Significance
It is currently thought that the transduction of bitter, sweet, and umami stimuli in taste cells depends on G protein-coupled receptor signaling with transient receptor potential melastatin 5 (TRPM5) acting as a common downstream component. However, in the absence of TRPM5, mice have a reduced, but not abolished, ability to detect these stimuli, suggesting that a TRPM5-independent pathway also contributes to taste transduction. Here, we identify a critical role for the TRPM4 channel in the detection of these taste qualities. Deletion of either TRPM4 or TRPM5 impairs sensitivity to bitter, sweet, and umami stimuli, and loss of both TRPM4 and TRPM5 abolishes taste responses to these stimuli. Our results show that both TRPM4 and TRPM5 are required and sufficient for the transduction of bitter, sweet, and umami stimuli.
Keywords: taste transduction, TRPM4, TRPM5
Abstract
Peripheral taste receptor cells use multiple signaling pathways to transduce taste stimuli into output signals that are sent to the brain. Transient receptor potential melastatin 5 (TRPM5), a sodium-selective TRP channel, functions as a common downstream component in sweet, bitter, and umami signaling pathways. In the absence of TRPM5, mice have a reduced, but not abolished, ability to detect stimuli, suggesting that a TRPM5-independent pathway also contributes to these signals. Here, we identify a critical role for the sodium-selective TRP channel TRPM4 in taste transduction. Using live cell imaging and behavioral studies in KO mice, we show that TRPM4 and TRPM5 are both involved in taste-evoked signaling. Loss of either channel significantly impairs taste, and loss of both channels completely abolishes the ability to detect bitter, sweet, or umami stimuli. Thus, both TRPM4 and TRPM5 are required for transduction of taste stimuli.
The detection of chemicals in potential food items relies on the activity of taste receptor cells housed in taste buds located in the oral cavity. Depending on the stimulus, tastants can directly interact with ion channels to cause cell depolarization or activate a G protein-coupled receptor (GPCR) signaling cascade. Taste buds are found in epithelial specializations called papillae and are composed of different cell types that vary in their anatomical features and in the signaling pathways that they express. Type I taste cells have glial-like properties and act primarily as support cells, while type II cells detect bitter, sweet, and umami stimuli through the activation of a GPCR pathway (1, 2). This GPCR pathway is composed of a phospholipase C (PLC) signaling cascade that causes calcium (Ca2+) release from internal stores and the subsequent activation of the monovalent selective transient receptor potential melastatin 5 (TRPM5) channel (3–10). Type II cells lack conventional synaptic specializations and, instead, activate TRPM5 to cause a cell depolarization (5, 7) which stimulates Calhm1 channels and causes release of ATP as a neurotransmitter (11, 12). Type III cells have conventional synapses, express voltage-gated Ca2+ channels (VGCCs) and synaptic proteins such as SNAP-25, and detect sour stimuli (13–15).
TRPM5 is a member of the transient receptor potential (TRP) superfamily of ion channels. All TRP channels have six transmembrane domains, and many of these channels are involved in sensory transduction (16). To date, only three TRP channels [TRPM5 (7), PKD1L3/PKD2L1 (17, 18), and TRPV1t (19)] have been identified in taste cells. Of these channels, TRPM5 has a well-defined role in the detection of bitter, sweet, and umami stimuli and is exclusively expressed in type II taste cells. TRPM5 is unique compared with most TRP channels because it is voltage-sensitive, monovalent cation-selective, and activated by elevated intracellular Ca2+ (5, 20, 21). Despite its importance in these processes, there is evidence that bitter, sweet, and umami stimuli are not exclusively transduced by TRPM5. While one study using Trpm5−/− mice reported that mice lacked all behavioral and nerve responses to bitter, sweet, and umami taste qualities (6), other studies using TRPM5-KO mice have shown reduced, but not abolished, bitter, sweet, and umami taste sensation (22–24). These data suggest the presence of a TRPM5-independent mechanism that is required for normal taste transduction.
TRPM4 shares key properties with TRPM5: It is voltage-sensitive, monovalent-selective, and activated by increased intracellular Ca2+ (25–28). TRPM4 mRNA is present in taste cells (29, 30), but its role in taste transduction has not been described. In our current study, we found that TRPM4 is expressed in both type II and III taste cells. Using live cell imaging in KO mice in conjunction with pharmacological blockers, we determined that both TRPM4 and TRPM5 contribute to taste-evoked signaling. Loss of both TRPM4 and TRPM5 eliminated the taste cell’s ability to detect bitter, sweet, or umami stimuli even though the upstream signaling pathway remained intact. Behavioral studies confirmed these physiological findings. Thus, the loss of either TRPM4 or TRPM5 impaired taste transduction, while the loss of both of these channels abolished the animal’s ability to detect these stimuli.
Results
TRPM4 Is Expressed in Type II and Type III Taste Cells but Not in Type I Cells.
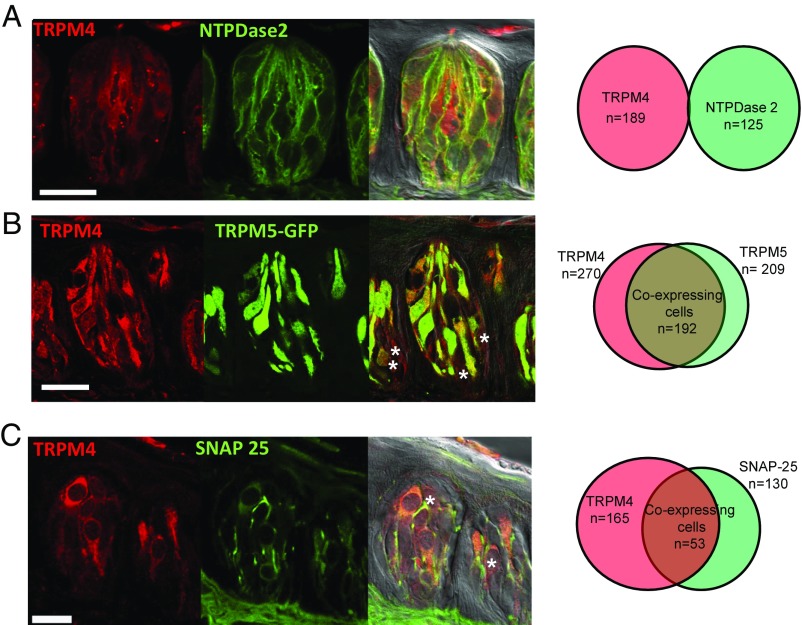
We used immunohistochemistry to evaluate the expression of TRPM4 and found it was widely expressed in taste cells. Colocalization studies with cell type-specific markers in C57BL/6 mice determined some specificity in its expression within the different taste cell types. We found no colocalization between TRPM4 and the type I taste cell marker nucleoside triphosphate diphosphohydrolase 2 (NTPDase2) (Fig. 1A; n = 189 TRPM4+ cells and n = 125 NTPDase2+ cells). Using the TRPM5-GFP mice to identify type II taste cells, immunohistochemical analysis showed a high level of colocalization between TRPM4 and TRPM5 (Fig. 1B; n = 270 TRPM4+ cells and n = 209 TRPM5-GFP cells). Approximately 90% of the TRPM5-GFP cells expressed TRPM4, while 70% of the TRPM4 expression was present in TRPM5-GFP taste cells.
Fig. 1.
TRPM4 is expressed in type II and III taste cells, but not in type I taste cells. (A) Immunocytochemical analyses of the CV papillae show the expression of TRPM4 (Left), the type I cell marker NTPDase2 (Center), and an overlay with a differential interference contrast (DIC) image (Right). (Far Right) Corresponding Venn diagram is shown representing the cell counting data. None of the TRPM4+ cells in the CV papillae expressed NTPDase2 (n = 3 mice). (B) Expression of TRPM4 (Left), expression of the type II cell marker TRPM5-GFP (Center), and an overlay with a DIC image (Right) are shown. The asterisks identify TRPM4-expressing cells that do not express TRPM5-GFP. (Far Right) Corresponding Venn diagram is shown representing the cell count data. A total of 71% of TRPM4+ cells were colocalized with TRPM5-GFP, while 92% of TRPM5-GFP cells expressed TRPM4 (n = 3 mice). (C) Expression of TRPM4 (Left), expression of the type III cell marker SNAP-25 (Center), and an overlay with a DIC image (Right) are shown. (Far Right) Corresponding Venn diagram representing the cell count data. A total of 32% of TRPM4+ taste cells expressed SNAP-25, while 41% of SNAP-25+ taste cells were positive for TRPM4 (n = 3 mice). The asterisks identify TRPM4+ cells that do not express SNAP-25. (Scale bars: 20 μm.)
