Abstract
Determining the correct enzymatic activity of putative glycosyltransferases (GTs) can be challenging as these enzymes can utilize multiple donor and acceptor substrates. Upon initial determination of the donor-sugar nucleotide(s), a GT utilizes various acceptor molecules that can then be tested. Here, we describe a quick method to screen sugar-nucleotide donor specificities of GTs utilizing a sensitive, nonradioactive, commercially available bioluminescent uridine diphosphate detection kit. This in vitro method allowed us to validate the sugar-nucleotide donor-substrate specificities of recombinantly expressed human, bovine, bacterial and protozoan GTs. Our approach, which is less time consuming than many traditional assays that utilize radiolabeled sugars and chromatographic separations, should facilitate discovery of novel GTs that participate in diverse biological processes.
Keywords: assays, enzymology, glycosyltransferase, methods, sugar-nucleotide
Introduction
Glycosyltransferases (GTs) are a large, diverse family of enzymes that catalyze the transfer of activated sugars, also termed donor substrates, to acceptor substrates that include a vast range of biomolecules such as a proteins, lipids, nucleic acids or other glycoconjugates (Lairson et al. 2008). Donor substrates are typically activated mono- or oligosaccharides in the form of nucleotide sugars, such as uridine diphosphate glucose (UDP-Glc), guanosine diphosphate mannose (GDP-Man), or cytidine monophosphate N-acetylneuraminic acid (CMP-NeuAc), but can also include dolichol-phosphate linked to mono- or oligosaccharides or other lipid-linked donor sugars (Lairson et al. 2008). Understanding the roles of GTs in biosynthetic pathways is key to understand many biological processes. Although GTs can be functionally predicted by genomic and primary amino acid sequence analyses, subtle changes in primary sequence, including single amino acid replacements (Ramakrishnan and Qasba 2002), can alter sugar nucleotide donor and/or acceptor utilization.
The characterization of putative GT activity can be hampered by the lack of information regarding both donor and acceptor substrate specificity. Traditional assays screening for GT activity have employed hydrophobic small molecule acceptors, such as para-nitrophenyl or 4-methylumbelliferyl-conjugated mono- or oligosaccharides, and the use of radiolabeled glycosyl donors by detecting the transfer of the radiolabeled sugar to the acceptor molecule (Wagner and Pesnot 2010). Fluorescence-based assays have been developed that utilize fluorescently modified glycosyl donors (Helm et al. 2003; Schweizer 2007; Pesnot and Wagner 2008), and mass spectrometric approaches have been used with success in functionally validating GT activity (Yang et al. 2005; Nagahori and Nishimura 2006). Assays for GT activity, which have been carefully reviewed by Wagner and Pesnot (2010) and Palcic and Sujino (2001), are extremely valuable tools for the glycobiologist. However, limitations may arise such as reagent availability, requirements for specialized or costly instrumentation in the case of mass spectrometry and working with hazardous radiolabeled sugars. In this report, we describe a simple, nonradioactive method to determine the specificity of the donor substrate in a hydrolysis assay in the absence of any acceptor substrate. A strategy similar to our approach has been reported using radiolabeled sugars in a hydrolysis assay, followed by anion-exchange chromatography to purify away the (uncharged) monosaccharide and liquid scintillation counting for detection of the radiolabeled sugar (Sethi et al. 2013). However, our strategy monitors the formation of the free nucleotide-phosphate leaving group from the activated donor instead of monitoring the sugar moiety, does not require any reaction product purification and utilizes a commercially available bioluminescent assay for GTs described in Materials and methods. An alternative malachite-green-based assay to screen for GT activity using phosphate-containing donors has also been previously reported; however, it requires the use of phosphatases specific for the type of nucleotide-phosphate to be detected, and the coupled reaction scheme requires that the specific phosphatases must remain active in the conditions used for the GT reactions (Wu et al. 2011).
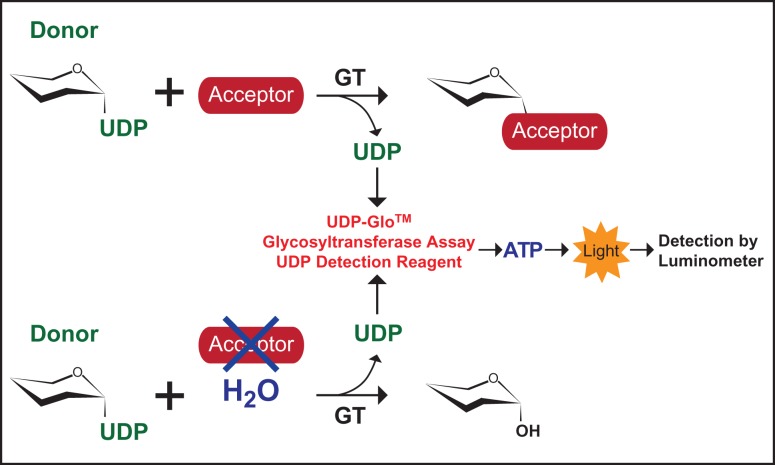
In a typical “Leloir type”, GT reaction that utilizes activated glycosyl donors in the form of covalently linked nucleotide sugars, the transfer of the sugar moiety from the nucleotide sugar is followed by the release of the nucleotide product (Figure 1). In the absence of an acceptor substrate, GTs often exhibit background hydrolysis of the donor substrate, which can be considered the enzymatically catalyzed transfer of the sugar moiety to a water molecule (Leemhuis and Dijkhuizen 2003; Sindhuwinata et al. 2010; Brockhausen 2014). Detection of the nucleotide product, such as UDP, can be achieved by the UDP-Glo™ Glycosyltransferase Assay kits recently developed by the Promega Corporation. The UDP-Glo™ assay detects GT activity based on the release of UDP, within the linear range of nM to µM concentrations, upon transfer of the carbohydrate to the acceptor molecule via a luciferase-based detection system allowing for detection of activity without radiolabeled sugars or product separation by high performance liquid chromatography (Figure 1). We hypothesized that we could use the UDP-GloTM assay to quickly screen for sugar-nucleotide donor specificities of GTs. To test our hypothesis, we assayed recombinantly expressed human, bovine, bacterial and protozoan GTs of known sugar-nucleotide specificity. Here, we report the feasibility of using a bioluminescent assay to screen for GT donor-substrate specificity.
