Abstract
Karyopherin beta-1/Importin beta-1 is a conserved nuclear transport receptor, acting in protein nuclear import in interphase and as a global regulator of mitosis. These pleiotropic functions reflect its ability to interact with, and regulate, different pathways during the cell cycle, operating as a major effector of the GTPase RAN. Importin beta-1 is overexpressed in cancers characterized by high genetic instability, an observation that highlights the importance of identifying its partners in mitosis. Here we present the first comprehensive profile of importin beta-1 interactors from human mitotic cells. By combining co-immunoprecipitation and proteome-wide mass spectrometry analysis of synchronized cell extracts, we identified expected (e.g., RAN and SUMO pathway factors) and novel mitotic interactors of importin beta-1, many with RNA-binding ability, that had not been previously associated with importin beta-1. These data complement interactomic studies of interphase transport pathways. We further developed automated proximity ligation assay (PLA) protocols to validate selected interactors. We succeeded in obtaining spatial and temporal resolution of genuine importin beta-1 interactions, which were visualized and localized in situ in intact mitotic cells. Further developments of PLA protocols will be helpful to dissect importin beta-1-orchestrated pathways during mitosis.
Introduction
Human karyopherin beta-1/importin beta-1 is a major effector of the GTPase RAN and a highly conserved member of the superfamily of nuclear transport receptors. As such, it regulates key cellular functions1, including i) protein import in interphase nuclei, underlying the fundamental processes of DNA replication, DNA repair, transcriptional and epigenetic control of gene expression2,3, ii) the localization and activity of factors implicated in mitotic spindle organization and function after nuclear envelope (NE) breakdown4, and, iii) the reorganization of the NE, nuclear pore complexes (NPCs) and nuclear structure at mitotic exit5,6.
Importin beta-1 has a well-characterized modular structure composed of HEAT repeats included in distinct functional domains7. Its interphase partners are both direct and indirect. Indirect partners include proteins carrying nuclear localization signals (NLS), recognized by an adaptor molecule belonging to the importin alpha subfamily. Using its C-terminal region, importin beta-1 interacts with importin alpha adaptor/NLS cargo, forming “classical” trimeric import complexes. In the nucleus, the GTPase RAN, loaded with GTP (RANGTP), binds importin beta-1 at its N-terminal region, thus destabilizing interactions at the C-terminus and triggering the cargo release from the import complex. Importin beta-1 also binds several proteins directly, including nucleoporins (NUPs) in their phenyl-glycine (FG)- and FxFG-rich regions, when crossing nuclear pores during nuclear import. Importin beta-1 also directly recognizes a variety of unrelated cargoes acting in different pathways2,8.
After NE breakdown, importin beta-1 takes on global regulatory roles in mitotic spindle organization and function4,9,10. In this mitotic mode, importin beta-1 generally inhibits factors with which it interacts2, and thus prevents their unscheduled activity and the premature onset of events during mitosis. Analogous to the mechanism operating in nuclear import, RANGTP binding to importin beta-1 releases mitotic “cargoes” in a free, biologically active form, thus enabling relevant mitotic event(s) to take place. When overexpressed in human cells, importin beta-1 overrides RANGTP regulation and keeps its mitotic partners in an inhibited state: this causes complex mitotic abnormalities, even under conditions under which nuclear import is not overtly affected11–14, indicating that mitosis is most sensitive to altered importin beta-1 levels. Regulated expression of importin beta-1 is therefore critical in control of cell division4. Importin beta-1 is overexpressed in many cancer types that generally display high genomic instability, e.g. cervical15 and gastric16 carcinoma, as well as diffuse large B-cell lymphoma17. Because many cancer types are dependent on importin beta-1 expression for their proliferation and survival15–20, importin beta-1 has been proposed as a novel therapeutic target21 and inhibitors are being developed, e.g. Importazole22 and INI-4323.
Interactomic analyses of importin-1 beta have been developed in interphase cultures to get insight into mechanisms of transport, based on stable isotope labeling by amino acids in cell cultures (SILAC)24, and, more recently, in a comparative study of twelve human importin family members to depict specificities in recognition of cargoes in distinct import pathways25. Studies of importin beta-1 mitotic interactors are less complete. In an early analysis of human importin beta-1 mitotic partners, we used MALDI-TOF mass spectrometry (MS) to examine importin beta-1 co-immunoprecipitates from mitotic-enriched cell cultures after thymidine arrest and release: we identified NUP358/ RAN-binding protein 2 (RANBP2), a large NUP with RAN-binding domains and E3 ligase activity for small ubiquitin-like modifier (SUMO) peptides, and RAN GTPase-activating protein 1 (RANGAP1) in the SUMO-conjugated form (RANGAP1-SUMO1)14. RANBP2 and RANGAP1-SUMO1 themselves form a stable complex throughout the cell cycle26. The complex (indicated as RRSU, for RANBP2/RANGAP1-SUMO1/UBC9) includes the SUMO E2 conjugating enzyme UBC9 and has enhanced SUMO ligase activity27. RANBP2 and RANGAP1-SUMO1 localize at the spindle microtubules (MTs) and accumulate to kinetochores (KTs) after MT attachment28. Importin beta-1 also localizes to the mitotic spindle MTs and poles12, and its interaction with RANBP2 prevents the RRSU premature localization at KTs prior to MT attachment14,29. Recent work has shown that this is essential to regulate the timing of SUMO conjugation of KT factors, and hence KT functions30,31. However, beyond individual studies of specific mitotic factors, a comprehensive view of importin beta-1 interactors in mitotic cells is currently lacking.
Here we present a comprehensive identification of importin beta-1 interactors from homogeneously synchronized mitotic cells using co-immunoprecipitation (co-IP) experiments hyphenated to a high resolution MS analysis. We identify expected partners, e.g. members of the RAN and SUMO pathways, as well as interactors that had not been previously associated with importin beta-1. Most mitotic protein interactions are highly dynamic in time and in space, and innovative methodologies are being developed to unravel specificities and variations. In situ proximity ligation assays (is-PLA) is an informative detection technique to visualize protein interactions at their intracellular site32. We have developed an automated PLA protocol to validate selected importin beta-1 interactions: that has enabled us to visualize the interactions of interest during stages of mitotic progression. Coupling the interactome analysis with automated is-PLA imaging provides valuable information on processes through which importin beta-1 can operate in mitosis.
Results
Co-immunoprecipitation of importin beta-1 and its partners from synchronized mitotic cells
To identify the interactions of importin beta-1 with protein partners during mitosis, we developed the workflow schematized in Fig. 1. Briefly, after an antibody selection step, large scale immunoprecipitates of importin beta-1 complexes from mitotic HeLa cells were analyzed by mass spectrometry (Step 2) and selected interactors were validated by is-PLA (Step 3).
Figure 1.
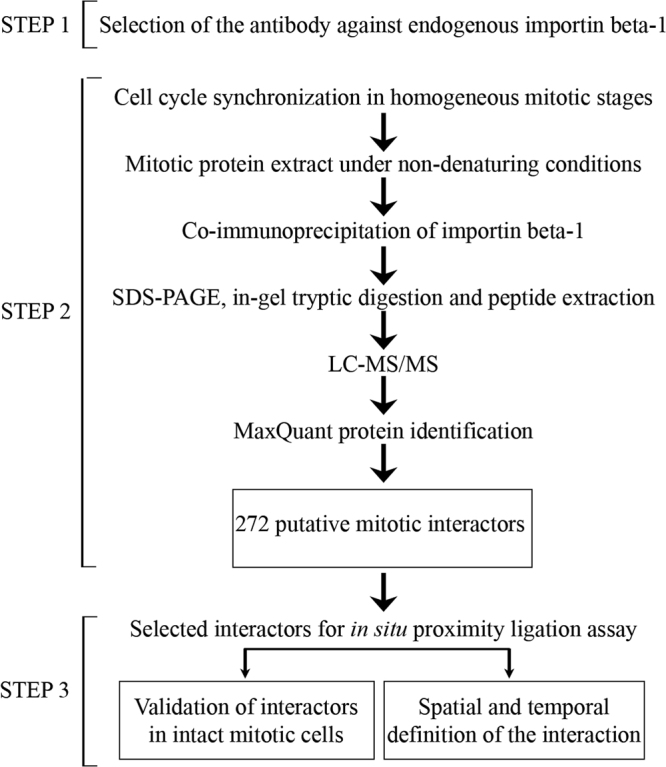
Schematics of the three-step interactomic workflow developed in this work. After selection of the importin beta-1 antibody (step 1), co-immunoprecipitating mitotic partners are identified (non denaturing conditions, low-stringency) and profiled in step 2; in step 3, selected partners are then validated by an automated is-PLA protocol.
Prior to developing affinity purification and mass spectrometry (AP/MS)33 analyses, we had to address two preliminary methodological issues, concerning the importin-beta-1 antibody bait and the purity of mitotic cell populations. Because importin beta-1 can engage various domains to interact with partners, different antibodies might be differentially hindered in co-IP experiments by importin beta-1 partners with which they may compete. We tested commercial antibodies against full-length or specific epitopes of importin beta-1 (map in Supplementary Fig. 1a). To compare with previous work14, protein extracts were prepared from thymidine-arrested (G1/S transition) and released cell cultures collected at mitotic round-up using the “shake-off” procedure. After cross-linking to beads and incubation with equal extract amounts, the antibodies showed differential effectiveness in their yield of immunoprecipitated importin beta-1 (Supplementary Fig. 1b). We compared the co-IP obtained with the three most effective antibodies (2, 4 and 5 in Supplementary Fig. 1a) by low-scale MALDI-TOF analysis. Antibodies against the full-length protein, 4 and 5, consistently identified RAN and RANBP2 in the importin beta-1 co-IP, as expected (Supplementary Fig. 1c). Antibody 5 proved particularly effective in the IP yield (Supplementary Fig. 1b) and was used thereafter.
