Significance
The genus of Aspergillus holds fungi relevant to plant and human pathology, food biotechnology, enzyme production, model organisms, and a selection of extremophiles. Here we present six whole-genome sequences that represent unexplored branches of the Aspergillus genus. The comparison of these genomes with previous genomes, coupled with extensive chemical analysis, has allowed us to identify genes for toxins, antibiotics, and anticancer compounds, as well as show that Aspergillus novofumigatus is potentially as pathogenic as Aspergillus fumigatus, and has an even more diverse set of secreted bioactive compounds. The findings are of interest to industrial biotechnology and basic research, as well as medical and clinical research.
Keywords: Aspergillus, fumigatus, comparative genomics, secondary metabolism
Abstract
The fungal genus of Aspergillus is highly interesting, containing everything from industrial cell factories, model organisms, and human pathogens. In particular, this group has a prolific production of bioactive secondary metabolites (SMs). In this work, four diverse Aspergillus species (A. campestris, A. novofumigatus, A. ochraceoroseus, and A. steynii) have been whole-genome PacBio sequenced to provide genetic references in three Aspergillus sections. A. taichungensis and A. candidus also were sequenced for SM elucidation. Thirteen Aspergillus genomes were analyzed with comparative genomics to determine phylogeny and genetic diversity, showing that each presented genome contains 15–27% genes not found in other sequenced Aspergilli. In particular, A. novofumigatus was compared with the pathogenic species A. fumigatus. This suggests that A. novofumigatus can produce most of the same allergens, virulence, and pathogenicity factors as A. fumigatus, suggesting that A. novofumigatus could be as pathogenic as A. fumigatus. Furthermore, SMs were linked to gene clusters based on biological and chemical knowledge and analysis, genome sequences, and predictive algorithms. We thus identify putative SM clusters for aflatoxin, chlorflavonin, and ochrindol in A. ochraceoroseus, A. campestris, and A. steynii, respectively, and novofumigatonin, ent-cycloechinulin, and epi-aszonalenins in A. novofumigatus. Our study delivers six fungal genomes, showing the large diversity found in the Aspergillus genus; highlights the potential for discovery of beneficial or harmful SMs; and supports reports of A. novofumigatus pathogenicity. It also shows how biological, biochemical, and genomic information can be combined to identify genes involved in the biosynthesis of specific SMs.
The Aspergillus genus is a diverse group of fungal species found worldwide in varying habitats. Several species are used in biotechnological industries for the production of enzymes and metabolites (commodity chemicals and pharmaceuticals), and as fermentation agents in food (1). Certain species, such as A. clavatus and A. fumigatus, are known food spoilers, mycotoxin producers, and opportunistic pathogens (1, 2). To study this diversity, it is important to have reference genomes of high assembly quality in all major clades of the genus. For this purpose, we selected four diverse Aspergillus species, A. campestris, A. novofumigatus, A. ochraceoroseus, and A. steynii, representing four phylogenetically very different sections in Aspergillus, for high-quality PacBio sequencing. The four selected genomes represent diverse and genomically unexplored sections of the Aspergillus genus: A. campestris is the first member of section Candidi to be sequenced, and likewise A. steynii is the first member of section Circumdati to be sequenced. A. ochraceoroseus, the first member of section Ochraceorosei, has recently been draft genome sequenced (3) and is available only in a large number of scaffolds. Here we also present a greatly improved assembly that may serve as a reference genome for this section. Furthermore, we have added a highly interesting member of section Fumigati, A. novofumigatus, which has a diverse secondary metabolite (SM) profile (4), as well as potentially being an opportunistic pathogen with close relation to the medically very important A. fumigatus (5). In addition, two strains from the Candidi section were Illumina sequenced to elucidate the chlorflavonin biosynthesis.
The four PacBio sequenced species explored in this study can act as a reference strain in their respective phylogenetic sections. The species may also be used to assess the natural variation within the Aspergillus genus via analysis of species-specific genes in comparison with other genome-sequenced species. Accordingly, we have compared our sequenced genomes with nine published Aspergillus reference genomes (from sections Nidulantes, Nigri, Fumigati, Flavi, Clavati, and Terrei) to serve as a compilation of reference strains for the genus.
In addition, the biosynthetic potential of these species is of interest. Filamentous fungi produce a diverse range of SMs, including bioactive compounds such as pharmaceuticals and toxins (6). SMs are not required for growth, but provide important benefits in the growth environment (7). Members of the Aspergillus genus are known to produce a wide variety of SMs with industrial, agricultural, medical, and economic importance (7, 8). The biosynthetic genes of SMs are located in clusters setting the stage for common gene regulation (9, 10). Clusters often span tens of kilobases (kbs) (11) and usually contain a gene or genes coding for one or more synthases (backbone enzyme) that define the product class of the cluster [i.e., polyketide synthases (PKS), nonribosomal peptide synthetases, and prenyltransferases or terpene cyclases (12)], in addition to tailoring enzymes such as transferases, hydroxylases, and regulatory proteins and transporters (11, 12).
With the increasing number of whole-genome sequences, the opportunity of performing analysis based on comparative genomics arises, which can give important insights and knowledge. With a focus on investigating bioactive and toxic compounds, we have here identified biosynthetic gene clusters responsible for interesting compounds from each of the PacBio sequences by combining genome analysis with knowledge of biochemical pathways and compound structure. We have identified candidates for the ochrindol cluster in A. steynii, and the chlorflavonin cluster in A. campestris.
A. novofumigatus was investigated on a genetic level, focusing on SMs. The secondary metabolic potential has been investigated, and biosynthetic gene clusters for three compounds (novofumigatonin, epi-aszonalenin, and ent-cycloechi) have been identified. Furthermore, the genomic differences and similarities of the closely related species A. novofumigatus and the pathogen A. fumigatus have been investigated, focusing on SMs, allergens, and virulence factors, and thereby addressing the potential pathogenicity of A. novofumigatus and how closely related the two morphologically similar species are.
In addition, the evolution of the aflatoxin (a highly carcinogenic compound) gene cluster from A. ochraceoroseus was investigated. The biosynthetic gene cluster was identified and studied earlier in several species, including A. flavus, A. parasiticus, and A. ochraceoroseus (13, 14). It has been seen that the synteny of the clusters are quite varying and that A. ochraceoroseus is missing some essential genes (aflQ and aflP) in the biosynthesis of aflatoxin known from A. flavus (14). With whole-genome sequences at hand, we have addressed some of these questions concerning the evolution of this biosynthetic gene cluster.
