Significance
Intermediate outcrossing rates are theoretically predicted to maintain effective selection against harmful alleles, but few studies have empirically tested this prediction with the use of genomic data. We used whole-genome resequencing data from alpine rock-cress to study how genetic variation and purifying selection vary with mating system. We find that populations with intermediate outcrossing rates have similar levels of genetic diversity as outcrossing populations, and that purifying selection against harmful alleles is efficient in mixed-mating populations. In contrast, self-fertilizing populations from Scandinavia have strongly reduced genetic diversity and accumulate harmful mutations, likely as a result of demographic effects of postglacial colonization. Our results suggest that mixed-mating populations can avoid some of the negative evolutionary consequences of high self-fertilization rates.
Keywords: self-fertilization, demographic history, bottleneck, fitness effects, genetic load
Abstract
Plant mating systems have profound effects on levels and structuring of genetic variation and can affect the impact of natural selection. Although theory predicts that intermediate outcrossing rates may allow plants to prevent accumulation of deleterious alleles, few studies have empirically tested this prediction using genomic data. Here, we study the effect of mating system on purifying selection by conducting population-genomic analyses on whole-genome resequencing data from 38 European individuals of the arctic-alpine crucifer Arabis alpina. We find that outcrossing and mixed-mating populations maintain genetic diversity at similar levels, whereas highly self-fertilizing Scandinavian A. alpina show a strong reduction in genetic diversity, most likely as a result of a postglacial colonization bottleneck. We further find evidence for accumulation of genetic load in highly self-fertilizing populations, whereas the genome-wide impact of purifying selection does not differ greatly between mixed-mating and outcrossing populations. Our results demonstrate that intermediate levels of outcrossing may allow efficient selection against harmful alleles, whereas demographic effects can be important for relaxed purifying selection in highly selfing populations. Thus, mating system and demography shape the impact of purifying selection on genomic variation in A. alpina. These results are important for an improved understanding of the evolutionary consequences of mating system variation and the maintenance of mixed-mating strategies.
Flowering plants show a great deal of variation in their reproductive modes, and variation in outcrossing rate is particularly common. Although ∼50% of flowering plants are predominantly outcrossing, a substantial proportion (35–40%) undergo intermediate levels of outcrossing, whereas only 10–15% are predominantly self-fertilizing (i.e., “selfing”) (1, 2). Whether mixed mating is evolutionarily stable or represents a transitional stage has long been debated (1, 3). Classic population genetic models predict that only high selfing and high outcrossing rates are evolutionarily stable strategies (4). However, mixed mating can be stable in ecologically more realistic models, such as those that account for reduced outcross pollen success with increased selfing (1). Moreover, population genetic models that incorporate linkage indicate that mixed-mating populations may avoid the reduced efficacy of selection associated with high selfing rates (5–7).
Although selfing can be favored because of its genetic transmission advantage and because it can confer reproductive assurance, it also has marked population genetic consequences that might contribute to the long-term demise of highly selfing lineages (6). For instance, highly selfing populations are expected to have a reduced effective population size (Ne) as a result of the direct effect of inbreeding (8, 9). Demographic processes such as frequent extinction and recolonization of local subpopulations (10) or founder events associated with the shift to selfing (6) can reduce genetic variation in selfers to an even greater extent. Self-fertilization is also expected to lead to elevated linkage disequilibrium, which means that background selection and other forms of linked selection can reduce genetic variation genome-wide, further reducing Ne in selfers (7, 11, 12).
Because the strength of selection scales with Ne (13), natural selection is expected to be less efficient in selfers than in outcrossers. Selfers are therefore expected to accumulate weakly deleterious mutations at a higher rate than outcrossers (14). However, low levels of outcrossing may be sufficient to prevent accumulation of mildly deleterious alleles (15). Furthermore, selfing also increases homozygosity, exposing recessive mutations to selection. This is expected to result in purging of recessive deleterious mutations unless selfing is associated with a strong reduction in Ne, which renders such purging ineffective (16–18). Theory predicts that purging should also be efficient in mixed-mating populations (17).
Although there is accumulating evidence of relaxed selection on weakly deleterious mutations in highly selfing lineages (5, 19–23), several fundamental questions about the evolutionary genomic consequences of mating systems remain unanswered. Specifically, it is unclear whether partial selfing generally results in purging of recessive deleterious alleles or this effect is possibly overridden by demographic effects, and there are few empirical genome-wide studies that have explicitly examined the selective consequences of mixed mating in plants (but see ref. 24). It is therefore unclear whether mixed-mating plants avoid the negative genetic effects associated with high selfing rates.
The broadly distributed arctic-alpine perennial herb Arabis alpina (Brassicaceae) is a promising plant system in which to address the impact of variation in outcrossing rates on genome-wide genetic variation and efficiency of selection. This species harbors populations that express a range of mating strategies from self-incompatible outcrossing (25) through mixed-mating to autonomous selfing (25–28). The colonization history of A. alpina has already begun to be characterized (26, 29–31), which facilitates interpretation of global patterns of polymorphism. Finally, the availability of a genome assembly of A. alpina (32) greatly facilitates population genomic studies.
In this study, we investigate the effects of mating system and demography on the efficacy of selection in A. alpina by population-genomic analyses of whole-genome resequencing data from outcrossing, mixed-mating, and highly selfing populations. We first investigate population structure and test whether populations with higher selfing rates have lower levels of genetic diversity, and then test whether higher selfing rates are associated with relaxed selection against weakly deleterious mutations and purging of strongly deleterious mutations. To do this, we use genome-wide allele frequency distributions at nonsynonymous and synonymous sites to estimate the fraction of weakly deleterious and strongly harmful new nonsynonymous mutations. We further test whether higher rates of selfing are associated with an increase in the frequency of derived alleles with major effects on gene integrity, which would suggest relaxed purifying selection. Finally, we compare two genomic proxies of genetic load, i.e., the reduction in mean fitness of a population caused by deleterious variation (33, 34), among outcrossing, mixed-mating, and highly self-fertilizing populations. Our results are important for an improved understanding of the population genetic consequences of mating system variation.
