Summary
Here, we present a large (N=107,207) genome-wide association study (GWAS) of general cognitive ability (g), further enhanced by combining results with a large-scale GWAS of educational attainment. We identified 70 independent genomic loci associated with GCA. Results showed significant enrichment for genes causing Mendelian disorders with an intellectual disability phenotype. Competitive pathway analysis implicated the biological processes of neurogenesis and synaptic regulation, as well as the gene targets of two pharmacologic agents: cinnarizine, a T-type calcium channel blocker; and LY97241, a potassium channel inhibitor. Transcriptome-wide and epigenome-wide analysis revealed that the implicated loci were enriched for genes expressed across all brain regions (most strongly in the cerebellum); enrichment was exclusive to genes expressed in neurons, but not oligodendrocytes or astrocytes. Finally, we report genetic correlations between cognitive ability and disparate phenotypes including psychiatric disorders, several autoimmune disorders, longevity, and maternal age at first birth.
Keywords: GWAS, general cognitive ability, nootropics, gene expression, neurodevelopment, synapse, calcium channel, potassium channel, cerebellum
Graphical abstract

Introduction
Genome-wide association studies (GWAS) have been highly successful at uncovering hundreds of genetic loci associated with heritable quantitative traits such as height (Wood et al., 2014) and weight/body mass index (Locke et al., 2015). However, identifying genetic loci underlying cognitive ability has been much more challenging, despite heritability of 0.5 or greater, as determined by both classical twin studies (Deary et al., 2009) and molecular genetic studies (Davies et al., 2011a). In part, the difficulty with cognitive GWAS may be caused by the relative heterogeneity in the measurement of the cognitive phenotype. Traditionally, general cognitive ability (g) has been defined as a latent trait underlying shared variance across multiple subdomains of cognitive performance, psychometrically obtained as the first principal component of several distinct neuropsychological test scores (Johnson et al., 2008). Using this approach, several cognitive GWAS with fewer than 20,000 subjects yielded no genome-wide significant (GWS) effects (Benyamin et al., 2013; Davies et al., 2011b; Lencz et al., 2014), while a few GWS loci were identified in larger GWAS of 35,298 (Trampush et al., 2017) and 53,949 (Davies et al., 2015) subjects, respectively. By contrast, two independent GWAS of height with sample sizes of approximately 30,000 subjects each yielded 20–30 GWS hits (Gudbjartsson et al., 2008; Weedon et al., 2008); allelic effect sizes were ~2–5 times larger than the largest obtained in cognitive GWAS (Trampush et al., 2015).
Very recently, a cognitive GWAS (Sniekers et al., 2017) was able to leverage a very brief measure of fluid intelligence, highly correlated with psychometrically defined g, obtained in over 50,000 subjects. In combination with several traditional cognitive GWAS cohorts, total sample size was 78,308. This sample size permitted discovery of 18 independent GWS allelic loci, as well as numerous additional loci from gene-based analysis. This report was critical in demonstrating that signal could be enhanced by combining data from cohorts with brief measures of intelligence with data from more traditional cognitive GWAS.
A further approach to enhancing power in cognitive GWAS has focused on educational attainment as a proxy phenotype (Rietveld et al., 2014). It is acknowledged that this phenotype is ‘noisy’, as it is influenced by non-cognitive genetic (Belsky et al., 2016) (e.g., personality) and environmental (Johnson et al., 2010) (e.g., socio-economic) factors; consequently, observed allelic effect sizes have been even smaller than those obtained for GWAS of g (Rietveld et al., 2013). However, by utilizing a single-item measure (years of education completed), obtained incidentally in large studies of other phenotypes, this approach has allowed investigators to obtain extremely large sample sizes. A recent study of educational attainment in nearly 300,000 individuals identified 74 independent GWS loci (Okbay et al., 2016). Moreover, a new technique called multi-trait analysis of GWAS (MTAG) (Turley et al., 2017) has been developed which permits integration of GWAS data across related traits, accounting for the possibility of overlapping samples across studies, and requiring only summary statistics. The developers of MTAG demonstrated its accuracy and utility in a study of traits (depression, neuroticism, and subjective well-being) that demonstrate genetic correlations in the range of ~.70–.75; importantly, the genetic correlation between cognitive performance and educational attainment has been consistently reported to be in the same range (Davies et al. 2015, 2016; Okbay et al. 2016; Trampush et al. 2017; Sniekers et al. 2017). MTAG is able to quantify the degree of “boost” to the signal of a single-trait GWAS, providing an estimate of observed sample size, and providing summary statistics (allelic weights) that can then be utilized in all downstream annotation pipelines available for GWAS output.
In the present study, we first utilized GWAS meta-analysis to combine our prior COGENT consortium GWAS (Trampush et al., 2017) of psychometrically defined g with the recently reported GWAS (Sniekers et al., 2017) relying primarily on the brief measure, resulting in a combined cohort of N=107,207 non-overlapping samples measured for cognitive performance. Next, we utilized MTAG to combine these results with the large-scale GWAS of educational attainment, resulting in further enhanced power. At each step, we performed both allelic and gene-based tests. We then performed downstream analyses on the resulting MTAG summary statistics, including: 1) competitive gene set analyses to identify key biological processes and potential drug targets implicated; 2) stratified linkage disequilibrium score regression (LDSC) to identify differential cell type expression; 3) transcriptome-wide association study (TWAS) methods, to identify specific effects of altered gene expression in the brain on cognition; and 4) LDSC to identify genetic correlations with other anthropometric and biomedical phenotypes.
Results
Meta-Analysis: Cognitive Performance GWAS
Meta-analysis of all non-overlapping cohorts from the two GWAS of cognitive performance (total N = 107,207) identified 28 independent genomic loci reaching genome-wide significance (GWS, P<5E-08), using default clumping parameters from the Functional Mapping and Annotation (FUMA) pipeline (Watanabe et al., 2017) pipeline (Figure 1a); this represents a 55.6% increase in loci compared to the previous GWAS (Sniekers et al., 2017) of cognitive performance. Two of these loci each contained two uncorrelated variants with independent effects, resulting in 30 independent lead SNPs. Evidence for spurious inflation of statistical tests was quite limited for a large study of a highly polygenic trait (λ=1.23; λ1000=1.001; LD score intercept=1.03; see also PP plot in Supplementary Figure 1), and overall SNP heritability was .168. Of the 28 GWS loci, 12 were not previously reported as GWS in published studies of cognitive or educational phenotypes (Supplementary Table 1). The majority of the 5,610 markers reaching a nominal significance threshold were intronic SNPs followed by those in the intergenic regions (Supplementary Table 2). As shown in Supplementary Table 3, several of the GWS loci overlap with loci related to schizophrenia, bipolar disorder, and other neuropsychiatric phenotypes, as well as obesity/body mass index and other traits.
Figure 1.
a) Manhattan plot depicting results of GWAS meta-analysis of cognitive performance. Dotted red line indicates threshold for genome-wide significance (P<5E-08). b) Manhattan plot depicting results of MTAG of cognitive performance with educational attainment. Dotted red line indicates threshold for genome-wide significance (P<5E-08).
The significant loci harbored 88 known protein coding genes (Supplementary Table 4), about half of which were in three large regions (Supplementary Figure 2), including two well-characterized regions: the distal 16p11.2 region, in which deletions have been associated with schizophrenia and other neuropsychiatric phenotypes (Guha et al., 2013), and the 17q21 region, in which inversions have been associated with neuropsychiatric disorders (Cooper et al., 2011). Using MAGMA (Multi-marker Analysis of GenoMic Annotation; de Leeuw et al., 2015) gene-based tests, 73 genes were genome-wide significant (Supplementary Table 5), of which 39 were overlapping with the 88 genes noted above, resulting in a total of 122 candidate genes with statistical evidence of association to cognitive performance.
MTAG: Combining Cognitive Performance and Educational Attainment GWAS
MTAG analysis combining the cognitive performance results obtained above with the large educational attainment GWAS previously reported (Okbay et al., 2016), resulted in a 75% enrichment of statistical power, effectively boosting the original sample size of N = 107,207 to a GWAS equivalent of N = 187,812. Default clumping procedures revealed that 70 independent genomic loci reached genome-wide significance, with 82 independent SNPs (Figure 1b). Similar to the GWAS results above, the PP plot (Supplementary Figure 3) demonstrated polygenicity without evidence for artifactual inflation of statistical tests (λ=1.28; λ1000=1.001; LD score intercept=0.91), and overall SNP heritability was 0.336. Of the 70 GWS loci, 34 were not previously reported as GWS in published studies of cognitive or educational phenotypes (Figure 2; Supplementary Table 1). All but two of the 30 loci identified in the meta-analysis remained genome-wide significant in the MTAG results; even these two loci showed the same direction of allelic effects between cognitive meta-analytic GWAS and the educational GWAS. The majority of the 13,549 SNPs reaching a nominal significance threshold in the MTAG analysis were intergenic or intronic (Supplementary Table 2; Supplementary Figure 4). GWAS catalog annotations are listed in Supplementary Table 3. Within the GWS loci, 265 protein coding genes were identified (Supplementary Table 4). Additionally, 256 genes were significant in MAGMA gene-based tests (Supplementary Table 6); of these, 85 genes were non-overlapping with the 265 genes within SNP GWS loci, resulting in a total of 350 genes receiving GWS support from the MTAG results.
Figure 2.
Venn diagram depicting overlap and independence of genome-wide significant SNP loci observed in three studies: the MTAG analysis of the present report; the cognitive performance GWAS reported by Sniekers et al. (2017); and the educational attainment GWAS of Okbay et al. (2016).
