Significance
B lymphocytes change the type of antibody they express to combat infections though a DNA breakage and joining process termed antibody class switch recombination (CSR). During CSR, DNA breaks are introduced into two specific regions of the antibody locus, and these two DNA breaks are joined to make the new form of the antibody gene. Like other genomic breaks, CSR DNA breaks activate a cellular DNA damage response pathway that helps ensure their proper repair. We now show that, when certain components of the DNA damage response pathway are inactivated, B-cell CSR junctions show different molecular signatures, indicating that they are repaired by a less-efficient alternative DNA repair pathway instead of the normal general cellular DNA break repair pathway.
Keywords: class switch recombination, DNA repair, microhomology-mediated end-joining, nonhomologous end-joining
Abstract
Ig heavy chain (IgH) class switch recombination (CSR) in B lymphocytes switches IgH constant regions to change antibody functions. CSR is initiated by DNA double-strand breaks (DSBs) within a donor IgH switch (S) region and a downstream acceptor S region. CSR is completed by fusing donor and acceptor S region DSB ends by classical nonhomologous end-joining (C-NHEJ) and, in its absence, by alternative end-joining that is more biased to use longer junctional microhomologies (MHs). Deficiency for DSB response (DSBR) factors, including ataxia telangiectasia-mutated (ATM) and 53BP1, variably impair CSR end-joining, with 53BP1 deficiency having the greatest impact. However, studies of potential impact of DSBR factor deficiencies on MH-mediated CSR end-joining have been technically limited. We now use a robust DSB joining assay to elucidate impacts of deficiencies for DSBR factors on CSR and chromosomal translocation junctions in primary mouse B cells and CH12F3 B-lymphoma cells. Compared with wild-type, CSR and c-myc to S region translocation junctions in the absence of 53BP1, and, to a lesser extent, other DSBR factors, have increased MH utilization; indeed, 53BP1-deficient MH profiles resemble those associated with C-NHEJ deficiency. However, translocation junctions between c-myc DSB and general DSBs genome-wide are not MH-biased in ATM-deficient versus wild-type CH12F3 cells and are less biased in 53BP1- and C-NHEJ−deficient cells than CSR junctions or c-myc to S region translocation junctions. We discuss potential roles of DSBR factors in suppressing increased MH-mediated DSB end-joining and features of S regions that may render their DSBs prone to MH-biased end-joining in the absence of DSBR factors.
Mature B cells undergo Ig heavy chain (IgH) class switch recombination (CSR) to change the constant region of IgH chains and, thereby, modulate antibody effector functions. In mice, six sets of CH exons lie 100 kb to 200 kb downstream of the initially expressed CH exon (Cμ) and are expressed in the context of CSR; these sets of downstream CH exons include Cγ3, Cγ1, Cγ2b, Cγ2a, Cε, and Cα (1). Each set of CH exons is flanked upstream by a long (1 kb to 10 kb), repetitive DNA Switch (S) region that contains large numbers of short DNA motifs that are targets for activation-induced cytidine deaminase (AID) (2). AID initiates CSR by introducing DNA cytidine deamination lesions in donor Sμ and into downstream acceptor S regions, which are then converted into DNA double-strand breaks (DSBs) by coopted activities of base excision and/or mismatch repair pathways (2). Productive CSR is completed by deletional joining of the upstream end of a donor Sμ DSB to the downstream end of an acceptor S-region DSB, in a CSR reaction that is mechanistically enforced to produce productive deletional joining CSR events at a much greater frequency (10- to 20-fold) than nonproductive inversional joining events (3).
Normal CSR DSB end-joining is carried out predominantly by classical nonhomologous end-joining (C-NHEJ) (1, 4). Consistent with C-NHEJ, many CSR junctions in normal B cells occur via direct (“blunt”) DSB end-joining (up to 30% depending on the S region), with the remainder involving joining of DSB ends via short (1 bp to 4 bp) overlaps of microhomology (MH) (4). B cells deficient for core C-NHEJ components (e.g., XRCC-4, ligase 4, Ku70, or Ku80) have moderately decreased (40 to 50% of normal) CSR and increased AID-dependent IgH chromosomal breaks; CSR junctions in such core C-NHEJ−deficient B cells are formed via alternative end-joining (A-EJ) and are mostly (90% or more) MH-mediated (4–7). CSR DSBs trigger the ataxia telangiectasia-mutated (ATM)-dependent DSB response (DSBR) in which the ATM kinase phosphorylates downstream substrates, including histone H2AX and 53BP1, which accumulate in foci surrounding DSBs (8). Deficiency for ATM or H2AX moderately impairs CSR (30 to 50% of normal) with a corresponding increased accumulation of IgH breaks, indicating a role for these proteins in CSR end-joining (1, 8). In contrast, 53BP1 deficiency more profoundly impairs CSR (5% of normal), also accompanied by increased IgH breaks (1, 8). The ATM DSBR likely promotes CSR end-joining by tethering isolated S-region DSBs for rejoining, thereby maintaining an intact chromosomal substrate for productive CSR joining (1, 3, 9). DSBR factors also function to prevent S-region DSB end resections that could generate long overhanging ends not suitable for C-NHEJ, but which may expose MH-rich substrates in the form of short S-region repeat sequences for A-EJ (3, 10, 11). In this regard, the greater impact of 53BP1 deficiency on CSR joining may reflect both impairment of end-tethering and substantial activation of increased DSB resection, due to inability to activate the associated Rif1 resection-blocking factor (3).
