Abstract
Stimulator of interferon genes (STING) is an endoplasmic reticulum transmembrane protein that serves as a molecular hub for activation of interferon and inflammatory cytokine response by multiple cellular DNA sensors. Not surprisingly, STING has been demonstrated to play an important role in host defense against microorganisms and pharmacologic activation of STING is considered as an attractive strategy to treat viral diseases and boost antitumor immunity. In light of this we established a HepAD38-derived reporter cell line that expresses firefly luciferase in response to the activation of cyclic GMP-AMP synthase (cGAS)-STING pathway for high throughput screening (HTS) of small molecular human STING agonists. This cell-based reporter assay required only 4 h treatment with a reference STING agonist to induce a robust luciferase signal and was demonstrated to have an excellent performance in HTS format. By screening 16,000 compounds, a dispiro diketopiperzine (DSDP) compound was identified to induce proinflammatory cytokine response in a manner dependent on the expression of functional human STING, but not mouse STING. Moreover, we showed that DSDP induced an interferon-dominant cytokine response in human skin fibroblasts and peripheral blood mononuclear cells, which in turn potently suppressed the replication of yellow fever virus, dengue virus and Zika virus. We have thus established a robust cell-based assay system suitable for rapid discovery and mechanistic analyses of cGAS-STING pathway agonists. Identification of DSDP as a human STING agonist enriches the pipelines of STING-targeting drug development for treatment of viral infections and cancers.
Keywords: high throughput assay, STING, Innate immune, antiviral
1. Introduction
The genomes of vertebrate animals encode an array of proteins called pattern recognition receptors (PRRs) that recognize pathogen associated molecular patterns upon infection of microorganisms to activate a proinflammatory cytokine response (Akira et al., 2006). This innate cytokine response not only inhibits the proliferation and limits the spread of microorganisms, but also orchestrates the induction of more powerful adaptive immune response to ultimately control the microorganism infections (Chang et al., 2012; Iwasaki and Medzhitov, 2015). Stimulator of interferon genes (STING) is a transmembrane protein localized in the endoplasmic reticulum (ER) membrane and serves as a PRR for cyclic dinucleotides produced by intracellular bacteria or synthesized by the cytoplasmic DNA sensor, cyclic GMP-AMP synthase (cGAS) (Sun et al., 2013; Wu et al., 2013). Binding of the cyclic dinucleotides to STING induces its dimerization and translocation from ER membrane to perinuclear vesicles and subsequently activates NFkB and TBK-1/IRF3 (Burdette et al., 2011; Yin et al., 2012). Activation of these signaling pathways induces the expression of type I and type III interferons as well as other proinflammatory cytokines (Tanaka and Chen, 2012). In addition, STING also serves as the adaptor for several other cytoplasmic and nuclear PRRs that recognize DNA to activate innate immune responses (Chen et al., 2016; Kondo et al., 2013). Therefore, STING is a molecular hub for DNA activation of innate immune response and has been demonstrated to play an essential role in host defense against the infection of DNA viruses, retroviruses, intracellular bacteria and protozoa (Cai et al., 2014). Moreover, accumulating evidence suggests that STING also plays an important role in host anti-tumor immunity (Corrales et al., 2016).
Recently, several studies showed that activation of STING with small molecular agonists can induce cellular processes that can efficiently inhibit the replication of many DNA and RNA viruses, boost host anti-tumor immune response and enhance the immunogenicity of vaccines (Chang and Guo, 2015; Corrales et al., 2016). These studies prove the concept that pharmacologic activation of STING is an attractive immunotherapeutic approach to treat viral infection and cancer (Corrales and Gajewski, 2015) (Chang and Guo, 2015; Woo et al., 2015). In addition, STING agonists have also been developed as vaccination adjuvants to break immune tolerance against cancer cells and viruses that establish chronic infections in humans (Li et al., 2013).
Currently, there are two classes of STING agonists, cyclic dinucleotides (CDNs) and non-nucleotide small molecules. Bacteria produced cyclic-di-GMP and cyclic-di-AMP are the first STING agonists identified (Burdette et al., 2011). With the discovery of cytosolic DNA sensor cGAS, its catalytic product 2′,3′-cGAMP was identified as an even more potent STING agonist (Zhang et al., 2013). Although the various formulations of CDNs have been demonstrated to facilitate the activation of antitumor immune response in mouse models (Fu et al., 2015), their poor cell membrane permeability and metabolic instability limit their biological activity and medical applications. Accordingly, medicinal chemistry efforts have been made to produce novel CDNs that are resistant to the degradation of cellular ecto-nucleotide pyrophosphatase/phosphodiesterase (ENPP1) (Li et al., 2014; Lioux et al., 2016). In addition, delivery of CDNs with nanoparticles or liposomes improved their antitumor activities in vivo (Hanson et al., 2015). So far, there are only two non-nucleotide small molecular STING agonists, DMXAA and G10. 5,6-dimethylxanthenone-4-acetic acid (DMXAA) was initially discovered and developed as a vascular disrupting agent with antitumor activity in various mouse models, but failed in phase III clinical trials for treatment of lung cancer (Conlon et al., 2013). It was recently identified to be a specific agonist of mouse STING and induced an interferon (IFN)-dominant cytokine response to potently inhibit the replication of influenza A virus, hepatitis B virus and also alphavirus in mice (Cavlar et al., 2013; Conlon et al., 2013; Guo et al., 2015). Interestingly, a genetic study revealed that a single amino acid substitution (S162A) in human STING confers DMXAA sensitivity, which provides a clue for the synthesis of DMXAA analogues as human STING agonists (Gao et al., 2013). G10, or 4-(2-chloro-6-fluorobenzyl)-N-(furan-2-ylmethyl)-3-oxo-3,4-dihydro-2H-benzo[b][1,4]thiazine-6-carboxamide, is a recently identified human STING-specific agonist by high throughput screening. G10 had been demonstrated to induce an antiviral response in human fibroblasts against alphaviruses, but its in vivo biological activity and pharmacological property remain to be determined (Sali et al., 2015).
In order to discover small molecular STING agonists with favorable pharmacological properties as the candidates of immunotherapeutics or vaccination adjuvants for viral diseases and cancers, we set out to establish a cell-based cGAS-STING pathway reporter assay and discovered a dispiro diketopiperzine (DSDP) compound that induces proinflammatory cytokine response in a human STING-dependent manner. We have thus demonstrated the robustness and usefulness of this assay as a platform for high throughput screening of the cGAS-STING pathway agonists. In addition, we have also developed a molecular and cellular tool kit for target validation and mechanistic analysis of the identified agonists.