Results and Discussion
Genome Statistics.
The genomes of A. campestris, A. novofumigatus, A. ochraceoroseus, and A. steynii were sequenced using PacBio RS, whereas A. taichungensis and A. candidus were sequenced using Illumina (see SI Appendix for details). Annotation of the genomes was completed using the JGI Annotation Pipeline (15). Table 1 lists genome sequence statistics for each of the six species. The four PacBio sequenced genomes have a relatively low number of scaffolds and do not contain internal gaps. For that reason, they are highly useful as references for comparative genomics, as well as for studies of the individual genomes. Of the sequenced genomes, A. steynii has the largest genome size and is comparable with that of A. oryzae (16). The genome of A. steynii is ∼27% larger than A. ochraceoroseus, which has the smallest genome in this set and has a genome size comparable with A. clavatus (17). The difference in genome size also reflects the numbers of predicted genes in the two species, which range from 13,211 to 8,924, respectively.
Table 1.
Overview of sequencing and annotation data for the four investigated PacBio-sequenced species, plus two additional Illumina-sequenced species
| A. campestris | A. novofumigatus | A. ochraceoroseus | A. steynii | A. candidus | A. taichungensis | |
| Genome size, Mbp | 28.3 | 32.4 | 27.7 | 37.8 | 27.3 | 27.12 |
| Number of proteins | 9,764 | 11,549 | 8,924 | 13,211 | 9,641 | 9,692 |
| Number of scaffolds | 62 | 62 | 34 | 37 | 268 | 310 |
| Number of scaffolds ≥2 kbp | 56 | 62 | 32 | 36 | 168 | 283 |
| Scaffold N50 | 6 | 4 | 4 | 4 | 23 | 47 |
| Scaffold L50 | 1,703,432 | 3,768,347 | 2,489,623 | 3,921,250 | 391,998 | 207,690 |
| Fraction of GC, % | 51.2 | 49.1 | 44.2 | 49.1 | 51.8 | 51.44 |
| Coverage of gaps, % | 0 | 0 | 0 | 0 | 0.0298 | 0.0155 |
| Coverage of InterPro, % | 68 | 67 | 67 | 66 | 75 | 25 |
Investigation of DNA Methylation.
Because the four Aspergillus genomes (A. steynii, A. campestris, A. novofumigatus, A. ochraceoroseus) have been sequenced using PacBio, it is possible to investigate the presence of N6-methyldeoxyadenine (6mA) (18). Previous attempts at validation of such low abundance of 6mA have proven challenging, making it difficult to conclude whether 6mA is present in these fungi and, if so, to discriminate between real 6mA sites and false-positives (18). The presence of 6mA was therefore explored across the four Aspergillus genomes (Table 2). Consistent with previous reports (18) of low levels of 6mA in the Dikarya, we detect very little 6mA in the Aspergilli, ranging from 0.012 (A. steynii) to 0.038 (A. campestris) percent adenines methylated compared with early-diverging fungi, in which up to 2.8% of all adenines were methylated (Table 2) (18). Furthermore, only a handful of 6mA sites were at ApT dinucleotides, and none was found symmetrically at ApTs, both of which are characteristic features of 6mA modification in early-diverging fungi (18). The results therefore suggest an absence or very low occurrence of 6mA methylation in Aspergilli.
Table 2.
Overview of the methylation pattern of A. campestris, A. novofumigatus, A. ochraceoroseus, and A. steynii
| Lineage | Percentage adenines methylated | Total number of sites | Percentage modifications at ApT sites |
| A. steynii | 0.012 | 6,753 | 0.054 |
| A. campestris | 0.038 | 9,156 | 0.041 |
| A. novofumigatus | 0.03 | 7,917 | 0.058 |
| A. ochraceoroseus | 0.021 | 7,355 | 0.027 |
Whole-Genome Phylogeny Confirms Species Found in Separate Clades.
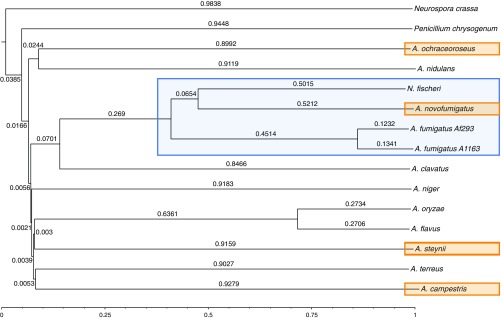
To provide an overview of the relationships among the sequenced species in the Aspergillus genus, we constructed a phylogenetic tree of the four PacBio sequenced species and the 11 reference strains, including Penicillium chrysogenum and Neurospora crassa as outgroups (Fig. 1).
Fig. 1.
Phylogenetic tree of 15 strains constructed from a composition vector approach of whole-proteome sequences using CVTree3 with a K’mer size of eight (19, 20). The time scale has been scaled to the root, thereby making the branch lengths relative to the distance between the root and the species. This time will therefore depend on what species are included in the set. The four PacBio-sequenced species are marked with orange, and the Fumigati section is marked with blue.
The constructed phylogenetic tree supports the results described earlier by Peterson (21), where a tree was constructed based on DNA sequences of four loci. A. campestris most closely resembles A. terreus of the reference genomes, whereas A. steynii relates closest to A. flavus and A. oryzae. Members of the Fumigati section are in a single clade (marked in blue on Fig. 1), with A. clavatus as a close relative. A. ochraceoroseus is placed next to A. nidulans, and both belong to the subgenus Nidulantes. All the species belonging to subgenus Circumdati (A. niger, A. oryzae, A. flavus, A. steynii, A. terreus, and A. campestris) are also placed in one clade. The tree further confirms that the three species A. ochraceoroseus, A. steynii, and A. campestris indeed represent distinct branches in the Aspergillus phylogram (22).
Unique Genes in the Genomes Often Encode Regulatory Proteins and Enzymes Involved in Secondary Metabolism.