Since TRPM4 was not completely colocalized with the type II cell marker TRPM5, we evaluated its potential expression in type III taste cells, using SNAP-25 as a marker. We found some colocalization between TRPM4 and SNAP-25 (n = 165 TRPM4+ cells and n = 130 SNAP-25+ cells). Of the 165 TRPM4+ cells, 53 cells (32%) also expressed SNAP-25 (Fig. 1C). These data were confirmed using PGP9.5, a protein that is expressed in neuronal cells, including type III cells. Fifty-four of the TRPM4+ taste cells (n = 169 total) also expressed PGP9.5 (Fig. S1A; n = 97, 32%). Thus, TRPM4 is present in a subset of type III cells, in addition to the majority of type II cells. To confirm that the GFP expression in the TRPM5-GFP mice was not misexpressed in type III taste cells, we did colocalization studies with the type III taste cell markers SNAP-25 and PGP9.5. TRPM5-GFP did not colocalize with either SNAP-25 (Fig. S1B; n = 153 TRPM5-GFP cells and n = 120 SNAP-25+ cells) or PGP9.5 (Fig. S1C; n = 140 TRPM5-GFP cells and n = 69 PGP9.5+ cells).
Further control experiments were performed to confirm the specificity of our transgenic mouse lines. Fig. S2 A and C shows representative images from each of the KO lines. None of the KO mice expressed the targeted protein for either single KOs (Fig. S2 A and C) or double KOs (DKOs) (Fig. S2E). We also confirmed that the TRPM5-GFP was specific to TRPM5-expressing cells (Fig. S2F). To ensure that the loss of both TRPM4 and TRPM5 did not impact the papillae structure or the signaling pathway upstream of TRPM5, we evaluated the expression of three upstream components of the pathway [PLCβ2, gustducin, and inositol 1,4,5-trisphosphate receptor type 3 (IP3R3)] in the taste cells from TRPM4/5-DKO mice. We did not find any structural deformities in the circumvallate (CV) papillae of the TRPM4/5-DKO mice, and the taste cells still expressed PLCβ2, gustducin, and IP3R3 (Fig. S2 G–I). Thus, the upstream components of the PLCβ signaling pathway remain intact in the TRPM4/5-DKO mice.
To determine if there was a change in expression of the TRPM channels in the KO mice, we evaluated TRPM4 expression in the TRPM5-KO mice and TRPM5 expression in the TRPM4-KO mice (Fig. S2 A–D). We quantitated the fluorescence intensity of our immunohistochemical images using ImageJ (NIH) to identify any changes in expression. We reasoned this approach would reveal any differences that were due to either an increased expression of the channel within the cell where it is normally expressed or any increase in expression because a greater than normal number of cells were now expressing the target protein. There was no change in TRPM4 expression in the CV of TRPM5-KO mice compared with wild-type mice (P = 1.0) (Fig. S2B) and no change in TRPM5 expression in the CV of TRPM4-KO mice compared with wild-type mice (P = 0.22024) (Fig. S2D). Thus, our data are not confounded by a nonspecific up-regulation of either TRPM4 or TRPM5 in the KO mice.
Using these transgenic mouse lines, our data found that TRPM5 was not solely responsible for transducing bitter, sweet, and umami stimuli. This finding is in contrast to a study by Zhang et al. (6) that concluded TRPM5 was solely responsible for the transduction of all these stimuli. To resolve this discrepancy, we analyzed the expression levels of TRPM4 in the Trpm5−/− mouse line, along with several of the upstream signaling components. We refer to the mice from the study by Zhang et al. (6) as Trpm5−/− and the mice used in our study as TRPM5-KO. These mice were originally described by Damak et al. (22). Our analysis of the Trpm5−/− mice identified some changes in this KO mouse line that indicate it was not a neutral mutation (Fig. S3). In the Trpm5−/− mouse, immunohistochemical analyses of the CV papillae identified a complete loss of TRPM5 (Fig. S3A) and normal PLCβ2 and gustducin expression (Fig. S3 B and C); however, the expression of IP3R3 and TRPM4 was severely reduced in the CV papillae of these mice (Fig. S3 D and E). Approximately 89% of IP3R3 expression was lost, while TRPM4 expression was reduced by 87% in the Trpm5−/− mice. Based on our data, this reduction in TRPM4 expression will have a significant impact on the ability to respond to bitter, sweet, or umami stimuli, and the loss of both TRPM4 and TRPM5 is expected to produce mice ageusic to these stimuli, as shown by Zhang et al. (6). The loss of IP3R3 will also severely reduce taste responsiveness as previously shown (31). It is unlikely that the Trpm5−/− mouse used by Zhang et al. (6) would be able to respond to bitter, sweet, or umami stimuli since the expression of all three of these proteins was compromised. Thus, the conclusions from this earlier study that TRPM5 is solely responsible for the transduction of all bitter, sweet, and umami stimuli did not account for the loss of these additional signaling proteins that occur in this strain.
Different Taste Stimuli Evoke Sodium Responses in Isolated Taste Cells.
We evaluated the contributions of TRPM4 and TRPM5 to taste-evoked signaling using live cell imaging on isolated taste cells taken from mice lacking TRPM4, TRPM5, or both TRPM4/5. Live cell imaging is a better approach for characterizing the relative contribution of a channel to taste transduction as we can measure responses from a large group of cells. This type of dataset provides a quantitative analysis of the role of a particular channel within the entire taste cell population. For this reason, we chose not to use patch-clamp analysis, which is limited by the number of cells that can feasibly be tested. Since TRPM4 and TRPM5 are sodium (Na+)-selective, we optimized our preparation to measure changes in taste-evoked Na+ responses. To our knowledge, Na+ imaging has not previously been used to measure evoked signals in taste receptor cells, so we used live cell imaging with both Ca2+- and Na+-selective dyes to validate that these Na+ signals were indeed taste-evoked responses.
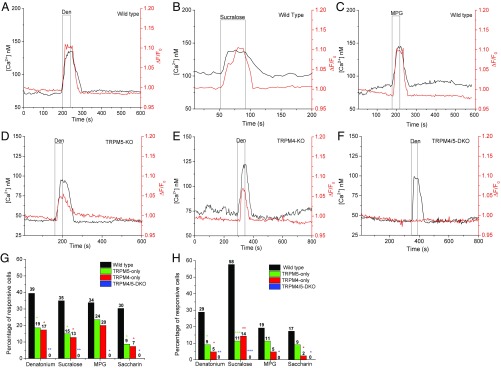
Isolated taste receptor cells from C57BL/6 mice were loaded with the Ca2+ indicator dye Fura-2 and the Na+ dye Asante NaTrium-2, and both the Ca2+ and Na+ responses to different taste stimuli were measured, including bitter [10 mM denatonium benzoate (Den)], sweet (20 mM sucralose), and umami [20 mM monopotassium glutamate (MPG)]. We chose these stimuli as representatives of bitter, sweet, or umami taste qualities, and we used maximal concentrations for each. We also included saccharin in some of our analyses. Previous studies in our laboratory determined that these concentrations of stimuli produce a maximal Ca2+ response and that higher concentrations do not increase the size of the taste-evoked response. We chose to use maximal concentrations to increase the likelihood that we would capture the responses from all taste cells that are sensitive to these stimuli. It also allows us to analyze the different responses to make comparisons. If we used less than maximal concentrations, it is possible that we would consider a cell as nonresponsive even though it could respond at higher stimulus concentrations. Our analyses of the responses could also be confounded if we were analyzing the responses at lower concentrations when the responses are expected to be more variable. We found a concurrent increase in intracellular concentrations of Ca2+ and Na+ in response to different taste stimuli (Fig. 2 A–C). This increase in cytosolic Na+ corresponds with the well-established increase in cytosolic Ca2+ that is routinely used to measure evoked responses in taste receptor cells.
Fig. 2.
