Significance
Subtype-selective modulation of ion channels is often important, but extremely difficult to achieve for drug development. Using Nav1.7 as an example, we show that this challenge could be attributed to poor design in ion channel assays, which fail to detect most potent and selective compounds and are biased toward nonselective mechanisms. By exploiting different drug binding sites and modes of channel gating, we successfully direct a membrane potential assay toward non–pore-blocking mechanisms and identify Nav1.7-selective compounds. Our mechanistic approach to assay design addresses a significant hurdle in Nav1.7 drug discovery and is applicable to many other ion channels.
Keywords: Nav1.7, N1742K, 1KαPMTX, veratridine, VSD4
Abstract
Many ion channels, including Nav1.7, Cav1.3, and Kv1.3, are linked to human pathologies and are important therapeutic targets. To develop efficacious and safe drugs, subtype-selective modulation is essential, but has been extremely difficult to achieve. We postulate that this challenge is caused by the poor assay design, and investigate the Nav1.7 membrane potential assay, one of the most extensively employed screening assays in modern drug discovery. The assay uses veratridine to activate channels, and compounds are identified based on the inhibition of veratridine-evoked activities. We show that this assay is biased toward nonselective pore blockers and fails to detect the most potent, selective voltage-sensing domain 4 (VSD4) blockers, including PF-05089771 (PF-771) and GX-936. By eliminating a key binding site for pore blockers and replacing veratridine with a VSD-4 binding activator, we directed the assay toward non–pore-blocking mechanisms and discovered Nav1.7-selective chemical scaffolds. Hence, we address a major hurdle in Nav1.7 drug discovery, and this mechanistic approach to assay design is applicable to Cav3.1, Kv1.3, and many other ion channels to facilitate drug discovery.
Ion channels play an essential role in physiological function and are among the most important therapeutic targets (1). Numerous drugs, such as local anesthetics, dihydropyridines, sulphonylureas, and benzodiazepines have been used to treat various human diseases. These early generation ion channel drugs were developed decades ago without knowledge of molecular targets or mechanisms of action. They often possess narrow therapeutic index due to inadequate potency or selectivity. In recent years, human genetics and ion channel research have linked more than 60 ion channels to specific human pathologies (2, 3). For example, the gain-of-function mutations in Nav1.7 channel cause excessive pain syndromes, whereas the loss-of-function mutations in Nav1.7 cause congenital insensitivity to pain while retaining normal responses to other sensory inputs (4–6). Additionally, mutations in Nav isoforms, such as Nav1.1, Nav1.4, and Nav1.5, are implicated in epilepsy, myotonia, and cardiac arrhythmia, respectively (7–9). Together, these studies suggest that selective block of Nav1.7 while sparing other Nav isoforms is an attractive strategy for pain management. Likewise, subtype-selective inhibition of Cav1.3 and Kv1.3 channels has been identified as viable approaches for treating Parkinson’s disease and autoimmune diseases, respectively (10–12).
Despite the recent progresses, few subtype-selective inhibitors exist for Nav1.7, Cav1.3, or Kv1.3 channels, a consequence of structural conservation within their respective channel families. For example, all Nav isoforms share the same overall architecture and high sequence homology (13). In the six transmembrane segments lining the ion conduction pore, the amino acid identity is >90%. Not surprisingly, most known sodium channel inhibitors, including local anesthetics, antiarrhythmics, and anticonvulsants, interact with a few conserved residues in the pore and therefore are nonselective (14). Recently, a group of arylsulfonamides was identified as selective Nav1.7 inhibitors (15, 16). These compounds were discovered empirically, before our current understanding that they bind to the voltage-sensing domain 4 (VSD4) instead of the central cavity. However, PF-771, the most advanced compound, was recently halted from clinical development, raising concerns about this chemical class.
To identify subtype-selective chemical scaffolds, large libraries of compounds, sometimes exceeding millions of individual compounds, are screened against the targets of interest (e.g., Nav1.7), followed by selectivity tests on other isoforms (e.g., Nav1.5). The effects of compounds are determined using in vitro assays such as electrophysiology (17–19) or fluorescence-based membrane potential assays (20–22). The major considerations for assay design have been signal robustness, throughput, and cost, whereas mechanisms of action, such as drug binding sites and molecular basis for selectivity, are often not taken into consideration. We hypothesize that this approach underlies the failure of ion channel drug discovery, and that mechanisms of action are critical and should be incorporated into assay design. We chose Nav1.7-membrane potential assay as a test case for several reasons. First, it is one of the most extensively utilized screening assays in modern drug discovery (20–24). Second, drug binding sites for both nonselective (e.g., local anesthetics and veratridine) and selective arylsulfonamide compounds (e.g., GX-936 and PF-771) have been determined, allowing rational assay design (14–16). Third, if successful, the approach could be extended to other assays and other ion channels such as Nav1.1, Cav1.3, and Kv1.3. The Nav1.7 membrane potential assay utilizes veratridine, an alkaloid from the plant Veratrum officinale, to stimulate channel conductance, and candidate compounds are identified based on their inhibition of veratridine-induced channel activities. The effect of veratridine is accounted for by a “foot-in-the-door” mechanism, whereby veratridine binds to the inner vestibule of the pore domain (25) and causes persistent opening by hindering inactivation and deactivation (26). We show that this assay is biased toward nonselective pore blockers and fails to detect highly potent and Nav1.7-selective compounds that act by different mechanisms. Guided by the knowledge of drug binding sites and mechanisms of action, we designed an assay capable of restricting the nonselective pore blockers and enriching compounds binding outside of the pore domains. We validated this approach in a high-throughput screen and identified unique Nav1.7-selective scaffolds.
Results
Evaluation of the Conventional Membrane Potential Assay.
Membrane potential assays for sodium channels have been reported by various groups (20–22). Consistent with previous findings, veratridine (100 μM) elicited robust fluorescence signals in Nav1.7-expressing cells loaded with the blue fluorescence membrane potential dye (Fig. 1A). To evaluate pharmacology of this assay, we tested several classic sodium channel blockers. TTX is a potent neurotoxin that binds to the outer vestibule of the channel, and tetracaine is a nonselective local anesthetic that binds to the inner pore domain (27–29). TTX and tetracaine inhibited veratridine-evoked signals in a concentration-dependent manner (Fig. 1 A and B). The IC50 was 0.034 ± 0.005 μM for TTX and 3.6 ± 0.4 μM for tetracaine (Fig. 1E; n = 6).