We have identified and investigated species-specific genes for the four newly sequenced species to examine the diversity within the Aspergillus genus. Genes that are unique to a species or a small group of species may be associated with phenotypic traits and adaptation of these species to specific environments. We define species-specific genes as those without any orthologs in other sequenced genomes. This definition makes the set of species-specific genes dependent on the strains included in the analysis. As more genomes are included, especially genomes from closely related species or strains, fewer species-specific genes will be identified. The species-specific genes for each genome were identified using a set consisting of the four PacBio sequenced genomes and 11 reference genomes (SI Appendix, Table S1). Two closely related strains will share most of their genes, and they will as such not be unique to the individual species. The unique genes are not expected to encode any key functions in the cell, as they are found in only one organism; instead, these genes might be involved in environmental adaptation and/or speciation. The strains have 22%, 15%, 21%, and 27% unique genes for A. campestris, A. novofumigatus, A. ochraceoroseus, and A. steynii, respectively, indicating the vast diversity found within the Aspergillus genus. Approximately one third of the species-specific genes could be associated with an InterPro sequence domain (SI Appendix, Table S1) (23), suggesting that these genes are not false annotations.
Comparative Analysis of the Genomes of A. novofumigatus and A. fumigatus.
A. novofumigatus and A. fumigatus are considered to be two closely related species, and A. novofumigatus has only been regarded a separate species since 2005 (24). The homology between A. novofumigatus and A. fumigatus has been investigated based on the number of A. novofumigatus proteins with BLASTP hits (≥50% identity ≥130% coverage of query plus hit) in A. fumigatus. Based on this, 8,385 of A. novofumigatus proteins have homologous proteins in A. fumigatus, corresponding to 73%. The synteny between the two species was also examined using NUCmer (Nucleotide Mummer) from the MUMmer 3.0 package to map A. novofumigtus genome to the reference genome of A. fumigatus (25–27). Based on these alignments, 23.1 Mbp of the A. novofumigatus genome can be mapped to A. fumigatus, corresponding to 71% of the A. novofumigatus genome. The maximum block size is 75 kbp, and the mean block size is 4.6 kbp.
To explore this difference genetically and functionally, we have explored the similarities and differences between these two species with a focus on allergens, genes involved in virulence, and production of SMs.
Secondary Metabolite Profile of A. novofumigatus Compared with A. fumigatus.
The extrolite production in A. fumigatus has been extensively studied, and an abundance of SMs have been identified (4). A. novofumigatus is also known to have a versatile secondary metabolism; however, there is very little overlap of extrolite production between the two closely related species, making it very interesting to compare their genetic potential for producing SMs (4).
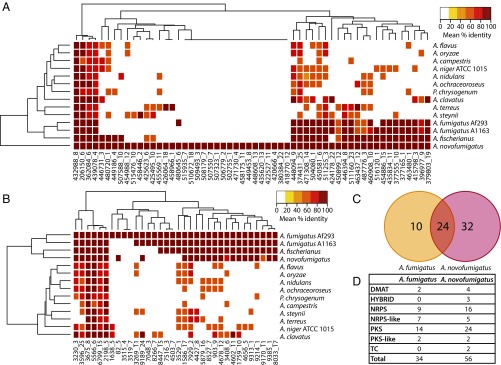
To investigate what type of SM gene clusters A. fumigatus and A. novofumigatus have in common, the SM gene clusters were predicted for each genome, using an implementation of SMURF (28). An overview of the predicted clusters and homologs in A. novofumigatus and A. fumigatus is presented in Fig. 2 A and B, respectively. Of the 34 predicted clusters in A. fumigatus and 56 predicted clusters in A. novofumigatus, 24 appear to be shared among the two species, based on bidirectional BLAST hits of the synthase (Fig. 2C). Of the 11 elucidated clusters from A. fumigatus [based on MIBiG (29)], homologs of seven (Gliotoxin, hexadehydro-astechrome, pseurotin A, fumagillin, endocrocin, helvolic acid, and trypacidin) can be found in A. novofumigatus (based on homology of the synthase). Several of these SMs are known to be involved in the virulence of A. fumigatus and are examined in more detail in the Comparative Genomics of Genes Encoding Allergens, Virulence, and Pathogenicity Factors.
Fig. 2.
(A) Overview of the SM gene clusters predicted in A. novofumigatus and their homologs in the reference species. (B) Overview of the SM gene clusters predicted in A. fumigatus and their homologs in the reference species. (C) A Venn diagram of the A. fumigatus and A. novofumigatus SM gene clusters. (D) The number and different types of SM gene clusters predicted in A. fumigatus and A. novofumigatus. DMATS, dimethylallyl tryptophan synthase; NRPS, nonribosomal peptide synthetase; PKS, polyketide synthase; TC, terpene cyclase.
The prediction of SM gene clusters also revealed considerable differences between the two closely related species. First, as seen in Fig. 2A, A. novofumigatus has 17 predicted clusters with no orthologs in any of the reference species. This is in contrast to A. fumigatus, which has only three clusters without orthologs in the reference species (Fig. 2B). Second, A. novofumigatus has 56 predicted SM clusters, whereas A. fumigatus has only 34 (Fig. 2D). Third, A. novofumigatus also has more different types of clusters. An overview of the cluster types and the number of clusters found in the two species can be seen in Fig. 2D. The diversity of SM gene clusters supports the identification of these two organisms as separate species.
SMs can present a competitive advantage in the battle for resources, but if the environment is stable, there is no need for a large arsenal of different metabolites. Thus, the large difference in SM potential between A. fumigatus and A. novofumigatus might be a reflection of the difference in natural environment and the competition in these environments, indicating that A. novofumigatus normally exists in a highly competitive environment and has a need for a larger repertoire of SMs. These results do not suggest in which conditions the metabolites are produced. Perhaps the differences are influenced by that fact that A. fumigatus Af293 is from a clinical isolate, whereas A. novofumigatus have been isolated from chamise chaparral soil after a bush fire in Southern California (2, 24). Indeed, earlier analyses have shown that clinical isolates produce fewer exometabolites and sporulate less (4, 30, 31).
