Significance
C1 is a large complex that triggers the destruction of invading pathogens via lysis or by stimulation of innate and adaptive immune processes. It is composed of C1q, a protein with a bouquet-like architecture, together with a tetramer assembled from two copies each of the serine proteases C1r and C1s, which activate when C1q binds to a pathogen surface. Here we describe detailed structures that show how C1r and C1s interact via an extensive interface encompassing the N-terminal regions of both proteases. Our findings reveal how the protease tetramer is organized and suggest a mechanism for the assembly and activation of C1.
Keywords: complement, structural biology, classical pathway, X-ray crystallography
Abstract
The multiprotein complex C1 initiates the classical pathway of complement activation on binding to antibody–antigen complexes, pathogen surfaces, apoptotic cells, and polyanionic structures. It is formed from the recognition subcomponent C1q and a tetramer of proteases C1r2C1s2 as a Ca2+-dependent complex. Here we have determined the structure of a complex between the CUB1-EGF-CUB2 fragments of C1r and C1s to reveal the C1r–C1s interaction that forms the core of C1. Both fragments are L-shaped and interlock to form a compact antiparallel heterodimer with a Ca2+ from each subcomponent at the interface. Contacts, involving all three domains of each protease, are more extensive than those of C1r or C1s homodimers, explaining why heterocomplexes form preferentially. The available structural and biophysical data support a model of C1r2C1s2 in which two C1r-C1s dimers are linked via the catalytic domains of C1r. They are incompatible with a recent model in which the N-terminal domains of C1r and C1s form a fixed tetramer. On binding to C1q, the proteases become more compact, with the C1r-C1s dimers at the center and the six collagenous stems of C1q arranged around the perimeter. Activation is likely driven by separation of the C1r-C1s dimer pairs when C1q binds to a surface. Considerable flexibility in C1s likely facilitates C1 complex formation, activation of C1s by C1r, and binding and activation of downstream substrates C4 and C4b-bound C2 to initiate the reaction cascade.
The classical pathway of complement activation triggers lysis and opsonization of invading pathogens and stimulates inflammatory and adaptive immune responses (1). It is initiated by a large multicomponent assembly, known as C1 (∼790 kDa), that binds to immune complexes, protein modulators (e.g., C-reactive protein), and polyanionic structures on pathogens and apoptotic cells. It is composed of a large recognition subcomponent, C1q (460 kDa), with a bouquet-like architecture consisting of six collagenous stems, each linked to a globular head, and four serine protease subcomponents, two C1r polypeptides (90 kDa) and two C1s polypeptides (80 kDa) that in the absence of C1q form a Ca2+-dependent heterotetramer. Binding to pathogens induces autoactivation in stepwise fashion: C1r autoactivates and then activates C1s (2, 3). C1s subsequently cleaves substrates C4 and C4b-bound C2 to form the C3 convertase (C4b2a), the next enzyme in the pathway.
C1r and C1s are modular proteases each with two N-terminal CUB domains (for complement C1r/C1s, Uegf and Bmp1), separated by an epidermal growth factor (EGF)-like domain, followed by two complement control modules (CCPs) and a C-terminal serine protease (SP) domain (4). In the absence of C1q, they form elongated S-shaped heterotetramers in electron micrographs (5, 6). The traditional explanation for this arrangement, first proposed in the 1980s, is that two central C1r polypeptides are linked via their catalytic domains (CCP1-CCP2-SP), each flanked by a C1s chain (6) (Fig. 1A). However, an alternative model (referred to herein as the stacked-tetramer model) has recently been proposed in which the CUB1-EGF-CUB2 domains of C1r and C1s stack to form an antiparallel ring-shaped tetramer, with the catalytic domains projecting out from opposite sides the ring (7) (Fig. 1B). Both models are compatible with small-angle X-ray scattering (SAXS) data, although the latter model does not explain the well-characterized interactions between the catalytic (CCP1-CCP2-SP) domains of C1r that are observed both in solution and in crystal structures (6, 8). Nevertheless, in the absence of high resolution structural information for the C1r–C1s interaction, it has not been possible to discriminate between these models.
Fig. 1.