2. Materials and Methods
2.1 Cell lines, viruses and reagents
Human hepatoblastoma cell line HepG2 was obtained from ATCC and maintained in Dulbecco’s modified minimal essential medium (DMEM)/F12 (Corning) supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin. HepAD38 is a HepG2-derived stable cell line supporting a tetracycline (tet)-inducible replication of hepatitis B virus (HBV) and was maintained in DMEM/F12 medium supplemented with 10% fetal bovine serum, 400 μg/ml of G418 and 1 μg/ml tetracycline (Ladner et al., 1997). HepAD38/cGAS-STING and HepAD38/cGAS-STINGΔC are HepAD38-derived cell line constitutively expressing human cGAS and STING or a mutant STING with deletion of 39 amino acid residues from carboxyl terminus (STINGΔC) and were maintained in DMEM/F12 medium supplemented with 10% fetal bovine serum, 400 μg/ml of G418, 2 μg/ml of puromycin and 1 μg/ml tetracycline (Guo et al., 2017). Vero (green monkey kidney) cells were maintained in DMEM (Corning) supplemented with 10% fetal bovine serum. THF cells are derived from primary human diploid foreskin fibroblasts (HFF) with extended passage life through expressing of a cDNA encoding the catalytic subunit of human telomerase and were maintained in DMEM with 10% fetal bovine serum (Bresnahan et al., 2000). Human Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Paque density gradient centrifugation (Miltenyi Biotech) from the whole blood of healthy donors (Biological Specialty). Freshly isolated PBMCs were cultured in RPMI-1640 medium (Corning) supplemented with L-Glutamine and 10% fetal bovine serum.
Sendai virus (SenV, strain 52) and human serum derived PRVABC59 strain of Zika virus (ZIKV) were purchased from ATCC. A plasmid containing the yellow fever virus (YFV) 17D complete genomic complementary DNA (cDNA), pACNR/FLYF-17Dx was a gift of Dr. Charles M. Rice at Rockefeller University (Bredenbeek et al., 2003; Rice et al., 1985). A plasmid containing the dengue virus (DENV) serotype 2 New Guinea C strain cDNA (pACYC177-NGC-DENV-2) was a gift of Dr. Pei-Yong Shi at University of Taxis Medical Branch in Galveston (Xie et al., 2013). YFV 17D and DENV-2 virus stocks were produced by electroporation of Huh7.5 cells with in vitro transcribed RNAs from the corresponding cDNA constructs, as described previously (Guo et al., 2016).
2′3′-cGAMP, Pam3CSK4, Poly I:C, LPS and Gardiquimod were purchased from Invivogen. G10, a small molecule human STING agonist (Sali et al., 2015), served as a control compound and was purchased from Aobious. Double strand DNA 90 (dsDNA-90) was prepared as previously described (Guo et al., 2017). Dispiro diketopiperzine (DSDP) was purchased from Maybridge. Mouse STING agonist 5,6-dimethylxanthenone-4-acetic acid (DMXAA) was purchased from AdooQ BioScience.
2.2 Establishment of reporter cell lines for high throughput screening
HepAD38/cGAS-STING-derived reporter cell line that expresses firefly luciferase under the control of an ISG54 promoter (HepAD38/cGAS-STING/ISG54Luc) were established by transduction of pGreenFire ISRE lentivector-based transcription reporter system following the manufacturer’s protocol (System Biosciences). The pGreenFire ISRE reporter lentiviral vector contains four copies of consensus interferon stimulated response element (ISRE) sequences derived from ISG54 ISRE1, which control the expression of both green fluorescent protein (GFP) and firefly luciferase in response to IRF3 activation and IFN stimulation (Levy et al., 1988). As a negative control, a HepAD38/cGAS-STINGΔC-derived ISG54 promoter reporter cell line (HepAD38/cGAS-STINGΔC/ISG54Luc) was also established using the same pGreenFire ISRE lentivector system.
2.3 High throughput screening assay
HitFinder library containing 16,000 small molecule compounds was screened (Maybridge). HepAD38/cGAS-STING cell line was originally established for investigation of HBV interaction with cGAS-STING DNA sensor pathway (Guo et al., 2017). However, for screening of cGAS and or STING agonists, the HepAD38/cGAS-STING/ISG54 reporter cell line was cultured in complete DMEM/F12 medium containing 1μg/ml tet throughout the experimental period, which completely prevents HBV replication (Ladner et al., 1997). Briefly, HepAD38/cGAS-STING/ISG54Luc cells were seeded in black wall/clear bottom 96-well plates at a density of 4×104/well in 1 mL of medium with 1μg/mL of tet overnight. For each plate, column 1 wells were mock-treated with 2% DMSO, and the column 12 wells were treated with 20 μM of G10 to serve as negative and positive controls, respectively. Each of the well in columns 2 to 11 was treated with a library compound at 20 μM with 2% DMSO. The firefly luciferase activities were determined at 4 h post treatment by using equal volume of Steady-Glo (Promega), followed by luminometry in a TopCounter (Perkin Elmer). Typically, forty to fifty plates containing 3,200 to 4,000 of library compounds were tested in a single experiment. In addition, a control plate, with half of the plate treated with 20 μM of G10 and half of the plate mock-treated, was included in each experiment to calculate Z′ value for that batch of experiment.
The compounds that induced luciferase by greater than 3-fold and Z score of greater than 5 were scored as primary “hits”. The primary “hits” were subjected to further evaluation of dose-dependent effect on luciferase activity in HepAD38/cGAS-STING/ISG54Luc and HepAD38/cGAS-STINGΔC/ISG54Luc cells. The compounds that dose-dependently enhanced luciferase expression with induction greater than 10-fold at the highest dose (100 μM) in HepAD38/cGAS-STING/ISG54Luc, but not in cGAS-STINGΔC/ISG54Luc cells were considered as confirmed agonist “hits” of cGAS-STING pathway.
2.4 Establishment of HepG2-derived cell lines stably expressing human and mouse STING
HepG2 cells stably expressing wild-type human STING (HepG2/STING), a mutant STING with deletion of 39 amino acid residues from carboxyl terminus (HepG2/STINGΔC), or mouse STING (HepG2/mSTING) were established by transduction of pCX4bsr retroviral vector (Addgene) containing the corresponding STING cDNA and selection with 10 μg/ml of blasticidin for two weeks. The HepG2/STING cells were also transduced by pGreenFire ISRE lentivector system (System Biosciences) to generate a reporter cell line HepG2/STING/ISG54Luc.
2.5 MTT assay
To determine the cell viability, a MTT assay (Sigma) was performed as described previously (Chang et al., 2009). Cells were treated with various concentrations of test compounds to determine the concentration that resulted in 50% cell death (CC50).
2.6 Analysis of cytokine gene expression by qRT-PCR assays
Total cellular RNAs were extracted using TRIzol (Invitrogen). IFN-β mRNA, TNFα mRNA, IL-28A, IL-29 mRNA and IL-6 mRNA were measured by quantitative one-step RT-PCR (qRT-PCR) assays using primers reported previously (Guo et al., 2017). β-actin mRNA served as internal control to normalize cytokine mRNAs.
2.7 YFV, DENV and ZIKV infection and antiviral assays
THF cells seeded in 96-well plate were mock-treated or pre-treated with indicated STING agonists for 8 h and then infected with YFV, DENV or ZIKV at a multiplicity of infection (MOI) of 0.1 for 1 h and cultured for 48 h. Total cellular RNA was extracted by using NucleoSpin 96 RNA kit (Macherey-Nagel). The amounts of YFV, DENV or ZIKV RNA were determined by one step qRT-PCR assays on LightCycler 480II (Roche). β-actin mRNA was quantified to normalize the levels of viral RNA. Primers for YFV were as reported previously (Guo et al., 2016). Primers for DENV are as the following: forward primer 5′-CAGGCTATGGCACTGTCACGAT-3′; reverse primer 5′-CCATTTGCAGCAACACCATCTC-3′. Primers for ZIKV are as the following: forward primer 5′-CCGCTGCCCAACACAAG-3′; reverse primer 5′-CCACTAACGTTCTTTTGCAGACAT-3′. Virus yield reduction assay was performed to measure YFV, DENV and ZIKV titers in culture media. Briefly, monolayers of Vero cells in 24-well plates were infected for 1 h with a serial of 10-fold dilutions of the culture media harvested from infected THF cells followed by overlay with media containing 0.75% methylcellulose and incubated at 37°C for 3 to 5 days. Plaques were either counted directly or after crystal violet staining (Chang et al., 2013).