As a formal validation that the MTAG methodology successfully predicts phenotype variance for cognitive performance, MTAG was re-analyzed, excluding the COGENT cohorts (i.e., the IQ GWAS of Sniekers et al. 2017 was combined with the educational GWAS of Okbay et al.2016). The ASPIS and GCAP datasets were held out as target cohorts used for calculation of polygenic risk score modelling for “g”. Despite the relatively small size of these hold-out cohorts, results show strongly significant polygenic prediction of “g” using MTAG-derived allele weights (Figure 3a and 3c), accounting for more than 4% of the variance in the GCAP cohort. For both cohorts, polygenic prediction began to drop at PT thresholds above 0.05, suggesting that there may be some degree of saturation of signal beyond the nominal 0.05 significance level at these sample sizes. Additional comparisons were made with IQ-only predictions (weights derived from Sniekers et al. 2017) and education-only predictions (weights derived from Okbay et al. 2016) for the same hold-out cohorts (Figure 3b and 3d), and we found that the MTAG-derived weights showed a 3.5-times and 3-times improvement in R2 variance explained in the ASPIS cohort, for IQ and Education respectively. For the GCAP cohort, there was a 5.1-times to 96-times improvement in R2 variance relative to IQ or education alone.
Figure 3.
a) Polygenic risk score prediction for MTAG results against held-out ASPIS cohort. b) Comparison of MTAG, cognitive (IQ) GWAS (Sniekers et al. 2017), and educational attainment (EDU) GWAS (Okbay et al. 2016) as source of weights for polygenic risk score prediction against held-out ASPIS cohort. c) Polygenic risk score prediction for MTAG results against held-out GCAP cohort. d) Comparison of MTAG, cognitive (IQ) GWAS (Sniekers et al. 2017), and educational attainment (EDU) GWAS (Okbay et al. 2016) as source of weights for polygenic risk score prediction against held-out GCAP cohort.
Overlap with Intellectual Disability Genes
We compared the list of 350 genes emerging from MTAG with a list of 621 genes known to cause autosomal dominant or autosomal recessive Mendelian disorders featuring intellectual disability (Harripaul et al., 2017; Vissers et al., 2016). As shown in Table 1, a total of 23 genes identified by MTAG appeared on this list, representing a 2-fold enrichment over chance (hypergeometric probability p=0.001). Examining autosomal dominant and recessive Mendelian genes demonstrated a somewhat stronger enrichment for autosomal dominant genes (p=.0017) than autosomal recessive genes (p=.054).
Table 1.
| GENE | CHR | START | MAGMA P | Min MTAG P | OMIM | Mode | Phenotype |
|---|---|---|---|---|---|---|---|
| AFF3 | 2 | 100152323 | 6.53E-12 | 6.8834E-15 | NA | AR | Nonsyndromal intellectual disability |
| AMT | 3 | 49444211 | 1.74E-09 | 8.5543E-09 | 605899 | AR | Glycine encephalopathy |
| ARFGEF2 | 20 | 47528427 | 7.28E-10 | 4.1558E-10 | 608097 | AR | Periventricular heterotopia with microcephaly |
| BCL11A | 2 | 60668302 | 8.5E-12 | 3.2174E-13 | 617101 | AD | Intellectual developmental disorder with persistence of fetal hemoglobin |
| C12orf65 | 12 | 123707463 | 1.48E-10 | 1.8088E-11 | 613559 | AR | Combined oxidative phosphorylation deficiency 7 |
| 615035 | AR | Spastic paraplegia 55 | |||||
| CLN3 | 16 | 28467983 | 2.31E-08 | 1.9502E-08 | 204200 | AR | Ceroid lipofuscinosis, neuronal 3 |
| DPYD | 1 | 97533299 | 0.005108 | 4.4603E-08 | 274270 | AR | Dihydropyrimidine dehydrogenase deficiency |
| 274270 | AR | 5-fluorouracil toxicity | |||||
| ERCC8 | 5 | 60159658 | 2.96E-07 | 5.5002E-7 | 216400 | AR | Cockayne syndrome, Type A |
| 614621 | AR | UV-sensitive syndrome 2 | |||||
| FOXP1 | 3 | 70993844 | 6.32E-07 | 3.5007E-09 | 613670 | AD | Mental retardation with language impairment and autistic features |
| GMPPB | 3 | 49744277 | 1.75E-14 | 6.6613E-16 | 613530 | AR | Muscular dystrophy-dystroglycanopathy (congenital w/ brain,eye anomalies), type A,14 |
| 615351 | AR | Muscular dystrophy-dystroglycanopathy (congenital with mental retardation), type B,14 | |||||
| 615352 | AR | Muscular dystrophy-dystroglycanopathy (limb--girdle), type C, 14 | |||||
| KANSL1 | 17 | 44097282 | 1.62E-08 | 5.0278E-12 | 610443 | AD | Koolen-De Vries syndrome |
| KCNH1 | 1 | 210846555 | 1.04E-06 | 5.2513E-08 | 135500 | AD | Zimmermann-Laband syndrome |
| KMT2D | 12 | 49402758 | 1.69E-07 | 4.3422E-08 | 147920 | AD | Kabuki syndrome, 1 |
| LARGE | 22 | 33548212 | 7.99E-07 | 5.4265E-07 | 613154 | AR | Muscular dystrophy-dystroglycanopathy (congenital w/ brain,eye anomalies), type A, 6 |
| 608840 | AR | Muscular dystrophy-dystroglycanopathy (congenital with mental retardation), type B, 6 | |||||
| MEF2C | 5 | 88003975 | 1.74E-13 | 1.1304E-12 | 613443 | AD | Mental retardation, stereotypic movements, epilepsy, and/or cerebral malformations |
| 613443 | AD | Chromosome 5q14.3 deletion syndrome | |||||
| NFIX | 19 | 13096422 | 2.45E-06 | 5.3017E-09 | 602535 | AD | Marshall-Smith syndrome |
| 614753 | AD | Sotos syndrome | |||||
| PDE4D | 5 | 58254865 | 9.13E-08 | 3.6537E-07 | 614613 | AD | Acrodysostosis 2 with or without hormone resistance |
| SHANK3 | 22 | 51102843 | 2.7E-10 | 8.0006E-08 | 606232 | AD | Phelan-McDermid syndrome |
| ST3GAL3 | 1 | 44161495 | 3.58E-13 | 1.6388E-10 | 611090 | AR | Mental retardation, autosomal recessive 12 |
| SUOX | 12 | 56380964 | 3.07E-05 | 4.1129E-08 | 272300 | AR | Sulfite oxidase deficiency |
| TCF4 | 18 | 52879562 | 1.02E-06 | 3.5713E-05 | 610954 | AD | Pitt-Hopkins syndrome |
| THRB | 3 | 24148651 | 0.000682 | 4.6883E-06 | 188570 | AD | Thyroid hormone resistance |
| 274300 | AR | Thyroid hormone resistance, autosomal recessive | |||||
| UBA7 | 3 | 49832640 | 2.11E-13 | 6.6613E-16 | NA | AR | Nonsyndromal intellectual disability |
AD=Autosomal Dominant; AR=Autosomal Recessive
Tissue Expression Enrichment and Competitive Pathway Analysis
Downstream MAGMA expression profiles and competitive pathway analysis were conducted as part of the FUMA pipeline. MAGMA tissue expression profile analysis revealed that genes emerging from the MTAG analysis were significantly enriched for expression in nearly all central nervous system tissues (except for substantia nigra and spinal cord), and that this enrichment was exclusive to neural tissues (Figure 4a). Notably, the strongest enrichment was observed for genes expressed in the cerebellum, followed by cortex, and slightly weaker (but still strongly significant) enrichment in subcortical and limbic structures. Competitive pathway analysis (based on gene ontology categories) for GWS MAGMA genes identified by MTAG revealed significant enrichment of neuronal and synaptic cellular components, as well as the biological processes of neurogenesis and regulation of synapse organization (Table 2, upper panel). Because three MTAG loci (at chromosome 3q21.31, 16p11.2, and 17q21.31) were unusually large, each containing 15 or more genes which may have disproportionately impacted enrichment results, we re-ran the above tissue expression and pathway analyses excluding these three regions. Results were substantively unchanged; all of the same neural tissues remained significantly enriched, in the same order of significance as shown in Figure 4a, and all of the same pathways remained significant (Bonferroni-corrected p<.05) as shown in Table 2, except for the cellular compartment “dendrite” (Bonferroni-corrected p=.089).
Figure 4.
a) Tissue expression profile analysis for genome-wide significant genes (as defined by MAGMA) emerging from the MTAG analysis. Gene results were significantly enriched for expression in nearly all central nervous system tissues (except for substantia nigra and spinal cord), but no tissues outside the CNS. b) Circular Manhattan Plot for MetaXcan results based on MTAG of cognitive performance with educational attainment. From inner circle out, GTEX tissue order is as follows: ACC: Anterior Cingulate Cortex; CDBG: Caudate – Basal Ganglia; CRBHM: Cerebellar Hemisphere; CRBLM: Cerebellum; CRTX: Cortex; FCTX: Frontal Cortex; HIPP: Hippocampus; HYPO: Hypothalamus; NACMB: Nucleus Accumbens; PUTM: Putamen. GWAS threshold is set at Bonferroni-corrected P < 0.05.