Results
Sequencing and SNPs.
We sampled 38 A. alpina individuals from 17 geographical sites, with targeted sampling of two to five individuals from 12 geographical sites harboring self-incompatible outcrossing populations (Greece and Italy), populations with intermediate outcrossing rates (France and Spain), and highly selfing populations (Scandinavia), in addition to a sample of single individuals from five additional geographic locations across the European range of A. alpina (SI Appendix, Table S1). Progeny array-based outcrossing estimates have shown that populations from Scandinavia are highly selfing (as much as ∼10% outcrossing), whereas intermediate outcrossing rates have been estimated for French and Spanish populations (∼19% and ∼18%, respectively; SI Appendix, SI Text) (28). We further verified that Greek and Italian individuals produced no offspring after forced self-pollination in the greenhouse. The distribution of genomic runs of homozygosity and decay of linkage disequilibrium supported mating system variation among the populations studied here (SI Appendix, Figs. S1 and S2).
Each individual was resequenced to high coverage (average, 26×; range, 16–45×) by using Illumina short-read technology. We called SNPs and applied stringent filtering criteria to identify a total of 1,514,615 high-quality SNPs, of which 98,564 were 0-fold degenerate nonsynonymous (i.e., sites at which any mutation will result in a nonsynonymous change), with a mean nucleotide diversity (π) of 0.0027, and 65,821 were fourfold degenerate synonymous (i.e., synonymous sites at which any mutation will result in a synonymous change), with a mean nucleotide diversity of 0.0102 (SI Appendix, Table S2).
Population Structure Has a Strong Geographic Component.
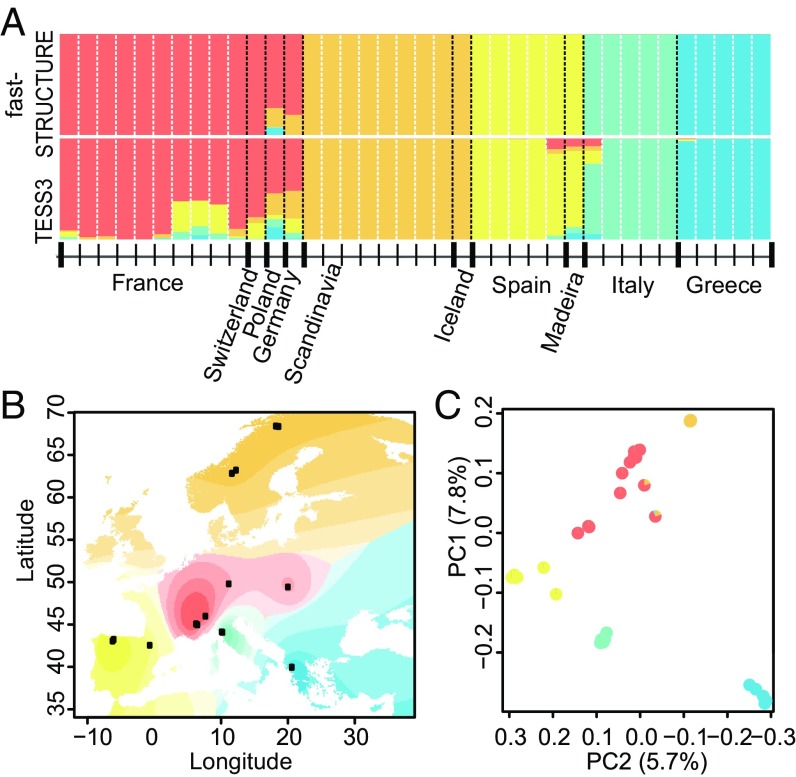
We analyzed population structure with two model-based Bayesian clustering approaches, implemented in the software fastSTRUCTURE (35) and TESS3 (36), based on 25,505 fourfold synonymous SNPs (Methods). Both methods, as well as principal component analysis (PCA), gave very similar results, and supported the presence of five clusters (Fig. 1) that were substantially differentiated (average pairwise fixation index, FST, 0.56; range, 0.39–0.82; SI Appendix, Table S3), in good agreement with previous analyses of population structure in A. alpina (30). These clusters correspond to a central European population of mixed-mating individuals from France, Germany, Poland, and Switzerland; a northern European population of highly self-fertilizing individuals from Sweden, Norway, and Iceland; a mixed-mating population containing individuals from Spain and Madeira; and two outcrossing populations representing individuals from Italy and Greece, respectively (Fig. 1). Subsequent population-genetic analyses are presented separately for regional population samples from each of these geographical regions.
Fig. 1.
Bayesian clustering analysis supports a strong geographic component to population structure in European A. alpina. (A) Ancestry proportions for K = 5 clusters correspond closely to geographical sampling locations. (B) Geographic interpolation of genetic structure across Europe based on TESS3 results for K = 5. (C) PCA of genetic data, with pie charts showing ancestry proportions for K = 5 and colors indicating geographic origin.
Genetic Diversity Is Maintained in Mixed-Mating but Not in Highly Selfing Populations.