Fig. 1.
GT reaction schematic. (Top) A simplified reaction where a GT catalyzes the transfer of a carbohydrate moiety from a specific donor substrate to an acceptor molecule. For screening of sugar-nucleotide donors, a GT is incubated with a panel of donor sugars in the absence of an acceptor substrate (Bottom), and the UDP released upon background hydrolysis of the UDP-sugar is detected using the UDP-Glo™ Glycosyltransferase Assay, where UDP is enzymatically converted to Adenosine triphosphate (ATP), followed by a luciferase reaction which emits light. The luminescence detected by the luminometer is directly proportional to UDP concentration. This figure is available in black and white in print and in color at Glycobiology online.
Results
Determination of purified recombinant GT UDP-sugar preference
In order to test whether the UDP-GloTM assay could be used to screen for nucleotide donor specificities of GTs without their target substrates, we recombinantly expressed one bacterial, one protozoan and four human GTs of known donor specificities. The enzyme glycosyltransferase A (GtfA) from Streptococcus pneumoniae was selected to assay for O-GlcNAc transferase (OGT) activity (Wu et al. 2010; Shi et al. 2014). The cytoplasmic, protozoan α-galactosyltransferase AgtA from the soil ameba Dictyostelium discoideum was selected to contribute to the diversity of species and cellular localization of our prototypic GTs (Ketcham et al. 2004; Schafer et al. 2014). With respect to human GTs, protein O-glucosyltransferase 1 (POGLUT1, Rumi in Drosophila) was chosen for its Glc- and xylose (Xyl)-transferase activities involved with O-β-glucosylation and O-β-xylosylation of specific epidermal growth factor (EGF) repeats (Takeuchi et al. 2011). Protein O-linked mannose β-1,2-N-acetylglucosaminyltransferase 1 (POMGNT1) catalyzes the transfer of GlcNAc from UDP-GlcNAc to O-linked Man on the protein α-dystroglycan to initiate Core M1- and M2-type structures [reviewed by Praissman and Wells (2014)], while the β-1,4-glucuronyltransferase (B4GAT1; formerly identified as B3GNT1) is responsible for adding β-1,4-linked glucuronic acid (GlcA) to Core M3-type O-Man glycans (Willer et al. 2014; Praissman et al. 2014), allowing for extension of the glycans by the bifunctional Like-acetylglucosaminyltransferases (LARGE1 or LARGE2), which synthesize a glycosaminoglycan-like repeating disaccharide (-Xylα1,3-GlcAβ1,3-) by utilizing UDP-GlcA and UDP-Xyl (Yoshida-Moriguchi and Campbell 2015). In addition, we selected the commercially available β-1,4-galactosyltransferase I (Bovine β4 Gal-T1) Y289L mutant that exhibits GalNAc-transferase activity (Ramakrishnan and Qasba 2002).
These GTs were selected based on their ability to use one or more UDP sugars within our panel of known UDP-containing glycosyl donors in humans, which includes UDP-Glucose (UDP-Glc), UDP-N-Acetylglucosamine (UDP-GlcNAc), UDP-Galactose (UDP-Gal), UDP-N-Acetylgalactosamine (UDP-GalNAc), UDP-glucuronic acid (UDP-GlcA) and UDP-Xylose (UDP-Xyl), all of which can be purchased or produced as Ultra Pure quality that have <0.005% UDP contamination as free UDP decreases assay sensitivity (see Materials and methods).
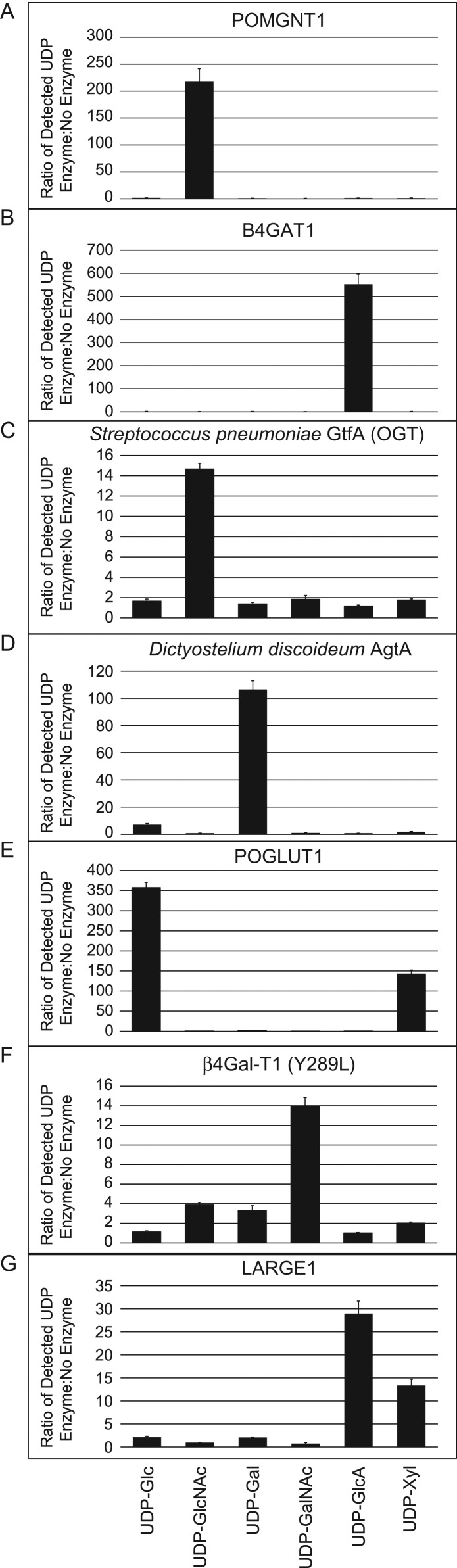
The assay conditions are fully described in Materials and methods, but essentially, each UDP-sugar was allowed to incubate overnight with and without each recombinantly expressed GT in separate pH-buffered reactions. The following day, detection of free UDP released by GT-mediated hydrolysis of the UDP sugars was performed according to the UDP-GloTM technical manual (see Materials and methods). After 1 hour of incubation with the UDP-GloTM Detection Reagent, luminescence was measured which is directly proportional to UDP concentration based on an UDP standard curve. Representative data from each assay are shown in Figure 2 and are represented as a ratio of the UDP measured from reactions containing the indicated GTs relative to the negative controls where no enzyme was added.