2.8 Western blot assay
Cells grown in 24-well plate were lysed with 100μL NuPAGE® LDS sample buffer (Thermo Fisher Scientific) supplemented with 2.5% 2-Mercaptoethanol (Sigma). Cell lysate was subjected to denaturing gel electrophoresis with NuPAGE 4–12% Bis-Tris Gel and NuPAGE MOPS SDS Running Buffer (Thermo Fischer Scientific). Proteins were transferred onto a PVDF membrane using iBlot 2 Dry Blotting System (Thermo Fischer Scientific). Membranes were blocked with TBST (50 mM Tris-HCl, pH 7.6; 150 mM NaCl and 0.1% Tween 20) containing 5% nonfat milk for 1 h and incubated with an antibody against cGAS, STING, IRF3, phosphorylated IRF3 or β-actin (Cell Signaling) for overnight at 4°C followed by incubation with LI-COR® IRDye® secondary antibodies and imaged with LI-COR Odyssey system (LI-COR Biotechnology).
2.9 Immunofluorescent assay
HepG2/STING and HepG2/STING C cells were mock-treated or treated with the indicated concentrations of G10 or DSDP for 2 h. The cells were fixed with PBS containing 4% paraformaldehyde followed by incubation with 0.1% Triton X-100 for 20 min. Cells were then blocked and incubated with an antibody against IRF3 (Cell Signaling). Bound primary antibody was visualized by Alexa Fluor 488-conjugated secondary antibody (Invitrogen) (Zhu et al., 2003). Cell nuclei were stained with DAPI.
2.10 Statistics
Z′ factor (Z′) was calculated using the following equation: Z′=1−((3δp+3δn)/|μp−μn|), where μp and μn represent mean values of positive control wells and negative control wells; δp and δn are the standard deviations (Zhang et al., 1999). Coefficient of variation (CV) was calculated using the following equation: CV(%)=δ/μ*100. Z score from each individual test compound was calculated using the following equation: Z score= (t−μn)/ δn where t represents value from the test compound. Ratio of signal to background, a parameter to represent signal dynamic was calculated as the following: S/B= μp/μn Ratio of signal to noise, a factor to represent signal strength was calculated as the following: S/N=(μp−μn)/ δn. The correlation of replicate plates was assessed by pair wise Pearson correlation analysis. P values were calculated using 2-tailed student’s t-test.
3 Results
3.1 Establishment and characterization of cGAS-STING reporter cell lines
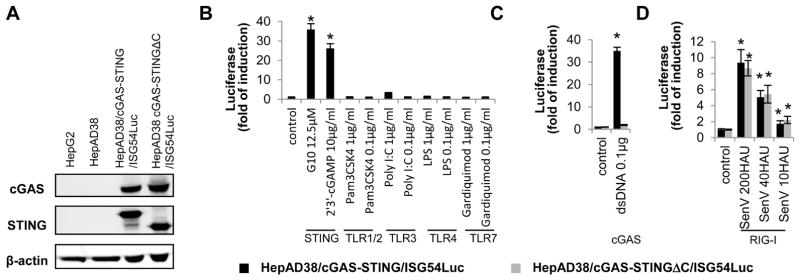
In order to establish a highly sensitive cell-based assay for high throughput screening of small molecular compounds that activate human STING, we first established a HepAD38-derived cell line that constitutively expresses human cGAS and STING, which successfully reconstituted a functional cytoplasmic DNA sensor pathway (Guo et al., 2017). Next, we introduced human ISG54 promoter-driven firefly luciferase expression cassette into the HepAD38/cGAS-STING cell line via lentiviral vector transduction and obtained the cell line HepAD38/cGAS-STING/ISG54Luc. Meanwhile, a reporter cell line, HepAD38/cGAS-STING C/ISG54Luc, that expresses a truncated STING with deletion of 39 amino acid residues from carboxyl terminus, was also established (Fig. 1A). Because the C-terminal domain of STING is essential for activation of IRF3 and subsequent induction of cytokines, this later reporter cell line serves as a negative control (Tanaka and Chen, 2012). Indeed, as shown in Fig. 1B, treatment with agonists of STING, such as 2′3′-cGAMP or G10, but not the agonists of TLR1/2, TLR3, TLR4 or TLR7, efficiently activated luciferase expression in HepAD38/cGAS-STING/ISG54Luc cells, but not in HepAD38/cGAS-STINGΔC/ISG54Luc cells. Furthermore, as anticipated, transfection of dsDNA90, a well known cGAS ligand (Abe et al., 2013), only induced luciferase expression in HepAD38/cGAS-STING/ISG54Luc, but not in HepAD38/cGAS-STINGΔC/ISG54Luc cells (Fig. 1C). The results thus indicate that ISG54 driven luciferase expression can report cGAS and/or STING activation in the HepG2 derived cell lines by their agonists. However, consistent with previous reports that hepatoma cells have an intact RIG-I/MDA5 mediated cytosolic RNA sensing pathway (Israelow et al., 2014), infection of the two reporter cell lines with Sendai virus, an RNA virus that is known to activate RIG-I, significantly induced luciferase expression in both reporter cell lines expressing wild-type or C-terminally truncated STING in a dose-dependent manner (Fig. 1D). In summary, these results indicate that the HepAD38/cGAS-STING/ISG54Luc cell line can be used for identification of compounds that activate cGAS-STING and RIG-I/MDA5 pathways, and the differential effects of the agonists in the two reporter cell lines expressing functional or dysfunctional human STING should distinguish which of the two innate immune pathways are targeted.
Figure 1. Activation of ISG54 promoter in HepAD38/cGAS-STING/ISG54Luc and HepAD38/cGAS-STINGΔC/ISG54Luc reporter cells by a panel of PRR agonists.
The expression of cGAS and STING in HepG2, HepAD38, HepAD38/cGAS-STING/ISG54Luc and HepAD38/cGAS-STINGΔC/ISG54Luc reporter cells were detected by Western blot assay using β-actin as loading control (A). The two reporter cells grown in 96-well plates were mock treated, or treated with indicated amount of STING, TLR1/2, TLR3, TLR4 or TLR7 agonists for 4 h (B), transfected with dsDNA 90 for 8 h (C), or infected with SenV for 24 h (D). ISG54 promoter activities were expressed as fold of induction of luciferase activity relative to mock treated controls (mean ± standard deviation, n≥3). * indicates p<0.05 compared to mock treated control.