The question of whether or not DSBR factor deficiencies promote increased MH usage in CSR has not been firmly resolved. A number of studies concluded that deficiency of ATM (12), H2AX (13), 53BP1 (11, 14), or Rif1 (15) did not result in altered MH usage in CSR junctions compared with that of WT B cells. On the other hand, other studies reported that ATM deficiency did lead to increased MH usage in mouse and human CSR junctions (16, 17). A major limitation with all prior studies of CSR end-joining in DSBR factor-deficient B cells is that, due to the technology then available for analysis, the long repetitive nature of S regions allowed assessment of only relatively short sequences at the peripheral edges of S regions lying between two PCR primers. As a result, such PCR-based CSR junction studies yielded a low number of junctions that do not come from the repetitive S-region cores where the majority of normal CSR junctions arise. In addition, conclusions of many prior studies were based on relatively small sample sizes, again based on limitations of technologies available at the time.
We have developed and refined high-throughput, genome-wide, translocation sequencing (HTGTS) to map, at nucleotide resolution, junctions between HTGTS “bait” DSBs and endogenous DSBs genome-wide (18–20). HTGTS bait DSBs may be introduced ectopically by designer nucleases or may be provided by recurrent endogenous DSBs, including those that initiate IgH CSR or V(D)J recombination (3, 20). In the latter context, we adapted HTGTS for use as a robust endogenous CSR assay, by using a series of AID-initiated bait DSBs in the 5′Sμ region to assay for AID-induced prey DSBs within downstream accepter S regions in CSR-activated normal primary B cells or CH12F3 B-lymphoma cells (3, 21). This HTGTS CSR assay offers many advantages compared with prior CSR end-joining assays, as it allows recovery of several orders of magnitude more CSR junctions and identifies CSR junctions from across the length of the repetitive acceptor S regions (3). To further elucidate the potential roles of DSBR factors in MH-mediated end-joining during CSR and chromosomal translocations, we analyzed junction patterns of large numbers of DSB junctions that occurred either during CSR or during translocations between Cas9/guide RNA (gRNA)-induced c-myc bait DSB to S-region DSBs or other general DSBs genome-wide in WT and DSBR factor-deficient primary B cells and CH12F3 cells activated for CSR.
Results
HTGTS Analysis of CSR Junction Profiles.
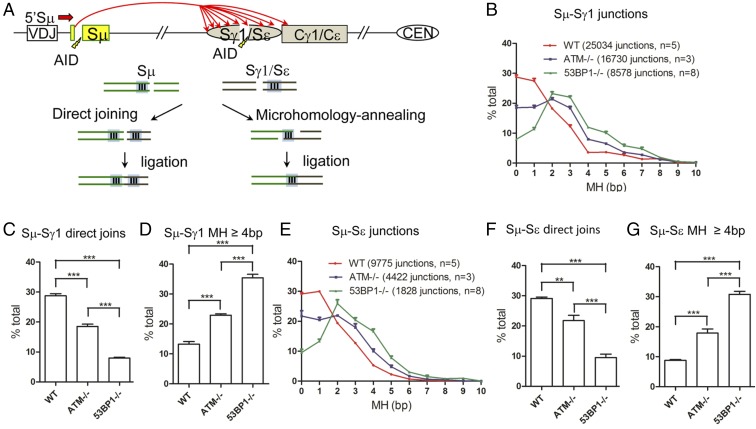
We studied the joining pattern of AID-initiated S-region breaks during CSR in mouse splenic B cells activated in culture with αCD40/IL-4 to stimulate CSR between Sμ and Sγ1 or Sε. We first analyzed tens of thousands of CSR junctions recovered in our prior HTGTS-based CSR orientation dependence studies that compared WT versus DSBR-deficient B cells (3). These HTGTS CSR assays were based on the use of 5′Sμ primer that allowed a series of AID-initiated DSBs in the very 5′ region of Sμ to serve as a combined HTGTS bait for capturing joins to AID-induced prey DSBs within the downstream core Sγ1 or Sε regions (Fig. 1A). CSR junctions analyzed in the current study can be broken into two general classes (4). The first is “direct” joins made up of bait sequences followed directly by prey sequences with no overlapping nucleotides. The second is “MH-mediated” joins that are made up of junctions with ≥1 bp of overlapping nucleotides that are most likely provided within overhanging flaps of resected bait and prey ends (Fig. 1A). We also recovered a significant number (about 20%) of junctions with ≥1 bp of inserted sequence, potentially resulting from PolQ activity (22) or unknown sources, from which direct versus MH-mediated joining mechanisms cannot be distinguished (SI Appendix, Table S1). We analyzed thousands of junctions and expressed the percentage of direct junctions and the number of junctions with various MH lengths for a given S region as a percentage of the total junctions to that S region (Fig. 1 B and E). Results for these experiments were highly reproducible (Fig. 1 B and E and SI Appendix, Fig. S1 and Table S2).
Fig. 1.