Fig. 2.
Comparison of glycosyl-donor specificities of different GTs. Recombinant human POMGNT1 (A), human B4GAT1 (B), SpGtfA (OGT) (C), DdAgtA (D), human POGLUT1 (E), bovine β4 Gal-T1 (Y289L) (F) and human LARGE1 (G) were incubated with 50 µM of each UDP-sugar (indicated at the bottom) for 16 h at 37°C, and the release of UDP was detected by the UDP-GloTM assay. Data are represented as the ratio of the UDP detected from the GT-containing reactions to the respective negative controls, which have all reaction components without GTs. Data are representative from three trials, and error bars represent standard deviation.
Optimization of GtfA hydrolysis reaction conditions
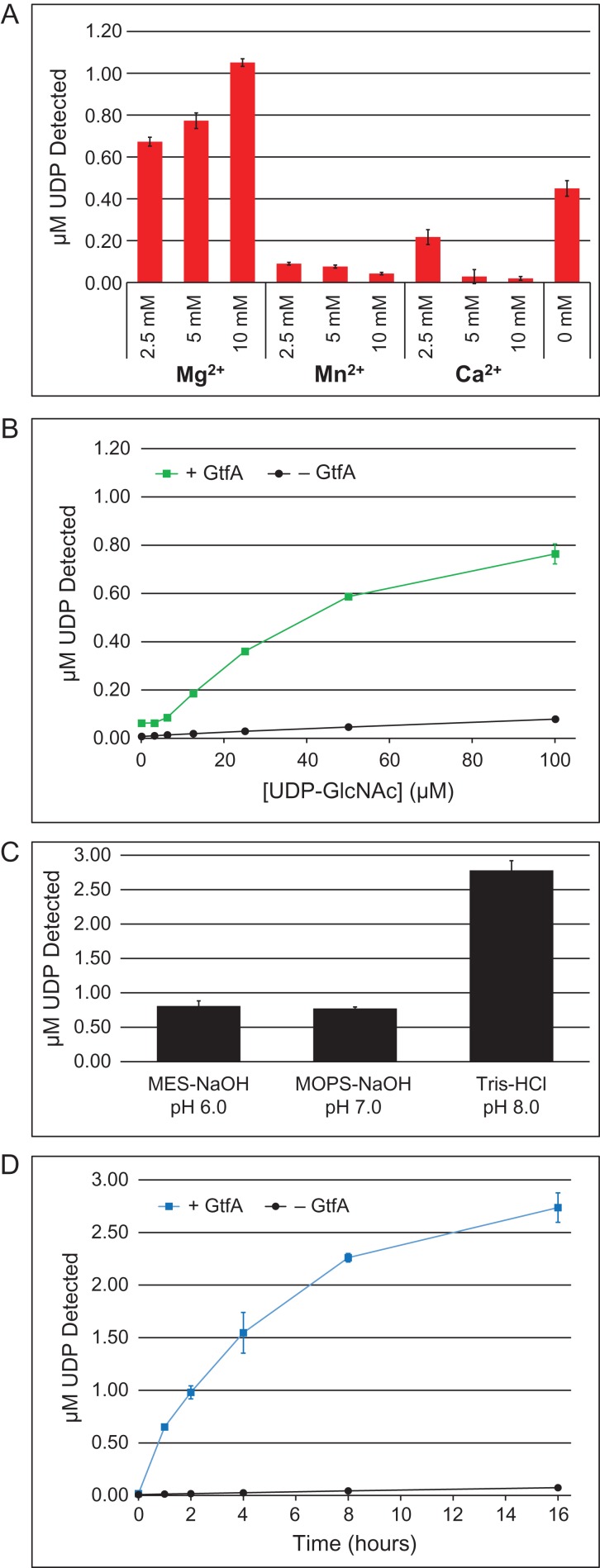
In order to determine if we could optimize the hydrolysis conditions using our methodology, we selected GtfA as a prototype GT since little is currently known about the enzymatic properties of this particular OGT from S. pneumoniae. Optimization of divalent cation supplementation in the hydrolysis reactions was performed using various concentrations of MgCl2, MnCl2 and CaCl2 in a reaction buffer containing 0.1 M MOPS-NaOH (pH 7.0) and 50 µM Ultra Pure UDP-GlcNAc (Figure 3A). Since the downstream analysis using the UDP-GloTM kit requires magnesium, the metal chelating agent, ethylenediaminetetraacetic acid (EDTA), could not be used to determine the metal-ion dependency, as directed by the manufacturer. GtfA displayed a concentration-dependent preference for Mg2+, whereas increasing concentrations of Mn2+ and Ca2+ inhibited hydrolase activity. A titration of UDP-GlcNAc in 0.1 M MOPS-NaOH (pH 7.0) and 10 mM MgCl2 was performed to evaluate the optimal sugar-nucleotide concentration for the hydrolysis assay, where the amount of UDP released began to reach a plateau in reactions containing 100 µM UDP-GlcNAc (Figure 3B). Three different buffers at pH 6, 7 and 8 were also evaluated and showed that GtfA had the greatest UDP-GlcNAc hydrolase activity in 0.1 M Tris–HCl (pH 8.0) out of the buffers tested (Figure 3C). Finally, using the optimized buffer conditions of 0.1 M Tris–HCl (pH 8.0), 10 mM MgCl2 and 100 µM UDP-GlcNAc, a time course experiment was performed to demonstrate that with this particular GT, the maximal amount of UDP-GlcNAc was hydrolyzed in 16 h (Figure 3D). In this case, product represented <3% of the total amount of input into the reaction, demonstrating that UDP-GlcNAc was not entirely consumed. This may imply that product (UDP) inhibition was occurring, which is not uncommon for GTs, including human OGT (Wang et al. 2014). Thus, our UDP-GloTM-coupled hydrolysis assay protocol can be used to quickly screen for optimal reaction conditions without any knowledge of an acceptor substrate.
Fig. 3.