Fig. 1.
A conventional Nav1.7 membrane potential assay detects robust channel inhibition by TTX and tetracaine but fails to detect PF-771 and GX-936. (A–D) Representative membrane potential traces for TTX, tetracaine, PF-771, and GX-936. A total of 100 μM veratridine was added (indicated by arrows) to evoke channel activity, as represented by an increase in fluorescence signals. Cells were incubated with compounds for 3 min before veratridine application. (E) Inhibition concentration–response for TTX and tetracaine. The IC50 was 0.034 ± 0.005 μM for TTX, and 3.6 ± 0.4 μM for tetracaine (n = 6). (F) Inhibition dose–response for PF-771 and GX-936. Only marginal inhibition was observed (n = 4). (G) Activities of 64,000 compounds from a membrane potential assay screen. Each dot represents one compound, numbered from 1 to 64,000. Yellow, 0.1% DMSO as negative control; blue, 1 μM TTX as positive control; red, four potent VSD4 blockers; and gray, library compounds. (H) Fold difference in IC50 between Nav1.5 and Nav1.7 for 21 most potent Nav1.7 hits. No compounds showed greater than twofold selectivity (dotted line).
Next, we tested PF-771 and GX-936, two of the most potent and selective Nav1.7 inhibitors belonging to the arylsulfonamide series (15, 16). PF-771 and GX-936 inhibited Nav1.7 with an IC50 of 0.011 and 0.001 μM, respectively, in voltage clamp assays (15, 16). However, in the membrane potential assay, PF-771 and GX-936 showed marginal effects (Fig. 1 C, D, and F). At concentrations that are several orders of magnitude over the electrophysiology IC50 values, PF-771 (5 μM) and GX-936 (3 μM) inhibited veratridine-induced response by less than 20% (Fig. 1F; n = 4).
The utility of membrane potential assay was further evaluated by a pilot screen of a chemical library of 64,000 compounds at 5 μM. The 0.1% DMSO, 1 μM TTX, and four potent blockers that bind to VSD4 domain (electrophysiology IC50 < 0.1 μM) were embedded in the screening sets. The average inhibitory effect of DMSO on veratridine responses was 0.4 ± 13.5% (n = 1,053) and the average inhibition by TTX was 99.7 ± 4.9% (n = 792). The mean inhibition was 6.1% for the 64,000-compound screen, with a SD of 30%. None of the VSD4 blockers showed >10% inhibition, hence these compounds were not identified as active by the screen (Fig. 1G, red dots). Among the 21 most potent Nav1.7 hits, no compound showed greater than twofold selectivity against Nav1.5 channels in patch-clamp experiments (Fig. 1H). In summary, the membrane potential assay fails to detect VSD4 inhibitors and is biased toward the nonselective compounds.
Characterization of N1742K Pore Mutant Channel.
We explored various ways of enhancing the sensitivity of the membrane potential assay for inhibitors that bind outside of the central cavity. We reasoned that by eliminating key pore binding sites, and by replacing veratridine (a pore binder) with a new activator of a different binding site, we could restrict pore binding and direct the assay toward other mechanisms. In rat Nav1.2 channel, an asparagine residue in the S6 segment of domain 4 (N1769) was critical for local anesthetics binding in the internal pore (30, 31). This asparagine residue is conserved in human Nav1.7 (N1742); therefore, we generated an asparagine-to-lysine mutant channel (N1742K). Whole-cell patch-clamp recordings using SyncroPatch 768 showed that N1742K channels were functional, despite a rightward shift in the voltage dependence of activation and inactivation (Fig. 2 A and B). The half-activation of N1742K was −9.80 ± 0.09 mV (n = 155; Fig. 2B) compared with −29.58 ± 0.14 mV for WT channels (n = 105), and the half inactivation of N1742K was −48.85 ± 0.07 mV (n = 155), compared with −62.85 ± 0.15 mV for WT channel (n = 105).
Fig. 2.
Biophysical and pharmacological characterization of Nav1.7 N1742K mutant channel. (A) Representative current traces for WT and N1742K channels. To assess voltage-dependent activation, currents were elicited by 20-ms test pulses (−80–15 mV in 5-mV increments) from a holding potential at −120 mV. Traces from 0-mV test pulses were highlighted. To determine voltage-dependent inactivation, peak currents at 0 mV were obtained after 500-ms conditioning prepulses varying from −120 to 15 mV. Traces obtained from test pulses (from −120 to 0 mV) were highlighted. Only selected voltage steps were shown in protocols. (B) Activation and inactivation curves for WT and N1742K mutant channels. Activation V1/2: −29.58 ± 0.14 mV (n = 105, WT); −9.80 ± 0.09 mV (n = 155, N1742K); inactivation V1/2: −62.85 ± 0.15 mV (n = 105, WT) and −48.85 ± 0.07 mV (n = 155, N1742K). (C) Pharmacology protocol and representative current traces. Cells were held at −50 mV, pulsed to 0 mV for 10 ms, followed by a 20-ms pulse at −150 mV and 10-ms pulse to 0 mV. Currents elicited at the second 0-mV pulse were used to derive inhibition at inactivated state. Current traces showed 20 nM TTX block of N1742K during a 10-min time period (from gray to black traces). (D) Fold change in IC50 between N1742K and WT channel. Compounds were grouped based on binding sites. Tetracaine, CNV-802, and AMG-52 bind to the inner pore; TTX binds to outer vestibule (OV); ProTx-II binds to VSD2; PF-771 and GX-936 bind to VSD4.
Next, we evaluated the pharmacology of N1742K channels by testing well-characterized sodium channel blockers using electrophysiology (Fig. 2 C and D). N1742K reduced the potency of tetracaine by ∼400-fold, consistent with the role of this asparagine residue in local anesthetics binding (30). N1742K also reduced the potency of AMG-52 (32) by 243-fold and the potency of CNV1014802 (raxatrigine) by 71-fold, suggesting their effects were mediated by interaction with the pore domain. TTX and ProTx-II are neurotoxins that bind to the outer vestibule and voltage sensor domain 2, respectively (25, 28); their potencies were not affected by N1742K. For compounds that bind to the VSD4 domain, N1742K changed the potency of PF-771 by 3.2-fold and the potency of GX-936 by 2.4-fold. Together, these data suggest that N1742K greatly reduces the sensitivity of compounds binding to the inner pore, and has relatively minimal effects on compounds binding outside of that region.