It is clear that the sequence of A. novofumigatus represents a significant number of unknown gene clusters, and thereby possibly interesting bioactive compounds. To start explore this treasure chest and to illustrate our approach of linking metabolites to their respective gene clusters, we have here identified four highly interesting compounds (novofumigatonin, ent-cycloechinulin, and epi-aszonalenin A and C) by liquid chromatography–mass spectrometry analysis (SI Appendix, Fig. S1), and we have identified the biosynthetic gene clusters by using comparative genomics. Our analysis targeted these four model compounds, as they represent major metabolites produced by A. novofumigatus and because we have them as pure standards in our in-house collection of fungal metabolites (32).
Novofumigatonin is chemically a very complex compound containing an orthoester and is at present only known to be produced by A. novofumigatus (33). It has been suggested that novofumigatonin is a meroterpenoid produced from the aromatic polyketide, 3,5-dimethylorsellinic acid, as an initial precursor. Terretonin is another fungal meroterpenoid derived from 3,5-dimethylorsellinic acid, and thus we hypothesized that the early-stage biosynthetic route for novofumigatonin would follow the same pathway as that for terretonin, found in A. terreus (34).
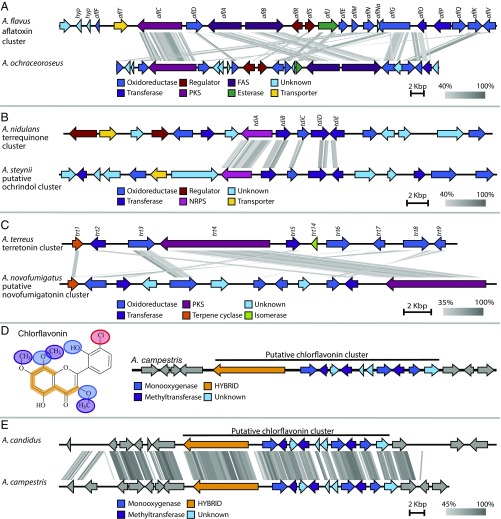
The terretonin biosynthetic genes in A. terreus were used to find homologs in A. novofumigatus, using BLASTP. Exactly one candidate cluster was identified (Fig. 3C), containing orthologs to all of the genes from the early stages of the terretonin pathway, with one exception: a terpene cyclase (trt1). However, we found this just upstream of the identified predicted cluster, and it was thus included in the putative cluster and in Fig. 3C.
Fig. 3.
Synteny plots of investigated clusters made using Easyfig tBLASTx. (A) The synteny of the predicted aflatoxin cluster in A. flavus NRRL3357 and the identified candidate aflatoxin cluster in A. ochraceoroseus (scaffold 2, 4,201,774–4,251,209 bp). (B) Synteny plot of the candidate cluster for ochrindol in A. steynii (scaffold 7, 2,783,445–2,824,507 bp) and terrequinone cluster in A. nidulans. The terrequinone cluster consists of a single-module nonribosomal peptide synthetase (tdiA), a prenyltransferase (tdiB), an oxidoreductase (tdiC), an aminotransferase (tdiD), and a gene of unknown function similar to a methyl transferase (tdiE). (C) Synteny plot of the known meroterpenoid cluster of terretonin in A. terreus and the candidate cluster of novofumigatonin in A. novofumigatus (scaffold 14, 103,246–136,450 bp). (D) The chemical structure of chlorflavonin and the candidate cluster for chlorflavonin in A. campestris (scaffold 1, 576,100–603,958 bp). The hydroxylation has been highlighted with blue, the O-methylation has been highlighted in purple, and the PKS backbone has been highlighted in orange. (E) Synteny plot of putative chlorflavonin clusters in A. candidus and A. campestris.
Novofumigatonin is closely related to fumigatonin, which has been reported to be produced by A. fumigatus (35). Interestingly, no similar cluster could be found in A. fumigatus. The most similar cluster in A. fumigatus (which is not a very strong hit: three proteins with amino acid identity <50%) is an already known cluster responsible for production of another meroterpenoid, pyripyropene A (36). This is very puzzling, and one could speculate that the report of fumigatonin might have been obtained from a misidentified isolate supposed to be A. fumigatus (35), but in reality an A. novofumigatus strain, which was only recently described as a separate species (24).
A putative cluster (scaffold 3, 33,642–53,133 bp) for ent-cycloechinulin (SI Appendix, Fig. S2) in A. novofumigatus was identified using the fumitremorgins (ftm) cluster from A. fumigatus as the starting point. Fumitremorgins are similar to ent-cycloechinulin (SI Appendix, Fig. S2), but with some important differences. Ent-cycloechinulin uses alanine instead of proline as a starter unit. Furthermore, the following prenylation occurs in a reverse manner. The genes responsible for these steps therefore have a low identity (≤35%). The following hydroxylation and O-methylation, however, are more similar, which is also reflected in the identity for the genes (≥65%). Even though the identity for the identified genes is low, they still represent the best hits in A. novofumigatus, supporting that this is the best candidate cluster.
In a similar fashion, a putative cluster for epi-aszonalenin A and C in A. novofumigatus was identified using a similar acetylaszonalenin cluster (SI Appendix, Fig. S2 for chemical structure) from A. terreus. Again, the acetylaszonalenin cluster proteins were used for a comparative genomics search in A. novofumigatus. This way, a very similar cluster was identified (scaffold 3, 1,663,448–1,719,848 bp) as a candidate for epi-aszonalenin A and C biosynthesis.
Comparative Genomics of Genes Encoding Allergens, Virulence, and Pathogenicity Factors.
A. fumigatus is known to be a common opportunistic human pathogen (37), whereas A. novofumigatus has only been reported as pathogenic in one instance (5). This difference in known pathogenicity offers the opportunity to identify and compare allergens and genes involved in the pathogenicity based on orthology, and to gather insights into the potential harmfulness of A. novofumigatus.
A list of currently accepted allergenic proteins from the well-studied A. fumigatus can be extracted from the Allergome database (www.allergome.org). The sequences of the allergenic proteins from this list were compared with the annotated A. novofumigatus protein list using BLAST+, with parameters set to report full-length sequence matches. Results shown in SI Appendix, Table S2 indicate that all A. fumigatus allergen proteins are represented in the A. novofumigatus genome. Of a total of 41 proteins, 34 proteins showed >90% identity, four showed 85–90% identity, and three showed 50–80% identity. As proteins with >50% identity are likely to cross-react to IgE (38), these results strongly indicate that A. novofumigatus possesses a strong allergen repertoire that will at least cross-react strongly with IgE to A. fumigatus and is likely to be able to provoke an immune response in the same manner as A. fumigatus. It is not possible to rule out the possibility that A. novofumigatus could be a more virulent pathogen or allergenic sensitizer than A. fumigatus.
