mice were used to reconstitute the immune system of lethally irradiated CD45.1+ B6 recipients to generate chimeric mice, which were analyzed 3 wk later. The analysis indicated that BM cells from CD45.2+ CD11c-cre/ALK5fl/fl mice were able to generate LCs in the oral epithelium, and some of these cells were able to migrate to the draining LNs (Fig. 5 C). Oral mucosal LCs were also detected directly in the oral epithelium of CD11c-cre/ALK5fl/fl mice, though at lower amounts than their littermate controls (Fig. S3 A). To further examine the contribution of ALK5 to mucosal LC differentiation, we reconstituted lethally irradiated adult CD45.1+ B6 mice with a mixture of BM cells consisting CD45.2+ CD11c-cre/ALK5fl/fl (3 × 106 cells) and CD45.1+ WT (1 × 106 cells) adult mice. Analysis of the BM of mixed chimeras revealed that in line with the 3:1 ratio of initial reconstitution, the frequencies of CD45.2+ pre-DCs and monocyte precursors were approximately three to four times higher than their CD45.1+ counterparts (Fig. 5 D). Nevertheless, despite the low frequencies of CD45.1+ WT precursors, the majority (>90%) of LCs in the oral mucosa were CD45.1+, whereas the remaining cells were of CD45.2+ origin. Examining LC frequencies in the draining LN yielded a picture similar to that in the oral epithelium, suggesting that the low percentages of CD45.2+ LCs in the oral mucosa cannot be attributed solely to their rapid migration to the LN. In agreement with this notion, when the mixed BM cells were tested in vitro, CD45.2+ and CD45.1+ cells contributed equally to the differentiation of LC-like cells, although the cultures contained three to four times less CD45.1+ than CD45.2+ precursors (Fig. S3 B). Collectively, these data suggest that signaling via ALK3 is required for epithelial transition of mucosal LC precursors, whereas signaling via ALK5 appears to be necessary for their final differentiation. ALK3 may also be involved in LC differentiation, because mucosal LC developed to some extent in the absence of ALK5.
Figure 5.
Signaling via both ALK3 and ALK5 is essential for the development of mucosal LCs. (A and B) A cohort of adult langerin-DTR mice were treated once with DT or left untreated; DT treated mice further received injections of dorsomorphin or vehicle only (three injections per day for 6 d). (A) Representative flow cytometry plots and graph present the frequencies and fold changes, respectively, of LCs in gingival and buccal epithelial tissues among the different group. Fold change was calculated based on absolute LC numbers and was normalized to untreated mice. (B) Graph indicates the fold change in total CD11c+MHCII+ cells in the oral epithelium between DT and DT + dorsomorphin groups (absolute LC numbers were normalized to DT-treated mice). Data present results pooled from two independent experiments shown as the mean. Each experiment included at least three separately analyzed mice. (C) Total BM cells were purified from adult CD11c-cre/ALK5fl/fl (CD45.2) mice and transplanted into lethally irradiated WT (CD45.1) hosts. Representative flow cytometry plots demonstrate the frequencies of CD45.2 versus CD45.1 among gingival and buccal LC 3 wk after BM reconstitution. (D) A mixture of total BM cells from adult CD11c-cre/ALK5fl/fl (CD45.2) mice and WT (CD45.1; a ratio of 3:1 KO/WT, respectively) was transplanted into lethally irradiated WT (CD45.1) hosts. 3 wk later, the frequencies of CD45.1+ versus CD45.2+ cells were examined in pre-DCs and monocytes derived from the reconstituted BM (left). The frequency of CD45.1+ versus CD45.2+ cells was also tested in the LC and non-LC population in the oral epithelium (middle) and cervical draining LNs (right). Representative data of one out of two independent experiments are presented. Each experiment included at least three separately analyzed mice. *, P < 0.05.
The microbiota shapes the differentiation of oral LCs
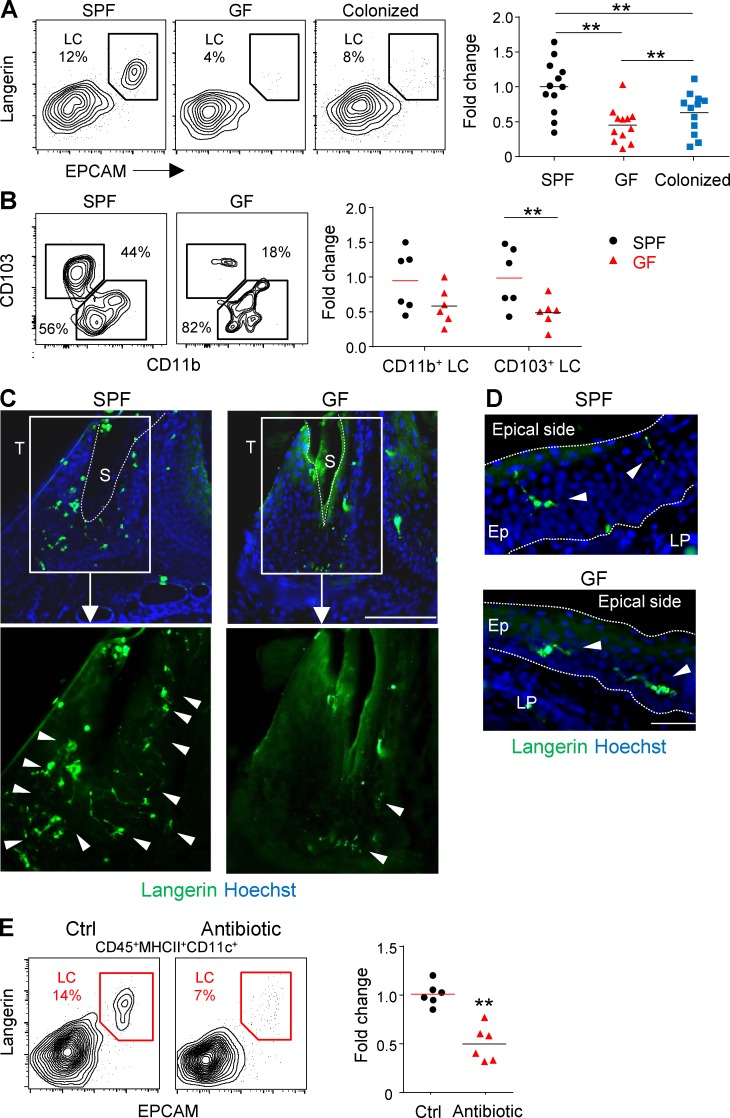
Because LCs develop in the epithelium, a tissue interfacing with external microorganisms, we asked whether the microbiota has any impact on LC differentiation. To address this issue, we analyzed LCs in the oral cavity of adult germ-free (GF) mice that lack microorganisms from birth. We found a significant reduction of ∼50% in the frequencies of LCs in the gingival epithelium of GF mice in comparison to specific pathogen–free (SPF) controls (Fig. 6 A). This reduction was also confirmed by counting langerin+ cells using immunofluorescence staining (Fig. S4 A). Further analysis revealed that the reduction in gingival LCs was attributed mainly to a decrease in the CD103+ LC subset (Fig. 6 B). Cohousing of GF with SPF mice for 2 mo restored LC frequencies in the gingiva, but not to the same levels seen in SPF mice (Fig. 6 A). Of note, a lesser but statistically significant reduction in LC numbers was also found in the buccal epithelium of GF mice, whereas no differences were observed between GF and SPF mice with regard to skin LCs (Fig. S4 B). The large reduction in gingival LCs led us to hypothesize that it might be related to the presence of the bacterial biofilm close to the gingiva. We thus generated gingival cross sections for immunofluorescence staining and focused our analysis on the sulcular and junctional epithelia surrounding the biofilm. As demonstrated in Fig. 6 C, in adult SPF mice, many langerin+ cells were situated in the junctional/sulcular epithelium and their large dendrites were aimed to the gingival sulcus where the biofilm is located (Curtis et al., 2011). In adult GF mice, however, only few langerin+ cells were observed, and their sizes as well as dendrites seemed to be very small as compared with their SPF equivalents. We also examined langerin+ cells located in gingival epithelial regions not facing directly to the biofilm. Whereas in SPF mice the dendrites of these cells were pointed to the apical end of the epithelium, in GF mice many of the cells failed to do so (Fig. 6 D; see also Fig. 4 C). We then investigated whether reducing the microbial load in adult mice by treating them with broad-spectrum antibiotics for 2 mo will also affect oral LCs. First, we confirmed that antibiotic treatment reduced the microbial load in the gut and oral cavity (Fig. S4 C). As a result, a significant decline in the frequencies of gingival LCs was detected, similar to GF mice (Fig. 6 E). A decrease in LCs was also observed in the buccal mucosa, whereas skin LCs remained unaffected (Fig. S4 D). Unlike GF mice, antibiotic treatment resulted in a significant decrease of both CD103+ and CD11b+ LC subsets (not depicted), which could be attributed to the reported capacity of antibiotics to decrease the monocyte population that gives rise to CD11b+ LCs (Bain et al., 2014).