Identification of 1KαPMTX as an Activator of N1742K Nav1.7 Channels.
Next, we sought to identify an activator alternative to veratridine, to develop an N1742K-based membrane potential assay. We focused on activators binding to the VSD4 domain, because this region exhibits the most sequence divergence among Nav channels and because several peptide toxins and small molecules binding to this region showed subtype selectivity (15, 16, 33–35). 1KαPMTX, a derivative of wasp venom toxin α-pompilidotoxin (PMTX) (36, 37), showed the highest TTX-sensitive signal, followed by αPMTX, 1KβPMTX, and βPMTX (Fig. 3 A and B). This group of activators all bind to the VSD4 domain, and remove fast inactivation through immobilizing VSD4 domain (36). N1742K mutant channels had diminished response to veratridine (39 ± 1%, n = 4, relative to 1KαPMTX response). Interestingly, Nav1.7 WT channels did not produce a robust response to 1KαPMTX (Fig. 3B), possibly due to the leftward shift in voltage-dependent inactivation for WT channels (Fig. 2B). Hence, a membrane potential assay using a combination of N1742K and 1KαPMTX was developed.
Fig. 3.
Development of a N1742K-based membrane potential assay using 1KαPMTX as an activator. (A) Activities of 12 Nav activators on N1742K mutant channels tested at 100 µM in the presence and absence of 5 µM TTX (n = 4). The fluorescence signals were normalized to peak fluorescence obtained with 1KαPMTX. (B) Time-course of fluorescence responses to 100 µM 1KαPMTX. Signals were normalized by peak response of 1KαPMTX. (C) 1KαPMTX did not evoked robust response in WT channels. Signals were normalized by peak response of veratridine. Arrows indicate addition of 1KαPMTX.
Pharmacological Evaluation of the N1742K-Based Membrane Potential Assay.
We tested the pharmacology of the N1742K-based membrane potential assay on TTX, tetracaine, ProTx-II, PF-771, and GX-936. TTX blocked 1KαPMTX activation of N1742K channels, with an IC50 comparable to its inhibition of veratridine-activated WT channels (0.048 ± 0.010 μM for N753K vs. 0.034 ± 0.005 μM for WT, n = 6; Fig. 4A). The potency of tetracaine on N1742K response was reduced dramatically (IC50: 488 ± 153 μM for N1742K vs. 3.6 ± 0.4 for WT, n = 6; Fig. 4B), consistent with the electrophysiology data (Fig. 2D). ProTx-II inhibits Nav1.7 channel by binding to VSD2 (38). ProTx-II blocked 1KαPMTX activation of N1742K and veratridine activation of WT channels with comparable potency (IC50: 0.762 ± 0.066 μM vs. 0.794 ± 0.037 μM, n = 4; Fig. 4 C and D. In contrast to their marginal effect on WT channel responses to veratridine, PF-771 and GX-936 exhibited robust inhibition of the 1KαPMTX-evoked N1742K response, with an IC50 of 0.205 ± 0.005 μM for PF-771 and 0.040 ± 0.007 μM for GX-936, respectively (n = 6; Fig. 4 E–H). Therefore, unlike the conventional WT channel-based assay, the N1742K–1KαPMTX assay is capable of detecting VSD4 inhibitors while reducing sensitivity to pore blockers.
Fig. 4.
Characterization of Nav1.7 blockers in N1742K-based membrane potential assay. A total of 100 μM 1KαPMTX and 100 μM veratridine were used to activate N1742K and WT Nav1.7, respectively. (A) Concentration–response relationships of TTX in N1742K (black line) and in WT (dotted line) membrane potential assays. TTX IC50 was 0.048 ± 0.010 μM for N1742K, 0.034 ± 0.005 μM for WT (n = 6). (B) Concentration–response relationships for tetracaine for N1742K (black line) and for WT (dotted line) membrane potential assays. IC50 was 488 ± 153 μM (n = 4) for N1742K and 3.6 ± 0.4 μM for WT (n = 6). (C) Representative kinetic traces for varying concentrations of ProTx-II in N1742K membrane potential assay. (D) Dose–response of ProTx-II in N1742K and WT membrane potential assays. IC50 was 0.762 ± 0.066 (n = 4) for N1742K and 0.794 ± 0.037 μM for WT (n = 4). (E) Representative kinetic traces for PF-771 in N1742K membrane potential assay. (F) Concentration dose–responses of PF-771. IC50 for PF-771 was 0.205 ± 0.005 μM for N1742K (n = 6); PF-771 only had marginal effect on WT (n = 4). (G) Representative kinetic traces for GX-936 in N1742K membrane potential assay. (H) Concentration–response relationship for GX-936. IC50 for GX-936 was 0.040 ± 0.007 μM for N1742K (n = 6); GX-936 only had marginal effect on WT (n = 4). Dose–responses for WT (dotted lines in A, B, F, and H) appeared in Fig. 1 but were included for comparison purpose. Arrows indicate addition of 1KαPMTX.
High-Throughput Screen Using N1742K Membrane Potential Assay.
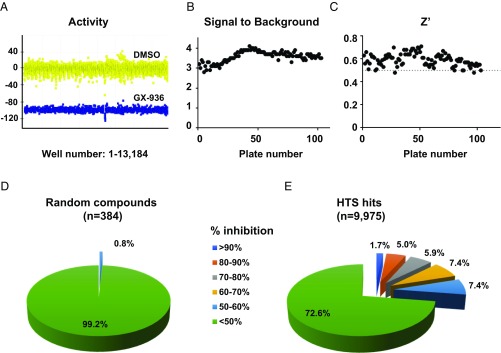
After assay optimization, we developed a fully automated, 1,536-well format membrane potential assay for N1742K channel, and screened a library of 1 million compounds at 5 µM concentration. We used 100 μM 1KαPMTX to activate N1742K mutant channel and 1 μM GX-936 as a control for channel inhibition. Assay robustness was demonstrated by the separation of activity between 0.1% DMSO and GX-936 controls (Fig. 5A). Based on analysis of 103 plates, the signal-to-background ratio was between 2.8 and 4.2 (Fig. 5B); Z′ value, an indication of assay quality, was above 0.5 for 101 of 103 plates; therefore, the assay was suitable for high-throughput screening (Fig. 5C). Mean inhibition for the screen was 16%, with a SD of 20%. Based on a selection criterion of inhibition greater than threefold of the SD, the hit rate was 1.3%. Further criteria were applied to remove potential activators, fluorescence quenchers, and toxic compounds, followed by cheminformatics pruning. These analyses generated a set of 9,975 compounds for follow-up studies.
