Abstract
miR408 is highly conserved among different plant species and targets transcripts encoding copper-binding proteins. The function of miR408 in reproductive development remains largely unclear despite it being known to play important roles during vegetative development in Arabidopsis. Here, we show that transgenic Arabidopsis plants overexpressing MIR408 have altered morphology including significantly increased leaf area, petiole length, plant height, flower size, and silique length, resulting in enhanced biomass and seed yield. The increase in plant size was primarily due to cell expansion rather than cell proliferation, and was consistent with higher levels of myosin gene expression and gibberellic acid (GA) measured in transgenic plants. In addition, photosynthetic rate was significantly increased in the MIR408-overexpressing plants, as manifested by higher levels of chloroplastic copper content and plastocyanin (PC) expression. In contrast, overexpression of miR408-regulated targets, Plantacyanin and Laccase 13, resulted in reduced biomass production and seed yield. RNA-sequencing revealed that genes involved in primary metabolism and stress response were preferentially enriched in the genes upregulated in MIR408-overexpressing plants. These results indicate that miR408 plays an important role in regulating biomass and seed yield and that MIR408 may be a potential candidate gene involved in the domestication of agricultural crops.
Keywords: miR408, biomass, seed yield, photosynthesis, copper
Introduction
miRNAs are a class of fundamental, sequence-specific regulatory small RNA molecules that repress target gene expression post-transcriptionally. In plants, these 20–24 nucleotide-long RNA species are processed from stem-loop structured precursors by a Dicer-like enzyme and are integrated into silencing complexes, where, in general, they function as gene repressors by directing cleavage of complementary mRNA transcripts (Llave et al., 2002; Brodersen et al., 2008; Voinnet, 2009; Rogers and Chen, 2013). Additionally, miRNAs can also mediate DNA methylation and histone modification to affect gene expression (Schramke and Allshire, 2004; Wu et al., 2010; Khraiwesh et al., 2012).
As regulatory molecules, miRNAs share a common regulatory logic with transcription factors (Chen and Rajewsky, 2007; Hobert, 2008). For instance, miRNAs recognize their target mRNAs based on short complementary sequence. A single miRNA can potentially regulate the expression of multiple target genes, and multiple miRNAs may act synergistically to regulate the same genes. This, together with the large number of miRNA genes, indicates that miRNAs may have a substantial impact on the transcriptome post-transcriptionally. Accumulating evidence has shown that miRNAs are involved in the control of almost all biological and metabolic processes in plants. A number of these processes are potentially responsible for some of the most challenging plant traits in agricultural production, such as regulation of plant development and plant architecture, and responses to environmental stresses and defense (Jones-Rhoades et al., 2006; Garcia and Frampton, 2008; Rubio-Somoza and Weigel, 2011; Comai and Zhang, 2012; Khraiwesh et al., 2012; Sun, 2012). Thus, advances in functional identification of miRNAs and miRNA networks will open up new avenues for understanding the genetic mechanisms underlying these complex traits (Sun, 2012). Moreover, it is believed that miRNAs themselves can be regarded as a reservoir of resourceful genes for modifying these challenging traits in agricultural production (Sun, 2012; Zhou et al., 2013).
MiR408 is one of the most conserved miRNA families and has, to date, been annotated in more than 30 plant species (Axtell and Bowman, 2008; Kozomara and Griffiths-Jones, 2011), implying that its function is fundamental to plants. It has been reported in various plant species that miR408 is differentially expressed in response to a variety of environmental cues, including copper, light, mechanical stress, dehydration, cold, and reactive oxygen species (Lu et al., 2005; Yamasaki et al., 2007; Abdel-Ghany and Pilon, 2008; Kantar et al., 2010; Li et al., 2010; Trindade et al., 2010; Mutum et al., 2013; Zhang and Li, 2013; Zhang et al., 2014; Ma et al., 2015). Moreover, Feng et al. (2013) reported that tae-miR408 was positively correlated with the resistance of host plants to abiotic stresses and stripe rust by regulating one of its target genes, chemocyanin-like protein gene (TaCLP1), in wheat. Thus, these observations indicate that miR408 is a key regulatory hub in abiotic and biotic stress signaling. Expression of MIR408 gene adapting to the diverse environmental stresses suggests that multiple different transcription factors may be involved in its regulation. However, our knowledge regarding the regulation of MIR408 is still relatively sparse, and just a few of transcription factors, known as LONG HYPOCOTYL 5 (HY5) and SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE 7 (SPL7), have been reported to directly regulate MIR408 transcription in response to varying light and copper conditions (Yamasaki et al., 2009; Zhang and Li, 2013; Zhang et al., 2014).
Previous studies have indicated that miR408 targets several genes for copper-binding proteins that belong to two distinct families of phytocyanin and laccase. Plastocyanin is a copper-binding protein and functions as a mobile electron carrier between the membrane-bound cytochrome b6f complex and P-700, the reaction center of photosystem I (PSI) in eukaryotic photosynthetic organisms (Raven et al., 1999; Joliot and Joliot, 2006). In Arabidopsis, there have two plastocyanin genes (PETE1 and PETE2), and PETE2 is the predominant isoform. Plants with mutations in the plastocyanin genes show impaired growth (Abdel-Ghany, 2009; Pesaresi et al., 2009). Laccases are also copper-containing oxidase enzymes and play a role in the formation of lignin by promoting the oxidative coupling of monolignols, such as LAC3, LAC4, LAC 12, and LAC13 (Zhao et al., 2013; Schuetz et al., 2014; Wang et al., 2014). Copper (Cu) plays a vital role in plant growth and development because it acts as a cofactor of many proteins and enzymes involved in number of physiological processes, such as photosynthesis, seed production, carbohydrate distribution, nitrogen fixation, antioxidant activity, cell wall metabolism and hormone perception (Pilon et al., 2006). The Cu-microRNAs are responsible for distribution of Cu allowing plants to coordinate Cu protein expression and properly development (Pilon, 2017). Taken together, these findings clearly demonstrate the importance of miR408 in copper homeostasis.
Our previous studies have revealed that miR408 accumulation promotes vegetative growth as well as biosynthesis of pigments in Arabidopsis young seedlings (Zhang et al., 2011, 2014; Zhang and Li, 2013). However, the underlying molecular mechanisms remain unclear. More importantly, it is not yet clear how miR408 affects reproductive development. Dong et al. (2005) demonstrated that overexpression of plantacyanin (ARPN) led to reduction of seed production in Arabidopsis. More recently, Zhao et al. (2016) reported that overexpression of tae-miR408 promoted heading time as well as increased transgenic plant height in wheat. These observations indicate that miR408 may play a role in reproduction in plants. To develop a better understanding of the biological function of miR408, further physiological characterization of miR408 transgenic plants and functional analysis of its targets at the molecular level become essential. In this work, we present evidence supporting the involvement of miR408 in different growth stages, further illustrating its functional regulatory role in biomass production and seed yield.
Materials and Methods
Plant Materials and Culture Conditions
Wild-type plants used were the Arabidopsis thaliana ecotype Columbia-0. The 35S:MIR408 (MIR408-OX) and amiR408 lines were previously described by Zhang and Li (2013). The T-DNA insertion lines of SALK_091945 for ARPN and SALK_023935 for LAC13 were from the Arabidopsis Biological Resource Center (ABRC). The sterilized seeds were sown on half-strength Murashige and Skoog (MS) medium after storage at 4°C for 4 days. The seedlings were grown in a growth chamber with the following settings: standard long-day (16 h of light/8 h of darkness) conditions at 23 ± 1°C, light intensity of 100–120 μmol m-2 s-1, and a relative humidity of approximately 50%. All phenotypic characterization experiments were conducted on multiple biological samples and repeated at least three times.
Generation of Transgenic Plants
To obtain the 35S:Plantacyanin (ARPN-OX) transgenic plants, the full length cDNA of Plantacyanin gene was amplified by PCR using primer pairs with restriction endonuclease cleavage site, and inserted into the pJim19 binary vector (Basta resistance; Zhang et al., 2008), then transformed into the wild type background. Transgenic plants were selected with 20 mg/L Basta. T3 generation homozygous lines were used for all experiments.
To obtain the 35S:LAC13 (LAC13-OX) transgenic plants, the full length cDNA of LAC13 was amplified by PCR using primer pairs with restriction endonuclease cleavage site, and inserted into the pJim19 binary vector (Hygromycin resistance; Zhang et al., 2008), then transformed into the wild type background. Transgenic plants were selected with 40 mg/L hygromycin. T3 generation homozygous lines were used for all experiments.
The ARPN-OX/LAC13-OX plants were generated by crossing ARPN-OX and LAC13-OX transgenic plants. F2 or F3 homozygous progenies were identified by growing the seedlings on MS medium containing both hygromycin and basta.
All the PCR products were obtained using Phusion high-fidelity DNA polymerase (NEB1) and the resultant constructs were sequenced by GENWIZE to ensure their integrity. The plasmids were electroporated into Agrobacterium tumefaciens GV3101, respectively. Arabidopsis transformation was performed by the floral dip method (Clough and Bent, 1998). All of the primers used to generate the above-mentioned constructs are listed in Supplementary Table S2.
