Significance
An expanding body of evidence argues that the Aβ and tau proteins share important characteristics of prion propagation to cause pathogenesis in Alzheimer’s disease (AD). Aβ and tau form a number of amyloids (β-sheet–rich structures) with distinct conformations (“strains”), some of which give rise to different diseases and associated pathologies. We develop new probes of amyloid structure and use these to identify conformational strains of Aβ in heritable and sporadic forms of AD patient samples. We demonstrate that distinct strains of Aβ can be discerned in different disease types, or in different brain compartments within a given patient. Our findings may potentially explain the spectrum of clinical and pathologic features observed in AD.
Keywords: Alzheimer’s disease, amyloid-β, conformational strains, spectral imaging, protein misfolding
Abstract
Point mutations in the amyloid-β (Aβ) coding region produce a combination of mutant and WT Aβ isoforms that yield unique clinicopathologies in familial Alzheimer’s disease (fAD) and cerebral amyloid angiopathy (fCAA) patients. Here, we report a method to investigate the structural variability of amyloid deposits found in fAD, fCAA, and sporadic AD (sAD). Using this approach, we demonstrate that mutant Aβ determines WT Aβ conformation through prion template-directed misfolding. Using principal component analysis of multiple structure-sensitive fluorescent amyloid-binding dyes, we assessed the conformational variability of Aβ deposits in fAD, fCAA, and sAD patients. Comparing many deposits from a given patient with the overall population, we found that intrapatient variability is much lower than interpatient variability for both disease types. In a given brain, we observed one or two structurally distinct forms. When two forms coexist, they segregate between the parenchyma and cerebrovasculature, particularly in fAD patients. Compared with sAD samples, deposits from fAD patients show less intersubject variability, and little overlap exists between fAD and sAD deposits. Finally, we examined whether E22G (Arctic) or E22Q (Dutch) mutants direct the misfolding of WT Aβ, leading to fAD-like plaques in vivo. Intracerebrally injecting mutant Aβ40 fibrils into transgenic mice expressing only WT Aβ induced the deposition of plaques with many biochemical hallmarks of fAD. Thus, mutant Aβ40 prions induce a conformation of WT Aβ similar to that found in fAD deposits. These findings indicate that diverse AD phenotypes likely arise from one or more initial Aβ prion conformations, which kinetically dominate the spread of prions in the brain.
The pathogenesis of Alzheimer’s disease (AD) is characterized by the formation and propagation of both amyloid-β (Aβ) and tau prions in the brain. Mutations in the Aβ precursor protein (APP) gene or the γ-secretase enzyme complex cause early-onset, familial AD (fAD) or cerebral amyloid angiopathy (fCAA) (1). Specifically, point mutations within the Aβ coding region determine the composition of Aβ isoforms (2, 3), the pattern of cerebral amyloid deposition, and the early onset of disease (4–6). In contrast with sporadic AD (sAD) cases, the extent of parenchymal amyloid deposition in fAD (7) and vascular amyloid in fCAA patients correlates with cognitive impairment (8, 9). Typically, familial mutations within the central coding region of Aβ decrease thermodynamic stability and accelerate fibrillogenesis in vitro (10–13). Because fAD and fCAA are autosomal dominant disorders, heterozygous carriers produce both mutant and WT Aβ. In vitro experiments suggest that the aggregation-prone mutant Aβ initially misfolds to form a prion that induces misfolding of both the WT and mutant peptides (11, 13). However, in vivo, it is not clear whether the mutant acts as a prion to induce misfolding of the WT or whether the plaques are composed of the more rapidly aggregating mutant peptide alone.
The current view of AD pathogenesis, like other neurodegenerative disorders involving protein aggregation, is that the formation of a pathogenic protein with a self-propagating structural state, a prion, is the underlying mechanism of disease (14, 15). Both Aβ and PrP prions have the capacity to propagate as distinct strains, which yield unique biochemical and histopathologic properties following transmission to animals (16–19). Strain-specific biological information is enciphered in the distinct prion conformation of the underlying protein (16, 17, 20–24). Alternative PrP prion conformations can emerge from environmental changes (25), therapeutic interventions (26–28), or mutations within the Prnp gene in mice (29, 30), leading to different incubation times and deposition patterns.
Similarly, we hypothesize that the unique pathology apparent in fAD and fCAA patients (with mutations in the Aβ coding region) is due to a distinct Aβ prion conformation defined by the Aβ mutation. Previous in vitro experiments have suggested that, following a dominant-negative mechanism, the mutant Aβ prion templates its structure onto WT Aβ, thereby inducing a conformational strain or strains not readily formed by the WT. The first clinical evidence suggesting a structural difference between Aβ deposits in sAD versus fAD was shown by positron emission tomography (PET)-based amyloid imaging in fAD patients with the Arctic [Aβ(E22G)] or Osaka [Aβ(E22∆)] mutation (31, 32). Surprisingly, these fAD patients tested negative for the amyloid-specific PET probe Pittsburgh compound B (PiB), which is commonly used to detect Aβ deposits in sAD cases (33), despite showing a severe cerebral amyloid burden postmortem (4). Thus, it would appear that these mutants adopt an alternative Aβ conformation (or strain) with a reduced binding affinity for PiB.
The accumulation of Aβ38 peptides in parenchymal and vascular deposits (2) is another hallmark of fAD and fCAA patients with mutations within the Aβ coding region. We recently demonstrated that brain tissue from a fAD [Aβ(E22G)] patient transmitted to mice induced a distinct pathological phenotype, which included Aβ38 deposition (19). These studies suggest that mutant Aβ may determine fAD pathogenesis. However, because the fAD patient-derived samples used in these studies contained a mixture of WT and mutant Aβ with isoform distributions different from those seen in sAD, it was not clear whether mutant, homogeneous Aβ peptides could act as prions to model pathogenic plaque formation in vivo.
To examine the mechanisms of mutant Aβ prion propagation, we investigated the interaction between mutant Aβ prions and WT Aβ in vitro and in transgenic (Tg) mice. We hypothesized that the mutant Aβ conformation determines unique conformational strain properties through a dominant-templating process. To test this hypothesis, we injected synthetic Aβ40(E22G) or Aβ40(E22Q) fibrils into the brains of Tg mice expressing only WT peptide. Mutant Aβ prions induced widespread deposition of WT Aβ in inoculated mice. Using multiple conformation-sensitive fluorescent probes (34) and high-resolution confocal spectral imaging in combination with principal component analysis (PCA), we demonstrate that WT Aβ deposits acquire a relatively homogeneous set of mutant-like conformations that are distinct from those seen in sAD, but closely resemble the conformations seen in fAD human brain samples. Furthermore, the plaque isoform composition induced in the mouse brain was rich in Aβ38, closely matching that seen in the corresponding fAD patients.
Our studies show that Aβ conformations associated with the mutant peptides are sufficient to induce major structural and pathological hallmarks of fAD in mice expressing only WT Aβ. Moreover, we found a dominant-templating process that likely explains the amyloid phenotype observed in fAD patients, although these patients have a heterogeneous mixture of WT and mutant Aβ peptides present in the brain. Importantly, our findings shape our understanding of the fundamental processes governing Aβ prion propagation. We show that the initial self-propagating Aβ conformation is the structure that defines the strain phenotype seen in AD patients.
Results
Amyloid Deposits from Distinct Aβ Prion Strains in Sporadic and fAD Patients Can be Differentiated with Structure-Sensitive Probes.
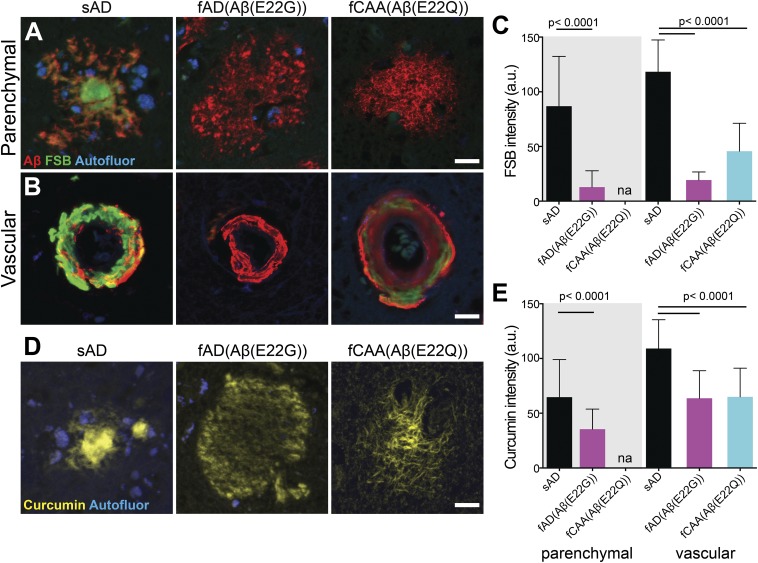
Amyloid-negative PET scans in fAD patients with abundant Aβ deposition suggest that distinct Aβ conformations are present in these individuals compared with sAD patients. Initially, we probed Aβ deposits using the amyloid-binding dye FSB [(E,E)-1-fluoro-2,5-bis(3-hydroxycarbonyl-4-hydroxy)styrylbenzene] (Figs. 1 and 2 and Fig. S1) in brain slices from 22 sAD patients, 5 fAD E22G (Arctic), and 8 fCAA E22Q (Dutch) cases (Table S1). Additionally, we examined six cases from fAD patients bearing the Swedish mutation (which occurs outside the Aβ coding region). This familial mutation provides an important control as it alters the β-secretase cleavage site N-terminal to the Aβ peptide region, which results in an increase in production of Aβ but not the amino acid sequences of the processed Aβ peptides. In each case, 40–70 plaques were imaged and their location (parenchymal versus vascular) was recorded. Using confocal imaging analysis, we observed bright staining of Aβ deposits from all of the sAD and Swedish cases. However, both parenchymal and vascular Aβ deposits in fAD Aβ(E22G) and fCAA Aβ(E22Q) brain samples showed a marked decrease in fluorescence intensity for FSB and other small-molecule amyloid dyes (Fig. 1 and Fig. S1). The low fluorescence intensity is not due to lower amounts of deposited Aβ as shown with colabeling by an N-terminal–specific Aβ antibody (Fig. 1A). These data clearly demonstrate that FSB intensity is a convenient indicator capable of differentiating E22G and E22Q mutant deposits from WT. Moreover, given the environment-sensitivity of this class of fluorophores, we suggest that the change in intensity reflects conformational differences between the mutants versus the WT, as previously shown in vitro using the dye thioflavin T (ThT) for position 22 mutants of Aβ peptides (11, 35).