Preliminarily we also needed to ensure homogeneity of mitotic synchronization. We realized that thymidine-arrested and released cultures tended to loose synchrony during release into S and G2 phases, reaching mitotic stages somewhat asynchronously (Supplementary Fig. 2a). We therefore compared protocols that synchronize the cell cycle closer to mitosis, using: i) the CDK1 inhibitor RO3306 (arrest at G2/M boundary), ii) the monastrol derivative STLC34, which inhibits the kinesin Eg5 and arrests cells in late G2/early prometaphase with unseparated centrosomes. By immunofluorescence (IF), STLC-arrested and released cell cultures, though being more synchronous compared to thymidine release, included many mitoses still exhibiting monopolar spindles 1 hour after wash-out (Supplementary Fig. 2c), suggesting that released cells progress through mitosis before they fully recover from Eg5 inhibition. RO3306-induced arrest and release (30–60 minutes) proved both effective and reversible, yielding cells that accomplished mitosis proficiently and synchronously (Supplementary Fig. 2b). We conclude that RO3306-mediated synchronization and release is suitable for large-scale proteomics in mitotic cell populations.
Profiling importin beta-1 mitotic interactors
In order to profile protein partners contacted by importin beta-1 in mitosis, we pulled down importin beta-1 from HeLa cells under non-denaturing conditions. Mitotic cell extracts were prepared from RO3306-synchronized and released cells collected by mechanical shake-off at mitotic round-up, then incubated with importin beta-1 antibody for preparative co-IP under low salt stringency, followed by Orbitrap MS analysis of mitotic partners. In parallel co-IP experiments, mouse IgG isotype were used as a negative control for non-specific interactors. An extended list of importin beta-1 interactors (272 protein groups) was obtained, which includes known as well as novel importin beta-1 interactors. The full list of all 272 hits is shown in Supplementary Table 1.
Among well-established importin beta-1 interactors we pulled down various classes of factors: i) the GTPase RAN and RANBP1, both with global roles in mitotic spindle organization and dynamics9,10,35,36; ii) importin alpha and importin-7, both of which cooperate with importin beta-1 in nuclear import of particular classes of cargoes in interphase1–3; and iii) components of the RRSU complex (RANBP2, SUMO-conjugated RANGAP1, and UBC9/UBE21), as expected from previous studies14,37.
The importin beta-1 co-IP also includes proteins with clear mitotic roles. Of note are tubulin beta (Supplementary Table 1), consistent with the evidence that importin beta-1 co-sediments with polymerized MTs in mitotic cells12, and NUSAP, a nucleolar protein in interphase that has tubulin-binding and MT-stabilizing activity in mitosis; through these activities NUSAP regulates the formation of asters and long MTs38,39. Interestingly, NUSAP is known to interact with both importin beta-1 and importin-7 (consistent with our detection of importin-7 in the importin beta-1 mitotic interactome); these two importins are reported to differentially inhibit NUSAP’s mitotic functions: importin beta-1 blocks aster assembly, while importin-7 enables the formation of small asters but prevents the assembly of long MTs38. Also of interest is the presence in the interactome of BUB3, a mitotic spindle checkpoint factor that promotes the stabilization of correct (“end-on”) MT attachments to KTs. BUB3 is required for BUB1 localization to KTs, and the BUB3/BUB1 complex cooperates with other spindle checkpoint factors to prevent premature anaphase onset. Importin-beta-1 was previously found to interact with BUB3, thus preserving it from the ubiquitin/proteasome system and regulating its stability40.
Besides these well-characterized factors, we wished to analyse the profile of importin beta-1 mitotic interactors. As a first approach, we examined the list of all 272 identified hits (Supplementary Table 1) by mining the Mitocheck database, built on a genome-wide siRNA-based screening and phenotypic analysis41. 195 of our 272 hits were annotated in Mitocheck. Of those, 86 (roughly 44%) were associated with specific mitotic phenotypes (e.g., mitotic arrest or delay; metaphase alignment problems or no metaphase; segregation problems with chromatin bridges or lagging chromosomes; “grape” phenotype, resulting from chromosome segregation defects at mitotic exit) when their coding genes were silenced. Of the remaining annotated genes, 7 (3.6%) gave a predominantly nuclear phenotype (polylobed or otherwise abnormally shaped nuclei, indicating aberrant nuclear reassembly), 10 (5%) affected cytokinesis, and 51 (26.15%) affected more general features (cell death, or, on the contrary, increased cell proliferation, or altered migration properties). With all possible technical reservations inherent to the Mitocheck siRNA-based methodology, these observations support the conclusion that many importin beta-1 interactors identified in our study are indeed required, directly or indirectly, for some aspect of mitosis.
We next inspected the full list of interactors more closely by text-mining in several resources (see Materials and Methods), in order to get more insight into their nature and functions. Table 1 shows a literature-driven selection of importin beta-1 interactors, most of them originally identified as members of other ontogenic pathways, which nevertheless play roles at various steps of mitotic progression and can thus be regarded as moonlighting proteins. A number of these proteins had not been previously associated with importin beta-1.
Table 1.
Importin beta-1 interacting factors with mitotic functions: literature-driven selection(a).
| Importin beta-1 interaction(b) | |||||
|---|---|---|---|---|---|
| Group/pathway | Protein ID | Gene name | Protein names | Pubmed | Biogrid |
| Nuclear import | Q14974 | KPNB1 | Importin subunit beta-1 | Yes | Yes |
| P52292 | KPNA2 | Importin subunit alpha-1 | Yes | Yes | |
| O95373 | IPO7 | Importin-7 | Yes | Yes | |
| RAN GTPase network | P62826 | RAN | GTP-binding nuclear protein Ran | Yes | Yes |
| P46060 | RANGAP1 | Ran GTPase-activating protein 1 | Yes | Yes | |
| P43487 | RANBP1 | Ran-specific GTPase-activating protein 1 | Yes | Yes | |
| SUMO modification | P49792 | RANBP2 | E3 SUMO-protein ligase RanBP2 | Yes | Yes |
| P63279 | UBE2I | SUMO-conjugating enzyme UBC9 | Yes | Yes | |
| Mitotic spindle | P07437 | TUBB | Tubulin beta chain | Yes | Yes |
| Q9BXS6 | NUSAP1 | Nucleolar spindle-associated protein 1 | Yes | Yes | |
| Mitotic checkpoint | O43684 | BUB3 | Mitotic checkpoint protein BUB3 | Yes | Yes |
| Signal transduction | P62258 | YWHAE | 14-3-3 protein epsilon | Yes | Yes |
| Chaperones | P08238 | HSP90AB1 | Heat shock protein HSP 90-beta | Yes | No |
| P14625 | HSP90B1 | Endoplasmin | Yes | No | |
| Nucleolus | P19338 | NCL | Nucleolin | Yes | Yes |
| Ribosome | P23396 | RPS3 | 40 S ribosomal protein S3 | Yes | No |
| Ribonucleoproteins | P07910 | HNRNPC | Heterogeneous nuclear ribonucleoproteins C1/C2 | Importin alpha-1 | No |
| P61978 | HNRNPK | Heterogeneous nuclear ribonucleoprotein K | Yes | No | |
| Q00839 | HNRNPU | Heterogeneous nuclear ribonucleoprotein U | Yes | No | |
| RNA binding | Q9UMS4 | PRPF19 | Pre-mRNA-processing factor 19 | No | No |
| Q01844 | EWSR1 | RNA-binding protein EWS | No | No | |
| Q96PK6 | RBM14 | RNA-binding protein 14 | No | No | |
| Q07666 | KHDRBS1 | KH domain-containing RNA-binding, signal transduction-associated protein 1 | Yes | No | |
| P84090 | ERH | Enhancer of Rudimentary Homolog | No | No | |
| Q86V81 | ALYREF | THO complex subunit 4 | No | No | |
| RNA helicases | P17844 | DDX5 | Probable ATP-dependent RNA helicase DDDX5 | No | No |
| Q92841 | DDX17 | Probable ATP-dependent RNA helicase DDDX17 | Yes | No | |
| Elongation factor | P13639 | EEF2 | Eukaryotic ElongationFactor 2 | No | No |
| Histones | Q92522 | H1FX | Histone H1x | No | No |
| Cytoskeleton | Q15149 | PLEC | Plectin | No | No |
| P63261 | ACTG1 | Actin, cytoplasmic 2 | No | No | |
| P60709 | ACTB | Actin, cytoplasmic 1 | No | No | |
| P46940 | IQGAP1 | Ras GTPase-activating-like protein IQGAP1 | No | No | |
| Vesicle transport | Q00610 | CLTC | Clathrin h)eavy chain 1 | Functional | No |
| Q92734 | TFG | Protein TFG (Trk-fused gene) | No | No | |
| Redox | Q06830 | PRDX1 | Peroxiredoxin-1 | No | No |
(a)The full list of all 272 hits in the mitotic importin beta-1 co-IP is shown in Supplementary Table 1.
(b)Interactions with imporitn beta-1 indicated as PubMed were retrieved from in-depth studies of specific proteins; all proteins reported in the list were also checked in Biogrid (https://thebiogrid.org/).
Some factors in the list have general roles in mitosis. Some of them, with signalling and chaperone activity, were known to bind importin beta-1. The 14–3–3 epsilon protein YWHAE regulates M entry by associating with and regulating the localization of cdc25B42, and also cooperates with dynein to maintain spindle orientation43. Heat shock proteins have roles in mitotic entry, progression and exit, often - though not exclusively - by regulating the stability of various mitotic proteins44. The HSP90B family members identified in our list were reported to interact with, and generally stabilize, G2/M checkpoint factors (e.g., the Chk1, Wee1, and greatwall kinases) and mitotic regulators (Plk1, topoisomearse II-alpha, survivin, and cyclin B1 itself). Some newly identified importin beta-1 interactors also exert global roles in mitosis, including the heterogeneous nuclear ribonucleoprotein C (HNRNPC)45, the translation elongation factor EEF246, and protein TFG (Trk-fused gene)47: all are involved in regulation of translation, which is essential in mitosis; consistent with this, all - when dysfunctional - yield abnormal mitoses or abrogation of mitosis.