Taste-evoked Na+ signals in isolated taste cells depend on the Ca2+-activated cation channels TRPM4 and TRPM5. Representative dual Ca2+ (black line) and Na+ (red line) imaging traces from wild-type mice showing cytosolic Ca2+ and Na+ responses to different taste stimuli: bitter (A; 10 mM Den), sweet (B; 20 mM sucralose), and umami (C; 20 mM MPG). (D) Representative dual Ca2+ and Na+ imaging from a TRPM5-KO cell showing the evoked Ca2+ and Na+ responses to a taste stimulus (10 mM Den). (E) Representative dual Ca2+ and Na+ imaging from a TRPM4-KO cell showing evoked responses to a taste stimulus (10 mM Den). (F) TRPM4/5-DKO cells lack a Na+ response but do generate a Ca2+ response to 10 mM Den. (G) χ2 Analysis was used to compare the percentage of responsive taste receptor cells from the CV papillae for wild-type, TRPM5-only, TRPM4-only, and TRPM4/5-DKO mice. Bar graphs (wild-type, black; TRPM5-only, green; TRPM4-only, red; TRPM4/5-DKO, blue) represent the percentages of evoked Na+ responses in taste cells isolated from the CV papillae. (H) Number of responsive taste cells isolated from Fun papillae to Den and sucralose was significantly reduced in the TRPM5-only, TRPM4-only, and TRPM4/5-DKO mice compared with wild-type mice. The number of responsive taste cells to saccharin and MPG was reduced for each of TRPM5-only, TRPM4-only, and TRPM4/5-DKO mice compared with wild-type mice (*P < 0.05; **P < 0.01; ***P < 0.001). Actual percentage values are shown on the graphs. No TRPM4/5-DKO cells had Na+ responses to any taste stimuli tested.
Taste Cells Lacking TRPM4 or TRPM5 Are Less Responsive to Different Taste Stimuli.
We then determined how the loss of either TRPM4 or TRPM5 affected the taste-evoked Na+ responses. Some taste cells that lacked TRPM5 could still generate a taste-evoked Na+ response as well as a normal Ca2+ response to the bitter, sweet, and umami stimuli tested (Fig. 2D and Fig. S4 A and B). Similar results were obtained from the TRPM4-KO mouse (Fig. 2E and Fig. S4 C and D). When both TRPM4 and TRPM5 were lost, taste cells were able to produce a normal increase in cytosolic Ca2+ in response to taste stimuli, but the ability to produce a taste-evoked Na+ signal was lost (Fig. 2F and Fig. S4 E and F). These data suggest that TRPM4 and TRPM5 both contribute to taste-evoked Na+ signals in taste cells and that these two channels are sufficient to generate these taste-evoked Na+ signals. Control experiments demonstrate that TTX-sensitive, voltage-gated Na+ channels are not contributing to these taste-evoked Na+ responses (Fig. S5 A–C). Our data also found that these taste-evoked Na+ signals are specific to Ca2+ release from stores because Ca2+ influx through VGCCs did not affect cytosolic Na+ levels (Fig. S5D).
Further control experiments demonstrated that the isolated taste cells from the TRPM5-KO, TRPM4-KO, and TRPM4/5-DKO animals could still generate Ca2+ responses to sour and salty stimuli, while no Na+ signals were produced (Fig. S5 E–L). These sour and salty responses were recorded in taste cells that also responded to high K, which indicates the presence of VGCCs and identifies them as type III taste cells (15, 32, 33). Thus, the loss of either TRPM4 or TRPM5 did not prevent taste cells from responding to sour or salty stimuli. Since we did not identify any other channels contributing to these taste-evoked Na+ responses, and in an effort to avoid confusion, we will refer to Na+ signals from TRPM4-KO mice as TRPM5-only, while the Na+ signals from TRPM5-KO mice will be called TRPM4-only.
We next analyzed the taste-evoked Na+ responses in wild-type, TRPM5-only (TRPM4-KO), TRPM4-only (TRPM5-KO), and TRPM4/5-DKO mice to determine the overall responsiveness of the cells to the different stimuli and to evaluate the characteristics of the remaining taste-evoked Na+ responses. Experiments on isolated taste cells from both CV and fungiform (Fun) papillae found that the number of bitter responsive taste cells was significantly reduced in either the TRPM5-only or TRPM4-only cells from both papillae compared with wild-type cells (Fig. 2 G and H). The overall responsiveness to Den and sucralose was significantly reduced in both CV and Fun taste cells from each of the KO mice, while saccharin responses were significantly reduced in both KO mouse lines for CV taste cells and TRPM4-only Fun cells. Saccharin responses were reduced in the Fun TRPM5-only taste cells, but were not significantly different (P = 0.3). While there were fewer MPG-sensitive taste cells in both TRPM5-only and TRPM4-only mice compared with controls, these values were not significantly different (Fig. 2 G and H). Taste cells from the TRPM4/TRPM5-DKO mice did not respond to any taste stimulus tested (Fig. 2 G and H).
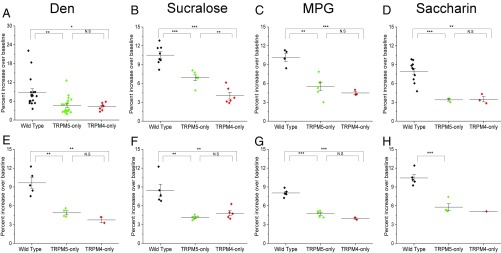
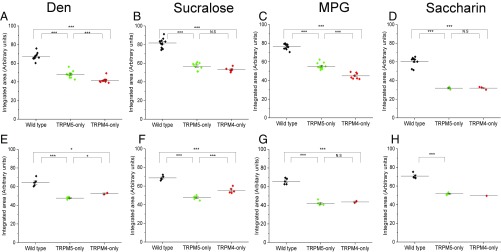
We characterized the remaining taste-evoked Na+ signals from the TRPM5-only and TRPM4-only mice and found that these responses had significantly reduced amplitudes (percent rise over baseline) for all of the taste stimuli tested compared with wild-type mice, in both the CV (Fig. 3 A–D) and the Fun taste cells (Fig. 3 E–H). We also evaluated the integrated area of the response to get a proportional measure of the taste-evoked Na+ signal when either TRPM4 or TRPM5 was absent. Compared with the responses from the wild-type mice, the overall size of the response was significantly reduced when either TRPM4 or TRPM5 was missing for both CV (Fig. 4 A–D) and Fun (Fig. 4 E–H) taste cells. Therefore, the loss of either TRPM4 or TRPM5 significantly reduces the ability of taste cells to respond to taste stimuli in comparable ways.
Fig. 3.
Loss of TRPM4 or TRPM5 reduces the amplitude of the evoked Na+ signals. Amplitudes of the taste-evoked Na+ responses were measured as a percent increase over baseline. One-way ANOVAs with a Bonferroni’s post hoc analysis and Student’s t test were used to determine significant changes in the amplitudes of taste-evoked Na+ responses. Amplitudes for CV taste cells were significantly reduced in both TRPM5-only (green) and TRPM4-only (red) mice for Den (A), sucralose (B), MPG (C), and saccharin (D) compared with wild-type cells (black). Amplitudes for Fun taste cells were significantly reduced in both TRPM5-only (green) and TRPM4-only (red) mice for Den (E), sucralose (F), MPG (G), and saccharin (H) compared with wild-type cells (black) (*P < 0.05; **P < 0.01; ***P < 0.001). There was only one recorded saccharin response in the TRPM4-only Fun taste cells, so no statistical analysis was performed. N.S, not significant.
Fig. 4.
Loss of TRPM4 or TRPM5 reduces the overall size of the evoked Na+ signals. The proportional sizes of the taste-evoked Na+ responses were measured as an integrated area under the curve. One-way ANOVAs with a Bonferroni’s post hoc analysis and Student’s t test were used to determine any significant changes in the sizes of taste-evoked Na+ responses. Integrated areas of the taste-evoked Na+ signals for CV taste cells were significantly reduced in both TRPM5-only (green) and TRPM4-only (red) mice for Den (A), sucralose (B), MPG (C), and saccharin (D) compared with responses from wild-type cells (black). Integrated areas of the taste-evoked Na+ signals for Fun taste cells were significantly reduced in both TRPM5-only (green) and TRPM4-only (red) mice for Den (E), sucralose (F), MPG (G), and saccharin (H) compared with wild-type cells (black) (*P < 0.05; **P < 0.01; ***P < 0.001). There was only one recorded saccharin response in the TRPM4-only Fun taste cells, so no statistical analysis was performed. N.S, not significant.
Taste-Evoked Na+ Signals Are Downstream of PLCβ Signal and Require Ca2+ Release from Endoplasmic Reticulum Ca2+ Stores.