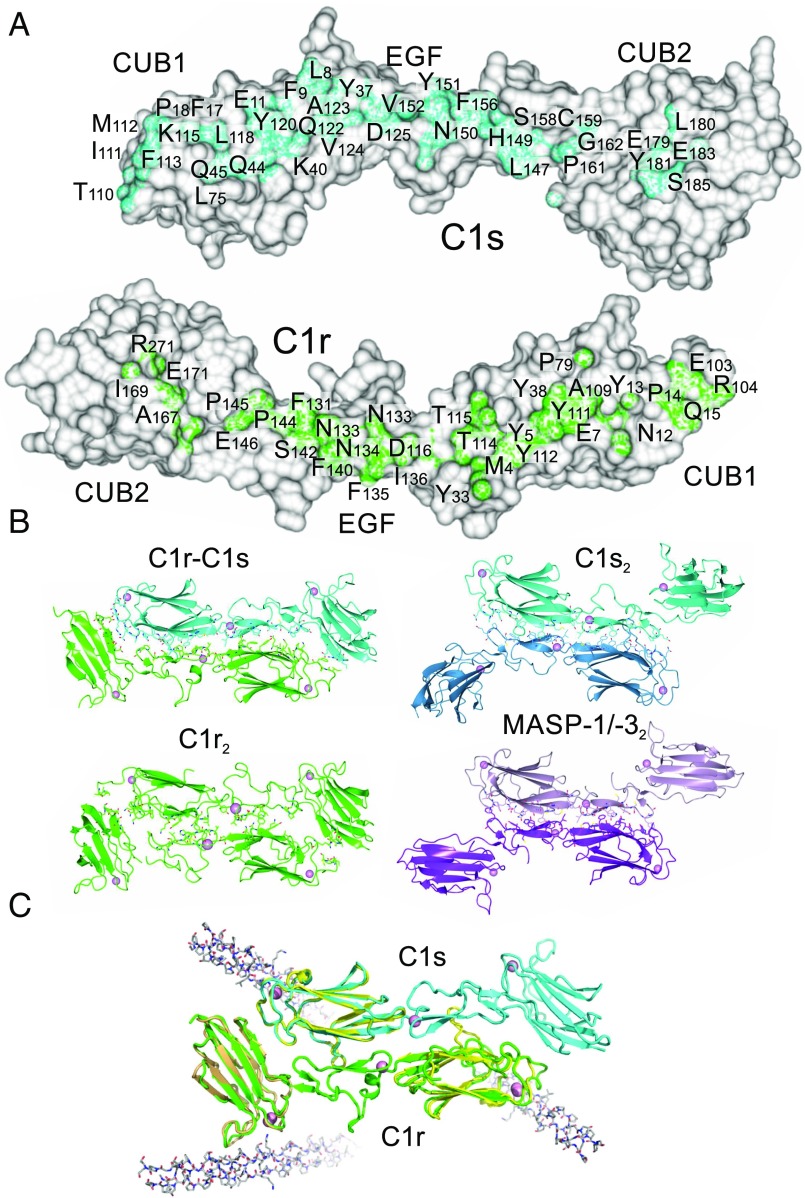
Structure of the C1r-C1s heterodimer. (A) The traditional model first proposed in ref. 6. (B) Stacked tetramer model (7). Black dots indicate the positions of the binding sites for the collagen-like domains of C1q (10). (C) Gel filtration of CUB1-EGF-CUB2 fragments of C1s, C1r, and an equimolar mixture of C1r and C1s fragments in Ca2+ (solid line) and EDTA (black dotted line). Two different loading concentrations of C1r and C1s fragments are shown (1 and 0.5 mg/mL), and 50 µL was loaded in each case. Samples were separated on a Superdex 200 column (10/30) equilibrated in 20 mM Tris pH 7.4, containing 150 mM NaCl. Elution positions of aldolase (158 kDa), conalbumin (75 kDa), and ovalbumin (43 kDa) are shown. (D) Structure of the C1r-C1s heterodimer formed from the CUB1-EGF-CUB2 fragment of each protease. Ca2+ is shown as pink spheres; carbohydrates, as white sticks.
The C1r2C1s2 tetramer folds up to form a more compact structure when it binds to C1q (5, 9). Each CUB domain of C1r and CUB1 of C1s binds to a separate collagenous stem, providing a total of six binding sites (10). Structures of CUB domains bound to collagen-like peptides, CUB1 of C1s [Protein Data Bank (PDB) ID code 4LOR] (11) and the CUB2 of the C1r/C1s homolog MASP-1 (PDB ID code 3POB) (12) show that a lysine side chain from the collagen penetrates the CUB domain to interact with Ca2+-coordinating residues. In this way, multiple weak interactions between C1q and C1r and C1s stabilize the complex.