Table 2.
| GO Category name | NGENES | BETA | BETA_STD | SE | P | Pbon |
|---|---|---|---|---|---|---|
| GO_cc:go_neuron_par t | 1204 | 0.155 | 0.0385 | 0.0304 | 1.84E-07 | 0.002008 |
| GO_cc:go_neuron_projection | 898 | 0.179 | 0.0388 | 0.0352 | 1.84E-07 | 0.002009 |
| GO_bp:go_neurogenesis | 1355 | 0.148 | 0.0388 | 0.0291 | 1.92E-07 | 0.002092 |
| GO_cc:go_synapse | 718 | 0.198 | 0.0386 | 0.0393 | 2.25E-07 | 0.002455 |
| GO_cc:go_synapse_part | 580 | 0.21 | 0.0369 | 0.0436 | 7.37E-07 | 0.008026 |
| GO_cc:go_dendrite | 430 | 0.229 | 0.0348 | 0.0501 | 2.49E-06 | 0.027087 |
| GO_bp:go_regulation_of_synapse_organization | 106 | 0.447 | 0.034 | 0.0987 | 2.94E-06 | 0.031982 |
| GO_bp:go_regulation_of_synapse_structure_or_activity | 223 | 0.291 | 0.032 | 0.0671 | 7.36E-06 | 0.080154 |
| GO_bp:go_regulation_of_nervous_system_development | 723 | 0.166 | 0.0325 | 0.0385 | 7.84E-06 | 0.085334 |
| GO_bp:go_modulation_of_synaptic_transmission | 291 | 0.253 | 0.0317 | 0.059 | 9.41E-06 | 0.102429 |
| GO_bp:go_calcium_dependent_cell_cell_adhesion_via_plasma_membrane_cell_adhesion_molecules | 26 | 1.06 | 0.0402 | 0.259 | 2.06E-05 | 0.224726 |
| GO_cc:go_postsynapse | 356 | 0.224 | 0.031 | 0.0553 | 2.64E-05 | 0.287583 |
| GO_cc:go_neuron_spine | 116 | 0.379 | 0.0302 | 0.0939 | 2.75E-05 | 0.299998 |
| GO_cc:go_cell_projection | 1710 | 0.103 | 0.0301 | 0.0258 | 3.36E-05 | 0.365381 |
| GO_bp:go_regulation_of_cell_development | 808 | 0.144 | 0.0297 | 0.0365 | 3.99E-05 | 0.434751 |
|
| ||||||
| Drug name | NGENES | BETA | BETA_STD | SE | P | Pbon |
|
| ||||||
| CINNARIZINE | 9 | 1.62 | 0.036 | 0.355 | 2.61E-06 | 0.007071 |
| LY97241 | 2 | 3.65 | 0.0382 | 0.842 | 7.59E-06 | 0.020535 |
| CELECOXIB | 45 | 0.632 | 0.0314 | 0.159 | 3.49E-05 | 0.094545 |
| ISRADIPINE | 8 | 1.59 | 0.0334 | 0.404 | 4.18E-05 | 0.11317 |
| NITRENDIPINE | 12 | 1.19 | 0.0305 | 0.323 | 1.19E-04 | 0.323151 |
| ABT-639;ML218;TTA-A2;Z944 | 3 | 2.31 | 0.0297 | 0.641 | 1.59E-04 | 0.429388 |
| NEUREGULIN-1;NEUREGULIN-2 | 2 | 2.39 | 0.0251 | 0.669 | 1.75E-04 | 0.473469 |
| FLUNARIZINE | 6 | 1.58 | 0.0287 | 0.457 | 2.67E-04 | 0.723503 |
| GLUCOCORTICOIDS | 2 | 3.68 | 0.0386 | 1.08 | 3.22E-04 | 0.872117 |
Competitive pathway analysis for drug pathways (Gaspar and Breen, 2017) revealed that the gene targets of two drugs were significantly enriched in the MTAG results (Table 2, lower panel): Cinnarizine, a T-type calcium channel blocker and LY97241, a potassium channel inhibitor. L-type calcium channel blockers and anti-inflammatories also showed suggestive evidence of enrichment. In a related analysis of drug classes, significant enrichment was observed for voltage-gated calcium channel subunits (p=9.28E-06, Bonferroni-corrected P=5.38E-04).
Stratified LD score regression (Finucane et al., 2017) also demonstrated an enrichment of cell type expression for neuronal tissues only. Notably, genes found in the neuronal expression list of Cahoy (Cahoy et al., 2008) were significantly enriched (p=.0129; Bonferroni-corrected p=.0386), whereas negative results were obtained for genes expressed in oligodendrocytes (p=.4997) and astrocytes (p=.9057). Additionally, using Roadmap annotations, epigenetic enrichment was strongest in fetal brain tissue DNase sites and H3K4me1 primed enhancers; followed by adult cortical H3K27ac active enhancer sites (see Supplementary Table 7 for further details). No enrichment was observed for any non-neuronal tissue. Again, results were not substantively changed when the three large loci were removed from these analyses.
Gene Expression Analyses
In order to derive specific biological insights from the broad association loci implicated by MTAG, we performed a series of analysis designed to identify individual gene expression changes associated with cognition. First, we performed transcriptome wide analysis (TWAS), using MetaXcan (Barbeira et al., 2016), on MTAG SNP results in order to identify transcripts for which up-regulation or down-regulation in specific neural compartments was associated with cognition. [Note that TWAS follows a similar logic to imputation, in that an external reference (in this case, publicly available GTEx eQTL data for 10 brain regions) is utilized to link SNP-based summary statistics to tissue-based expression levels]. As shown in Figure 4b (and detailed in Supplementary Table 8), most of the significant TWAS results are expressed across all neural tissues, involving genes such as AMIGO3, RNF123, and RBM6. Moreover, no individual tissue compartment was much more strongly enriched for associations compared to the others. However, a few strong transcriptomic associations were specific to individual brain regions. For example, the strongest result in hippocampus was with DAG1; TWAS demonstrated that greater expression of this gene in hippocampus was associated with higher cognitive scores. However, this gene was not expressed in other neural tissue types in the GTEx database. Similarly, lower levels of ACTR1A were significantly associated with better cognition, but this transcript was observed only in frontal cortex.
Second, we applied a Bayesian fine-mapping approach (CAVIAR-BF, Chen et al. 2015) to identify putative causal SNPs within each associated locus, as defined in Supplementary Table 9. CAVIAR-BF revealed that there was strong evidence (BF = 3.71e+2) for at least 1 causal SNP within each of the 70 independent MTAG loci. There is also evidence that there is at least 2 causal SNPs in 65 of the loci (BF = 3e+6) and at least 3 causal SNPs in 47 of the loci (BF = 2.86e+6). In the extended region analysis, there was evidence for at least 1 causal SNP (BF = 3.45e+2) and 2 causal SNPs (BF = 2.89e+6) for 70 and 63 loci respectively. Model search revealed that there were 386 putative causal SNPs within the 70 independent loci (Supplementary Table 10). Lookups of these SNPs in two brain eQTL databases (BrainEAC (Ramasamy et al., 2014) and CommonMind(Hauberg et al., 2017)) revealed several additional SNP-eQTL relationships that can explain variance in the cognitive phenotype (Supplementary Tables 11 and 12); the most notable eQTL effect was observed for rs3809912 on chromosome 18. This SNP, which was GWS in the MTAG results (p=7.06E-09), was a strong eQTL for CEP192 (p = 5.1e-38, FDR < 0.01). This eQTL was confirmed in the CommonMind database (FDR<.01), which demonstrated that expression of 44 independent transcripts in frontal cortex were significantly associated with MTAG SNPs at the FDR<.01 level. Combining annotation information from the Mendelian gene analysis, MetaXcan TWAS, Braineac and CommonMind databases, we found supporting functional evidence for 112 of the 350 candidate genes nominated by MTAG (Supplementary Table 13). The remaining 238 genes without functional support had statistical evidence for association to cognition, but are considered to be ‘candidate genes’ requiring further functional or experimental support.
Genetic Correlations with Other Phenotypes
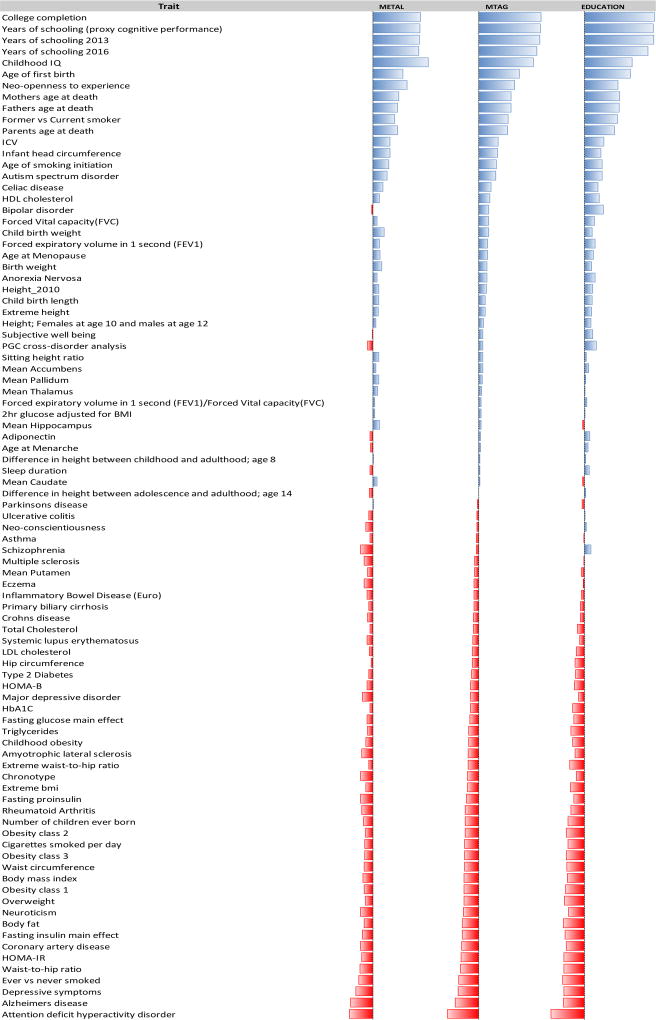
LD-score regression was carried out across 89 traits in 15 broad phenotypic categories in LD-hub (Zheng et al., 2017): 1) aging, 2) anthropometric, 3) autoimmune, 4) brain volume, 5) cardiometabolic, 6) education, 7) glycemic, 8) lipids, 9) lung function, 10) neurological, 11) personality, 12) psychiatric, 13) reproductive behavior, 14) sleep, and 15) smoking behavior (Figure 5; Supplementary Table 14). We performed LD-score regression separately for the results of our initial meta-analysis and for the MTAG results. For comparison, we also present LD-score regression results for the educational attainment GWAS of Okbay et al. (2016); it should be noted that only 14 phenotypes were examined for genetic correlation in that publication.