A set of 35 potential virulence genes was assembled from recent literature, as well as genes responsible for biosynthesis of the SMs melanin, fumagillin, fumitremorgins, gliotoxin, and helvolin, which are reported to play a direct role in virulence (4, 39, 40). The results are shown in SI Appendix, Table S3. The majority of the potential virulence genes are shared between A. fumigatus and A. novofumigatus with high similarity (>85% identity); only arp2 and gel2 had identity just below 50%. The fumitremorgins cluster consists of nine genes, six of which have identity <50%, including the synthase indicating that A. novofumigatus is unable to produce fumitremorgins. The two SM gene clusters for gliotoxin and fumagillin in A. fumigatus both have highly similar matches in A. novofumigatus. The cluster for helvolic acid has three genes of nine with low BLASTP identity of 42–48%. However, A. novofumigatus has been reported to produce helvolic acid, indicating that a high amino acid similarity of these genes is not required (4).
It is likely that different combinations of virulence factors among the species affect pathogenicity (31). It has been suggested that species unable to produce some metabolites may be able to produce proxy-exometabolites that can serve the same function. This could indicate that species producing many different kinds of exometabolites are potentially pathogenic (4).
A. novofumigatus possesses the full range of allergen proteins expressed by A. fumigatus, in addition to the majority of virulence factors including several SMs. Furthermore, A. novofumigatus has an extensive potential for SM production with 56 predicted gene clusters compared with 34 for A. fumigatus. Together, these results indicate that A. novofumigatus has a considerable potential to be pathogenic. The observation of only a single instance of invasive infection by A. novofumigatus (5) may result from the recent development of methods to identify this species, which has previously not been distinguishable from A. fumigatus. It has been found that ∼4–5% of A. fumigatus isolated from patients later turned out to be closely related species (41). Thus, the true pathogenic potential of A. novofumigatus might be underestimated. Similarly, allergen sensitization to A. novofumigatus is not currently tested, and this species may also have potential to contribute to the burden of fungal allergy.
Investigation and Evolution of the Aflatoxin Gene Cluster in A. ochraceoroseus.
It is well known that A. ochraceoroseus can produce aflatoxin, and the biosynthetic cluster has been identified (14). Also, it has been noted that the aflatoxin gene cluster in A. ochraceoroseus is missing homologs to the aflP and aflQ gene involved in the conversion of sterigmatocystin (ST) to aflatoxin.
Here we have compared the aflatoxin gene cluster from the whole-genome-sequenced A. flavus NRRL3357 with A. ochraceoroseus. The clusters were identified in both species by using the aflatoxin genes identified in A. flavus AF70 (AY510453) (42).
Comparing the two clusters from A. flavus NRRL3357 with A. ochraceoroseus, it is evident that the synteny is characterized by gene shuffling (Fig. 3A). The identified cluster in A. ochraceoroseus is more similar to the ST gene cluster known from A. nidulans in the organization of genes, which was also the result of previous findings (14). This is evolutionary very interesting, as the clusters producing the same compound are quite different in their synteny, suggesting cluster dynamics or distant evolutionary origin.
As found by Cary et al. (14) it was seen that the A. flavus aflP and aflQ genes are missing in the A. ochraceoroseus aflatoxin cluster. These genes are important for the biosynthesis of aflatoxin. The whole-genome sequence was searched for orthologs to the aflP and aflQ genes from A. flavus, using BLASTP. The best hit for aflQ was JGI protein 547596, with identity 56.3% and coverage of 95.3%. The best hits for aflP were JGI proteins 430163, 506769, and 427152, with identity ranging from 40.5% to 36.6% and coverage between 31.4% and 37.7%. All the potential genes are located on a different scaffold than the aflatoxin cluster. The genes identified here are possible candidates for the A. ochraceoroseus version of the aflP and aflQ genes. With this information, it is not possible to determine exactly which genes are responsible for the conversion from ST to aflatoxin, but based on homology, these are the best candidates. Another possibility could be that the aflP and aflQ genes in A. ochraceoroseus have arisen via convergent evolution, and would thus not be found via homology analysis.
In summary, the identified aflatoxin gene cluster in this A. ochraceoroseus genome shows that A. ochraceoroseus and A. flavus most likely represent various stages of the aflatoxin cluster evolution. However, to get the full picture and truly understand the evolution of the clusters, more aflatoxin and sterigmatocystin producers need to be sequenced to be able to make bigger comparisons and get a better idea of where and when the different variations were created.
Identifying the Ochrindol Cluster in A. steynii.
Ochrindoles are prenylated bisindolyl benzoid/quinone metabolites (SI Appendix, Fig. S2) that have shown anti-insectant properties (43), one reason that A. steynii is an interesting species. Ochrindol is produced by A. steynii, and the chemical structure is known, but the biosynthetic pathway is unknown (44). However, the biosynthesis of a similar compound, terrequinone (SI Appendix, Fig. S2), produced by A. nidulans, is known, and so are the five biosynthetic genes tdiA–tdiE (45). It has been shown that ochrindol D is produced as an intermediate during biosynthesis of terrequinone. We therefore hypothesize that the genes for the biosynthesis would be partly similar, and could thus be used to identify the ochrindol cluster.
First, the five tdi genes were identified in the A. nidulans genome in a predicted cluster consisting of 17 genes. Significantly, five genes similar to A. nidulans tdiA–tdiE were identified in a predicted cluster of 17 genes, with the synteny of the tdiA–tdiE orthologs conserved (Fig. 3B and SI Appendix, Table S4). However, none of the genes next to the five tdi genes showed any homology or synteny, suggesting that the size of the cluster is overpredicted, at least in A. nidulans. In A. steynii, some of the extra genes could be involved in ochrindol production.
Identifying the Chlorflavonin Cluster in A. campestris.