Here we describe the structure of the C1r–C1s interaction in the form of a complex between the CUB1-EGF-CUB2 fragments of each protease. The fragments form Ca2+-dependent heterodimers both in solution and in the crystals. The interface is extensive and spans all three domains of each protease. The data are incompatible with the stacked-tetramer model of C1r2C1s2, but instead support the traditional arrangement in which C1r-C1s heterodimers are linked via interactions between the catalytic domains of C1r. On association with C1q, the C1r-C1r contacts would prevent autoactivation of C1r as the proteases fold up with the C1r-C1s dimers at the center. Disruption of the C1r contacts when C1 binds to an activating surface very likely triggers autoactivation of C1r and subsequent activation of C1s. Activation is likely facilitated through hyperflexibility at the C1s EGF-CUB2 junction, enabling considerable movement of the catalytic domains.
Results
Structure of the Complex Between the CUB1-EGF-CUB2 Domains of C1r and C1s.
The CUB-EGF-CUB2 fragments of C1r and C1s were produced in Chinese hamster ovary cells and purified by affinity chromatography and gel filtration (13). Analytical gel filtration showed that the C1s fragment is a Ca2+-dependent dimer (Fig. 1C). The C1r fragment was also dimeric in Ca2+, but aggregated in EDTA. Analysis of an equimolar mixture of C1r and C1s fragments revealed a new dimer peak that eluted slightly earlier than either of the C1s or C1r homodimers, implying the formation of heterodimers (Fig. 1C). No peak corresponding to a tetrameric species (expected to be ∼150 kDa in size for the N-terminal domains) was observed at loading concentrations up to 1 mg/mL (>10-fold the normal serum concentration of C1r2C1s2), and the position of the heterodimer peak did not change over the concentration range examined, indicating a stable complex. To obtain sufficient material for structural analysis, preparations were scaled up, and fractions were collected across the heterodimer peak and concentrated for crystallization trials.
Crystals were grown in imidazole buffer at pH 8.0 with PEG 8000 as the precipitant. Data were collected for multiple crystals and could be classified into two groups. Two representative structures were determined using the best dataset from each group. The phases were solved by molecular replacement, using the structure of the CUB1-EGF-CUB2 (PDB ID code 4LMF) (11) domains of C1s as a search model, and the structures were refined to 4.2 Å and 4.5 Å resolution. The asymmetric unit of each crystal contains two C1r-C1s heterodimers. Each heterodimer binds a total of six Ca2+, with a single Ca2+ in each domain. The Ca2+ in the EGF-like domains are located near the dimer interface, explaining the Ca2+ dependence of the interaction. Ca2+ in both CUB domains of C1r and in the CUB1 domain of C1s form the binding sites for the collagenous stems of C1q (10, 11). The C1r and C1s fragments adopt similar conformations in both crystal forms, with the main difference between the structures in the relative orientations of the heterodimer pairs. Density for carbohydrate was observed at all of the potential N-linked glycosylation sites in C1r (two sites) and C1s (one site), with multiple sugar residues observed at some sites (Fig. 1D).
The C1r-C1s Interface.
The C1r and C1s fragments are both L-shaped and interlock with their binding partner in an antiparallel arrangement (Fig. 1D). Contacts span all three domains of each polypeptide, creating an extensive dimer interface (Fig. 2A). C1r and the CUB1 and EGF domains of C1s overlay closely in each C1r-C1s pair within the asymmetric unit of the crystal. However, the orientation of C1s CUB2 is different, indicating flexibility at the EGF-CUB2 junction (see below). Interestingly, although positioned differently, C1s CUB2 is packed against C1s in both conformations, with interfaces of 1,268 Å2 for C1r and 1,304 Å2 for C1s in one C1r-C1s pair and of 1,342.4 Å2 of C1r and 1,309.1 Å2 of C1s in the other.
Fig. 2.
The C1r-C1s interface. (A) Residues buried at the heterodimer interface. (B) Dimers formed from the CUB1-EGF-CUB2 domains of C1r, C1s (PDB ID code 4LMF) and MASP-1/-3 (PDB ID code 3DEM). (C) The C1r-C1s dimers superposed with CUB1-collagen (PDB ID code 4LOR; yellow) and CUB2 collagen structures (PDB ID code 3POB; light brown), showing the position of the collagen-binding sites. Collagen is shown in gray.