Fig. 5.
A high-throughput screen using N1742K membrane potential assay. (A) Separation of activity between 0.1% DMSO (yellow, 9,476 wells) and 1 µM GX-936 treated group (blue, 3,708 wells). Activity was plotted as percent effect on 100 µM 1KαPMTX-evoked responses. Data were obtained from 103 plates as described in Materials and Methods. (B) Distribution of signal-to-background ratios for 103 plates. The signal was obtained as fluorescence responses to 100 µM 1KαPMTX, and the background was defined as responses to 1KαPMTX with 1 µM GX-936 treatment; 103 plates were included. (C) Distribution of Z′ values, reflecting robustness of the assay. Z′ > 0.5 is desired for high throughput screening (48). (D) Electrophysiological characterization of 384 randomly selected compounds from Roche library. Compounds were tested at 6.6 µM on N1742K mutant channel using patch-clamp protocols shown in Fig. 2C. Compounds were binned based on percentage of inhibition. (E) Electrophysiological characterization of 9,975 membrane potential assay hits. Compounds were binned based on percentage of inhibition.
To validate the N1742K-based membrane potential assay, we characterized the 9,975 screening hits and 384 randomly selected compounds by using SyncroPatch 768 electrophysiology (18). All compounds were tested at a concentration of 6.6 µM on WT Nav1.7 channels. Among the 384 randomly selected compounds, only three compounds (0.8%) showed 50–60% inhibition, and no compounds showed >60% inhibition (Fig. 5D). Among the screening hits, 1.7% of the compounds showed inhibition >90%; 5.0% of compounds showed 80–90% inhibition; 5.9% of compounds showed 70–80% inhibition; 7.4% of compounds showed 60–70% inhibition; and 7.4% compounds showed 50–60% inhibition (Fig. 5E). Therefore, N1742K-based membrane potential assay was efficient in enriching for Nav1.7 inhibitors.
Identification of Novel Nav1.7-Selective Blockers.
The 2,740 compounds with inhibition >50% in SyncroPatch electrophysiology assays were further selected for concentration-response studies on Nav1.7 to confirm activity, and on Nav1.5 to determine subtype selectivity. Additionally, compounds were tested against various Nav1.7 mutant channels to reveal drug binding sites and mechanisms of actions. Through this effort, we identified compounds of high interest, as exemplified by GNE-0439 (Fig. 6A).
Fig. 6.
The identification of GNE-0439 as a selective inhibitor of Nav1.7. (A) Chemical structure of GNE-0439. (B) GNE-0439 (5 μM) inhibited 1KαPMTX response in N1742K-based assay but had no effect on WT–veratridine assay. Fluorescence signals were normalized against peak response; faint dotted lines indicate baseline. Arrows indicate 1KαPMTX or veratridine addition. (C) Inhibition dose–response of GNE-0439 on Nav1.7, Nav1.5, N1742K, and R1608A channels. IC50 value was 0.34 μM for Nav1.7 (95% confidence: 0.29–0.73 μM); 38.3 μM for Nav1.5 (95% confidence: 21.43–70.92 μM); and 0.37 μM for N1742K (95% confidence: 0.46–0.76 μM). GNE-0439 (66 μM) inhibited R1608A by 41.0 ± 12.8% (n = 6). The dotted line indicates 50% inhibition.
In the N1742K-based membrane potential assay, GNE-0439 (5 μM) nearly completely blocked responses to 1KαPMTX (Fig. 6B). However, in the conventional WT-based membrane potential assay, GNE-0439 did not inhibit the veratridine-evoked responses (Fig. 6B). Therefore, similar to PF-771 and GX-936 (Fig. 1), GNE-0439 would not have been discovered with the conventional membrane potential screen. In electrophysiology experiments (Fig. 6C), GNE-0439 inhibited Nav1.7 with an IC50 of 0.34 μM (95% confidence: 0.29–0.73 μM) and inhibited Nav1.5 with an IC50 of 38.3 μM (95% confidence: 21.43–70.92 μM). Therefore, GNE-0439 is highly selective against Nav1.5. GNE-0439 also inhibited N1742K channels (IC50 = 0.37 μM, 95% confidence: 0.46–0.76 μM), suggesting this pore residue is not involved in compound binding. R1608A, a mutation of the fourth charged arginine residue in the S4 segment of VSD4 domain, was reported to reduce arylsulfonamide binding by >1,000-fold (15). R1608A greatly reduced the potency of GNE-0439 (41.0 ± 12.8% inhibition at 66 μM, n = 6; Fig. 6C), strongly suggesting that GNE-0439 binds to the VSD4 domain.
The structure of GNE-0439 is unique compared with known selective VSD4 binders such as the arylsulfonamides (GX-936 and PF-771) (15, 16) and acylsulfonamides (39). It is structurally homologous to a class of VSD4 binders with amino acid-like constituents that we discovered through a structure-based drug design approach (40). GNE-0439 and this new class of inhibitor possess a carboxylic acid group that is predicted to form a favorable electrostatic interaction with R1608, similar to the interaction between GX-936 (an acidic arylsulfonamide) and R1608 observed in a cocrystal structure of a Nav1.7 chimeric channel (15). The discovery of GNE-0439 demonstrates that our assay is capable of identifying Nav1.7-selective inhibitors that bind outside of the channel pore.
Discussion
Membrane potential assays have been used to screen tens of millions of compounds for Nav1.7 blockers (20–24). Despite the tremendous efforts, few Nav1.7-selective chemical scaffolds have been identified. This paucity of results reflects the high sequence identity of residues in the pore domain of Nav channels and, particularly, the conservation of several key residues (e.g., F1737, N1742, and Y1744) that afford promiscuous binding to diverse chemicals (14). Here we show that the lack of productivity is further aggravated by poor assay design. In the conventional membrane potential assays, compounds are incubated with Nav1.7-expressing cells first, and veratridine is applied subsequently to stimulate channel activity. As veratridine binds to the pore domain (25), pore blockers (e.g., tetracaine) may preclude veratridine binding, and are therefore preferentially identified in the screening campaign (Figs. 1 and 7).