For each regional population, we quantified nucleotide diversity at three categories of sites—synonymous sites, nonsynonymous sites, and intergenic sites—in regions of low gene density and high recombination rate (Table 1 and SI Appendix, Table S4). At all three categories of sites, levels of nucleotide diversity (π) varied by an order of magnitude or more among regional populations (Table 1 and SI Appendix, Table S4). The outcrossing Greek population was the most genetically diverse (synonymous diversity πS, 0.008), whereas the highly self-fertilizing Scandinavian populations had very low nucleotide diversity (πS, 0.0002; Table 1). Levels of nucleotide diversity were intermediate and of a similar magnitude in outcrossing Italian and mixed-mating French and Spanish populations (Table 1), which also have similar demographic histories (SI Appendix, Fig. S7). These results thus suggest that, in A. alpina, mixed-mating populations maintain similar levels of genetic diversity as outcrossing populations. Similar patterns were seen for nonsynonymous sites, although the relative reduction in diversity in Scandinavia was less severe for nonsynonymous than for synonymous sites (SI Appendix, Table S4). This resulted in a markedly elevated ratio of nonsynonymous to synonymous nucleotide diversity (πN/πS) in Scandinavia (Table 1).
Table 1.
Population genetic summary statistics for regional populations
| Populations (n) | SS* | πS† | πN/πS‡ |
| Greece (5) | 30,920 | 0.0080 | 0.2740 |
| Italy (5) | 17,971 | 0.0054 | 0.2820 |
| Spain (5) | 15,852 | 0.0046 | 0.2911 |
| France (10) | 19,661 | 0.0052 | 0.2764 |
| Scandinavia (8) | 589 | 0.0002 | 0.3936 |
Segregating fourfold synonymous SNPs.
Mean fourfold synonymous nucleotide diversity.
Mean 0-fold degenerate nonsynonymous/fourfold degenerate synonymous polymorphism.
Selfing, but Not Mixed Mating, Is Associated with a Genomic Signature of Relaxed Purifying Selection.
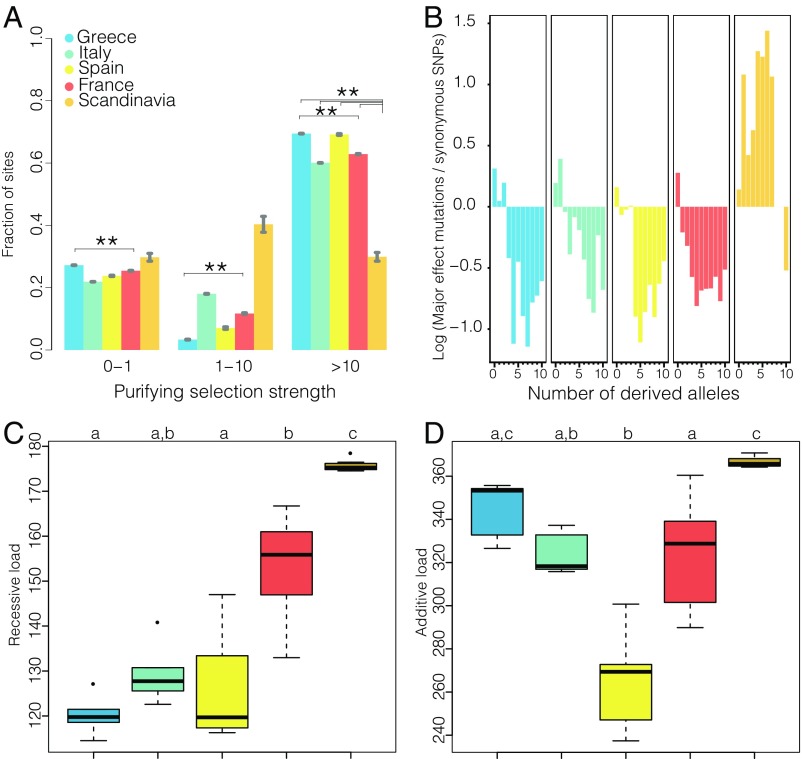
To test whether the impact of purifying selection varied with mating system, we estimated the distribution of negative fitness effects (DFE) of new nonsynonymous mutations using the software DFE-α (37) (further details provided in Methods). This method can detect changes in selection in association with plant mating system shifts, including purging of strongly deleterious mutations, which should be visible as an increase in the proportion of new mutations under strong purifying selection (38), and corrects for effects of demographic changes on allele frequency spectra by using a simple population size change model. We summarized the strength of purifying selection, defined as the product of the effective population size Ne and the selection coefficient s, in three bins, ranging from nearly neutral to strongly deleterious (0 < Nes < 1; 1 < Nes < 10; Nes > 10). Although the DFE of the mixed-mating Spanish and outcrossing Greek populations differed, there were no significant differences between the DFE of the outcrossing Italian population and the mixed-mating Spanish and French populations (Fig. 2A). The inferred proportion of strongly deleterious new nonsynonymous mutations was not generally higher in the mixed-mating French and Spanish populations than in the outcrossing Greek and Italian populations (Fig. 2A). Thus, A. alpina populations undergoing as much as 80% selfing do not show population-genomic evidence of relaxed purifying selection or increased purging of recessive deleterious mutations.
Fig. 2.
The impact of mating system on purifying selection in A. alpina. (A) The DFE in bins of Nes for new nonsynonymous mutations, estimated under a model with a stepwise population size change. Error bars show ±1 SE. Asterisks indicate significant differences (false discovery rate < 0.05) among populations. (B) Scandinavian A. alpina shows an increase in the frequency of derived major effect polymorphism relative to synonymous polymorphism. The figure shows the log ratio of major effect-derived allele frequencies to fourfold synonymous allele frequencies, down-sampled to 10 derived alleles in all populations. (C) Recessive genetic load for major effect alleles (number of derived homozygous genotypes). (D) Additive genetic load for major effect alleles (number of derived alleles). In C and D, letters indicate groups with statistically significant differences (P < 0.05, Kruskal–Wallis test, post hoc Dunn test).