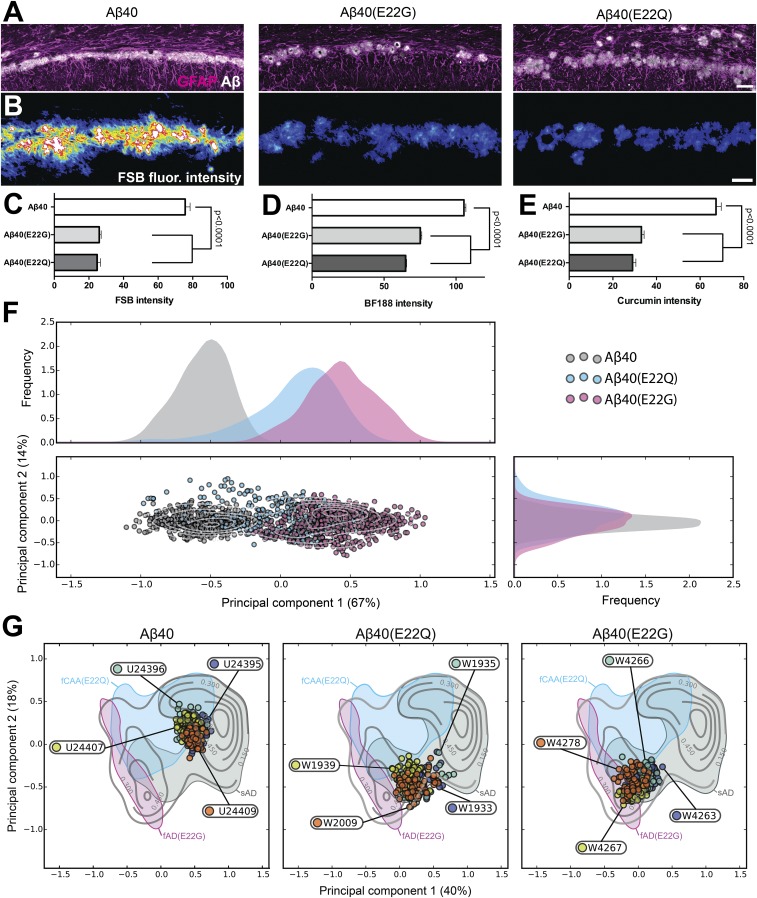
Fig. 1.
Reduced emission intensity of fluorescent β-sheet binding dyes in fAD and fCAA brain samples. (A and B) Confocal images show Aβ deposits in the brain gray matter (“parenchymal”) and along the walls of the cerebrovasculature (“vascular”), also known as CAA, in sporadic, fAD [Aβ(E22G)] (Arctic), and fCAA [Aβ(E22Q)] (Dutch) samples labeled with pan-Aβ antibody (Aβ1–16; red) and the fluorescent β-sheet binding small-molecule dye FSB (green). Autofluorescent structures (i.e., lipofuscin granules) were acquired in a separate channel (blue) to contrast these structures from labeled deposits. (C) Quantitation of FSB fluorescence intensity in Aβ-immunolabeled parenchymal and vascular Aβ deposits. Diffuse plaques in the Aβ(E22Q) (Dutch) cases were sparse and not sufficient in number for quantitative analyses (reported as “na” in graph). Sampling for FSB: n = 492 parenchymal/76 vascular deposits from five sAD cases; 296 parenchymal/40 vascular deposits from three fAD [Aβ(E22G)] cases; 210 vascular deposits from four fCAA [Aβ(E22Q)] cases. (D) Confocal images showing curcumin labeling of parenchymal deposits in sAD and fAD cases also reveal the diffuse plaques that are not visible when using the probes in A and B. (E) Quantitation of curcumin fluorescence intensity in parenchymal and vascular Aβ deposits. Sampling for curcumin: n = 520 parenchymal/96 vascular deposits from five sAD cases; 598 parenchymal/112 vascular deposits from three fAD [Aβ(E22G)] cases; 206 vascular deposits from four fCAA [Aβ(E22Q)] cases. Data shown as mean ± SD. A one-way ANOVA with Tukey’s post hoc statistical test was used for multiple comparisons. (Scale bars, 15 μm.)
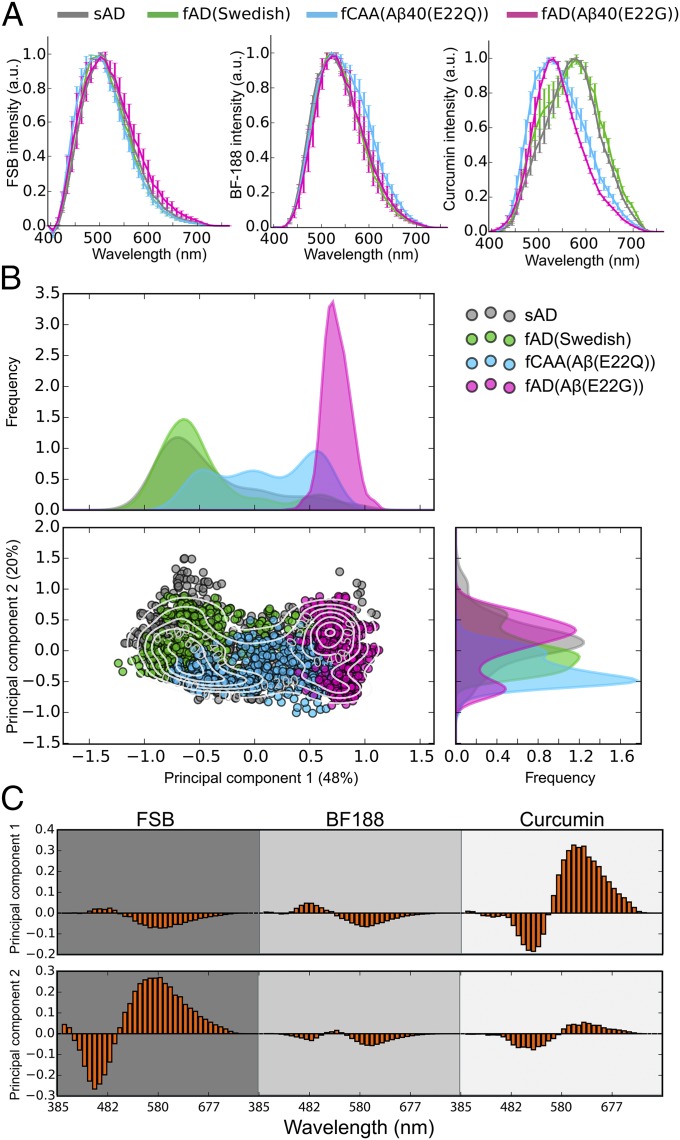
Fig. 2.
Spectral discrimination of distinct Aβ conformations in sAD and fAD patients. (A) Confocal microscopy-based emission spectra from curcumin-, FSB-, and BF-188–labeled Aβ parenchymal and vascular deposits for one representative patient of each disease type. Data shown as mean ± SD. (B) Projection of the combined spectra into the eigenspace defined by the first two principal components (see Methods). The sAD, fAD(Swedish), fCAA [Aβ(E22Q)], and fAD [Aβ(E22G)] are, respectively, shown in dark gray, green, cyan, and magenta. The contour levels indicate density of spectra, and the density distribution along each axis is reported on the top and to the right of the plot. (C) Multispectral vectors from representative patients are projected into the eigenspace defined in B, and their magnitudes reveal the wavelength-dependent contributions from the various dyes to the variation between the samples.