A group of importin beta-1 interactors regulate centrosome function or organization. For example, peroxiredoxin-1 (Prdx1) regulates H2O2 at the centrosome level and is crucial for centrosome function48. Prdx1 is inactivated by Cdk1-dependent phosphorylation in early mitosis, thus unshielding mitotic centrosomes from high H2O2 concentrations, and facilitating the inactivation of centrosomal phosphatases. Dephosphorylation and reactivation of Prdx1 in late mitosis then shields back the centrosome from H2O2, allowing the local reactivation of phosphatases. The abundance of importin beta-1 at spindle poles12, and its ability to interact with Prdx1 detected here, might provide an additional mechanism to regulate the timing of Prdx1 activity/inactivation at centrosomes. Interestingly, the importin beta-1 mitotic interactome also contains catalase (Supplementary Table 1), strengthening the possibility that importin beta-1 might control the redox status during mitosis. RNA-binding protein 14 (RBM14), also acts on centrosome and spindle functions; its inactivation induces ectopic centrosome formation and multipolar spindles49, a phenotype also induced by importin beta-1 overepxression. Plectin regulates centrosome positioning in association with BRCA250.
Many identified interactors localize to the spindle, the main site of importin beta-1 localization in mitosis. Clathrin, a component of the endocytic pathway with RNA-binding capacity, and a recognized example of a moonlighting protein, localizes to MTs and regulates spindle functions, in part by localizing other proteins to the spindle51. Interestingly, clathrin targets the MT-regulatory factor TACC-3 to the spindle MTs52 in an importin-beta-1-sensitive manner: importin beta-1 inhibits clathrin/TACC3 binding, while RANGTP restores it. Despite of this regulatory connection, a direct physical interaction between clathrin and importin beta-1 had not been shown before, suggesting the possibility that importin beta-1 regulates clathrin availability for TACC-3 by binding to it.
In addition, both the ribosomal protein RPS3 and the ribonucleoprotein HNRPU/SAF-A localize at spindles in mitosis and their inactivation yields mitotic delay, spindle defects and chromosome misalignment53,54. Interestingly, HNRPU/SAF-A co-immunoprecipitates with Aurora-A and TPX2, which are required for HNRPU/SAF-A association with the spindle; in turn, HNRPU/SAF-A contributes to targeting of Aurora-A to the spindle MTs55. Importin beta-1 interacts with and negatively regulates TPX2 activity; the identification of HNRPU/SAF-A in the interactome suggests a possible interplay between importin beta-1, HNRPU/SAF-A, TPX2, and Aurora-A in steps of spindle assembly. Among newly identified importin beta-1 interactors localizing to the spindle, the RNA processing and transport protein EWSR1 is required for mitotic entry and, in cells that do enter mitosis, for proper spindle organization56,57. The PRP19 pre-mRNA processing factor affects the mitotic spindle MT density and chromosome alignment58. These newly identified interactors might hint at additional mechanisms through which importin beta-1 can regulate the mitotic spindle.
We also identified regulators of MT/KT interactions in the importin beta-1 interactome, the depletion of which generally results in chromosome misalignment and missegregation. A known example is nucleolin, the major nucleolar component, which interacts with the chromosomal periphery in mitosis, including around outer KTs, where it contributes to regulate MT interactions59. Noteworthily, nucleolin associates with Hsp90, which contributes to stabilize it60, and co-immunoprecipitates with HNRPU/SAF-A55: the importin beta-1 interactome contains all these components. Several newly identified importin beta-1 interactors perform similar function. The spliceosome component ERH (Drosophila Enhancer of Rudimentary Homolog), which also interacts with the interactome member RPS3, regulates the levels of the CENP-E kinetochore-associated motor protein61, and hence KT interactions with MTs. The THO complex subunit 4 (ALYREF), involved in export of spliced mRNA, is also required for proper chromosome alignment62. The RNA helicases (DDX5 and DDX17) regulate the stability of Aurora-B mRNA, and hence the overall Aurora-B protein abundance63. The identification of diverse interactors that play roles in chromosome congression and MT/KT interactions thus expands the range of mechanisms that importin beta-1 might regulate.
Finally, the list of importin beta-1 interactors includes factors acting at mitotic exit. Here we identify KHDRBS1/SAM68, known to play roles in mitotic exit64. In addition, IQGAP recruits several proteins to the cleavage furrow in cytokinesis65; importantly, it functions in recruitment of NUP98 and other NUPs (MAB414 antibody substrates) during NE reassembly at mitotic exit66, a process in which importin beta-1 plays crucial roles6. Together, the data identify a variety of factors and pathways potentially regulated by importin beta-1 at various steps during mitosis.
Automation of a PLA-based method for the validation of interactions
Experimental validation of protein partners after interactomic screening often relies on Western blotting assays of co-immunoprecipitating complexes, or other cell extract-based methods that fail to provide spatial or temporal information on dynamic interactions. We decided to develop an in situ proximity ligation assays (is-PLA) protocol to validate true importin beta-1 interactors in mitotic cells and depict where and when the interactions occur. Is-PLA uses ordinary primary antibodies to the proteins of interest, that are revealed by secondary antibodies conjugated to DNA oligonucleotides of complementary sequence: when the target proteins interact, or are in close proximity, the oligonucleotides can be ligated and amplified, and the amplification products are visualized at the site(s) and temporal window in which the interactions occur. We reasoned that the is-PLA method, if developed to be automated, could advance the validation of the interactomic results and add temporal and spatial information on the identified interactions.
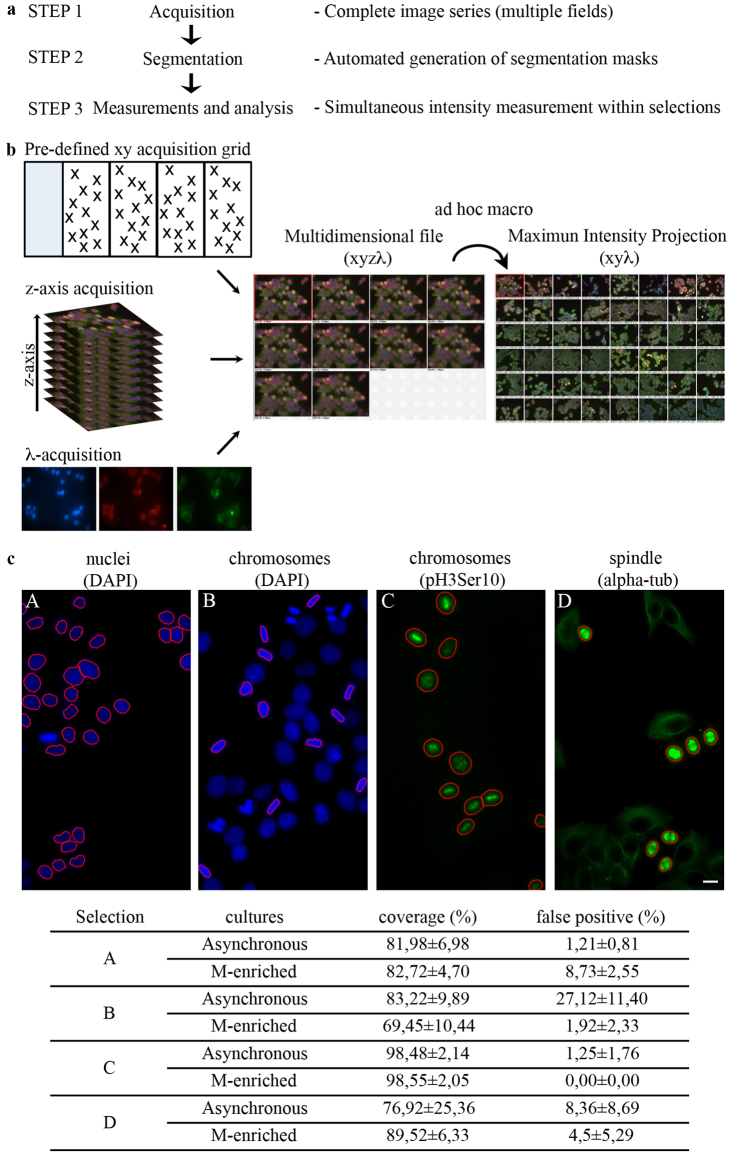
Figure 2a shows a schematics of the is-PLA automation. We combined is-PLA with distinctive labelling markers for mitotic figures, i.e. chromosome labelling (DAPI staining of the DNA, or IF with phospho-Ser10-H3 antibody to the mitotic-specific modification of histone H3), and mitotic spindle visualization (alpha-tubulin IF). Automated image acquisition is schematized in Fig. 2b. Segmentation procedures are used to define single objects within a digital image. We defined intensity, size and circularity thresholds for reproducible image segmentation (see Methods). Panels in Fig. 2c display mitotic cells, automatically distinguished from interphases for their higher DAPI fluorescence intensity and loss of rounded nuclear shape (panel B; compare with interphase nuclei in A), H3 phosphorylation (panel C), and bright stained mitotic spindles (panel D: with the set thresholds for IF intensity and object “roundness”, spindles distinctively stand out from the cytoskeleton of adherent interphase cells). Experiments were set up in both non-synchronized cultures, to control the abundance and localization of the interactions of interest in the absence of checkpoint activation after drug-induced cell cycle arrest, and RO-synchronized cultures to reproduce the conditions used for the interactomic profiling. For all markers, over 70% (non-synchronized cultures) and 80% (mitoses-enriched cultures) of the objects were recognized, with >75% or >90% confidence, respectively (Fig. 2c). Thus, our image acquisition and segmentation workflow yields rapid and accurate detection of mitotic structures in an automated manner.
Figure 2.