3.2 Performance of HepAD38/cGAS-STING/ISG54Luc cell-based high throughput screening assay
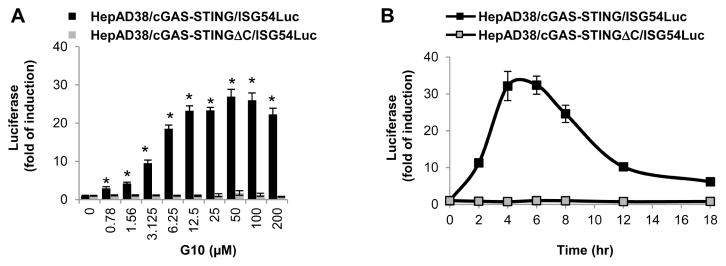
Having demonstrated that HepAD38/cGAS-STING/ISG54Luc cells could sensitively report the activation of cGAS-STING pathway by small molecules (Fig. 1B), we first determined the activation dynamics of the cell line by STING agonists in a high throughput format. As shown in Fig. 2A, under the condition of 4 h treatment, the highest induction of luciferase by G10 was achieved between concentrations of 12.5 to 50 μM. A time-course study indicated that 20 μM G10 treatment induced the maximum level of luciferase expression between 4 to 6 h (Fig. 2B). As expected, no significant induction of luciferase expression by G10 in HepAD38/cGAS-STINGΔC/ISG54luc cells was observed. Accordingly, we determined that the compound concentration and treatment duration for HTS assay should be 20 μM and 4 h, respectively. Such a short term treatment protocol not only allows for completion of the screening procedure within one day, but also limits the potential false negative or positive results due to long-term treatment associated cytotoxicity.
Figure 2. Optimization of high throughput assay.
HepAD38/cGAS-STING/ISG54Luc and HepAD38/cGAS-STINGΔC/ISG54Luc cells grown in 96-well plates were mock treated or treated with a serial of doses of G10 for 4 h (A). Alternatively, the two cell lines were treated with 20 μM of G10. The luciferase activities were measured before (0 h) or at the indicated time post treatment (B). ISG54 promoter activities were expressed as fold of induction of luciferase activity relative to 0 h (mean ± standard deviation, n=6). * indicates p<0.05 compared to mock treated control.
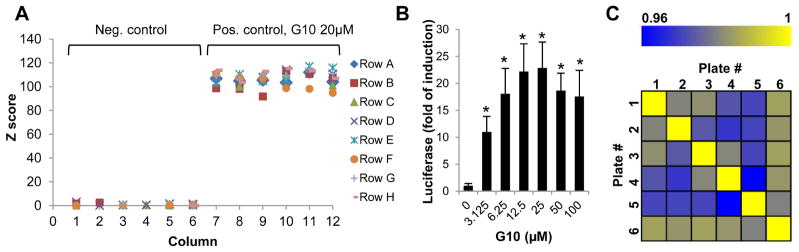
We next determined the key performance parameters of HepAD38/cGAS-STING/ISG54Luc reporter assay in a 96-well plate HTS format. Treatment of one-half plate of the reporter cells with 20 μM of reference compound G10 or 2% DMSO for 4 h and luciferase activities were determined at 5, 30 and 60 min post addition of luciferase substrate. At different time of plate reading, no significant difference in signal-to-background ratio (S/B), signal to noise ratio (S:N), CV% or Z′ was observed (Table 1). As shown in Fig. 3A, using data obtained at 60 min post substrate addition, the positive control wells treated with G10 had a Z score value of greater than 100. The Z′ of the assay is ~0.8, indicating its high signal dynamic and low variability.
Table 1.
Effect of plate reading time on high throughput assay performance
| Plate reading time | S/B | S/N | %CV neg.* | %CV pos.# | Z′ |
|---|---|---|---|---|---|
| 5 min | 19.8 | 103.8 | 18.1 | 6.3 | 0.77 |
| 30 min | 20.0 | 105.8 | 17.9 | 6.0 | 0.78 |
| 60 min | 19.8 | 107.5 | 17.5 | 5.1 | 0.81 |
CV% of mock treated control wells
CV% of G10 treated wells
Figure 3. Characterization of high throughput assay using control plates and hypothetical replicate plates.
(A) In each batch of experiment, a plate of HepAD38/cGAS-STING/ISG54Luc cells was used as control plate with 6 columns of the wells treated with 2% DMSO (n=48) and 6 columns of the wells treated with 20μM of reference compound G10 (n=48). The firefly luciferase activities were measured in parallel with all the other test plates at 4 h post treatment. The Z scores were calculated and shown for a representative control plate. (B and C) HepAD38/cGAS-STING/ISG54Luc cells were seeded in 6 of 96-well plate. Each plate had 6 columns of the wells treated with 2% DMSO (n=48 per plate) and the other columns of the wells treated with 6 increasing concentrations of G10 (n=8 per dose, per plate). The average luciferase value in treated wells relative to control wells (fold of induction) were calculated from 6 plates, and expressed as mean ± standard deviation. * indicates p<0.05 compared to mock treated control (B). Pairwise Pearson’s coefficient of data on 6 hypothetical replicate plates was shown in a correlation heat map (C).
In order to further determine the assay reproducibility, in each of six replicate plates seeded with HepAD38/cGAS-STING/ISG54Luc reporter cells, half of the wells were mock treated with 2% DMSO and half of the wells were treated with a serial concentrations of G10 for 4 h. Fig. 3B showed the average and standard deviation of the luciferase induction by G10 from the six replicate plates. Pearson’s coefficient was determined ranging from 0.96 to 1.0 among the replicate plates (Fig. 3C), suggesting a high reproducibility of the assay under the selected experimental condition.
Taken together, HepAD38/cGAS-STING/ISG54Luc reporter cell line can be used for high throughput discovery of cGAS-STING agonists. The assay is quite robust and can be completed within 4 h of treatment with acceptable performance in HTS format.