Previously, we found that three dyes were necessary to fully discriminate the conformations of closely related tau amyloids (36), so we similarly examined a series of dyes for the detection of Aβ deposits. Curcumin is an environment-sensitive dye (34), which we found to be particularly useful because of its higher relative fluorescence intensity for fAD [Aβ(E22G)] and fCAA [Aβ(E22Q)] deposits compared with traditional histochemical dyes (Fig. 1 C and E). Indeed, the diffuse parenchymal plaques in the E22G and E22Q brain samples can be discriminated from WT based on both the intensity and the shape of the emission spectrum (Figs. 1 C and E and 2A and Fig. S1). We also examined the benzimidazole BF-188 as a third probe, which has been reported to be a conformation-sensitive amyloid dye (37).
Fig. 2 shows significant differences in the intensity-normalized spectra averaged over each patient group. While the averages are informative, the variations between individual spectra provide even more valuable information about the homogeneity and conformational differences between individuals and regions in the brain. To facilitate this comparison, we used PCA (Methods), which allows one to project the variations in the thousands of spectra into a few dimensions. Indeed, 68% of the spectral variation can be captured using only two components (Fig. 2B). The first component is dominated by contributions from curcumin, while the second component primarily reflects contributions from FSB. The resolving power realized by PCA of the three fluorophores can be appreciated by comparing the simple averaged spectra (Fig. 2A) to the plot of the first two principal components (Fig. 2B). The distributions for WT and Swedish variants are indistinguishable, as would be expected from their identical sequence in the Aβ region. This finding confirms our earlier qualitative conclusions based on FSB intensity and the shape of the curcumin spectra. Furthermore, the E22G mutant distribution is now very well separated from that of WT. Finally, the E22Q mutant shows some overlap but is clearly differentiable by this metric.
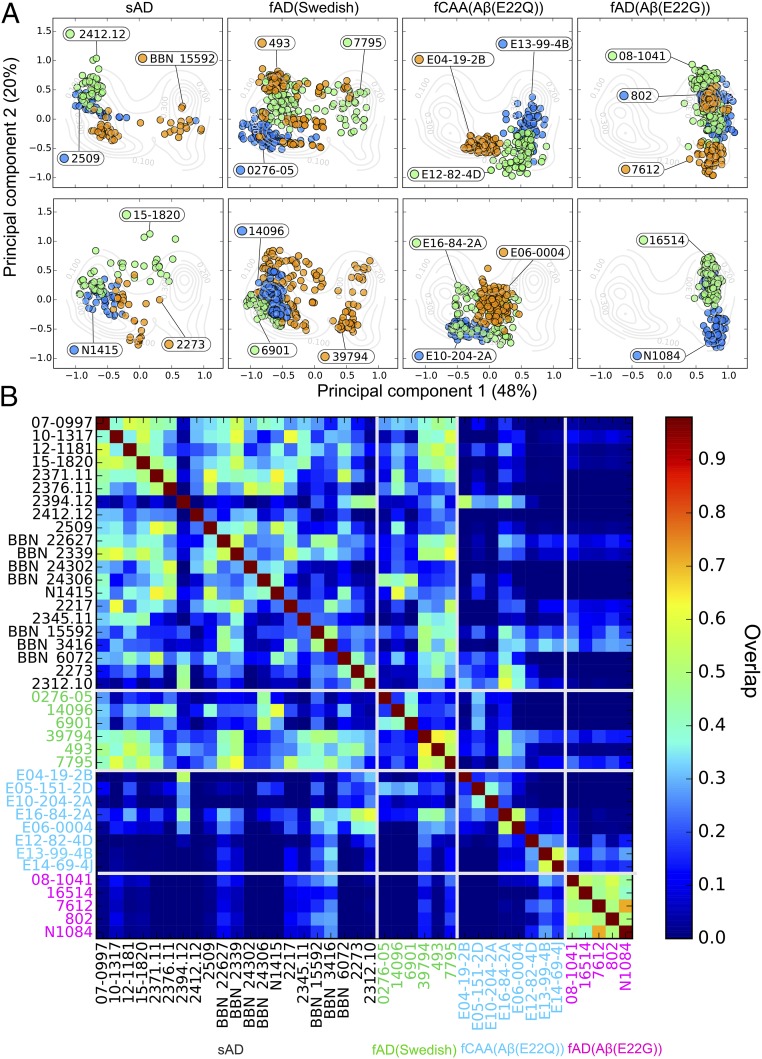
Recent solid-state NMR data suggest that there are a large number of biologically relevant conformers of Aβ amyloids in different individuals and disease states, but that only one or two conformational forms are found within a single brain (38). We therefore asked whether our PCA might identify intersubject variability within sAD and fAD subjects. Thus, we examined the distribution of points for the 40–70 amyloid plaques observed for each patient relative to the distribution for plaques from all patients. If the amyloid deposits in a given brain are identical with respect to conformation and isoform composition, then the plots for a given patient would match the overall distribution seen for the corresponding set of sAD or fAD brain samples. In contrast, if the overall distribution reflects a set of conformational types that vary between individuals, the points for each individual would be more tightly clustered than that of the overall population average.
In each case, the distributions observed for a given patient were more tightly clustered than the corresponding distributions for the pooled samples of a given genotype (WT, Swedish, Dutch, or Arctic). These data support the hypothesis that the conformational variability within a patient is much smaller than in the overall population. However, as shown for representative samples in Fig. 2C, the degree of homogeneity varied from one patient to the next (Fig. 3A). To facilitate a pairwise comparison of each sample, we constructed a 2D heat map that quantifies the similarity between plaques from each patient (Fig. 3B). This analysis clearly shows that, although there is a spread in the degree of similarity between patients of a given type, the Swedish and sporadic populations are highly similar, while the E22Q and E22G patients are clearly differentiated from each of the other samples. Interestingly, fAD [Aβ(E22G)] patients showed the lowest intersubject variability and were the most well separated from the other patient populations, possibly because the Glu22 to Gly replacement in E22G represents a greater perturbation than the more conservative Glu22 to Gln mutation in fCAA [Aβ(E22Q)] patients.
Fig. 3.
Comparison of interpatient variability between sAD and different fAD samples. (A) Two-dimensional PCA plots for selected patients. (B) The heat map shows the spectral similarity between the patients, computed as the overlap between patient spectral distribution (see Methods). The labels of sAD, fAD (Swedish) (Swedish AD), fCAA [Aβ(E22Q)], and fAD [Aβ(E22G)] patients are, respectively, shown in black, green, cyan, and magenta.
Finally, we asked whether the intersubject variability might be a result of a number of variables including brain region, age at time of death, sex, identity of the brain repository, or the location of the plaques within the gray matter (parenchymal) versus areas surrounding the vasculature (Fig. 4 and Fig. S2B). Of these variables, there was little correlation with sex or the brain repository. There did appear to be a potentially interesting clustering of patients according to age and brain region, although the number of patients in each group precluded an in-depth analysis. Therefore, we focused on the location of the plaques within the parenchymal versus the vasculature, as most brain slices had large numbers of each type of plaque. Thus, the staining properties of the parenchymal versus vascular plaques could be directly compared within each individual, without confounding influences associated with the other variables.
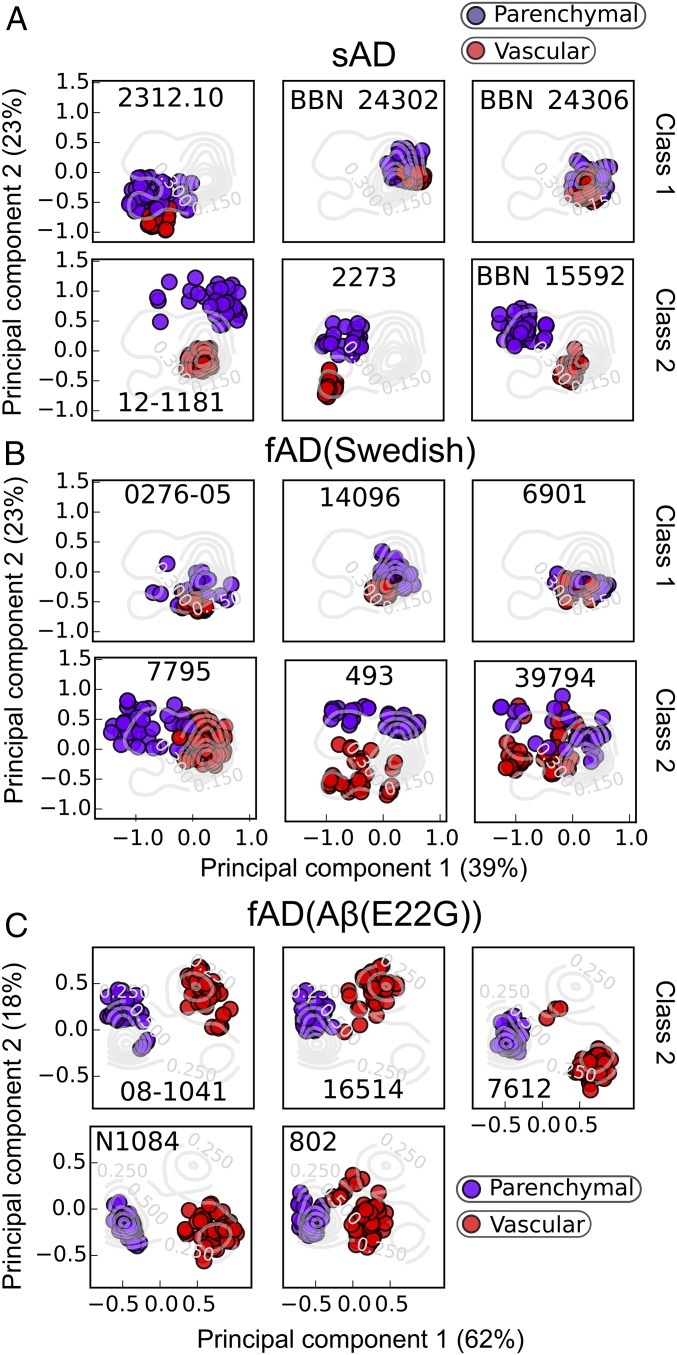
Fig. 4.