RNA Analyses
Total RNA was extracted from Arabidopsis seedlings using the RNeasy plant mini kit (Qiagen) and quantified by NanoDrop 2000, and the integrity was examined by agarose gel electrophoresis. For real-time qRT-PCR, 5 μg of DNaseI treated RNA was reverse transcribed using the SuperScript II reverse transcriptase (Invitrogen), and the resultant cDNA was analyzed using the Power SYBR Green PCR Master Mix (Takara) with a Bio-Rad CFX96 real-time PCR detection system in triplicate. A total volume of 20 μl reaction system was set up according to the manufacture’s instruction, including 10 μl of 2x SYBR Green Master Mix, 0.2 μM of forward primer, 0.2 μM of reverse primer, 50 ng of cDNA template, ddH2O up to 20 μl. The Actin2 amplicon was used for normalization. Determination of relative gene expression level was carried out using the standard 2-ΔΔC(T) method. All experiments were performed on three independent biological samples with each including three technical replicates. Statistical significance was calculated using the two-tailed Student’s t-test. For RNA gel blot analysis, 20 μg of total RNA were loaded per lane and blotting was performed as described previously. Fragments of ARPN and LAC13 used for probe labeling were generated by PCR. Primer sequences are listed in Supplementary Table S2.
Glucose Measurement
Measurement of glucose content was performed using the Glucose and Sucrose Assay Kit (Biovision) according to the manufacturer’s instructions as described previously (Zhang et al., 2014). Rosette leaves from the plants grown in soil under standard long-day (16 h of light/8 h of darkness) conditions for 2 weeks were homogenized in a glucose assay buffer, and the supernatant was collected after centrifuging at 12,000 rpm for 10 min. The reaction system was set up in a 100-μL total volume including 50 μL of sample in glucose assay buffer and 50 μL of glucose assay mix (46 μL of glucose assay buffer, 2 μL of glucose probe, and 2 μL of glucose enzyme mix) and incubated at 37°C for 30 min. The A570 was collected, and glucose concentrations of the test samples were calculated based on the standard curve. The experiments were performed on three independent biological samples.
Immunoblotting
Proteins were extracted from the rosette leaves of plants grown in soil under standard long-day (16 h of light/8 h of darkness) conditions for 2 weeks, and immunoblotting was performed as described previously (Feng et al., 2004). After electroblotting on a nitrocellulose membrane, protein gel blot analysis was performed using antibodies against plastocyanin (Acris Antibodies, AS06141). Detection was performed using goat anti-rabbit IgG (H+L) horseradish peroxidase conjugate secondary antibodies (Sigma). RPT5 (Abcam, ab22676) was used as loading control.
Dry Weight and Cell Size Measurements
Dry weight was measured by drying aerial parts of 2-week-old plants grown in soil at 80°C for 10 h. For cell size measurements, a fully expanded first rosette leaf was fixed in FAA [formalin:glacial acetic acid:ethanol (70%) = 1:1:18)]. The epidermal cells of the leaves or the petioles were peeled off by hand, and then photographed using a Leica DM5500 microscope. The epidermal cell area and mesophyll cell length were measured using ImageJ analysis software.
RNA Sequencing
Total RNA from three biological replicates was extracted with an RNeasy plant mini kit (Qiagen) from the aerial parts of 10-day-old plants grown on MS plate under continuous light condition. Library construction and sequencing on the HiSeq 2000 platform were performed according to the manufacturer’s instructions (Illumina) by BioMarker cooperation (Beijing, China). Bowtie (Langmead et al., 2009) was used to map the sequence reads to the Arabidopsis genome (TAIR 10) and only uniquely mapped reads were used in subsequent analyses. Reads mapped to exonic regions of annotated gene models were normalized against the length of the transcript. On average, 74.4% of the total reads mapped to the Arabidopsis reference genome sequence (Supplementary Table S3), with most of the reads mapping to exons and only a small portion mapping to introns or intergenic regions. To compare gene expression between wild type and MIR408-OX genotypes, length-normalized read density was quantile-normalized. Relevant genes were those identified as differing in expression by at least 1.5-fold (p < 0.01) in the test and control samples. The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Wang et al., 2017a) in BIG Data Center (Wang et al., 2017b), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number CRA000553 that are publicly accessible at http://bigd.big.ac.cn/gsa.
Measurement of Photosynthesis and Contents of Cellular Copper and Gibberellins
The net photosynthesis was measured using a portable photosynthesis system (CIRAS-1, PP Systems, United Kingdom) by an open system. Measurements were made by attaching a light source to the leaf chamber window under saturating photosynthetic photon flux densities (1500 μmol m-2 s-1), ambient CO2 concentration (Ca) at 290 μmol mol-1, and relative humidity of 50–60%. Leaf temperature and vapor pressure deficit (VPD) were maintained at 28°C and 0.99 ± 0.2 kPa, respectively. Data were determined at least in six leaves from six different plants of each genotype grown in soil for 2 weeks in a growth chamber.
For cellular copper content analysis, 10-day-old seedlings grown on half MS medium under continuous light were harvested and weighted, and then washed twice with 1 mM EDTA and once with double-distilled water. The following treatment and measurement were performed as previously described (Zhang et al., 2014).
The endogenous hormone content was measured using the Agilent 1100 high-performance liquid chromatography (HPLC) system (Agilent, Palo Alto, CA, United States). The aerial parts from more than fifteen individual plants of wild type and MIR408-OX plants grown in soil for 2 weeks were cold-dried under vacuum. 0.2 g dried leaves were added to 10 mL 80% aqueous methanol and immediately homogenized on ice, then kept at 4°C overnight in darkness with continuous shaking. The homogenates were centrifuged for 10 min at 4500 g at 4°C, and the supernatant was collected and evaporated under vacuum. Dry residue was re-dissolved in 5 mL of ammonium acetic buffer (0.1 M, pH 9.0) and centrifuged at 14 000 rpm for 20 min, and the supernatant was collected and purified sequentially through Polyvinylpolypyrrolidone (PVPP) column and DEAE Sephadex A225 column. Before HPLC analysis, the elution with 50% aqueous methanol was concentrated by Sep-Pak C18 column (Waters Chromatography). Standard gibberellins were purchased from Fluka Co. (Switzerland), and all solvents and buffers were HPLC-quality. Two biologically independent replicates were performed.
Results
Phenotypic Effects of Increased miR408 Expression in Arabidopsis
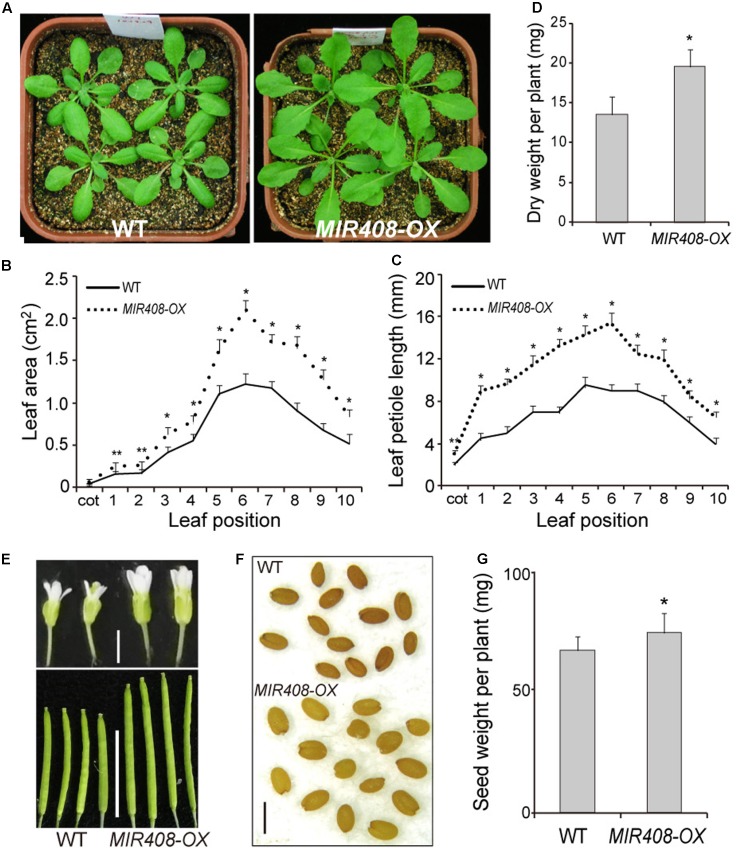
Constitutive expression of miR408 in Arabidopsis implies that it has the potential to act throughout the growth and development, and its importance at the seedling stage is well known (Zhang and Li, 2013). To determine the phenotypic impact of increased miR408 expression during the later development, transgenic plants overexpressing miR408 (MIR408-OX; Zhang and Li, 2013) were examined. The MIR408-OX transgenic plants grew rapidly and were morphologically larger than wild type (Figure 1A). In plants grown in soil for 2 weeks after stratification, the areas of rosette leaves of MIR408-OX transgenic plants were significantly increased compared with the ones of wild type (Figure 1B). The average petiole length from the cotyledon to the tenth leaves of the MIR408-OX transgenic plants also becomes much longer than that of wild type (Figure 1C and Supplementary Figures S1a,b). In addition, both fresh weight and dry weight of the MIR408-OX plants were dramatically increased compared with wild type (Supplementary Figure S1c and Figure 1D). While grown in soil for 6 weeks, the transgenic plants were much taller than wild type (Supplementary Figure S1d). In the plants at the reproductive stage, a comparison of flower size and silique length indicated significant increase for the MIR408-OX transgenic plants (Figure 1E and Supplementary Figure S1e). Furthermore, wild type plants exhibited a darker seed coat color than the transgenic ones (Figure 1F), and the seed yield of the MIR408-OX plants significantly increased (Figure 1G). In summary, the data show that constitutive expression of miR408 in wild type background leads to changes in morphology as well as increases in biomass and seed yield.