In contrast to results for mixed-mating populations, several lines of evidence suggest that selection against deleterious alleles is compromised in highly self-fertilizing Scandinavian A. alpina. First, there was a strong difference in the DFE of nonsynonymous mutations, with a strong, significant reduction in selection against strongly deleterious nonsynonymous mutations (i.e., Nes > 10) in Scandinavia compared with all other regional populations (Fig. 2A). Thus, DFE analyses suggest that the Scandinavian population could be accumulating strongly deleterious nonsynonymous mutations.
In line with our inference of accumulation of deleterious alleles in Scandinavian A. alpina, we found that derived alleles with a major effect on gene integrity (Methods) were at a markedly higher frequency relative to derived synonymous alleles in Scandinavia compared with all other regional populations (Fig. 2B). Because selected and neutral allele frequency spectra can be differently affected by demographic changes even in the absence of an actual change in selection (39–41), we considered additional proxies for genetic load. First, we estimated the average number of homozygous derived major-effect genotypes (Fig. 2C), which is expected to be proportional to genetic load if deleterious mutations act recessively, and then we estimated the average number of derived major-effect alleles (Fig. 2D), which is proportional to genetic load if deleterious mutations have additive effects (42). According to both statistics, the highly selfing Scandinavian regional population had an elevated genetic load compared with mixed-mating populations, and outcrossing and mixed-mating populations were not different from each other (Fig. 2 C and D). Estimates of additive load were lower in mixed-mating than in outcrossing populations, but the difference was not statistically significant (Fig. 2D). In addition, the number of fixed derived major-effect variants in Scandinavia was highly inflated (∼70% increase) compared with the outcrossing and mixed-mating populations (SI Appendix, Table S5). A similar pattern was seen for derived nonsynonymous variants as well as derived nonsynonymous variants at evolutionarily constrained sites, and when considering each geographical population separately (SI Appendix, Figs. S3 and S4 and Table S5). Overall, this suggests that the highly selfing A. alpina from Scandinavia are accumulating deleterious mutations, whereas we observe no statistically significant evidence of purging of genetic load in mixed-mating populations.
A Recent Bottleneck and Selfing Explain Polymorphism in the Scandinavian Population.
Theory predicts that high levels of self-fertilization should result in effective purging of recessive deleterious variation unless selfing is associated with strong reductions in Ne (17). Given that we observed no evidence for purging and a strong reduction in genetic variation in Scandinavian A. alpina, we asked whether this could have resulted from a bottleneck, or if increased background selection caused by selfing is sufficient to explain the reduction of diversity.
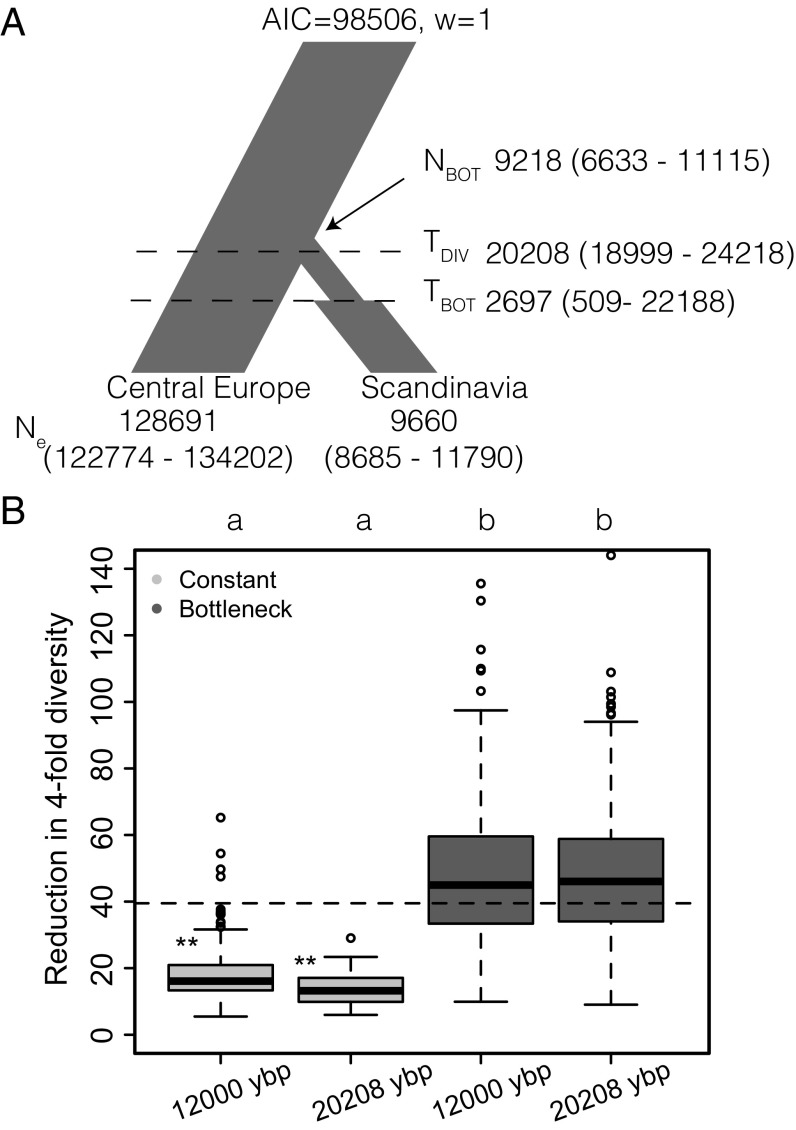
Previous phylogeographic analyses have shown that A. alpina likely originated in Asia Minor and subsequently spread westward approximately 500 kya (29, 31). Population-genetic analyses have identified Central European populations as the most likely source for Scandinavian A. alpina populations (30), but no explicit demographic model has yet been fit to estimate the timing or demographics of colonization of Scandinavia by A. alpina. For this purpose, we used a maximum likelihood-based approach to estimate the parameters of a demographic model of colonization of Scandinavia by using a 2D site frequency spectrum (2D-SFS; SI Appendix, Fig. S5) based on a scattered sample of individuals from Central Europe and the highly selfing Scandinavian A. alpina (Methods). For analyses of demographic history, we used a set of 12,967 SNPs in intergenic regions with low gene density and high recombination rate, which are expected to be less affected by linked selection and thus useful for demographic inference (12, 43). According to our best-fit model, the split between Central Europe and Scandinavia occurred ∼20 kya and was associated with a prolonged bottleneck (Fig. 3A and SI Appendix, Figs. S6 and S7 and Table S6).