Effect of the localization of the deposits on the Aβ conformations in sAD and fAD patients. The multispectral vectors from representative sAD (A), fAD (Swedish) (B), and fAD [Aβ(E22G)] (C) patients are projected into the eigenspace defined by the first two components of the PCA. Data are colored according to the localization of the aggregates in the brain tissue. Parenchymal deposits are shown in purple and vascular deposits in red.
Fig. 4 illustrates data exploring the variation of parenchymal and vascular localization of deposits in sAD, fAD Swedish, and fAD [Aβ(E22G)] patients. In one class representing about a third of sAD patients and half of fAD Swedish patients, the points are relatively tightly clustered in the same position within the plot for both parenchymal and vascular plaques (Fig. 4 A and B, Upper). Thus, these “class 1” amyloids have a single conformation (within the resolution of this method) that can deposit indiscriminately between parenchymal gray matter and the vasculature. Furthermore, although the class 1 patients have highly homogeneous amyloid staining within an individual, significant variation is seen between individuals. Quite distinct “class 2” behavior can be seen for another third of the sAD and half of the fAD Swedish patients, which show two discrete clusters within a given patient: one located in the parenchymal area and the other surrounding the vasculature (Fig. 4 A and B, Lower). Thus, these patients appear to have a different Aβ conformational form surrounding the vasculature versus in the gray matter. The remaining sAD samples either had too few vascular plaques to allow a significant analysis, or had multiple conformations within both their parenchymal and vascular deposits (Fig. S3).
In contrast to the differing behavior seen between patients with sAD and fAD (Swedish), all of the fAD [Aβ(E22G)] mutants showed class 2 behavior (Fig. 4C). Two clusters were seen within a given patient, one in the parenchyma and one surrounding the vasculature. Furthermore, for each patient sample, the parenchymal plaques populated an elongated region of the plot, the first component of which reported on the location of the plaques, while the second component reported on the variation between individuals. There were too few fCAA [Aβ(E22Q)] parenchymal plaques present in the brains to permit a similar analysis.
Synthetic WT and Mutant Aβ Amyloid Propagates in Vivo.
Previous studies with synthetic peptides have indicated that the position 22 mutants aggregate more rapidly than WT (11, 13, 35) and serve as a seed to induce rapid aggregation of WT (11). We therefore asked whether synthetic mutants could act as prions to template fAD [Aβ(E22G)]- and fCAA [Aβ(E22Q)]-like conformations of WT protein in vitro and in vivo. For these studies, we used synthetic Aβ40 (the most abundant isoform produced in humans and in Tg mice) and methods of in vitro fibril formation and characterization similar to earlier studies (Fig. S4). Briefly, kinetic analysis of amyloid formation using the ThT fibrillization assay (Fig. S4C) or under label-free conditions using solution 1D-1H NMR (Fig. S5) confirmed that mutant Aβ40 peptides aggregate more rapidly than WT Aβ40. Moreover, the mutant fibrils reached a lower absolute ThT fluorescence intensity at the end of the amyloid formation process (Fig. S4D), indicating a different conformation, which is consistent with earlier studies (11, 35), and matching our above observations using clinically derived samples. All samples contained fully formed fibrils, so the low observed intensity was not a result of less complete fibril formation (Fig. S4E).
To investigate conformational differences, we adapted the confocal spectral imaging approach to discriminate WT and mutant synthetic fibrils (Fig. S6). We immobilized WT or mutant fibrils in agarose gel and stained them with the panel of fluorescent amyloid dyes used on human brain slices (Fig. S6 A–C). Performing PCA of curcumin, FSB, and BF-188 spectra, we identified distinct conformations of synthetic WT and mutant fibrils (Fig. S6 D and E).
To determine whether mutant Aβ fibrils could template WT Aβ to fold into a mutant-like conformation, we performed a series of in vitro seeding experiments that combined mutant Aβ40 and WT Aβ40 as the substrate, followed by fluorescence intensity measurements using ThT. We exposed monomeric WT Aβ40 to preformed mutant Aβ40(E22G) and Aβ40(E22Q) fibrils in a ratio of either 90:10 (10%) or 99:1 (1%) (Fig. S4 E and F). Mutant Aβ led to templated amyloid formation of WT Aβ40 in a low ThT-binding conformation, consistent with the reduced fluorescence intensity seen in fAD brain samples (Fig. 1) and spontaneously formed synthetic fibrils (Fig. S4C). Thus, the sequence changes in these mutants encipher altered conformations that resemble the pathogenic plaques found in fAD.
Encouraged by these in vitro templating results, we modeled the interaction between mutant Aβ40 fibrils and WT Aβ in vivo. We inoculated Tg(APP23) mice expressing WT Aβ with synthetic fibrils composed of either WT Aβ40, Aβ40(E22G), or Aβ40(E22Q) (Fig. 5 and Fig. S7). All three fibril preparations induced widespread Aβ deposition throughout the corpus callosum region bordering the CA1 area of the hippocampus, a signature pattern following intrathalamic inoculation in our experimental paradigm (Fig. 5 A–C and Fig. S7 A and B). We observed different fluorescence intensities for amyloid plaques induced by mutant and WT synthetic Aβ40 prions (Fig. 5 D–F). Tg(APP23) mice inoculated with WT Aβ fibrils displayed relatively bright fluorescence intensity of FSB-, BF-188–, or curcumin-labeled plaques, while inoculation with mutant Aβ prions showed a marked reduction in signal (Fig. 5 D–F), similar to our findings in vitro and in patient tissue. It is important to note that these mice express only WT Aβ and that these changes in deposit phenotype are solely based on a distinct amyloid conformation induced by structurally distinct synthetic Aβ40 prions used for propagation in vivo.
Fig. 5.
Synthetic mutant Aβ40 prions transmitted to mice expressing WT Aβ induce a distinct conformation. (A) Brain-slice staining for Aβ (Aβ1–16 epitope) and GFAP-immunolabeled astrocytes reveal the stereotypical deposition pattern induced in the corpus callosum above the CA1 region of the hippocampus following thalamic injection, but with strikingly different FSB fluorescence signal (B). (Scale bars, 50 μm in A; 25 μm in B.) (C–E) Tg mice inoculated with synthetic mutant Aβ40 prions show reduced fluorescence intensity of (C) FSB, (D) BF-188, and (E) curcumin signals in induced plaque deposits compared with control mice inoculated with synthetic WT Aβ40 prions. Quantitation of (C) FSB plaque intensity (n = 4 animals per group/292 plaques), (D) BF-188 plaque intensity (n = 4–5 animals per group/1,336 plaques), or (E) curcumin plaque intensity (n = 4 animals per group/195 plaques) is shown. Data shown as mean ± SE of mean. A one-way ANOVA with Tukey’s post hoc statistical test was used for multiple comparisons. (F) Spectral analysis using a combination (see Methods) of curcumin, FSB, and BF-188 spectra datasets. The density distribution along each axis is reported on the top and to the right of the plot. (G) Projections of the spectra from individual mice into the eigenspace defined by human patients.
To test the molecular conformation of the induced Aβ deposits, we analyzed the fluorescence spectral profiles of bound amyloid dyes and found a similar trend in curcumin spectral shifts (Fig. S7C) compared with Aβ deposits found in the patient samples (Fig. 2A and Fig. S1B). Using a combination of curcumin, FSB, and BF-188 spectra to perform PCA further differentiated the mutant and WT Aβ40 prion-induced plaques (Fig. 5G). Therefore, inoculation of the 40-residue isoform of synthetic mutant Aβ was sufficient to model key aspects of fAD and fCAA strains in mice expressing WT Aβ. PCA of FSB-, BF-188–, or curcumin-labeled plaques provided additional information about the conformations induced by synthetic fibrils in mouse brains and how these compared with the conformations seen in human brains (Fig. 2B). Comparing the distributions seen for human brains to mouse brains inoculated with a synthetic sample shows several interesting features (Fig. 5G). First, relatively sharp distributions are apparent for a given synthetic peptide with very little intermouse variability, indicating that a given synthetic peptide induces a relatively homogeneous conformational form in the WT Aβ expressed in a given mouse. Second, there is a significant separation between the distributions induced by WT, E22Q, and E22G mutants, similar to the separation seen when comparing sAD with fAD mutants. Finally, the WT Aβ40-seeded mouse has a distribution located near the central region observed for sAD patients, while the E22G-seeded materials were located near the corresponding region seen in patients bearing the Arctic mutation. The distributions for the E22Q-inoculated brains were also close to the most populated region in patients bearing the E22Q mutation, although the overlap was not as good as for WT and E22G. Taken together, these data clearly show that the synthetic Aβ40 mutant peptides act as prions to induce distinct conformational strains of amyloid in mice expressing WT Aβ40, and these prion-templated conformational strains show many similarities to those seen in the corresponding fAD and fCAA patients.