The C1r-C1s interface is more extensive than that seen between homodimers of C1s (PDB ID code 4LMF; buried surfaces of 966 Å2 and 960 Å2) (11), C1r (see below), or MASP-1/-3 (PDB ID code 3DEM; 1,036 Å2 and 1,051 Å2) (14) and MASP-2 (PDB ID codes 5CIS, 5CKM, and 5CKN; 1,083 Å2 from each partner) (15), the homologous serine proteases of the lectin pathway of complement activation (Fig. 2B). It is predominantly hydrophobic in nature, although potential hydrogen bond donors and acceptors are also present (but could not be unequivocally assigned at the resolution obtained). Reciprocal interactions between the CUB1 and EGF-like domains of C1r and C1s are similar to those previously observed in C1s and MASP homodimers, with many conserved contacts and with equivalent areas of buried surface (886 Å2 from C1r and 914 Å2 from C1s). The main difference is the contacts between the CUB2 of one protease and the CUB1 of its partner in the heterodimer, which do not occur in C1s or MASP homodimers. The larger buried surface would explain why C1r and C1s preferentially form heterodimers rather than homodimers.
Flexibility at the EGF-CUB2 Junction of C1s.
The orientation of the CUB2 domain of C1s differs by ∼60° in the two C1r-C1s dimers within the asymmetric unit of the crystal, with rotation around the long axis of the L-shaped polypeptide. Rotation by ∼140° is observed in comparison with previous structures of C1s (PDBs ID codes 4LOR and 4LFM) (11), indicating that the EGF-CUB2 junction of C1s permits considerable flexibility (Fig. 3). The overall effect of this flexibility is to allow huge movements of the catalytic domains of C1s, by as much as 200 Å relative to the CUB1-EGF-CUB2 core. Such movements are likely to facilitate assembly of C1, activation of C1s by C1r, and binding and activation of the downstream substrates of C1s, C4 and C4b-bound C2, during complement activation.
Fig. 3.
Flexibility in C1s. Top (A) and side (B) views of the C1r-C1s dimer showing three different orientations of the CUB2 domain of C1s. (1) is from PDB ID code 4LMF. (2) and (3) show the two conformations of the C1s fragment in the two heterodimers of the asymmetric unit in the C1r-C1s crystals. The CUB1-EGF-CUB2 structures are overlaid with structures of the CUB2-CCP1 (PDB ID code 4LOS) and CCP1-CCP2-SP (PDB ID code 4J1Y) regions of C1s (white), to show the effects of flexibility at the EGF-CUB2 junction on the full-length C1s polypeptide.
Structure of the C1r Homodimer.
We obtained crystals of the C1r fragment alone under similar crystallization conditions as those that were successful for the heterodimer complex, but at 4 °C. Although these crystals diffracted relatively weakly, by analyzing data with a high multiplicity of measurements, it was possible to determine the structure to 5.8 Å resolution. A single antiparallel dimer was present in the asymmetric unit of the crystal, with each C1r fragment adopting a similar L-shaped structure to that observed in the C1r-C1s heterodimer (Fig. 2B). The CUB2 domain of C1r was offset by ∼30° about the long axis compared with the C1r-C1s complex, suggesting at least limited rotation at the EGF-CUB2 junction of C1r (Fig. S1). Although the CUB1-EGF-CUB2 fragments of C1r dimerize in solution, it is unclear whether these contacts form in the full-length protein or whether they are blocked by the interactions between the CCP1-CCP2-SP domains.
Buried surfaces in the C1r homodimer were smaller than those in the heterodimer, with 946 Å2 from one polypeptide and 901 Å2 from its partner. Although the binding interface spans all three domains in each complex, the overall shape complementarity is better in the heterodimer (Fig. S2 A and B). Many of the residues buried at the interface of the heterodimer also form contacts in the homodimer (25 out of 38); differences are mainly in CUB1 and CUB2 (Fig. S2C). Interestingly, previous studies have shown that smaller fragments of C1r that lack the intact CUB2 domain are monomeric in solution, indicating that CUB2 is necessary for dimerization (16, 17). Consistent with this finding, the CUB2 domain contributes >200 Å toward the buried surface of the homodimer.
Assembly of C1.