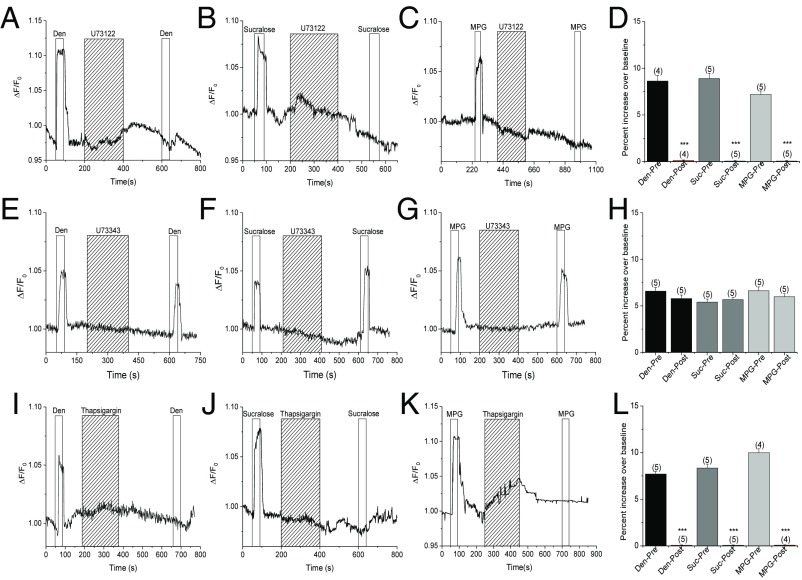
Since the TRPM5-only, TRPM4-only, and TRPM4/5-DKO cells retained an intact Ca2+ response, we hypothesized that the taste-evoked Na+ signals are downstream of the PLCβ signaling pathway that transduces bitter, sweet, and umami stimuli (6). To test this hypothesis, we applied the irreversible PLC inhibitor U73122 and measured the effect of blocking PLC activity on the taste-evoked Na+ signals. We found that the bitter-, sweet-, and umami-evoked Na+ signals were all abolished by U73122 (Fig. 5 A–D). The inactive analog of U73122, U73433, did not affect the taste-evoked Na+ responses (Fig. 5 E–H). Further, thapsigargin, which inhibits the sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) pump and prevents the refilling of endoplasmic reticulum Ca2+ stores, also eliminated the bitter, sweet, and umami taste-evoked Na+ signals (Fig. 5 I–L). Based on these data, we conclude that the taste-evoked Na+ signals are downstream of the PLCβ signaling pathway and depend on Ca2+ release from internal stores.
Fig. 5.
Taste-evoked Na+ signals are downstream of the PLCβ signaling pathway. Application of the specific PLC blocker U73122 (1 μM) abolished the taste-evoked Na+ responses to Den (A), sucralose (B), and MPG (C). (D) U73122 significantly inhibited the taste-evoked Na+ signals to Den, sucralose (Suc), and MPG. Application of the inactive analog of U73122, U73433 (1 μM), did not affect the taste-evoked Na+ responses to Den (E), sucralose (F), and MPG (G). (H) U73343 did not affect the taste-evoked Na+ signals to Den, Suc, and MPG. Application of 3 μM thapsigargin abolished the taste-evoked Na+ responses to Den (I), sucralose (J), and MPG (K). (L) Thapsigargin significantly inhibited the taste-evoked Na+ signals to Den, Suc, and MPG (***P < 0.001) (n = 4–5 cells collected from three mice for all experiments).
Taste-Evoked Na+ Signals Are Solely Mediated by TRPM4 and TRPM5.
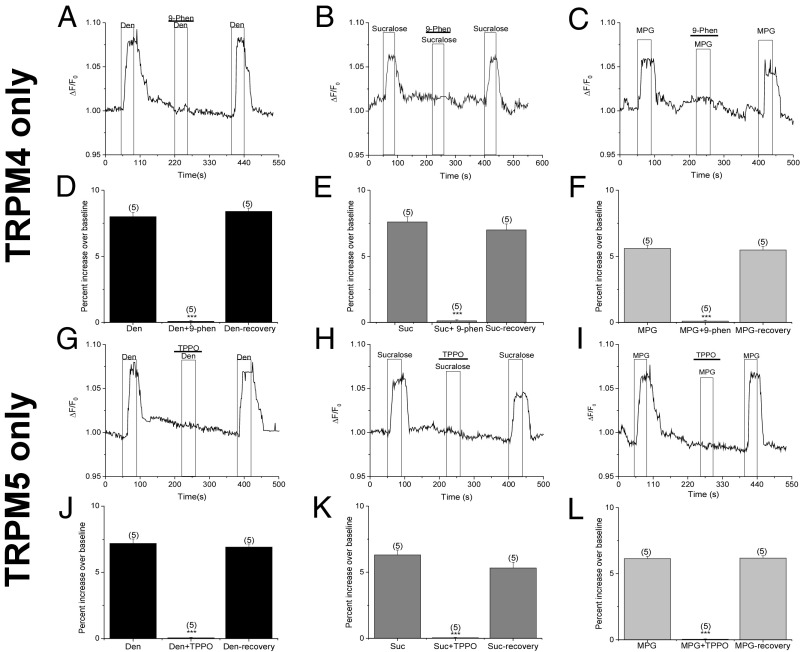
Our imaging experiments using the TRPM4/TRPM5-DKO mice indicate that these two channels are solely responsible for generating the taste-evoked Na+ signals. To ensure this was the case, we used specific TRPM4 and TRPM5 inhibitors and the TRPM5-only (TRPM4-KO) and TRPM4-only (TRPM5-KO) mice to evaluate the taste-evoked Na+ signals. The TRPM4 inhibitor 9-phenanthrol is a reversible, selective blocker of the TRPM4 channel (34–36). When we applied 9-phenanthrol (50 μM) to taste cells from the TRPM4-only mice, the bitter, sweet, and umami taste-evoked Na+ responses were abolished, indicating that the remaining taste-evoked Na+ signal are due to TRPM4 (Fig. 6 A–F). After washout of 9-phenanthrol, the taste-evoked Na+ signals were restored. Triphenylphosphine oxide (TPPO) is a reversible, selective blocker for the TRPM5 channel (37). Application of TPPO (50 μM) eliminated the remaining taste-evoked Na+ responses in TRPM5-only mice, indicating that these taste-evoked Na+ signals are solely mediated by TRPM5. Responses were restored after washout of TPPO (Fig. 6 G–L). To test the specificity of our pharmacological blockers, we applied the TRPM5 blocker TPPO in the TRPM4-only mice (Fig. S6 A–C) and the TRPM4 blocker 9-phenanthrol in the TRPM5-only mice (Fig. S6 D–F). These antagonists had no effect on the taste-evoked signals in these experiments. Based on these data and our data from the TRPM4/TRPM5-DKO mice (Fig. 2), we conclude that both TRPM4 and TRPM5 are responsible for the taste-evoked Na+ signals and that no other channel contributes to these responses.
Fig. 6.
Taste-evoked Na+ responses are due entirely to TRPM4 and TRPM5. Application of 9-phenanthrol (50 μM), a specific blocker for TRPM4, abolished the remaining taste-evoked Na+ responses in TRPM4-only mice for Den (A and D), sucralose (Suc; B and E), and MPG (C and F). Taste-evoked Na+ responses returned upon washout of 9-phenanthrol (9-Phen). Application of TPPO (50 μM), a specific blocker of TRPM5, abolished the remaining taste-evoked Na+ responses in TRPM5-only mice for Den (G and J), Suc (H and K), and MPG (I and L) (***P < 0.001). Evoked Na+ responses returned upon washout of TPPO (n = 4–5 cells collected from three mice for all experiments).
TRPM4 and TRPM5 Have Different Sensitivities in Taste Cells.
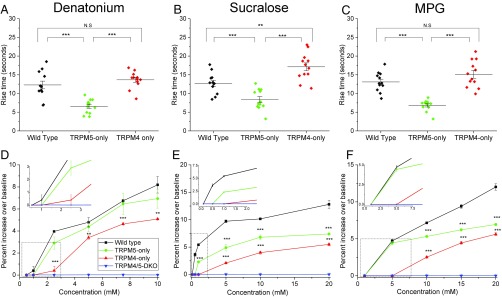
Analysis of the peak rise time in wild-type, TRPM5-only, and TRPM4-only taste cells revealed significant differences in the activation of the TRPM4 and TRPM5 channels during taste-evoked signaling (Fig. 7 A–C). We used maximal stimulus concentrations for these analyses to reduce any variability in the responses. Our data were initially analyzed separately for the CV and Fun taste cells, but no differences were found between these groups and the data were combined. When TRPM5 was the only functional channel present, the rise time was significantly faster for all of the stimuli tested compared with either the wild-type or TRPM4-only responses (Fig. 7 A–C). The rise time for TRPM4 channels was comparable to the rise time from wild-type cells for both Den and MPG (Fig. 7 A and C), while it was significantly slower than the rise time from wild-type cells for sucralose (Fig. 7B).
Fig. 7.