Increased MH usage in Sμ−Sγ1 and Sμ−Sε CSR junctions from DSBR factor-deficient primary B cells. (A) Illustration of joining outcomes of Sμ region DSBs to downstream Sγ1 and Sε region breaks undergoing direct or MH-mediated joining. WT, ATM−/,−and 53BP1−/− primary splenic B cells were stimulated with αCD40/IL4 for 96 h. (B and E) MH usage from junctions with direct and up to 10-bp MH for 5′Sμ (B) to Sγ1 and (E) to Sε DSBs are plotted as percentage of total junctions. (C, D, F, and G) Percentages of direct joins of 5′Sμ to (C) Sγ1 and (F) Sε and percentage of junctions with 4-bp or longer MH in (D) Sγ1 or (G) Sε in different genetic backgrounds are compared. Unpaired two-tailed t test was used to calculate P values for significant difference between samples (quantitative data = average ± SEM; **P ≤ 0.01, ***P ≤ 0.001). At least three independent samples were used for each experiment. SI Appendix, Table S2 lists detailed information, including junction numbers and percentage of direct versus MH-mediated joins found for each of the nonsized matched libraries analyzed to generate the data described in this figure. We also obtained highly similar results upon analyzing three replicate libraries for each genotype in which Sμ−Sγ1 or Sμ−Se junctional sequence numbers from each experimental sample were normalized via random selection to that of the experimental sample with the smallest number of recovered junctions (see SI Appendix, Fig. S1 for details).
Direct Versus MH-Mediated CSR Junctions in WT Versus DSBR-Deficient Splenic B Cells.
Total (combined deletional and inversional) Sμ–Sγ1 and Sμ–Sε CSR junctions in WT anti-CD40 plus IL-4–treated B cells were ∼30% direct, with most of the others using 1 bp to 3 bp of MH and a small fraction using longer MHs (Fig. 1 B and E). In contrast, ATM-deficient B cells had a marked increase in MH-mediated Sμ–Sγ1 and Sμ–Sε junctions, with about 20% of each being direct (Fig. 1 C and F) and a correspondingly increased fraction of MH-mediated junctions (Fig. 1 B and E). Notably, the severely CSR-impaired 53BP1-deficient B cells had the greatest decrease in direct junctions, with less than 10% of Sμ–Sγ1 and Sμ–Sε CSR junctions being direct (Fig. 1 C and F) and a significantly increased fraction of MH-mediated junctions (Fig. 1 B and E). H2AX-deficient and Rif1-deficient B cells displayed ∼20% direct joins and an overall pattern of CSR junctions very similar to that of ATM-deficient B cells and distinct from that of 53BP1-deficient B cells (SI Appendix, Fig. S2). In this regard, WT cells had an average MH length of 1.67 bp for Sμ–Sγ1 and 1.48 bp for Sμ–Sε CSR junctions, while, at the other extreme, 53BP1-deficient B cells had average MH length of 3.17 bp and 2.8 bp for these two sets of junctions (SI Appendix, Table S3). The ATM, H2AX, and Rif1-deficient B cells had CSR junctions with increased average MH lengths that were intermediated between WT and 53BP1-deficient B cells (SI Appendix, Table S3). Finally, we also assessed relative MH usage in deletional versus inversional Sμ–Sγ1 and Sμ–Sε junctions in WT versus DSBR-deficient genotypes and observed the same general patterns, indicating that the usage of direct versus MH-mediated end-joining does not markedly contribute to inversional versus deletional CSR recombination (SI Appendix, Table S3).
To further elucidate the influence of the various DSB response factor deficiencies on end-joining used to form CSR junctions, we assessed the relative percentage of Sμ–Sγ1 and/or Sμ–Sε CSR junctions for each genotype that utilized MH of 4 bp or longer, a length considered not favorable for joining via C-NHEJ (23). Consistent with predominant joining of CSR junctions via C-NHEJ in WT cells, only about 12% and 8% of Sμ–Sγ1 and Sμ–Sε junctions, respectively, reached the 4-bp or longer length (Fig. 1 D and G). In contrast, relative levels of these longer MH-mediated junctions from ATM-deficient B cells were increased to about 24% and 20%, respectively. for Sμ–Sγ1 and Sμ–Sε junctions (Fig. 1 D and G), and H2AX- and Rif1-deficient B cells showed similar increases (SI Appendix, Fig. S2 C, F, and I). Notably, however, the levels of these long MH-mediated junctions were highest for the CSR junctions in 53BP1-deficient B cells, reaching 35% and 33%, respectively, for Sμ–Sγ1 and Sμ–Sε junctions, levels about 3 to 4 times greater than those observed for WT B cells (Fig. 1 D and G).
CSR Junctions in WT Versus DSBR- or Ligase 4-Deficient CH12F3 Cells.