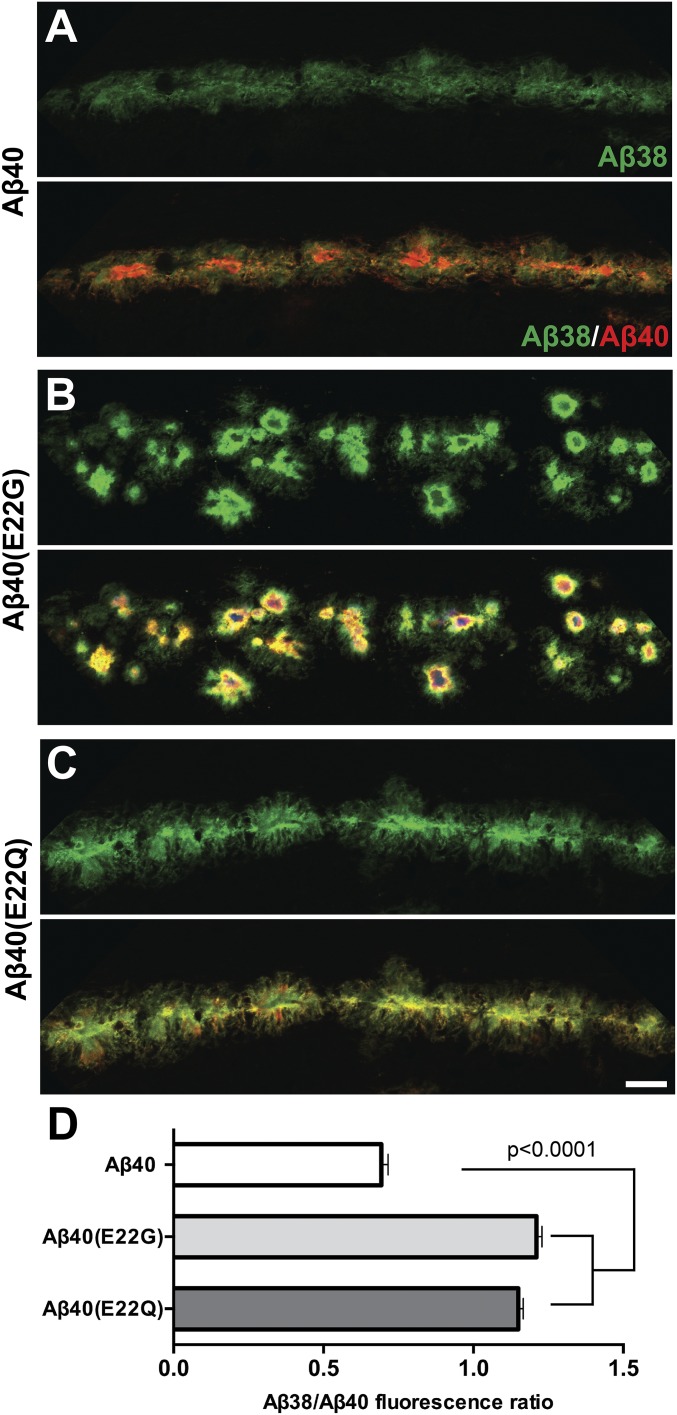
Inoculation of Mutant Aβ40 Fibrils Leads to Enhanced Deposition of WT Aβ38 in Mouse Brains, Reproducing a Key Feature of fAD.
We investigated whether inoculation of Tg mice with mutant Aβ40 prions also affected the amount of Aβ38 deposition. Brain slices of the inoculated mice were immunolabeled with isoform-specific antibodies. Quantitative confocal imaging revealed that both Aβ40(E22G) and Aβ40(E22Q) synthetic prions induced a significant increase in the levels of Aβ38 isoform recruited to induced plaques (Fig. 6). This finding is entirely consistent with both neuropathological observations in fAD brain samples (Fig. S8A) as well as histochemical observations following transmission of fAD and fCAA brain extracts to Tg(APP23) mice (Fig. S8 B, D, and E). Together, these data demonstrate that a mutant conformation of a single Aβ isoform is sufficient to induce the unique deposit characteristics in Tg mice expressing WT Aβ.
Fig. 6.
Synthetic mutant Aβ40 fibrils induce recruitment of the Aβ38 isoform in vivo. Tg (APP23) mice inoculated with synthetic mutant Aβ40 prions have differential levels of accumulated Aβ isoforms within the induced plaque deposits found in the corpus callosum/CA1 hippocampal region compared with control mice inoculated with WT Aβ40. (A–C) Confocal images show Aβ38 (green) and Aβ40 (red) immunopositive plaques in mice inoculated with WT Aβ40, Aβ40 (E22G) (Arctic), or Aβ40 (E22Q) (Dutch) fibrils. (D) Quantification of Aβ38 and Aβ40 levels is shown (n = 4 animals per group/307 plaques, data shown as mean ± SE or mean). A one-way ANOVA with Tukey’s post hoc statistical test was used. (Scale bar, 25 μm.)
Characterization of Mutant Aβ38 Fibrils in Vitro Provides Insight into the Robust Aβ38 Deposition in fAD.
To gain further insight into the enriched deposition of Aβ38 in fAD and fCAA patients (2) and in patient brain-inoculated mice (19), we characterized the intrinsic properties of synthetic Aβ38 and Aβ38(E22G) in vitro (Fig. S9). WT Aβ38 displayed a very low aggregation propensity, reaching the half-maximum of ThT fluorescence [T(ThTmax-1/2)] in ∼20 h compared with ∼3 h for WT Aβ40 (Fig. S9 A and B). In contrast, mutant Aβ38(E22G) aggregated almost immediately, albeit with low ThT fluorescence (Fig. S9A), as observed with all mutant Aβ peptides in this study (Fig. S3). Synthetic Aβ38 and Aβ38(E22G) fibrils were probed with a panel of conformation-sensitive fluorescent dyes (Fig. S10A). Conformational probing by PCA revealed that WT Aβ38 and Aβ40 fibrils occupy a distinct conformational space that is in good agreement with their different aggregation propensities (Fig. S10B). In contrast, when Aβ38 and Aβ40 shared the E22G mutation, fibrils produced overlapping clusters, which suggests a very similar conformation.
The significant difference in aggregation kinetics between mutant and WT Aβ38 explains the accumulation of mutant Aβ38 in fAD cases and its absence in sAD samples. Moreover, elevated levels of plaque-associated WT Aβ38 in our in vivo experiments suggest a preferred recruitment of Aβ38 by mutant Aβ(E22G/Q) prions. Taken together, the data show that the high aggregation propensity of mutant Aβ38 in combination with preferred templating of WT Aβ38 by mutant Aβ prions (Fig. 6) are likely the main causes for the robust deposition pattern of Aβ38 in fAD and fCAA.
Discussion
Here, we develop a confocal fluorescence microscopy technique with multiple fluorescent dyes to assess the degree of homogeneity and similarity in amyloid plaques of diverse origins, spanning from in vivo- to in vitro-derived materials. Deposits from fAD [Aβ(E22G)] and fCAA [Aβ(E22Q)] patients can be distinguished from sAD patients based on the fluorescence intensity of several amyloid-staining dyes. Furthermore, additional fine-tuned structural differences in amyloid plaques can be distinguished from the shape of the emission spectra of multiple dyes; PCA of these data provides 2D fingerprints with considerable resolving power. These fingerprints are influenced by the conformational strain of the amyloids, and possibly also the relative ratios of the different Aβ peptide isoforms forming the plaques. At the grossest level, they provide a second metric that readily distinguishes amyloid plaques from sAD versus fAD [Aβ(E22G)] and fCAA [Aβ(E22Q)] patients.
The 2D fingerprints generated in this work can be used to probe conformational homogeneity and similarity in much the same way as chemical shifts are used to provide a conformational fingerprint from solid-state NMR (35, 38). The major advantage, however, is that this method can be used directly to interrogate biological samples, and it does not require purification of the amyloids or seeding of large amounts of isotopically labeled peptides in vitro. We first use the method to examine inter- and intrasubject heterogeneity in sAD brains. In each case, intrasubject variability is less than intersubject variability, but the degree of intrasubject homogeneity is not uniform among all patients. In class 1 patients, it appears that one conformational form arises and then spreads as a prion throughout both the gray matter and the region surrounding the vasculature. In contrast, in class 2 patients, structurally different amyloids appear to deposit in these two anatomically distinct regions.
This method might also prove to be a useful strategy to differentiate amyloid conformations that predispose the brain toward disease. There is a clear separation between rapidly progressing E22Q and E22G mutants versus the sAD cases. We also see intriguing separations based on patient age at time of death, which suggests it might be possible to differentiate patients for whom disease progresses most rapidly. However, it will be necessary to conduct additional experiments in which other variables, such as sex and brain region, are more carefully controlled before making firm conclusions along these lines.