Automated image acquisition. (a) The diagram shows the development of a three-step protocol for automated microscopy acquisition and analysis: 1) acquisition of complete image series, giving one multidimensional file as the output; 2) segmentation step, enabling the automated generation of the regions of interest; 3) simultaneous measurements of segmented images in one step. (b) Image acquisition protocol. For each PLA combination, a single file is composed of 60 xy points (acquired using a pre-defined xy acquisition grid), each containing the information from 10 z-stacks and 3 wave-lengths (λ: DAPI; FITC; TRITC). The generated multidimensional file (xyzλ) is processed using an ad-hoc macro for the automated generation of the Maximum intensity projection (xyλ), which is then analyzed as described in step 2 and 3. (c) Segmentation set-up for automated is-PLA: interphase vs. mitotic figures. Exemplifying segmented images are shown for each selection. The % (± standard deviation) coverage (detected objects over the total number of real objects, determined by manual counting of the same images) and false positives (determined by a manual check of the segmentation process) are indicated in the Table below. At least 3 replicates per condition were analyzed. Bar, 10 μm.
Among importin beta-1 interactors to test in PLA assays, we selected RAN, RANBP1 and RANBP2 as established importin beta-1 interactors in all cell cycle stages, and NUSAP and BUB3 as mitotic factors. Two approaches were used to control the specificity of the PLA automated mode. First, as a negative control, native HeLa cells, not expressing GFP, were incubated with importin beta-1 and GFP-specific antibodies, followed by oligonucleotide-conjugated secondary antibodies: no PLA signal was detected, indicating that no aspecific ligation or amplification occurred (Fig. 3a). Second, for each tested reaction, we set up parallel PLAs in cultures treated with importazole, which disrupts importin beta-1 interactions22. Results are shown in Fig. 3. Briefly, importin beta-1/RAN interactions (Fig. 3b, upper panel) were detected within the mitotic spindle selection. Mitotic PLA products were similarly detected when testing importin beta-1 in combination with RANBP1 (Fig. 3b, middle panel) and, more abundantly, with RANBP2 (Fig. 3b, lower panel).
Figure 3.
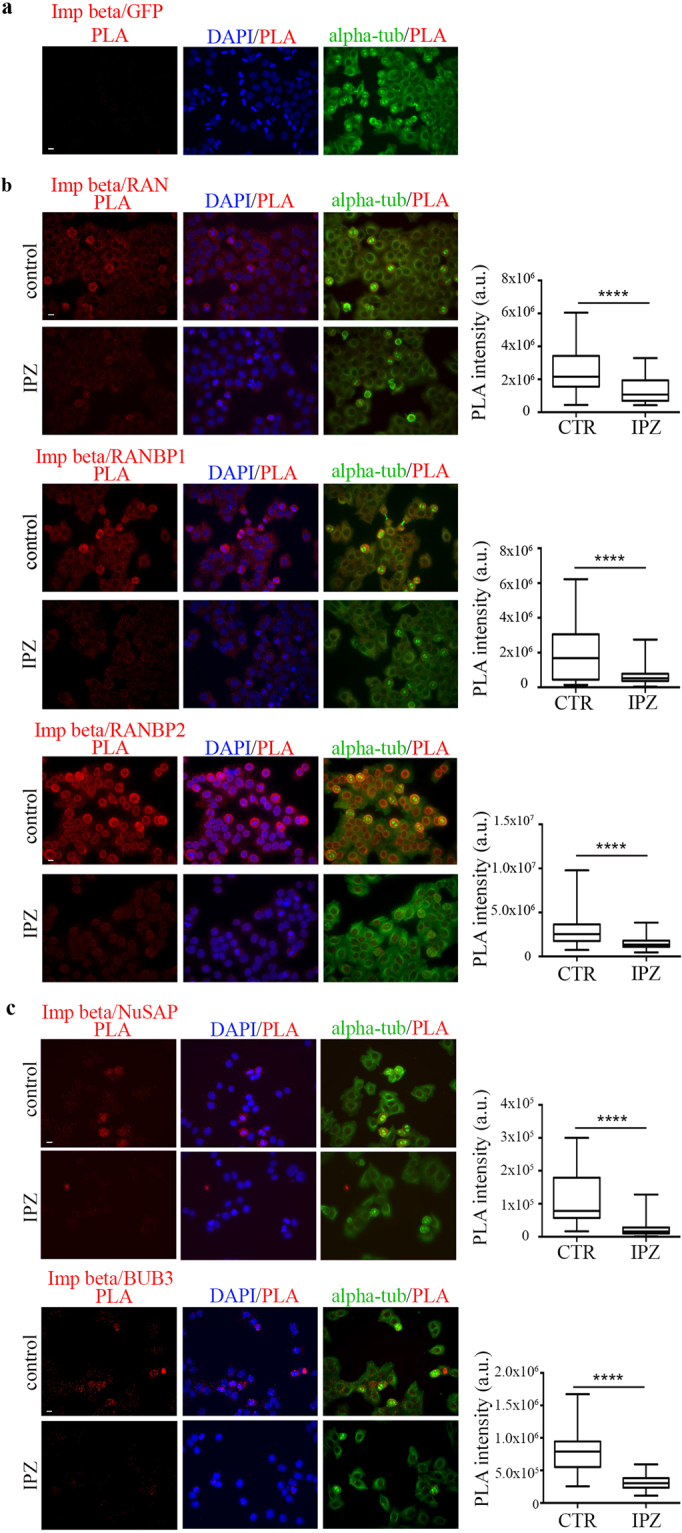
Automated detection of importin beta-1-containing PLA products. (a) Negative control for is-PLA experiments: note the absence of PLA signals in non-transfected HeLa cells processed for amplification after incubation with antibodies for importin beta-1 and GFP (not expressed in HeLa cells). (b) Fluorescence panels on the left show PLA-positive cells for the interaction between importin beta-1 and well-characterized interactors: RAN, RANBP1 and RANBP2, in untreated cells (control, CTR) and after importazole treatment (IPZ); the box-plot panels on the right represent the distribution of PLA signal intensities (arbitrary units, a.u.) in individual cells. (c) PLA-positive cells for the reaction between importin beta-1 and NUSAP or BUB3. Exemplifying panels used for automated measures are shown on the left; on the right, PLA signals are quantified as described for section b. 40 × objective, bar, 10 μm. n counted cells, without and with IPZ were: RAN, 40–35; RANBP1, 190-150; RANBP2, 140-105; NuSAP, 35-30; BUB3, 55-40. ****0.0001, Mann-Whitney test.
The is-PLA protocol also visualized importin-beta-1 interaction products with the spindle-associated protein NUSAP and the spindle checkpoint component BUB3 (Fig. 3c): positive PLA signals were detected for both interactions around the areas of chromosome selection, suggesting an increased abundance in the spindle regions that make contacts with chromosomes. All five PLA reactions were prevented by treatment with importazole. The results demonstrate that the automated PLA mode is reliable and depicts genuine interactions between importin beta-1 and its partners in mitotic cells.
Localization of importin beta-1 PLA interactions with partners in mitotic cells
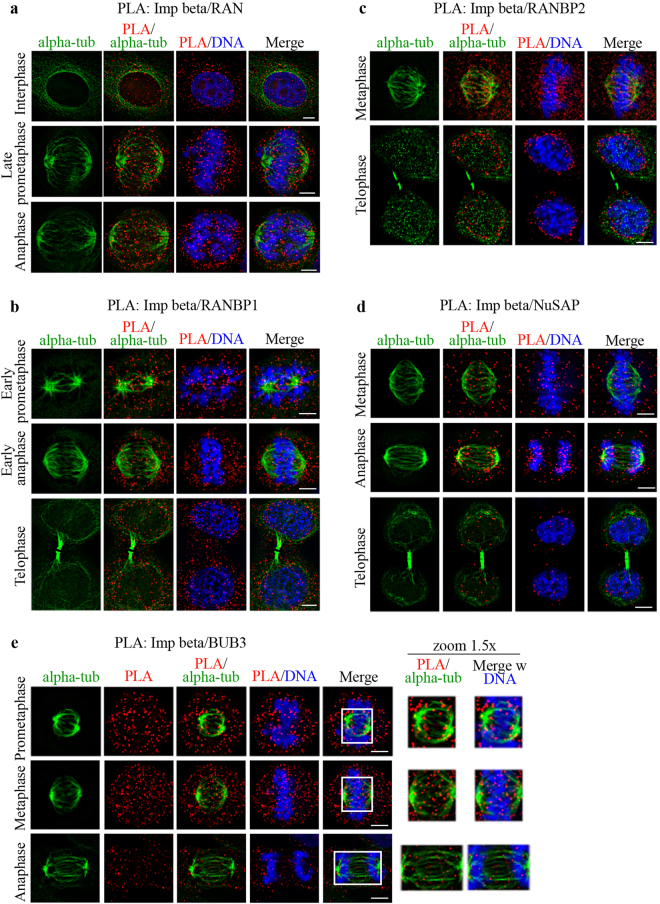
During the unfolding of mitosis, importin beta-1 can use different mechanisms to repress its interactors. It inhibits certain MT-regulatory factors, for example TPX2, which it binds via an importin alpha molecule, by co-localizing with them and rendering them inactive in the interaction. In other cases, importin beta-1 displaces factors away from their site of action. For example, it prevents the unscheduled onset of KT functions by localizing certain regulatory factors (e.g., RANBP2) away from KTs14,29,30. Depicting the spatial map of importin beta-1 interactions with its partners can help to understand its modes of action in mitotic cells. To fully exploit the informative content of the PLA method, we carried out PLA assays in asynchronous HeLa cultures, where all stages of mitosis are present with physiological frequency, and examined the PLA interaction signals in higher magnification to characterize their localization and abundance with spatiotemporal resolution.
RAN is a highly mobile protein. Previous studies depicted RANGTP enrichment around chromosomes using an indirect method based on FRET reporters for RANGTP-rich territories13,67. RANGTP-specific conformational antibody68 also detected fractions at centrosomes69 and spindle MTs35. In PLA assays, RAN/importin beta-1 interactions were visualized around and within the spindle region (Fig. 4a), particularly along MTs (late prometaphase, middle row). PLA signals remained visible at MTs during spindle elongation and chromosome segregation in anaphase (lower row).
Figure 4.