Figure 6.
Commensal microbiota regulates the frequencies of mucosal LC. (A) Gingival mucosal tissue from adult Swiss Webster SPF mice, GF mice, and GF mice cohoused with SPF mice for 2 mo was processed and analyzed by flow cytometry to examine the percentages of LCs. Representative flow cytometry plots present the frequencies of gingival LCs (CD45+CD11c+MHCII+langerin+EpCAM+). Graph presents the fold change in absolute numbers of LCs normalized to SPF mice and (mean). Results pooled from four independent experiments are shown; each experiment included three separately analyzed mice. (B) Representative plots illustrate the percentages of CD103+LCs and CD11b+LCs in SPF and GF mice. Graph presents the fold change in absolute numbers of the two LC subsets normalized to SPF mice presented as the mean. Results pooled from two independent experiments are shown, each experiment included three separately analyzed mice. (C and D) Representative immunofluorescence images of gingival cross section prepared from adult SPF and GF mice, stained for langerin (green) and Hoechst (blue). Confocal microscopy images from one out of three independent experiments are presented. Arrowheads indicate the location of LCs, and the dotted line specifies the sulcus region or the basal and epical ends of the epithelium. S, sulcus; T, tooth. (E) Adult SPF B6 mice were treated with a broad-spectrum antibiotic cocktail in the drinking water for 2 mo. Representative flow cytometry plots demonstrate the percentages of LCs in gingival tissues of antibiotic-treated mice and control group. Graph presents the fold change in the absolute numbers of LCs normalized to SPF mice and presented as the mean. Results pooled from two independent experiments are shown, each experiment included three separately analyzed mice. Bars, 50 µm. **, P < 0.001.
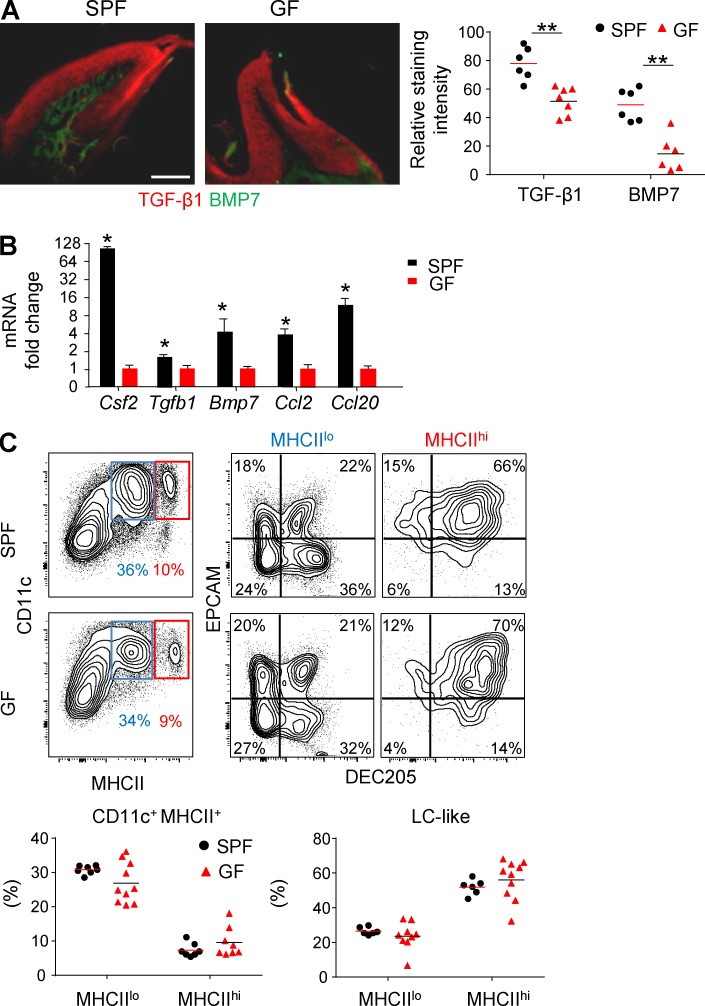
We next quantified in GF mice the expression of molecules involved in LC differentiation. Immunofluorescence staining revealed that TGF-β1 and BMP7 protein levels were significantly lower in GF mice than SPF mice (Fig. 7 A). These results were further confirmed on the transcription level using quantitative RT-PCR (Fig. 7 B). Expression of Ccl20, Ccl2, and Csf2 in the gingiva was also lower in GF mice than SPF mice. Because it was reported that the microbiota has an impact on steady-state hematopoiesis of BM cells (Khosravi et al., 2014), we asked whether this might explain the decrease in gingival LCs. BM cells were purified from adult GF or SPF mice, and their capability to differentiate into LC-like cells was examined as described in Fig. 3. No differences were found in the ability of BM cells from the two groups to generate LC-like cells (Fig. 7 C). Collectively, these data suggest that the oral microbiota modulate the development of mucosal LCs by regulating the expression of cytokines and chemokines involved in LC differentiation.
Figure 7.
The impact of microbiota on mucosal LC differentiation is mediated locally rather than systemically. (A) Immunofluorescence confocal microscopy images of gingival cross sections from adult Swiss Webster SPF and GF mice stained for TGF-β1 (red), BMP7 (green), and Hoechst (blue). Graph indicates the relative staining intensity of the two cytokines using ImageJ software (n = 6–7). One representative experiment out of three independent experiments is shown. (B) Total RNA was extracted from gingival tissues of adult SPF and GF mice, and expression levels of the indicated genes were determined by quantitative RT-PCR. Bar graph presents the fold change in gene expression normalized to GF mice presented as the mean ± SEM. Data were pooled from three independent experiments, and each experiment included four separately analyzed mice. (C) Total BM cells from adult SPF or GF mice were incubated for 5 d in serum containing SM in the presence of GM-CSF and TGF-β1. Representative flow cytometry plots demonstrate the differentiation of CD11c-expressing MHCIIlo and MHCIIhi subsets as well as LC-like cells based on further expression of CD205 and EpCAM. Graphs present results pooled from three independent experiments shown as the mean values (n = 6 SPF and 9 GF mice). Bar, 50 µm. *, P < 0.05; **, P < 0.001.