Chlorflavonin was the first fully characterized flavone with fungal origin, and it is also the first naturally occurring flavone discovered to be chlorinated. It has been shown to have antifungal properties against specific species (46). The chemical structure of chlorflavonin (SI Appendix, Fig. S2) is known, and a biosynthetic pathway has been proposed, but no genes associated with the biosynthesis have been identified (47). With the whole-genome sequence for A. campestris at hand, we started exploring the genetic potential to identify the biosynthetic gene cluster responsible for producing this interesting compound.
Initially, looking at the chemical structure of this fungal flavonoid, an obvious idea for the biosynthesis would be that the backbone structure is created by a type III PKS, as the compound is so similar to plant flavonoids produced by type III PKS (48, 49). However, no type III PKS were found in A. campestris, suggesting a fungal-specific mode of biosynthesis. Next, investigating the chemical structure and proposed general biosynthesis for chlorflavonin (47), it could be deduced that the cluster must contain at least a PKS/hybrid backbone, three monooxygenases, three methyltransferases, and a chlorinating enzyme. Only one cluster met the requirements of three monooxygenases and three methyltransferases (Fig. 3D). The only concern with this candidate cluster is the lack of the essential chlorinating enzyme (SI Appendix, Table S5, Part 3).
First, sequences of known chlorinating enzymes (SI Appendix, Table S5, Part 1) were used to search for similar proteins in A. campestris, using BLASTP, but no genes were found (51–54). Second, relevant possible chlorinating InterPro domains were identified and found in four genes (SI Appendix, Table S5, Part 2), although it was not possible to pinpoint the best candidate of the chlorinating enzyme with these methods. However, the identified cluster is currently the best candidate cluster for chlorflavonin in A. campestris. Verification of this by knock-out experiments or heterologous expression could verify the candidate clusters as being responsible for the production of chlorflavonin, but this organism is not currently genetically engineerable, and the gene cluster is too large to transfer.
We therefore set out to support our prediction by sequencing and comparing genomes of several closely related species from section Candidi. A. candidus is a known chlorflavonin producer, whereas A. taichungensis is not (50). These species were therefore whole-genome sequenced to compare the pattern of the producers with the predicted clusters. A. campestris, A. candidus, and A. taichungensis each have 48, 45, and 43 predicted clusters. Based on the backbone, A. campestris and A. candidus share 35 clusters and A. campestris and A. taichungensis share 31 (BLASTP ≥50% identity and ≥130% hit+query coverage).
Comparing the genes found in the putative chlorflavonin cluster in A. campestris with the whole-genome sequences of A. candidus and A. taichungensis, A. candidus was homologous to genes in the putative chlorflavonin cluster (Fig. 3E). Moreover, this cluster is also the only cluster in A. candidus that has three methyltransferases and three monooxygenases. A. taichungensis, in contrast, does not have any significant hits of the predicted biosynthesis genes, as would be expected.
In addition, the chlorinating potential was investigated in these species. As with A. campestris, there were no BLASTP hits in A. candidus and A. taichungensis from the known chlorinating proteins (SI Appendix, Table S5, Part 1) (51–54).
Also, the possible chlorinating InterPro domains were investigated in the genomes of A. candidus and A. taichungensis. The number of hits were similar; however, A. campestris had one more hit for IPR001568, and both A. campestirs and A. candidus had one more hit for IPR008775, but none of the hits is found in SMURF-predicted clusters (SI Appendix, Table S5, Part 2).
These investigations further support that the identified cluster in A. campestris is the best candidate for chlorflavonin biosynthesis.
Conclusion
In this study, high-quality PacBio genome sequence data were generated for four Aspergillus species (A. campestris, A. novofumigatus, A. ochraceoroseus, and A. steynii) and investigated using comparative genomics. Furthermore, we have prepared draft genome sequences for two additional species: A. taichungensis and A. candidus. These six species are diverse and represent various sections of the Aspergillus genus, and thereby provide insight into the genomic and biochemical diversity and potential of the genus.
The four PacBio sequenced species have been compared with a group of already whole-genome-sequenced Aspergillus species to determine the level of genetic diversity. A phylogram was constructed on the basis of the whole-genome proteomes, and the resulting tree supports the taxonomy of the genus and fits with a phylogenetic tree constructed by Peterson SW (21) and Kocsubé S, et al. (22), based on four loci or nine loci (21, 22). The tree confirms that A. campestris, A. ochraceoroseus, and A. steynii indeed represent sections of the Aspergillus genus, which have not been genome sequenced before. Analysis of the genomes show that these genomes represent a large number of species-specific genes, particularly within secondary metabolism.
Investigation of the presence of N6-methyldeoxyadenine of the four presented species shows very low levels of 6mA. Moreover, no 6mA sites were found symmetrically at ApTs, which has been found to be a characteristic feature of 6mA modification in early-diverging fungi (18), thus confirming previous suggestions that 6mA methylation is not significant in Aspergilli.
A. novofumigatus has been compared with a close relative, the pathogenic species A. fumigatus, to better understand the mechanism of pathogenicity and virulence. The predicted SM gene clusters were found to be very different for the two close relatives, with A. novofumigatus containing 65% more clusters than A. fumigatus.
All allergens known from A. fumigatus are also present in A. novofumigatus, and the majority of the virulence factors are shared between the two species. The major difference is that A. novofumigatus lacks the fumitremorgin cluster. However, it has been suggested that proxy-exometabolites may serve the same function and A. novofumigatus has an extensive arsenal of additional SM gene clusters. It is thus highly likely that A. novofumigatus is a highly capable pathogen.
Furthermore, we have, with multiple examples, demonstrated that it is possible to identify the respective gene cluster using whole-genome sequences if one has a well-established structure of a SM and biological and chemical insights to the pathway. This way we have reidentified the aflatoxin gene cluster in A. ochraceoroseus; the epi-aszonalenins, novofumigatonin, and ent-cycloechinulin gene clusters in A. novofumigatus; the ochrindol cluster in A. steynii; and finally, the chlorflavonin cluster in A. campestris, backed by additional info from sequencing the A. taichungensis and A. candidus genomes.
In summary, the six genome sequences presented in this study illustrate the large diversity found in the Aspergillus genus and highlight the potential for discovery of structurally diverse SMs. As our project of sequencing +300 species progresses along with other fungal genome sequencing projects (e.g., the 1K fungal genomes project 1000.fungalgenomes.org/home/), the potential for applying comparative genomics to get evolutionary insights and discover interesting SMs will only increase.