Spatiotemporal resolution of PLA interactions between importin beta-1 and the indicated partners during mitotic progression. PLA panels (100 × objective) depict the sites of interaction of importin beta-1 with partners validated in Fig. 3, relative to the mitotic apparatus (alpha-tubulin), chromosomes or reforming nuclei (DAPI): (a) RAN; (b) RANBP1; (c) RANBP2; (d) NUSAP; (e) BUB3. Representative stages for each combination are shown and details are given in the text. Bar, 5 μm.
We then assessed the mitotic localization of importin beta-1/ RANBP1 PLA products. In early prometaphase, RANBP1/importin beta-1 PLA products were found to distribute throughout the cell volume, yet showed a discrete fraction localizing around asters from which MTs project (Fig. 4b, top row), then persisted in the cytoplasm and at spindle MTs (middle row). In anaphase/early telophase (recognized from the thick midbody), PLA products relocated around the segregating chromosomes (Fig. 4b, bottom row). In more advanced telophase (elongated midbody, decondensed chromatin) PLA products decreased (data not shown), consistent with the notion that RANBP1 is down-regulated at mitotic exit, concomitant with importin beta-1’s resumption of function in nuclear and NE reorganization70.
We next analyzed importin beta-1/RANBP2 PLA products. PLA signals localized along the spindle MTs and at the interface between MT plus-ends and KTs in metaphase (Fig. 4c, top row); chromosomal regions were devoid of PLA signals, consistent with the notion that the KT-associated RANBP2 pool is free of importin beta-130. In telophase, importin beta-1/RANBP2 PLA products relocalized around the decondensing chromatin, in the region where the NE and NPCs reassemble around the reforming nuclei (Fig. 4c, lower row).
PLA analysis of the importin beta-1/NUSAP pair showed a temporally regulated pattern (Fig. 4d). In early mitosis, PLA signals were distributed throughout the cytoplasm, with a small fraction along MTs, suggesting transient dispersal after NE breakdown and nucleolus disassembly (data not shown). The signals then progressively concentrated at MTs until metaphase (Fig. 4d, upper row); most PLA products then remained associated to MTs, while the spindle poles moved apart during anaphase, resulting in the gradual distancing of the PLA product sets during chromosome segregation (middle row). At mitotic exit, PLA signals were visible within nuclei, indicating the resumption of importin beta-1-dependent nuclear import of NUSAP (bottom row). We noticed that intranuclear PLA products have low abundance, suggesting that, once they enter the nucleus, RANGTP readily disassembles them.
Lastly we examined importin beta-1/BUB3 PLA products. As mentioned, importin beta-1 interaction with BUB3 was shown to protect the latter from ubiquitination, and hence proteasome targeting, ultimately modulating the timing of the metaphase-to-anaphase transition40. Most studies of BUB3 used GFP-tagged overexpressed chimaeras, and so the endogenous localization is not extensively documented during mitotic progression. We therefore first assessed the precise localization of endogenous, untagged BUB3 in mitotic cells by IF. After mild solubilization, BUB3 was depicted at prometaphase and metaphase KTs (visualized by CREST); its amount increased in cells treated with nocodazole (NOC), which prevented MT assembly and induced sustained activation of the spindle assembly checkpoint (Supplementary Fig. 3). In PLA images (Fig. 4e), many importin beta-1/BUB3 PLA signals are dispersed in the cell volume, with a fraction associated with MTs in prometaphase (Fig. 4e, top row), and in the vicinity of the MT/KT interface in metaphase (middle row). The signals decreased in anaphase (compare the signal abundance during mitotic progression in the PLA column), consistent with the timing at which BUB3 is reported to become accessible to the degradation machinery39.
Together, the PLA results for five selected interactions provide an effective validation tool for the co-immunoprecipitating mitotic partners identified in our proteome-wide approach. In addition, they depict spatial and dynamic information for specific pairs of importin beta-1 and its partners, which could not have been obtained otherwise.
Discussion
Here we report novel information on the versatile ability of importin beta-1 to interact with mitotic partners, which extends and complements studies of interphase transport. We have defined an extensive list of importin beta-1 interacting partners in human mitotic cells. Many proteins identified here are implicated in control of mitosis at various steps and had not been previously associated with importin beta-1. The identification of novel mitotic interactors is relevant, given the implication of importin beta-1 in the generation of mitotic abnormalities in contexts in which it is expressed in a deregulated manner, as in many cancer types15–20.
We have identified both known and new targets of importin beta-1. Thirty-five proteins were previously identified as importin beta-1 partners using stable isotope labeling by amino acids in cell cultures (SILAC) in a study of transport pathways24, and included essentially ribosomal proteins, nuclear RNPs, and a few proteins acting as nuclear import cargoes in interphase, e.g. nucleolin. Some actually act in mitosis, indicating “moonlighting” roles. We also depicted all components of the RRSU SUMO ligase complex, which is emerging as a temporal regulator of KT functions28,30,31, consistent with the established ability of importin beta-1 to interact with RANBP214,37. Another interesting group comprises tubulin beta, the tubulin-binding protein NUSAP and importin-7, with which NUSAP also interacts. These data provide reciprocal evidence to previous studies that identified importin beta-1 in the mitotic MT interactome71 and, more recently, in the co-IP of NUSAP, together with the MT-regulatory kinesin MCAK72, and demonstrate the reliability of our identification method.
We have searched several resources to obtain specific information on importin beta-1 mitotic interactors. Cross-comparison of our full interactome list (Supplementary Table 1) vs. the Mitocheck database confirms that most importin beta-1 interactors identified here actually play roles in mitosis, as their inactivation is associated with aberrant mitotic phenotypes. Importin beta-1 generally inhibits partners with which it interacts. Our data, therefore, may hint at novel potential pathways of importin beta-1-dependent mitotic control. A number of novel importin beta-1 mitotic interactors (Table 1) include ribonucleoproteins, ribosomal proteins, RNA-binding, splicing and processing factors, and even components of the “translasome” (elongation factors). Mitotic roles are growingly emerging for many proteins of these groups, consistent with the finding that RNAs are part of the mitotic apparatus73. Elucidating the precise mechanism by which importin beta-1 acts on these factors, and their regulated pathways, will be important to uncover yet poorly investigated pathways of control of specific mitotic events by importin beta-1.
Cell cycle synchronization protocols were optimised prior to developing our interactomic analysis; nevertheless, due to the highly dynamic nature of mitosis and the rapidity of the prometaphase-metaphase transition, interactions that vary in abundance before or after MT attachment to KTs, such as that involving BUB3 for example, may be over- or under-represented in our list. It was therefore important to validate the interactions identified in the proteomic screening with spatiotemporal resolution in mitotic cells. We report an is-PLA-based automated method that has enabled us to visualize the actual timing and sites at which importin beta-1-containing complexes localize in intact mitotic cells. Our assays show that is-PLA provides a highly informative tool, yielding information that complements that derived from biochemical assays, where detection methods depict the average behaviour of a bulk cell population or cell extract. The integration of these approaches in automated pipelines may represent a key step in high-content screening of protein interactions74,75.
High resolution is-PLA assays illustrate the potential of the method to provide visual evidence for functional mechanisms of importin beta-1 with specific partners during mitosis. For example, importin beta-1/NUSAP interactions show variations both in localization and in abundance in mitosis, which may differentially modulate NUSAP activity before, and after, MT attachments to kinetochores. Similarly, we visualized BUB3/importin beta-1 PLA products in early mitosis, which became down-regulated after metaphase, consistent with models for the accessibility of importin beta-1-free BUB3 to the ubiquitination/proteasome machinery past metaphase. More generally, subcellular “territories” in which particular interactions occur can be identified, and predictions for modes of mitotic regulation may be drawn.
In conclusion, the information presented in this work extend our knowledge of importin beta-1 functions available from interphase transport studies and can assist the systematic dissection of the global regulatory roles of importin beta-1 during mitosis.
Materials and Methods
Cell cultures
Human HeLa cervix carcinoma epithelial cells were cultured in DMEM supplemented with 10% fetal calf serum, 2% L-glutamine, and 2% penicillin/streptomycin in a humidified atmosphere at 37 °C in 5% CO2. Preliminary comparisons of cell cycle synchronization protcols are described in detail in Supplementary Materials and Methods. For proteomic analysis, 5 × 106 HeLa cells were grown in 150 cm2 flasks in the presence of RO3306 (9 μM; SML0569, Sigma) for 20 hours, then released in RO3306-free medium, monitored under an inverted microscope and collected at round-up by mechanical shake-off. For automated image acquisition, cells were grown on 4-chamber Culture Slides (Falcon) (8 × 104 cells per 1.7 cm2 well), and either left to cycle asynchronously, or synchronized using RO3306 as above and fixed 60 minutes after RO3306 wash-out. Where indicated, cells were treated with 50 μM Importazole (Sigma) for the last 2–3 hours of culture before harvesting the cells.
Preparation of sepharose-antibody immunoconjugate
To covalently bind antibodies to Protein G-conjugated sepharose beads (Protein G-Sepharose 4 Fast Flow; GE Healthcare), dimethylpimelimidate (DMP; Sigma-Aldrich) was used as crosslinker. Importin beta-1 antibody Sigma I2534 (roughly, 2.5 μg equivalent), or mouse IgG (Santa Cruz Biotechnology sc-2025), were diluted in 1 ml of phosphate buffered saline (PBS), mixed with 15 μl of bead pellet, and incubated overnight at 4 °C on a rotating wheel. The mixture was then centrifuged (3000 rpm, 4 °C, 5 minutes), the pellet was washed with 10 volumes of 0.2 M sodium borate (pH 9.0) and suspended in 100 volumes of 0.2 M sodium borate (pH 9.0)/PBS, to which 20 mM DMP was added just before use. After rotation for 1 hour at room temperature (RT), the beads were washed in 0.2 M ethanolamine (pH 9.0). The remaining reactive amino groups were quenched by adding 0.2 M ethanolamine (pH 9.0) (RT, 2 hours). The bead pellet was then centrifuged (3000 rpm, RT, 5 minutes), washed with 0.1 M glycine (pH 2.5) to remove non-crosslinked antibodies, washed further 3 × in 10 volumes PBS, and stored in PBS at 4 °C.