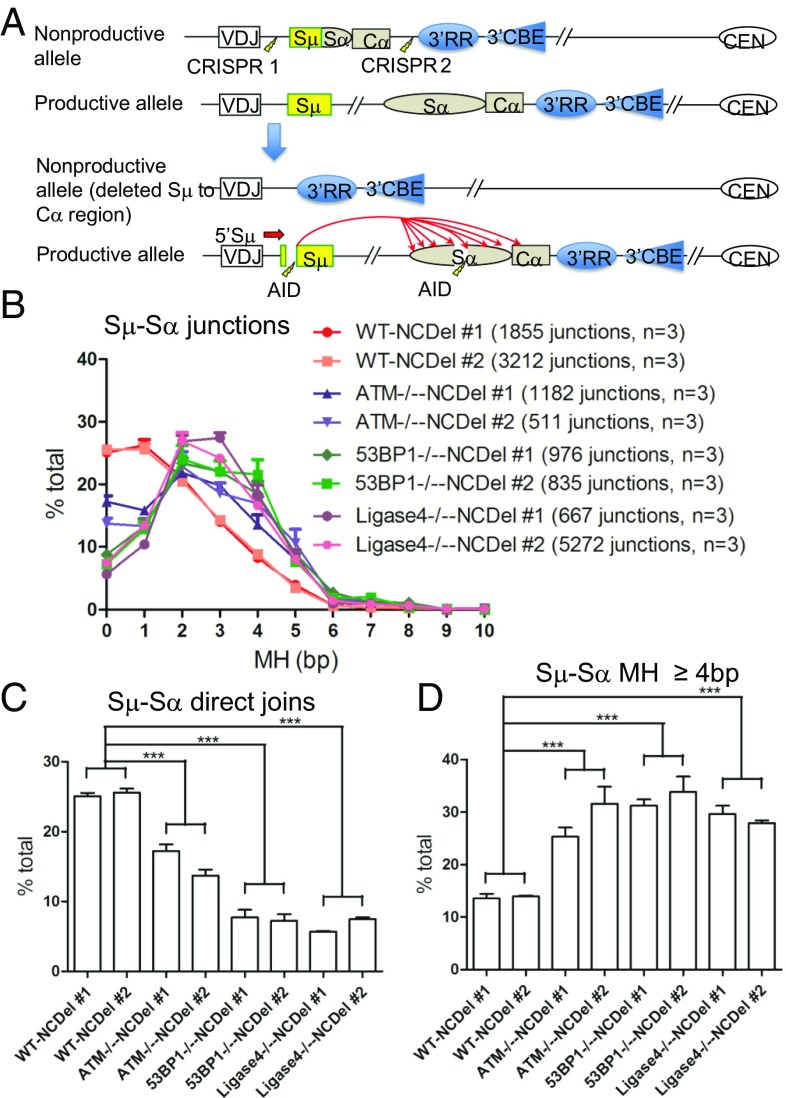
We examined relative levels of direct and MH-mediated CSR junctions in Sα through studies of mouse CH12F3 B-lymphoma cells which can be stimulated to undergo robust CSR to IgA upon stimulation with αCD40/IL4/TGF-β in culture (24). For this purpose, we generated ATM-deficient and 53BP1-deficient CH12F3 cells by CRISPR-Cas9–mediated gene deletion (SI Appendix, Fig. S3 A–D). To obviate potentially confounding effects of the previously IgA switched nonproductive IgH allele on assessment of CSR junctions, we also deleted the preswitched Sμ−Sα region of nonproductive IgH allele in all genotypes (Fig. 2A and SI Appendix, Fig. S3E). To account for clonal variation in CH12F3 cells, we assayed two independently derived cell lines for each genotype. Following 72 h of αCD40/IL4/TGF-β stimulation, the ATM- and 53BP1-deficient CH12F3 cells showed CSR levels that, respectively, were on, average, ∼35% and 25% of WT IgA class-switching levels (SI Appendix, Fig. S3F). Consistent with known clonal variation of CSR in CH12F3 cell lines (25), we observed significant variation in overall IgH class switching levels between two independent clones of each genotype (SI Appendix, Fig. S3F).
Fig. 2.
Increased MH usage in Sμ−Sα CSR junctions from WT, ATM-, 53BP1-, and ligase 4-deficient CH12F3 cells. (A) Illustration of the construction of nonproductive allele-deleted CH12F3 cells and the HTGTS strategy to assay the Sμ−Sα junctions with 5′Sμ bait after 72-h stimulation with αCD40/IL4/TGF-β. (B) MH usage in junctions from joining AID-induced 5′Sμ breaks to Sα breaks in nonproductive allele-deleted WT, ATM−/−, 53BP1−/−, and ligase 4−/− CH12F3 cells were plotted as percentage of junctions with indicated length of MH over the total number of junctions in Sα region. (C and D) Percentage of (C) direct junctions and (D) junctions with 4-bp and longer MH in Sμ−Sα joining in nonproductive allele-deleted WT, ATM−/−, 53BP1−/−, and ligase 4−/− CH12F3 cells was plotted. Two-tailed t test was used for statistical analysis (quantitative data = average ± SEM; ***P ≤ 0.001). Three independent samples were used for each experiment, and details are provided in SI Appendix, Table S4.
We used the HTGTS CSR assay to recover 5′Sμ to Sα junctions from the two sets of stimulated WT and mutant CH12F3 cells and assayed MH usage in recovered junctions from three independent experiments for each line; these experiments gave consistent results between independent clones of each genotype (Fig. 2 B–D and SI Appendix, Table S4). Total Sμ−Sα CSR junctions in WT CH12F3 cells were ∼25% direct, with most others using 1 bp to 4 bp of MH (Fig. 2B). In contrast, ATM-deficient CH12F3 cells had a marked increase in MH-mediated Sμ−Sα junctions, with only about 15% direct junctions (Fig. 2C). The 53BP1-deficient CH12F3 cells, as observed for 53BP1-deficient primary B cells, had the greatest decrease in direct Sμ−Sα junctions, with less than 10% being direct (Fig. 2C). ATM-deficient and 53BP1-deficient CH12F3 cells also had corresponding increases in the average MH length of the 5′Sμ to Sα junctions (SI Appendix, Table S5) and substantially increased frequency of 5′Sμ to Sα junctions that utilized MH of 4 bp or longer (Fig. 2D).