Materials and Methods
The materials include a list of sequenced strains. Methods include strain cultivation; genome sequencing, assembly, and annotation; DNA-methylation analysis; details for comparative genomics analysis; phylogeny; and chemical analysis of secondary metabolism. Details for all methods are found in SI Appendix, SI Text. In particular, we provide a detailed protocol for efficient, reproducible, and scalable DNA and RNA extraction from fungi.
Supplementary Material
Acknowledgments
M.R.A. and T.C.V. gratefully acknowledge funding from the Villum Foundation, Grant VKR023437. Genome sequencing was kindly supported by Joint BioEnergy Institute and Joint Genome Institute. The work conducted by the US Department of Energy Joint Genome Institute, a US Department of Energy Office of Science User Facility, is supported by the Office of Science of the US Department of Energy under Contract DE-AC02-05CH11231. The US Department of Energy Joint BioEnergy Institute (www.jbei.org) is supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, through Contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the US Department of Energy.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All the sequencing data are available at the JGI Genome Portal (genome.jgi.doe.gov). A. campestris (accession no. MSFM00000000) genome.jgi.doe.gov/Aspcam1/Aspcam1.home.html A. novofumigatus (accession no. MSZS00000000) genome.jgi.doe.gov/Aspnov1/Aspnov1.home.html A. ochraceoroseus (accession no. MSFN00000000) genome.jgi.doe.gov/Aspoch1/Aspoch1.home.html A. steynii (accession no. MSFO00000000) genome.jgi.doe.gov/Aspste1/Aspste1.home.html A. candidus (accession no. PKFS00000000) genome.jgi.doe.gov/Aspcand1/Aspcand1.home.html A. taichungensis (accession no. PKFW00000000) genome.jgi.doe.gov/Asptaic1/Asptaic1.home.html.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715954115/-/DCSupplemental.
References
- 1.Samson RA, et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud Mycol. 2014;78:141–173. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nierman WC, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 3.Moore G, Mack B, Beltz S. Draft genome sequences of two closely-related aflatoxigenic Aspergillus species obtained from the Ivory Coast. Genome Biol Evol. 2015;8:729–732. doi: 10.1093/gbe/evv246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frisvad JC, Larsen TO. Extrolites of Aspergillus fumigatus and other pathogenic species in Aspergillus section Fumigati. Front Microbiol. 2016;6:1485. doi: 10.3389/fmicb.2015.01485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peláez T, et al. Invasive aspergillosis caused by cryptic Aspergillus species: A report of two consecutive episodes in a patient with leukaemia. J Med Microbiol. 2013;62:474–478. doi: 10.1099/jmm.0.044867-0. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmeister D, Keller NP. Natural products of filamentous fungi: Enzymes, genes, and their regulation. Nat Prod Rep. 2007;24:393–416. doi: 10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- 7.Inglis DO, et al. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol. 2013;13:91. doi: 10.1186/1471-2180-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frisvad JC, Larsen TO. Chemodiversity in the genus Aspergillus. Appl Microbiol Biotechnol. 2015;99:7859–7877. doi: 10.1007/s00253-015-6839-z. [DOI] [PubMed] [Google Scholar]
- 9.Perrin RM, et al. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007;3:e50. doi: 10.1371/journal.ppat.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer JM, Keller NP. Secondary metabolism in fungi: Does chromosomal location matter? Curr Opin Microbiol. 2010;13:431–436. doi: 10.1016/j.mib.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brakhage AA, Schroeckh V. Fungal secondary metabolites: Strategies to activate silent gene clusters. Fungal Genet Biol. 2011;48:15–22. doi: 10.1016/j.fgb.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Osbourn A. Secondary metabolic gene clusters: Evolutionary toolkits for chemical innovation. Trends Genet. 2010;26:449–457. doi: 10.1016/j.tig.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Yu J, et al. Clustered pathway genes in aflatoxin biosynthesis. Appl Environ Microbiol. 2004;70:1253–1262. doi: 10.1128/AEM.70.3.1253-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cary JW, Ehrlich KC, Beltz SB, Harris-Coward P, Klich MA. Characterization of the Aspergillus ochraceoroseus aflatoxin/sterigmatocystin biosynthetic gene cluster. Mycologia. 2009;101:352–362. doi: 10.3852/08-173. [DOI] [PubMed] [Google Scholar]
- 15.Grigoriev IV, Martinez DA, Salamov AA. Fungal genomic annotation. Appl Mycol Biotechnol. 2006;6:123–142. [Google Scholar]
- 16.Machida M, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 17.Wortman JR, et al. Whole genome comparison of the A. fumigatus family. Med Mycol. 2006;44:S3–S7. doi: 10.1080/13693780600835799. [DOI] [PubMed] [Google Scholar]
- 18.Mondo SJ, et al. Widespread adenine N6-methylation of active genes in fungi. Nat Genet. 2017;49:964–968. doi: 10.1038/ng.3859. [DOI] [PubMed] [Google Scholar]
- 19.Qi J, Luo H, Hao B. CVTree: A phylogenetic tree reconstruction tool based on whole genomes. Nucleic Acids Res. 2004;32:W45–W47. doi: 10.1093/nar/gkh362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuo G, Li Q, Hao B. On K-peptide length in composition vector phylogeny of prokaryotes. Comput Biol Chem. 2014;53:166–173. doi: 10.1016/j.compbiolchem.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 21.Peterson SW. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia. 2008;100:205–226. doi: 10.3852/mycologia.100.2.205. [DOI] [PubMed] [Google Scholar]