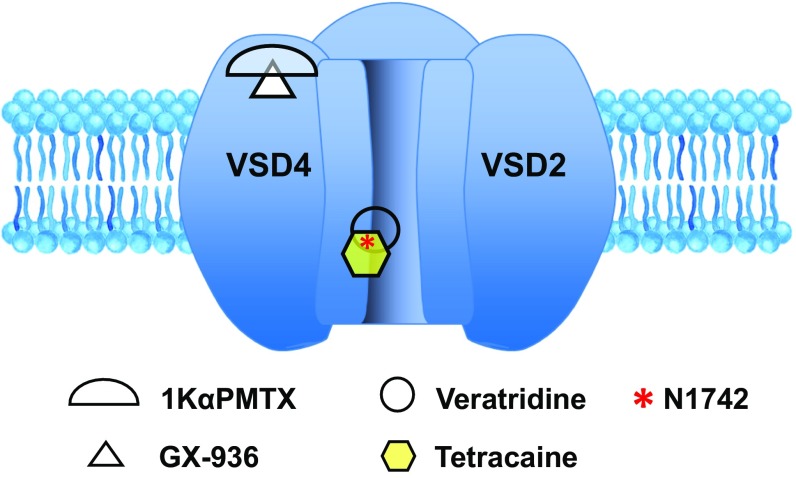
Fig. 7.
Drug binding sites for Nav1.7 activators and inhibitors underlie membrane potential assay performance. Veratridine and tetracaine bind to the inner pore, in the vicinity of residue N1742. In WT membrane potential assay, tetracaine affects veratridine binding and therefore inhibits response to veratridine; in N1742K mutant assay, tetracaine binding to N1742K channel is reduced. GX-936 and 1KαPMTX bind to the vicinity of VSD4 domain. In the N1742K mutant assay, GX-936 binding inhibits N1742K response to 1KαPMTX.
Besides the bias toward pore binders, the conventional membrane potential assay failed to detect the most potent and selective VSD4 compounds (Fig. 1 C, D, F, and G). At concentrations several orders of magnitude greater than their IC50 values, PF-771 and GX-936 had minimal effect on veratridine-induced response. The current data are consistent with a previous report of an assay using voltage-sensor probes (21), where arylsulfonamides failed to inhibit WT response to veratridine. In the assay using blue fluorescence membrane potential dye, arylsulfonamides delayed the onset of fluorescence but failed to inhibit peak signals. This lack of sensitivity to arylsulfonamides could be related to their strong preference for inactivated state block. For example, PF-771 and GX-936 are ∼1,000-fold more potent on the inactivated state compared with the closed state of the channel (15, 16). In the conventional membrane potential assay, veratridine removes inactivation and therefore compromises the effects of these compounds. Regardless of the mechanisms, the conventional membrane potential assay is not capable of detecting VSD4 blockers.
Based on mechanisms of action and drug binding sites (Fig. 7), we modified the conventional assay by replacing WT Nav1.7 with the N1742K mutant, and by replacing veratridine with 1KαPMTX as an alternative activator. The N1742K mutation disrupted a key binding site, thus reducing sensitivity to pore binders. 1KαPMTX was derived from a family of wasp toxins known for removing fast inactivation and increasing steady-state currents of several sodium channels (36, 37). PMTXs bind to the extracellular S3–S4 linker of VSD4 domain (Fig. 7) (41). Among the four VSDs of Nav channels, VSD4 is crucial for mediating fast inactivation (42, 43). Specifically, the intramembrane charge displacement associated with movement of VSD4 is necessary for the development and recovery from fast inactivation (44). Presumably, the binding of 1KαPMTX impedes VSD4 movement and thereby removes fast inactivation. In addition, due to the low sequence conservation in VSD4 domain, binding to VSD4 confers subtype selectivity for peptide toxins and small molecule inhibitors (15, 16, 33–35). By combining the use of N1742K pore mutant and 1KαPMTX as an activator, we were able to direct the assay toward non–pore-binding mechanism and facilitate the detection of subtype-selective VSD4 binders, such as PF-771, GX-936, and GNE-0439. Most recently, antillatoxin, a lipopeptide from pantropical marine cyanobacteria, was used to activate Nav1.7 channel in a membrane potential assay (45). Similar to veratridine, antillatoxin activates sodium channels through binding to the inner pore domain (25, 46). The antillatoxin-based assay could detect TTX and local anesthetics, but its sensitivity to VSD4 binders was not reported (45).
Theoretically, the N1742K-based assay could lead to the identification of false positives by introducing a potential new binding site residue (K1742). However, such compounds would gain potency on the N1742K mutant channel and could be eliminated by comparing potency on WT and K1742 mutant channels using electrophysiology (as in Figs. 5E and 6C). Of note, the N1742K-based assay reduced but did not completely eliminate detection of pore binders. Although N1742 has been implicated in the binding of many pore blockers (30, 31), N1742 is not the only critical residue, and thus N1742K mutation can reduce, but may not abolish, pore binding. In fact, several other residues, including F1737 and Y1744, are also implicated in binding of pore blockers (14). Channels harboring two (e.g., F1737A/Y1744A) or three (F1737A/N1742K/Y1744A) mutations would potentially restrict a larger subset of pore binders. Unfortunately, the response of double- or triple-mutant channels to 1KαPMTX was not robust enough for compound screening. Of note, we observed a significant potency shift between the membrane potential assay and electrophysiology experiments (e.g., IC50: 0.205 μM vs. 0.011 μM for PF-771). There could be several explanations for this shift in potency. First, the membrane potential assay is not a linear readout of channel function. Second, to obtain robust signals for compound screening, a high concentration of 1KαPMTX (100 μM) was used, which may potentially affect IC50 values. Third, inhibition of Nav1.7 by arylsulfonamides is strongly state dependent (15, 16); electrophysiology data were obtained from channels in inactivated states (Fig. 2C). The membrane potential assay did not control voltage and the 1KαPMTX responses might be obtained from different channel states. Interestingly, both the N1742K–1KαPMTX assay and the WT–veratridine assay could detect ProTx-II, a VSD2 binder without state dependency, though its potency (IC50: 0.762 μM for N1742K and 0.794 μM for WT; Fig. 4D) was significantly reduced compared with electrophysiology measurements (IC50: 0.6 nM for N1742K and 0.3 nM for WT) (38). This decrease in potency might be partially caused by the slow kinetics of ProTx-II and by its promiscuous binding to compound plates and pipette tips during membrane potential experiments. It is possible that our assay is biased toward VSD4 ligands, and its utility for detecting small molecules with other binding sites (e.g., VSD2) remains to be determined. Despite these limitations, targeting VSD4 has been the most successful strategy for achieving subtype-specific modulation (15, 16, 33–35), and our assay represents a valuable tool for Nav1.7 ligand discovery. A high-throughput screen using this assay successfully enriched sodium channel blockers, leading to Nav1.7-selective hits (Figs. 5D and 6).