FIGURE 1.
Effects of miR408 overexpression on Arabidopsis development. (A) Wild type (left) and miR408-overexpressing plants (MIR408-OX; right) grown in soil for 2 weeks. (B,C) Leaf area (B) petiole length (C) of individual leaves of the wild type and MIR408-OX plants grown in soil for 2 weeks (n = 15). (D) Dry weight was determined for 2-week-old plants grown in soil (n = 20). (E) Comparison of flowers (upper, scale bar = 2 mm) and silique (lower, scale bar = 1 cm) between the wild type and MIR408-OX plants. (F) Seed morphology of the wild type and MIR408-OX plants, scale bar = 500 μm. (G) Seeds from more than 15 plants were collected and quantified. Data are means ± SD from n biological repeats. ∗p < 0.01, ∗∗p < 0.05 vs. wild type, by two-tailed Student’s t-test.
Cell Expansion and Elongation Likely Contributes to the Larger Phenotypes
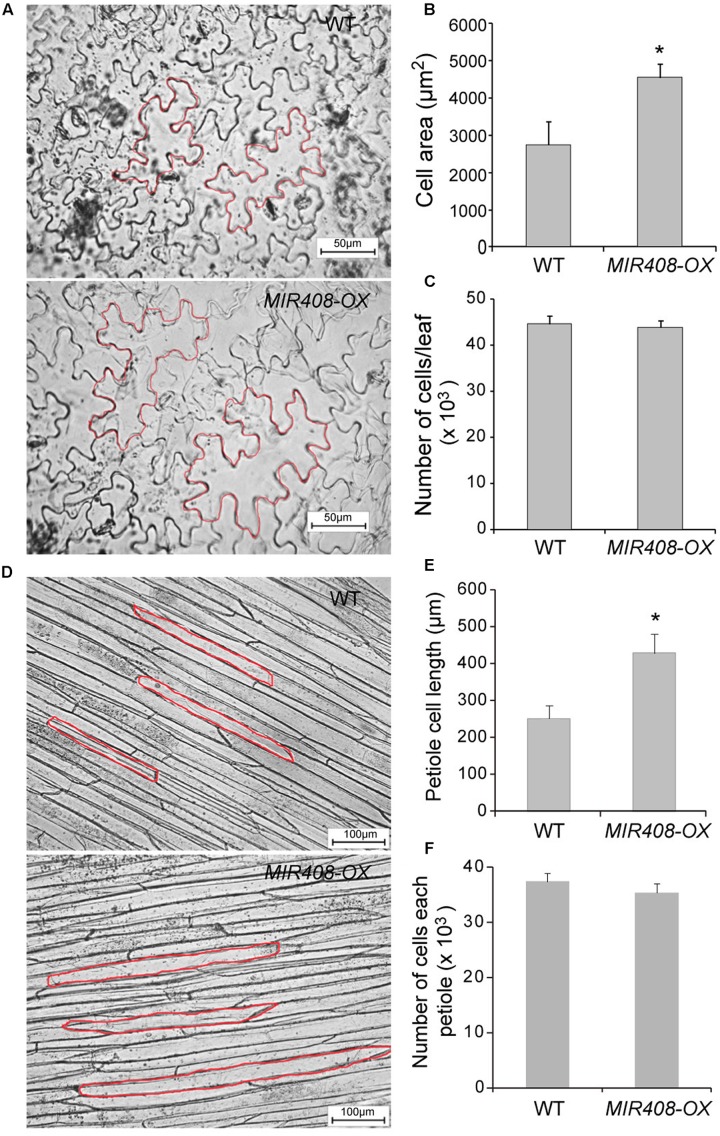
Either increased cell number or cell length could lead to larger leaf size (Tsukaya et al., 2002). To investigate whether enhanced leaf area in the MIR408-OX plants was primarily a result of cell division or cell expansion, the lengths of cells in the middle of the first rosette leaf were measured. Microscopic analysis revealed that the epidermal cell size of the transgenic plants was about 165% that of wild type (Figures 2A,B). However, the total number of epidermal cells showed no obvious changes in these two genotypes (Figure 2C). Consistent with the markedly enhanced leaf petiole elongation, the dramatic increase in the epidermal cell length was observed in the transgenic plants (Figures 2D,E) meanwhile the cell number showed no difference between wild type and MIR408-OX transgenic plants (Figure 2F). Together, increased leaf size was in good agreement with the increase in the epidermal cell length, suggesting that enhanced leaf size or leaf petiole elongation in the MIR408-OX plants primarily resulted from an increase in cell expansion rather than cell proliferation.
FIGURE 2.
Cell size and number. (A) Epidermal cells of the first rosette leaf from the wild type (upper) and MIR408-OX plants (lower). The scale bars represent 50 μm. (B) Epidermal cell area of the first rosette leaf (n = 50). (C) The number of cells per leaf (n = 10). (D) Epidermal cells of the first leaf petiole from the wild type (upper) and MIR408-OX (lower) plants. The scale bars represent 50 μm. (E) Cell length of the first leaf petiole epidermal cell (n = 50). (F) The number of cell from the first leaf petiole (n = 10). Data are means ± SD from n biological repeats. ∗p < 0.01 vs. the wild type, by two-tailed Student’s t-test.
Enhanced Expression of Genes Involved in Cytoplastic Growth and GA Biosynthesis in the MIR408-OX Plants
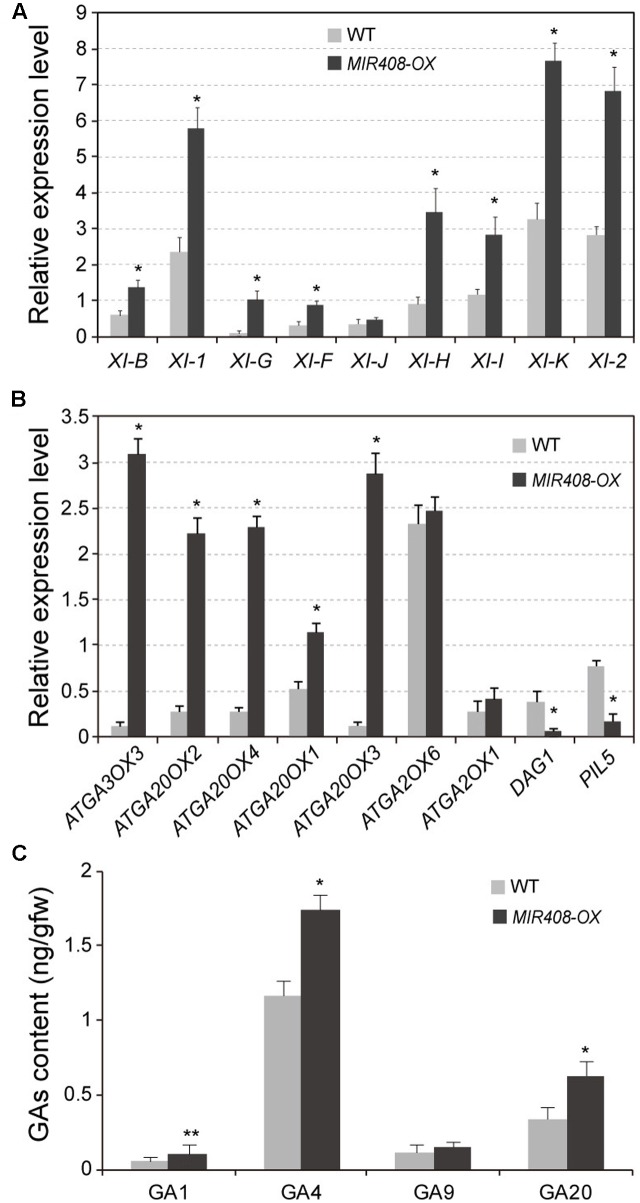
Previous reports indicated that cytoplasmic streaming, a key determinant of plant size, is generated by organelle associated myosin XI moving (Shimmen and Yokota, 2004; Ojangu et al., 2012; Tominaga et al., 2013). Gene knockout analyses of several myosin XI members lead to growth defects concomitant with reduction in cell size (Prokhnevsky et al., 2008; Peremyslov et al., 2010; Ojangu et al., 2012). Therefore, we examined the expression of nine myosin XI genes, including XI-B, XI-1, XI-G, XI-F, XI-J, XI-H, XI-I, XI-K, and XI-2. Compared to wild type, eight of the tested genes except for XI-J exhibited dramatically increased transcript levels in the MIR408-OX transgenic plants (Figure 3A). Among these myosin members, XI-K and XI-2 transcripts were the most abundant, which is consistent with the conclusion that XI-2 and XI-K are considered the major myosins providing the motive force for cytoplasmic streaming (Peremyslov et al., 2008; Ueda et al., 2010). Thus, it is likely that high level accumulation of myosin in the MIR408-OX transgenic plants might drive cytoplasmic growth, thereby promoting cell expansion and thus leading to the larger morphology.