Fig. 3.
A recent bottleneck and selfing explain the reduction of polymorphism in Scandinavia. (A) The best-fit demographic model of the colonization of Scandinavia from a Central European population. Estimated times are given in years before present (ybp). (B) Background selection alone does not explain the reduction in diversity in Scandinavian A. alpina. Boxplots show the ratio of synonymous polymorphism between an outcrossing population and a 90% selfing population experiencing a constant population size or a 10-fold bottleneck, with the two populations diverging 12,000 ybp or 20,208 ybp. The dashed line indicates the observed ratio of synonymous polymorphism in Central Europe to that in Scandinavia. Letters indicate significant difference between models (Mann–Whitney test, P < 0.001). Asterisks indicate an observed neutral diversity reduction significantly greater than that expected based on 300 simulations.
We conducted population-genetic simulations to assess whether the reduction in diversity in Scandinavia could be explained by a stronger impact of background selection caused by selfing, without additional demographic changes. These simulations, which used realistic settings for A. alpina genome structure, including variation in recombination rate, gene density, and mutation rates, and a realistic distribution of negative fitness effects (Methods), show that models that incorporate a transition to selfing but no demographic change cannot explain the reduction in diversity we observe in the Scandinavian population (P = 0.013 based on 300 simulations), whereas a model that includes a 10-fold bottleneck and a transition to selfing is consistent with the observed reduction in genetic diversity (P = 0.58 based on 300 simulations; Fig. 3B). These conclusions are robust if we assume a more recent split between the Central European and the Scandinavian population, as late as 12 kya (Fig. 3B). The results also hold under a different DFE (Methods and SI Appendix, Fig. S9). These demographic modeling results suggest that a postglacial colonization bottleneck reduced diversity and affected the impact of natural selection in Scandinavian A. alpina.
Discussion
We have used population-genomic analyses to investigate effects of mating system and demographic history on genetic variation and purifying selection in the arctic-alpine crucifer species A. alpina. Our results show that populations with intermediate levels of self-fertilization maintain genetic variation and experience similar levels of purifying selection as outcrossing populations. We further find that highly selfing Scandinavian A. alpina experience relaxed purifying selection, most likely as a result of having undergone a severe bottleneck, the timing of which is consistent with postglacial recolonization of Northern Europe. Our results suggest a strong effect of demography on the impact of purifying selection in selfing populations of this species and demonstrate that intermediate levels of outcrossing can allow populations to avoid the negative population genetic consequences of self-fertilization.
The fact that mixed-mating populations maintained similar levels of genetic diversity as outcrossing populations suggests that the loss of self-incompatibility in A. alpina was not associated with a recent and strong bottleneck. Empirical studies in other plant species have frequently found higher diversity in outcrossing and mixed-mating species relative to highly self-fertilizing species (7, 44), in good agreement with our results. Taken together, these findings agree well with the expectation that levels of diversity should be higher in outcrossing than in self-fertilizing populations (6).
We detected no strong evidence for purging of deleterious alleles in mixed-mating A. alpina populations based on analyses of the DFE. This suggests that, in contrast to expectations from single-locus theory (17), strongly deleterious alleles may not be rapidly purged as a result of partial selfing in A. alpina, perhaps as a result of selective interference (5). Although our power to detect purging may be limited if strongly deleterious recessive mutations are rare, a recent simulation-based study showed that the analysis method used here can detect the signal of purging following shifts to higher selfing rates (38). Indeed, no strong differences between outcrossing and mixed-mating populations were found for the DFE of new nonsynonymous mutations, or with respect to derived allele frequencies of major-effect alleles. Estimates of additive load were lower for mixed-mating than for outcrossing populations, which suggests that nonrandom mating could result in some purging of deleterious alleles (17); however, these differences were not statistically significant. Our results thus suggest that the genome-wide impact of purifying selection is similar in outcrossing and mixed-mating populations of A. alpina.
Although there is a dearth of genome-wide studies contrasting purifying selection in outcrossing and mixed-mating plants, one previous study also found modest differences in purifying selection between outcrossing and mixed-mating species (24). These observations agree with simulation-based results that suggest a that low degree of outcrossing is sufficient to maintain efficient purifying selection (15). Thus, mixed-mating populations may avoid some of the negative evolutionary genetic effects associated with high selfing rates, which could imply that population genetic effects contribute to the stable maintenance of intermediate outcrossing rates (5). However, ultimately, direct estimates of inbreeding depression are needed to fully elucidate the maintenance of mating system variation in A. alpina.