Langerin-expressing cells are required for oral mucosal homeostasis
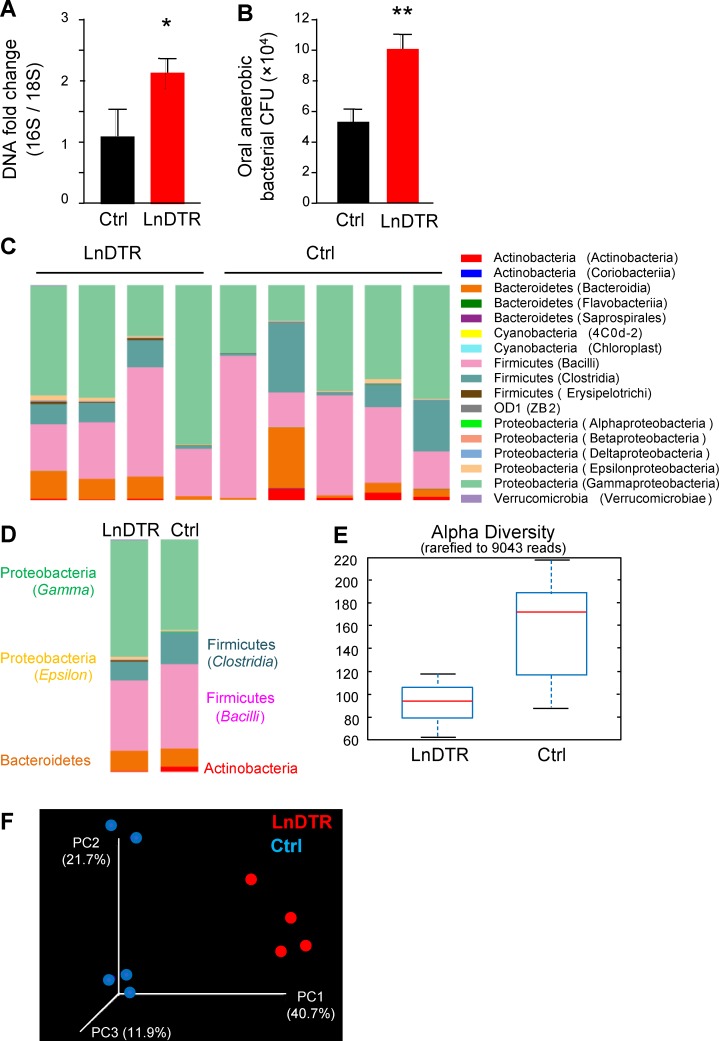
Because the microbiota exert a major influence on the development of oral LCs, we next examined whether LCs in turn regulate microbial/immunological homeostasis in the oral mucosa. To address this question, we treated adult Langerin-DTR mice and littermate controls with DT for 4 mo, which resulted in long-term ablation of langerin-expressing cells (Bennett and Clausen, 2007; Kautz-Neu et al., 2011). Of note, unlike the skin, where langerin is expressed by both LCs and dermal CD103+ DCs, in the oral mucosa, langerin-expressing cells are exclusively LCs (Fig. S1 B), as was previously reported in the vaginal mucosa (Hervouet et al., 2010). To probe the oral microbiota in the presence or absence of LCs, DNA was extracted from oral swabs, and the ratio of ribosomal 16S/18S genes was calculated to compare the total bacterial load. As depicted in Fig. 8 A, upon prolonged DT treatment, the oral cavity of Langerin-DTR mice contained an elevated oral bacterial load in comparison to littermate controls. We further quantified the burden of anaerobic bacteria by plating oral swab samples on blood agar under anaerobic conditions. Significantly more oral anaerobic bacteria could be cultivated from Langerin-DTR mice than from littermate controls (Fig. 8 B). To examine whether these alterations in DT-treated Langerin-DTR mice affect bacterial diversity, we performed a taxonomic analysis of the oral microbiota. Indeed, significant changes in the composition of oral microbiota were detected in DT-treated Langerin-DTR mice when compared to controls (Fig. 8, C and D; and Fig. S5, A and B). Specifically, elevated relative frequencies of bacteria of the Proteobacteria and Bacteroidetes phyla, as well as a reduction in Clostridia class (Firmicutes phylum), were detected in the absence of LCs. In addition, α diversity analysis indicated a lower taxa richness in DT-treated Langerin-DTR mice than in controls (Fig. 8 E), whereas the taxa present in these mice varied, as indicated by an unweighted β diversity (Fig. 8 F).
Figure 8.
Prolonged ablation of langerin-expressing cells induces oral microbial dysbiosis. 8-wk-old langerin-DTR (LnDTR) mice and littermate controls (Ctrl) were injected intraperitoneally with DT on a weekly basis for 4 mo. (A) Total oral bacterial load was determined in oral swabs taken from the mice by quantitative RT-PCR of the 16S rRNA gene. Bar graphs present the 16S/18S ratio in each group and represent the mean values ± SEM (n = 12). (B) Enumeration of cultivable oral anaerobic bacteria was performed by plating serial dilutions of oral swabs samples on blood agar under anaerobic conditions. Graph presents the calculated CFU values in each group and represent the mean values ± SEM (n = 6). Representative bacterial load data of one out of three independent experiments are presented. (C–F) Relative abundance of taxa in oral swabs sampled from adult LnDTR mice and littermate controls upon prolonged DT treatment. Histograms represent the distribution of sequences in operational taxonomic units (OTUs) assigned to each class. Histograms for individual mice (C) as well as a histogram representing the mean distribution of OTU (D) are provided. (E) Alpha diversity plot representing taxa richness in samples of both groups of mice. Data represent the mean ± SEM (n = 4–5). (F) Principal coordinates (PCs) analysis of weighted UniFrac distances based on 16S rRNA of LnDTR mice versus littermate control (P < 0.05). Taxonomic data from one out of two independent experiments are shown. *, P < 0.05, **, P < 0.001.
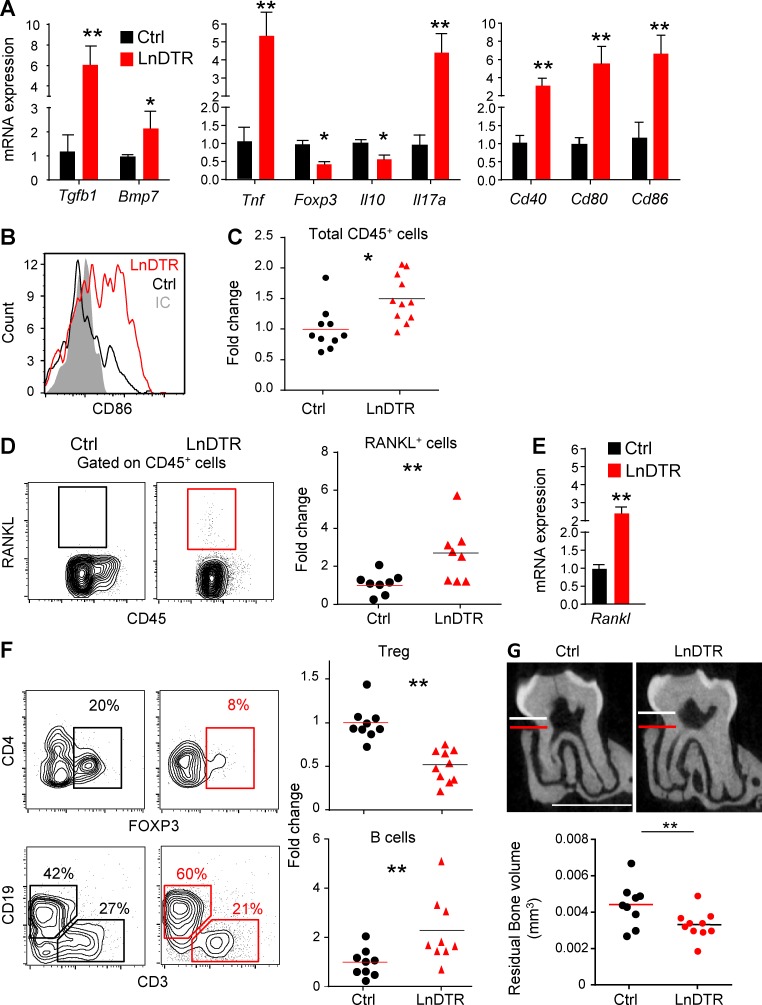
To further study the impact of the absence of LCs on oral mucosal homeostasis, we examined gingival immune responses. First, expression of TGF-β1 and BMP7 increased significantly in DT-treated Langerin-DTR mice as compared with littermate controls (Fig. 9 A). Quantitative RT-PCR analysis also revealed elevated expression of the proinflammatory cytokines TNF-α and IL-17, whereas the regulatory T cell (T reg cell) marker Foxp3 and antiinflammatory cytokine IL-10 were reduced (Fig. 9 A). Increased expression levels of CD40, CD80 and CD86 were detected in the gingiva, and the increment in CD86 expression was verified on gingival APCs using flow cytometry (Fig. 9, A and B). Moreover, ∼1.5 times more of CD45+ cells were found in the gingiva of DT-treated Langerin-DTR mice than in littermate controls (Fig. 9 C). These CD45+ cells expressed high amount of receptor activator of nuclear factor kappa-B ligand (RANKL), a molecule known to play a detrimental role in gingiva-associated pathologies (Fig. 9, D and E). In agreement with the aforementioned quantitative RT-PCR analysis, we detected reduced frequencies of CD4+Foxp3+ T reg cells in Langerin-DTR mice by flow cytometry (Fig. 9 F). In addition, LC ablation resulted in considerably higher percentages of B cells in the gingiva, whereas no change occurred in the frequencies of neutrophils, T cells, and circulating or inflammatory monocytes (Fig. S5 C). Because these data indicate that ablation of LCs resulted in an inflammatory state in the gingiva, we next examined the production of proinflammatory cytokines by splenocytes as a representative of systemic inflammation. Indeed, upon restimulation, an increased secretion of TNF-α and IFN-γ was measured in splenocytes from Langerin-DTR as compared with controls (Fig. S5 D). Finally, it has been reported that elevated inflammation and RANKL expression, together with a decrease in oral microbial diversity, are associated with periodontal pathology (Cochran, 2008). Concurringly, we detected a significant alveolar bone loss in Langerin-DTR mice (Fig. 9 G). These data suggest that oral LCs regulate T cell polarization and are crucial for maintaining microbial and immunological homeostasis in the oral mucosa.