FIGURE 3.
Expression levels of myosin and GA biosynthetic genes in the wild type and MIR408-OX transgenic plants. (A,B) Quantitative RT-PCR analysis of mRNA levels for myosin genes (A) and GA biosynthetic genes (B) using the rosette leaves from the plants grown in soil for 2 weeks. Actin2 was used as an internal control. Data are means ± SD of three biological experiments. (C) Levels of endogenous GAs in wild type and MIR408-OX plants. Data are means ± SD of three technical repeats. ∗p < 0.01, ∗∗p < 0.05 vs. the wild type, by two-tailed Student’s t-test.
Previous studies showed that alteration of Gibberellin (GA) metabolism or disruption of the GA signal transduction plays a critical role in regulation of plant dwarfism (Hedden and Phillips, 2000; Monna et al., 2002; Sasaki et al., 2002). Overexpression or repression of genes for enzymes in GA biosynthesis could lead to the alteration of GA levels and thereby result in dwarf or tall phenotypes. Thus, we determined the expression levels of genes involved in GA biosynthesis. As shown in Figure 3B, the transcript levels of biosynthetic genes for GA20-oxdases (GA20OX) and GA3-oxidases (GA3OX) exhibited significantly induction in the MIR408-OX transgenic plants compared with wild type. On the contrary, transcripts levels of the biosynthetic negative regulators of PIL5 and DAG1 were dramatically decreased in the transgenic plants. Expression level of GA2-oxidase (GA2OX) for GA deactivation did not show obvious difference between the wild type and the MIR408-OX transgenic plants. Moreover, GA contents were examined in the rosette leaves of wild type and miR408 over-expressing plants grown in soil for 2 weeks. The levels of GA4, the major endogenous bioactive GA in Arabidopsis, were significantly increased in the MIR408-OX plants although that of GA9, the immediate precursor of GA4, showed a slight increase (Figure 3C). In addition, the contents of GA1 as well as its immediate precursor GA20 were dramatically increased in the MIR408-OX plants. Taken together, these observations suggest that accumulation of miR408 might directly or indirectly modulate the cytoplasmic growth and/or GA biosynthesis to promote cell expansion and thus leading to the larger phenotypes.
Increased Photosynthesis and Chloroplastic Copper Content by miR408 Overexpression
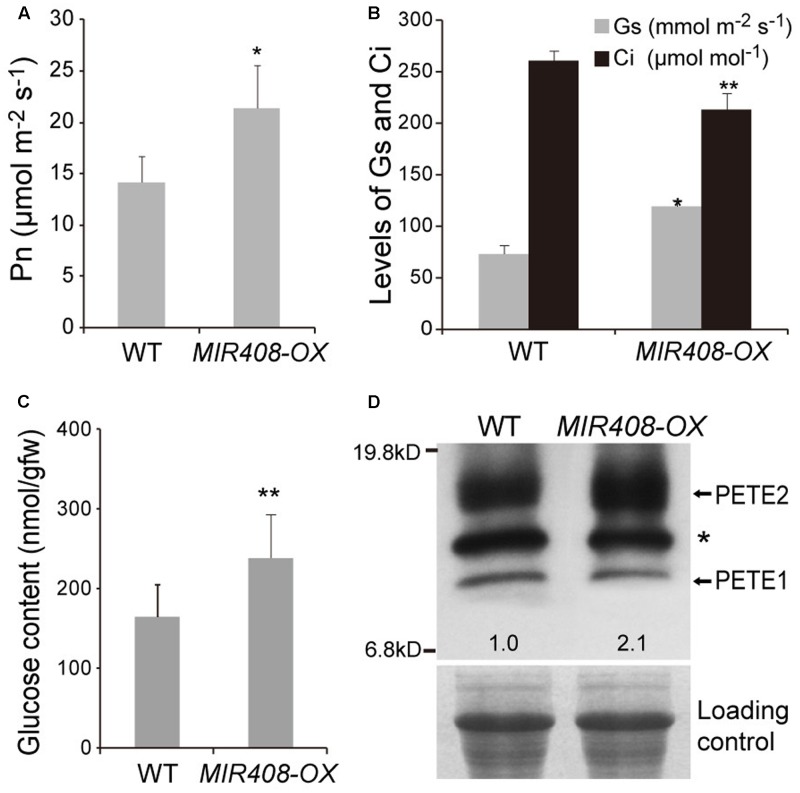
A key element to increased growth and reproduction is photosynthesis. To investigate the impact of increased expression of miR408 on photosynthesis, the rates were measured using a portable photosynthesis system (CIRAS-1, PP Systems) in the fifth rosette leaves of wild type and MIR408-OX plants grown in soil for 2 weeks. The net photosynthetic rate was significantly increased in the MIR408-OX transgenic plants compared to that of wild type (Figures 4A,B). Consequently, the contents of glucose were found to be significantly increased in the rosette leaves of the transgenic plants (Figure 4C). In the final step of the linear electron transport of photosynthesis, ferredoxin-NADP+-oxidoreductase (FNR) catalyzes the reduction of NADP+ by ferredoxin (Fd) and provides the reducing power for CO2 fixation in the Calvin cycle (Carrillo and Ceccarelli, 2003). FNR is supposed to be one of the limiting factors in photosynthetic electron transport, and the amount of FNR has been shown to correlate with photosynthetic activity (Hajirezaei et al., 2002). We found that the transcript levels of genes for leaf-type FNRs (ATLFNR1 and ATLFNR2) showed significant increase in the MIR408-OX plants (Supplementary Figure S2a).
FIGURE 4.
Enhanced photosynthesis by overexpressing MIR408. (A) Comparison of net photosynthetic rate between the wild type and MIR408-OX transgenic plants grown in soil for 2 weeks (n = 5). (B) The net photosynthetic rate is positively correlated with stomatal conductance (Gs) and negatively correlated with internal CO2 concentration (Ci) (n = 5). (C) Glucose content in the shoots of plants grown in soil for 2 weeks (n = 3). (D) Immunoblot analysis of PETE (PC) protein levels in the wild type and MIR408-OX transgenic plants. Values represent PETE2 levels normalized against the loading control RPT5 using Image J software and set to one for wild type. Data are means ± SD from n biological repeats. ∗p < 0.01, ∗∗p < 0.05 vs. the wild type, by two-tailed Student’s t-test.
Plastocyanin (PC), which is encoded by two paralogous genes, PETE1 (less abundant) and PETE2 (more abundant) is a copper-binding protein that functions as electron carrier in the thylakoid lumen of the chloroplast (Weigel et al., 2003; Abdel-Ghany, 2009; Pesaresi et al., 2009). Previous studies have indicated that miR408 repress several genes encoding for copper-binding proteins with non-photosynthetic usage (Yamasaki et al., 2007; Abdel-Ghany and Pilon, 2008). It is, thus, plausible that high level of miR408 repress non-photosynthesis related copper-binding proteins and increases copper’s availability for proteins such as PC. To address this question, we first measured copper content in the whole seedlings, and found that there were no differences between wild type and the MIR408-OX plants in overall copper content. The MIR408-OX plants, however, had higher chloroplastic copper than wild type plants (Table 1). Consistent with this observation, transcript level of PAA1 which specifically delivers copper from cytosol to chloroplast (Shikanai et al., 2003; Abdel-Ghany et al., 2005) was significantly increased in the MIR408-OX plants (Supplementary Figure S2b). Thus, elevated miR408 promotes the copper allocation to chloroplast from the cytosol. We next compared the PC levels between wild type and the MIR408-OX transgenic plants because PC is one of the major destinations of chloroplastic copper (Ramshaw et al., 1973). PETE2 was significantly increased in the MIR408-OX plants at both the mRNA and protein levels, though PETE1 was unchanged (Figure 4D and Supplementary Figure S2c). Taken together, these results suggest that over-accumulation of miR408 promotes photosynthesis by modulating the distribution of copper in plant cells.
Table 1.
Copper content (μg.g-1 fresh weight).
| Genotype | Total | Chloroplast | Percentage |
|---|---|---|---|
| WT | 0.92 ± 0.07 | 0.27 ± 0.09 | 29.34% |
| MIR408-OX | 1.18 ± 0.06 | 0.48 ± 0.11∗ | 40.67% |
Values are from three biological repeats, and star represents significant difference compared with the wild type (two-tailed Student’s t-test, p < 0.05).