3.3 Discovery of a dispiro diketopiperzine compound as cGAS-STING pathway activator
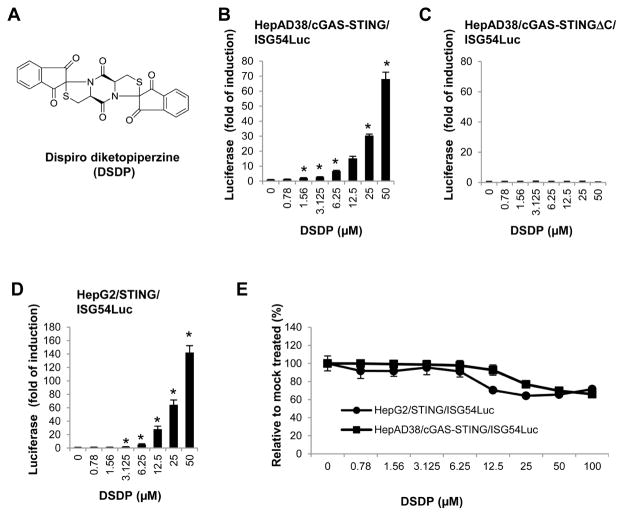
Screening a library of 16,000 small molecule compounds was carried out under the established HTS experimental condition described above. Raw luciferase values reporting the activation of ISG54 promoter activity were normalized as fold of induction relative to mock treated controls (n=8) within each assay plate. In addition, a Z score value, which indicates how many standard deviations a test value is from the mean of mock treated controls, was also calculated. Ten compounds that induced luciferase expression by greater than 3-fold and Z score of greater than 5 were scored as primary “hits”. The primary “hits” were subjected to further evaluation of dose-dependent effect (from 0.78 to 100μM) on luciferase expression in HepAD38/cGAS-STING/ISG54Luc and HepAD38/cGAS-STINGΔC/ISG54Luc cells. Five compounds that dose-dependently induced luciferase expression with peak induction greater than 10-fold in HepAD38/cGAS-STING/ISG54, but not in cGAS-STINGΔC/ISG54Luc cells were considered as confirmed STING pathway agonists. The potency of these compounds was ranked by their concentrations required for 5- and 10-fold induction of luciferase activity. Among these five confirmed “hits”, a dispiro diketopiperzine compound, 2,7,2″,2″-dispiro[indene-1″,3″-dione]-tetrahydrodithiazolo[3,2-a:3′,2′-d]pyrazine-5,10(5aH,10aH)-dione (DSDP) (Fig. 4A), most potently induced the ISG54 promoter activity in HepAD38/cGAS-STING/ISG54 cells and was selected for further investigation. As shown in Fig. 4B and C, DSDP treatment induced a concentration-dependent luciferase expression and reached 70-fold induction at 50μM concentration in HepAD38/cGAS-STING/ISG54Luc cells, but failed to induce luciferase expression at any of the tested concentrations in cGAS-STINGΔC/ISG54Luc cells. The results thus suggested that DSDP is most likely a cGAS/STING pathway activator.
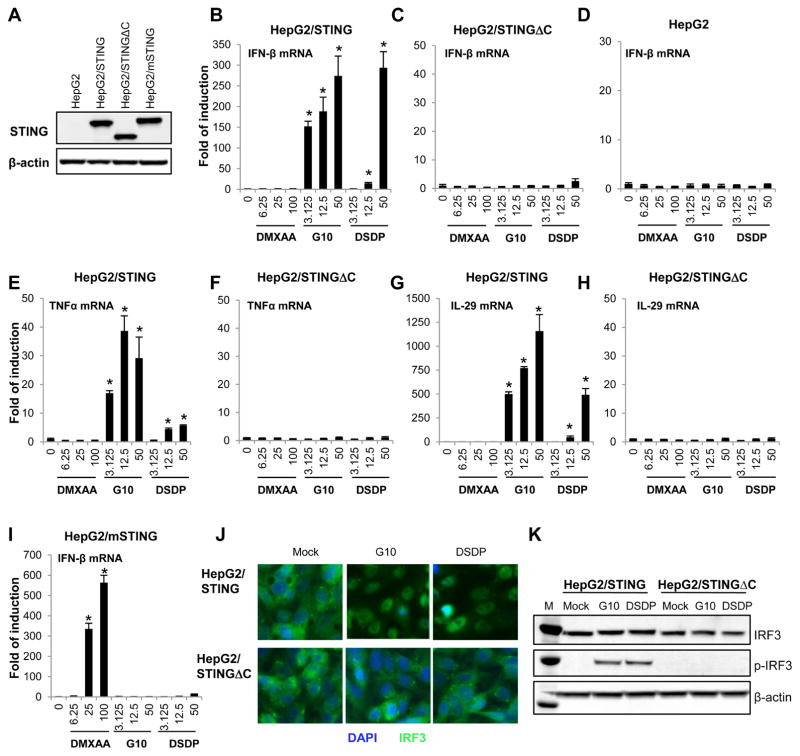
Figure 4. DSDP activated ISG54 promoter activity in HepG2 based cell lines with reconstituted human STING, but not the truncated STING.
(A) Chemical structure of DSDP. Dose-dependent activation of ISG54 promoter activities by DSDP were measured in HepAD38/cGAS-STING/ISG54Luc cells (B) HepAD38/cGAS-STINGΔC/ISG54Luc cells (C) and HepG2/STING/ISG54Luc cells (D). ISG54 promoter activities were expressed as fold of induction of luciferase activity relative to mock treated controls (mean ± standard deviation, n=3). * indicates p<0.05 compared to mock treated control. (E) An MTT assay was performed in HepAD38/cGAS-STING/ISG54Luc and HepG2/STING/ISG54Luc cells. Values represented absorbance relative to mock treated controls (mean ± standard deviation, n=3).
To further investigate whether DSDP activation of the ISG54 promoter depends on cGAS, HepG2/STING/ISG54 reporter cells lacking cGAS were treated with a serial concentration of DSDP. As shown in Fig. 4D, DSDP dose-dependently induced ISG54 driven luciferase expression in a similar fashion as that in HepAD38/cGAS-STING/ISG54 reporter cells, with peak induction of 140-fold at 50μM. In both the reporter cell lines, the CC50 values of DSDP were greater than 100μM. These results thus suggest that DSDP activated a cellular component that is downstream of cGAS, but at or upstream of STING.
3.4 DSDP activation of interferon-dominant cytokine expression requires a functional human STING
To further validate the biological activity of DSDP, its activity on induction of inflammatory cytokines was investigated in parental HepG2 and HepG2-derived cells lines expressing wild-type or C-terminally truncated human STING, or mouse STING (Fig. 5A). Consistent with results obtained with ISG54 reporter assays, both G10 and DSDP dose-dependently induced IFN-β, IL29 (IFN-λ1) and TNF-α mRNA expression in HepG2/STING cells, but not in parental HepG2 and HepG2/STINGΔC cells (Fig. 5B to H). As anticipated, the mouse STING agonist DMXAA did not induce noticeable IFN-β, IL29 and TNF-α mRNA expression in parental HepG2, HepG2/STING or HepG2/STINGΔC cells (Fig. 5B to H), but induced a robust IFN response in HepG2/mSTING cells (Fig. 5I). However, Similar to G10, a human STING specific agonist, DSDP failed to activate IFN-β gene expression in HepG2/mSTING cells (Fig. 5I). Hence, our results suggest that like G10, DSDP induces an IFN-dominant proinflammatory cytokine response in human STING-dependent manner.
Figure 5. DSDP induced the expression of cytokines in HepG2 based cell lines with reconstituted human STING, but not human STING with C-terminal truncation or mouse STING.
The expression of STING in HepG2, HepG2/STING, HepG2/STINGΔC and HepG2/mSTING cells were detected by Western blot assay using an antibody cross-reactive with both human and mouse STING. β-actin served as loading control (A). The induction of IFN-β mRNA were detected after 6 h treatment with indicated concentrations of compounds (DMXAA, G10 and DSDP in μM) in HepG2/STING cells expressing wild-type human STING (B), HepG2/STINGΔC cells expressing human STING with C-terminal truncation (C) and wild-type HepG2 cells (D). Similarly, the induction of TNFα and IL-29 mRNA were measured in HepG2/STING cells (E and G) or HepG2/STINGΔC cells (F and H). The induction of IFN-β mRNA was detected in HepG2/mSTING expressing mouse STING (I). The mRNA levels of the cytokines were measured by qRT-PCR assay using β-actin as internal control and expressed as fold of induction relative to mock treated control (mean ± standard deviation, n=4). * indicates p<0.05 compared to mock treated control. (J and K) HepG2/STING and HepG2/STINGΔC cells were mock treated or treated with 50μM of either DSDP or G10 for 2 h, followed by immune staining of IRF3 () (J) and Western blot assay to detect the total and phosphorylated IRF3(K).