Figure 5.
Genetic correlations (rg) between cognitive phenotypes and other publicly available GWAS results, based on LD score regression. The first and second columns (labelled METAL and MTAG, respectively) refer to results of the cognitive meta-analyses in the present report. The third column displays correlations for the educational attainment GWAS of Okbay et al. (2016).
Cognition appeared to be strongly associated at the genetic level with aging, education, personality, neuropsychiatric disorders, reproductive behavior, and smoking behavior. Strong association with parental age at death was observed for both the GWAS meta-analysis and MTAG results. Meanwhile, moderate associations with anthropometric traits were observed, although associations with brain volumes were surprisingly modest, except for total intracranial volume (rg for MTAG results = 0.31, p=7.37E-19). While many of these correlations have been described previously (Hagenaars et al., 2016; Okbay et al., 2016; Sniekers et al., 2017; Trampush et al., 2017), two results observed in the present study were not reported in those prior publications. First, we report a strong positive genetic correlation between cognitive performance and maternal age at first birth (rg for MTAG results = 0.63, p=2.36E-163) and inverse correlation with parental number of children ever born (rg for MTAG results = −0.22; p=6.91E-13). It is possible that these effects are mediated by years of higher education, insofar as correlations were even stronger with educational attainment (rg for parental age at first birth=0.72, p=2.24E-244; rg for number of children= −0.26, p=3.34E-18). As with any other regression relationship, a role for unmeasured mediators, such as propensity for delayed gratification, cannot be ruled out. Second, we observed modest, yet nominally significant, inverse correlations between cognition and autoimmune diseases such as eczema and Crohn’s disease, attaining Bonferroni significance for rheumatoid arthritis (rg for MTAG results = −0.2086; p=1.60E-08); there was also a Bonferroni-significant positive genetic correlation with celiac disease (rg for MTAG results = 0.1922; p=0.0001). While results of cross-trait analyses were largely consistent using either the GWAS results, the MTAG results, or the previously-published educational attainment datasets, there were notable divergences in correlations with psychiatric phenotypes, especially schizophrenia and bipolar disorder.
Discussion
Uncovering the molecular genetic basis of individual differences in cognitive performance can have a significant impact on our understanding of neuropsychiatric disorders, which are both phenotypically (Burdick et al., 2011; Ferreri et al., 2011; Keefe and Harvey, 2012; Snyder, 2013) and genetically (Lencz et al., 2014; Smeland et al., 2017; Stergiakouli et al., 2017) correlated with cognition, as well as numerous non-psychiatric health-relevant phenotypes (Hagenaars et al., 2016) which also demonstrate significant genetic correlations with cognitive function. Here, we have presented the largest GWAS of cognition to date, with 107,207 individuals phenotypically characterized for performance on standardized tests measuring general cognitive ability. Results were further enhanced by utilizing a relatively new approach to allow meta-analysis with a large-scale GWAS of educational attainment, which is highly (though not perfectly) correlated with cognitive ability at the genetic level. With this approach, we were able to identify 70 genomic loci significantly associated with cognition, implicating 350 candidate genes underlying cognitive ability. In total, we found that common SNPs were able to account for roughly half of the overall heritablility of the phenotype as determined by prior family studies (Plomin and Deary, 2015).
Downstream analysis confirmed an important role for neurodevelopmental processes in cognitive ability, consistent with implications from the education GWAS (Okbay et al., 2016). Significant genes were more strongly enriched for expression in fetal brain tissue than adult tissue; results were also enriched for genes implicated in early neurodevelopmental disorders; and neurogenesis was the most strongly enriched GO biological process. At the same time, it is important to emphasize that adult neural tissues were also strongly represented in the results, and multiple synaptic components were significant in the pathway analysis. In this context, it is noteworthy that many cellular processes necessary for early neurodevelopment are also involved in adult synaptic plasticity. This duality is represented by several significant genes emerging from our analysis: CELSR3 encodes an atypical cadherin plasma membrane protein involved in long-range axon guidance in neurodevelopment through planar cell polarity signaling (Chai et al., 2015), but is also necessary for adult formation of hippocampal glutamatergic synapses (Thakar et al., 2017). Similarly SEMA3F is a negative regulator of dendritic spine development in adult hippocampus (Tran et al., 2009), but embryonically serves as an endogenous chemorepellent, guiding septohippocampal fibers away from non-limbic regions of developing cortex (Pascual et al., 2005).
While synaptic mechanisms were strongly implicated by our results, it is noteworthy that there was no statistical evidence for enrichment of genes expressed in oligodendrocytes or astrocytes. While developmental disorders primarily affecting oligodendrocytes, such as metachromatic leukodystrophy, are marked by cognitive impairment (Faust et al. 2010), it is possible that individual variation in cognitive ability within the normal range is less directly under genetic control via white matter mechanisms. By contrast, strong evidence was provided for the involvement of genes expressed in the cerebellum. Converging evidence from functional imaging studies, lesion studies, structural connectivity, and evolutionary considerations strongly implicate a role for cerebellum in higher cognitive functions (Buckner, 2013), possibly through the mechanism of prediction and error-based learning (Sokolov et al. 2017).
By utilizing TWAS methodology, we were able to isolate expression effects of specific genes within some of our broad GWAS loci. For example, ACTR1A, which lies near the GWAS peak at chromosome 10q24, encodes a microtubular dynactin protein involved in retrograde axon transport (Moughamian et al., 2013); other genes at this locus were not significant in the TWAS analysis (although a role in cognition cannot be ruled out, given the limited sample size in the reference brain expression datasets in GTEx). However, most of the genes implicated by TWAS were clustered in a few “hot” genomic loci, which may represent topologically associated domains (TADs) under the control of a shared 3-dimensional chromatin structure (Gonzalez-Sandoval and Gasser, 2016). Whether effects on cognition are driven by all differentially expressed genes within such loci, or if specific effects can be disentangled through experimental means, remains to be determined.
The overlap of 23 genes from our results with known genes for Mendelian disorders characterized by intellectual disability has several implications. First, this statistically significant enrichment provides partial validation of our MTAG results. Second, genes with known mutations of large effect, when combined with our data demonstrating SNPs with smaller regulatory effects on the same phenotype (cognition), can be considered an “allelic series” (Plenge et al., 2013) – a natural set of experiments powerfully demonstrating directional information (in the form of a dose-response curve) regarding gene function. Such information can be leveraged for the identification of novel drug targets. Third, converging evidence across the Mendelian and GWAS lists can aid interpretation of specific pathways and molecular processes that are necessary to normal neuronal function, and vice versa. For example, two genes on both the Mendelian and GWAS lists (GMPPB and LARGE) are associated with dystroglycanopathies with mental retardation. This information provides context for the observation that DAG1, which encodes dystroglycan 1, is the strongest TWAS result in the hippocampus. DAG1 is necessary for GABAergic signaling in hippocampal interneurons (Früh et al., 2016), While dystroglycanopathies are most prominently characterized by muscular dystrophy and retinal abnormalities, it is possible that all of these genes play a role in hippocampal synapse formation that is relevant to normal cognitive ability.
As noted above, one of the most important aims of GWAS studies is the identification of novel drug targets, and it has been suggested that targets with supporting GWAS evidence may be twice as successful in clinical development compared to those without such evidence (Nelson et al. 2015). Our drug set enrichment analysis pointed to several potential nootropic mechanisms. Most notably, the strongest signal was for cinnarizine, a T-type calcium channel inhibitor typically prescribed for seasickness. In the present study, we discovered an association of cognition to CACNA1I, which encodes one component of the voltage-dependent T-Type Cav3.3 channel, and has been previously associated with schizophrenia (PGC2-SCZ, 2014). While cinnarizine has strong antihistamine activity and may be inappropriate for general cognitive enhancement, a novel agent targeting Cav3.3 has shown nootropic activity in preclinical models (Moriguchi et al., 2012). In addition to gene set results suggesting a potential role for calcium and potassium channel regulation, single-gene results also point towards a potential role for the metabotropic glutamate receptor encoded by GRM3. This gene is also implicated in schizophrenia (PGC-SCZ, 2014), and drugs targeting GRM3 have been suggested as a potential treatment (Lencz & Malhotra, 2015) however, a large-scale trial of one such agent was unsuccessful in treating psychotic symptoms (Downing et al. 2014). Based on the present results, future studies may seek to examine a role for such compounds in cognitive remediation. It is also noteworthy that the present study identified genome-wide significant evidence implicating three phosphodiesterase genes: PDE1C, PDE2A, and PDE4D. In particular, there is growing interest in PDE2A inhibitors as potential agents for cognitive enhancement (Trabanco et al. 2016), and evidence suggests that these agents may enhance synaptic plasticity via presynaptic modulation of cAMP hydrolysis (Fernández-Fernández et al. 2015). PDE4D inhibition is also under investigatation as a potential therapy for neurodegenerative disease (Ricciarelli et al. 2017).