Transcriptome Changes in MIR408-OX Plants
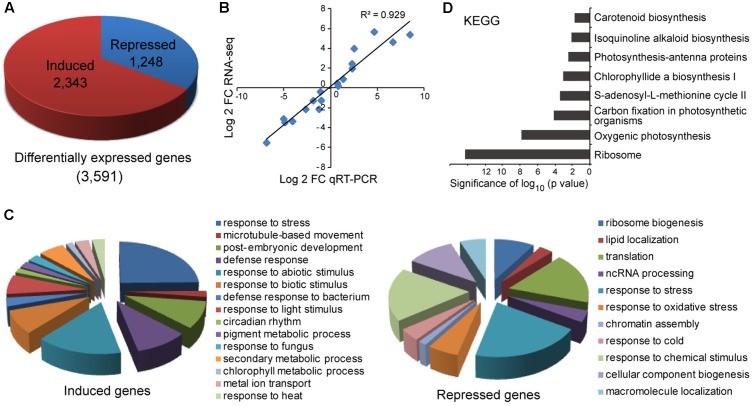
To gain more insight into the molecular changes associated with miR408 overexpression, total RNA was extracted from whole seedlings of wild type and the MIR408-OX seedlings grown on MS plate for 10 days under continuous light, and subjected to high throughput sequencing. We identified 3,591 differentially expressed genes, of which 2,343 were induced and 1,248 repressed in the MIR408-OX transgenic plants, respectively (Figure 5A and Supplementary Table S1). To further confirm the RNA sequencing data, we performed RT-qPCR to monitor the transcripts levels on a handful of copper-responsive as well as randomly selected genes. As shown in Figure 5B, the results generated by the two methods agreed well. Gene Ontology (GO) analysis revealed that the genes induced in MIR408-OX transgenic plants preferentially associated with GO terms such as response to stimulus and stress, microtubule-based movement, chlorophyll metabolic process, pigment biosynthetic process, and metal ion transport (Figure 5C). Biological pathways responsive to ribosome biogenesis, lipid location, response to oxidative stress, and RNA processing were greatly enriched among the downregulated genes (Figure 5C). Regarding annotated pathways, ribosome, photosynthesis, carbon fixation, pigments biosynthetic processes are among the most significantly enriched terms (Figure 5D). Thus, we hypothesize that these transcriptomic changes may contribute to the photosynthesis, myosin-mediated cytoplasmic growth, abiotic stress responses, and better protection of cells against reactive oxygen species (ROS) especially from the higher photosynthesis of the MIR408-OX transgenic plants. Overall, these results suggest that overexpression of miR408 influences multiple metabolic processes directly or indirectly and thus impacts on multiple aspects of plant growth and development.
FIGURE 5.
Characterization of genes with altered levels of expression in the MIR408-OX transgenic plants. (A) Differentially expressed genes between the wild type and MIR408-OX plants. (B) Correlation between RNA-sequencing and qRT-PCR data on the selected genes. Pearson correlation was calculated using data points representing Log2 transformed transcript level ratios of wild type and MIR408-OX plants. (C) Enriched representative GO terms (p < 0.001) in the biological category that associated with induced (left) and repressed (right) genes in the MIR408-OX transgenic plants. (D) Enriched pathways associated with the differentially expressed genes by KEGG analysis.
Effect of miR408 on Reproduction via Modulating Its Target Genes
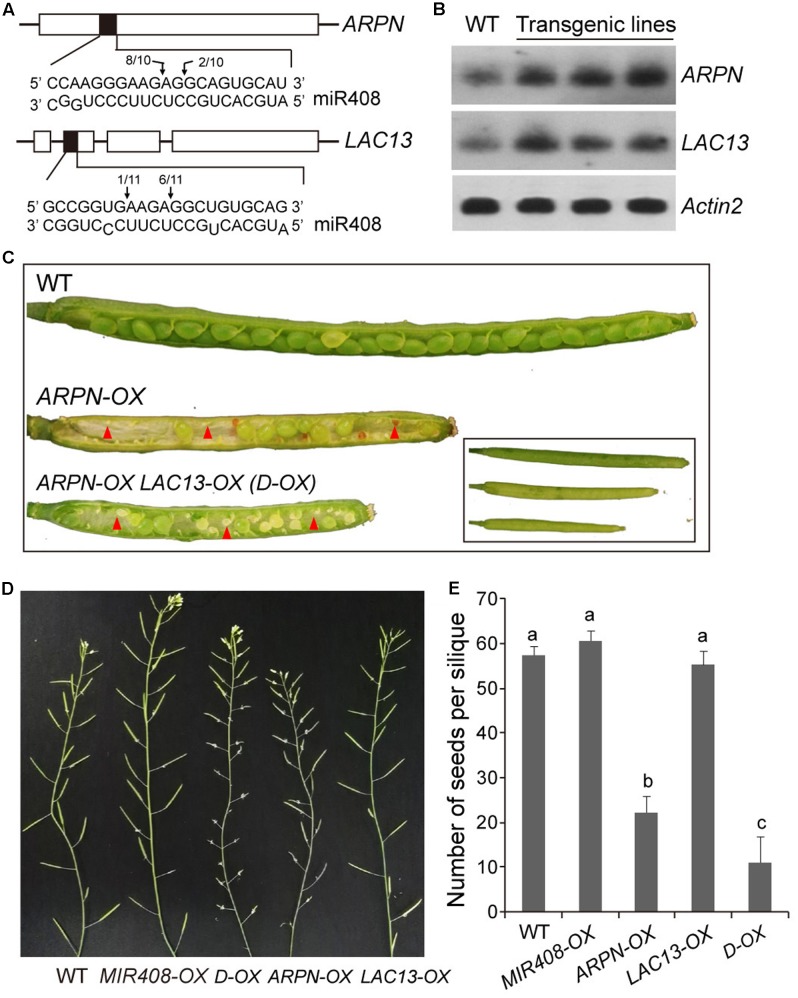
Identification and functional analysis of the targets of miRNA is crucial for understanding the biological function of miRNA. To further dissect the function of miR408, two of its target genes, Plastacyanin (ARPN) and LAC13 (Figure 6A), were analyzed. We obtained two T-DNA insertion lines of SALK_091945 for ARPN and SALK_023935 for LAC13 from the Arabidopsis Biological Resource Center (ABRC). Unfortunately, we did not see any visible phenotypes for both mutants under normal growth conditions, which are most likely due to the functional redundancy of the miR408 target genes, especially for those laccase members. Thus, we resorted to the overexpression approach to determine their functions. In order to create ARPN or LAC13 overexpressors, we used the CaMV35S promoter to drive protein expression in Arabidopsis. As shown in Figure 6B, homozygous transgenic plants contained higher transcript levels of ARPN or LAC13 compared with the wild type. Compared to wild type, the ARPN-OX transgenic lines showed shorter siliques and increased silique number of inflorescence (Figures 6C,D). Seed formation was dramatically decreased when siliques were randomly picked for seed counts (Figure 6E) and thus led to less seed yield although the fresh and dry weight showed no obvious differences from the wild type (Table 2). For the LAC13-OX transgenic plants, we did not observe any obvious phenotypic changes at either vegetative or reproductive stage (Table 2). However, the F1 progenies of ARPN-OX and LAC13-OX plants showed much shorter siliques and significantly reduction of seeds number and seed yield although the silique number of inflorescence increased (Figures 6C–E). In addition, the F1 progenies exhibited significant reduction of fresh and dry weight compared with the wild type (Table 2). Meanwhile, the transgenic plants (amiR408) with decreased level of endogenous miR408 generated by the artificial miRNA approach reported in the previous study (Zhang and Li, 2013) showed similar phenotypes to the ARPN-OX/LAC13-OX plants (Supplementary Figure S3 and Table 2), suggesting that miR408 may play a role in control of reproductive development by regulating its target genes. Taken together, these results demonstrate that miR408 appears to have pleotropic effects on plant growth and development.
FIGURE 6.
Biomass and seed yield traits of Plantacyanin (ARPN) and LAC13 overexpressors. (A) Confirmation of miR408 targeting on ARPN and LAC13 by 5′-RACE. The complementary mRNA and miRNA sequences are shown with shaded boxes. Vertical arrows mark the sequenced cleavage sites with the frequency of clones shown. (B) RNA gel blot analysis for wild type and ARPN-OX or LAC13-OX transgenic lines. Actin2 served as loading control. (C) Stereomicroscopy images of siliques obtained from self-pollinated wild type, ARPN-OX, and ARPN-OX/LAC13-OX (D-OX) parental plants. Red arrowheads indicate abnormal ovules. (D) Comparison of silique development on 6-week-old plants of the wild type, MIR408-OX, ARPN-OX, LAC13-OX, and D-OX genotypes. (E) Seeds from different genotypes were collected and quantified (n = 15). Data are means ± SD. Genotypes labeled with the same letters have no statistical difference, while different letters denote groups with significant differences (ANOVA, p < 0.01).
Table 2.
Comparison of biomass and seed yield among various genotypic plants.
| WT | ARPN-OX | LAC13-OX | D-OX | AmiR408 | |
|---|---|---|---|---|---|
| Fresh weight (g) | 0.18 ± 0.06 | 0.16 ± 0.03 | 0.19 ± 0.05 | 0.11 ± 0.05∗ | 0.12 ± 0.03∗ |
| Dry weight (mg) | 13.4 ± 2.3 | 12.9 ± 3.6 | 13.7 ± 2.8 | 10.2 ± 3.3∗ | 9.8 ± 1.4∗ |
| Seed yield (mg) | 105.3 ± 16.1 | 38.7 ± 5.2∗ | 99.6 ± 8.4 | 14.5 ± 4.2∗ | 23.1 ± 3.6∗ |
Values are means ± SD from n biological replicates, where n = 20 for fresh weight or dry weight, and n = 10 for seed yield. Stars represent significant difference compared with the wild type (two-tailed Student’s t-test, p < 0.05).