Although the neurovascular toxicity of mutant Aβ (39–41) and the efficiency of its clearance from the brain (42) have been extensively studied in vivo, the role of mutations in propagating unique Aβ amyloid-deposition phenotypes in the AD brain remains poorly understood. Particularly, the precise mechanisms underlying the interaction between different Aβ isoforms and the distinct structural characteristics of Aβ deposits in fAD are unknown. Molecular characterization of AD has been traditionally performed in human postmortem samples or in Tg mouse models with a defined pathological phenotype. However, this changed when investigators exploited the prion mechanism by exogenously administering different self-propagating Aβ conformations into Tg mice (17–19, 43–45).
Importantly, we previously demonstrated that transmitting a single, synthetic Aβ isoform, artificially formed into a different conformation, is sufficient to catalyze distinct patterns of cerebral amyloidosis in recipient mice (17). Alternatively, distinct Aβ fibril conformations can be induced by familial mutations at residue 22, which represents a structural switch for distinct amyloid conformations (35, 46). In the present study, we inoculated mice with synthetic Aβ40 fibrils containing fAD-associated substitutions at this position to test our hypothesis that mutant Aβ directs WT Aβ to misfold into a prion. Remarkably, mutant Aβ induced plaques in these mice that showed reduced intensity for amyloid dyes and a distinct 2D spectral fingerprint (Fig. 4). These characteristics are consistent with the strain features that we observed in the human fAD brain and following transmission of AD patient samples to Tg mice (Figs. 1 and 2 and Fig. S7A).
Experimental propagation of mutant Aβ prions in vitro and in vivo has demonstrated that the mutants have a dominant-templating effect on WT Aβ, which adopts a mutant prion structure. Bigenic mice expressing human WT and mutant Aβ exhibit brain deposits that shift to a morphologically distinct cerebrovascular phenotype (47, 48). Furthermore, it has been postulated that the Δ22E (Osaka) mutant of Aβ acts in a dominant-negative manner; the high propensity of this mutant to aggregate leads to the formation of seeds that can induce rapid fibrillization of WT Aβ (11). Similarly, we find that the mutants studied here aggregate very rapidly and seed WT Aβ in vitro. In animal models, these variants (35) induce novel phenotypes, which model key aspects of fAD [Aβ(E22G)] and fCAA [Aβ(E22Q)], including the ability to induce the incorporation of WT Aβ38 within the plaques (2). In contrast to fAD, there is typically little to no Aβ38 deposition in most sAD cases, with the exception of a report describing a low to moderate level of Aβ38 deposition in the cerebrovasculature (3). It is known, however, that increased Aβ38 deposition occurs in other forms of early-onset fAD that are caused by point mutations in APP near the γ-secretase cleavage site (49, 50) or in the presenilin genes (PSEN1 and PSEN2), which encode the catalytic subunit of γ-secretase (51–53). Altered trimming of APP can produce a higher proportion of Aβ42 and Aβ38 in the total Aβ pool (54–56). Thus, it has been unclear how fAD cases caused by a point mutation within the Aβ coding region (which does not directly alter Aβ production) could increase the proportion of Aβ38. Our finding that synthetic mutant Aβ40 prions are sufficient to induce the preferential incorporation of WT Aβ38 in Tg mice expressing only WT Aβ isoforms (Fig. 5D) strongly suggests that preferred templating of Aβ38 is an important mechanism for its elevated deposition in the fAD patient brain. Together, our in vitro and in vivo experiments show that WT Aβ38 is preferentially templated by mutant Aβ prions and that mutated Aβ38 has an increased propensity to aggregate. These two independent mechanisms act synergistically to cause Aβ38 deposition in fAD cases.
In summary, position 22 mutants of Aβ adopt a dominant prion conformation defining the fAD phenotype. By studying Aβ prion self-propagation in vivo using novel conformation-specific tools, new strains of WT Aβ prions may be identified that explain rapidly progressive sAD (57, 58) or the pathologic heterogeneity among patients with similar cognitive function (59–62). Indeed, WT Aβ misfolds into unique prion strains in different environmental conditions, and these strains cause distinct pathologies when transmitted to mice (17), indicating that structural variation of the Aβ prion conformation may similarly occur in different AD patients (23, 38). Taken together, our findings argue that the deposition pattern within specific regions of AD patients is likely defined by the first stable Aβ prion formed through a kinetically dominant self-propagation process. Indeed, in some cases only a single conformational form within an individual is detected using the methods developed in this work. Using the prion paradigm to characterize the many sets of patient-derived Aβ prion strains will further elucidate the molecular spectrum of AD diversity. Indeed, insights from our earlier studies (17) suggest that even WT Aβ40 and Aβ42 possess a structural plasticity to form distinct prion strains. This characteristic may shed light on the more common forms of sAD where a subset of pathologically confirmed cases do not present with a positive PiB-PET scan antemortem (63). Importantly, our studies show the immediate need for clinical PET probes that can detect a broader spectrum of pathological Aβ conformations, as shown for newly developed amyloid tracers (64, 65). Probes that are more sensitive will result in earlier AD detection, providing a longer time frame for therapeutic interventions in mild, or even subclinical, AD patients.
Methods
Neuropathology.
All comparisons were conducted on brains from age- and sex-matched animals. Animals were maintained in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International in accordance with the Guide for the Care and Use of Laboratory Animals (66). All procedures for animal use were approved by the University of California, San Francisco’s Institutional Animal Care and Use Committee. Samples were immersion-fixed in 10% buffered formalin and then embedded in paraffin using standard procedures. Sections (8 µm) were cut, deparaffinized, and then processed for immunohistochemistry. Sections to be stained with anti-Aβ antibodies were pretreated by incubation in formic acid followed by heated sodium citrate buffer as an epitope retrieval step. Following blocking with 10% normal goat serum, sections were incubated with primary antibody overnight at room temperature. The following antibodies were used: anti-Aβ (Aβ1–16 epitope; BioLegend) and anti-GFAP (Dako) to detect astrocytic gliosis. Antibody binding was performed using goat secondary antibodies with conjugated AlexaFluor 488 and AlexaFluor 647 (A-21245; Life Technologies).
For immunofluorescence, double-staining with specific antibodies targeting Aβ38 (clone 7-14-4; BioLegend 808601), Aβ40 (Millipore AB5074), and Aβ1–16 (BioLegend 802901), brain sections were pretreated by incubation in formic acid followed by heated sodium citrate buffer as an epitope retrieval step. Sections were blocked with 10% normal goat serum and then incubated with primary antibodies at a 1:250 dilution overnight at room temperature. The following day, sections were thoroughly washed and incubated with the fluorescent secondary antibodies, goat anti-mouse AlexaFluor 488 (A-11029; Life Technologies), and goat anti-rabbit AlexaFluor 647 (A-21245; Life Technologies). All immunohistochemistry stained sections were subsequently stained with 2.5 µM FSB (Congo red derivative) (67) (Santa Cruz) in 1× PBS and washed before mounting. Fluorescence intensity and spectral measurements also used BTA-1 (“PiB” precursor; ThT derivative) (68) (Sigma), BF-188 (37) (custom synthesis), and curcumin (Sigma #08511).
Samples were visualized using a 40× water-immersion lens (1.1 NA) in sequential scan mode on a Leica SP8 confocal microscope equipped with three HyD detectors and an acousto-optical beam splitter. Using control (spontaneous disease) brains to establish standardized acquisition settings for all experimental groups, 8-bit image z-stacks (1.5-µm steps) were collected at 1,024 × 1,024 pixel resolution with the full dynamic range of fluorescence intensity.
Confocal-Based Spectral Profiling of Fluorescent Amyloid-Binding Dyes in Brain Slices and Gel-Embedded Synthetic Fibrils.
Curcumin-, BF-188–, and FSB-labeled Aβ deposits were imaged in the spectral (Lambda) scan mode of a Leica SP8 confocal microscope using a 40× water immersion lens (1.1 NA), a 405-nm laser for fluorescent amyloid-dye excitation, and a HyD detector at 512- × 512-pixel resolution. For each field-of-view in the brain slice experiments, the optical plane was moved to the center of the z-stack volume for a given Aβ deposit, and fluorescence emission was acquired from a series of 40-image steps spanning 385- to 765-nm wavelengths using a 15-nm-wide detection window at each interval. For each field-of-view in the gel-embedded synthetic fibril experiments, confocal z-stack at multiple random positions in a well was acquired with identical spectral parameters as described above (XYλZ mode) using a 63× water immersion lens (1.2 NA). The size of the z-stack ranged from 100- to 200-µm thick depending on the density of aggregates in a given well. Raw spectral data were analyzed with Leica LASAF and NIH ImageJ software.
Principal Component Analysis.
The signal-processing algorithm for the analysis of the spectra was implemented in Python using NumPy and sklearn packages. Sudden spikes in each spectrum (a result of occasional noise in the photodetector) were first removed using a five-element sliding window spline function, and all spectra were rescaled to a [0, 1] interval. To combine the spectral information from the different dyes, a new dataset of multispectral vectors was built. For a given patient and localization, a multispectral vector was composed by randomly concatenating an emission spectrum from each dye. Forty to 70 vectors were assembled for each patient. A PCA was then performed on the obtained dataset, and each vector was projected into the eigenspace defined by the two first components (covering about 60–75% of the variance).