Optimization of GtfA-catalyzed hydrolysis of UDP-GlcNAc. (A) GtfA (3 µg) was incubated with 50 µM of UDP-GlcNAc in 0.1 M MES-NaOH (pH 7.0) and the indicated concentration of divalent cation (Mg2+, Mn2+ or Ca2+) for 16 h at 37°C. (B) Same as in Panel A ± GtfA (3 µg), except with 10 mM MgCl2 and varying concentration of UDP-GlcNAc (0–100 µM). (C) Same as in Panel A, except with 10 mM MgCl2 and different pH buffers as indicated. Data are represented as the ratio of the UDP detected from the GT-containing reactions to the respective negative controls, which have all reaction components without GTs. (D) Time course experiment with ±GtfA (3 µg) in the optimized buffer consisting of 0.1 M Tris–HCl (pH 8.0), 10 mM MgCl2 and 100 µM UDP-GlcNAc. In all panels, the release of UDP was detected by the UDP-GloTM assay. Data are representative from three trials, and error bars represent standard deviation. This figure is available in black and white in print and in color at Glycobiology online.
Discussion
As anticipated, each GT displayed specific hydrolysis activity toward the expected UDP sugars. In particular, human POMGNT1, human B4GAT1, S. pneumoniae OGT and D. discoideum AgtA were all selective for a single UDP-sugar (Figure 2A–D). These results demonstrate the applicability of our assay toward different GTs originating from various species and indiscriminate of the anomeric configuration of the sugar moiety after transfer. POGLUT1 utilizes both UDP-Glc and UDP-Xyl, and specific amino acid consensus sequences of acceptor substrates have been defined and reported to influence whether POGLUT1 transfers Glc or Xyl to specific EGF sequences (Takeuchi et al. 2011). In our experiments, in the absence of a substrate, POGLUT1 can hydrolyze both sugar-donors, however, at ~2.3-fold lower efficiency for UDP-Xyl with respect to UDP-Glc at the 50 µM donor concentrations used (Figure 2E). As expected, the bovine β4 Gal-T1 (Y289L) mutant exhibits strong GalNAc-transferase activity (Figure 2F); however, it also demonstrated broader specificity, though minor hydrolysis, toward other UDP sugars due to the opening of the catalytic pocket as a result of the Y289L substitution (Ramakrishnan and Qasba 2002). LARGE1 utilizes UDP-GlcA at an ~3-fold greater level of hydrolysis activity over UDP-Xyl at the pH tested (pH 6.0, Figure 2G). This result is entirely consistent with a previous report of an optimal pH of 5.0 for Xyl-transferase activity (with ~30% of maximal activity at pH 6.0) and broader pH range (pH 5.5–8.0) for GlcA-transferase activity (Inamori et al. 2013). Although the relative utilization of donor substrates in the cell will depend on their concentrations, the buffer and the respective Km values of the GT of interest, these in vitro reactions can provide important indications of enzyme potential. In total, we were able to identify the donor substrate for three retaining activities and five inverting activities. Thus, regardless of whether the GTs utilized a retaining or inverting mechanism, the hydrolysis assay was able to identify the preferred UDP-sugar(s) (Table I).
Table I.