Discussion
MiR408 is among the most conserved miRNA families and has so far been annotated in more than 30 plant species, suggesting that its role is fundamental to plant development and function (Axtell and Bowman, 2008; Kozomara and Griffiths-Jones, 2011). It has shown in the earlier studies that miR408 is involved in photomorphorgenetic development, copper-light signaling pathway, and biotic stress responses as well as vegetative biomass in Arabidopsis seedlings (Zhang and Li, 2013; Zhang et al., 2014; Ma et al., 2015). In our present work, constitutive expression of miR408 influences various developmental stages, and promotes vigorous plants growth and seed yield by increasing photosynthetic efficiency. Therefore, miR408 likely have pleiotropic effects on plant growth and development.
Transgenic plants overexpressing MIR408 exhibited vigorous growth phenotypes such as increased leaf area, petiole length, plant height, and biomass yield. Tsukaya et al. (2002) demonstrated that both increased cell number and increased cell length lead to larger size of leaf or petiole length. Our results showed that increased cell size but not cell number leads to the larger phenotype of the MIR408-OX transgenic plants. Interestingly, a previous study demonstrated that myosin-mediated cytoplasmic streaming is a key determinant of plant size (Tominaga et al., 2013). In Arabidopsis, there have 13 myosins in the class XI family (Reddy, 2001). Among the nine tested myosin genes, eight of them exhibited significantly increased levels in the MIR408-OX transgenic plants compared with wild type (Figure 3A), implying that induction of myosin might be an explanation for the larger morphology in the transgenic plants. Apart from copper, expression levels of MIR408, as well as its target genes, were investigated in different plant species in response to various environmental conditions, such as light, cold, salinity, oxidative stress, mechanical stress, drought and osmotic stress (Sunkar and Zhu, 2004; Lu et al., 2005; Kantar et al., 2010; Li et al., 2010; Trindade et al., 2010; Sunkar et al., 2012; Zhang et al., 2014; Rajwanshi et al., 2014). These observations may suggest a new adaptation strategy to environmental fluctuations for plants survival such as flood and canopy shade by manipulating miR408 abundance.
Leaf position plays a vital role for photosynthesis, and the control of leaf petiole elongation is an important mechanism to ensure plant leaves at appropriate positions. Several key regulators that control leaf petiole elongation had been identified (Tsukaya et al., 2002). Mutation of the ROTUNDIFOLIA 3 (ROT3) gene encoding a protein of the cytochrome P450 family results in short leaf petioles, and overexpression of the ROT3 gene in transgenic Arabidopsis produces elongated leaf petioles (Tsuge et al., 1996; Kim et al., 1999). In addition, knock-out of ACAULIS 2 gene (ACL2) suppresses leaf petiole elongation as well as flower stalks (Tsukaya et al., 1995). Recently, three related receptor-like kinases encoded by HERCULES1 (HERK1), THESEUS1 (THE1) and FERONIA (FER) that regulate the brassinosteroid signaling pathway were reported to control petiole growth, which mutants showed short leaf petioles phenotypes (Guo et al., 2009). Furthermore, previous studies have revealed that cell extensibility regulated by cell-wall loosening proteins is one of important factors controlling cell elongation in plants (Cosgrove, 2000; Ookawara et al., 2005). Consistently, expression levels of ACL2, ROT3, HERK1, and several of cell-wall loosening genes were significantly changed between the MIR408-OX and wild type plants (Supplementary Table S1). These observations suggest that overexpression of miR408 indirectly influences the cell elongation genes and thus leads to elongated petiole. Additionally, hormones, such as gibberellins (GAs), are well known regulators of cell elongation in many species, such as Arabidopsis, maize and rice (Luo et al., 2006; Achard et al., 2009; Bolduc and Hake, 2009). In our study, transcript levels of genes for GA biosynthesis were dramatically increased in the MIR408-OX transgenic plants (Figure 2B), and an increased production of hormone could be expected (Figure 2C), implying that GA-enhanced cell elongation could play a role in the larger phenotypes as well.
In addition to miR408, there are several copper-regulated miRNAs that respond to external copper concentrations in Arabidopsis, e.g., miR397, miR398, and miR857, which together repress the levels of mRNA transcripts for a number of copper-containing proteins under Cu-limited conditions (Abdel-Ghany and Pilon, 2008; Yamasaki et al., 2009). Constitutive expression of miR397a or miR397b in rice could promote panicle branching and increase grain size and yield (Zhang and Li, 2013). Furthermore, compared to wild type Arabidopsis, the transgenic plants overexpressing miR397b developed more inflorescence shoots and showed increased silique number and silique length, which resulted in higher seed numbers (Wang et al., 2014). In contrast, transgenic plants overexpressing its target gene, LAC4, were severely dwarfed with small rosette leaves, short inflorescence stems, short silique, and less seeds, a completely opposite phenotype to that observed for the miR397b-OX plants. Previous studies have indicated that under copper deficient conditions, elevated miR408 level can lead to inhibition of mRNA transcripts for copper-containing proteins, thus promoting the preferential delivery of copper from cytoplasm to chloroplast, which in turn ensures photosynthesis (Weigel et al., 2003; Burkhead et al., 2009). Thus, it is not surprising that constitutive expression of miR408 results in higher accumulation of copper in the chloroplast of the MIR408-OX transgenic plants compared with the wild type and obviously increased photosynthetic efficiency (Table 1 and Figures 4A,B). Our data presented here may suggest that overexpression of miR408 results in vigorous growth and enlarged morphology as well as increases biomass and seeds yield through promoting photosynthetic efficiency. In terms of silique size and seeds yield, the MIR408-OX transgenic plants are similar to the miR397b-OX ones in Arabidopsis. However, the LAC13-OX Arabidopsis showed no obvious difference from wild type although both LAC4 and LAC13 belong to the laccase family. These observations indicate that regulatory mechanism may be distinctive for individual copper miRNAs.
It should be noted that actual plants’ photosynthetic efficiency is rather low from 0.1 to 8% because of inefficient conversion of solar light to electric energy (Zhu et al., 2010). To overcome this limitation, genetically modify plants can be developed to achieve greater efficiencies and enhanced growth. To data, genetic engineering has been widely used to develop new agricultural crops with stress tolerance against degradation of global environment. However, genetically modified plants with desirable agricultural traits by manipulating related regulatory effectors frequently show growth limitation and yield penalties for the interplay between developmental and stress responsive signaling networks (Cabello et al., 2014). In this regard, it becomes desirable to explore candidate genes that can confer stress tolerance without restricting the plant growth and yield. Our results demonstrate that the transgenic plants overexpressing MIR408 showed significantly increased photosynthetic rates and produced higher biomass and seed yield. More recently, Ma et al. (2015) reported that overexpression of miR408 led to improved tolerance to abiotic stresses in Arabidopsis, such as salinity, cold, and oxidative stress. Together with a previous study that overexpression of miR408 significantly increased drought tolerance in chickpea (Hajyzadeh et al., 2015), it is plausible that the MIR408-OX transgenic plants could be resistant to stress conditions. Thus, miR408 may provide an important cross-link between plant growth, development and stress response, and play a central role in plant survival. Furthermore, it is well known that miR408 targets three laccase genes, LAC3, LAC12, and LAC13, which polymerize monolignols into lignin (Bao et al., 1993; Mayer and Staples, 2002). Thus, a reduction of lignin deposition could be expected in the MIR408-OX plants as well. This information may have potential to improve a wide range of plant species for use as bioenergy feedstocks. Taken together, our results suggest that MIR408 can be of potential interest as a candidate gene in developing new agricultural crops. Further exploration of miR408 in various plant species including crops will thus provide much needed insight as to the coordinated control of the superior phenotypes.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MIR408 (At2g47015), Plantacyanin (At2g02850), LAC13 (At5g07130), PETE1 (At1g76100), PETE2 (At1g20340), Myosin XI-B (At1g04160), Myosin XI-1 (At1g17580), Myosin XI-1 (At5g43900), Myosin XI-G (At2g20290), Myosin XI-F (At2g31900), Myosin XI-J (At3g58160), Myosin XI-H (At4g28710), Myosin XI-I (At4g33200), Myosin XI-K (At5g20490), ATGA3OX3 (At4g21690), ATGA20OX2 (At5g51810), ATGA20OX4 (At1g60980), ATGA20OX1 (At4g25420), ATGA20OX3 (At5g07200), ATGA2OX1 (At1g78440), ATGA2OX6 (At1g02400), DAG1 (At3g61850), PIL5 (At2g20180), LFNR1 (At5g66190), LFNR2 (At1g20020), RFNR1 (At4g05390), RFNR2 (At1g30510), HMA1 (At4g37270), PAA1(At5g35590), PAA2 (At5g21930), ACTIN2 (At3g18780). The T-DNA insertion lines are SALK_091945 for Plantacyanin and SALK_023935 for LAC13.
Author Contributions
Project design: ZS, YW, and HZhang. Data cultivation and collection: ZS, YW, LZ, SL, HZhang, HL, and HZhao. Data analysis: ZS, YW, LZ, SL, and HZhang. Writing: ZS and HZhang.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant 31670288 to HZhang) and the Innovation Special Program of Henan Agricultural University for Science and Technology (Grant 30600764 to HZhang).