The structure of C1r-C1s heterodimers has important implications with respect to assembly of C1r2C1s2 and of the C1 complex. In particular, our data are incompatible with the recent stacked-tetramer model for C1r2C1s2, because the CUB1-EGF-CUB2 fragments do not form tetramers, even at higher concentrations than found in serum. Instead, the data support the traditional model in which the C1r2C1s2 tetramer is composed of two C1r-C1s pairs linked by interactions between the catalytic domains of C1r. To bind to C1q, the extended S-shaped tetramer must fold up to become more compact, with the CUB1-EGF-CUB2 domains of each C1r-C1s pair at the center, held in place by the collagenous stems of C1q (Fig. 4A). The interactions between the catalytic domains of C1r would not only link the two C1r-C1s dimers together during assembly, but also prevent one C1r polypeptide activating its partner.
Fig. 4.
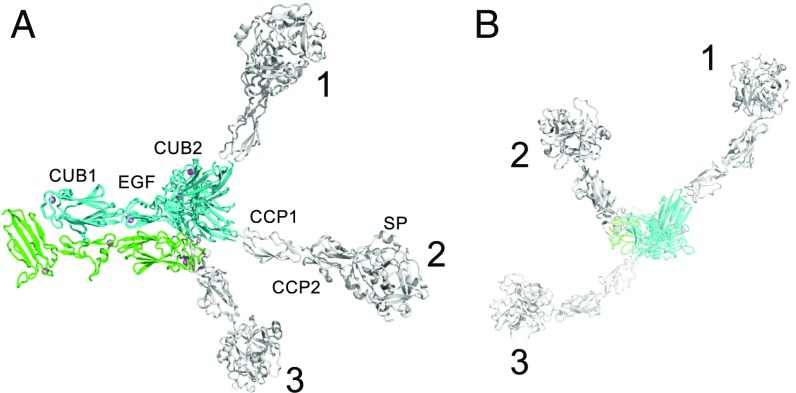
Proposed mechanism of assembly of C1. (A) C1r2C1s2 adopts an extended structure in solution (Left), in which the two C1r-C1s dimers are linked by a central interaction between the catalytic domains of C1r. It folds up (Middle) to form a more compact structure to bind to the six collagenous stems of C1q (Right). Contacts between the catalytic domains of C1r prevent one C1r polypeptide from activating its partner. Black dots show the positions of the binding sites for the collagen-like domains of C1q (10). (B) Model of C1 generated by rigid-body fitting to SAXS data. C1q is in gray, C1r is in green, and C1s is in blue. (C) Rigid-body fit to scattering data SASDB38. I(s) is the intensity, and s is the scattering vector. The χ2 value for the fit is 2.9. The fit is shown as a solid line, and the residuals to the fit are shown below with a scale of ±0.5.
To test the feasibility of the proposed arrangement, we created a model of the C1 complex using known crystal structures of C1r, C1s, collagen-like peptides (PDB ID code 1CAG) (18) and the globular domains of C1q (PDB ID code 1PK6) (19). In this model, two CUB1-EGF-CUB2 heterodimers are positioned at the center with the C1s polypeptides innermost. The dimer formed by the catalytic domains of C1r (PDB ID code 1GPZ) (8) is fitted between the N-terminal fragments. The six collagenous stems of C1q are arranged around the outside of C1r and C1s at positions corresponding to the known binding sites of C1q. The collagen stems converge close to an interruption in the collagen-like domain (the C1q kink), creating the characteristic bouquet-like architecture observed by electron microscopy. With this structure as a template, we performed rigid-body modeling using previously obtained SAXS data for C1 (Small Angle Scattering Biological Data Bank ID: SASDB38) (7) using Coral, part of the ATSAS package (20). Flexibility was permitted at the C1q kink and at the junction between the collagen-like domains and the globular heads of C1q. Although there is no direct evidence of flexibility in C1q, flexibility has been observed at collagen junctions in other proteins (21). In addition, collagen peptides themselves have been shown to have limited flexibility (22), and interruptions in collagen-like domains, such as occurs at the kink region of C1q, are often associated with considerable flexibility (23, 24). Flexibility was also allowed at EGF-CUB2 and CUB2-CCP1 and CCP1-CCP2 junctions of C1s, and between the EGF-CUB2 and CUB2-CCP1 domains of C1r, but the catalytic domains of C1r were fixed as dimers according to the crystal structure (PDB ID code 1GPZ) (8). Notably, flexibility has previously been observed at all of these junctions in overlapping crystal structures of C1r (in this work) and C1s (11), with the exception of the CUB2-CCP1 junction of C1r, for which no structure is available.