TRPM4 and TRPM5 have different sensitivities in taste cells. The rise times of the responses were compared by one-way ANOVA with a Bonferroni’s post hoc analysis and Student’s t test between wild-type and KO mice to determine any differences in the time to peak response when either TRPM4 or TRPM5 was the only channel present. The rise times for TRPM5-only to Den (A), sucralose (B), and MPG (C) were significantly faster than the rise times for either wild-type or TRPM4-only. TRPM4 was not different from wild-type for Den or MPG (A and C), but was significantly slower than wild-type for sucralose (B). The rise time analysis for Den, sucralose, and MPG is based on n = 12 cells for wild-type, TRPM5-only, and TRPM4-only mice. A concentration gradient analysis of a bitter stimulus (D; Den), a sweet stimulus (E; sucralose), and an umami stimulus (F; MPG) revealed reduced amplitudes for TRPM4-only and TRPM5-only mice compared with wild-type mice at all concentrations (**P < 0.01; ***P < 0.001). N.S., not significant. Graphs plot the average ± SE of the responses from five cells taken from at least three mice. These experiments (raw data are shown in Fig. S7) identified different responsive ranges for TRPM4 and TRPM5. Amplitudes were compared by one-way ANOVA with a Bonferroni’s post hoc analysis and Student’s t test between wild-type, TRPM4-only, and TRPM5-only mice. (D–F, Insets) Average ± SE of the responses for wild-type (black), TRPM5-only (green), TRPM4-only (red), and TRPM4/5-DKO (blue) mice at the lowest stimulus concentrations tested.
We also evaluated the responsiveness of each TRPM channel to a range of stimulus concentrations (Fig. 7 D–F and Fig. S7). At lower stimulus concentrations, the taste cells had smaller responses that increased in size with increasing stimulus concentration. Taste cells from the wild-type mice were the most sensitive and responded to the lowest concentrations of stimuli presented. Taste cells from the TRPM4/TRPM5-DKO mice did not respond to any stimulus concentration presented (Fig. 7 D–F and Fig. S7 J–L). The TRPM5 channel was also activated at lower stimulus concentrations compared with TRPM4, although not always comparably to wild type. Wild-type taste cells had a measurable response to 1 mM Den, while the taste cells that only expressed TRPM5 or TRPM4 did not respond until the concentration was 2.5 mM (Fig. 7D, Inset). TRPM4-dependent signals were not robust until the Den concentration was 5 mM. There was a similar response profile for sucralose responses (Fig. 7E, Inset). For MPG, the wild-type cells had a small response to 1 mM, while the TRPM5-only cells did not respond until 5 mM MPG was applied. Interestingly, this response was comparable to the wild-type response (Fig. 7F, Inset); however, the responses in the TRPM5-only taste cells were significantly smaller than wild-type responses at higher concentrations. TRPM4-only taste cells required 10 mM MPG to generate a measurable response. Clearly, TRPM4 and TRPM5 have different sensitivities to concentrations of different stimuli, but neither can solely mediate the wild-type response.
Loss of TRPM4 and TRPM5 Impairs Behavioral Preference for Bitter, Sweet, and Umami Tastants.
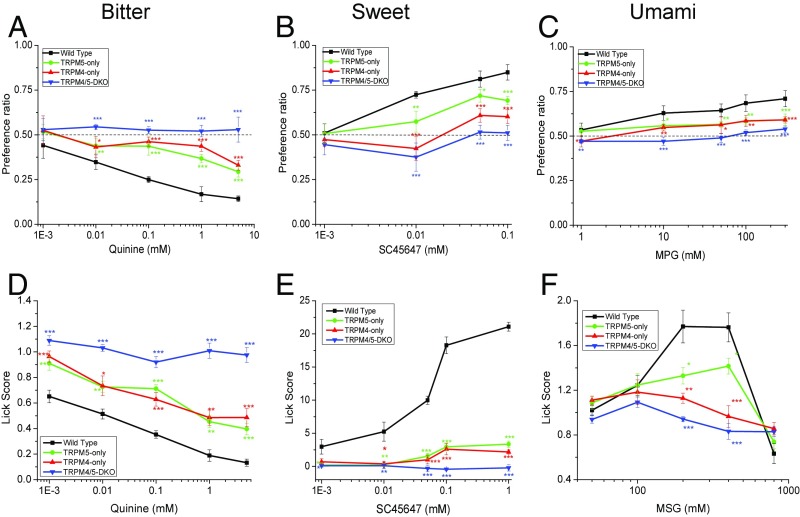
We performed behavioral experiments to determine if the loss of taste-evoked Na+ signals correlated with loss of taste sensitivity. We used two-bottle preference tests to analyze the preference ratios for TRPM5-only, TRPM4-only, and TRPM4/5-DKO mice compared with wild-type mice. We tested two bitter, two sweet, and two umami stimuli (Fig. 8 A–C and Fig. S8) and found that loss of either TRPM4 or TRPM5 significantly changed the preference ratios for all of the tested taste stimuli. The TRPM4/TRPM5-DKO mice did not show any concentration-dependent changes in preference; in effect, they did not treat any of the taste stimuli presented as different from water. TRPM4/TRPM5-DKO mice did prefer the highest concentrations of sucrose (Fig. S8B; 300 and 500 mM). Since these were long-term exposures to the stimuli (48 h) and sucrose has well-established postingestive effects (38), we concluded that their increased intake at these concentrations is most likely due to the positive postingestive effects associated with sucrose and not due to their ability to taste this stimulus.
Fig. 8.
Loss of TRPM4 or TRPM5 affects behavioral responses to taste stimuli. Mean preference ratios (±SD) from two-bottle preference tests compare the responses of TRPM5-only (green line), TRPM4-only (red line), and TRPM4/5-DKO (blue line) mice with the responses of wild-type mice (C57BL/6, black line) for the bitter stimulus quinine hydrochloride (A), the sweet stimulus SC45647 (B), and the umami stimulus MPG (C) (n = 5 mice of each genotype for all experiments). (D) Lick scores (stimulus/water) of the wild-type mice for the bitter stimulus quinine hydrochloride were significantly different from those of TRPM4/5-DKO, TRPM5-only, and TRPM4-only mice. (E) Lick scores (stimulus–water) of the wild-type mice for the sweet stimulus SC45647 were significantly different from those of TRPM4/5-DKO, TRPM5-only, and TRPM4-only mice. (F) Lick scores (stimulus/water) of the wild-type mice for umami stimulus monosodium glutamate (MSG) + 10 μM amiloride were significantly different from those of the TRPM4/5-DKO, TRPM5-only, and TRPM4-only mice (*P < 0.05; **P < 0.01; ***P < 0.001).
The behavioral responses of mice in the two-bottle preference test depend on peripheral nerve input, postingestive factors, hedonics, and central integration. To ensure that these behavioral effects were not a result of some nonspecific effect due to using long-term behavioral assays, brief-access lick assays were also performed. The wild-type mice showed decreased licking of quinine at a concentration of 0.01 mM and higher, while TRPM4/5-DKO mice maintained a lick ratio close to 1.0 for all quinine concentrations (i.e., they did not treat any concentration of quinine tested as different from water). TRPM4-only (TRPM5-KO) and TRPM5-only (TRPM4-KO) mice showed intermediate licking behavior (Fig. 8D). Because SC45647 is hedonically positive under wild-type conditions, the lick score is calculated as licks to the stimulus minus licks to water; therefore, a score close to zero resembles water. In this case, the TRPM5-only, TRPM4-only, and TRPM4/5-DKO mice showed no concentration-dependent effect on licking to SC45647 across the entire range of concentrations tested, but remained close to zero. In contrast, the wild-type mice showed strong concentration-dependent increases in licking to SC45647 concentrations (Fig. 8E). In the umami brief-access test, the wild-type mice showed increases in unconditioned licking for MSG up to 400 mM and then decreased licking at the highest concentration (800 mM). The TRPM4-only and TRPM5-only mice had fewer licks compared with the wild-type mice, and the TRPM4/5-DKO mice showed no dose-dependent change in licking behavior but, instead, maintained a lick ratio close to 1.0, suggesting the DKO mice treat the stimulus as similar to water. These data clearly show that both TRPM4 and TRPM5 are required for normal taste perception of bitter, sweet, and umami taste qualities.