In contrast to the lack of differences in purifying selection between outcrossing and mixed-mating populations, we found strong evidence for accumulation of deleterious mutations in highly self-fertilizing Scandinavian A. alpina. Indeed, our DFE analyses indicate less efficient selection against strongly deleterious alleles, and we observe an increase in the relative allele frequency and fixed derived major-effect mutations, as well as elevated additive and expressed genetic load in Scandinavian A. alpina. These results suggest that deleterious alleles are not efficiently selected against and have been able to increase in frequency in these populations. In the context of human population history, theory and simulations have shown that range expansions (45) or strong and extended bottlenecks (39, 41, 42) can lead to increased genetic load. It has previously been shown that genetic variation is strongly reduced over a vast geographical area representing the northern part of the distribution of A. alpina (30). Our demographic inference and population-genetic simulations suggest that this reduction of genetic variation is most likely a result of postglacial colonization bottlenecks. Thus, bottlenecks associated with range expansion appear to have strongly reduced Ne and increased the impact of drift in the Northern part of the species range, causing accumulation of deleterious variants in Scandinavian A. alpina. The impact of demographic history on genetic load is currently a strongly debated topic in human population genetics, in which studies on the effect of the out-of-Africa bottleneck on genetic load have come to different conclusions depending on the statistics they applied (39, 46–48). Here, we extend these studies to a plant species and document increased accumulation of deleterious mutations in bottlenecked Scandinavian A. alpina.
Increased background selection can lead to sharply reduced genetic diversity in highly selfing populations (7, 12), but our forward population genetic simulations show that this effect alone cannot explain the reduction in diversity in Scandinavia, whereas a model with selfing and a population size reduction is consistent with the observed reduction in diversity. At present, however, we cannot rule out a contribution of positive selection, or other changes in selection during range expansion, to reduced diversity in the Scandinavian A. alpina population. Indeed, reciprocal transplant experiments have documented adaptive differentiation between Spanish and Scandinavian populations of A. alpina (49). Although elevated load can be associated with enhanced local adaptation following a range expansion (50), this raises the possibility that some of the increase in frequencies of major-effect variants in Scandinavia may have been directly or indirectly driven by positive selection. However, we believe it is unlikely to result in the genome-wide signature of relaxed purifying selection that we observe in Scandinavian A. alpina. Empirical identification of genomic regions responsible for local adaptation and population genomic studies using larger sample sizes will be needed to explore the genomic impact of positive selection.
Here, we have investigated the impact of demographic history and mating system on genomic patterns of variation in A. alpina. Our results show that mixed-mating populations maintain genetic variation and purifying selection at similar levels as outcrossing populations. In contrast, we find an increase in genetic load in highly self-fertilizing populations, most likely as a result of demographic effects associated with postglacial range expansion. Our results are important for a more general understanding of the impact of mating system and demographic history on genomic variation and selection in plants.
Methods
Data and Sequencing.
We performed paired-end (100-bp) whole-genome resequencing of 38 A. alpina individuals from Europe (SI Appendix, Table S1) by using libraries with an insert size of 300–400 bp (SI Appendix, SI Text) on an Illumina HiSeq 2000 instrument (Illumina). We obtained a total of 10,079 Gbp (quality score QC > 30) with a mean coverage of 26×, ranging from 16× to 45×.
Quality Assessment, Trimming, Genotype Calling, and Filtering.
Adapter-trimmed reads were mapped to the A. alpina V4 reference genome assembly (32) by using BWA-MEM v0.7.8 (51), and duplicate-free BAM alignment files were further processed by using the Genome Analysis Toolkit (52) (v.3.4.0; SI Appendix, SI Text). The A. alpina genome assembly is enriched for repetitive elements relative to Brassicaceae relatives (32), so we employed a variety of hard and custom filtering techniques (SI Appendix, SI Text) to avoid calling SNPs in regions of the genome that putatively represent copy number variants. After applying all filters, the dataset contained 1,514,615 SNPs and 43,209,020 invariant sites.
Inference of Population Structure and Population-Genetic Analyses of Selection.
Population structure was inferred by using a combination of PCA and Bayesian clustering analysis with the use of fastSTRUCTURE v1.0 (35) and TESS v3 (36) (SI Appendix, SI Text).
For each of the five regional populations identified, we estimated summary statistics (S, π, Tajima’s D) at fourfold degenerate synonymous sites, 0-fold degenerate nonsynonymous sites, and intergenic sites in regions with high recombination and low gene density (Table 1 and SI Appendix, SI Text). We estimated the DFE by using DFE-α v2.15 (37) on folded fourfold and 0-fold SFS under a stepwise population size change model (SI Appendix, SI Text and Table S7). We compared the DFE of regional populations based on 200 bootstrap replicates.
Major Effect Mutations and Genetic Load.
Presence and frequency of major effect mutations, i.e., loss of start and stop codons, gain of stop codons, and changes in splice sites, were calculated per population by using snpEFF v4.2 (53). To avoid reference biases, we polarized the SNPs using the Arabis montbretiana genome assembly (ASM148412v1) (54) as an outgroup (SI Appendix, SI Text). As a proxy for genetic load, we estimated the average number of derived nonsynonymous and major-effect homozygous genotypes per individual (41, 46), and the average number of derived alleles for nonsynonymous and major-effect alleles per individual (42) for each regional population. We repeated these analyses for nonsynonymous variants at highly constrained sites (SI Appendix, SI Text). In addition, we counted the total number of fixed derived nonsynonymous and major-effect alleles for each regional population.
Demographic Modeling and Simulations.
We conducted demographic inference in the software fastsimcoal2 v2.5.2.21 (55). To estimate parameters associated with the origin of Scandinavian A. alpina, we compared three demographic models (SI Appendix, Fig. S5) using 2D joint SFS based on a scattered sample from central Europe and the Scandinavian population (SI Appendix, SI Text and Fig. S6). We used 12,967 intergenic sites, a mutation rate of 7 × 10−9, and a generation time of 1.5 y. We used forward simulation in SLiM2 v2.1 (56) to assess the impact of demography and selection associated with a shift to selfing on genetic diversity in the Scandinavian population under four demographic models with varying bottleneck severity and population split time (SI Appendix, SI Text).