It is important to emphasize that uncovering genetic variation underlying general cognitive ability in the healthy population does not have deterministic implications. As has been previously explicated in similar studies (e.g., Trampush et al. 2015), effect sizes for each allele are extremely small (R2<0.1% for even the strongest effects), and the combined effects genome-wide predict only a small proportion of the total variance in hold-out samples (Figure 3). Thus, results of the present study do not hold the potential for individual prediction or classification. Nevertheless, the results may still have substantial impact on our understanding of molecular mechanisms underlying cognitive ability.
Experimental Procedures
Subject Details
The cohorts included in the current study were described in detail in two prior reports on cognitive performance (Sniekers et al., 2017; Trampush et al., 2017) and one prior report on educational attainment (Okbay et al., 2016). Sample sizes for these three studies were N=78,308, N=35,298, and N = 328,917, respectively. For the present study, two cohorts reported in Trampush et al., 2017 were excluded, so that cohorts included will be independent from those reported in Sniekers et al., 2017: i) Minnesota Center for Twin and Family Research (MCTFR) and ii) Lothian Birth Cohort 1936 Study. As a result, sample sizes decreased from the originally reported N = 35,298 to N = 28,899. All phenotypes included were as reported originally in the respective publications. All subjects provided written, informed consent to procedures that were approved by local review boards for the institutions at which each cohort was collected. Further details are available in the supplementary materials to those three publications.
GWAS Quality Control
Markers reported in the prior COGENT study (Trampush et al., 2017) were updated to build 37 coordinates, but were originally imputed against the HRC reference panel (McCarthy et al., 2016) via the Sanger imputation server. To ensure that markers, allele frequencies, and alleles were aligned to the 1000 genomes phase 3 reference panel (The 1000 Genomes Project Consortium, 2015), the COGENT summary statistics (Trampush et al., 2017) were checked using the EasyQC pipeline (Winkler et al., 2014) which allows summary statistics to be aligned and checked against a reference panel of choice. We used the default 1000 genomes phase 3 reference panel (The 1000 Genomes Project Consortium, 2015), provided along with the EasyQC package. Markers were inspected for allele frequency outliers, presence of duplicated markers, and allele mismatches with the 1000 genomes reference panel. Quality control filters for INFO < .6 and N < 10000 were additionally implemented. After EasyQC quality control, 8,040,131 SNPs were available for analysis. Only 87 SNPs were excluded due to allele mismatches, 13,276 SNPs were excluded due to allele frequency mismatches from the 1000 genomes phase 3 reference panel, 283,163 were found to be duplicates and excluded, 104 SNPs were found on the HRC reference panel, but not on the 1000 genomes phase 3 reference panel, and 2,723,493 SNPs had sample sizes less 10000 individuals. None of the SNPs failed the INFO < .6 cutoff. The same set of SNPs was utilized for subsequent reduced sample meta-analysis without the overlapping LBC1936 and MCTFR cohorts in Trampush et al., 2017. As the other prior studies of cognitive performance (Sniekers et al., 2017) and education (Okbay et al., 2016) were imputed to the 1000 genomes phase 3 reference panel, summary statistics were used as provided (URL: https://ctg.cncr.nl/software/summary_statistics; https://www.thessgac.org/data).
GWAS Meta-Analysis
Fixed-effect meta-analysis was conducted between Sniekers et al., 2017 and independent cohorts reported in Trampush et al., 2017 using the METAL package (Willer et al., 2010). To ensure that results of the meta-analysis were contributed by both studies, markers present only in Sniekers et al., 2017 or Trampush et al., 2017 but not in both were excluded for further analysis. The number of available markers after QC filtering was 7,357,080. Because the GWAS of Sniekers et al. (2017) utilized the sample-size weighted method to perform meta-analysis across its own cohorts, and did not report variance terms, our meta-analysis was conducted using the sample-size weighted method.
Multi-Trait Analysis for GWAS (MTAG)
To further enrich genetic signals, we employed a newly developed methodology that integrates LD-score regression and meta-analysis techniques across related traits: MTAG (Turley et al., 2017). MTAG (v0.9.0) was applied to the METAL results described immediately above, combined with summary statistics from the recent, large-scale education GWAS (Okbay et al., 2016). MTAG analysis allows the boosting of genetic signals across related traits, and has been found to be effective in resolving unknown sample overlaps, generating trait-specific effect estimates weighted by bivariate genetic correlation. The MTAG QC pipeline aligned all alleles across both sets of summary statistics, and ensured that SNPs were present across all datasets. SNPs that were not present in either dataset were removed. The final SNP count for MTAG was 7,333,576. The MTAG methodology proceeds by i) estimating the variance-covariance matrix of the GWAS estimation error, by using a series of LD score regressions, of which, under the known properties of LD score regression captures relevant sources of estimation error, incorporating population stratification, unknown sample overlap and cryptic relatedness ii) estimating the variance-covariance of SNP effects using the maximum likelihood procedure reported in (Turley et al., 2017) and iii) computes the MTAG estimator for each SNP and each trait. Summary statistics consisting of SNP, CHR, BP, per SNP sample size, BETA and SE for each trait were entered to the MTAG python command line. The resulting effect estimates and p-values are interpreted in the same as single-trait GWAS, which allows standard downstream follow-up analysis on the summary statistics. The python code for MTAG is available at https://github.com/omeed-maghzian/mtag.
Functional Mapping and Annotation for GWAS
GWAS summary statistics from the METAL meta-analysis and MTAG analysis were separately entered into the Functional Mapping and Annotation (FUMA) pipeline (Watanabe et al., 2017). The FUMA pipeline enables fast prioritization of genomic variants and genes, and permits interactive visualization of genomic results with respect to state-of-art bioinformatics resources. Manhattan and QQ plots are produced, and MAGMA gene-based analysis is performed, accounting for gene size and LD structure32. FUMA was also utilized to perform competitive gene-set analyses for GO cell compartment and biological process categories using the Molecular Signature Database (MsigDB 5.2). A separate competitive gene-set analysis was also conducted for the drug-based pathways previously described by Gaspar & Breen (2017). The pipeline also generates aggregated statistics for independent loci, lead SNPs, tagged genes, and supplementary plots – including SNP and locus annotations. Default clumping parameters are GWAS p-value < 5E-08; r2 threshold to define LD structure of independent SNPs > 0.1; maximum P-value cutoff < 0.05; population for clumping = EUR; minor allele frequency filter > 0.01; maximum distance between LD blocks to merge into a single locus: 250kb. Follow-up queries were then made for independent loci of the cognitive performance meta-analysis as well as the MTAG results and compared against summary statistics for the prior cognitive and education GWAS. For purposes of comparison, loci in which the lead SNPs were within 500kb of each other were considered overlapping.
We compared the list of genes resulting from the MTAG analysis (including all genes within GWS SNP loci, as well as GWS genes identified with MAGMA) with a list of 621 genes known to cause autosomal dominant or autosomal recessive Mendelian disorders featuring intellectual disability; this list is primarily derived from a recent comprehensive review (Vissers et al., 2016), supplemented by a subsequent large-scale study of consanguineous multiplex families (Harripaul et al., 2017). A total of 193 autosomal dominant genes were identified, and a total of 413 autosomal recessive genes were identified. Fifteen genes were annotated as causing both autosomal dominant and autosomal recessive disorders with intellectual disability. Statistical significance was determined by probabilities derived according to the hypergeometric distribution. For this purpose, the total pool of autosomal genes was set to 19,011 (per Gencode).
Polygenic Risk Prediction for Independent Datasets
To validate that the genetic architecture elucidated via the MTAG methodology, we attempted to predict the phenotypic variance of general cognitive function in two of the independent COGENT cohorts (ASPIS and GCAP). MTAG analysis was conducted as above, but removing the COGENT cohorts. Polygenic score prediction across multiple thresholds of PT was conducted using PRSice (Euesden et al., 2015). To compare the effectiveness of MTAG, we also conducted polygenic risk prediction using IQ only and Education only summary statistics. Finally, R2 across SNP thresholds are compared to obtain the degree of improvement in terms of the ratio of MTAG PRS R2 values versus those of IQ or Education PRS R2.
Stratified LD regression: Cell type Expression and Epigenomics
Functional characterization of GWAS summary statistics was carried out via stratified LD regression to investigate if heritability of cognitive performance is enriched in specific tissue or cell types. Summary statistics were first subjected to baseline partitioned heritability and thereafter passed through a cell type-specific functional characterization pipeline (Finucane et al., 2017). Cell type characterization includes the DEPICT tissue expression database, GTEX tissue expression, IMMGEN immune cell types, CAHOY brain level cell types, and the ROADMAP cell epigenomic marks.
Transcriptome Wide Analysis and Brain Expression lookups
Transcriptome wide analysis was carried out via MetaXcan (Barbeira et al., 2016), which allows for GTEx brain expression data to be integrated with GWAS summary statistics. MetaXcan computes downstream phenotypic associations of genetic regulation of molecular traits, using elastic, adjustment for model uncertainty and colocalization of of GWAS and eQTL signals (Barbeira et al., 2016). GTEx Version 6, brain tissue expression profiles/sample sizes include the Anterior Cingulate Cortex (N=72); Caudate – Basal Ganglia (N=100); Cerebellar Hemisphere (N=89); Cerebellum (N=103); Cortex (N=96); Frontal Cortex (N=92); Hippocampus (N=81); Hypothalamus (N=81); Nucleus Accumbens (N=93); and Putamen (N=82).