To directly compare the MH usage between DSBR factor-deficient and C-NHEJ−deficient B cells, we also recovered Sμ−Sα junctions from CH12F3 cells deficient for the core c-NHEJ ligase 4 factor in which we also deleted the preswitched Sμ−Sα region of the nonproductive IgH allele (SI Appendix, Fig. S3E). Compared with WT “matched” (i.e., preswitched Sμ−Sα deleted) CH12F3 cells, the ligase 4-deficient CH12F3 cells showed greatly increased MH usage, increased average MH usage, and increased frequency of Sμ−Sα junctions with more than 4 bp of MH (Fig. 2 B–D and SI Appendix, Table S5). For each measured parameter, values were remarkably similar to those of matched 53BP1-deficient CH12F3 cells (Fig. 2 B–D and SI Appendix, Table S5). Finally, compared with WT or ATM-deficient cells, the 53BP1- and ligase 4-deficient CH12F3 cells also both had a significant fraction of directional junctions mapping to nonrepetitive 5′ and 3′ regions adjacent to Sα (SI Appendix, Fig. S4A), which we previously have concluded occur in the context of long resections of Sα DSBs (3). As shown previously (3), WT, ATM-, and 53BP1-deficient CH12F3 cell CSR junctions, respectively, showed an increasing contribution of long CSR resections and corresponding loss of orientation-dependent joining bias (SI Appendix, Fig. S4 B and C and Tables S6 and S7). Notably, despite similarity to 53BP1-deficient cells in MH-mediated CSR joins, ligase 4-deficient CH12F3 cells showed only intermediately increased long resections and a more modest impact on orientation-dependent joining (SI Appendix, Fig. S4 B and C and Tables S6 and S7).
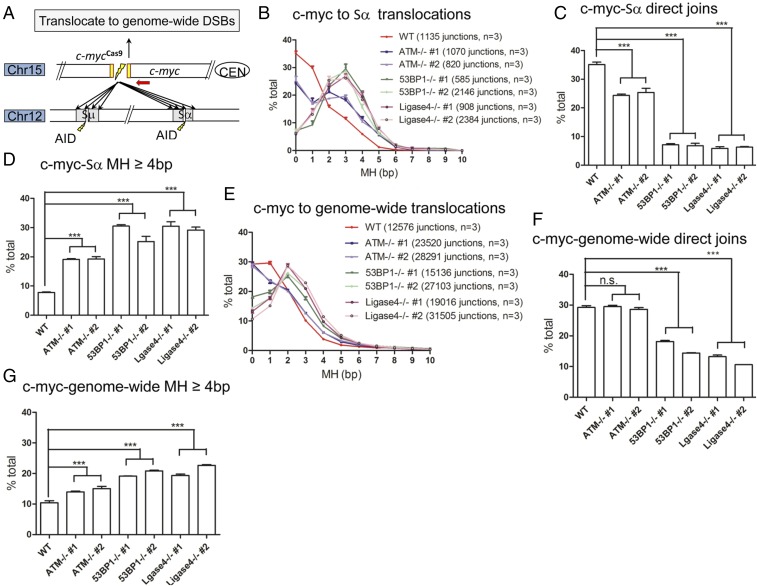
Impact of DSBR Deficiency on Translocation Junctions to Genome-Wide Versus CSR DSBs.
To determine whether increased MH usage in CSR junctions observed in DSBR factors-deficient CH12F3 cells extends to other genome-wide DSBs, we introduced a CRISPR-Cas9−mediated DSB at c-myc locus on chromosome 15 and employed HTGTS to assay translocation junctions to Sμ and Sα DSBs and other DSBs genome-wide. We performed these studies in αCD40/IL4/TGF-β−stimulated WT CH12F3 cells and two independently derived clones each of ATM-, 53BP1-, and ligase 4-deficient CH12F3 cells, all with the nonproductive Sμ−Sα allele intact (Fig. 3A). Junctions between the c-myc bait DSB to AID-initiated Sμ and Sα DSBs were remarkably similar, for each assayed genotype, to those of Sμ−Sα CSR junctions in terms of overall junctional profiles, frequency of direct joins, average MH usage, and frequency of junctions with more than 4 bp of MH (Fig. 3 B–D and SI Appendix, Fig. S5 and Tables S8 and S9). In all characteristics of joining in this set of assays, translocation junctions of 53BP1- and ligase 4-deficient CH12F3 cells were very similar to each other (Fig. 3 B–D and SI Appendix, Fig. S5). Similar to CSR junctions in 53BP1- and ligase 4-deficient cells, we found that a significant fraction of translocations between c-myc bait DSBs to the Sμ and, particularly, Sα locales, compared with those in other backgrounds, involved DSBs into nonrepetitive resection regions beyond the S regions (SI Appendix, Fig. S6).