Summary of GTs used in this study
| UniProtKB entry IDa | Protein name | GT activity | Species | Mechanism | Donor substrate |
|---|---|---|---|---|---|
| Q8WZA1 | POMGNT1 | β-1,2-N-acetylglucosaminyltransferase | Homo sapiens | Inverting | UDP-GlcNAc |
| O43505 | B4GAT1 | β-1,4-Glucuronyltransferase | H. sapiens | Inverting | UDP-GlcA |
| A0A0H2URG7 | GtfA | Protein O-N-acetylglucosaminyltransferase | S. pneumoniae | Retaining | UDP-GlcNAc |
| Q54RP0 | AgtA | α-1,3-Galactosyltransferase | D. discoideum | Retaining | UDP-Gal |
| Q8NBL1 | POGLUT1 | Protein O-β-glucosyltransferase, protein O-β-xylosyltransferase | H. sapiens | Inverting | UDP-Glc, UDP-Xyl |
| P08037 | β4 Gal-T1 (Y289L) | β-1,4-N-acetylgalactosaminyltransferase, β-1,4-galactosyltransferase | Bos taurus | Inverting | UDP-GalNAc, UDP-Gal, UDP-GlcNAc |
| O95461 | LARGE1 | Bifunctional, β-1,3-glucuronyltransferase, α-1,3-xylosyltransferase | H. sapiens | Retaining, Inverting | UDP-GlcA, UDP-Xyl |
aUniProtKB: http://www.uniprot.org/
Once sugar-donor specificity of a GT has been identified, our methodology can also be used to screen different buffering conditions and components such as pH, salt and divalent cation selectivity for hydrolysis. We selected GtfA for further optimization and successfully optimized UDP-GlcNAc hydrolase conditions in the absence of any exogenous acceptor substrate (Figure 3). Attempts to assay desalted Escherichia coli cell extracts expressing GtfA, without purification, were unsuccessful, presumably due to endogenous hydrolases (data not shown). Thus, it is imperative to assay purified GTs using our methodology. As an example of the utility of looking at hydrolysis, a parasite polypeptide UDP-GlcNAc:Thr α-GlcNAc transferase, whose UDP-GlcNAc hydrolysis activity was competitive with its transferase activity, a hydrolysis assay was important for its detection and purification, and allowed accurate determination of its Km for UDP-GlcNAc (Heise et al. 2009). Further, different acceptor substrates can then be tested using the same assay and validated by radioactivity transfer assays and/or mass spectrometry. Recently, we have applied the methodology described in this report to characterize the Xyl-transferase activity of transmembrane protein 5 (Praissman et al. 2016). The assays are not restricted to only UDP-based sugars as CMP/UMP- and GDP-based GloTM assays are currently under development by Promega (personal communication) and can be utilized to assay for CMP/UMP- and GDP-containing sugar-nucleotide donors which would account for all 9 sugar nucleotides known for mammals [or 10, including the recently identified activated reduced sugar, cytidine diphosphate-ribitol (Riemersma et al. 2015; Gerin et al. 2016; Kanagawa et al. 2016; Praissman et al. 2016)]. In conclusion, this straightforward method is a new valuable tool in the glycobiologist's toolbox for assisting in the assignment of GT activities.
Materials and methods
Enzyme expression and purification
Expression and purification of full length His6-DdAgtA was performed exactly as previously described (Schafer et al. 2014) and stored at −80°C in a buffer containing 50 mM Tris–HCl (pH 8.0), 0.2 mM EDTA and 5 mM β-mercaptoethanol.
Expression and purification of human POGLUT1 containing a C-terminal myc-His6 tag was performed as previously described (Takeuchi et al. 2011) and stored at −80°C in 20% glycerol in Tris-buffered saline pH 7.4.
The coding region of S. pneumoniae GtfA (Gene ID: SP_1758, UniProt A0A0H2URG7) lacking a stop codon was obtained in Gateway clone sets from Biodefense and Emerging Infections Research Resources Repository (BEI Resources). An LR-clonase reaction was performed to insert the gene into the pET-DEST42 (Thermo Fisher Scientific, Waltham, MA, USA) destination vector for the expression of a carboxy-terminal His6-tagged fusion protein in E. coli BL21 cells. BL21 cells transformed with the pET-DEST42-SP1758 plasmid were grown in LB medium supplemented with 100 µg/mL ampicillin at 37°C, and cell density was monitored by absorbance at 600 nm (A600). Once the A600 reached 0.6, the cells were transferred into 25°C, protein expression was induced by the addition of Isopropyl β-D-1-thiogalactopyranoside to a final concentration of 1 mM and the cell culture was allowed to incubate for 4 h (until A600 reached ~1.1). Cells were harvested by centrifugation and stored at −80°C until ready for lysis and purification. Cells were then resuspended in phosphate-buffered saline (PBS, pH 7.2) with 1 mg/mL lysozyme for 20 min at 30°C, probe sonicated for 2 min (four cycles of 20 s on, 10 s off), clarified by centrifugation at 17,000 × g for 1 h at 4°C, and passed through a 0.45 µm syringe filter. The enzyme (SpGtfA-His6, or simply, GtfA) was purified by Ni2+-NTA chromatography at 4°C, eluted with 300 mM imidazole and buffer exchanged into PBS pH 7.2. Protein concentration was determined by bicinchoninic acid assay.