Co-immunoprecipitation experiments
The importin beta-1 interactome was captured from mitotic HeLa cells using anti-Importin beta-1 antibody (Sigma-Aldrich I2534). Mouse IgG antibodies (Santa Cruz Biotechnology sc-2025) were used as a control for non-specific binding proteins. RO336-released mitotic cells were centrifuged at low speed, washed twice at 4 °C in cold PBS/0.5 mM PMSF and resuspended in lysis buffer (50 mM Tris HCl (pH 8.0), 150 mM NaCl, 1% NP40) containing protease and phosphatase inhibitors (aprotinin, leupeptin, and pepstatin, all 1 μg/ml; 1 mM PMSF, 50 mM sodium fluoride, 2 mM sodium orthovanadate, 10 mM sodium pyrophosphate). After incubation on ice for 30 minutes, cells were gently homogenized using a Dounce homogenizer and the extent of mechanical disruption of the cells was checked by microscopic observation. After centrifugation (14000 rpm, 4 °C, 20 minutes), the supernatant was quantified by Bradford assay and 1 mg of lysate was immediately precleared with Protein G-Sepharose at 4 °C for 1 hour (15 μl of resin slurry per mg total protein). Importin beta-1 interacting partners were co-immunoprecipitated overnight on a rotating wheel at 4 °C from the precleared mitotic supernatant with anti-importin-beta-1 antibody covalently coupled to Protein G-Sepharose (1 mg protein sample: 2 μg covalent immune-conjugate, home made as described above). For control, the same amount of precleared mitotic extract was incubated with mouse non-specific IgG covalently coupled with Protein G-Sepharose. The mixture was then centrifuged at 3000 rpm, 4 °C for 15 minutes and the pellet was washed 5 × in lysis buffer and again centrifuged as above. After the last centrifugation step, protein complexes were eluted from the beads by a gentle 3 × washing cycle in 15 μl of 0.1 M glycine buffer (pH 2.5) at RT. After centrifugation (3000 rpm, RT, 5 minutes), the pH of the eluted proteins was neutralized by adding 1.5 M Tris HCl pH 8.0.
Proteomics and data analysis
Aliquots of co-immunoprecipitates were diluted in Laemmli buffer and fractionated through SDS-PAGE on 4–20% Mini-PROTEAN TGX™ gel (Bio-Rad). After colloidal Coomassie staining76, gel slices were processed as described77 and tryptic peptide mixtures extracted and desalted according to ref.78. LC-MS/MS analyses were performed by reverse phase chromatography (C18, 5 μm particle size, 200 Å pore size; Magic C18 AQ, Michrom), on an Ultimate3000 system (Dionex, Sunnyvale, CA, USA). Peptides eluted by a two-step gradient of ACN containing 0.1% FA (5–40% in 120 minutes, and 40–85% in 15 minutes) at 300 nl/min flow rate, were directly injected into an LTQ-Orbitrap XL mass spectrometer (Thermo-Fisher Scientific). Data-dependent tandem MS were performed with full precursor ion scans (MS1) collected at 30,000 resolution, with an automatic gain control (AGC) of 1 × 106 ions, and maximal injection time of 1000 ms. The five most intense (>200 counts) ions with charge states ≥ +2 were selected for collision-induced dissociation (CID). Dynamic exclusion was active with 90 ms exclusion for ions selected twice within a 30 ms window. For MS/MS scanning, the minimum MS signal was set to 500, activation time to 30 ms, target value to 10,000 ions, and injection time of 100 ms. All MS/MS spectra were collected using a normalized collision energy of 35% and an isolation window of 2 Th. Spectra were searched against the Homo sapiens UniProtKB database (release 2016–07–09, 20154 sequences) and common contaminant proteins using the software package MaxQuant (version 1.5.5.1, Max Planck Institute of Biochemistry, Martinsried, Germany; 64). We set oxidation (methionine) and acetylation (protein N-terminus) as variable modifications, carbamidomethylation (cysteine) as fixed modification, mass tolerance of 20 ppm for the precursor ion (MS) and of 0.5 Da for the fragment ions (MSMS). High-confidence peptide-spectral matches were filtered at <1% false discovery rate. Due to the high sensitivity and resolving power of the mass spectrometric platform used here, together with the high power of the Andromeda search engine in assigning peptide sequences from tandem MS data, the MaxQuant platform yielded a large set of starting data. As a first step of our established proteomic pipeline in data analysis of the original outputs, we discarded identifications for all annotated potential laboratory contaminants (i.e. keratins from experimenters, proteins from reagents etc.), and for hits that the search engine identified in a reverse decoy database used as a false negative control. To extract meaningful information from this cleaned data, a higher confidence matrix was set up by filtering out protein groups recognized with a low confidence level. In particular, we removed proteins which the Andromeda algorithm identified based on an extremely limited number of assigned peptide sequences (i.e.: i) number of unique peptides ≤0, ii) number of peptides ≤1, and iii) only by a peptide carrying a post-translational modification, or iv) through less than 3 MS/MS spectra), or v) presenting an ambiguous association, e.g. associated with multiple genes; see Supplementary Table 1 header legend). Finally, mass spectra intensity difference values between the pulled-down proteins by anti-importin beta-1 and by a non-specific mouse IgG were statistically compared using the Perseus software package (version 1.5.5.3, Max Planck Institute of Biochemistry, Martinsried, Germany79) (p-value < 0.05). The resultant list of the 272 high confidence hits was analyzed by data mining against the interphase list24, the Mitocheck database version 2.1 (based on EnsEMBL v89, http://www.mitocheck.org/gene.shtml), and the latest releases of UNiProt http://www.uniprot.org/, Biogrid https://thebiogrid.org/, MiCroKITS http://microkit.biocuckoo.org/ and Pubmed https://www.ncbi.nlm.nih.gov/pubmed.
Immunofluorescence
Cells grown on coverlisps were fixed in 3.7% paraformaldehyde/30 mM sucrose (10 min), permeabilized in 0.1% Triton X-100, then blocked in 3% bovine serum albumin in PBS/0.05% Tween-20 (1 hour, RT). Antibodies used for image segmentation were: chicken anti-alpha-tubulin (Abcam, Ab89984, 1:50), mouse anti-phospho-Ser10-histoneH3 (Millipore, 1:5000). Secondary antibodies were: anti-chicken FITC (Jackson Immunoresearch Laboratories, 1:100), anti-mouse FITC (Jackson Immunoresearch Laboratories, 1:200) and anti-rabbit Cy3 (Jackson Immunoresearch Laboratories, 1:1000). DNA was stained with 0.1 μg/ml 4,6-diamidino-2-phenylindole (DAPI, Sigma Aldrich). Slides were mounted in Vectashield (Vector Laboratories).
Proximity ligation assay
Cells grown on 4-chamber Culture Slides (Falcon) were processed for PLA in four steps: i) incubation of fixed cell slides with primary specific antibodies for the proteins under examination; ii) incubation with secondary antibodies conjugated with complementary oligonucleotide tails (PLA probes, conventionally called PLUS and MINUS); iii) ligase addition; when the target proteins interact or are very close, the ligation step will produce a DNA circle; iv) rolling circle amplification. All steps were performed using the Duolink® In Situ Detection Reagents Red DUO92008 (Sigma-Aldrich). Cells were fixed, blocked, and incubated with primary antibodies as for conventional IF; we used combination of mouse and rabbit primary antibodies for each protein pair (Supplementary Fig. 3). Anti-mouse MINUS and anti-rabbit PLUS PLA probes (PLADuolink® In Situ PLA® Probe Anti-Rabbit PLUS, Affinity purified Donkey anti-Rabbit IgG (H + L), DUO92002, and Duolink® In Situ PLA® Probe Anti-Mouse MINUS, Affinity purified Donkey anti-Mouse IgG (H + L) DUO92004, respectively, Sigma-Aldrich) were then added (diluted 1:5 in PBS containing 0.05% Tween-20 and 3% bovine serum albumin) and incubated in a pre-heated humidity chamber (60 minutes, 37 °C). Subsequent ligation (30 minutes, 37 °C) and amplification (70 minutes, 37 °C) steps were performed following the Olink Bioscience protocol. To localize PLA signals, cells were co-stained using DAPI and chicken anti-alpha tubulin antibody, followed by FITC-conjugated anti-chicken IgG.
Image segmentation and measurements
Segmentation is used to partition a digital image to identify objects or extract relevant information. After a background subtraction step, image segmentation on Maximum Intensity Projections was performed using Nis-Elements HC 4.2 (Nikon). We used the Threshold function of the Nis-Elements HC 4.2 software to define regions in the images where we intended to perform the analysis (nuclei, chromosomes, spindles), based on signal intensity, size and circularity. Segmentation masks for chromosomes and spindles were saved and automatically reloaded for each new image set. All steps were performed on a single.nd2 file for the whole experiment acquisition. To evaluate the effectiveness of segmentation, manual analysis of the same images was performed in parallel. For the automated analysis of PLA signals, the “Automated measurements results” function of the software was used: Sum Intensity values for PLA signals within the selected masks were obtained and exported to Excel. Data were statistically analyzed using Graph Pad Prism 6.
Automated image acquisition
Images from cell samples processed for PLA in 4-chamber culture slides were acquired under a Nikon Eclipse Ti microscope equipped with the Perfect Focus System, a Nikon DS-Qi1 Cooled Digital Monochrome Camera and a 40 × objective (PlanFuor, N.A. 0,75, Nikon), using the Nis-Elements AR 3.2 software. A grid (15 fields per well) was generated, saved and re-loaded for all acquisitions, with a focus adjustment step performed for each new slide. In order to collect the whole spindle-associated signal from rounded-up mitoses, images were acquired over an 8 μm range along the z-dimension (z-step: 0.8 μm). An ad-hoc macro was then applied for the automated generation of 2D projection (Maximum Intensity Projection) of all acquired images within a single.nd2 file at the end of the acquisition process.