It is conceivable that our current assay could be further improved, or designed toward specific mechanisms or drug binding sites by using various combinations of mutant channels and activators. The mechanism-specific assay design can also be extended to other assay formats (e.g., electrophysiology), other sodium channel isoforms (e.g., Nav1.1), and other ion channel families. For example, we now routinely use electrophysiology to screen compounds using mutant channels for specific mechanisms (e.g., pore and VSD4; Fig. 6C). Besides Nav1.7, achieving subtype selectivity is challenging for many other ion channel targets such as Nav1.1, Cav1.3, and Kv1.3 (10, 35, 47). Nav1.1 possesses the same local anesthetics binding sites in the pore domain. By combining Nav1.1 pore mutations with Hm1a, a selective activator binding to VSD4 (35), assays could be developed to identify Nav1.1-selective compounds. Similarly, the promiscuous drug binding sites for Cav1.3 (dihydropyridine) and Kv1.3 (tetraethylammonium) reside in the highly conserved pore domains. Point mutations that disrupt these sites could be exploited to direct compound screening toward non–pore-mediated mechanisms.
In summary, we revealed the intrinsic limitations of a conventional membrane potential assay that have confounded Nav1.7 drug discovery. By using a rational assay design, we directed the assay toward desired mechanisms and identified unique Nav1.7-selective scaffolds. The mechanism-specific assay design can be applied to other ion channels to facilitate future drug discovery.
Materials and Methods
Cells.
Human Nav1.7, N1742K and R1608A, and Nav1.5 were cloned in the tetracycline (Tet)-inducible system (pcDNA6/TR), and stably expressed in HEK293 cells. Each cell line was grown in DMEM high-glucose medium supplemented with 10% FBS (Tet-free), 2 mM l-glutamine (Gln), blasticidin at 5 μg/mL, Zeocin at 400 μg/mL, in 5% CO2 and 37 °C. When cell cultures reached ∼70% confluency, expression was induced with doxycycline (1 µg/mL) in DMEM + 2% FBS + l-Glu, in the absence of antibiotics for 16 h. Cells were harvested with Accutase, spun down at 100 × g (Allegra 6R; Beckman Coulter) for 10 min, and resuspended in DMEM + 2% FBS + l-Glu at a density of 5 × 106 cells per milliliter.
Reagents.
Blue membrane potential dye (R8034) was obtained from Molecular Devices. Tet-free FBS was obtained from Clontech (631101), and other cell culture reagents were from Life Technologies. TTX was obtained from Enzo Life Sciences; 1KαPMTX and voltage-gated sodium channel activator explorer kit were obtained from Alomone Labs; Veratridine and tetracaine were from Sigma Aldrich; PF-771, GX-936, and GNE-0439 were synthesized at Genentech.
Membrane Potential Assays for WT and N1742K Mutant Channels.
Assays were run in the 1,536-well format. BioRAPTR (Beckman Coulter) was used to dispense cells and membrane potential dye. ECHO (Labcyte) was used for dispensing of library compounds. Multidrop Combi (Thermo Fisher) was used to dilute compounds in 1,536 plates. FDSS7000 (Hamamatsu) was used for compound addition and detection of fluorescent signals. Cells were dispensed at 2,000 cells per well in 4 µL total volume into Aurora Kalypsys, 1,536 black, clear-bottom plates (CLS3833-100EA; Corning). For Nav1.7 WT cells, a 2-h attachment period at 37 °C preceded membrane potential dye addition. For N1742K cells, membrane potential dye was added at the same time as the cells. Membrane potential dye was diluted into buffer A (157.5 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM Hepes, 10 mM glucose, pH 7.4) and transferred by BioRAPTR to the plates at 2 µL per well. Cells and dye were incubated for 1 h at 37 °C, then 15 min at room temperature. Plates were then transferred to FDSS7000. Compound plates (1,536, 782270-1B; Greiner) were generated on ECHO and diluted with buffer A (see above) on multidrop.
For high-throughput screening, compounds were tested at a concentration of 5 µM. After 3-min incubation, veratridine was added to activate wild-type Nav1.7, and 1KαPMTX was used to activate N1742K. Veratridine and 1KαPMTX were each dissolved in buffer B (115 mM NaCl, 45 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM Hepes, 10 mM glucose, pH 7.4), and 2 µL per well was added to a final 100 µM concentration. Fluorescence signals were collected every second for 4.5 min, starting at 10 s before compound addition. Each compound plate contained 92 wells for negative control (0.1% DMSO) and 36 wells for positive control (1 µM TTX for WT–veratridine assay, or 1 µM GX-936 for N1742K–1KαPMTX assay).
Membrane potential assay results were processed by evaluating changes in fluorescence from the average fluorescence of initial 10-s vs. peak fluorescence upon activator addition. Data were normalized to basal levels. Genedata and Vortex (Dotmatics) software programs were used to process high-throughput screening data. GraphPad Prism software was used for generating inhibition concentration responses.
Electrophysiology.
Automated patch-clamp recordings were performed using the SyncroPatch 768PE (Nanion). Chips with single-hole medium resistance (5∼8 MΩ) were used. Pulse generation and data collection were performed with PatchController384 V1.4.1 and DataController384 V1.3.3. Whole-cell recordings were conducted according to Nanion’s procedure. Briefly, cells were stored at 10 °C in a cell hotel (shake speed at 60 rpm). Cell catching, sealing, whole-cell formation, liquid application, recording, and data acquisition were performed sequentially. The voltage protocol consists of 220 ms leak pulse to obtain seal resistance, series resistance, and cell capacitance. Series resistance compensation was set to 80% and currents were sampled at 10 kHz. All data were corrected for leak currents. After establishing baseline currents (IBaseline), compounds were applied and currents were obtained after 5-min incubation of compounds (IEND), then followed by 2 µM TTX treatment to obtain full block (). Compound effect after 5 min was calculated using the following formula:
IC50 values were calculated by fitting dose–responses to four-parameter Hill equation using GraphPad Prism with constrained bottom at 0 and top at 1. For high-throughput screen data analysis, the Genedata screener was used.