Figure 9.
Extended depletion of langerin-expressing cells causes inflammation-driven alveolar bone loss. (A) Gingival tissues were collected from DT-treated adult LnDTR mice and littermate controls. mRNA levels of the indicated genes were quantified by quantitative RT-PCR. Graphs present the fold change in transcript levels normalized to littermate controls depicted as mean ± SEM (n = 6–12). Data pooled from at least three independent experiments are shown. (B) FACS plot demonstrate the mean fluorescence intensity (MFI) levels of CD86 on live CD45+CD11c+CD11b+ cells in each group, IC; isotype antibody control. (C) Frequencies of total CD45+ leukocytes in the gingiva of adult LnDTR mice and littermate controls 4 mo after DT treatment. (D) Representative flow cytometry plots and graph demonstrate the percentages of RANKL-expressing leukocytes in the gingiva. Graph presents the fold change in RANKL-expressing cells normalized to littermate controls presented as mean (n = 8). (E) Fold change in gingival Rankl mRNA levels normalized to littermate controls presented as mean ± SEM (n = 8). (F) Frequencies of live gingival T reg cells (CD45+CD3+CD4+FOXP3+) and B cells (CD45+CD3−CD19+). Graphs present the fold change in the frequencies of noted cells that was normalized to littermate control and represent the mean (n = 9). Data pooled from three independent experiments are shown. (G) Representative μCT sections of the second upper molar indicate the distance between the cementoenamel junction and alveolar bone crest. Graph demonstrates 3D quantification of the residual alveolar bone volume 4 mo after DT treatment, and data represent the mean (n = 9). Bone loss data of one out of two independent experiments are shown. Bars, 1 mm. *, P < 0.05; **, P < 0.001.
Discussion
This study reveals the unique mechanisms of LC development in the murine mucosa, highlighting their distinct ontogeny in comparison to skin LCs. The compartmentalized expression of TGF-β1 and BMP7 in the mucosa indicates that both cytokines have distinct roles in mucosal LC differentiation. We provide a conclusive line of evidence for a role for BMP7 in promoting the transition of LC precursors to the mucosal epithelium. First, BMP7 expression is restricted to the lamina propria/basal membrane, whereas it is not expressed in the epithelium, where the precursors acquire their final LC phenotype. This observation is in line with studies demonstrating that BMP7 exhibits high-affinity binding to the basement membrane and type IV collagen that is confined to the basal lamina (Jiang et al., 1994; Vukicevic et al., 1994). Second, BMP7 fails to induce the differentiation of LC-like cells in BM cultures. Third, postnatal expression kinetics of TGF-β1, rather than BMP7, correlates with the postnatal differentiation of LCs. Fourth, blocking of ALK3, the receptor of BMP7, upon ablation of LCs reduced total numbers of CD11c+MHCII+ cells in the epithelium. Fifth, BMP7 up-regulates the expression of cytokines and chemokine receptors that promote transition to the epithelium. In this regard, the differential expression of CCL2 and CCL20 in the epithelium might imply that CCL20 leads the LC precursors to where TGF-β1 is most expressed, which fits with the reduced differentiation of oral LCs reported in CCR6-deficient mice (Capucha et al., 2015). We thus propose that BMP7 induces MET-like programming in LC precursors during mucosal LC development.
In contrast to TGF-β1, BMP7 was not able to generate LC-like cells from murine BM cells. These results oppose a recent study demonstrating the superiority of BMP7 over TGF-β1 in generating LC-like cells from human CD34+ hematopoietic progenitor cells (HPCs; Yasmin et al., 2013). This discrepancy is unlikely to be related to the more primitive stage of human CD34+ HPCs because BMP7 also failed to induce LC-like cells from murine fetal liver cells. Nevertheless, the advantage of BMP7 in the human CD34+ HPC system is probably attributed to its capability to induce proliferation of these primitive precursors because TGF-β1 and BMP7 equally differentiate human blood CD1c+ DCs into LC-like cells (Milne et al., 2015). Whereas it remains unclear why BMP7 fails to generate LC-like cells in murine BM cultures, TGF-β1 can induce the differentiation of these cells from both mouse and human precursors (Chopin et al., 2013; Bigley et al., 2015; Milne et al., 2015). This capacity of TGF-β1 in human cultures is mediated by ALK3 signaling, whereas signaling via ALK5 impaired the differentiation of human LC-like cells (Yasmin et al., 2013; Bigley et al., 2015). In the current study, we demonstrate that ALK5 is essential for murine mucosal LC development both in vitro and in vivo, in agreement with a previous study on skin LCs (Kel et al., 2010). ALK3, on the other hand, was found to affect the transition of LC precursors to the mucosal epithelium in vivo by binding to BMP7. Nevertheless, it is possible that in the epithelium, ALK3/TGF-β1 signaling is also involved in LC differentiation, including langerin expression because blocking ALK3 signaling resulted in accumulation of EpCAM+ Langerin− cells. Thus, both ALK3 and ALK5 are required for BM-derived LCs, supporting a model of sequential signaling by BMP7 (ALK3) and TGF-β1 (ALK5 and ALK3).
The significant reduction in mucosal LCs in GF mice can be attributed to the decrease in CD103+ LCs. In the intestine, CD103+CD11b− DCs play a crucial role in mucosal homeostasis because they induce an antiinflammatory response in epithelial cells, T cell homing, and T reg cell differentiation (Coombes et al., 2007; Ruane and Lavelle, 2011; Muzaki et al., 2016). Similar to CD103+CD11b− DCs, mucosal CD103+ LCs originate from pre-DCs and depend on the transcription factors IRF8 and Id2 for their development, suggesting that both cell types might share some immunological functions (Ginhoux et al., 2009; Capucha et al., 2015). It is thus possible that whereas basal levels of mucosal LCs develop in the oral epithelium postnatally independently of local microbiota, exposure to microbiota increases the population of CD103+ LCs that facilitates mucosal homeostasis. This effect of the microbiota on oral LCs is mediated locally rather than systemically as recently suggested (Khosravi et al., 2014) because the microbiota up-regulated expression of cytokines and chemokines capable of attracting and instructing LC differentiation. The remarkable reduction of LCs observed in the gingiva of GF mice, particularly around the sulcus, supports these findings because this barrier tissue deals with a unique and substantial microbial challenge. Notably, although a 2-mo period of cohousing did not completely restore LC frequencies in GF mice, some of them exhibited LC levels similar to SPF mice. This strongly suggests that over time, the frequencies of LCs will be normal in cohoused mice, most likely because of an increase of CD103+ LCs. Such adaptation of oral LCs to the microbiota appears to be a dynamic process rather than a unique postnatal event because broad-spectrum antibiotic treatment also reduced LC levels in adult SPF mice. As a reflection of such a dynamic process, ablation of oral LCs resulted in considerable microbial changes, particularly elevated load of oral anaerobic bacteria of the Proteobacteria and Bacteroidetes phyla. We recently reported that bacteria of this phyla can expand in the oral mucosa by consuming inflammatory byproducts by anaerobic respiration (Nassar et al., 2017). Such bacterial dysbiosis accompanied by elevated inflammation was shown to induce periodontal pathology upon infection with an oral pathogen (Hajishengallis, 2015). Here, we report periodontal damage without infection, probably reflecting an accelerated bone loss occurring spontaneously with age (Liang et al., 2010). Of note, the frequencies of gingival LCs are reduced in elderly humans and mice (Rittman et al., 1987; Zavala and Cavicchia, 2006), suggesting that an age-associated decrease in LCs might cause, at least in part, periodontal pathology.