Image acquisition for high-resolution PLA analysis
Fixed samples on microscope slides were analyzed under a Nikon Eclipse 90i microscope equipped with a Qicam Fast CCD camera (Qimaging). Single-cell images were taken using an immersion oil 100 × objective (NA 1.3). Images (.nd2 format) were acquired using NIS-Elements AR 3.2 software (Nikon) along the z-axis (0.4 μm stacks, range: 4.4 μm); out of focus planes were uniformly removed before processing. After a background subtraction step, images were deconvolved using the ‘AutoQuant’ deconvolution module of NIS-Element HC (latest release, HC 5.02). Image projections were created using the Extended Depth of Focus (EDF) function of NIS-Element HC 5.02. Focused images were uniformly processed using the LUT (Look Up Table) tool of NIS-Element HC 5.02, and exported in TIFF format for Figure composition using Photoshop CS6.
Data Availability
The data that support the findings of this study are available from the authors upon reasonable request.
Electronic supplementary material
Acknowledgements
This work was supported by grants from Consiglio Nazionale delle Ricerche InterOmics Flagship Project to PL and MES (grant SYNLETHER), Associazione Italiana per la Ricerca sul Cancro to PL (AIRC IG14534) and Fondi di Ateneo (Sapienza Università di Roma) to MES. IAA was supported by a fellowship from Fondazione Italiana per la Ricerca sul Cancro (FIRC grant 18313). We are grateful to Gianni Colotti and Stefano Gianni for suggestions and advice. We would also like to acknowledge Erica Di Cesare, Andrea Ilari and Veronica Morea for many discussions and support throughout this work.
Author Contributions
L.D.F. designed and performed proteomics experiments; A.V. designed and performed cell cycle and cell biology experiments, contributed to write the paper; I.A.A. designed and performed automated imaging analyses; P.R. performed fluorescence experiments and microscopy analyses; P.C. developed ad-hoc macros and software tools for image automation; G.G. supervised image automation and microscopy; M.E.S. supervised interactomics studies and wrote the paper; P.L. supervised cell biology studies and wrote the paper. All authors reviewed the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Laura Di Francesco and Annalisa Verrico contributed equally to this work.
Maria Eugenia Schininà and Patrizia Lavia jointly supervised this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-19351-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Maria Eugenia Schininà, Email: eugenia.schinina@uniroma1.it.
Patrizia Lavia, Email: patrizia.lavia@uniroma1.it.
References
- 1.Harel A, Forbes DJ. Importin beta-1: conducting a much larger cellular symphony. Mol. Cell. 2004;16:319–30. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Chook YM, Süel KE. Nuclear import by karyopherin-βs: recognition and inhibition. Biochim. Biophys. Acta. 2011;1813:1593–606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kimura M, Imamoto N. Biological significance of the importin-β family-dependent nucleocytoplasmic transport pathways. Traffic. 2014;15:727–48. doi: 10.1111/tra.12174. [DOI] [PubMed] [Google Scholar]
- 4.Forbes DJ, Travesa A, Nord MS, Bernis C. Nuclear transport factors: global regulation of mitosis. Curr. Opin. Cell Biol. 2015;35:78–90. doi: 10.1016/j.ceb.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schellhaus AK, De Magistris P, Antonin W. Nuclear reformation at the end of mitosis. J. Mol. Biol. 2016;428:1962–85. doi: 10.1016/j.jmb.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 6.Ungricht R, Kutay U. Mechanisms and functions of nuclear envelope remodelling. Nat. Rev. Mol. Cell Biol. 2017;18:229–45. doi: 10.1038/nrm.2016.153. [DOI] [PubMed] [Google Scholar]
- 7.Christie M, et al. Structural biology and regulation of protein import into the nucleus. J. Mol. Biol. 2016;428:2060–90. doi: 10.1016/j.jmb.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 8.Marfori M, et al. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta. 2011;1813:1562–77. doi: 10.1016/j.bbamcr.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 9.Ciciarello M, Mangiacasale R, Lavia P. Spatial control of mitosis by the GTPase Ran. Cell. Mol. Life. Sci. 2007;6:1891–914. doi: 10.1007/s00018-007-6568-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalab P, Heald R. The RanGTP gradient - a GPS for the mitotic spindle. J. Cell Sci. 2008;121:1577–86. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nachury MV, et al. Importin beta-1 is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 2001;104:95–106. doi: 10.1016/S0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- 12.Ciciarello M, et al. Importin beta-1 is transported to spindle poles during mitosis and regulates Ran-dependent spindle assembly factors in mammalian cells. J. Cell Sci. 2004;117:6511–22. doi: 10.1242/jcs.01569. [DOI] [PubMed] [Google Scholar]
- 13.Kaláb P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- 14.Roscioli E, et al. Importin-β negatively regulates multiple aspects of mitosis including RANGAP1 recruitment to kinetochores. J. Cell Biol. 2012;196:435–50. doi: 10.1083/jcb.201109104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Watt PJ, et al. The Karyopherin proteins, Crm1 and Karyopherinbeta1, are overexpressed in cervical cancer and are critical for cancer cell survival and proliferation. Int. J. Cancer. 2009;124:1829–40. doi: 10.1002/ijc.24146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu J, et al. Upregulation of KPNβ1 in gastric cancer cell promotes tumor cell proliferation and predicts poor prognosis. Tumour Biol. 2016;37:661–72. doi: 10.1007/s13277-015-3839-7. [DOI] [PubMed] [Google Scholar]
- 17.He S, et al. Upregulation of nuclear transporter, Kpnβ1, contributes to accelerated cell proliferation- and cell adhesion-mediated drug resistance (CAM-DR) in diffuse large B-cell lymphoma. J. Cancer Res. Clin. Oncol. 2016;142:561–72. doi: 10.1007/s00432-015-2057-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martens-de Kemp SR, et al. Functional genetic screens identify genes essential for tumor cell survival in head and neck and lung cancer. Clin. Cancer Res. 2013;19:1994–2003. doi: 10.1158/1078-0432.CCR-12-2539. [DOI] [PubMed] [Google Scholar]
- 19.Kuusisto HV, Jans DA. Hyper-dependence of breast cancer cell types on the nuclear transporter Importin β1. Biochim. Biophys. Acta. 2015;1853:1870–8. doi: 10.1016/j.bbamcr.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Yang L, et al. Suppression of the nuclear transporter-KPNβ1 expression inhibits tumor proliferation in hepatocellular carcinoma. Med. Oncol. 2015;32:128. doi: 10.1007/s12032-015-0559-1. [DOI] [PubMed] [Google Scholar]
- 21.van der Watt PJ, Stowell CL, Leaner VD. The nuclear import receptor Kpnβ1 and its potential as an anticancer therapeutic target. Crit. Rev. Eukaryot. Gene Expr. 2013;23:1–10. doi: 10.1615/CritRevEukarGeneExpr.2013004845. [DOI] [PubMed] [Google Scholar]
- 22.Soderholm JF, et al. Importazole, a small molecule inhibitor of the transport receptor importin-β. ACS Chem. Biol. 2011;6:700–8. doi: 10.1021/cb2000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Watt PJ, et al. Targeting the nuclear import receptor Kpnβ1 as an anticancer therapeutic. Mol. Cancer Ther. 2016;15:560–73. doi: 10.1158/1535-7163.MCT-15-0052. [DOI] [PubMed] [Google Scholar]
- 24.Kimura M, Okumura N, Kose S, Takao T, Imamoto N. Identification of cargo proteins specific for importin-β with importin-α applying a stable isotope labeling by amino acids in cell culture (SILAC)-based in vitro transport system. J. Biol. Chem. 2013;288:24540–9. doi: 10.1074/jbc.M113.489286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura M, et al. Extensive cargo identification reveals distinct biological roles of the 12 importin pathways. Elife. 2017;6:e21184. doi: 10.7554/eLife.21184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swaminathan S, et al. RanGAP1*SUMO1 is phosphorylated at the onset of mitosis and remains associated with RanBP2 upon NPC disassembly. J. Cell Biol. 2004;164:965–71. doi: 10.1083/jcb.200309126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Werner A, Flotho A, Melchior F. The RanBP2/RanGAP1*SUMO1/Ubc9 complex is a multisubunit SUMO E3 ligase. Mol. Cell. 2012;46:287–98. doi: 10.1016/j.molcel.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 28.Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 2004;14:611–7. doi: 10.1016/j.cub.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 29.Torosantucci L, De Luca M, Guarguaglini G, Lavia P, Degrassi F. Localized RanGTP accumulation promotes microtubule nucleation at kinetochores in somatic mammalian cells. Mol. Biol. Cell. 2008;19:1873–82. doi: 10.1091/mbc.E07-10-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilistro E, et al. P. Importin beta and CRM1 control a RANBP2 spatiotemporal switch essential for mitotic kinetochore function. J. Cell Sci. 2017;130:2564–2578. doi: 10.1242/jcs.197905. [DOI] [PubMed] [Google Scholar]
- 31.Vecchione L, et al. A vulnerability of a subset of colon cancers with potential clinical utility. Cell. 2016;165(2):317–30. doi: 10.1016/j.cell.2016.02.059. [DOI] [PubMed] [Google Scholar]
- 32.Weibrecht I, et al. Proximity ligation assays: a recent addition to the proteomics toolbox. Expert. Rev. Proteomics. 2010;7:401–9. doi: 10.1586/epr.10.10. [DOI] [PubMed] [Google Scholar]
- 33.Pardo M, Choudhary JS. Assignment of protein interactions from affinity purification/mass spectrometry data. J. Proteome Res. 2012;11:1462–74. doi: 10.1021/pr2011632. [DOI] [PubMed] [Google Scholar]
- 34.Skoufias DA, et al. S-trityl-L-cysteine is a reversible, tight binding inhibitor of the human kinesin Eg5 that specifically blocks mitotic progression. J. Biol. Chem. 2006;281:17559–69. doi: 10.1074/jbc.M511735200. [DOI] [PubMed] [Google Scholar]