To further confirm the mechanism underlying the G10 and DSDP activation of innate immune response, subcellular localization of IRF3 was determined by immune fluorescence assay. As shown in Fig. 5J and K, both G10 and DSDP treatment induced IRF3 nuclear translocation and phosphorylation in HepG2/STING cells, but not in HepG2/STINGΔC cells. The results thus indicate that G10 and DSDP activate IRF3 in a functional STING dependent manner.
3.5 DSDP induced the expression of cytokines in human fibroblasts and peripheral blood mononuclear cells
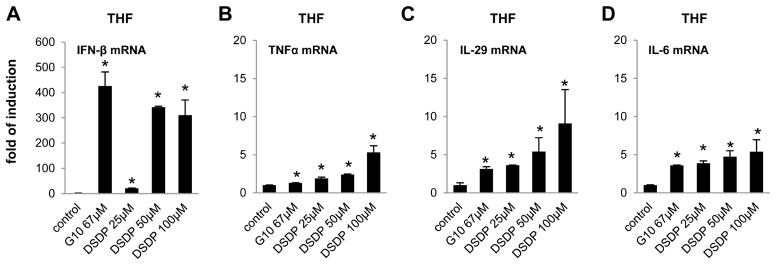
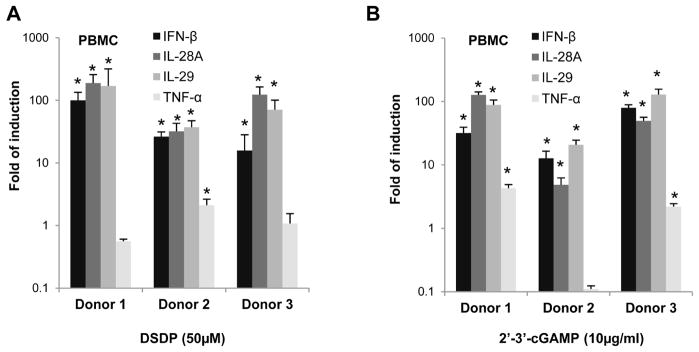
In order to determine whether DSDP can activate STING pathway in cells expressing physiologically relevant levels of STING, we examined its effects on proinflammatory gene expression in THF cells, a cell line derived from primary human diploid foreskin fibroblasts (HFF) with extended passage life, as well as human peripheral blood mononuclear cells (PBMCs). As shown in Fig. 6, similar to G10, DSDP treatment of THF cells for 6 h dose-dependently induced the mRNA expression of IFN-β, and to a lesser extent, IL-29, TNF-α, and IL-6. Also similar to 2′3′-cGAMP, the endogenous ligand of STING, DSDP treatment of PBMCs derived from three different donors efficiently induced mRNA expression of both type I (IFN-β) and type III (IL-28A and IL-29) IFNs, but not TNF-α (Fig. 7).
Figure 6. DSDP induced the expression of cytokines in human fibroblast cell line THF.
THF cells were seeded in 12 well plate, and left mock treated or treated with serial of doses of DSDP or control compound G10 for 6 h. Total cellular RNAs were extracted to detect the mRNA of IFN-β (A), TNFα (B), IL-29 (C), and IL-6 (D) by qRT-PCR. Values represented fold of induction relative to mock treated control (mean ± standard deviation, n=4). * indicates p<0.05 compared to mock treated control.
Figure 7. DSDP induced the expression of cytokines in human PBMCs.
2×106 of PBMCs freshly isolated from human blood of three healthy donors were seeded in 12-well plate, and left mock treated or treated with 50μM of DSDP (A) or 10μg/ml of 2′3′-cGAMP (B) for 6 h. Total cellular RNAs were extracted to detect the mRNA of IFN-β, IL-28A, IL-29 and TNFα by qRT-PCR. Values represented fold of induction relative to mock treated control (mean ± standard deviation, n=4). * indicates p<0.05 compared to mock treated control.
3.6 DSDP inhibited the replication of multiple flaviviruses in THF cells
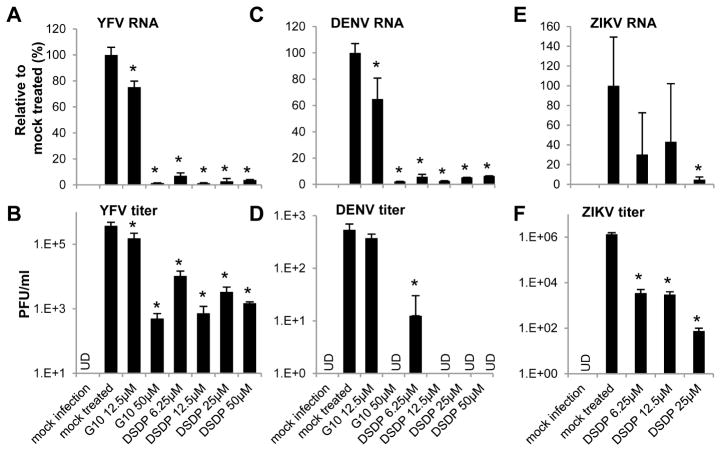
Finally, to investigate whether DSDP activation of human STING pathway induces an antiviral response, THF cells were mock-treated or pre-treated with the indicated concentrations of DSDP for 8 h. The cells were then infected with YFV, DENV or ZIKV at a MOI of 0.1. Forty eight hours post infection, intracellular viral RNA and virus yields in culture media were determined by qRT-PCR and plaque assays, respectively. As shown in Fig. 8, DSDP treatment significantly reduced the intracellular YFVand DENV RNA levels at concentration as low as 6.25 μM or at 25 μM for ZIKV (Fig. 8A, C and E). Consistent with the inhibition of viral RNA replication, the virus titers were reduced by at least 1 log at 6.25 μM, and 2 to 4 logs at higher concentrations of DSDP treatment (Fig. 8B, D and F).
Figure 8. DSDP inhibited the replication of Yellow fever, Dengue and Zika viruses in THF cells.
THF cells were left untreated or treated with indicated concentrations of DSDP or control compound G10 for 8 h. Cells were then infected with YFV, DENV, or ZIKV at MOI of 0.1 for 1 h followed by culture for additional 48 h. Total cellular RNAs were extracted to detect viral genomic RNA by qRT-PCR using β-actin as internal control (A, C and E). Values represented percentage to mock treated controls (mean ±standard derivations, n=3). Culture media were harvested and the virus titers (B, D and F) were determined by plaque assay and presented as number of plaque forming units per milliliter medium (PFU/ml) (mean ±standard derivations, n=3). UD, under detection limit. * indicates p<0.05 compared to mock treated control.