Supplementary Material
Acknowledgments
We thank Tiina Mattila for advice on population genetic analyses and Cindy Canton for help with plant care and extractions. Computations were performed on resources provided by the Swedish National Infrastructure for Computing (SNIC) through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Projects snic2014-1-194, b2013022, and b2013237. This work was funded by grants from the Swedish Research Council (to J.Å. and T.S.), a grant from the Deutsche Forschungsgemeinschaft through SPP1529 (to G.C.), and SciLifeLab (to T.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the European Bioinformatics Institute database (accession no. PRJEB20772).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707492115/-/DCSupplemental.
References
- 1.Goodwillie C, Kalisz S, Eckert C. The evolutionary enigma of mixed mating systems in plants: Occurrence, theoretical explanations, and empirical evidence. Annu Rev Ecol Evol Syst. 2005;36:47–79. [Google Scholar]
- 2.Igic B, Kohn JR. The distribution of plant mating systems: Study bias against obligately outcrossing species. Evolution. 2006;60:1098–1103. [PubMed] [Google Scholar]
- 3.Igic B, Busch JW. Is self-fertilization an evolutionary dead end? New Phytol. 2013;198:386–397. doi: 10.1111/nph.12182. [DOI] [PubMed] [Google Scholar]
- 4.Lande R, Schemske DW. The evolution of self-fertilization and inbreeding depression in plants. I. Genetic models. Evolution. 1985;39:24–40. doi: 10.1111/j.1558-5646.1985.tb04077.x. [DOI] [PubMed] [Google Scholar]
- 5.Hartfield M. Evolutionary genetic consequences of facultative sex and outcrossing. J Evol Biol. 2016;29:5–22. doi: 10.1111/jeb.12770. [DOI] [PubMed] [Google Scholar]
- 6.Wright SI, Kalisz S, Slotte T. Evolutionary consequences of self-fertilization in plants. Proc Biol Sci. 2013;280:20130133. doi: 10.1098/rspb.2013.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrett SCH, Arunkumar R, Wright SI. The demography and population genomics of evolutionary transitions to self-fertilization in plants. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130344. doi: 10.1098/rstb.2013.0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pollak E. On the theory of partially inbreeding finite populations. I. Partial selfing. Genetics. 1987;117:353–360. doi: 10.1093/genetics/117.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nordborg M. Linkage disequilibrium, gene trees and selfing: An ancestral recombination graph with partial self-fertilization. Genetics. 2000;154:923–929. doi: 10.1093/genetics/154.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ingvarsson PK. A metapopulation perspective on genetic diversity and differentiation in partially self-fertilizing plants. Evolution. 2002;56:2368–2373. doi: 10.1111/j.0014-3820.2002.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 11.Charlesworth D, Wright SI. Breeding systems and genome evolution. Curr Opin Genet Dev. 2001;11:685–690. doi: 10.1016/s0959-437x(00)00254-9. [DOI] [PubMed] [Google Scholar]
- 12.Slotte T. The impact of linked selection on plant genomic variation. Brief Funct Genomics. 2014;13:268–275. doi: 10.1093/bfgp/elu009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge Univ Press; Cambridge, UK: 1983. [Google Scholar]
- 14.Ohta T. Slightly deleterious mutant substitutions in evolution. Nature. 1973;246:96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- 15.Kamran-Disfani A, Agrawal AF. Selfing, adaptation and background selection in finite populations. J Evol Biol. 2014;27:1360–1371. doi: 10.1111/jeb.12343. [DOI] [PubMed] [Google Scholar]
- 16.Bataillon T, Kirkpatrick M. Inbreeding depression due to mildly deleterious mutations in finite populations: Size does matter. Genet Res. 2000;75:75–81. doi: 10.1017/s0016672399004048. [DOI] [PubMed] [Google Scholar]
- 17.Glémin S. How are deleterious mutations purged? Drift versus nonrandom mating. Evolution. 2003;57:2678–2687. doi: 10.1111/j.0014-3820.2003.tb01512.x. [DOI] [PubMed] [Google Scholar]
- 18.Glémin S. Mating systems and the efficacy of selection at the molecular level. Genetics. 2007;177:905–916. doi: 10.1534/genetics.107.073601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu S, Zeng K, Slotte T, Wright S, Charlesworth D. Reduced efficacy of natural selection on codon usage bias in selfing Arabidopsis and Capsella species. Genome Biol Evol. 2011;3:868–880. doi: 10.1093/gbe/evr085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ness RW, Siol M, Barrett SCH. Genomic consequences of transitions from cross- to self-fertilization on the efficacy of selection in three independently derived selfing plants. BMC Genomics. 2012;13:611. doi: 10.1186/1471-2164-13-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brandvain Y, Slotte T, Hazzouri KM, Wright SI, Coop G. Genomic identification of founding haplotypes reveals the history of the selfing species Capsella rubella. PLoS Genet. 2013;9:e1003754. doi: 10.1371/journal.pgen.1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slotte T, et al. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat Genet. 2013;45:831–835. doi: 10.1038/ng.2669. [DOI] [PubMed] [Google Scholar]
- 23.Douglas GM, et al. Hybrid origins and the earliest stages of diploidization in the highly successful recent polyploid Capsella bursa-pastoris. Proc Natl Acad Sci USA. 2015;112:2806–2811. doi: 10.1073/pnas.1412277112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salcedo A, Kalisz S, Wright SI. Limited genomic consequences of mixed mating in the recently derived sister species pair, Collinsia concolor and Collinsia parryi. J Evol Biol. 2014;27:1400–1412. doi: 10.1111/jeb.12384. [DOI] [PubMed] [Google Scholar]