Spectral Overlap Analysis.
A kernel density estimate was used to estimate the 2D spectral distribution for each patient after projection into the eigenspace defined by the two first principal components. The similarity between two patients was computed as the integral of the union of their respective distributions.
Supplementary Material
Acknowledgments
We thank the staff at the Hunters Point animal facility for their assistance with the maintenance of our mouse colony; visiting Prof. George A. Carlson [University of California, San Francisco (UCSF)] for editing the manuscript; and the UCSF Neurodegenerative Disease Brain Bank and Prof. William W. Seeley (UCSF Memory and Aging Center) for providing brain tissue for this study. Autopsy tissue obtained from the University of Washington Neuropathology Core is supported by the Alzheimer’s Disease Research Center (AG005136), the Adult Changes in Thought Study (AG006781), and the Morris K. Udall Center of Excellence for Parkinson’s Disease Research (NS062684). Autopsy tissue was also supplied by the University of Edinburgh (Department of Neuropathology) and the Manchester Brain Bank (University of Manchester), which is part of the Brains for Dementia Research Initiative, jointly funded by Alzheimer’s Society and Alzheimer’s Research UK. Human brain tissue was provided by the Brain Bank at Karolinska Institutet, Stockholm, Sweden, which has received financial support from StratNeuro at the Karolinska Institutet, Swedish Brain Power and Stockholm County Council. This work was supported by the National Institutes of Health Grants AG002132 and AG031220, the UCSF Program for Breakthrough Biomedical Research, as well as by support from Daiichi Sankyo Co., Ltd., the Sherman Fairchild Foundation, the Schott Foundation for Public Education, and the Rainwater Charitable Foundation. C.C. was supported by a research supplement to a National Institute on Aging Grant AG010770-19S1. T.L. was supported by the Swiss National Science Foundation Grant PA164691. J.S. was supported by a fellowship from the Glenn Foundation and the New Investigator Research Grant of the Alzheimer’s Association.
Footnotes
Conflict of interest statement: The Institute for Neurodegenerative Diseases has a research collaboration with Daiichi Sankyo (Tokyo, Japan). S.B.P. is the chair of the Scientific Advisory Board of Alzheon, Inc., which has not contributed financial or any other support to these studies.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714966115/-/DCSupplemental.
References
- 1.Bertram L, Lill CM, Tanzi RE. The genetics of Alzheimer disease: Back to the future. Neuron. 2010;68:270–281. doi: 10.1016/j.neuron.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 2.Moro ML, et al. APP mutations in the Aβ coding region are associated with abundant cerebral deposition of Aβ38. Acta Neuropathol. 2012;124:809–821. doi: 10.1007/s00401-012-1061-x. [DOI] [PubMed] [Google Scholar]
- 3.Reinert J, et al. Aβ38 in the brains of patients with sporadic and familial Alzheimer’s disease and transgenic mouse models. J Alzheimers Dis. 2014;39:871–881. doi: 10.3233/JAD-131373. [DOI] [PubMed] [Google Scholar]
- 4.Kalimo H, et al. The Arctic AβPP mutation leads to Alzheimer’s disease pathology with highly variable topographic deposition of differentially truncated Aβ. Acta Neuropathol Commun. 2013;1:60. doi: 10.1186/2051-5960-1-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Philipson O, et al. The Arctic amyloid-β precursor protein (AβPP) mutation results in distinct plaques and accumulation of N- and C-truncated Aβ. Neurobiol Aging. 2012;33:1010.e1–1010.e13. doi: 10.1016/j.neurobiolaging.2011.10.022. [DOI] [PubMed] [Google Scholar]
- 6.Farrer LA, et al. Transmission and age-at-onset patterns in familial Alzheimer’s disease: Evidence for heterogeneity. Neurology. 1990;40:395–403. doi: 10.1212/wnl.40.3_part_1.395. [DOI] [PubMed] [Google Scholar]
- 7.Wang F, et al. Dominantly Inherited Alzheimer Network Cerebral amyloidosis associated with cognitive decline in autosomal dominant Alzheimer disease. Neurology. 2015;85:790–798. doi: 10.1212/WNL.0000000000001903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Natté R, et al. Dementia in hereditary cerebral hemorrhage with amyloidosis-Dutch type is associated with cerebral amyloid angiopathy but is independent of plaques and neurofibrillary tangles. Ann Neurol. 2001;50:765–772. doi: 10.1002/ana.10040. [DOI] [PubMed] [Google Scholar]
- 9.Bugiani O, et al. Hereditary cerebral hemorrhage with amyloidosis associated with the E693K mutation of APP. Arch Neurol. 2010;67:987–995. doi: 10.1001/archneurol.2010.178. [DOI] [PubMed] [Google Scholar]
- 10.Vandersteen A, et al. A comparative analysis of the aggregation behavior of amyloid-β peptide variants. FEBS Lett. 2012;586:4088–4093. doi: 10.1016/j.febslet.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 11.Cloe AL, Orgel JPRO, Sachleben JR, Tycko R, Meredith SC. The Japanese mutant Aβ (ΔE22-Aβ(1-39)) forms fibrils instantaneously, with low-thioflavin T fluorescence: Seeding of wild-type Aβ(1-40) into atypical fibrils by ΔE22-Aβ(1-39) Biochemistry. 2011;50:2026–2039. doi: 10.1021/bi1016217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulic L, et al. Early accumulation of intracellular fibrillar oligomers and late congophilic amyloid angiopathy in mice expressing the Osaka intra-Aβ APP mutation. Transl Psychiatry. 2012;2:e183. doi: 10.1038/tp.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lashuel HA, et al. Mixtures of wild-type and a pathogenic (E22G) form of Aβ40 in vitro accumulate protofibrils, including amyloid pores. J Mol Biol. 2003;332:795–808. doi: 10.1016/s0022-2836(03)00927-6. [DOI] [PubMed] [Google Scholar]
- 14.Walker LC, Jucker M. Neurodegenerative diseases: Expanding the prion concept. Annu Rev Neurosci. 2015;38:87–103. doi: 10.1146/annurev-neuro-071714-033828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prusiner SB. Cell biology. A unifying role for prions in neurodegenerative diseases. Science. 2012;336:1511–1513. doi: 10.1126/science.1222951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Telling GC, et al. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science. 1996;274:2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 17.Stöhr J, et al. Distinct synthetic Aβ prion strains producing different amyloid deposits in bigenic mice. Proc Natl Acad Sci USA. 2014;111:10329–10334. doi: 10.1073/pnas.1408968111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heilbronner G, et al. Seeded strain-like transmission of β-amyloid morphotypes in APP transgenic mice. EMBO Rep. 2013;14:1017–1022. doi: 10.1038/embor.2013.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watts JC, et al. Serial propagation of distinct strains of Aβ prions from Alzheimer’s disease patients. Proc Natl Acad Sci USA. 2014;111:10323–10328. doi: 10.1073/pnas.1408900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colby DW, et al. Design and construction of diverse mammalian prion strains. Proc Natl Acad Sci USA. 2009;106:20417–20422. doi: 10.1073/pnas.0910350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanders DW, et al. Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron. 2014;82:1271–1288. doi: 10.1016/j.neuron.2014.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paravastu AK, Qahwash I, Leapman RD, Meredith SC, Tycko R. Seeded growth of beta-amyloid fibrils from Alzheimer’s brain-derived fibrils produces a distinct fibril structure. Proc Natl Acad Sci USA. 2009;106:7443–7448. doi: 10.1073/pnas.0812033106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen ML, et al. Rapidly progressive Alzheimer’s disease features distinct structures of amyloid-β. Brain. 2015;138:1009–1022. doi: 10.1093/brain/awv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 25.Lasmézas CI, et al. BSE transmission to macaques. Nature. 1996;381:743–744. doi: 10.1038/381743a0. [DOI] [PubMed] [Google Scholar]
- 26.Berry DB, et al. Drug resistance confounding prion therapeutics. Proc Natl Acad Sci USA. 2013;110:E4160–E4169. doi: 10.1073/pnas.1317164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giles K, et al. Different 2-aminothiazole therapeutics produce distinct patterns of scrapie prion neuropathology in mouse brains. J Pharmacol Exp Ther. 2015;355:2–12. doi: 10.1124/jpet.115.224659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts BE, et al. A synergistic small-molecule combination directly eradicates diverse prion strain structures. Nat Chem Biol. 2009;5:936–946. doi: 10.1038/nchembio.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Telling GC, et al. Interactions between wild-type and mutant prion proteins modulate neurodegeneration in transgenic mice. Genes Dev. 1996;10:1736–1750. doi: 10.1101/gad.10.14.1736. [DOI] [PubMed] [Google Scholar]