4. Discussion
Pharmacologic activation of STING has been shown to induce interferon and cytokine responses that efficiently inhibited virus replication and attenuated viral disease progress in mouse models (Guo et al., 2015; Sali et al., 2015; Shirey et al., 2011). It has also been demonstrated that cGAS-STING pathway is important for intrinsic antitumor immunity and treatment of solid tumors with variety of STING agonist formulations activated dendritic cells and enhanced cross-presentation of tumor associated antigens to CD8 T cells, which resulted in the suppression of tumor growth and improvement of tumor-bearing animal survival (Corrales et al., 2015). Moreover, cGAS-STING pathway-mediated immune activation also plays an important role in gamma-radiation treatment of cancers (Deng et al., 2014). Interestingly, it was shown recently that cGAS is indispensable for the antitumor effect of immune checkpoint blockade in mice. Wild-type, but not cGAS-deficient, mice exhibited slower growth of B16 melanomas in response to a PD-L1 antibody treatment. Consistently, the combination of cGAMP and PD-L1 antibody exerted stronger antitumor effects than did either treatment alone (Corrales et al., 2015). In addition to their antiviral and anti-tumor activities, activation of cGAS and STING also promotes vaccination induced immune response (Wang et al., 2017). Hence, the agonists of cGAS-STING pathway hold a great promise for treatment of viral infections and cancers, either as immunotherapeutics to activate intrinsic host antiviral and antitumor immune response or adjuvants of therapeutic vaccinations to break host immune tolerance against chronically infecting viruses and tumor cells.
However, despite the great promise, there are only very limited chemotypes of STING agonists in the development pipelines and discovery of novel chemotypes of cGAS-STING pathway agonists with improved pharmacological property should facilitate the development of this therapeutic strategy toward its clinical use. In this study, we established a HepAD38-based luciferase reporter assay for HTS discovery of novel agonists of cGAS-STING pathway. Systematic evaluation of the assay’s performance in HTS format demonstrated its high signal dynamic range in response to STING agonist treatment and low variability. The assay is robust and only 4 h of compound treatment is needed to induce a significant reporter luciferase signal. It is worth noting that in the reporter cells the ISG54 ISRE1 promoter controls the expression of both GFP and firefly luciferase, and therefore, although not relevant to the current screening, should preclude any GFP-based readouts in parallel. In addition to cGAS-STING pathway, the HepAD38-derived reporter cell line also has a functional RIG-I/MDA5 pathway, but is not responsive to all the TLR agonists tested. The primary screening with HepAD38/cGAS-STING/ISG54Luc cells should not only identify the agonists of cGAS-STING pathway, but also of the RLR pathway. The later are also valuable candidates of immunotherapeutics for viral infections.
In order to categorize the primary “hits” and identify their molecular targets, all the primary “hits” were counter-screened with HepAD38/cGAS-STINGΔC/ISG54Luc cells. The compounds only active in HepAD38/cGAS-STING/ISG54Luc cells most likely activate STING or cellular components upstream of STING in the cGAS-STING pathway. On the contrary, the compounds active in both cell lines most likely activate RLR or downstream shared signaling components by STING and RLR pathways. To further determine if the compounds activate cGAS or cellular components downstream of cGAS, we also evaluated their activity to induce interferons and cytokines in HepG2-derived cells expressing human STING or mouse STING. A compound that only induces the cytokines in human STING-expressing cells strongly indicates its direct targeting of human STING. Through screening 16,000 compounds, one compound, DSDP, fits this criterion and is thus a specific human STING agonist. Our results indicate that the primary screening assay is highly specific and has very low “hit” rate and false positive rate. This greatly reduced the burden of “primary hit” validation and categorization.
Finally, because our primary screening and “hit” validation/categorization were performed with hepatoma cells ectopically expressing cGAS and/or wild-type and mutant STING, the validated cGAS-STING pathway agonists should be further confirmed in cells that express physiological levels of STING. In this study, the ability of DSDP to activate interferon and cytokine response and induce antiviral state was validated in human fibroblasts and peripheral blood mononuclear cells derived from three individuals. The slight difference in cytokine induction profiles between HepG2/STING and THF cells could simply due to the difference in the levels of cGAS/STING expression or other cellular components in the cGAS-STING signaling pathway between the two cell lines. The quantitative difference in cytokine induction and kinetics between DSDP and a reference compound G10 could be due to many reasons. Most possibly, the two compounds may bind and activate STING via distinct mechanisms. Taken together, we have developed a robust assay system for HTS discovery and validation of the agonists of human cGAS-STING pathway. Identification of DSDP as a human STING agonist enriches the pipeline of STING-targeting drug development and provides a chemical probe for investigating the molecular mechanism of STING activation and biological functions.
Highlights.
A robust cell-based reporter assay was established for high throughput screening of cGAS-STING pathway agonists
A serial of molecular and cellular tools were developed for target validation and mechanistic analysis of the identified agonists
A dispiro diketopiperzine compound DSDP was identified as human STING agonist from screening of 16,000 compounds
DSDP induced an interferon-dominant cytokine response and suppressed the replication of flaviviruses
Acknowledgments
We thank Wade Bresnahan at University of Minnesota for providing THF cells and Eain Murphy at Forge Life Science for providing the early passage of HFF cells. A cDNA clone of serotype 2 dengue virus, pACYC177-NGC-DENV-2, was a gift from Pei-Yong Shi at University of Texas Medical Branch. A cDNA clone of Yellow fever virus 17D strain, pACNR/FLYF-17Dx, was a gift from Charles Rice at Rockefeller University. This work was supported by grant from the National Institutes of Health, USA (AI113267), Arbutus Biopharma Inc. and the Commonwealth of Pennsylvania through the Hepatitis B Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe T, Harashima A, Xia T, Konno H, Konno K, Morales A, Ahn J, Gutman D, Barber GN. STING recognition of cytoplasmic DNA instigates cellular defense. Mol Cell. 2013;50:5–15. doi: 10.1016/j.molcel.2013.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Bredenbeek PJ, Kooi EA, Lindenbach B, Huijkman N, Rice CM, Spaan WJ. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J Gen Virol. 2003;84:1261–1268. doi: 10.1099/vir.0.18860-0. [DOI] [PubMed] [Google Scholar]