- 35.Tedeschi A, et al. RANBP1 localizes a subset of mitotic regulatory factors on spindle microtubules and regulates chromosome segregation in human cells. J. Cell Sci. 2007;120:3748–61. doi: 10.1242/jcs.009308. [DOI] [PubMed] [Google Scholar]
- 36.Zhang MS, Arnaoutov A, Dasso M. RanBP1 governs spindle assembly by defining mitotic Ran-GTP production. Dev. Cell. 2014;31:393–404. doi: 10.1016/j.devcel.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hashizume C, Kobayashi A, Wong RW. Down-modulation of nucleoporin RanBP2/Nup358 impaired chromosomal alignment and induced mitotic catastrophe. Cell Death. Dis. 2013;4:e854. doi: 10.1038/cddis.2013.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ribbeck K, et al. NuSAP, a mitotic RanGTP target that stabilizes and cross-links microtubules. Mol. Biol. Cell. 2006;17:2646–60. doi: 10.1091/mbc.E05-12-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ribbeck K, Raemaekers T, Carmeliet G, Mattaj IW. A role for NuSAP in linking microtubules to mitotic chromosomes. Curr. Biol. 2007;17:230–6. doi: 10.1016/j.cub.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 40.Jiang H, He X, Feng D, Zhu X, Zheng Y. RanGTP aids anaphase entry through Ubr5-mediated protein turnover. J. Cell Biol. 2015;211:7–18. doi: 10.1083/jcb.201503122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neumann B, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–7. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cui C, et al. 14-3-3 epsilon prevents G2/M transition of fertilized mouse eggs by binding with CDC25B. BMC Dev. Biol. 2014;14:33. doi: 10.1186/s12861-014-0033-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahale S, et al. The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic Spindle Orientation. Sci. Rep. 2016;6:22. doi: 10.1038/s41598-016-0030-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Regan L, Sampson J, Fry AM. Hsp70 proteins in mitosis and disease. Oncotarget. 2015;6:32293–4. doi: 10.18632/oncotarget.5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aviner R, et al. Proteomic analysis of polyribosomes identifies splicing factors as potential regulators of translation during mitosis. Nucleic Acids Res. 2017;2(45):5945–5957. doi: 10.1093/nar/gkx326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith EM, Proud CG. cdc2-cyclin B regulates eEF2 kinase activity in a cell cycle- and amino acid-dependent manner. EMBO J. 2008;27:1005–16. doi: 10.1038/emboj.2008.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Möller A, et al. Proteomic analysis of mitotic RNA polymerase II reveals novel interactors and association with proteins dysfunctional in disease. Mol. Cell Proteomics. 2012;11(M111):011767. doi: 10.1074/mcp.M111.011767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim JM, Lee KS, Woo HA, Kang D, Rhee SG. Control of the pericentrosomal H2O2 level by peroxiredoxin I is critical for mitotic progression. J. Cell Biol. 2015;6(210):23–33. doi: 10.1083/jcb.201412068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shiratsuchi G, Takaoka K, Ashikawa T, Hamada H, Kitagawa D. RBM14 prevents assembly of centriolar protein complexes and maintains mitotic spindle integrity. EMBO J. 2015;34:97–114. doi: 10.15252/embj.201488979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niwa T, et al. BRCA2 interacts with the cytoskeletal linker protein plectin to form a complex controlling centrosome localization. Cancer Sci. 2009;100:2115–25. doi: 10.1111/j.1349-7006.2009.01282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rao SR, et al. The Clathrin-dependent Spindle Proteome. Mol. Cell Proteomics. 2016;15:2537–53. doi: 10.1074/mcp.M115.054809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu W, et al. Clathrin recruits phosphorylated TACC3 to spindle poles for bipolar spindle assembly and chromosome alignment. J. Cell Sci. 2010;123:3645–51. doi: 10.1242/jcs.075911. [DOI] [PubMed] [Google Scholar]
- 53.Jang CY, Kim HD, Zhang X, Chang JS, Kim J. Ribosomal protein S3 localizes on the mitotic spindle and functions as a microtubule associated protein in mitosis. Biochem. Biophys. Res. Commun. 2012;429:57–62. doi: 10.1016/j.bbrc.2012.10.093. [DOI] [PubMed] [Google Scholar]
- 54.Douglas P, et al. Phosphorylation of SAF-A/hnRNP-U serine 59 by polo-like kinase 1 is required for mitosis. Mol. Cell Biol. 2015;35:2699–713. doi: 10.1128/MCB.01312-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma N, et al. The nuclear scaffold protein SAF-A is required for kinetochore-microtubule attachment and contributes to the targeting of Aurora-A to mitotic spindles. J. Cell Sci. 2011;124:394–404. doi: 10.1242/jcs.063347. [DOI] [PubMed] [Google Scholar]
- 56.Embree LJ, Azuma M, Hickstein DD. Ewing sarcoma fusion protein EWSR1/FLI1 interacts with EWSR1 leading to mitotic defects in zebrafish embryos and human cell lines. Cancer Res. 2009;69:4363–71. doi: 10.1158/0008-5472.CAN-08-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang YL, et al. EWSR1 regulates mitosis by dynamically influencing microtubule acetylation. Cell Cycle. 2016;15:2202–15. doi: 10.1080/15384101.2016.1200774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hofmann JC, et al. The Prp19 complex directly functions in mitotic spindle assembly. PLoS One. 2013;8:e74851. doi: 10.1371/journal.pone.0074851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma N, et al. Nucleolin functions in nucleolus formation and chromosome congression. J. Cell Sci. 2007;120:2091–105. doi: 10.1242/jcs.008771. [DOI] [PubMed] [Google Scholar]
- 60.Wang SA, et al. Heat shock protein 90 stabilizes nucleolin to increase mRNA stability in mitosis. J. Biol. Chem. 2011;3(286):43816–29. doi: 10.1074/jbc.M111.310979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fujimura A, Kishimoto H, Yanagisawa J, Kimura K. Enhancer of rudimentary homolog (ERH) plays an essential role in the progression of mitosis by promoting mitotic chromosome alignment. Biochem. Biophys. Res. Commun. 2012;423:588–92. doi: 10.1016/j.bbrc.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 62.Yamazaki T, et al. The closely related RNA helicases, UAP56 and URH49, preferentially form distinct mRNA export machineries and coordinately regulate mitotic progression. Mol. Biol. Cell. 2010;21:2953–65. doi: 10.1091/mbc.E09-10-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jung E, Seong Y, Seo JH, Kwon YS, Song H. Cell cycle-dependent regulation of Aurora kinase B mRNA by the Microprocessor complex. Biochem. Biophys. Res. Commun. 2014;28(446):241–7. doi: 10.1016/j.bbrc.2014.02.104. [DOI] [PubMed] [Google Scholar]
- 64.Pillay I, Nakano H, Sharma SV. Radicicol inhibits tyrosine phosphorylation of the mitotic Src substrate Sam68 and retards subsequent exit from mitosis of Src-transformed cells. Cell Growth Differ. 1996;7:1487–99. [PubMed] [Google Scholar]
- 65.Adachi M, Kawasaki A, Nojima H, Nishida E, Tsukita S. Involvement of IQGAP family proteins in the regulation of mammalian cell cytokinesis. Genes Cells. 2014;9:803–20. doi: 10.1111/gtc.12179. [DOI] [PubMed] [Google Scholar]
- 66.Lian AT, Hains PG, Sarcevic B, Robinson PJ, Chircop M. IQGAP1 is associated with nuclear envelope reformation and completion of abscission. Cell Cycle. 2015;14:2058–74. doi: 10.1080/15384101.2015.1044168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hasegawa K, Ryu SJ, Kaláb P. Chromosomal gain promotes formation of a steep RanGTP gradient that drives mitosis in aneuploid cells. J. Cell Biol. 2013;200:151–61. doi: 10.1083/jcb.201206142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Richards SA, Lounsbury KM, Macara IG. The C-terminus of the nuclear RAN/TC4 GTPase stabilizes the GDP-bound state and mediates interactions with RCC1, RAN-GAP, and HTF9A/RANBP1. J. Biol. Chem. 1995;270:14405–11. doi: 10.1074/jbc.270.24.14405. [DOI] [PubMed] [Google Scholar]
- 69.Kéryer G, et al. Part of Ran is associated with AKAP450 at the centrosome: involvement in microtubule-organizing activity. Mol. Biol. Cell. 2003;14:4260–71. doi: 10.1091/mbc.E02-11-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ciciarello M, et al. Nuclear reformation after mitosis requires downregulation of the Ran GTPase effector RanBP1 in mammalian cells. Chromosoma. 2010;119:651–68. doi: 10.1007/s00412-010-0286-5. [DOI] [PubMed] [Google Scholar]
- 71.Hughes JR, et al. A microtubule interactome: complexes with roles in cell cycle and mitosis. PLoS Biol. 2008;6:e98. doi: 10.1371/journal.pbio.0060098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li C, et al. NUSAP modulates the dynamics of kinetochore microtubules by attenuating MCAK depolymerisation activity. Sci. Rep. 2016;6:18773. doi: 10.1038/srep18773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alliegro MC. The centrosome and spindle as a ribonucleoprotein complex. Chromosome Res. 2011;19:367–76. doi: 10.1007/s10577-011-9186-7. [DOI] [PubMed] [Google Scholar]
- 74.Aitchison JD, Rout MP. The interactome challenge. J. Cell Biol. 2015;211:729–32. doi: 10.1083/jcb.201510108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lobingier BT, et al. An approach to spatiotemporally resolve protein interaction networks in living cells. Cell. 2017;169(350–360):e12. doi: 10.1016/j.cell.2017.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Candiano G, et al. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25:1327–33. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- 77.Di Francesco L, et al. 14-3-3εmarks the amyloid-stimulated microglia long-term activation. Proteomics. 2012;12:124–34. doi: 10.1002/pmic.201100113. [DOI] [PubMed] [Google Scholar]
- 78.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 79.Tyanova S, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–40. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the authors upon reasonable request.