Discussion
Bitter, sweet, and umami taste qualities are detected by taste receptor cells that share a common signaling pathway in which the TRPM5 channel is a critical component (6, 7, 22). Only one of these studies (6) reported that mice with a partial deletion of TRPM5 lost all behavioral and nerve responses to bitter, sweet, and umami compounds, while other studies reported reduced, but not abolished, responses to bitter, sweet, and umami stimuli when TRPM5 is lost (22–24). Our analysis of the Trpm5−/− mouse used in the study by Zhang et al. (6) suggests that the TRPM5 mutation in this line is not a neutral mutation. In this mouse line, the construct retained the gene’s upstream promoter along with exons 1–14 (6). Our immunohistochemical analysis of this mouse line (Fig. S3) shows loss of TRPM5 with normal PLCβ2 and gustducin expression. However, we also found reduced IP3R3 and TRPM4 expression. The loss of these proteins, in addition to the loss of TRPM5, would explain why the mice in the study by Zhang et al. (6) were not able to detect any bitter, sweet, and umami stimuli. Studies using a different TRPM5-KO mouse line, which was constructed by removing the promoter and exons 1–4 of the TRPM5 gene, including the translation start site, reported that loss of TRPM5 caused a significant impairment in the transduction of bitter, sweet, and umami stimuli but that these TRPM5-KO mice retained some sensitivity to these taste qualities (22, 23). This mouse line was the one we used in our studies, and our control experiments reveal no change in the upstream components (gustducin, PLCβ2, or IP3R3) when TRPM4 and TRPM5 are lost. We also found no change in the expression of TRPM4 when TRPM5 is lost or in TRPM5 expression when TRPM4 is lost (Fig. S2). These earlier studies indicate the presence of a TRPM5-independent pathway that is also required for normal transduction of bitter, sweet, and umami stimuli.
Until now, the identity of this TRPM5-independent component in taste-evoked signaling has remained elusive. Earlier reports identified a Ca2+-activated cation current in taste receptor cells that the authors speculated, but did not conclude, may be carried through the TRPM4 channel (39, 40). A separate study reported that TRPM4 mRNA is present in taste cells (29), while RNA sequencing analysis revealed high levels of mRNA for both TRPM4 and TRPM5 in isolated taste cells (30) (Fig. S2J). Our data have clearly identified that TRPM4 has an important role in the transduction of bitter, sweet, and umami stimuli and that both TRPM4 and TRPM5 are required for normal signaling in taste cells.
The primary difference between TRPM4 and TRPM5 in other cell types is their sensitivity to intracellular Ca2+. Analyses in HEK-293 cells found that the Ca2+ concentration for half-maximal current activation (EC50) for TRPM4 (20.2 ± 4.0 μM) is much larger than the EC50 of TRPM5 (0.70 ± 0.1 μM) (41). If these EC50 values are comparable in taste cells, then we predict that during cell stimulation, TRPM5 would be activated first when cytosolic Ca2+ levels are lower. As Ca2+ concentrations increase, TRPM4 would also contribute to the taste-evoked signal. While we do not know the EC50 values for TRPM4 or TRPM5 in taste cells, our data suggest that these channels do have different Ca2+ sensitivities. We evaluated the taste-evoked responses when either TRPM4 or TRPM5 was absent and found that TRPM5 channels responded to lower stimulus concentrations compared with TRPM4 channels. These findings suggest that TRPM5 requires a smaller increase in cytosolic Ca2+ for activation compared with TRPM4.
Our rise time analyses also demonstrate that TRPM5 channels open significantly faster than TRPM4 channels for all of the taste-evoked responses tested, further supporting the idea that these channels have different Ca2+ sensitivities for activation. If TRPM4 requires more cytosolic Ca2+ for activation, then the rise time for this channel will be slower since the channel will not be opening very efficiently until Ca2+ levels are sufficiently higher. The rise times of the responses when both channels were contributing to the signal (wild-type cells) were between the rise times for the TRPM4-only and TRPM5-only responses. When both of these channels contribute to the taste-evoked Na+ influx, the cell depolarization will be larger. Perhaps a larger depolarization is needed to sufficiently activate the voltage-dependent ATP release from the Calhm1 channel. It has been postulated that TRPM5 activity alone is unlikely sufficient to activate ATP release consistently enough to mediate rapid taste responses (42). The contribution of TRPM4 will increase this initial depolarization, and it appears that both channels are needed to depolarize the taste cells sufficiently to reliably activate ATP release in response to taste stimuli.
Approximately 90% of TRPM5+ cells also express TRPM4, so it is feasible that these channels are working together in taste signaling. At higher stimulus concentrations, both TRPM4-only and TRPM5-only responses are required to generate the wild-type response. At the lower stimulus concentrations, both TRPM4-only and TRPM5-only cells were not responsive, even though the wild-type cells responded (Fig. 7 D–F, Insets). Analyses of the overall size of the taste-evoked responses revealed that the size of the Na+ response when only one channel was present was significantly reduced for all taste stimuli tested (Fig. 4). While significantly smaller, the taste-evoked signals in either TRPM5-only or TRPM4-only taste cells were not half the size of the wild-type response. If the channels were independently contributing to the wild-type response, we would predict that adding the responses together would approximate the size of the wild-type response. However, if we added the actual individual responses together, they would be larger than the wild-type response. Taken together, these data suggest there is a functional coupling of TRPM4 and TRPM5 that is needed to generate the appropriate cellular response. However, if this functional coupling exists, it is not required for the channels to function, as they were able to independently respond to higher stimulus concentrations. Perhaps there is some coupling of TRPM4 and TRPM5 that conveys a greater sensitivity to Ca2+ changes. TRP channels have been shown to functionally couple in other systems (43–45), so it is possible that this is also occurring in taste cells.
Our data demonstrate that the taste-evoked Na+ responses are solely mediated by TRPM4 and TRPM5 channels and require Ca2+ release from intracellular stores through activation of the PLC signaling cascade. The activation of TRPM4 by PLC signaling and Ca2+ release from stores is well established in other systems (46, 47). While a recent study reported that the PLC inhibitor U73122 directly activates TRPM4 (48), U73122 application abolished taste-evoked TRPM4-dependent Na+ responses in our study. This earlier work was done with heterologous cell lines and a high concentration of U73122, which likely caused nonspecific effects. We also found that the Na+ responses in taste cells were not activated by a high K+-dependent Ca2+ influx (Fig. S5D). Our data are consistent with the previous findings that TRPM4 is activated by oscillatory changes in intracellular Ca2+, but not by a large influx of Ca2+ (28).
Finally, our behavior experiments clearly show that both TRPM4 and TRPM5 are required for normal responses to bitter, sweet, and umami stimuli. In both two-bottle preference tests and brief-access lick tests, TRPM4-only and TRPM5-only mice had impaired behavioral responses compared with wild-type mice. TRPM4/5-DKO mice did not demonstrate behavioral responses to any taste stimuli tested, suggesting that the loss of both TRPM4 and TRPM5 abolished their ability to detect the bitter, sweet, or umami stimuli that we tested. Taken with the live cell imaging data, we conclude that both TRPM4 and TRPM5 are required, and sufficient, for the normal transduction of bitter, sweet, and umami taste stimuli.
Materials and Methods
Mice.
Animals were cared for in compliance with the University at Buffalo Institutional Animal Care and Use Committee. We used the following transgenic mice for the experiments: TRPM5-GFP (49), TRPM4-KO (50), TRPM5-KO (22), and TRPM4/5-DKO mice, which are all in the C57BL/6 background. All transgenic mice were generously provided by Robert Margolskee, Monell Chemical Senses Center, Philadelphia. All mice used in the experiments were between 2 and 6 mo of age, and both sexes were used for imaging experiments and immunohistochemical analyses. Animals were kept on a 12-h dark/12-h light cycle. No more than three mice were kept per cage. Mice were only used for a single experiment and were not subjected to any prior analysis.
Immunohistochemistry.
Mice were transcardially perfused, and tongues were collected. Forty-micrometer sections of the CV papillae were cut and then subjected to immunohistochemical analysis. Further details are provided in SI Materials and Methods. Images were obtained with a Zeiss LSM 510 Meta NLO Confocal Microscope and were only adjusted for brightness and contrast.
The following primary antibodies were used with their research resource identifier (RRID) provided: anti–SNAP-25 (1:250, catalog no. 20-783-70323; GenWay Biotech, Inc.; RRID: AB-1024914), anti-TRPM4 (1:50, catalog no. OST00027W; Invitrogen; RRID: AB-1091055), anti-TRPM4 (1:50, catalog no. ab63080; Abcam; RRID: AB-956418), anti-PGP9.5 (1:100, catalog no. ab108986; Abcam; RRID: AB-10891773), anti-NTPDase2 [1:100 (51); RRID: AB-2314986], anti-PLCβ2 (1:250, catalog no. SC-206; Santa Cruz Biotechnology; RRID: AB-632197), anti-gustducin (1:100, catalog no. SC-395; Santa Cruz Biotechnology; RRID: AB-10177605), anti-IP3R3 (1:50, catalog no. 610313; BD Biosciences; RRID: AB-397705), and anti-TRPM5 [1:200 (40)]. Secondary antibodies were purchased from Jackson ImmunoResearch and used at a dilution of 1:250.