- Bresnahan WA, Hultman00 GE, Shenk T. Replication of wild-type and mutant human cytomegalovirus in life-extended human diploid fibroblasts. J Virol. 2000;74:10816–10818. doi: 10.1128/jvi.74.22.10816-10818.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Chiu YH, Chen ZJ. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol Cell. 2014;54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- Cavlar T, Deimling T, Ablasser A, Hopfner KP, Hornung V. Species-specific detection of the antiviral small-molecule compound CMA by STING. EMBO J. 2013;32:1440–1450. doi: 10.1038/emboj.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Block TM, Guo JT. The innate immune response to hepatitis B virus infection: implications for pathogenesis and therapy. Antiviral Res. 2012;96:405–413. doi: 10.1016/j.antiviral.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Chang J, Guo JT. Treatment of chronic hepatitis B with pattern recognition receptor agonists: Current status and potential for a cure. Antiviral Res. 2015;121:152–159. doi: 10.1016/j.antiviral.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Wang L, Ma D, Qu X, Guo H, Xu X, Mason PM, Bourne N, Moriarty R, Gu B, Guo JT, Block TM. Novel imino sugar derivatives demonstrate potent antiviral activity against flaviviruses. Antimicrob Agents Chemother. 2009;53:1501–1508. doi: 10.1128/AAC.01457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Warren TK, Zhao X, Gill T, Guo F, Wang L, Comunale MA, Du Y, Alonzi DS, Yu W, Ye H, Liu F, Guo JT, Mehta A, Cuconati A, Butters TD, Bavari S, Xu X, Block TM. Small molecule inhibitors of ER alpha-glucosidases are active against multiple hemorrhagic fever viruses. Antiviral Res. 2013;98:432–440. doi: 10.1016/j.antiviral.2013.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol. 2016;17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- Conlon J, Burdette DL, Sharma S, Bhat N, Thompson M, Jiang Z, Rathinam VA, Monks B, Jin T, Xiao TS, Vogel SN, Vance RE, Fitzgerald KA. Mouse, but not human STING, binds and signals in response to the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid. Journal of immunology. 2013;190:5216–5225. doi: 10.4049/jimmunol.1300097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales L, Gajewski TF. Molecular Pathways: Targeting the Stimulator of Interferon Genes (STING) in the Immunotherapy of Cancer. Clin Cancer Res. 2015;21:4774–4779. doi: 10.1158/1078-0432.CCR-15-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales L, Glickman LH, McWhirter SM, Kanne DB, Sivick KE, Katibah GE, Woo SR, Lemmens E, Banda T, Leong JJ, Metchette K, Dubensky TW, Jr, Gajewski TF. Direct Activation of STING in the Tumor Microenvironment Leads to Potent and Systemic Tumor Regression and Immunity. Cell reports. 2015;11:1018–1030. doi: 10.1016/j.celrep.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales L, McWhirter SM, Dubensky TW, Jr, Gajewski TF. The host STING pathway at the interface of cancer and immunity. J Clin Invest. 2016;126:2404–2411. doi: 10.1172/JCI86892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, Li XD, Mauceri H, Beckett M, Darga T, Huang X, Gajewski TF, Chen ZJ, Fu YX, Weichselbaum RR. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity. 2014;41:843–852. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Kanne DB, Leong M, Glickman LH, McWhirter SM, Lemmens E, Mechette K, Leong JJ, Lauer P, Liu W, Sivick KE, Zeng Q, Soares KC, Zheng L, Portnoy DA, Woodward JJ, Pardoll DM, Dubensky TW, Jr, Kim Y. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci Transl Med. 2015;7:283ra252. doi: 10.1126/scitranslmed.aaa4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA, Gaffney BL, Shuman S, Jones RA, Deng L, Hartmann G, Barchet W, Tuschl T, Patel DJ. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell. 2013;154:748–762. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Han Y, Zhao X, Wang J, Liu F, Xu C, Wei L, Jiang JD, Block TM, Guo JT, Chang J. STING agonists induce an innate antiviral immune response against hepatitis B virus. Antimicrob Agents Chemother. 2015;59:1273–1281. doi: 10.1128/AAC.04321-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Tang L, Shu S, Sehgal M, Sheraz M, Liu B, Zhao Q, Cheng J, Zhao X, Zhou T, Chang J, Guo JT. Activation of STING in hepatocytes suppresses the replication of hepatitis B virus. Antimicrob Agents Chemother. 2017 doi: 10.1128/AAC.00771-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Wu S, Julander J, Ma J, Zhang X, Kulp J, Cuconati A, Block TM, Du Y, Guo JT, Chang J. A Novel Benzodiazepine Compound Inhibits Yellow Fever Virus Infection by Specifically Targeting NS4B Protein. J Virol. 2016 doi: 10.1128/JVI.01253-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MC, Crespo MP, Abraham W, Moynihan KD, Szeto GL, Chen SH, Melo MB, Mueller S, Irvine DJ. Nanoparticulate STING agonists are potent lymph node-targeted vaccine adjuvants. J Clin Invest. 2015;125:2532–2546. doi: 10.1172/JCI79915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelow B, Narbus CM, Sourisseau M, Evans MJ. HepG2 cells mount an effective antiviral interferon-lambda based innate immune response to hepatitis C virus infection. Hepatology. 2014;60:1170–1179. doi: 10.1002/hep.27227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R. Control of adaptive immunity by the innate immune system. Nat Immunol. 2015;16:343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Kobayashi J, Saitoh T, Maruyama K, Ishii KJ, Barber GN, Komatsu K, Akira S, Kawai T. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc Natl Acad Sci U S A. 2013;110:2969–2974. doi: 10.1073/pnas.1222694110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner SK, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT, Seeger C, King RW. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother. 1997;41:1715–1720. doi: 10.1128/aac.41.8.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DE, Kessler DS, Pine R, Reich N, Darnell JE., Jr Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. 1988;2:383–393. doi: 10.1101/gad.2.4.383. [DOI] [PubMed] [Google Scholar]
- Li L, Yin Q, Kuss P, Maliga Z, Millan JL, Wu H, Mitchison TJ. Hydrolysis of 2′3′-cGAMP by ENPP1 and design of nonhydrolyzable analogs. Nat Chem Biol. 2014;10:1043–1048. doi: 10.1038/nchembio.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioux T, Mauny MA, Lamoureux A, Bascoul N, Hays M, Vernejoul F, Baudru AS, Boularan C, Lopes-Vicente J, Qushair G, Tiraby G. Design, Synthesis, and Biological Evaluation of Novel Cyclic Adenosine-Inosine Monophosphate (cAIMP) Analogs That Activate Stimulator of Interferon Genes (STING) J Med Chem. 2016;59:10253–10267. doi: 10.1021/acs.jmedchem.6b01300. [DOI] [PubMed] [Google Scholar]
- Rice CM, Lenches EM, Eddy SR, Shin SJ, Sheets RL, Strauss JH. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985;229:726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- Sali TM, Pryke KM, Abraham J, Liu A, Archer I, Broeckel R, Staverosky JA, Smith JL, Al-Shammari A, Amsler L, Sheridan K, Nilsen A, Streblow DN, DeFilippis VR. Characterization of a Novel Human-Specific STING Agonist that Elicits Antiviral Activity Against Emerging Alphaviruses. PLoS Pathog. 2015;11:e1005324. doi: 10.1371/journal.ppat.1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirey KA, Nhu QM, Yim KC, Roberts ZJ, Teijaro JR, Farber DL, Blanco JC, Vogel SN. The anti-tumor agent, 5,6-dimethylxanthenone-4-acetic acid (DMXAA), induces IFN-beta-mediated antiviral activity in vitro and in vivo. J Leukoc Biol. 2011;89:351–357. doi: 10.1189/jlb.0410216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Science signaling. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hu S, Chen X, Shi H, Chen C, Sun L, Chen ZJ. cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc Natl Acad Sci U S A. 2017;114:1637–1642. doi: 10.1073/pnas.1621363114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo SR, Corrales L, Gajewski TF. The STING pathway and the T cell-inflamed tumor microenvironment. Trends Immunol. 2015;36:250–256. doi: 10.1016/j.it.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Gayen S, Kang C, Yuan Z, Shi PY. Membrane topology and function of dengue virus NS2A protein. J Virol. 2013;87:4609–4622. doi: 10.1128/JVI.02424-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, Wu H. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol Cell. 2012;46:735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Guo JT, Seeger C. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J Virol. 2003;77:9204–9210. doi: 10.1128/JVI.77.17.9204-9210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]