- 25.Tedder A, Ansell SW, Lao X, Vogel JC, Mable BK. Sporophytic self-incompatibility genes and mating system variation in Arabis alpina. Ann Bot. 2011;108:699–713. doi: 10.1093/aob/mcr157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ansell SW, Grundmann M, Russell SJ, Schneider H, Vogel JC. Genetic discontinuity, breeding-system change and population history of Arabis alpina in the Italian Peninsula and adjacent Alps. Mol Ecol. 2008;17:2245–2257. doi: 10.1111/j.1365-294X.2008.03739.x. [DOI] [PubMed] [Google Scholar]
- 27.Buehler D, Graf R, Holderegger R, Gugerli F. Contemporary gene flow and mating system of Arabis alpina in a central European alpine landscape. Ann Bot. 2012;109:1359–1367. doi: 10.1093/aob/mcs066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toräng P, et al. Evolution of the selfing syndrome: Anther orientation and herkogamy together determine reproductive assurance in a self-compatible plant. Evolution. 2017;71:2206–2218. doi: 10.1111/evo.13308. [DOI] [PubMed] [Google Scholar]
- 29.Koch MA, et al. Three times out of Asia Minor: The phylogeography of Arabis alpina L. (Brassicaceae) Mol Ecol. 2006;15:825–839. doi: 10.1111/j.1365-294X.2005.02848.x. [DOI] [PubMed] [Google Scholar]
- 30.Ehrich D, et al. Intrabiodiv Consortium Genetic consequences of Pleistocene range shifts: Contrast between the Arctic, the Alps and the East African mountains. Mol Ecol. 2007;16:2542–2559. doi: 10.1111/j.1365-294X.2007.03299.x. [DOI] [PubMed] [Google Scholar]
- 31.Ansell SW, et al. The importance of Anatolian mountains as the cradle of global diversity in Arabis alpina, a key arctic-alpine species. Ann Bot. 2011;108:241–252. doi: 10.1093/aob/mcr134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willing E-M, et al. Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nat Plants. 2015;1:14023. doi: 10.1038/nplants.2014.23. [DOI] [PubMed] [Google Scholar]
- 33.Morton NE, Crow JF, Muller HJ. An estimate of the mutational damage in man from data on consanguineous marriages. Proc Natl Acad Sci USA. 1956;42:855–863. doi: 10.1073/pnas.42.11.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimura M, Maruyama T, Crow JF. Mutation load in small populations. Genetics. 1963;48:1303–1312. doi: 10.1093/genetics/48.10.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raj A, Stephens M, Pritchard JK. fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics. 2014;197:573–589. doi: 10.1534/genetics.114.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caye K, Deist TM, Martins H, Michel O, François O. TESS3: Fast inference of spatial population structure and genome scans for selection. Mol Ecol Resour. 2016;16:540–548. doi: 10.1111/1755-0998.12471. [DOI] [PubMed] [Google Scholar]
- 37.Keightley PD, Eyre-Walker A. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics. 2007;177:2251–2261. doi: 10.1534/genetics.107.080663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arunkumar R, Ness RW, Wright SI, Barrett SCH. The evolution of selfing is accompanied by reduced efficacy of selection and purging of deleterious mutations. Genetics. 2015;199:817–829. doi: 10.1534/genetics.114.172809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gravel S. When is selection effective? Genetics. 2016;203:451–462. doi: 10.1534/genetics.115.184630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brandvain Y, Wright SI. The limits of natural selection in a nonequilibrium world. Trends Genet. 2016;32:201–210. doi: 10.1016/j.tig.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 41.Pedersen CT, et al. The effect of an extreme and prolonged population bottleneck on patterns of deleterious variation: Insights from the greenlandic inuit. Genetics. 2017;205:787–801. doi: 10.1534/genetics.116.193821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henn BM, Botigué LR, Bustamante CD, Clark AG, Gravel S. Estimating the mutation load in human genomes. Nat Rev Genet. 2015;16:333–343. doi: 10.1038/nrg3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Messer PW, Petrov DA. Frequent adaptation and the McDonald-Kreitman test. Proc Natl Acad Sci USA. 2013;110:8615–8620. doi: 10.1073/pnas.1220835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamrick JL, Godt MJW. Effects of life history traits on genetic diversity in plant species. Philos Trans R Soc Lond B Biol Sci. 1996;351:1291–1298. [Google Scholar]
- 45.Peischl S, Dupanloup I, Kirkpatrick M, Excoffier L. On the accumulation of deleterious mutations during range expansions. Mol Ecol. 2013;22:5972–5982. doi: 10.1111/mec.12524. [DOI] [PubMed] [Google Scholar]
- 46.Henn BM, et al. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc Natl Acad Sci USA. 2016;113:E440–E449. doi: 10.1073/pnas.1510805112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lohmueller KE, et al. Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451:994–997. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simons YB, Turchin MC, Pritchard JK, Sella G. The deleterious mutation load is insensitive to recent population history. Nat Genet. 2014;46:220–224. doi: 10.1038/ng.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toräng P, et al. Large-scale adaptive differentiation in the alpine perennial herb Arabis alpina. New Phytol. 2015;206:459–470. doi: 10.1111/nph.13176. [DOI] [PubMed] [Google Scholar]
- 50.Gilbert KJ, et al. Local adaptation interacts with expansion load during range expansion: Maladaptation reduces expansion load. Am Nat. 2017;189:368–380. doi: 10.1086/690673. [DOI] [PubMed] [Google Scholar]
- 51.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kiefer C, et al. Divergence of annual and perennial species in the Brassicaceae and the contribution of cis-acting variation at FLC orthologues. Mol Ecol. 2017;26:3437–3457. doi: 10.1111/mec.14084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa VC, Foll M. Robust demographic inference from genomic and SNP data. PLoS Genet. 2013;9:e1003905. doi: 10.1371/journal.pgen.1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haller BC, Messer PW. SLiM 2: Flexible, interactive forward genetic simulations. Mol Biol Evol. 2017;34:230–240. doi: 10.1093/molbev/msw211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.