A recent study using Langerin-DTA mice suggests that LCs are not involved in alveolar bone loss upon infection with an oral pathogen (Bittner-Eddy et al., 2016). In contrast, we have previously established that LCs protect mice from infection-driven bone loss by inducing regulatory T cells (Arizon et al., 2012), and in the current study, we demonstrate that LCs regulate spontaneous bone loss. Although the reason for these contradicting findings remains elusive, they may be explained by the distinct mouse models used by both studies to ablate LCs in vivo, as was also reported in other experimental settings (Clausen and Kel, 2010; Clausen and Stoitzner, 2015).
Collectively, this study reveals the unique mechanisms of LC development in mucosal tissues, highlighting the intimate interplay between LCs and the mucosa. Mucosal LCs are considered to be involved in various pathologies of the oral and vaginal mucosa (Solano et al., 2000; Miller and Shattock, 2003; Upadhyay et al., 2013). Therefore, besides increasing basic knowledge regarding these cells, the present study will stimulate clinical research dissecting the role of LCs in mucosal pathologies and facilitate the development of novel therapeutic approaches to treat them.
Materials and methods
Mice
CD45.2+ C57BL/6 (B6) mice were purchased from Harlan. Langerin-DTR mice were obtained from B.E. Clausen (Bennett et al., 2005). The animals were maintained under SPF conditions and analyzed between 8 and 12 wk of age unless stated otherwise in the text. All animal protocols were approved by the Hebrew University Institutional Animal Care and Use Committee (IACUC). CD11c-Cre/ALK5fl/fl mice (Kel et al., 2010) were maintained under SPF conditions at the central animal facility of the University Medical Center Mainz. Experiments were performed in accordance with guidelines of the IACUC of the University of Mainz. GF or SPF adult male outbred Swiss Webster mice (a widely used mouse strain for GF experiments) were maintained in sterile isolators at the Weizmann Institute of Science. For cohousing experiments, GF and SPF mice were housed together for 2 mo. All GF studies were approved by the IACUC of the Weizmann Institute of Science.
Antibodies and reagents
The following fluorochrome-conjugated monoclonal antibodies and the corresponding isotype controls were purchased from BioLegend: I-A/I-E (M5/114.15.2), CD4 (GK1.5), CD8α (53–6.7), RANKL (IK22/5), CD205 (NLDC-145), CD45.2 (104), B220 (RA3-6B2), CD86 (PO3), CD115 (AFS98), Flt3 (A2F10), Ly6G (1A8), Ly6C (HK1.4), CD3 (17A2), CD19 (6D5), NK1.1 (PK136), Ter119 (TER-119), EpCAM (G8.8), CD11b (M1/70), (53–6.7), CD135 (A2F10), CD11c (N418), E-cadherin (DECMA-1), and c-Kit (2B8). CD172a/SIRP-α (P84) was obtained from BD. Anti-FOXP3 (MF-14) was purchased from BioLegend and the T reg cell staining kit from Invitrogen. Anti–CD207/langerin antibody (929F3.01) was purchased from Imgenex and used for intracellular staining, whereas clone 4C7 (BioLegend) was used for extracellular staining.
Immunofluorescence staining
For whole-mount staining, gingival tissues were fixed in ice-cold 95% ethanol for 40 min and subsequently digested in 1 M ammonium acetate in water for 90 min at 37°C or alternatively with 2 mg/ml Dispase II (Roche Diagnostics) in PBS for 45 min at 37°C. For staining frozen sections, the maxilla was fixed overnight at 4°C in 4% paraformaldehyde in PBS solution and then washed for 1 wk in 0.5 M EDTA in PBS that was changed every other day. The tissue was cryopreserved in 30% sucrose (overnight at 4°C), embedded in optimal cutting temperature medium, and cryosectioned into 10-µm-thick sections. The cross sections and separated epithelium were washed three times in PBS, blocked in blocking buffer (5% FCS and 0.1% Triton X-100 in PBS) for 1 h at room temperature, and incubated overnight at 4°C with the following primary antibodies: goat anti-langerin (E17; Santa Cruz Biotechnology), rat anti–I-AI-E, clone M5/114.15.2 (BioLegend), rabbit anti–TGFβ1 (Abcam), mouse anti–BMP-7 (Abcam), goat anti–Laminin-5α (Santa Cruz Biotechnology), rabbit anti-CCL2 (Abcam), and rabbit anti-CCL20 (Abcam). After three washing steps in PBS, the samples were incubated with a secondary antibody (donkey anti–goat IgG, donkey anti–rat IgG, or donkey anti–rabbit IgG; Jackson ImmunoResearch) diluted 1:100 in blocking buffer for 1 h at room temperature, washed three times, stained with Hoechst, and mounted. As a negative staining control, primary antibody was omitted and replaced with blocking buffer. Signals were visualized and digital images were obtained with an Olympus BX51 fluorescence microscope.
Flow cytometry analysis of mucosal and skin LCs
Gingival, buccal, or skin (ear) tissues were excised and pooled from two to five mice per mucosal sample so that a sufficient numbers of LCs were obtained for flow cytometry analysis, and these pooled tissues were considered n = 1. In some experiments, epithelial sheets were prepared by predigestion of tissue with 2 mg/ml Dispase II in PBS for 30–40 min (oral and skin tissues) at 37°C. The epithelium and subepithelium (dermis or lamina propria) were carefully separated with forceps under a binocular microscope. Mucosal tissues were then minced and treated with 2 mg/ml collagenase type II (Worthington Biochemicals) and 1 mg/ml DNase I (Roche) solution in PBS plus 2% FCS for 25 min at 37°C in a shaker bath. A total of 20 µl of 0.5 M EDTA per 2-ml sample was added to the digested tissues and incubated for an additional 10 min. Ear skin was excised, separated into two halves, and treated similarly, except that 1 mg/ml collagenase was used. The cells were washed, passed through a 70-µM filter, stained and run in LSR II (BD Biosciences) flow cytometer, and further analyzed by FlowJo software (Tree Star).
Western blot
Gingival tissues were isolated and homogenized in ice-cold lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.5% Nonidet P-40, 0.1% SDS, and 0.5 mM EDTA) supplemented with a protease inhibitor mixture (Sigma). After lysate incubation for 30 min on ice, tissue debris were removed by centrifugation (15,000 × g for 30 min at 4°C), and protein concentration in the supernatant was determined using the Pierce BSA Protein Assay kit (Thermo Scientific). Samples of 5–20 µg total protein were loaded onto 10% acrylamide gel and subject to SDS-PAGE. Proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore), the membrane was blocked in 5% skim milk for 30 min at room temperature, and reacted with primary antibody overnight at 4°C. The primary antibodies used are rabbit anti–TGFβ1 (Abcam), mouse anti–BMP-7 (Abcam), and rabbit polyclonal anti-Actin (I-19; Santa Cruz Biotechnology). The membrane was then washed three times in 1× TBST for 10 min at room temperature, incubated with secondary HRP-conjugated antibody (Abcam) in blocking buffer for 1 h at room temperature and washed three times in TBST before the blots were reacted with ECL substrate (Western blot detection kit; Advansta). Images were captured using a ChemiDoc MP Image System (Bio-Rad), and band quantification was done using ImageJ (National Institutes of Health).
RNA extraction
For RNA isolation from the maxilla, soft or both soft and hard tissues (as indicated in the text) were homogenized in 1 ml TRI reagent (Sigma) using an electric homogenizer (IKA labortechnik), and RNA was extracted according to the manufacturer’s instructions. Extraction of RNA from cell cultures was executed using 200 μl TRI reagent, and RNA was extracted according to the manufacturer’s instructions. cDNA synthesis was performed using the qScript cDNA Synthesis kit (Quanta-BioSciences). Quantitative RT-PCR reactions (20 µl volume) were performed using Power SYBR Green PCR Master Mix (Quanta-BioSciences). The following reaction conditions were used: 10 min at 95°C, 40 cycles of 15 s at 95°C, and 60 s at 60°C. The samples were normalized to the TBP (TATA box–binding protein) or Actin genes as control mRNA, by the change in cycling threshold (ΔCT) method, and calculated based on 2−ΔΔCT.
Isolation of LC precursors
Pre-DCs were isolated from BM cells stained with linage specific antibodies (CD19, CD3, NK1.1, B220, Ter119, CD11b, and MHCII) and with antibodies against CD11c, Flt3, and Sirpα. The cells were then sorted using FACS Aria III to obtain pre-DC cells as previously described (Capucha et al., 2015). Monocytes were preenriched from BM cells using the EasySep Mouse Monocyte Enrichment kit (STEMCELL Technologies). The eluted fraction was then stained with CD11b and Ly6G antibodies and sorted using FACS AriaIII instrument to obtain CD11b+ ly6G− cells as previously described (Capucha et al., 2015).