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02114/full#supplementary-material
References
- Abdel-Ghany S. E. (2009). Contribution of plastocyanin isoforms to photosynthesis and copper homeostasis in Arabidopsis thaliana grown at different copper regimes. Planta 229 767–779. 10.1007/s00425-008-0869-z [DOI] [PubMed] [Google Scholar]
- Abdel-Ghany S. E., Muller-Moule P., Niyogi K. K., Pilon M., Shikanai T. (2005). Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17 1233–1251. 10.1105/tpc.104.030452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Ghany S. E., Pilon M. (2008). MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 283 15932–15945. 10.1074/jbc.M801406200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P., Gusti A., Cheminant S., Alioua M., Dhondt S., Coppens F., et al. (2009). Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr. Biol. 19 1188–1193. 10.1016/j.cub.2009.05.059 [DOI] [PubMed] [Google Scholar]
- Axtell M. J., Bowman J. L. (2008). Evolution of plant microRNAs and their targets. Trends Plant Sci. 13 343–349. 10.1016/j.tplants.2008.03.009 [DOI] [PubMed] [Google Scholar]
- Bao W., O’Malley D. M., Whetten R., Sederoff R. R. (1993). A laccase associated with lignification in loblolly pine xylem. Science 260 672–674. 10.1126/science.260.5108.672 [DOI] [PubMed] [Google Scholar]
- Bolduc N., Hake S. (2009). The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21 1647–1658. 10.1105/tpc.109.068221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y. Y., Sieburth L., et al. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320 1185–1190. 10.1126/science.1159151 [DOI] [PubMed] [Google Scholar]
- Burkhead J. L., Reynolds K. A., Abdel-Ghany S. E., Cohu C. M., Pilon M. (2009). Copper homeostasis. New Phytol. 182 799–816. 10.1111/j.1469-8137.2009.02846.x [DOI] [PubMed] [Google Scholar]
- Cabello J. V., Lodeyro A. F., Zurbriggen M. D. (2014). Novel perspectives for the engineering of abiotic stress tolerance in plants. Curr. Opin. Biotechnol. 26 62–70. 10.1016/j.copbio.2013.09.011 [DOI] [PubMed] [Google Scholar]
- Carrillo N., Ceccarelli E. A. (2003). Open questions in ferredoxin-NADP+ reductase catalytic mechanism. Eur. J. Biochem. 270 1900–1915. 10.1046/j.1432-1033.2003.03566.x [DOI] [PubMed] [Google Scholar]
- Chen K., Rajewsky N. (2007). The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 8 93–103. 10.1038/nrg1990 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Comai L., Zhang B. (2012). MicroRNAs: key gene regulators with versatile functions. Plant Mol. Biol. 80:1. 10.1007/s11103-012-9947-5 [DOI] [PubMed] [Google Scholar]
- Cosgrove D. J. (2000). Loosening of plant cell walls by expansins. Nature 407 321–326. 10.1038/35030000 [DOI] [PubMed] [Google Scholar]
- Dong J., Kim S. T., Lord E. M. (2005). Plantacyanin plays a role in reproduction in Arabidopsis. Plant Physiol. 138 778–789. 10.1104/pp.105.063388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H., Zhang Q., Wang Q., Wang X., Liu J., Li M., et al. (2013). Target of tae-miR408, a chemocyanin-like protein gene (TaCLP1), plays positive roles in wheat response to high-salinity, heavy cupric stress and stripe rust. Plant Mol. Biol. 83 433–443. 10.1007/s11103-013-0101-9 [DOI] [PubMed] [Google Scholar]
- Feng S., Shen Y., Sullivan J. A., Rubio V., Xiong Y., Sun T. P., et al. (2004). Arabidopsis CAND1 an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. Plant Cell 16 1870–1882. 10.1105/tpc.021949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia P., Frampton J. (2008). Hematopoietic lineage commitment: miRNAs add specificity to a widely expressed transcription factor. Dev. Cell 14 815–816. 10.1016/j.devcel.2008.05.011 [DOI] [PubMed] [Google Scholar]
- Guo H., Li L., Ye H., Yu X., Algreen A., Yin Y. (2009). Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 106 7648–7653. 10.1073/pnas.0812346106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajirezaei M. R., Peisker M., Tschiersch H., Palatnik J. F., Valle E. M., Carrillo N., et al. (2002). Small changes in the activity of chloroplastic NADP+-dependent ferredoxin oxidoreductase lead to impaired plant growth and restrict photosynthetic activity of transgenic tobacco plants. Plant J. 29 281–293. 10.1046/j.0960-7412.2001.01209.x [DOI] [PubMed] [Google Scholar]
- Hajyzadeh M., Turktas M., Khawar K. M., Unver T. (2015). miR408 overexpression causes increased drought tolerance in chickpea. Gene 555 186–193. 10.1016/j.gene.2014.11.002 [DOI] [PubMed] [Google Scholar]
- Hedden P., Phillips A. L. (2000). Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 5 523–530. 10.1016/S1360-1385(00)01790-8 [DOI] [PubMed] [Google Scholar]
- Hobert O. (2008). Gene regulation by transcription factors and microRNAs. Science 319 1785–1786. 10.1126/science.1151651 [DOI] [PubMed] [Google Scholar]
- Joliot P., Joliot A. (2006). Cyclic electron flow in C3 plants. Biochim. Biophys. Acta 1757 362–368. 10.1016/j.bbabio.2006.02.018 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M. W., Bartel D. P., Bartel B. (2006). MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 57 19–53. 10.1146/annurev.arplant.57.032905.105218 [DOI] [PubMed] [Google Scholar]
- Kantar M., Unver T., Budak H. (2010). Regulation of barley miRNAs upon dehydration stress correlated with target gene expression. Funct. Integr. Genomics 10 493–507. 10.1007/s10142-010-0181-4 [DOI] [PubMed] [Google Scholar]
- Khraiwesh B., Zhu J. K., Zhu J. (2012). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 1819 137–148. 10.1016/j.bbagrm.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G. T., Tsukaya H., Saito Y., Uchimiya H. (1999). Changes in the shapes of leaves and flowers upon overexpression of cytochrome P450 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 96 9433–9437. 10.1073/pnas.96.16.9433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2011). miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39 D152–D157. 10.1093/nar/gkq1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Li H., Zhang Y. X., Liu J. Y. (2010). Identification and analysis of seven HO-responsive miRNAs and 32 new miRNAs in the seedlings of rice (Oryza sativa L. ssp. indica). Nucleic Acids Res. 39 2821–2833. 10.1093/nar/gkq1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C., Xie Z., Kasschau K. D., Carrington J. C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297 2053–2056. 10.1126/science.1076311 [DOI] [PubMed] [Google Scholar]
- Lu S., Sun Y. H., Shi R., Clark C., Li L., Chiang V. L. (2005). Novel and mechanical stress-responsive MicroRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17 2186–2203. 10.1105/tpc.105.033456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo A., Qian Q., Yin H., Liu X., Yin C., Lan Y., et al. (2006). EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol. 47 181–191. 10.1093/pcp/pci233 [DOI] [PubMed] [Google Scholar]
- Ma C., Burd S., Lers A. (2015). miR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 84 169–187. 10.1111/tpj.12999 [DOI] [PubMed] [Google Scholar]
- Mayer A. M., Staples R. C. (2002). Laccase: new functions for an old enzyme. Phytochemistry 60 551–565. 10.1016/S0031-9422(02)00171-1 [DOI] [PubMed] [Google Scholar]
- Monna L., Kitazawa N., Yoshino R., Suzuki J., Masuda H., Maehara Y., et al. (2002). Positional cloning of rice semidwarfing gene, sd-1: rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res. 9 11–17. 10.1093/dnares/9.1.11 [DOI] [PubMed] [Google Scholar]
- Mutum R. D., Balyan S. C., Kansal S., Agarwal P., Kumar S., Kumar M., et al. (2013). Evolution of variety-specific regulatory schema for expression of osa-miR408 in indica rice varieties under drought stress. FEBS J. 280 1717–1730. 10.1111/febs.12186 [DOI] [PubMed] [Google Scholar]
- Ojangu E. L., Tanner K., Pata P., Jarve K., Holweg C. L., Truve E., et al. (2012). Myosins XI-K, XI-1, and XI-2 are required for development of pavement cells, trichomes, and stigmatic papillae in Arabidopsis. BMC Plant Biol. 12:81. 10.1186/1471-2229-12-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ookawara R., Satoh S., Yoshioka T., Ishizawa K. (2005). Expression of alpha-expansin and xyloglucan endotransglucosylase/hydrolase genes associated with shoot elongation enhanced by anoxia, ethylene and carbon dioxide in arrowhead (Sagittaria pygmaea Miq.) tubers. Ann. Bot. 96 693–702. 10.1093/aob/mci221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov V. V., Prokhnevsky A. I., Avisar D., Dolja V. V. (2008). Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis. Plant Physiol. 146 1109–1116. 10.1104/pp.107.113654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov V. V., Prokhnevsky A. I., Dolja V. V. (2010). Class XI myosins are required for development, cell expansion, and F-Actin organization in Arabidopsis. Plant Cell 22 1883–1897. 10.1105/tpc.110.076315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P., Scharfenberg M., Weigel M., Granlund I., Schroder W. P., Finazzi G., et al. (2009). Mutants, overexpressors, and interactors of Arabidopsis plastocyanin isoforms: revised roles of plastocyanin in photosynthetic electron flow and thylakoid redox state. Mol. Plant 2 236–248. 10.1093/mp/ssn041 [DOI] [PubMed] [Google Scholar]