Bayesian Fine-mapping Analysis and functional annotations
To identify potential causal variants in each of the independent loci, CAVIAR-BF is implemented to a region +/− 50KB of a lead SNP identified in the MTAG analysis. We followed similar procedures setting prior effect distribution σa to 0.1 in the model, which was recommended for GWAS studies (Chen et al., 2015; https://bitbucket.org/Wenan/caviarbf). The prior probability of being causal for each SNP is set to 1/m, where m is the number of SNPs. Bayes factor was calculated for three model sets for independent loci, which modelled for 1,2, and up to 3 causal SNPs within each independent regions. After which a model search algorithm searches and identifies the putative causal SNPs. These SNPs were then annotated using the Ensembl Variant Effect Predictor (McLaren et al., 2016). The analysis was repeated for extended regions taking into account the length of the independent loci identified by earlier FUMA procedures modelling for either 1 or 2 causal SNPs. SNPs identified by the two stage CAVIARBF analysis were then examined for potential gene expression in the BrainEAC (Ramasamy et al., 2014) and CommonMind (Hauberg et al., 2017) databases. BrainEAC top SNP lookups were for the following tissue expression: aveALL: All area combined; CRBL: cerebellum; FCTX: frontal cortex; HIPP: Hippocampus; MEDU: medulla; OCTX: occipital cortex; PUTM: putamen; SNIG: substantia nigra; TCTX: temporal cortex; THAL: thalamus; and WHMT: white matter across N=134 individuals. Finally, the prefrontal cortex lookup was included as part of the CommonMind consortium brain expression profile in n=467 genetically-inferred Caucasian samples.
Linkage Disequilibrium Score Regression
LD score regression allows genetic correlations to be computed across traits (Bulik-Sullivan et al., 2015a, 2015b), which allows further insights to be drawn from understanding the degree to which genetic architecture are shared across traits. To further examine potential traits that overlap with the cognitive architecture from the cognition meta-analysis results and MTAG results, LD score regression was conducted via the LD-hub pipeline, a centralized trait database (Zheng et al. 2017). LD-score regression was carried out across 89 traits in 15 broad phenotypic categories: 1) aging, 2) anthropometric, 3) autoimmune, 4) brain volume, 5) cardiometabolic, 6) education, 7) glycemic, 8) lipids, 9) lung function, 10) neurological, 11) personality, 12) psychiatric, 13) reproductive behavior, 14) sleep, and 15) smoking behavior. Very recent reported GWAS summary statistics for attention deficit hyperactivity disorder (ADHD, Demontis et al., 2017) and intracranial volume (ICV, Adams et al., 2016) were included as additional phenotypes. For comparison, we also present LD-score regression results for the educational attainment GWAS of Okbay et al. (2016); it should be noted that only 14 phenotypes were examined for genetic correlation in that publication. It should be noted that the MHC region was redacted from all datasets prior to LD score regression analysis, as per standard protocol at LD-Hub.
Supplementary Material
Acknowledgments
This work has been supported by grants from the National Institutes of Health (R01 MH079800 and P50 MH080173 to AKM; R01 MH080912 to DCG; K23 MH077807 to KEB; K01 MH085812 to MCK). Data collection for the TOP cohort was supported by the Research Council of Norway, South-East Norway Health Authority, and KG Jebsen Foundation. The NCNG study was supported by Research Council of Norway Grants 154313/V50 and 177458/V50. The NCNG GWAS was financed by grants from the Bergen Research Foundation, the University of Bergen, the Research Council of Norway (FUGE, Psykisk Helse), Helse Vest RHF and Dr Einar Martens Fund. The Helsinki Birth Cohort Study has been supported by grants from the Academy of Finland, the Finnish Diabetes Research Society, Folkhälsan Research Foundation, Novo Nordisk Foundation, Finska Läkaresällskapet, Signe and Ane Gyllenberg Foundation, University of Helsinki, Ministry of Education, Ahokas Foundation, Emil Aaltonen Foundation. For the LBC1936 cohort, phenotype collection was supported by The Disconnected Mind project. Genotyping was funded by the UK Biotechnology and Biological Sciences Research Council (BBSRC grant No. BB/F019394/1). The work was undertaken by The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross council Lifelong Health and Wellbeing Initiative, which is funded by the Medical Research Council and the Biotechnology and Biological Sciences Research Council (MR/K026992/1). The CAMH work was supported by the CAMH Foundation and the Canadian Institutes of Health Research. The Duke Cognition Cohort (DCC) acknowledges K. Linney, J.M. McEvoy, P. Hunt, V. Dixon, T. Pennuto, K. Cornett, D. Swilling, L. Phillips, M. Silver, J. Covington, N. Walley, J. Dawson, H. Onabanjo, P. Nicoletti, A. Wagoner, J. Elmore, L. Bevan, J. Hunkin and R. Wilson for recruitment and testing of subjects. DCC also acknowledges the Ellison Medical Foundation New Scholar award AG-NS-0441-08 for partial funding of this study as well as the National Institute of Mental Health of the National Institutes of Health under award number K01MH098126. The UCLA Consortium for Neuropsychiatric Phenomics (CNP) study acknowledges the following sources of funding from the NIH: Grants UL1DE019580 and PL1MH083271 (RMB), RL1MH083269 (TDC), RL1DA024853 (EL) and PL1NS062410. The ASPIS study was supported by National Institute of Mental Health research grants R01MH085018 and R01MH092515 to Dr. Dimitrios Avramopoulos. Support for the Duke Neurogenetics Study was provided the National Institutes of Health (R01 DA033369 and R01 AG049789 to ARH) and by a National Science Foundation Graduate Research Fellowship to MAS. Recruitment, genotyping and analysis of the TCD healthy control samples were supported by Science Foundation Ireland (grants 12/IP/1670, 12/IP/1359 and 08/IN.1/B1916).
Data access for several cohorts used in this study was provided by the National Center for Biotechnology Information (NCBI) database of Genotypes and Phenotypes (dbGaP). dbGaP accession numbers for these cohorts were:
Cardiovascular Health Study (CHS): phs000287.v4.p1, phs000377.v5.p1, and phs000226.v3.p1 Framingham Heart Study (FHS): phs000007.v23.p8 and phs000342.v11.p8
Multi-Site Collaborative Study for Genotype-Phenotype Associations in Alzheimer’s Disease (GENADA): phs000219.v1.p1
Long Life Family Study (LLFS): phs000397.v1.p1
Genetics of Late Onset Alzheimer’s Disease Study (LOAD): phs000168.v1.p1 Minnesota Center for Twin and Family Research (MCTFR): phs000620.v1.p1 Philadelphia Neurodevelopmental Cohort (PNC): phs000607.v1.p1 The acknowledgment statements for these cohorts are found below:
Framingham Heart Study: The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (Contract No. N01-HC-25195 and HHSN268201500001I). This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or NHLBI. Funding for SHARe Affymetrix genotyping was provided by NHLBI Contract N02-HL-64278. SHARe Illumina genotyping was provided under an agreement between Illumina and Boston University.
Cardiovascular Health Study: This research was supported by contracts HHSN268201200036C, HHSN268200800007C, N01-HC-85079, N01-HC-85080, N01-HC-85081, N01-HC-85082, N01-HC-85083, N01-HC-85084, N01-HC-85085, N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, and N01-HC-85239; grant numbers U01 HL080295 and U01 HL130014 from the National Heart, Lung, and Blood Institute, and R01 AG-023629 from the National Institute on Aging, with additional contribution from the National Institute of Neurological Disorders and Stroke. A full list of principal CHS investigators and institutions can be found at https://chs-nhlbi.org/pi. This manuscript was not prepared in collaboration with CHS investigators and does not necessarily reflect the opinions or views of CHS, or the NHLBI. Support for the genotyping through the CARe Study was provided by NHLBI Contract N01-HC-65226. Support for the Cardiovascular Health Study Whole Genome Study was provided by NHLBI grant HL087652. Additional support for infrastructure was provided by HL105756 and additional genotyping among the African-American cohort was supported in part by HL085251, DNA handling and genotyping at Cedars-Sinai Medical Center was supported in part by National Center for Research Resources grant UL1RR033176, now at the National Center for Advancing Translational Technologies CTSI grant UL1TR000124; in addition to the National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
Multi-Site Collaborative Study for Genotype-Phenotype Associations in Alzheimer’s Disease: The genotypic and associated phenotypic data used in the study were provided by the GlaxoSmithKline, R&D Limited. Details on data acquisition have been published previously in: Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, Hosford D, Barnes MR, Briley JD, Borrie M, Coletta N, Delisle R, Dhalla D, Ehm MG, Feldman HH, Fornazzari L, Gauthier S, Goodgame N, Guzman D, Hammond S, Hollingworth P, Hsiung GY, Johnson J, Kelly DD, Keren R, Kertesz A, King KS, Lovestone S, Loy-English I, Matthews PM, Owen MJ, Plumpton M, Pryse-Phillips W, Prinjha RK, Richardson JC, Saunders A, Slater AJ, St George-Hyslop PH, Stinnett SW, Swartz JE, Taylor RL, Wherrett J, Williams J, Yarnall DP, Gibson RA, Irizarry MC, Middleton LT, Roses AD. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol., Jan;65(1):45–53, 2008 (PMID: 17998437). Filippini N, Rao A, Wetten S, Gibson RA, Borrie M, Guzman D, Kertesz A, Loy-English I, Williams J, Nichols T, Whitcher B, Matthews PM. Anatomically-distinct genetic associations of APOE epsilon4 allele load with regional cortical atrophy in Alzheimer’s disease. Neuroimage, Feb 1;44(3):724-8, 2009. (PMID: 19013250).