- 30.McLean CA, et al. The D178N (cis-129M) “fatal familial insomnia” mutation associated with diverse clinicopathologic phenotypes in an Australian kindred. Neurology. 1997;49:552–558. doi: 10.1212/wnl.49.2.552. [DOI] [PubMed] [Google Scholar]
- 31.Schöll M, et al. Low PiB PET retention in presence of pathologic CSF biomarkers in Arctic APP mutation carriers. Neurology. 2012;79:229–236. doi: 10.1212/WNL.0b013e31825fdf18. [DOI] [PubMed] [Google Scholar]
- 32.Tomiyama T, et al. A new amyloid β variant favoring oligomerization in Alzheimer’s-type dementia. Ann Neurol. 2008;63:377–387. doi: 10.1002/ana.21321. [DOI] [PubMed] [Google Scholar]
- 33.Jack CR, Jr, et al. 11C PiB and structural MRI provide complementary information in imaging of Alzheimer’s disease and amnestic mild cognitive impairment. Brain. 2008;131:665–680. doi: 10.1093/brain/awm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Condello C, Yuan P, Schain A, Grutzendler J. Microglia constitute a barrier that prevents neurotoxic protofibrillar Aβ42 hotspots around plaques. Nat Commun. 2015;6:6176. doi: 10.1038/ncomms7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elkins MR, et al. Structural polymorphism of Alzheimer’s β-amyloid fibrils as controlled by an E22 switch: A solid-state NMR study. J Am Chem Soc. 2016;138:9840–9852. doi: 10.1021/jacs.6b03715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stöhr J, et al. A 31-residue peptide induces aggregation of tau’s microtubule-binding region in cells. Nat Chem. 2017;9:874–881. doi: 10.1038/nchem.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harada R, et al. Use of a benzimidazole derivative BF-188 in fluorescence multispectral imaging for selective visualization of tau protein fibrils in the Alzheimer’s disease brain. Mol Imaging Biol. 2014;16:19–27. doi: 10.1007/s11307-013-0667-2. [DOI] [PubMed] [Google Scholar]
- 38.Qiang W, Yau WM, Lu JX, Collinge J, Tycko R. Structural variation in amyloid-β fibrils from Alzheimer’s disease clinical subtypes. Nature. 2017;541:217–221. doi: 10.1038/nature20814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park L, et al. Age-dependent neurovascular dysfunction and damage in a mouse model of cerebral amyloid angiopathy. Stroke. 2014;45:1815–1821. doi: 10.1161/STROKEAHA.114.005179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miao J, et al. Cerebral microvascular amyloid β protein deposition induces vascular degeneration and neuroinflammation in transgenic mice expressing human vasculotropic mutant amyloid β precursor protein. Am J Pathol. 2005;167:505–515. doi: 10.1016/s0002-9440(10)62993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu W, et al. Cerebral microvascular rather than parenchymal amyloid-β protein pathology promotes early cognitive impairment in transgenic mice. J Alzheimers Dis. 2014;38:621–632. doi: 10.3233/JAD-130758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davis J, et al. Deficient cerebral clearance of vasculotropic mutant Dutch/Iowa double Aβ in human A betaPP transgenic mice. Neurobiol Aging. 2006;27:946–954. doi: 10.1016/j.neurobiolaging.2005.05.031. [DOI] [PubMed] [Google Scholar]
- 43.Stöhr J, et al. Purified and synthetic Alzheimer’s amyloid beta (Aβ) prions. Proc Natl Acad Sci USA. 2012;109:11025–11030. doi: 10.1073/pnas.1206555109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyer-Luehmann M, et al. Exogenous induction of cerebral beta-amyloidogenesis is governed by agent and host. Science. 2006;313:1781–1784. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- 45.Morales R, Duran-Aniotz C, Castilla J, Estrada LD, Soto C. De novo induction of amyloid-β deposition in vivo. Mol Psychiatry. 2012;17:1347–1353. doi: 10.1038/mp.2011.120. [DOI] [PubMed] [Google Scholar]
- 46.Schütz AK, et al. Atomic-resolution three-dimensional structure of amyloid β fibrils bearing the Osaka mutation. Angew Chem Int Ed Engl. 2015;54:331–335. doi: 10.1002/anie.201408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herzig MC, Eisele YS, Staufenbiel M, Jucker M. E22Q-mutant Abeta peptide (AbetaDutch) increases vascular but reduces parenchymal Abeta deposition. Am J Pathol. 2009;174:722–726. doi: 10.2353/ajpath.2009.080790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu F, et al. Cerebral vascular amyloid seeds drive amyloid β-protein fibril assembly with a distinct anti-parallel structure. Nat Commun. 2016;7:13527. doi: 10.1038/ncomms13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muratore CR, et al. The familial Alzheimer’s disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons. Hum Mol Genet. 2014;23:3523–3536. doi: 10.1093/hmg/ddu064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suárez-Calvet M, et al. Autosomal-dominant Alzheimer’s disease mutations at the same codon of amyloid precursor protein differentially alter Aβ production. J Neurochem. 2014;128:330–339. doi: 10.1111/jnc.12466. [DOI] [PubMed] [Google Scholar]
- 51.Page RM, et al. Generation of Abeta38 and Abeta42 is independently and differentially affected by familial Alzheimer disease-associated presenilin mutations and gamma-secretase modulation. J Biol Chem. 2008;283:677–683. doi: 10.1074/jbc.M708754200. [DOI] [PubMed] [Google Scholar]
- 52.Czirr E, et al. Independent generation of Aβ42 and Aβ38 peptide species by gamma-secretase. J Biol Chem. 2008;283:17049–17054. doi: 10.1074/jbc.M802912200. [DOI] [PubMed] [Google Scholar]
- 53.Okochi M, et al. γ-Secretase modulators and presenilin 1 mutants act differently on presenilin/γ-secretase function to cleave Aβ42 and Aβ43. Cell Rep. 2013;3:42–51. doi: 10.1016/j.celrep.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 54.Chávez-Gutiérrez L, et al. The mechanism of γ-secretase dysfunction in familial Alzheimer disease. EMBO J. 2012;31:2261–2274. doi: 10.1038/emboj.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szaruga M, et al. Qualitative changes in human γ-secretase underlie familial Alzheimer’s disease. J Exp Med. 2015;212:2003–2013. doi: 10.1084/jem.20150892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Szaruga M, et al. Alzheimer’s-causing mutations shift Aβ length by destabilizing γ-secretase-Aβn interactions. Cell. 2017;170:443–456.e14. doi: 10.1016/j.cell.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 57.Schmidt C, et al. Rapidly progressive Alzheimer’s disease: A multicenter update. J Alzheimers Dis. 2012;30:751–756. doi: 10.3233/JAD-2012-120007. [DOI] [PubMed] [Google Scholar]
- 58.Chitravas N, et al. Treatable neurological disorders misdiagnosed as Creutzfeldt-Jakob disease. Ann Neurol. 2011;70:437–444. doi: 10.1002/ana.22454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cupidi C, et al. Neocortical variation of Abeta load in fully expressed, pure Alzheimer’s disease. J Alzheimers Dis. 2010;19:57–68. [Google Scholar]
- 60.Bird TD, et al. Phenotypic heterogeneity in familial Alzheimer’s disease: A study of 24 kindreds. Ann Neurol. 1989;25:12–25. doi: 10.1002/ana.410250104. [DOI] [PubMed] [Google Scholar]
- 61.Armstrong RA, Nochlin D, Bird TD. Neuropathological heterogeneity in Alzheimer’s disease: A study of 80 cases using principal components analysis. Neuropathology. 2000;20:31–37. doi: 10.1046/j.1440-1789.2000.00284.x. [DOI] [PubMed] [Google Scholar]
- 62.Lam B, Masellis M, Freedman M, Stuss DT, Black SE. Clinical, imaging, and pathological heterogeneity of the Alzheimer’s disease syndrome. Alzheimers Res Ther. 2013;5:1. doi: 10.1186/alzrt155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosen RF, et al. Deficient high-affinity binding of Pittsburgh compound B in a case of Alzheimer’s disease. Acta Neuropathol. 2010;119:221–233. doi: 10.1007/s00401-009-0583-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sundaram GSM, et al. Fluselenamyl: A novel benzoselenazole derivative for PET detection of amyloid plaques (Aβ) in Alzheimer’s disease. Sci Rep. 2016;6:35636. doi: 10.1038/srep35636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sehlin D, et al. Antibody-based PET imaging of amyloid beta in mouse models of Alzheimer’s disease. Nat Commun. 2016;7:10759. doi: 10.1038/ncomms10759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Committee for the Update of the Guide for the Care and Use of Laboratory Animals IfLAR; Division of Earth and Life Sciences; National Research Council of the National Academies . Guide for the Care and Use of Laboratory Animals. 8th Ed The National Academies Press; Washington, DC: 2011. [Google Scholar]
- 67.Velasco A, et al. Detection of filamentous tau inclusions by the fluorescent Congo red derivative FSB [(trans,trans)-1-fluoro-2,5-bis(3-hydroxycarbonyl-4-hydroxy)styrylbenzene] FEBS Lett. 2008;582:901–906. doi: 10.1016/j.febslet.2008.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bacskai BJ, et al. Four-dimensional multiphoton imaging of brain entry, amyloid binding, and clearance of an amyloid-β ligand in transgenic mice. Proc Natl Acad Sci USA. 2003;100:12462–12467. doi: 10.1073/pnas.2034101100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.