LC-like cell differentiation cultures
Total BM cells were isolated as previously described (Nassar et al., 2017). BM cells (5 × 105 cells) were cultured in 24-well plates (Nunc) in RPMI media (450 ml RPMI 1640, 5 ml l-glutamine, 50 µM β-mercaptoethanol, 100 U/ml penicillin,100 µg/ml streptomycin, and 50 µg/ml gentamicin) supplemented with 20 ng/ml SCF, 50 ng/ml FLT3L, and 2.5 ng/ml TNF-α (SM). The SM was further supplemented with 100 ng/ml GM-CSF with or without 10 ng/ml TGF-β1 or 200 ng/ml BMP7 for 5 d to induce their differentiation into LC-like cells. In some experiments, including experiments in which pre-DCs (5 × 104 cells) or monocytes (5 × 104 cells) were cultured, 5% FCS was added to the culture to facilitate cell survival. The cells were then washed with PBS plus 2% FCS and stained with the noted antibodies to analyze their differentiation by flow cytometry. To examine gene expression, total mRNA was extracted from the culture and analyzed by quantitative RT-PCR as described in the RNA extraction section.
Generation of chimeric mice
Adult CD45.1+ B6 mice were lethally irradiated with 950 rad, and 24 h later, the mice were injected intravenously with 3 × 106 BM cells obtained from congenic CD45.2+ CD11c-cre/ALK5fl/fl animals to allow identification of donor-derived cells. To generate mix BM chimeras, lethally irradiated CD45.1+ B6 mice received a mixture of BM cells purified from CD45.2+ CD11c-cre/ALK5fl/fl mice (3 × 106 cells) and CD45.1+ WT mice (1 × 106 cells). The chimeric mice were analyzed 3 wk after BM reconstitution.
In vivo administration of dorsomorphin
Adult Langerin-DTR mice were injected intraperitoneally with 1 µg DT (Sigma-Aldrich) per mouse in 150 µl PBS or PBS only as a control. Part of the mice received also intraperitoneal injection 2 µM dorsomorphin (Tocris) per mouse three times a day for 6 d.
Cohousing experiment
Cohousing was performed using 6-wk-old Swiss Webster GF females with age-matched colonized SPF mice. The GF and SPF mice were cohoused in new cages at a 1:1 ratio for 8 wk to allow microbial transfer and colonization of the GF mice.
Broad-spectrum antibiotic treatment
Adult mice were given a combination of 0.5 g/liter vancomycin, 1 g/liter ampicillin, 1 g/liter kanamycin, and 1 g/liter metronidazole in the drinking water for 2 mo as previously described (Levy et al., 2015). All antibiotics were obtained from Sigma-Aldrich.
Microbiota analysis
Oral microbiota of age- and sex-matched mice was collected from a single cage to exclude any effects that are not related to the depletion of LCs. The oral cavities of anesthetized individual mice were swabbed for 30 s, and DNA was extracted from the swabs using the UltraClean Tissue & Cells DNA Isolation kit according to the manufacturer’s instructions (MO BIO). After confirming high DNA purity, DNA samples were analyzed by quantitative RT-PCR as follows. Total bacterial load was determined using universal primers against the r16S gene after normalization based on the levels of r18S gene copies in each sample. For taxonomic microbiota analysis we performed r16S amplicon pyrosequencing. PCR amplification was performed spanning the V1/2 region of the 16S rRNA gene and subsequently sequenced using 500-bp paired-end sequencing (Illumina MiSeq). Reads were then processed using the QIIME (quantitative insights into microbial ecology) analysis pipeline with USEARCH against the Greengenes Database.
Cultivation of oral microbiota
Oral cavity of anesthetized individual DT-treated and untreated control adult mice were swabbed for 30 s, and the swabs were then inserted into Eppendorf tubes containing 100 µl Wilkins-Chalgren medium (Oxoid). The samples were serially diluted and plated on blood agar for 2 d under anaerobic conditions at 37°C, and colonies were enumerated to determine the colony-forming units of total cultivatable oral anaerobic bacteria.
Ablation of langerin-expressing cells in vivo
The progeny of heterozygous Langerin-DTR breeding pairs (Langerin-DTR+/− and +/+ littermate control) were cohoused from birth to adulthood and separated into two new cages before DT administration. Both groups of mice received a single intraperitoneal injection of 1 µg DT (Sigma-Aldrich) in 150 µl PBS per mouse. For long-term depletion, mice received additional injections of 0.6 µg DT in 150 µl PBS every 2 wk for 4 mo.
Bone loss quantification
4 mo after constitutive ablation of LCs in langerin-DTR mice, the hemimaxillae were harvested from the mice and littermate controls and alveolar bone loss was quantified using μCT (Scanco Medical). In brief, the sagittal plan of the specimens was set parallel to the x-ray beam axis. The specimens were scanned at a resolution of 12 µm in all three spatial dimensions. The scans were Gaussian-filtered and segmented using a multilevel global thresholding procedure for the segmentation of enamel, dentin, and bone. Residual alveolar bone volume was determined separately for either root (buccomesial and buccodistal) using a direct 3D approach. The apical basis of the measured volume was set mesio-distally parallel to the cementoenamel junction and buccopalatinally parallel to the occlusal plane. The results represented the residual bone above the reference plane in millimeters cubed.
Ki67 staining
Whole-mount gingival samples were incubated in 50% formamide for 2 h at 65°C, rinsed in 2 × SSC for 15 min at room temperature. DNA was denatured in 2N HCl for 30 min at 37°C and neutralized with 0.1M boric acid, followed by three washes in TBS (5 M NaCl and 1 M Tris in double-distilled water) and blocked in blocking buffer (3% FBS and 0.25% Triton X-100 in TBS) for 1 h at room temperature, followed by incubation with primary antibody for 72 h at 4°C. The next day, the sections were washed three times in TBS and blocked in blocking buffer for 15 min at room temperature, followed by incubation with fluorescently conjugated secondary antibody in blocking buffer for 1 h at room temperature.
Statistics
Data are expressed as means ± SEM. Statistical tests were performed using one-way ANOVA and a Student t test.
Online supplemental material
Fig. S1 depicts the analysis of pre-DCs and LCs in the lamina propria postnatally and the steady-state proliferation of mucosal LCs. Fig. S2 shows further characterization of LC-like cell differentiation from BM and murine fetal liver cells. Fig. S3 indicates that ALK5 is essential for LC differentiation. Fig. S4 illustrates that the microbiota regulates the levels of oral mucosa, but not skin, LCs. Fig. S5 demonstrates that prolonged depletion of langerin-expressing cells alters oral microbiota and augments secretion of proinflammatory cytokines by splenocytes.
Supplementary Material
Acknowledgments
This work was supported by the Israel Science Foundation (grant 766/16 to A.-H. Hovav).
The authors declare no competing financial interests.
Author contributions: T. Capucha, N. Koren, and A.-H. Hovav designed research. T. Capucha and N. Koren analyzed data. T. Capucha, N. Koren, M. Nassar, O. Heyman, T. Nir, L. Eli-Berchoer, and K. Zelentova performed experiments. G. Zilberman-Schapira, M. Levy, and E. Elinav contributed to GF mice experiments and microbiota analysis. A. Wilensky, T. Hieronymus, M. Zenke, and H. Bercovier contributed to manuscript preparation. T. Capucha, B.E. Clausen, and A.-H. Hovav wrote the manuscript.