Acknowledgments
We thank Saundra Clausen, Jennifer Perry-Ruzic, Rebecca Turincio, Timothy Dawes, Margaret Scott, Cristina Lewis, Moulay Hicham Alaoui Ismaili, and other colleagues for their support; and Dr. Michael Sanguinetti for comments on the manuscript.
Footnotes
Conflict of interest statement: H.V., K.K., and C.J.C. are employees of Xenon Pharmaceuticals; and T.C.-R., T.L., G.L., S.W.J., M.H.B., C.L., D.F.O., S.J.M., D.H.H., D.S., and J.C. are employees of Genentech.
This article is a PNAS Direct Submission.
References
- 1.Wickenden A, Priest B, Erdemli G. Ion channel drug discovery: Challenges and future directions. Future Med Chem. 2012;4:661–679. doi: 10.4155/fmc.12.4. [DOI] [PubMed] [Google Scholar]
- 2.Dib-Hajj SD, Yang Y, Black JA, Waxman SG. The Na(V)1.7 sodium channel: From molecule to man. Nat Rev Neurosci. 2013;14:49–62. doi: 10.1038/nrn3404. [DOI] [PubMed] [Google Scholar]
- 3.George AL., Jr Inherited disorders of voltage-gated sodium channels. J Clin Invest. 2005;115:1990–1999. doi: 10.1172/JCI25505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox JJ, et al. An SCN9A channelopathy causes congenital inability to experience pain. Nature. 2006;444:894–898. doi: 10.1038/nature05413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fertleman CR, et al. SCN9A mutations in paroxysmal extreme pain disorder: Allelic variants underlie distinct channel defects and phenotypes. Neuron. 2006;52:767–774. doi: 10.1016/j.neuron.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Yang Y, et al. Mutations in SCN9A, encoding a sodium channel alpha subunit, in patients with primary erythermalgia. J Med Genet. 2004;41:171–174. doi: 10.1136/jmg.2003.012153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett PB, Yazawa K, Makita N, George AL., Jr Molecular mechanism for an inherited cardiac arrhythmia. Nature. 1995;376:683–685. doi: 10.1038/376683a0. [DOI] [PubMed] [Google Scholar]
- 8.Escayg A, et al. A novel SCN1A mutation associated with generalized epilepsy with febrile seizures plus–And prevalence of variants in patients with epilepsy. Am J Hum Genet. 2001;68:866–873. doi: 10.1086/319524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasaki R, et al. A novel mutation in the gene for the adult skeletal muscle sodium channel alpha-subunit (SCN4A) that causes paramyotonia congenita of von Eulenburg. Arch Neurol. 1999;56:692–696. doi: 10.1001/archneur.56.6.692. [DOI] [PubMed] [Google Scholar]
- 10.Cahalan MD, Chandy KG. The functional network of ion channels in T lymphocytes. Immunol Rev. 2009;231:59–87. doi: 10.1111/j.1600-065X.2009.00816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang S, et al. Structure-activity relationship of N,N′-disubstituted pyrimidinetriones as Ca(V)1.3 calcium channel-selective antagonists for Parkinson’s disease. J Med Chem. 2013;56:4786–4797. doi: 10.1021/jm4005048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitz A, et al. Design of PAP-1, a selective small molecule Kv1.3 blocker, for the suppression of effector memory T cells in autoimmune diseases. Mol Pharmacol. 2005;68:1254–1270. doi: 10.1124/mol.105.015669. [DOI] [PubMed] [Google Scholar]
- 13.Catterall WA. Forty Years of sodium channels: Structure, function, pharmacology, and epilepsy. Neurochem Res. 2017;42:2495–2504. doi: 10.1007/s11064-017-2314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ragsdale DS, McPhee JC, Scheuer T, Catterall WA. Common molecular determinants of local anesthetic, antiarrhythmic, and anticonvulsant block of voltage-gated Na+ channels. Proc Natl Acad Sci USA. 1996;93:9270–9275. doi: 10.1073/pnas.93.17.9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahuja S, et al. Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist. Science. 2015;350:aac5464. doi: 10.1126/science.aac5464. [DOI] [PubMed] [Google Scholar]
- 16.Alexandrou AJ, et al. Subtype-selective small molecule inhibitors reveal a fundamental role for Nav1.7 in nociceptor electrogenesis, axonal conduction and presynaptic release. PLoS One. 2016;11:e0152405. doi: 10.1371/journal.pone.0152405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chambers C, Witton I, Adams C, Marrington L, Kammonen J. High-throughput screening of Na(V)1.7 modulators using a giga-seal automated patch clamp instrument. Assay Drug Dev Technol. 2016;14:93–108. doi: 10.1089/adt.2016.700. [DOI] [PubMed] [Google Scholar]
- 18.Li T, et al. High-throughput electrophysiological assays for voltage gated ion channels using SyncroPatch 768PE. PLoS One. 2017;12:e0180154. doi: 10.1371/journal.pone.0180154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerne R, Wakulchik M, Krambis MJ, Burris KD, Priest BT. IonWorks Barracuda assay for assessment of state-dependent sodium channel modulators. Assay Drug Dev Technol. 2016;14:84–92. doi: 10.1089/adt.2015.677. [DOI] [PubMed] [Google Scholar]
- 20.Felix JP, et al. Functional assay of voltage-gated sodium channels using membrane potential-sensitive dyes. Assay Drug Dev Technol. 2004;2:260–268. doi: 10.1089/1540658041410696. [DOI] [PubMed] [Google Scholar]
- 21.Finley M, et al. Kinetic analysis of membrane potential dye response to NaV1.7 channel activation identifies antagonists with pharmacological selectivity against NaV1.5. J Biomol Screen. 2016;21:480–489. doi: 10.1177/1087057116629669. [DOI] [PubMed] [Google Scholar]
- 22.Trivedi S, et al. Cellular HTS assays for pharmacological characterization of Na(V)1.7 modulators. Assay Drug Dev Technol. 2008;6:167–179. doi: 10.1089/adt.2007.090. [DOI] [PubMed] [Google Scholar]