Fig. 3.
Comparison of translocation junctions to S region or general DSBs in WT and mutant CH12F3 cells. WT, ATM-, 53BP1-, and ligase 4-deficient CH12F3 cells were stimulated with αCD40/IL4/TGF-β, and cmyc-Cas9 bait DSB was introduced 12 h after stimulation. HTGTS libraries were made with c-myc-Cas9 DSB as bait, with genomic DNA harvested 72 h poststimulation. (A) Translocation outcomes of c-myc-Cas9 DSB to genome-wide or Sμ/Sα breaks are depicted. (B and E) MH usage in junctions from c-myc-Cas9 break joining to (B) Sα breaks and (E) genome-wide breaks were plotted as percentage of junctions with indicated length of MH over the total number of junctions in the respective regions. (C, D, F, and G) Percentages of direct joins from c-myc-Cas9 break joining to (C) Sα breaks and (F) genome-wide breaks, and percentages of junctions with 4 bp and longer MH from c-myc-Cas9 break to (D) Sα breaks and (G) genome-wide breaks in different genetic backgrounds are shown. Unpaired two-tailed t test was used for statistical analysis (quantitative data = average ± SEM; n.s. means P > 0.05; ***P ≤ 0.001). Three independent samples were used for each experiment, and details are provided in SI Appendix, Table S9.
For analysis of junction profiles of c-myc bait DSB translocations to general genome-wide DSBs in this same set of CH12F3 WT and mutant cells, we excluded both junctions mapping in the IgH locus to avoid S-region DSBs, as well as those in the 20-mb region surrounding the c-myc break site, to avoid break site junctions mediated mostly by resection (18, 19). We also excluded sequence at known AID hotspot junctions. These studies revealed that translocation junctions between the c-myc bait DSBs and general DSBs genome-wide in WT and ATM-deficient CH12F3 cells were indistinguishable with respect to frequency of direct joins (Fig. 3 E and F). In contrast, 53BP1- and ligase 4-deficient CH12F3 cells had significantly reduced direct genome-wide translocation junctions compared with those of WT and ATM-deficient cells (Fig. 3 E and F). Despite similar levels of direct joins with WT cells, ATM-deficient cells had a slightly increased frequency of c-myc to general DSB translocation junctions with greater than 4 bp MH, and this class of junctions was further increased in 53BP1- and ligase 4-deficient cells (Fig. 3G). Notably, the relative level of direct c-myc to general DSB translocation junctions versus c-myc to Sα translocation junctions, compared with respective WT controls, was increased and the frequency of junctions with greater than 4 bp MH was decreased in the ATM-, 53BP1-, and ligase 4-deficient backgrounds (compare Fig. 3 C and D with Fig. 3 F and G). Finally, in all characteristics of joining in this set of assays, translocation junctions of 53BP1- and ligase 4-deficient CH12F3 cells again were very similar (Fig. 3 E–G).
Discussion
Prior studies reported that DSBR factor deficiencies had essentially no impact on MH usage in CSR junctions (12–15). However, we now demonstrate that deficiencies for various ATM-dependent DSBR factors in activated mouse primary B cells and/or the mouse CH12F3 line, compared with WT counterparts, lead to a significant overall increase in MH-mediated versus direct CSR junctions between Sμ and all tested downstream S regions (Sγ1, Sε, and Sα). In this context, 53BP1 deficiency had the most dramatic increase, while ATM, H2AX, and Rif1 deficiencies had intermediate increases. The relative impacts of these different DSBR-deficient backgrounds in level of MH usage in CSR junctions also correlated well with recovery of junctions with long MHs (greater than 4 bp), suggesting increased use of A-EJ (4). Several factors may contribute to the differences between our current findings and those of prior studies. One is that most prior studies, due to technical limitations, analyzed small numbers of CSR junctions, whereas our current HTGTS-based CSR junction studies recovered two to three orders of magnitude more (3). A second factor is that many prior studies only analyzed junctions lying at the peripheral edges of S regions, due to restraints on primers that could be used for the PCR-based assays employed (26). In contrast, our HTGTS-based CSR assay recovers junctions across the repetitive core of downstream acceptor S regions, where most CSR occurs (3). Our clear-cut finding of increased use of MH in CSR junctions from ATM-deficient mouse B cells supports the conclusion of prior studies, with more-limited junctions, that found increased MH usage in CSR junctions in the context of ATM deficiency in mice and humans (16, 17).