References
- Arizon M., Nudel I., Segev H., Mizraji G., Elnekave M., Furmanov K., Eli-Berchoer L., Clausen B.E., Shapira L., Wilensky A., and Hovav A.H.. 2012. Langerhans cells down-regulate inflammation-driven alveolar bone loss. Proc. Natl. Acad. Sci. USA. 109:7043–7048. 10.1073/pnas.1116770109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asiedu M.K., Ingle J.N., Behrens M.D., Radisky D.C., and Knutson K.L.. 2011. TGFbeta/TNF(alpha)-mediated epithelial-mesenchymal transition generates breast cancer stem cells with a claudin-low phenotype. Cancer Res. 71:4707–4719. 10.1158/0008-5472.CAN-10-4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain C.C., Bravo-Blas A., Scott C.L., Perdiguero E.G., Geissmann F., Henri S., Malissen B., Osborne L.C., Artis D., and Mowat A.M.. 2014. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat. Immunol. 15:929–937. 10.1038/ni.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer T., Zagórska A., Jurkin J., Yasmin N., Köffel R., Richter S., Gesslbauer B., Lemke G., and Strobl H.. 2012. Identification of Axl as a downstream effector of TGF-β1 during Langerhans cell differentiation and epidermal homeostasis. J. Exp. Med. 209:2033–2047. 10.1084/jem.20120493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C.L., and Clausen B.E.. 2007. DC ablation in mice: promises, pitfalls, and challenges. Trends Immunol. 28:525–531. 10.1016/j.it.2007.08.011 [DOI] [PubMed] [Google Scholar]
- Bennett C.L., van Rijn E., Jung S., Inaba K., Steinman R.M., Kapsenberg M.L., and Clausen B.E.. 2005. Inducible ablation of mouse Langerhans cells diminishes but fails to abrogate contact hypersensitivity. J. Cell Biol. 169:569–576. 10.1083/jcb.200501071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigley V., McGovern N., Milne P., Dickinson R., Pagan S., Cookson S., Haniffa M., and Collin M.. 2015. Langerin-expressing dendritic cells in human tissues are related to CD1c+ dendritic cells and distinct from Langerhans cells and CD141high XCR1+ dendritic cells. J. Leukoc. Biol. 97:627–634. 10.1189/jlb.1HI0714-351R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner-Eddy P.D., Fischer L.A., Kaplan D.H., Thieu K., and Costalonga M.. 2016. Mucosal Langerhans Cells Promote Differentiation of Th17 Cells in a Murine Model of Periodontitis but Are Not Required for Porphyromonas gingivalis-Driven Alveolar Bone Destruction. J. Immunol. 197:1435–1446. 10.4049/jimmunol.1502693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobr A., Olvera-Gomez I., Igyarto B.Z., Haley K.M., Hogquist K.A., and Kaplan D.H.. 2010. Acute ablation of Langerhans cells enhances skin immune responses. J. Immunol. 185:4724–4728. 10.4049/jimmunol.1001802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkowski T.A., Letterio J.J., Farr A.G., and Udey M.C.. 1996. A role for endogenous transforming growth factor β 1 in Langerhans cell biology: The skin of transforming growth factor β 1 null mice is devoid of epidermal Langerhans cells. J. Exp. Med. 184:2417–2422. 10.1084/jem.184.6.2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capucha T., Mizraji G., Segev H., Blecher-Gonen R., Winter D., Khalaileh A., Tabib Y., Attal T., Nassar M., Zelentsova K., et al. 2015. Distinct Murine Mucosal Langerhans Cell Subsets Develop from Pre-dendritic Cells and Monocytes. Immunity. 43:369–381. 10.1016/j.immuni.2015.06.017 [DOI] [PubMed] [Google Scholar]
- Chopin M., Seillet C., Chevrier S., Wu L., Wang H., Morse H.C. III, Belz G.T., and Nutt S.L.. 2013. Langerhans cells are generated by two distinct PU.1-dependent transcriptional networks. J. Exp. Med. 210:2967–2980. 10.1084/jem.20130930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorro L., Sarde A., Li M., Woollard K.J., Chambon P., Malissen B., Kissenpfennig A., Barbaroux J.B., Groves R., and Geissmann F.. 2009. Langerhans cell (LC) proliferation mediates neonatal development, homeostasis, and inflammation-associated expansion of the epidermal LC network. J. Exp. Med. 206:3089–3100. 10.1084/jem.20091586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen B.E., and Kel J.M.. 2010. Langerhans cells: critical regulators of skin immunity? Immunol. Cell Biol. 88:351–360. 10.1038/icb.2010.40 [DOI] [PubMed] [Google Scholar]
- Clausen B.E., and Stoitzner P.. 2015. Functional Specialization of Skin Dendritic Cell Subsets in Regulating T Cell Responses. Front. Immunol. 6:534 10.3389/fimmu.2015.00534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran D.L. 2008. Inflammation and bone loss in periodontal disease. J. Periodontol. 79(8 Suppl.):1569–1576. 10.1902/jop.2008.080233 [DOI] [PubMed] [Google Scholar]
- Coombes J.L., Siddiqui K.R., Arancibia-Cárcamo C.V., Hall J., Sun C.M., Belkaid Y., and Powrie F.. 2007. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β and retinoic acid-dependent mechanism. J. Exp. Med. 204:1757–1764. 10.1084/jem.20070590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.A., Zenobia C., and Darveau R.P.. 2011. The relationship of the oral microbiotia to periodontal health and disease. Cell Host Microbe. 10:302–306. 10.1016/j.chom.2011.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fainaru O., Woolf E., Lotem J., Yarmus M., Brenner O., Goldenberg D., Negreanu V., Bernstein Y., Levanon D., Jung S., and Groner Y.. 2004. Runx3 regulates mouse TGF-beta-mediated dendritic cell function and its absence results in airway inflammation. EMBO J. 23:969–979. 10.1038/sj.emboj.7600085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Liu K., Helft J., Bogunovic M., Greter M., Hashimoto D., Price J., Yin N., Bromberg J., Lira S.A., et al. 2009. The origin and development of nonlymphoid tissue CD103+ DCs. J. Exp. Med. 206:3115–3130. 10.1084/jem.20091756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker C., Kirsch R.D., Ju X.S., Hieronymus T., Gust T.C., Kuhl C., Jorgas T., Kurz S.M., Rose-John S., Yokota Y., and Zenke M.. 2003. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat. Immunol. 4:380–386. 10.1038/ni903 [DOI] [PubMed] [Google Scholar]
- Hajishengallis G. 2015. Periodontitis: from microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 15:30–44. 10.1038/nri3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helft J., Böttcher J., Chakravarty P., Zelenay S., Huotari J., Schraml B.U., Goubau D., and Reis e Sousa C.. 2015. GM-CSF Mouse Bone Marrow Cultures Comprise a Heterogeneous Population of CD11c(+)MHCII(+) Macrophages and Dendritic Cells. Immunity. 42:1197–1211. 10.1016/j.immuni.2015.05.018 [DOI] [PubMed] [Google Scholar]
- Hervouet C., Luci C., Rol N., Rousseau D., Kissenpfennig A., Malissen B., Czerkinsky C., and Anjuère F.. 2010. Langerhans cells prime IL-17-producing T cells and dampen genital cytotoxic responses following mucosal immunization. J. Immunol. 184:4842–4851. 10.4049/jimmunol.0901695 [DOI] [PubMed] [Google Scholar]
- Hieronymus T., Zenke M., Baek J.H., and Seré K.. 2015. The clash of Langerhans cell homeostasis in skin: Should I stay or should I go? Semin. Cell Dev. Biol. 41:30–38. 10.1016/j.semcdb.2014.02.009 [DOI] [PubMed] [Google Scholar]
- Hoeffel G., Wang Y., Greter M., See P., Teo P., Malleret B., Leboeuf M., Low D., Oller G., Almeida F., et al. 2012. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J. Exp. Med. 209:1167–1181. 10.1084/jem.20120340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D.J., Wilson D.F., Smith P.S., Pierce A.M., and Wiebkin O.W.. 1994. Distribution of basal lamina type IV collagen and laminin in normal rat tongue mucosa and experimental oral carcinoma: ultrastructural immunolocalization and immunogold quantitation. Eur. J. Cancer B Oral Oncol. 30B:237–243. 10.1016/0964-1955(94)90004-3 [DOI] [PubMed] [Google Scholar]