Recombinant expression of soluble, secreted versions of green fluorescent protein (GFP)-B4GAT1 and GFP-LARGE were performed as previously described (Praissman et al. 2014). The catalytic domain of human POMGNT1 (amino acid residues 60–660, UniProt Q8WZA1) was expressed as a soluble, secreted fusion protein by transient transfection of HEK293 suspension cultures (Meng et al. 2013). The coding regions were amplified from Mammalian Gene Collection (Gerhard et al. 2004) clones using primers that appended a tobacco etch virus (TEV) protease cleavage site (Phan et al. 2002) to the NH2-terminal end of the coding region and attL1 and attL2 Gateway adaptor sites to the 5′- and 3′-terminal ends of the amplimer products. The amplimers were recombined via BP clonase reaction into the pDONR221 vector, and the DNA sequences were confirmed. The pDONR221 clones were then recombined via LR-clonase reaction into a custom Gateway adapted version of the pGEn2 mammalian expression vector (Barb et al. 2012; Meng et al. 2013) to assemble a recombinant coding region comprises a 25 amino acid NH2-terminal signal sequence from the Trypanosoma cruzi lysosomal α-mannosidase (Vandersall-Nairn et al. 1998) followed by an 8xHis tag, 17 amino acid AviTag (Beckett et al. 1999), ‘superfolder’ GFP (Pedelacq et al. 2006), the nine amino acid sequence encoded by attB1 recombination site, followed by the TEV protease cleavage site and the respective GT catalytic domain coding region. Suspension culture HEK293F cells (Life Technologies, Grand Island, NY) were transfected as previously described (Meng et al. 2013) and the culture supernatant was subjected to Ni-NTA superflow chromatography (Qiagen, Valencia, CA). Enzyme preparations eluted with 300 mM imidazole were concentrated to ∼1 mg/mL using an ultrafiltration pressure cell membrane (Millipore, Billerica, MA) with a 10 kDa molecular weight cutoff and buffer exchanged into PBS pH 7.2. The β1,4-galactosyltransferase I mutant [β4 Gal-T1 (Y289L)] was purchased from Invitrogen, Waltham, MA, USA.
UDP hydrolysis reactions
Ultra Pure UDP-Glc, UDP-GlcNAc, UDP-Gal, UDP-GalNAc and UDP-GlcA were purchased from Promega Corporation, Madison, WI, USA. Ultra Pure UDP-Xyl was prepared as previously described (Sheikh and Wells 2016). Specificity of each recombinant enzyme's sugar-nucleotide hydrolysis activity was performed by incubation of 3 μg of either POGLUT1-myc-His6, His6-DdAgtA, GFP-POMGNT1, GFP-B4GAT1 or β4 Gal-T1 (Y289L) with 50 μM of a single UDP-sugar (Ultra Pure UDP-Glc, UDP-GlcNAc, UDP-Gal, UDP-GalNAc, UDP-GlcA or UDP-Xyl) in the absence of any acceptor substrate in separate 20 µL reactions containing 0.1 M MOPS-NaOH pH 7.0, 10 mM MgCl2 and 10 mM MnCl2 at 37°C for 16 h. GFP-LARGE (3 μg) was assayed in the same way, except in a buffer containing 0.1 M MES-NaOH pH 6.0 instead of 0.1 M MOPS-NaOH pH 7.0 due to the optimal pH of the enzyme (Inamori et al. 2013). GtfA (3 μg) was initially assayed in the same way, except in a buffer containing 0.1 M Tris–HCl pH 7.4, followed by optimization of the reaction conditions as described in GtfA reaction optimization. Negative controls consisted of all reaction components, except for the enzymes, for each of the respective UDP sugars. Following the incubation period, reactions were allowed to equilibrate to room temperature for 10 min. Hydrolysis reactions were stopped by the addition of the UDP-GloTM Detection Reagent, and detection of free UDP was performed as described in UDP-GloTM Glycosyltransferase Assay.
UDP-GloTM Glycosyltransferase Assay
Detection of free UDP after hydrolysis of the sugar nucleotide was performed using the UDP-GloTM Glycosyltransferase Assay Kit (Promega Corporation), which detects UDP after UDP-sugar hydrolysis or transfer by converting UDP to light (measured in Relative Luminescence Units) in a luciferase type reaction. A standard curve using 0–25 µM UDP was performed, and the range of measurements was determined to be in the linear range of detection (in all cases, UDP-sugar consumption was <10% of the starting amount), where the luminescence detected is directly proportional to UDP concentration. Following the manufacturer's protocol, each sugar-nucleotide hydrolysis reaction was combined in a ratio of 1:1 (5 µL:5 µL) with the UDP-GloTM Detection Reagent in independent wells of a white, flat bottom 384-well assay plate (Corning) and allowed to incubate at ambient temperature. After 1 h of incubation, luminescence was measured in triplicate using a GloMax®-Multi+ Microplate Luminometer (Promega Corporation).
GtfA reaction optimization
The reaction conditions for GtfA-mediated hydrolysis of UDP-GlcNAc were optimized by first performing the hydrolysis assay using 3 µg of GtfA per reaction as described in UDP hydrolysis reactions in a buffer containing 0.1 M MOPS-NaOH (pH 7.0), 50 µM Ultra Pure UDP-GlcNAc and either MgCl2, MnCl2 or CaCl2 at various concentrations (all at 0, 2.5, 5 and 10 µM) incubated at 37°C for 16 h to determine divalent cation preference. UDP-GlcNAc titrations from 0 to 100 µM were then carried out in a buffer containing 0.1 M MOPS-NaOH (pH 7.0), 10 mM MgCl2 and 50 µM Ultra Pure UDP-GlcNAc, incubated at 37°C for 16 h. Buffer pH optimization was performed using 10 mM MgCl2, 50 µM Ultra Pure UDP-GlcNAc and either 0.1 M MES-NaOH (pH 6.0), 0.1 M MOPS-NaOH (pH 7.0) or 0.1 M Tris–HCl (pH 8.0), incubated at 37°C for 16 h. Finally, a time course hydrolysis experiment (0–16 h) was performed in the optimized buffer containing 0.1 M Tris–HCl (pH 8.0), 10 mM MgCl2 and 100 µM Ultra Pure UDP-GlcNAc.
Acknowledgments
The authors thank all members of the Wells, Moremen, Avci, Haltiwanger and West laboratories for helpful discussions.
Funding
This work was supported in part by grants from the National Institute of Health [R01GM111939 to L.W., P41GM103490 to K.W.M. and L.W., P01GM107012 to K.W.M. and L.W., R21AI123161 to C.M.W. and L.W., R01AI123383 to F.Y.A., R21AI115451 to F.Y.A., R01GM061126 to R.S.H. and R01GM037539 to C.M.W.].