- 22.Kocsubé S, et al. Aspergillus is monophyletic: Evidence from multiple gene phylogenies and extrolites profiles. Stud Mycol. 2016;85:199–213. doi: 10.1016/j.simyco.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell A, et al. The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong S-B, Go S-J, Shin H-D, Frisvad JC, Samson RA. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia. 2005;97:1316–1329. doi: 10.3852/mycologia.97.6.1316. [DOI] [PubMed] [Google Scholar]
- 25.Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delcher AL, et al. Alignment of whole genomes. Nucleic Acids Res. 1999;27:2369–2376. doi: 10.1093/nar/27.11.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khaldi N, et al. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medema MH, et al. Minimum information about a biosynthetic gene cluster. Nat Chem Biol. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frisvad JC, Samson RA. Modern Concepts in Penicillium Apergillus Classification. 1990. Chemotaxonomy and morphology of Aspergillus fumigatus and related species; pp. 201–208. [Google Scholar]
- 31.Tamiya H, et al. Secondary metabolite profiles and antifungal drug susceptibility of Aspergillus fumigatus and closely related species, Aspergillus lentulus, Aspergillus udagawae, and Aspergillus viridinutans. J Infect Chemother. 2015;21:385–391. doi: 10.1016/j.jiac.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen KF, Månsson M, Rank C, Frisvad JC, Larsen TO. Dereplication of microbial natural products by LC-DAD-TOFMS. J Nat Prod. 2011;74:2338–2348. doi: 10.1021/np200254t. [DOI] [PubMed] [Google Scholar]
- 33.Rank C, et al. Novofumigatonin, a new orthoester meroterpenoid from Aspergillus novofumigatus. Org Lett. 2008;10:401–404. doi: 10.1021/ol7026834. [DOI] [PubMed] [Google Scholar]
- 34.Guo C-J, et al. Molecular genetic characterization of a cluster in A. terreus for biosynthesis of the meroterpenoid terretonin. Org Lett. 2012;14:5684–5687. doi: 10.1021/ol302682z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okuyama E, Yamazaki M, Katsube Y. Fumigatonin, a new meroterpenoid from Aspergillus fumigatus. Tetrahedron Lett. 1984;25:3233–3234. [Google Scholar]
- 36.Itoh T, et al. Reconstitution of a fungal meroterpenoid biosynthesis reveals the involvement of a novel family of terpene cyclases. Nat Chem. 2010;2:858–864. doi: 10.1038/nchem.764. [DOI] [PubMed] [Google Scholar]
- 37.Latgé JP. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aalberse RC, Akkerdaas J, van Ree R. Cross-reactivity of IgE antibodies to allergens. Allergy. 2001;56:478–490. doi: 10.1034/j.1398-9995.2001.056006478.x. [DOI] [PubMed] [Google Scholar]
- 39.Valiante V, Macheleidt J, Föge M, Brakhage AA. The Aspergillus fumigatus cell wall integrity signaling pathway: Drug target, compensatory pathways, and virulence. Front Microbiol. 2015;6:325. doi: 10.3389/fmicb.2015.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rementeria A, et al. Genes and molecules involved in Aspergillus fumigatus virulence. Rev Iberoam Micol. 2005;22:1–23. doi: 10.1016/s1130-1406(05)70001-2. [DOI] [PubMed] [Google Scholar]
- 41.Hong S-B, et al. Re-identification of Aspergillus fumigatus sensu lato based on a new concept of species delimitation. J Microbiol. 2010;48:607–615. doi: 10.1007/s12275-010-0084-z. [DOI] [PubMed] [Google Scholar]
- 42.Ehrlich KC, Yu J, Cotty PJ. Aflatoxin biosynthesis gene clusters and flanking regions. J Appl Microbiol. 2005;99:518–527. doi: 10.1111/j.1365-2672.2005.02637.x. [DOI] [PubMed] [Google Scholar]
- 43.de Guzman FS, et al. Ochrindoles A-D: New bis-indolyl benzenoids from the sclerotia of Aspergillus ochraceus NRRL 3519. J Nat Prod. 1994;57:634–639. doi: 10.1021/np50107a011. [DOI] [PubMed] [Google Scholar]
- 44.Frisvad JC, Frank JM, Houbraken JAMP, Kuijpers AFA, Samson RA. New ochratoxin A producing species of Aspergillus section Circumdati. Stud Mycol. 2004;50:23–43. [Google Scholar]
- 45.Balibar CJ, Howard-Jones AR, Walsh CT. Terrequinone A biosynthesis through L-tryptophan oxidation, dimerization and bisprenylation. Nat Chem Biol. 2007;3:584–592. doi: 10.1038/nchembio.2007.20. [DOI] [PubMed] [Google Scholar]
- 46.Richards M, Bird AE, Munden JE. Chlorflavonin, a new antifungal antibiotic. J Antibiot. 1969;22:388–389. [Google Scholar]
- 47.Burns MK, Coffin JM, Kurobane I, Vining LC. Biosynthesis of chlorflavonin in Aspergillus candidus : A novel fungal route to flavonoids. J Chem Soc Chem Commun. 1979:426–427. [Google Scholar]
- 48.Hashimoto M, Nonaka T, Fujii I. Fungal type III polyketide synthases. Nat Prod Rep. 2014;31:1306–1317. doi: 10.1039/c4np00096j. [DOI] [PubMed] [Google Scholar]
- 49.Austin MB, Noel JP. The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep. 2003;20:79–110. doi: 10.1039/b100917f. [DOI] [PubMed] [Google Scholar]
- 50.Varga J, Frisvad JC, Samson RA. Polyphasic taxonomy of Aspergillus section Candidi based on molecular, morphological and physiological data. Stud Mycol. 2007;59:75–88. doi: 10.3114/sim.2007.59.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaillancourt FH, Yeh E, Vosburg DA, O’Connor SE, Walsh CT. Cryptic chlorination by a non-haem iron enzyme during cyclopropyl amino acid biosyn. Nature. 2005;436:1191–1194. doi: 10.1038/nature03797. [DOI] [PubMed] [Google Scholar]
- 52.Kirner S, et al. Functions encoded by pyrrolnitrin biosynthetic genes from Pseudomonas fluorescens. J Bacteriol. 1998;180:1939–1943. doi: 10.1128/jb.180.7.1939-1943.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu X, et al. Identification of the first diphenyl ether gene cluster for pestheic acid biosynthesis in plant endophyte Pestalotiopsis fici. ChemBioChem. 2014;15:284–292. doi: 10.1002/cbic.201300626. [DOI] [PubMed] [Google Scholar]
- 54.Fullone MR, et al. Insight into the structure-function relationship of the nonheme iron halogenases involved in the biosynthesis of 4-chlorothreonine–Thr3 from Streptomyces sp. OH-5093 and SyrB2 from Pseudomonas syringae pv. syringae B301DR. FEBS J. 2012;279:4269–4282. doi: 10.1111/febs.12017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.