ImageJ (NIH) was used to measure the corrected immunofluorescence intensity of TRPM4 and TRPM5 in Fig. S2 and the corrected fluorescence intensity of TRPM5, gustducin, PLCβ2, IP3R3, and TRPM4 in Fig. S3. The area, mean gray value, and immunofluorescence intensity were measured in the area of interest and in five small areas selected as the background signal. The corrected integrated density was calculated as follows:
The two-tailed Student’s t test was done to determine the statistical difference between the immunofluorescence intensity of different mouse lines.
Live Cell Imaging.
Na+ imaging.
All measurements of intracellular Na+ were performed in isolated taste receptor cells. Taste cells were isolated and then loaded for 20 min with the Na+ indicator dye Asante NaTrium-2 (TEFLabs, Inc.) containing the nonionic dispersing agent Pluronic F-127 (Invitrogen, ThermoFisher Scientific). Loaded taste cells were visualized using an Olympus IX73 microscope with a 40× oil immersion lens, and images were captured using a Hamamatsu ORCA-03G camera. Cells were excited at 488 nm, and images were captured at 540 nm. During experiments, the cells were kept under constant perfusion and images were collected every 1 s using Imaging Workbench (Indec Biosystems). Experiments were graphed and analyzed using Origin 2016 software. Further details are provided in SI Materials and Methods.
Peak responses and response magnitudes were averaged and plotted with SEMs. Rise times to the peak responses were also averaged and plotted with SEMs. Data were analyzed for statistically significant differences using either a two-sided Student’s t test or a one-way analysis of variance (ANOVA) with a Bonferroni’s post hoc analysis as appropriate. Taste cell responsiveness was compared using a χ2 analysis to determine if there were any significant differences between wild-type, TRPM4-KO, TRPM5-KO, and TRPM4/5-DKO mice. For all analyses, a significance level of P < 0.05 was used and SEMs were reported. In all figures, statistical significance is labeled as follows: *P < 0.05, **P < 0.01, and ***P < 0.001. Sample numbers varied between experiments due to the variability in the responsiveness of the cells to particular stimuli. Before analysis, data were evaluated for normal distribution using the Shapiro–Wilk test and were determined to be normally distributed at P < 0.05.
Dual Ca2+ and Na+ imaging.
Cells were collected according to the procedures described above and were loaded simultaneously with Fura 2-AM (Invitrogen) and Asante NaTrium-2. The cells were excited at 340-, 380-, and 488-nm excitation wavelengths. The dual Ca2+ and Na+ images were captured every 4 s using a multiedge dichroic beam-splitter that captures emission at both 510 and 540 nm and Imaging Workbench (Indec Biosystems). Experiments were graphed and analyzed using Origin 2016 software.
Behavioral Experiments.
Two-bottle preference test.
Mice from C57BL/6 (wild-type), TRPM5-only, TRPM4-only, and TRPM4/5-DKO lines were subjected to two-bottle preference tests in blind conditions in which each mouse tested was not identified by genotype during the analysis. Each test concentration was presented simultaneously with water for a total of 48 h. Test solutions were switched with water every 24 h to control for side preferences. Preference ratios were calculated as volume of taste solution intake/total volume intake (taste solution + water). Five male mice from each background were used, for a total of 20 mice. Two taste stimuli for each taste quality were tested: sweet [sucrose (10, 50, 100, 300, and 500 mM) and SC45647 (0.001, 0.01, 0.05, and 0.1 mM)], bitter [Den (0.1, 0.5, 1, 5, 10, and 20 mM) and quinine hydrochloride (0.001, 0.01, 0.1, 1, and 5 mM)], and umami [MPG (1, 10, 50, 100, and 300 mM) and inosine monophosphate (0.01, 0.1, 1, 10, and 50 mM)]. Preference ratios between different groups of mice were compared using repeated-measures two-way ANOVA, with a Bonferroni’s post hoc analysis and two-sided Student’s t test used to determine significant differences for each concentration. Average values were plotted with the SD for each stimulus concentration. The significance level was set at P < 0.05.
Analysis of licking behavior.
We recorded the unconditioned licking responses to varying concentrations of taste stimuli in a test chamber designed to measure brief-access licking (Davis MS80 Rig; Dilog Instruments and Systems). Each individual lick was detected by a contact lickometer and recorded on a computer via DavisPro collection software (Dilog Instruments and Systems). Mice were tested on varying concentrations of SC45647, quinine hydrochloride, and MSG in 10 μM amiloride, in that order. Animals were water-deprived for 22 h for all testing except SC45647, for which water-replete animals were tested. Once an animal began licking the tube, it was allowed 10 s of access before the shutter closed. Further details are provided in SI Materials and Methods. Lick scores and licks relative to water are compared by repeated-measures ANOVA, with genotype as a between-factor variable and concentration as a repeated-measures within-factor variable. Significant interaction terms were followed by Tukey’s honestly significant difference tests. Statistical analyses were performed in Statistica.
Supplementary Material
Acknowledgments
We thank Dr. Robert Margolskee for generously providing the transgenic mice and comments on the manuscript. We also thank Dr. Stefan G. E. Roberts for comments on the manuscript and Angela Wirth, Amy R. Nelson, Folake Olaleye, and Tianyi Zhou for technical assistance. Generation of the TRPM4-KO mouse was supported by Deutsche Forschungsgemeinschaft grants, including Grants SFB 1118 (German Centre for Cardiovascular Research and German Ministry of Education and Research), TR-SFB 152, and FOR 2289 (FR1638/3-1) (to M.F.). This work was supported by National Science Foundation Grant 1256950 (to K.F.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718802115/-/DCSupplemental.
References
- 1.Roper SD. Signal transduction and information processing in mammalian taste buds. Pflugers Arch. 2007;454:759–776. doi: 10.1007/s00424-007-0247-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liman ER, Zhang YV, Montell C. Peripheral coding of taste. Neuron. 2014;81:984–1000. doi: 10.1016/j.neuron.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rössler P, Kroner C, Freitag J, Noè J, Breer H. Identification of a phospholipase C beta subtype in rat taste cells. Eur J Cell Biol. 1998;77:253–261. doi: 10.1016/s0171-9335(98)80114-3. [DOI] [PubMed] [Google Scholar]
- 4.McLaughlin SK, McKinnon PJ, Margolskee RF. Gustducin is a taste-cell-specific G protein closely related to the transducins. Nature. 1992;357:563–569. doi: 10.1038/357563a0. [DOI] [PubMed] [Google Scholar]
- 5.Liu D, Liman ER. Intracellular Ca2+ and the phospholipid PIP2 regulate the taste transduction ion channel TRPM5. Proc Natl Acad Sci USA. 2003;100:15160–15165. doi: 10.1073/pnas.2334159100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, et al. Coding of sweet, bitter, and umami tastes: Different receptor cells sharing similar signaling pathways. Cell. 2003;112:293–301. doi: 10.1016/s0092-8674(03)00071-0. [DOI] [PubMed] [Google Scholar]
- 7.Pérez CA, et al. A transient receptor potential channel expressed in taste receptor cells. Nat Neurosci. 2002;5:1169–1176. doi: 10.1038/nn952. [DOI] [PubMed] [Google Scholar]
- 8.Clapp TR, Stone LM, Margolskee RF, Kinnamon SC. Immunocytochemical evidence for co-expression of type III IP3 receptor with signaling components of bitter taste transduction. BMC Neurosci. 2001;2:6. doi: 10.1186/1471-2202-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernhardt SJ, Naim M, Zehavi U, Lindemann B. Changes in IP3 and cytosolic Ca2+ in response to sugars and non-sugar sweeteners in transduction of sweet taste in the rat. J Physiol. 1996;490:325–336. doi: 10.1113/jphysiol.1996.sp021147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spielman AI, et al. Rapid kinetics of second messenger production in bitter taste. Am J Physiol. 1996;270:C926–C931. doi: 10.1152/ajpcell.1996.270.3.C926. [DOI] [PubMed] [Google Scholar]
- 11.Finger TE, et al. ATP signaling is crucial for communication from taste buds to gustatory nerves. Science. 2005;310:1495–1499. doi: 10.1126/science.1118435. [DOI] [PubMed] [Google Scholar]