Conflict of interest statement
None declared.
Abbreviations
β4 Gal-T1 (Y289L), β-1,4-galactosyltransferase I mutant Y289L; B4GAT1, β-1,4-glucuronyltransferase; CMP, cytidine monophosphate; Dd, Dictyostelium discoideum; EDTA, ethylenediaminetetraacetic acid; EGF, epidermal growth factor; Gal, galactose; GalNAc, N-acetylgalactosamine; GDP, guanosine diphosphate; Glc, glucose; GlcA, glucuronic acid; GtfA, glycosyltransferase A; GT, glycosyltransferase; GFP, green fluorescent protein; LARGE1, Like-acetylglucosaminyltransferase 1; Man, mannose; OGT, O-GlcNAc transferase; PBS, phosphate-buffered saline; POGLUT1, protein O-glucosyltransferase 1; POMGNT1, protein O-linked mannose β-1,2-N-acetylglucosaminyltransferase 1; Sp, Streptococcus pneumoniae; UDP, uridine diphosphate; Xyl, xylose.
References
- Barb AW, Meng L, Gao Z, Johnson RW, Moremen KW, Prestegard JH. 2012. NMR characterization of immunoglobulin G Fc glycan motion on enzymatic sialylation. Biochemistry. 51:4618–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett D, Kovaleva E, Schatz PJ. 1999. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 8:921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhausen I. 2014. Crossroads between bacterial and mammalian glycosyltransferases. Front Immunol. 5:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard DS, Wagner L, Feingold EA, Shenmen CM, Grouse LH, Schuler G, Klein SL, Old S, Rasooly R, Good P et al. 2004. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC). Genome Res. 14:2121–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerin I, Ury B, Breloy I, Bouchet-Seraphin C, Bolsee J, Halbout M, Graff J, Vertommen D, Muccioli GG, Seta N et al. 2016. ISPD produces CDP-ribitol used by FKTN and FKRP to transfer ribitol phosphate onto alpha-dystroglycan. Nat Commun. 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise N, Singh D, van der Wel H, Sassi SO, Johnson JM, Feasley CL, Koeller CM, Previato JO, Mendonca-Previato L, West CM. 2009. Molecular analysis of a UDP-GlcNAc:polypeptide alpha-N-acetylglucosaminyltransferase implicated in the initiation of mucin-type O-glycosylation in Trypanosoma cruzi. Glycobiology. 19:918–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm JS, Hu YN, Chen L, Gross B, Walker S. 2003. Identification of active-site inhibitors of MurG using a generalizable, high-throughput glycosyltransferase screen. J Am Chem Soc. 125:11168–11169. [DOI] [PubMed] [Google Scholar]
- Inamori K, Hara Y, Willer T, Anderson ME, Zhu Z, Yoshida-Moriguchi T, Campbell KP. 2013. Xylosyl- and glucuronyltransferase functions of LARGE in alpha-dystroglycan modification are conserved in LARGE2. Glycobiology. 23:295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagawa M, Kobayashi K, Tajiri M, Manya H, Kuga A, Yamaguchi Y, Akasaka-Manya K, Furukawa J, Mizuno M, Kawakami H et al. 2016. Identification of a post-translational modification with ribitol-phosphate and its defect in muscular dystrophy. Cell Rep. 14:2209–2223. [DOI] [PubMed] [Google Scholar]
- Ketcham C, Wang F, Fisher SZ, Ercan A, van der Wel H, Locke RD, Sirajud-Doulah K, Matta KL, West CM. 2004. Specificity of a soluble UDP-galactose: fucoside alpha1,3-galactosyltransferase that modifies the cytoplasmic glycoprotein Skp1 in Dictyostelium. J Biol Chem. 279:29050–29059. [DOI] [PubMed] [Google Scholar]
- Lairson LL, Henrissat B, Davies GJ, Withers SG. 2008. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 77:521–555. [DOI] [PubMed] [Google Scholar]
- Leemhuis H, Dijkhuizen L. 2003. Engineering of hydrolysis reaction specificity in the transglycosylase cyclodextrin glycosyltransferase. Biocatal Biotransfor. 21:261–270. [Google Scholar]
- Meng L, Forouhar F, Thieker D, Gao Z, Ramiah A, Moniz H, Xiang Y, Seetharaman J, Milaninia S, Su M et al. 2013. Enzymatic basis for N-glycan sialylation: structure of rat alpha2,6-sialyltransferase (ST6GAL1) reveals conserved and unique features for glycan sialylation. J Biol Chem. 288:34680–34698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahori N, Nishimura SI. 2006. Direct and efficient monitoring of glycosyltransferase reactions on gold colloidal nanoparticles by using mass spectrometry. Chem-Eur J. 12:6478–6485. [DOI] [PubMed] [Google Scholar]
- Palcic MM, Sujino K. 2001. Assays for glycosyltransferases. Trends Glycosci Glyc. 13:361–370. [Google Scholar]
- Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 24:79–88. [DOI] [PubMed] [Google Scholar]
- Pesnot T, Wagner GK. 2008. Novel derivatives of UDP-glucose: concise synthesis and fluorescent properties. Org Biomol Chem. 6:2884–2891. [DOI] [PubMed] [Google Scholar]
- Phan J, Zdanov A, Evdokimov AG, Tropea JE, Peters HK IIIrd, Kapust RB, Li M, Wlodawer A, Waugh DS. 2002. Structural basis for the substrate specificity of tobacco etch virus protease. J Biol Chem. 277:50564–50572. [DOI] [PubMed] [Google Scholar]