A striking result from our current findings is that 53BP1 deficiency and deficiency for ligase 4 lead to remarkably similar impacts on MH usage in Sμ to Sα CSR junctions recovered from activated CH12F3 B-lymphoma cells. Thus, for CSR junctions in these two backgrounds, the frequency of MH-mediated CSR junctions, overall usage of MH, and frequency of MHs over 4 bp are quite comparable. This striking correlation between MH usage for end-joining between these two genotypes also applies to translocation junctions between bait c-myc Cas9/gRNA-generated DSBs and Sμ/Sα and to more general genome-wide translocation junctions, supporting the notion that 53BP1 contributes to promoting C-NHEJ end-joining during CSR and, potentially, other classes of genome-wide DSBs (11). Thus, in the absence of 53BP1, MH-mediated A-EJ may predominate. A significant fraction of CSR-associated DSBs that are joined in the absence of 53BP1 arise from S regions that have undergone long resections that could expose long single-stranded regions that inhibit C-NHEJ and, thereby, contribute to the junctional MH phenotype (3). Notably, however, ligase 4 deficiency has a much less severe impact on CSR than 53BP1 deficiency and does not promote nearly the same degree of long S resections. Thus, the dramatic effect of 53BP1 deficiency on CSR and orientation of recovered CSR joins clearly reflects additional factors beyond the dominant use of an MH-biased A-EJ pathway for CSR, potentially including impaired end-tethering (3). The relative increase in MH-biased CSR junctions in other backgrounds also could more strongly reflect relative influences of end resection (3), but more work is required to clarify this possibility, since our assay only reliably detects long resections (3). Finally, the moderate impact of ligase 4 deficiency on orientation-specific CSR also implicates factors beyond junctional MH usage in the much more severe impact of 53BP1 deficiency on CSR orientation specificity.
In WT, ATM-, 53BP1-, and ligase 4-deficient CH12F3 cells, we find that the frequency of MH usage in translocation junctions, overall MH patterns and usage, and use of long homologies in c-myc bait to Sα translocation junctions are remarkably similar to those of Sμ to Sα CSR junctions. Thus, the distinctive MH usage phenotypes with respect to junctional MH used for CSR in WT and various mutant backgrounds is not limited to junctions between two repetitive S-region sequences or to localization within an IgH locus domain. On the other hand, the MH patterns of junctions between c-myc bait DSBs and general DSBs genome-wide in the WT and different mutant backgrounds tested had less pronounced differences compared with those observed for Sμ to Sα CSR or c-myc bait DSB to Sα junctions. The most extreme example of such less-pronounced differences was junctions between the c-myc bait DSB and general DSBs in WT and ATM-deficient CH12F3 cells in which the level of MH usage, as evidenced by level of direct joins, was essentially indistinguishable. To test the generality of this surprising latter finding, we also assayed translocation junctions in WT and ATM-deficient primary neural stem and progenitor cells and made similar findings with respect to similarity in MH usage (SI Appendix, Fig. S7). Why S-region DSBs are significantly more prone to use MH for DSB end-joining in the absence of ATM, and, to a lesser extent, other DSBR factors, than many other DSBs in the genome will require further investigation. However, distinctive candidate features that may contribute include S-regions DSBs being AID-initiated, occurring in very repetitive sequences, and undergoing increased resection in DSBR-deficient backgrounds.
Experimental Procedures
Mice.
ATM−/−, H2AX−/−, 53BP1−/−, and Rif1f/fCD19cre lines have been reported previously (3). Mouse work was performed under protocols approved by the Boston Children’s Hospital.
Plasmids and Oligonucleotides.
All gRNA oligonucleotides for CRISPR/Cas9 were cloned into pX330 vector (Addgene plasmid ID 42230). All of the oligonucleotides are listed in SI Appendix, Table S10.
Cell Culture.
Primary splenic B cells were isolated by a CD43-negative selection kit and cultured in medium R15 (RPMI1640, 15% FBS, l-glutamate, 1× penicillin and streptomycin). Primary B-cell stimulation was performed with anti-CD40 (1 μg/mL; eBioscience) plus IL4 (20 ng/mL; PeproTech) for 96 h. CH12F3 cells were cultured in medium R15 and stimulated with anti-CD40 (1 μg/mL; eBioscience) and IL4 (20 ng/mL; PeproTech) plus TGF-β (0.5 ng/mL; R&D Systems) for 72 h. Nucleofection of the c-myc Cas9 plasmid was carried out 12 h poststimulation.
Cell Lines.
Generation of noncoding allele-deleted CH12F3 cell lines was described previously (3). To obtain ATM−/− cells, WT CH12F3 cells were first nucleofected using the 4D-nucleofector X (solution SF, protocol CA-137; Lonza) with the two pX330 vectors with gRNA sequence targeting the ATM gene. Single-cell subclones were seeded into 96-well plates 12 h postnucleofection, and the resulting clones were screened by PCR and Southern blot. Two independent clones used in this study were confirmed by Western blot with α-ATM antibody [(2C1) sc23921; SantaCruz Biotechnology]. The 53BP1−/− CH12F3 cells were obtained similarly as above, and two independent clones generated were confirmed by Southern blot and Western blot with α-53BP1 antibody (#4908; Cell Signaling Technology). Ligase 4-deficient CH12F3 (clone 2) was derived in a similar manner previously (21).
HTGTS and Data Analysis.
HTGTS junctions from the various CH12F3 cell genetic backgrounds were isolated as described previously (3). Isolation of primary B-cell HTGTS junctions has been described (3). HTGTS data analyses, including junctional MH analyses (21), of all of the HTGTS libraries were performed and characterized using the bioinformatics pipeline (v2) described previously (3), and raw data have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus (GEO). MH is defined as the longest region at the switch junction of perfect uninterrupted donor/acceptor identity. Sequences with a gap between donor and acceptor sequences are considered as insertions and were excluded from calculations. Direct joins were defined as junctions with no MH and no insertions (MH = 0). To plot the MH pattern, direct joins and junctions with MH were pooled and sorted by length, and the number of junctions with indicated length of MH were counted and calculated as percentage of the total number of junctions mapped to the region of interest. The coordinates of each S region for the calculation are described previously (3).