- 23.Liu CJ, et al. A high-capacity membrane potential FRET-based assay for NaV1.8 channels. Assay Drug Dev Technol. 2006;4:37–48. doi: 10.1089/adt.2006.4.37. [DOI] [PubMed] [Google Scholar]
- 24.Huang CJ, et al. Characterization of voltage-gated sodium-channel blockers by electrical stimulation and fluorescence detection of membrane potential. Nat Biotechnol. 2006;24:439–446. doi: 10.1038/nbt1194. [DOI] [PubMed] [Google Scholar]
- 25.Stevens M, Peigneur S, Tytgat J. Neurotoxins and their binding areas on voltage-gated sodium channels. Front Pharmacol. 2011;2:71. doi: 10.3389/fphar.2011.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leibowitz MD, Sutro JB, Hille B. Voltage-dependent gating of veratridine-modified Na channels. J Gen Physiol. 1986;87:25–46. doi: 10.1085/jgp.87.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fozzard HA, Sheets MF, Hanck DA. The sodium channel as a target for local anesthetic drugs. Front Pharmacol. 2011;2:68. doi: 10.3389/fphar.2011.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fozzard HA, Lipkind GM. The tetrodotoxin binding site is within the outer vestibule of the sodium channel. Mar Drugs. 2010;8:219–234. doi: 10.3390/md8020219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noda M, Suzuki H, Numa S, Stühmer W. A single point mutation confers tetrodotoxin and saxitoxin insensitivity on the sodium channel II. FEBS Lett. 1989;259:213–216. doi: 10.1016/0014-5793(89)81531-5. [DOI] [PubMed] [Google Scholar]
- 30.Ragsdale DS, McPhee JC, Scheuer T, Catterall WA. Molecular determinants of state-dependent block of Na+ channels by local anesthetics. Science. 1994;265:1724–1728. doi: 10.1126/science.8085162. [DOI] [PubMed] [Google Scholar]
- 31.Wright SN, Wang SY, Wang GK. Lysine point mutations in Na+ channel D4-S6 reduce inactivated channel block by local anesthetics. Mol Pharmacol. 1998;54:733–739. [PubMed] [Google Scholar]
- 32.Bregman H, et al. Identification of a potent, state-dependent inhibitor of Nav1.7 with oral efficacy in the formalin model of persistent pain. J Med Chem. 2011;54:4427–4445. doi: 10.1021/jm200018k. [DOI] [PubMed] [Google Scholar]
- 33.McCormack K, et al. Voltage sensor interaction site for selective small molecule inhibitors of voltage-gated sodium channels. Proc Natl Acad Sci USA. 2013;110:E2724–E2732. doi: 10.1073/pnas.1220844110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bosmans F, Martin-Eauclaire MF, Swartz KJ. Deconstructing voltage sensor function and pharmacology in sodium channels. Nature. 2008;456:202–208. doi: 10.1038/nature07473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osteen JD, Sampson K, Iyer V, Julius D, Bosmans F. Pharmacology of the Nav1.1 domain IV voltage sensor reveals coupling between inactivation gating processes. Proc Natl Acad Sci USA. 2017;114:6836–6841. doi: 10.1073/pnas.1621263114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konno K, et al. Molecular determinants of binding of a wasp toxin (PMTXs) and its analogs in the Na+ channels proteins. Neurosci Lett. 2000;285:29–32. doi: 10.1016/s0304-3940(00)01017-x. [DOI] [PubMed] [Google Scholar]
- 37.Schiavon E, et al. Voltage-gated sodium channel isoform-specific effects of pompilidotoxins. FEBS J. 2010;277:918–930. doi: 10.1111/j.1742-4658.2009.07533.x. [DOI] [PubMed] [Google Scholar]
- 38.Schmalhofer WA, et al. ProTx-II, a selective inhibitor of NaV1.7 sodium channels, blocks action potential propagation in nociceptors. Mol Pharmacol. 2008;74:1476–1484. doi: 10.1124/mol.108.047670. [DOI] [PubMed] [Google Scholar]
- 39.DiMauro EF, et al. Application of a parallel synthetic strategy in the discovery of biaryl acyl sulfonamides as efficient and selective NaV1.7 inhibitors. J Med Chem. 2016;59:7818–7839. doi: 10.1021/acs.jmedchem.6b00425. [DOI] [PubMed] [Google Scholar]
- 40.Andrez J-C, et al. 2016. Therapeutic Compounds and Methods of Use Thereof. US Patent WO2016007534.
- 41.Kinoshita E, et al. Novel wasp toxin discriminates between neuronal and cardiac sodium channels. Mol Pharmacol. 2001;59:1457–1463. [PubMed] [Google Scholar]
- 42.Horn R, Ding S, Gruber HJ. Immobilizing the moving parts of voltage-gated ion channels. J Gen Physiol. 2000;116:461–476. doi: 10.1085/jgp.116.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheets MF, Kyle JW, Kallen RG, Hanck DA. The Na channel voltage sensor associated with inactivation is localized to the external charged residues of domain IV, S4. Biophys J. 1999;77:747–757. doi: 10.1016/S0006-3495(99)76929-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capes DL, Goldschen-Ohm MP, Arcisio-Miranda M, Bezanilla F, Chanda B. Domain IV voltage-sensor movement is both sufficient and rate limiting for fast inactivation in sodium channels. J Gen Physiol. 2013;142:101–112. doi: 10.1085/jgp.201310998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao F, et al. Development of a rapid throughput assay for identification of hNav1.7 antagonist using unique efficacious sodium channel agonist, antillatoxin. Mar Drugs. 2016;14:E36. doi: 10.3390/md14020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao Z, Gerwick WH, Murray TF. Antillatoxin is a sodium channel activator that displays unique efficacy in heterologously expressed rNav1.2, rNav1.4 and rNav1.5 α subunits. BMC Neurosci. 2010;11:154. doi: 10.1186/1471-2202-11-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang S, et al. CaV1.3-selective L-type calcium channel antagonists as potential new therapeutics for Parkinson’s disease. Nat Commun. 2012;3:1146. doi: 10.1038/ncomms2149. [DOI] [PubMed] [Google Scholar]
- 48.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]