A range of different models were obtained with χ2 values usually between 2 and 4; an example is shown in Fig. 4B. Viewed from above, the six globular heads of C1q were typically arranged in two almost parallel lines (rather than around the points of a regular hexagon), with the catalytic domains of C1s projecting between the collagenous stems and with the catalytic domains of C1r at the center. The models of C1 are compatible with negative-stain EM images of cross-linked C1 in which a central mass is visible between the C1q stalks (the CUB1-EGF-CUB2 domains of C1r and C1s and the catalytic domains of C1r) (5). They are also compatible with recent cryoEM data for C1 (7), in which images appear to show between six and nine peripheral globular structures (six C1q heads, two C1s catalytic domains, and the central collagen hub and/or density from C1r polypeptides). An additional globular structure observed in some negatively stained images could be caused by disruption of the central C1r–C1r interaction, either as a result of spontaneous dissociation and separation or as a result of induced dissociation under the harsher experimental conditions.
Discussion
The structure of the C1r–C1s interaction provides insight into the assembly of C1r2C1s2 and the C1 complex, in addition to the mechanism of C1 activation. C1r and C1s polypeptides bind via an extensive interface involving all three N-terminal domains. Additional buried surface explains why heterodimers form preferentially to C1r or C1s homodimers. A Ca2+-binding site in the EGF domain of each subcomponent forms part of the binding interface, and explains the Ca2+-dependence of the interaction. Additional Ca2+ sites present in each CUB domain of C1r and in CUB1 of C1s form the binding sites for the collagen-like domains of C1q. Overall, the CUB1-EGF-CUB2 domains of the C1r-C1s heterodimer adopt a more compact arrangement than in equivalent structures of MASP homodimers, with the CUB2 domain of one polypeptide folded back against its partner.
Assembly of C1r2C1s2 has been shown to confer protection of the N-linked glycans at positions Asn159 in CUB2 of C1s and Asn204 in CUB2 of C1r against peptide:N-glycosidase F (25). Both residues are relatively close to the C1r-C1s interface, so complex formation might sterically hinder access of the glycosidase or alter the local structure of the polypeptide, to prevent cleavage of the glycan.
The data reported here are incompatible with the recently proposed stacked-tetramer model for C1r2C1s2 (7). Instead, each C1r-C1s pair must be held together by contacts between the catalytic domains of C1r to form the elongated S-shaped structures observed in EM images (5). This arrangement would prevent autoactivation of C1r, because the SP domain of one C1r polypeptide packs against the CCP domains of its partner (8). Rigid body modeling implies that these interactions can be maintained as the tetramer folds up during formation of the C1 complex. Autoactivation of the complex would require disruption of the C1r–C1r interaction, followed by alignment of the catalytic site of one polypeptide with the cleavage site of the other. This intramolecular autoactivation mechanism is compatible with the first-order kinetics observed for spontaneous C1 activation (26). Our favored model for activation is one in which binding to a surface perturbs the structure of C1 to favor separation of the SP domains of C1r (8, 27); subsequent repositioning of the CUB2 domains of C1r (to the conformation observed in the crystal structure) would help to align the SP domains for autocatalysis (11). Flexibility in C1s as demonstrated here would then allow activation of C1s by C1r and subsequent cleavage and activation of downstream substrates.
Based on an alternative model of C1 generated by rigid-body modeling of SAXS data (7), it was recently suggested that C1 cross-activates by interacting with neighboring complexes rather than by autoactivation within individual C1 complexes. In this model, the proteases are almost fully extended, and the catalytic regions project from opposite sides of the cone created by the collagenous stems of C1q. Such a model is difficult to reconcile with the first-order kinetics observed for spontaneous activation of C1 (26), which favor an intramolecular activation mechanism. In addition, the extended arrangement of C1r required to permit cross-activation is incompatible with the structures of the heterodimer, in which the C1r and C1s fragments adopt more compact, folded-back (L-shaped) configurations. Furthermore, the proposed complex would require disruption of the interaction between the catalytic domains of C1r during assembly of the C1 complex, which is likely to lead to spontaneous autoactivation of one C1r by its partner. Notably, this C1 model was generated using the stacked-tetramer model of C1r2C1s2 as a starting point, in which the catalytic domains of C1r are on opposite sides of the tetramer, thereby preventing autoactivation (Fig. 1A). However, the data reported herein, together with the previous characterization of C1r (6, 8), show that these domains must be linked together at the center of the heterotetramer. Thus, autoactivation of C1r likely occurs as soon as contacts between the catalytic domains are broken.