Joining Orientation Bias and Acceptor S-Region Resection Calculation.
Acceptor S-region junctions mapped to Sα (SI Appendix, Fig. S4A) were divided into six regions (denoted by a through f) on either (−) or (+) orientation as shown below:
Junction numbers denoted in each of the above regions specified in three replicates of a given genotype (SI Appendix, Fig. S4A) were used to calculate inversion (INV):deletion (DEL) bias ratio as follows: Bias ratio = (d + e)/(b + c). Percentage of junctions with long resection was calculated by c/(b + c) × 100.
Statistical Analysis.
Unpaired two-tailed Student t test was used to examine the significant difference between samples. A P value of <0.05 was considered significant.
Data Deposition.
HTGTS sequencing data analyzed here have been deposited in the GEO database under accession nos. GSE71005, GSE106922, and GSE74356 (SI Appendix, Fig. S7).
Supplementary Material
Acknowledgments
We thank Dr. Kefei Yu for providing the ligase 4−/− CH12F3 cell line (designated clone 1). This work was supported by National Institutes of Health Grant R01AI077595 (to F.W.A.). J.D. is supported by the Program for Guangdong Introducing Innovative and Entrepreneurial Teams (Grant 2016ZT06S252). F.W.A. is an investigator and Z.D. is a postdoctoral associate of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE71005, GSE106922, and GSE74356).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719988115/-/DCSupplemental.
References
- 1.Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Methot SP, Di Noia JM. Molecular mechanisms of somatic hypermutation and class switch recombination. Adv Immunol. 2017;133:37–87. doi: 10.1016/bs.ai.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Dong J, et al. Orientation-specific joining of AID-initiated DNA breaks promotes antibody class switching. Nature. 2015;525:134–139. doi: 10.1038/nature14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 5.Soulas-Sprauel P, et al. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J Exp Med. 2007;204:1717–1727. doi: 10.1084/jem.20070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han L, Yu K. Altered kinetics of nonhomologous end joining and class switch recombination in ligase IV-deficient B cells. J Exp Med. 2008;205:2745–2753. doi: 10.1084/jem.20081623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 8.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franco S, et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol Cell. 2006;21:201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Bothmer A, et al. Mechanism of DNA resection during intrachromosomal recombination and immunoglobulin class switching. J Exp Med. 2013;210:115–123. doi: 10.1084/jem.20121975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bothmer A, et al. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med. 2010;207:855–865. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reina-San-Martin B, Chen HT, Nussenzweig A, Nussenzweig MC. ATM is required for efficient recombination between immunoglobulin switch regions. J Exp Med. 2004;200:1103–1110. doi: 10.1084/jem.20041162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reina-San-Martin B, et al. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J Exp Med. 2003;197:1767–1778. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reina-San-Martin B, Chen J, Nussenzweig A, Nussenzweig MC. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1-/- B cells. Eur J Immunol. 2007;37:235–239. doi: 10.1002/eji.200636789. [DOI] [PubMed] [Google Scholar]
- 15.Di Virgilio M, et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339:711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lumsden JM, et al. Immunoglobulin class switch recombination is impaired in Atm-deficient mice. J Exp Med. 2004;200:1111–1121. doi: 10.1084/jem.20041074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan Q, et al. Alternative end joining during switch recombination in patients with ataxia-telangiectasia. Eur J Immunol. 2002;32:1300–1308. doi: 10.1002/1521-4141(200205)32:5<1300::AID-IMMU1300>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 18.Chiarle R, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frock RL, et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu J, et al. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat Protoc. 2016;11:853–871. doi: 10.1038/nprot.2016.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar V, Alt FW, Frock RL. PAXX and XLF DNA repair factors are functionally redundant in joining DNA breaks in a G1-arrested progenitor B-cell line. Proc Natl Acad Sci USA. 2016;113:10619–10624. doi: 10.1073/pnas.1611882113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood RD, Doublié S. DNA polymerase θ (POLQ), double-strand break repair, and cancer. DNA Repair (Amst) 2016;44:22–32. doi: 10.1016/j.dnarep.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yousif AS, Stanlie A, Mondal S, Honjo T, Begum NA. Differential regulation of S-region hypermutation and class-switch recombination by noncanonical functions of uracil DNA glycosylase. Proc Natl Acad Sci USA. 2014;111:E1016–E1024. doi: 10.1073/pnas.1402391111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura M, et al. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. Int Immunol. 1996;8:193–201. doi: 10.1093/intimm/8.2.193. [DOI] [PubMed] [Google Scholar]
- 25.Kim A, Han L, Santiago GE, Verdun RE, Yu K. Class-switch recombination in the absence of the IgH 3′ regulatory region. J Immunol. 2016;197:2930–2935. doi: 10.4049/jimmunol.1600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stavnezer J, Björkman A, Du L, Cagigi A, Pan-Hammarström Q. Mapping of switch recombination junctions, a tool for studying DNA repair pathways during immunoglobulin class switching. Adv Immunol. 2010;108:45–109. doi: 10.1016/B978-0-12-380995-7.00003-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.