- Pilon M. (2017). The copper microRNAs. New Phytol. 213 1030–1035. 10.1111/nph.14244 [DOI] [PubMed] [Google Scholar]
- Pilon M., Abdel-Ghany S. E., Cohu C. M., Gogolin K. A., Ye H. (2006). Copper cofactor delivery in plant cells. Curr. Opin. Plant Biol. 9 256–263. 10.1016/j.pbi.2006.03.007 [DOI] [PubMed] [Google Scholar]
- Prokhnevsky A. I., Peremyslov V. V., Dolja V. V. (2008). Overlapping functions of the four class XI myosins in Arabidopsis growth, root hair elongation, and organelle motility. Proc. Natl. Acad. Sci. U.S.A. 105 19744–19749. 10.1073/pnas.0810730105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajwanshi R., Chakraborty S., Jayanandi K., Deb B., Lightfoot D. A. (2014). Orthologous plant microRNAs: microregulators with great potential for improving stress tolerance in plants. Theor. Appl. Genet. 127 2525–2543. 10.1007/s00122-014-2391-y [DOI] [PubMed] [Google Scholar]
- Ramshaw J. A., Brown R. H., Scawen M. D., Boulter D. (1973). Higher plant plastocyanin. Biochim. Biophys. Acta 303 269–273. 10.1016/0005-2795(73)90357-7 [DOI] [PubMed] [Google Scholar]
- Raven J. A., Evans M. C. W., Korb R. E. (1999). The role of trace metals in photosynthetic electron transport in O2-evolving organisms. Photosynth. Res. 60 111–149. 10.1023/A:1006282714942 22544768 [DOI] [Google Scholar]
- Reddy A. S. (2001). Molecular motors and their functions in plants. Int. Rev. Cytol. 204 97–178. 10.1016/S0074-7696(01)04004-9 [DOI] [PubMed] [Google Scholar]
- Rogers K., Chen X. (2013). Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25 2383–2399. 10.1105/tpc.113.113159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I., Weigel D. (2011). MicroRNA networks and developmental plasticity in plants. Trends Plant Sci. 16 258–264. 10.1016/j.tplants.2011.03.001 [DOI] [PubMed] [Google Scholar]
- Sasaki A., Ashikari M., Ueguchi-Tanaka M., Itoh H., Nishimura A., Swapan D., et al. (2002). Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416 701–702. 10.1038/416701a [DOI] [PubMed] [Google Scholar]
- Schramke V., Allshire R. (2004). Those interfering little RNAs! Silencing and eliminating chromatin. Curr. Opin. Genet. Dev. 14 174–180. 10.1016/j.gde.2004.02.006 [DOI] [PubMed] [Google Scholar]
- Schuetz M., Benske A., Smith R. A., Watanabe Y., Tobimatsu Y., Ralph J., et al. (2014). Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol. 166 798–807. 10.1104/pp.114.245597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T., Muller-Moule P., Munekage Y., Niyogi K. K., Pilon M. (2003). PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell 15 1333–1346. 10.1105/tpc.011817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmen T., Yokota E. (2004). Cytoplasmic streaming in plants. Curr. Opin. Cell Biol. 16 68–72. 10.1016/j.ceb.2003.11.009 [DOI] [PubMed] [Google Scholar]
- Sun G. (2012). MicroRNAs and their diverse functions in plants. Plant Mol. Biol. 80 17–36. 10.1007/s11103-011-9817-6 [DOI] [PubMed] [Google Scholar]
- Sunkar R., Li Y. F., Jagadeeswaran G. (2012). Functions of microRNAs in plant stress responses. Trends Plant Sci. 17 196–203. 10.1016/j.tplants.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Sunkar R., Zhu J. K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16 2001–2019. 10.1105/tpc.104.022830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga M., Kimura A., Yokota E., Haraguchi T., Shimmen T., Yamamoto K., et al. (2013). Cytoplasmic streaming velocity as a plant size determinant. Dev. Cell 27 345–352. 10.1016/j.devcel.2013.10.005 [DOI] [PubMed] [Google Scholar]
- Trindade I., Capitao C., Dalmay T., Fevereiro M. P., Santos D. M. (2010). miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 231 705–716. 10.1007/s00425-009-1078-0 [DOI] [PubMed] [Google Scholar]
- Tsuge T., Tsukaya H., Uchimiya H. (1996). Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122 1589–1600. [DOI] [PubMed] [Google Scholar]
- Tsukaya H., Inaba-Higano K., Komeda Y. (1995). Phenotypic characterization and molecular mapping of an acaulis2 mutant of Arabidopsis thaliana with flower stalks of much reduced length. Plant Cell Physiol. 36 239–246. 10.1093/oxfordjournals.pcp.a078755 [DOI] [Google Scholar]
- Tsukaya H., Kozuka T., Kim G. T. (2002). Genetic control of petiole length in Arabidopsis thaliana. Plant Cell Physiol. 43 1221–1228. 10.1093/pcp/pcf147 [DOI] [PubMed] [Google Scholar]
- Ueda H., Yokota E., Kutsuna N., Shimada T., Tamura K., Shimmen T., et al. (2010). Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc. Natl. Acad. Sci. U.S.A. 107 6894–6899. 10.1073/pnas.0911482107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136 669–687. 10.1016/j.cell.2009.01.046 [DOI] [PubMed] [Google Scholar]
- Wang C. Y., Zhang S., Yu Y., Luo Y. C., Liu Q., Ju C., et al. (2014). MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis. Plant Biotechnol. J. 12 1132–1142. 10.1111/pbi.12222 [DOI] [PubMed] [Google Scholar]
- Wang Y., Song F., Zhu J., Tang B., Yang Y., Chen T. (2017a). The BIG Data Center: from deposition to integration to translation. Nucleic Acids Res. 45 D18–D24. 10.1093/nar/gkw1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Song F., Zhu J., Zhang S., Yang Y., Chen T., et al. (2017b). GSA: genome sequence archive. Genomics Proteomics Bioinformatics 15 14–18. 10.1016/j.gpb.2017.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel M., Varotto C., Pesaresi P., Finazzi G., Rappaport F., Salamini F., et al. (2003). Plastocyanin is indispensable for photosynthetic electron flow in Arabidopsis thaliana. J. Biol. Chem. 278 31286–31289. 10.1074/jbc.M302876200 [DOI] [PubMed] [Google Scholar]
- Wu L., Zhou H., Zhang Q., Zhang J., Ni F., Liu C., et al. (2010). DNA methylation mediated by a microRNA pathway. Mol. Cell 38 465–475. 10.1016/j.molcel.2010.03.008 [DOI] [PubMed] [Google Scholar]
- Yamasaki H., Abdel-Ghany S. E., Cohu C. M., Kobayashi Y., Shikanai T., Pilon M. (2007). Regulation of copper homeostasis by micro-RNA in Arabidopsis. J. Biol. Chem. 282 16369–16378. 10.1074/jbc.M700138200 [DOI] [PubMed] [Google Scholar]
- Yamasaki H., Hayashi M., Fukazawa M., Kobayashi Y., Shikanai T. (2009). SQUAMOSA promoter binding protein-Like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21 347–361. 10.1105/tpc.108.060137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., He H., Wang X., Wang X., Yang X., Li L., et al. (2011). Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65 346–358. 10.1111/j.1365-313X.2010.04426.x [DOI] [PubMed] [Google Scholar]
- Zhang H., Li L. (2013). SQUAMOSA promoter binding protein-like7 regulated microRNA408 is required for vegetative development in Arabidopsis. Plant J. 74 98–109. 10.1111/tpj.12107 [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhao X., Li J., Cai H., Deng X. W., Li L. (2014). MicroRNA408 is critical for the HY5-SPL7 gene network that mediates the coordinated response to light and copper. Plant Cell 26 4933–4953. 10.1105/tpc.114.127340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Feng S., Chen F., Chen H., Wang J., McCall C., et al. (2008). Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell 20 1437–1455. 10.1105/tpc.108.058891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Nakashima J., Chen F., Yin Y., Fu C., Yun J., et al. (2013). LACCASE is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25 3976–3987. 10.1105/tpc.113.117770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. Y., Hong P., Wu J. Y., Chen X. B., Ye X. G., Pan Y. Y., et al. (2016). The tae-miR408-mediated control of TaTOC1 genes transcription is required for the regulation of heading time in wheat. Plant Physiol. 170 1578–1594. 10.1104/pp.15.01216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Li D., Li Z., Hu Q., Yang C., Zhu L., et al. (2013). Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 161 1375–1391. 10.1104/pp.112.208702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. G., Long S. P., Ort D. R. (2010). Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 61 235–261. 10.1146/annurev-arplant-042809-112206 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.