- 12.Taruno A, et al. CALHM1 ion channel mediates purinergic neurotransmission of sweet, bitter and umami tastes. Nature. 2013;495:223–226. doi: 10.1038/nature11906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medler KF, Margolskee RF, Kinnamon SC. Electrophysiological characterization of voltage-gated currents in defined taste cell types of mice. J Neurosci. 2003;23:2608–2617. doi: 10.1523/JNEUROSCI.23-07-02608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeFazio RA, et al. Separate populations of receptor cells and presynaptic cells in mouse taste buds. J Neurosci. 2006;26:3971–3980. doi: 10.1523/JNEUROSCI.0515-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richter TA, Caicedo A, Roper SD. Sour taste stimuli evoke Ca2+ and pH responses in mouse taste cells. J Physiol. 2003;547:475–483. doi: 10.1113/jphysiol.2002.033811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venkatachalam K, Montell C. TRP channels. Annu Rev Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishimaru Y, et al. Transient receptor potential family members PKD1L3 and PKD2L1 form a candidate sour taste receptor. Proc Natl Acad Sci USA. 2006;103:12569–12574. [Google Scholar]
- 18.LopezJimenez ND, et al. Two members of the TRPP family of ion channels, Pkd1l3 and Pkd2l1, are co-expressed in a subset of taste receptor cells. J Neurochem. 2006;98:68–77. doi: 10.1111/j.1471-4159.2006.03842.x. [DOI] [PubMed] [Google Scholar]
- 19.Lyall V, et al. The mammalian amiloride-insensitive non-specific salt taste receptor is a vanilloid receptor-1 variant. J Physiol. 2004;558:147–159. doi: 10.1113/jphysiol.2004.065656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofmann T, Chubanov V, Gudermann T, Montell C. TRPM5 is a voltage-modulated and Ca(2+)-activated monovalent selective cation channel. Curr Biol. 2003;13:1153–1158. doi: 10.1016/s0960-9822(03)00431-7. [DOI] [PubMed] [Google Scholar]
- 21.Prawitt D, et al. TRPM5 is a transient Ca2+-activated cation channel responding to rapid changes in [Ca2+]i. Proc Natl Acad Sci USA. 2003;100:15166–15171. doi: 10.1073/pnas.2334624100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Damak S, et al. Trpm5 null mice respond to bitter, sweet, and umami compounds. Chem Senses. 2006;31:253–264. doi: 10.1093/chemse/bjj027. [DOI] [PubMed] [Google Scholar]
- 23.Ohkuri T, et al. Multiple sweet receptors and transduction pathways revealed in knockout mice by temperature dependence and gurmarin sensitivity. Am J Physiol Regul Integr Comp Physiol. 2009;296:R960–R971. doi: 10.1152/ajpregu.91018.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eddy MC, et al. A conditioned aversion study of sucrose and SC45647 taste in TRPM5 knockout mice. Chem Senses. 2012;37:391–401. doi: 10.1093/chemse/bjr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clapham DE. TRP channels as cellular sensors. Nature. 2003;426:517–524. doi: 10.1038/nature02196. [DOI] [PubMed] [Google Scholar]
- 26.Guinamard R, Demion M, Launay P. Physiological roles of the TRPM4 channel extracted from background currents. Physiology (Bethesda) 2010;25:155–164. doi: 10.1152/physiol.00004.2010. [DOI] [PubMed] [Google Scholar]
- 27.Guinamard R, Sallé L, Simard C. The non-selective monovalent cationic channels TRPM4 and TRPM5. Adv Exp Med Biol. 2011;704:147–171. doi: 10.1007/978-94-007-0265-3_8. [DOI] [PubMed] [Google Scholar]
- 28.Launay P, et al. TRPM4 is a Ca2+-activated nonselective cation channel mediating cell membrane depolarization. Cell. 2002;109:397–407. doi: 10.1016/s0092-8674(02)00719-5. [DOI] [PubMed] [Google Scholar]
- 29.Liu P, Shah BP, Croasdell S, Gilbertson TA. Transient receptor potential channel type M5 is essential for fat taste. J Neurosci. 2011;31:8634–8642. doi: 10.1523/JNEUROSCI.6273-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shandilya J, Gao Y, Nayak TK, Roberts SG, Medler KF. AP1 transcription factors are required to maintain the peripheral taste system. Cell Death Dis. 2016;7:e2433. doi: 10.1038/cddis.2016.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hisatsune C, et al. Abnormal taste perception in mice lacking the type 3 inositol 1,4,5-trisphosphate receptor. J Biol Chem. 2007;282:37225–37231. doi: 10.1074/jbc.M705641200. [DOI] [PubMed] [Google Scholar]
- 32.Sukumaran SK, et al. Whole transcriptome profiling of taste bud cells. Sci Rep. 2017;7:7595. doi: 10.1038/s41598-017-07746-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewandowski BC, Sukumaran SK, Margolskee RF, Bachmanov AA. Amiloride-insensitive salt taste is mediated by two populations of type III taste cells with distinct transduction mechanisms. J Neurosci. 2016;36:1942–1953. doi: 10.1523/JNEUROSCI.2947-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guinamard R, Hof T, Del Negro CA. The TRPM4 channel inhibitor 9-phenanthrol. Br J Pharmacol. 2014;171:1600–1613. doi: 10.1111/bph.12582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sagredo AI, et al. TRPM4 regulates Akt/GSK3-beta activity and enhances beta-catenin signaling and cell proliferation in prostate cancer cells. Mol Oncol. 2017 doi: 10.1002/1878-0261.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grand T, et al. 9-phenanthrol inhibits human TRPM4 but not TRPM5 cationic channels. Br J Pharmacol. 2008;153:1697–1705. doi: 10.1038/bjp.2008.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmer RK, et al. Triphenylphosphine oxide is a potent and selective inhibitor of the transient receptor potential melastatin-5 ion channel. Assay Drug Dev Technol. 2010;8:703–713. doi: 10.1089/adt.2010.0334. [DOI] [PubMed] [Google Scholar]
- 38.Sclafani A, Marambaud P, Ackroff K. Sucrose-conditioned flavor preferences in sweet ageusic T1r3 and Calhm1 knockout mice. Physiol Behav. 2014;126:25–29. doi: 10.1016/j.physbeh.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sato T, Nishishita K, Okada Y, Toda K. The receptor potential of frog taste cells in response to cold and warm stimuli. Chem Senses. 2010;35:491–499. doi: 10.1093/chemse/bjq039. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Z, Zhao Z, Margolskee R, Liman E. The transduction channel TRPM5 is gated by intracellular calcium in taste cells. J Neurosci. 2007;27:5777–5786. doi: 10.1523/JNEUROSCI.4973-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ullrich ND, et al. Comparison of functional properties of the Ca2+-activated cation channels TRPM4 and TRPM5 from mice. Cell Calcium. 2005;37:267–278. doi: 10.1016/j.ceca.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Taruno A, Matsumoto I, Ma Z, Marambaud P, Foskett JK. How do taste cells lacking synapses mediate neurotransmission? CALHM1, a voltage-gated ATP channel. BioEssays. 2013;35:1111–1118. doi: 10.1002/bies.201300077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chubanov V, et al. Disruption of TRPM6/TRPM7 complex formation by a mutation in the TRPM6 gene causes hypomagnesemia with secondary hypocalcemia. Proc Natl Acad Sci USA. 2004;101:2894–2899. doi: 10.1073/pnas.0305252101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lintschinger B, et al. Coassembly of Trp1 and Trp3 proteins generates diacylglycerol- and Ca2+-sensitive cation channels. J Biol Chem. 2000;275:27799–27805. doi: 10.1074/jbc.M002705200. [DOI] [PubMed] [Google Scholar]
- 45.Xu XZ, Li HS, Guggino WB, Montell C. Coassembly of TRP and TRPL produces a distinct store-operated conductance. Cell. 1997;89:1155–1164. doi: 10.1016/s0092-8674(00)80302-5. [DOI] [PubMed] [Google Scholar]
- 46.Cheng H, et al. TRPM4 controls insulin secretion in pancreatic beta-cells. Cell Calcium. 2007;41:51–61. doi: 10.1016/j.ceca.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Launay P, et al. TRPM4 regulates calcium oscillations after T cell activation. Science. 2004;306:1374–1377. doi: 10.1126/science.1098845. [DOI] [PubMed] [Google Scholar]
- 48.Leitner MG, et al. Direct modulation of TRPM4 and TRPM3 channels by the phospholipase C inhibitor U73122. Br J Pharmacol. 2016;173:2555–2569. doi: 10.1111/bph.13538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clapp TR, Medler KF, Damak S, Margolskee RF, Kinnamon SC. Mouse taste cells with G protein-coupled taste receptors lack voltage-gated calcium channels and SNAP-25. BMC Biol. 2006;4:7. doi: 10.1186/1741-7007-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vennekens R, et al. Increased IgE-dependent mast cell activation and anaphylactic responses in mice lacking the calcium-activated nonselective cation channel TRPM4. Nat Immunol. 2007;8:312–320. doi: 10.1038/ni1441. [DOI] [PubMed] [Google Scholar]
- 51.Bartel DL, Sullivan SL, Lavoie EG, Sévigny J, Finger TE. Nucleoside triphosphate diphosphohydrolase-2 is the ecto-ATPase of type I cells in taste buds. J Comp Neurol. 2006;497:1–12. doi: 10.1002/cne.20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.