Genetics of Late Onset Alzheimer’s Disease Study: Funding support for the “Genetic Consortium for Late Onset Alzheimer’s Disease” was provided through the Division of Neuroscience, NIA. The Genetic Consortium for Late Onset Alzheimer’s Disease includes a genome-wide association study funded as part of the Division of Neuroscience, NIA. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by Genetic Consortium for Late Onset Alzheimer’s Disease. A list of contributing investigators is available at https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000168.v1.p1 Long Life Family Study: Funding support for the Long Life Family Study was provided by the Division of Geriatrics and Clinical Gerontology, National Institute on Aging. The Long Life Family Study includes GWAS analyses for factors that contribute to long and healthy life. Assistance with phenotype harmonization and genotype cleaning as well as with general study coordination, was provided by the Division of Geriatrics and Clinical Gerontology, National Institute on Aging. Support for the collection of datasets and samples were provided by Multicenter Cooperative Agreement support by the Division of Geriatrics and Clinical Gerontology, National Institute on Aging (UO1AG023746; UO1023755; UO1023749; UO1023744; UO1023712). Funding support for the genotyping which was performed at the Johns Hopkins University Center for Inherited Disease Research was provided by the National Institute on Aging, National Institutes of Health.
Minnesota Center for Twin and Family Research: This project was led by William G. Iacono, PhD. And Matthew K. McGue, PhD (Co-Principal Investigators) at the University of Minnesota, Minneapolis, MN, USA. Co-investigators from the same institution included: Irene J. Elkins, Margaret A. Keyes, Lisa N. Legrand, Stephen M. Malone, William S. Oetting, Michael B. Miller, and Saonli Basu. Funding support for this project was provided through NIDA (U01 DA 024417). Other support for sample ascertainment and data collection came from several grants: R37 DA 05147, R01 AA 09367, R01 AA 11886, R01 DA 13240, R01 MH 66140.
Philadelphia Neurodevelopmental Cohort: Support for the collection of the data sets was provided by grant RC2MH089983 awarded to Raquel Gur, MD, and RC2MH089924 awarded to Hakon Hakonarson, MD, PhD. All subjects were recruited through the Center for Applied Genomics at The Children’s Hospital in Philadelphia.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
T.L. designed the study and supervised the data analyses. M.L. performed the primary data analyses, and J.T.W., J.Y., and E.K provided additional statistical input. A.K.M., D.C.G., I.J.D., K.E.B., and G.D. provided the initial conceptual framework for the COGENT consortium. M.L. and T.L. drafted the manuscript. All other authors were involved in ascertainment, assessment, and analysis of individual cohorts, provided conceptual input to study design, and critically reviewed the manuscript.
Web Resources
METAL http://genome.sph.umich.edu/wiki/METAL_Documentation
MTAG https://github.com/omeed-maghzian/mtag
LD-HUB http://ldsc.broadinstitute.org/
LDSC https://github.com/bulik/ldsc
METAXCAN https://github.com/hakyimlab/MetaXcan
PRSice http://prsice.info/
BRAINEAC http://www.braineac.org/
CommonMind https://www.synapse.org//#!Synapse:syn2759792/wiki/69613
Conflict of Interest
The authors declare no conflict of interest.
References
- Adams HHH, Hibar DP, Chouraki V, Stein JL, Nyquist PA, Rentería ME, Trompet S, Arias-Vasquez A, Seshadri S, Desrivières S, et al. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat. Neurosci. 2016;19:1569–1582. doi: 10.1038/nn.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeira A, Shah KP, Torres JM, Wheeler HE, Torstenson ES, Edwards T, Garcia T, Bell GI, Nicolae D, Cox NJ, et al. MetaXcan: Summary Statistics Based Gene-Level Association Method Infers Accurate PrediXcan Results. BioRxiv 2016 [Google Scholar]
- Belsky DW, Moffitt TE, Corcoran DL, Domingue B, Harrington H, Hogan S, Houts R, Ramrakha S, Sugden K, Williams BS, et al. The Genetics of Success: How Single-Nucleotide Polymorphisms Associated With Educational Attainment Relate to Life-Course Development. Psychol. Sci. 2016;27:957–972. doi: 10.1177/0956797616643070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyamin B, Pourcain Bs, Davis OS, Davies G, Hansell NK, Brion M-J, Kirkpatrick RM, Cents RaM, Franić S, Miller MB, et al. Childhood intelligence is heritable, highly polygenic and associated with FNBP1L. Mol. Psychiatry. 2013 doi: 10.1038/mp.2012.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL. The cerebellum and cognitive function: 25 years of insight from anatomy and neuroimaging. Neuron. 2013;80:807–15. doi: 10.1016/j.neuron.2013.10.044. [DOI] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, ReproGen Consortium; Psychiatric Genomics Consortium; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3. Duncan L, Perry JR, Patterson N, Robinson EB, Daly MJ, Price AL, Neale BM. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015a;47:1236–41. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium. Patterson N, Daly MJ, Price AL, Neale BM. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015b;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick KE, Goldberg TE, Cornblatt BA, Keefe RS, Gopin CB, DeRosse P, Braga RJ, Malhotra AK. The MATRICS Consensus Cognitive Battery in Patients with Bipolar I Disorder. Neuropsychopharmacology. 2011;36:1587–1592. doi: 10.1038/npp.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. Off. J. Soc. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai G, Goffinet AM, Tissir F. Celsr3 and Fzd3 in axon guidance. Int. J. Biochem. Cell Biol. 2015;64:11–14. doi: 10.1016/j.biocel.2015.03.013. [DOI] [PubMed] [Google Scholar]
- Chen W, Larrabee BR, Ovsyannikova IG, Kennedy RB, Haralambieva IH, Poland GA, Schaid DJ. Fine Mapping Causal Variants with an Approximate Bayesian Method Using Marginal Test Statistics. Genetics. 2015;200:719–736. doi: 10.1534/genetics.115.176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, et al. A copy number variation morbidity map of developmental delay. Nat. Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, Ke X, Le Hellard S, Christoforou A, Luciano M, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol. Psychiatry. 2011a;16:996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, Ke X, Le Hellard S, Christoforou A, Luciano M, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol. Psychiatry. 2011b;16:996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Armstrong N, Bis JC, Bressler J, Chouraki V, Giddaluru S, Hofer E, Ibrahim-Verbaas CA, Kirin M, Lahti J, et al. Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N=53 949) Mol. Psychiatry. 2015;20:183–192. doi: 10.1038/mp.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Marioni RE, Liewald DC, Hill WD, Hagenaars SP, Harris SE, Ritchie SJ, Luciano M, Fawns-Ritchie C, Lyall D. Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N= 112 151) Mol. Psychiatry. 2016 doi: 10.1038/mp.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deary IJ, Johnson W, Houlihan LM. Genetic foundations of human intelligence. Hum. Genet. 2009;126:215–232. doi: 10.1007/s00439-009-0655-4. [DOI] [PubMed] [Google Scholar]
- Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, Belliveau R, Bybjerg-Grauholm J, Bækved-Hansen M, Cerrato F, et al. Discovery Of The First Genome-Wide Significant Risk Loci For ADHD. BioRxiv 2017 [Google Scholar]
- Downing AM, Kinon BJ, Millen BA, Zhang L, Liu L, Morozova MA, Brenner R, Rayle TJ, Nisenbaum L, Zhao F, Gomez JC. A Double-Blind, Placebo-Controlled Comparator Study of LY2140023 monohydrate in patients with schizophrenia. BMC Psychiatry. 2014;14:351. doi: 10.1186/s12888-014-0351-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euesden J, Lewis CM, O’Reilly PF. PRSice: Polygenic Risk Score software. Bioinforma. Oxf. Engl. 2015;31:1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust PL, Kaye EM, Powers JM. Myelin lesions associated with lysosomal and peroxisomal disorders. Expert Rev Neurother. 2010;10:1449–66. doi: 10.1586/ern.10.127. [DOI] [PubMed] [Google Scholar]
- Fernández-Fernández D, Rosenbrock H, Kroker KS. Inhibition of PDE2A, but not PDE9A, modulates presynaptic short-term plasticity measured by paired-pulse facilitation in the CA1 region of the hippocampus. Synapse. 2015;69:484–96. doi: 10.1002/syn.21840. [DOI] [PubMed] [Google Scholar]
- Ferreri F, Lapp LK, Peretti C-S. Current research on cognitive aspects of anxiety disorders. Curr. Opin. Psychiatry. 2011;24:49–54. doi: 10.1097/YCO.0b013e32833f5585. [DOI] [PubMed] [Google Scholar]
- Finucane H, Reshef Y, Anttila V, Slowikowski K, Gusev A, Byrnes A, Gazal S, Loh P-R, Genovese G, Saunders A, et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. BioRxiv. 2017 doi: 10.1038/s41588-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Früh S, Romanos J, Panzanelli P, Bürgisser D, Tyagarajan SK, Campbell KP, Santello M, Fritschy J-M. Neuronal Dystroglycan Is Necessary for Formation and Maintenance of Functional CCK-Positive Basket Cell Terminals on Pyramidal Cells. J. Neurosci. Off. J. Soc. Neurosci. 2016;36:10296–10313. doi: 10.1523/JNEUROSCI.1823-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar HA, Breen G. Pathways analyses of schizophrenia GWAS focusing on known and novel drug targets. Scientific Reports, Sci Rep. 2017;7:12460. doi: 10.1038/s41598-017-12325-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Sandoval A, Gasser SM. On TADs and LADs: Spatial Control Over Gene Expression. Trends Genet. 2016;32:485–495. doi: 10.1016/j.tig.2016.05.004. [DOI] [PubMed] [Google Scholar]
- Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, Sulem P, Thorlacius S, Gylfason A, Steinberg S, et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- Guha S, Rees E, Darvasi A, Ivanov D, Ikeda M, Bergen SE, Magnusson PK, Cormican P, Morris D, Gill M, et al. Implication of a rare deletion at distal 16p11.2 in schizophrenia. JAMA Psychiatry. 2013;70:253–260. doi: 10.1001/2013.jamapsychiatry.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenaars SP, Harris SE, Davies G, Hill WD, Liewald DCM, Ritchie SJ, Marioni RE, Fawns-Ritchie C, Cullen B, Malik R, et al. Shared genetic aetiology between cognitive functions and physical and mental health in UK Biobank (N=112 151) and 24 GWAS consortia. Mol. Psychiatry. 2016;21:1624–1632. doi: 10.1038/mp.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harripaul R, Vasli N, Mikhailov A, Rafiq MA, Mittal K, Windpassinger C, Sheikh TI, Noor A, Mahmood H, Downey S, et al. Mapping autosomal recessive intellectual disability: combined microarray and exome sequencing identifies 26 novel candidate genes in 192 consanguineous families. Mol. Psychiatry. 2017 doi: 10.1038/mp.2017.60. [DOI] [PubMed] [Google Scholar]
- Hauberg ME, Zhang W, Giambartolomei C, Franzén O, Morris DL, Vyse TJ, Ruusalepp A, Fromer M, Sieberts SK, Johnson JS, et al. Large-Scale Identification of Common Trait and Disease Variants Affecting Gene Expression. Am. J. Hum. Genet. 2017;100:885–894. doi: 10.1016/j.ajhg.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W, Nijenhuis te J, Bouchard TJ., Jr Still just 1 g: Consistent results from five test batteries. Intelligence. 2008;36:81–95. [Google Scholar]
- Johnson W, Deary IJ, Silventoinen K, Tynelius P, Rasmussen F. Family background buys an education in Minnesota but not in Sweden. Psychol. Sci. 2010;21:1266–1273. doi: 10.1177/0956797610379233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefe RS, Harvey PD. Cognitive impairment in schizophrenia. In: Geyer MA, Gross G, editors. Handbook of Experimental Pharmacology. Berlin, Heidelberg: Springer; 2012. pp. 11–37. [DOI] [PubMed] [Google Scholar]
- de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Comput. Biol. 2015;11 doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencz T, Knowles E, Davies G, Guha S, Liewald DC, Starr JM, Djurovic S, Melle I, Sundet K, Christoforou A, et al. Molecular genetic evidence for overlap between general cognitive ability and risk for schizophrenia: a report from the Cognitive Genomics consorTium (COGENT) Mol. Psychiatry. 2014;19:168–174. doi: 10.1038/mp.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencz T, Malhotra AK. Targeting the schizophrenia genome: a fast track strategy from GWAS to clinic. Mol Psychiatry. 2015;20:820–6. doi: 10.1038/mp.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriguchi S, Shioda N, Yamamoto Y, Tagashira H, Fukunaga K. The T-type voltage-gated calcium channel as a molecular target of the novel cognitive enhancer ST101: enhancement of long-term potentiation and CaMKII autophosphorylation in rat cortical slices. J. Neurochem. 2012;121:44–53. doi: 10.1111/j.1471-4159.2012.07667.x. [DOI] [PubMed] [Google Scholar]
- Moughamian AJ, Osborn GE, Lazarus JE, Maday S, Holzbaur ELF. Ordered recruitment of dynactin to the microtubule plus-end is required for efficient initiation of retrograde axonal transport. J. Neurosci. Off. J. Soc. Neurosci. 2013;33:13190–13203. doi: 10.1523/JNEUROSCI.0935-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, Floratos A, Sham PC, Li MJ, Wang J, Cardon LR, Whittaker JC, Sanseau P. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–60. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA, Turley P, Chen G-B, Emilsson V, Meddens SFW. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–542. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual M, Pozas E, Soriano E. Role of class 3 semaphorins in the development and maturation of the septohippocampal pathway. Hippocampus. 2005;15:184–202. doi: 10.1002/hipo.20040. [DOI] [PubMed] [Google Scholar]
- PGC2-SCZ. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov. 2013;12:581–594. doi: 10.1038/nrd4051. [DOI] [PubMed] [Google Scholar]
- Plomin R, Deary IJ. Genetics and intelligence differences: five special findings. Mol. Psychiatry. 2015;20:98–108. doi: 10.1038/mp.2014.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, De T, UK Brain Expression Consortium, North American Brain Expression Consortium. Coin L, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17:1418–1428. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciarelli R, Brullo C, Prickaerts J, Arancio O, Villa C, Rebosio C, Calcagno E, Balbi M, van Hagen BT, Argyrousi EK, Zhang H, Pronzato MA, Bruno O, Fedele E. Memory-enhancing effects of GEBR-32a, a new PDE4D inhibitor holding promise for the treatment of Alzheimer’s disease. Sci Rep. 2017;7:46320. doi: 10.1038/srep46320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld CA, Medland SE, Derringer J, Yang J, Esko T, Martin NW, Westra H-J, Shakhbazov K, Abdellaoui A, Agrawal A, et al. GWAS of 126,559 Individuals Identifies Genetic Variants Associated with Educational Attainment. Science. 2013;340:1467–1471. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld CA, Esko T, Davies G, Pers TH, Turley P, Benyamin B, Chabris CF, Emilsson V, Johnson AD, Lee JJ, et al. Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proc. Natl. Acad. Sci. 2014;111:13790–13794. doi: 10.1073/pnas.1404623111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeland OB, Frei O, Kauppi K, Hill WD, Li W, Wang Y, Krull F, Bettella F, Eriksen JA, Witoelar A, et al. Identification of Genetic Loci Jointly Influencing Schizophrenia Risk and the Cognitive Traits of Verbal-Numerical Reasoning, Reaction Time, and General Cognitive Function. JAMA Psychiatry. 2017 doi: 10.1001/jamapsychiatry.2017.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniekers S, Stringer S, Watanabe K, Jansen PR, Coleman JRI, Krapohl E, Taskesen E, Hammerschlag AR, Okbay A, Zabaneh D, et al. Genome-wide association meta-analysis of 78,308 individuals identifies new loci and genes influencing human intelligence. Nat. Genet. 2017;49:1107–1112. doi: 10.1038/ng.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder HR. Major depressive disorder is associated with broad impairments on neuropsychological measures of executive function: a meta-analysis and review. Psychol. Bull. 2013;139:81–132. doi: 10.1037/a0028727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov AA, Miall RC, Ivry RB. The Cerebellum: Adaptive Prediction for Movement and Cognition. Trends Cogn Sci. 2017;21:313–332. doi: 10.1016/j.tics.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiakouli E, Martin J, Hamshere ML, Heron J, St Pourcain B, Timpson NJ, Thapar A, Davey Smith G. Association between polygenic risk scores for attention-deficit hyperactivity disorder and educational and cognitive outcomes in the general population. Int. J. Epidemiol. 2017;46:421–428. doi: 10.1093/ije/dyw216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakar S, Wang L, Yu T, Ye M, Onishi K, Scott J, Qi J, Fernandes C, Han X, Yates JR, et al. Evidence for opposing roles of Celsr3 and Vangl2 in glutamatergic synapse formation. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E610–E618. doi: 10.1073/pnas.1612062114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabanco AA, Buijnsters P, Rombouts FJ. Towards selective phosphodiesterase 2A (PDE2A) inhibitors: a patent review (2010 - present) Expert Opin Ther Pat. 2016;26:933–46. doi: 10.1080/13543776.2016.1203902. [DOI] [PubMed] [Google Scholar]
- Trampush JW, Yang MLZ, Yu J, Knowles E, Davies G, Liewald DC, Starr JM, Djurovic S, Melle I, Sundet K, et al. GWAS meta-analysis reveals novel loci and genetic correlates for general cognitive function: a report from the COGENT consortium. Mol. Psychiatry. 2017;22:336–345. doi: 10.1038/mp.2016.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TS, Rubio ME, Clem RL, Johnson D, Case L, Tessier-Lavigne M, Huganir RL, Ginty DD, Kolodkin AL. Secreted semaphorins control spine distribution and morphogenesis in the postnatal CNS. Nature. 2009;462:1065–1069. doi: 10.1038/nature08628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turley P, Walters RK, Maghzian O, Okbay A, Lee JJ, Fontana MA, Nguyen-Viet TA, Furlotte NA, Team, 23andMe Research Consortium SSGAC et al. MTAG: Multi-Trait Analysis of GWAS. Nature Genetics. 2017 in press. [Google Scholar]
- Vissers LELM, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat. Rev. Genet. 2016;17:9–18. doi: 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Taskesen E, van Bochoven A, Posthuma D. FUMA: Functional mapping and annotation of genetic associations. BioRxiv. 2017 doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, Freathy RM, Perry JRB, Stevens S, Hall AS, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinforma. Oxf. Engl. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler TW, Day FR, Croteau-Chonka DC, Wood AR, Locke AE, Mägi R, Ferreira T, Fall T, Graff M, Justice AE, et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 2014;9:1192–1212. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, Hemani G, Tansey K, Laurin C, Pourcain BS, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.