- Jurkin J., Krump C., Köffel R., Fieber C., Schuster C., Brunner P.M., Borek I., Eisenwort G., Lim C., Mages J., et al. 2017. Human skin dendritic cell fate is differentially regulated by the monocyte identity factor Kruppel-like factor 4 during steady state and inflammation. J. Allergy Clin. Immunol. 139:1873–1884. 10.1016/j.jaci.2016.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D.H., Li M.O., Jenison M.C., Shlomchik W.D., Flavell R.A., and Shlomchik M.J.. 2007. Autocrine/paracrine TGFβ1 is required for the development of epidermal Langerhans cells. J. Exp. Med. 204:2545–2552. 10.1084/jem.20071401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautz-Neu K., Noordegraaf M., Dinges S., Bennett C.L., John D., Clausen B.E., and von Stebut E.. 2011. Langerhans cells are negative regulators of the anti-Leishmania response. J. Exp. Med. 208:885–891. 10.1084/jem.20102318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kel J.M., Girard-Madoux M.J., Reizis B., and Clausen B.E.. 2010. TGF-beta is required to maintain the pool of immature Langerhans cells in the epidermis. J. Immunol. 185:3248–3255. 10.4049/jimmunol.1000981 [DOI] [PubMed] [Google Scholar]
- Khosravi A., Yáñez A., Price J.G., Chow A., Merad M., Goodridge H.S., and Mazmanian S.K.. 2014. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 15:374–381. 10.1016/j.chom.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konradi S., Yasmin N., Haslwanter D., Weber M., Gesslbauer B., Sixt M., and Strobl H.. 2014. Langerhans cell maturation is accompanied by induction of N-cadherin and the transcriptional regulators of epithelial-mesenchymal transition ZEB1/2. Eur. J. Immunol. 44:553–560. 10.1002/eji.201343681 [DOI] [PubMed] [Google Scholar]
- Levy M., Thaiss C.A., Zeevi D., Dohnalová L., Zilberman-Schapira G., Mahdi J.A., David E., Savidor A., Korem T., Herzig Y., et al. 2015. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell. 163:1428–1443. 10.1016/j.cell.2015.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Gao X.H., and Mi Q.S.. 2016. TGF-β1-Smad signaling pathways are not required for epidermal LC homeostasis. Oncotarget. 7:15290–15291. 10.18632/oncotarget.8167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S., Hosur K.B., Domon H., and Hajishengallis G.. 2010. Periodontal inflammation and bone loss in aged mice. J. Periodontal Res. 45:574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Manz M.G., Karsunky H., Wagers A., Peters W., Charo I., Weissman I.L., Cyster J.G., and Engleman E.G.. 2002. Langerhans cells renew in the skin throughout life under steady-state conditions. Nat. Immunol. 3:1135–1141. 10.1038/ni852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C.J., and Shattock R.J.. 2003. Target cells in vaginal HIV transmission. Microbes Infect. 5:59–67. 10.1016/S1286-4579(02)00056-4 [DOI] [PubMed] [Google Scholar]
- Milne P., Bigley V., Gunawan M., Haniffa M., and Collin M.. 2015. CD1c+ blood dendritic cells have Langerhans cell potential. Blood. 125:470–473. 10.1182/blood-2014-08-593582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzaki A.R., Tetlak P., Sheng J., Loh S.C., Setiagani Y.A., Poidinger M., Zolezzi F., Karjalainen K., and Ruedl C.. 2016. Intestinal CD103(+)CD11b(-) dendritic cells restrain colitis via IFN-γ-induced anti-inflammatory response in epithelial cells. Mucosal Immunol. 9:336–351. 10.1038/mi.2015.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao K., Ginhoux F., Leitner W.W., Motegi S., Bennett C.L., Clausen B.E., Merad M., and Udey M.C.. 2009. Murine epidermal Langerhans cells and langerin-expressing dermal dendritic cells are unrelated and exhibit distinct functions. Proc. Natl. Acad. Sci. USA. 106:3312–3317. 10.1073/pnas.0807126106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S., Bouladoux N., Wilhelm C., Molloy M.J., Salcedo R., Kastenmuller W., Deming C., Quinones M., Koo L., Conlan S., et al. 2012. Compartmentalized control of skin immunity by resident commensals. Science. 337:1115–1119. 10.1126/science.1225152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S., Bouladoux N., Linehan J.L., Han S.J., Harrison O.J., Wilhelm C., Conlan S., Himmelfarb S., Byrd A.L., Deming C., et al. 2015. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 520:104–108. 10.1038/nature14052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar M., Tabib Y., Capucha T., Mizraji G., Nir T., Pevsner-Fischer M., Zilberman-Schapira G., Heyman O., Nussbaum G., Bercovier H., et al. 2017. GAS6 is a key homeostatic immunological regulator of host-commensal interactions in the oral mucosa. Proc. Natl. Acad. Sci. USA. 114:E337–E346. 10.1073/pnas.1614926114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouchi T., Kubo A., Yokouchi M., Adachi T., Kobayashi T., Kitashima D.Y., Fujii H., Clausen B.E., Koyasu S., Amagai M., and Nagao K.. 2011. Langerhans cell antigen capture through tight junctions confers preemptive immunity in experimental staphylococcal scalded skin syndrome. J. Exp. Med. 208:2607–2613. 10.1084/jem.20111718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittman B.R., Hill M.W., Rittman G.A., and Mackenzie I.C.. 1987. Age-associated changes in Langerhans cells of murine oral epithelium and epidermis. Arch. Oral Biol. 32:885–889. 10.1016/0003-9969(87)90102-6 [DOI] [PubMed] [Google Scholar]
- Romani N., Clausen B.E., and Stoitzner P.. 2010. Langerhans cells and more: langerin-expressing dendritic cell subsets in the skin. Immunol. Rev. 234:120–141. 10.1111/j.0105-2896.2009.00886.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruane D.T., and Lavelle E.C.. 2011. The role of CD103+ dendritic cells in the intestinal mucosal immune system. Front. Immunol. 2:25 10.3389/fimmu.2011.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seré K., Baek J.H., Ober-Blöbaum J., Müller-Newen G., Tacke F., Yokota Y., Zenke M., and Hieronymus T.. 2012. Two distinct types of Langerhans cells populate the skin during steady state and inflammation. Immunity. 37:905–916. 10.1016/j.immuni.2012.07.019 [DOI] [PubMed] [Google Scholar]
- Solano T., España A., Sola J., and López G.. 2000. Langerhans’ cell histiocytosis on the vulva. Gynecol. Oncol. 78:251–254. 10.1006/gyno.2000.5836 [DOI] [PubMed] [Google Scholar]
- Takahashi E., Nagano O., Ishimoto T., Yae T., Suzuki Y., Shinoda T., Nakamura S., Niwa S., Ikeda S., Koga H., et al. 2010. Tumor necrosis factor-alpha regulates transforming growth factor-beta-dependent epithelial-mesenchymal transition by promoting hyaluronan-CD44-moesin interaction. J. Biol. Chem. 285:4060–4073. 10.1074/jbc.M109.056523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay J., Upadhyay R.B., Agrawal P., Jaitley S., and Shekhar R.. 2013. Langerhans cells and their role in oral mucosal diseases. N. Am. J. Med. Sci. 5:505–514. 10.4103/1947-2714.118923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukicevic S., Latin V., Chen P., Batorsky R., Reddi A.H., and Sampath T.K.. 1994. Localization of osteogenic protein-1 (bone morphogenetic protein-7) during human embryonic development: high affinity binding to basement membranes. Biochem. Biophys. Res. Commun. 198:693–700. 10.1006/bbrc.1994.1100 [DOI] [PubMed] [Google Scholar]
- Xu Y.P., Shi Y., Cui Z.Z., Jiang H.H., Li L., Wang X.F., Zhou L., and Mi Q.S.. 2012. TGFβ/Smad3 signal pathway is not required for epidermal Langerhans cell development. J. Invest. Dermatol. 132:2106–2109. 10.1038/jid.2012.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasmin N., Bauer T., Modak M., Wagner K., Schuster C., Köffel R., Seyerl M., Stöckl J., Elbe-Bürger A., Graf D., and Strobl H.. 2013. Identification of bone morphogenetic protein 7 (BMP7) as an instructive factor for human epidermal Langerhans cell differentiation. J. Exp. Med. 210:2597–2610. 10.1084/jem.20130275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala W.D., and Cavicchia J.C.. 2006. Deterioration of the Langerhans cell network of the human gingival epithelium with aging. Arch. Oral Biol. 51:1150–1155. 10.1016/j.archoralbio.2006.06.008 [DOI] [PubMed] [Google Scholar]
- Zeisberg M., Shah A.A., and Kalluri R.. 2005. Bone morphogenic protein-7 induces mesenchymal-to-epithelial transition in adult renal fibroblasts and facilitates regeneration of injured kidney. J. Biol. Chem. 280:8094–8100. 10.1074/jbc.M413102200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.