Materials and Methods
The CUB1-EGF-CUB2 fragments of C1r and C1s (each with a C-terminal hexahistidine tag) were produced in Chinese hamster ovary cells and purified by affinity chromatography on a nickel-Sepharose affinity column as described previously (13). Protein was purified further by gel filtration on a Superdex 200 16/60 column (GE Healthcare) in 20 mM Tris pH 7.5, 50 mM NaCl, and 2 mM CaCl2, and was then concentrated by filtration using a 10-kDa molecular mass cutoff membrane (Amicon) before crystallization. Heterocomplexes were formed by mixing molar equivalent amounts of C1r and C1s dimers, followed by gel filtration on a Superdex 200 16/60 column in the same buffer.
All crystals were grown using the sitting-drop vapor diffusion method by mixing equal volumes (1.2 + 1.2 μL) of protein and reservoir solution. Crystals of the C1r-C1s complex (3–4 mg/mL) were grown in 12–18% PEG 8000 and 100 mM imidazole at pH 8.0 and 25 °C. Similar conditions were used to crystallize C1r alone, except that the crystals were grown at 4 °C. All crystals were transferred to reservoir solution containing 20% glucose before being stored in liquid nitrogen, and were maintained at 100 K during data collection.
Diffraction data were collected at Diamond Light Source and were processed with iMosflm. Phases were determined by molecular replacement with Phaser (28) using the C1s CUB-EGF-CUB (29) structure as a search model. Models were optimized using cycles of manual refinement with Coot and refinement in Refmac5 (30), part of the CCP4 software suite (31), and in Phenix (32). A structure of the CUB2 domain of C1r (at 1.95 resolution; Table S1 and Fig. S3) was determined independently and used as a reference for the C1r-C1s structures during refinement (Supporting Information).
Supplementary Material
Acknowledgments
We thank Diamond Light Source (DLS) for access to beamlines I02, 103, and I04-1 and the UK Midlands Block Allocation Group mx14692, mx10369, and mx8359. We also thank beamline scientists at DLS for their help with data collection. Funding for this work was provided by the Medical Research Council (Grant G1000191/1, to R.W., P.C.E.M., and W.J.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6F1C, 6F1H, 6F39, and 6F1D).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718709115/-/DCSupplemental.
References
- 1.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: A key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaboriaud C, Ling WL, Thielens NM, Bally I, Rossi V. Deciphering the fine details of C1 assembly and activation mechanisms: “Mission impossible”? Front Immunol. 2014;5:565. doi: 10.3389/fimmu.2014.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallis R, Mitchell DA, Schmid R, Schwaeble WJ, Keeble AH. Paths reunited: Initiation of the classical and lectin pathways of complement activation. Immunobiology. 2010;215:1–11. doi: 10.1016/j.imbio.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forneris F, Wu J, Gros P. The modular serine proteases of the complement cascade. Curr Opin Struct Biol. 2012;22:333–341. doi: 10.1016/j.sbi.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Strang CJ, Siegel RC, Phillips ML, Poon PH, Schumaker VN. Ultrastructure of the first component of human complement: Electron microscopy of the crosslinked complex. Proc Natl Acad Sci USA. 1982;79:586–590. doi: 10.1073/pnas.79.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villiers CL, Arlaud GJ, Colomb MG. Domain structure and associated functions of subcomponents C1r and C1s of the first component of human complement. Proc Natl Acad Sci USA. 1985;82:4477–4481. doi: 10.1073/pnas.82.13.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mortensen SA, et al. Structure and activation of C1, the complex initiating the classical pathway of the complement cascade. Proc Natl Acad Sci USA. 2017;114:986–991. doi: 10.1073/pnas.1616998114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Budayova-Spano M, et al. The crystal structure of the zymogen catalytic domain of complement protease C1r reveals that a disruptive mechanical stress is required to trigger activation of the C1 complex. EMBO J. 2002;21:231–239. doi: 10.1093/emboj/21.3.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perkins SJ. Molecular modelling of human complement subcomponent C1q and its complex with C1r2C1s2 derived from neutron-scattering curves and hydrodynamic properties. Biochem J. 1985;228:13–26. doi: 10.1042/bj2280013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bally I, et al. Identification of the C1q-binding sites of human C1r and C1s: A refined three-dimensional model of the C1 complex of complement. J Biol Chem. 2009;284:19340–19348. doi: 10.1074/jbc.M109.004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkatraman Girija U, et al. Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation. Proc Natl Acad Sci USA. 2013;110:13916–13920. doi: 10.1073/pnas.1311113110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gingras AR, et al. Structural basis of mannan-binding lectin recognition by its associated serine protease MASP-1: Implications for complement activation. Structure. 2011;19:1635–1643. doi: 10.1016/j.str.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Phillips AE, et al. Analogous interactions in initiating complexes of the classical and lectin pathways of complement. J Immunol. 2009;182:7708–7717. doi: 10.4049/jimmunol.0900666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teillet F, et al. Crystal structure of the CUB1-EGF-CUB2 domain of human MASP-1/3 and identification of its interaction sites with mannan-binding lectin and ficolins. J Biol Chem. 2008;283:25715–25724. doi: 10.1074/jbc.M803551200. [DOI] [PubMed] [Google Scholar]
- 15.Nan R, et al. Flexibility in mannan-binding lectin-associated serine proteases-1 and -2 provides insight on lectin pathway activation. Structure. 2017;25:364–375. doi: 10.1016/j.str.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thielens NM, Aude CA, Lacroix MB, Gagnon J, Arlaud GJ. Ca2+ binding properties and Ca2+-dependent interactions of the isolated NH2-terminal alpha fragments of human complement proteases C1-r and C1-s. J Biol Chem. 1990;265:14469–14475. [PubMed] [Google Scholar]
- 17.Thielens NM, et al. The N-terminal CUB-epidermal growth factor module pair of human complement protease C1r binds Ca2+ with high affinity and mediates Ca2+-dependent interaction with C1s. J Biol Chem. 1999;274:9149–9159. doi: 10.1074/jbc.274.14.9149. [DOI] [PubMed] [Google Scholar]
- 18.Bella J, Eaton M, Brodsky B, Berman HM. Crystal and molecular structure of a collagen-like peptide at 1.9 Å resolution. Science. 1994;266:75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- 19.Gaboriaud C, et al. The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties. J Biol Chem. 2003;278:46974–46982. doi: 10.1074/jbc.M307764200. [DOI] [PubMed] [Google Scholar]
- 20.Petoukhov MV, et al. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Cryst. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Resnick D, Chatterton JE, Schwartz K, Slayter H, Krieger M. Structures of class A macrophage scavenger receptors: Electron microscopic study of flexible, multidomain, fibrous proteins and determination of the disulfide bond pattern of the scavenger receptor cysteine-rich domain. J Biol Chem. 1996;271:26924–26930. doi: 10.1074/jbc.271.43.26924. [DOI] [PubMed] [Google Scholar]
- 22.Walker KT, et al. Non-linearity of the collagen triple helix in solution and implications for collagen function. Biochem J. 2017;474:2203–2217. doi: 10.1042/BCJ20170217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brodsky B, Thiagarajan G, Madhan B, Kar K. Triple-helical peptides: An approach to collagen conformation, stability, and self-association. Biopolymers. 2008;89:345–353. doi: 10.1002/bip.20958. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann H, Voss T, Kühn K, Engel J. Localization of flexible sites in thread-like molecules from electron micrographs: Comparison of interstitial, basement membrane and intima collagens. J Mol Biol. 1984;172:325–343. doi: 10.1016/s0022-2836(84)80029-7. [DOI] [PubMed] [Google Scholar]
- 25.Aude CA, Lacroix MB, Arlaud GJ, Gagnon J, Colomb MG. Differential accessibility of the carbohydrate moieties of Cls-Clr-Clr-Cls, the catalytic subunit of human Cl. Biochemistry. 1988;27:8641–8648. doi: 10.1021/bi00423a020. [DOI] [PubMed] [Google Scholar]
- 26.Ziccardi RJ. Spontaneous activation of the first component of human complement (C1) by an intramolecular autocatalytic mechanism. J Immunol. 1982;128:2500–2504. [PubMed] [Google Scholar]
- 27.Kardos J, et al. Revisiting the mechanism of the autoactivation of the complement protease C1r in the C1 complex: Structure of the active catalytic region of C1r. Mol Immunol. 2008;45:1752–1760. doi: 10.1016/j.molimm.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 28.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feinberg H, et al. Crystal structure of the CUB1-EGF-CUB2 region of mannose-binding protein associated serine protease-2. EMBO J. 2003;22:2348–2359. doi: 10.1093/emboj/cdg236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 31.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 32.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.