- Praissman JL, Live DH, Wang S, Ramiah A, Chinoy ZS, Boons GJ, Moremen KW, Wells L. 2014. B4GAT1 is the priming enzyme for the LARGE-dependent functional glycosylation of alpha-dystroglycan. eLife. 3:e03943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praissman JL, Wells L. 2014. Mammalian O-mannosylation pathway: glycan structures, enzymes, and protein substrates. Biochemistry. 53:3066–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praissman JL, Willer T, Sheikh MO, Toi A, Chitayat D, Lin YY, Lee H, Stalnaker SH, Wang S, Prabhakar PK et al. 2016. The functional O-mannose glycan on α-dystroglycan contains a phospho-ribitol primed for matriglycan addition. eLife. 5:e14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan B, Qasba PK. 2002. Structure-based design of beta 1,4-galactosyltransferase I (beta 4 Gal-T1) with equally efficient N-acetylgalactosaminyltransferase activity: point mutation broadens beta 4 Gal-T1 donor specificity. J Biol Chem. 277:20833–20839. [DOI] [PubMed] [Google Scholar]
- Riemersma M, Froese DS, van Tol W, Engelke Udo F, Kopec J, van Scherpenzeel M, Ashikov A, Krojer T, von Delft F, Tessari M et al. 2015. Human ISPD is a cytidyltransferase required for dystroglycan O-mannosylation. Chem Biol. 22:1643–1652. [DOI] [PubMed] [Google Scholar]
- Schafer CM, Sheikh MO, Zhang D, West CM. 2014. Novel regulation of Skp1 by the dictyostelium AgtA alpha-galactosyltransferase involves the Skp1-binding activity of Its WD40 repeat domain. J Biol Chem. 289:9076–9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer F. 2007. Synthesis of fluorescently labelled and internally quenched UDP-Gal probes. Carbohydr Res. 342:1831–1840. [DOI] [PubMed] [Google Scholar]
- Sethi MK, Buettner FF, Ashikov A, Bakker H. 2013. In vitro assays of orphan glycosyltransferases and their application to identify Notch xylosyltransferases. Methods Mol Biol. 1022:307–320. [DOI] [PubMed] [Google Scholar]
- Sheikh MO, Wells L. 2016. Preparation of low background sugar-nucleotide donors for use in the UDP-Glo™ glycosyltransferase assay. Promega Corporation, Madison, WI, USA. [Google Scholar]
- Shi WW, Jiang YL, Zhu F, Yang YH, Shao QY, Yang HB, Ren YM, Wu H, Chen YX, Zhou CZ. 2014. Structure of a novel O-linked N-acetyl-D-glucosamine (O-GlcNAc) transferase, GtfA, reveals insights into the glycosylation of pneumococcal serine-rich repeat adhesins. J Biol Chem. 289:20898–20907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindhuwinata N, Munoz E, Munoz FJ, Palcic MM, Peters H, Peters T. 2010. Binding of an acceptor substrate analog enhances the enzymatic activity of human blood group B galactosyltransferase. Glycobiology. 20:718–723. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Fernandez-Valdivia RC, Caswell DS, Nita-Lazar A, Rana NA, Garner TP, Weldeghiorghis TK, Macnaughtan MA, Jafar-Nejad H, Haltiwanger RS. 2011. Rumi functions as both a protein O-glucosyltransferase and a protein O-xylosyltransferase. Proc Natl Acad Sci USA. 108:16600–16605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandersall-Nairn AS, Merkle RK, O'Brien K, Oeltmann TN, Moremen KW. 1998. Cloning, expression, purification, and characterization of the acid a-mannosidase from Trypanosoma cruzi. Glycobiology. 8:1183–1194. [DOI] [PubMed] [Google Scholar]
- Wagner GK, Pesnot T. 2010. Glycosyltransferases and their assays. ChemBioChem. 11:1939–1949. [DOI] [PubMed] [Google Scholar]
- Wang S, Shen DL, Lafont D, Vercoutter-Edouart AS, Mortuaire M, Shi Y, Maniti O, Girard-Egrot A, Lefebyre T, Pinto BM et al. 2014. Design of glycosyltransferase inhibitors targeting human O-GlcNAc transferase (OGT). MedChemComm. 5:1172–1178. [Google Scholar]
- Willer T, Inamori K, Venzke D, Harvey C, Morgensen G, Hara Y, Beltran Valero de Bernabe D, Yu L, Wright KM, Campbell KP. 2014. The glucuronyltransferase B4GAT1 is required for initiation of LARGE-mediated alpha-dystroglycan functional glycosylation. eLife. 3:e0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Zhou M, Wu H. 2010. Purification and characterization of an active N-acetylglucosaminyltransferase enzyme complex from Streptococci. Appl Environ Microbiol. 76:7966–7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZL, Ethen CM, Prather B, Machacek M, Jiang WP. 2011. Universal phosphatase-coupled glycosyltransferase assay. Glycobiology. 21:727–733. [DOI] [PubMed] [Google Scholar]
- Yang M, Brazier M, Edwards R, Davis BG. 2005. High-throughput mass-spectrometry monitoring for multisubstrate enzymes: determining the kinetic parameters and catalytic activities of glycosyltransferases. ChemBioChem. 6:346–357. [DOI] [PubMed] [Google Scholar]
- Yoshida-Moriguchi T, Campbell KP. 2015. Matriglycan: a novel polysaccharide that links dystroglycan to the basement membrane. Glycobiology. 25:702–713. [DOI] [PMC free article] [PubMed] [Google Scholar]