Abstract
Grain size and weight are important determinants of rice yield. The identification of beneficial genes from wild rice that have been lost or weakened in cultivated rice has become increasingly important for modern breeding strategies. In this study, we constructed a set of chromosome segment substitution lines (CSSLs) of wild rice, Oryza rufipogon with the indica cultivar 9311 genetic background. Four grain-related traits, i.e., grain length (GL), grain width (GW), length-width ratio (LWR), and thousand grain weight (TGW), were screened across six environments. A total of 37 quantitative trait loci (QTLs) were identified in these environments and mapped to 12 chromosomes. Sixteen QTLs were detected in at least two environments, and two QTL clusters were observed on Chr. 4 and Chr. 8. Based on a comparative analysis with QTLs identified in previous studies, the CSSLs between Oryza rufipogon accessions and 9311 had high genetic diversity. Among the sixteen stable QTLs, seven for TGW, LWR, GL, and GW were not previously identified, indicating potentially novel alleles from wild rice. These CSSLs provide powerful tools for functional studies and the cloning of essential genes in rice; furthermore, we identified elite germplasm for rice variety improvement.
Keywords: Oryza rufipogon Griff., chromosome segment substitution lines, grain-related traits, quantitative trait loci
Introduction
Rice (Oryza sativa L.) is one of the most important staple foods in the world; half of the world population depends on rice as the main food resource. The Food and Agriculture Organization predicted that the human population will reach 9 billion in 2050, requiring a 60% increase in food production to feed the population (Alexandratos and Bruinsma 2012). At present, rice breeding is challenged by a yield plateau owing to the narrow genetic background. Common wild rice (O. rufipogon Griff.), which is more genetically diverse than O. sativa (Sun et al. 2001, Wang et al. 1992), is considered the direct ancestor of cultivated rice (Wei et al. 2012). Transferring genes that control desirable traits from wild relatives to cultivated rice has proven to be an important strategy in rice breeding (McCouch et al. 2007, Xiao et al. 1996).
Most agronomic traits of rice are controlled by multiple quantitative trait loci (QTL) (Yamamoto et al. 2009, Yano and Sasaki 1997). Genetic populations such as the F2, BC1F2 generations, and some permanent populations, including doubled haploid (Yoshida et al. 2002) and recombinant inbred lines, could be used for QTL analyses (Onishi et al. 2007). Major QTLs are easily detected from those populations, but minor QTLs are not. Moreover, it is difficult to make repeated observations using temporary populations. Recently, chromosome segment substitution lines (CSSLs) have been developed in rice (Ando et al. 2008, Ebitani et al. 2005, Kubo et al. 2002, Shim et al. 2010) for the detection of functionally important QTLs and the identification of elite genetic resources. Each CSSL consists of one or a few homozygous chromosome segments derived from a donor parent in the genetic background of the recurrent parent, and the effect of each substituted segment on a trait can be evaluated without genetic interactions among QTLs. The same CSSL can be simultaneously and repeatedly planted in various locations or in different years for diverse analyses.
The identification of QTLs that control grain production is a primary step in rice cultivar improvement. Nearly 400 QTLs have been detected for grain traits (Huang et al. 2013), some of which have been fine mapped (Bai et al. 2010, Wan et al. 2006, Xie et al. 2008). Additionally, some genes related to grain size and weight have been cloned, including GS3 (Fan et al. 2006), GS5 (Li et al. 2011), GL3.1 (Qi et al. 2012), GW2 (Song et al. 2007), and GW6a (Song et al. 2015).
QTLs that impact breeding must be stable across different environments and in different genetic backgrounds. However, grain-related QTL analyses performed in multiple environments have indicated that environmental conditions contribute substantially to changes in rice grain-related traits. Although the phenotypic characters of wild rice are inferior to those of cultivated rice, modern molecular biology studies have revealed that some genes hidden in wild rice are essential to yield-related trait improvement (Tian et al. 2005). Some QTLs associated with rice grain traits have been detected using wild rice introgression lines (Tan et al. 2008, Yuan et al. 2009). Therefore, the identification of stable QTLs associated with grain size and weight in wild rice is critical for rice breeding.
In this study, we evaluated a set of CSSLs that carry the chromosome segments of O. rufipogon from Hainan province of China in the genetic background of the elite indica cultivar 9311 to detect QTLs for rice grain size and weight that were not specific to a particular environment and to identify elite genetic resources for yield-related traits. Our results will provide elite materials for rice breeding and map-based cloning of desirable genes.
Materials and Methods
Plant materials
The high-yield indica rice variety 9311, which has a sequenced genome and is used as the recipient parent for hybrid rice, was used as the recurrent parent. Wild rice (O. rufipogon, laboratory preserved accession number CWR274) from Hainan province, China was used as the donor parent. The F1 hybrid was backcrossed several times consecutively to 9311. An advanced backcrossed population was generated after three or four rounds of self-pollination. Molecular markers that were polymorphic between the two parents were used to select target lines to construct CSSLs from the BC3F1 plants and the subsequent backcross and self-pollination generations.
Field experiments
The parental variety 9311 and 133 CSSLs were grown in six environments (two years × three locations) (Table 1) using the randomized complete block design with two replications. Each plot consisted of rows with 10 plants. Forty plants of each genotype in each plot were planted with a spacing of 10 × 25 cm. Crop management and the control of diseases and insect pests were performed as locally recommended.
Table 1.
Test environments in which the CWR274 × 9311 CSSLs populations were evaluated
| Environment | Replication | Crop location | Cropping season | Seeding date | Transplanting date |
|---|---|---|---|---|---|
| E1 | 2 | Changping, Beijing N40.20°, E115.51° | May–Oct. 2014 | April 22, 2014 | May 26, 2014 |
| E2 | 2 | Nanjing, Jiangsu N32.03°, E118.47° | May–Oct. 2014 | May 15, 2014 | June 16, 2014 |
| E3 | 2 | Sanya, Hainan N18.15°, E109.3° | Dec. 2013–May 2014 | Dec. 04, 2013 | Jan. 05, 2014 |
| E4 | 2 | Changping, Beijing N40.20°, E115.51° | May–Oct. 2015 | April 28, 2015 | June 02, 2015 |
| E5 | 2 | Nanjing, Jiangsu N32.03°, E118.47° | May–Oct. 2015 | May 11, 2015 | June 13, 2015 |
| E6 | 2 | Sanya, Hainan N18.15°, E109.3° | Dec. 2014–May 2015 | Dec. 01, 2014 | Dec. 30, 2015 |
Phenotypic evaluation
At maturity, the grains of five plants of each CSSL were harvested and all fully filled grains were selected and dried in an oven at 40°C for 48 h. Then, 200–300 fully filled seeds per CSSL were measured using an automatic seed analyzer (Wanshen Detection Technology Co., Ltd., Hangzhou, China, www.wseen.com). The automatic seed analyzer instrument contained a 5 million pixel resolution camera and a backlight source grass plate. The grain length (GL), grain width (GW), grain length and width ratio (LWR) were automatically calculated via the automatic instrument. Seed weights were detected using an analytical balance (Botoo Electronic Technology Co., Ltd., Shenzhen, China) linked with the automatic seed analyzer, and values were automatically converted to thousand grain weights (TGW), all traits were measured with three replications.
DNA extraction and SSR analysis
DNA was extracted from fresh leaves according to the procedure described by Zheng et al. (1995). A polymorphism survey between the parents was conducted using simple sequence repeat (SSR) markers, which were synthesized according to published sequences (McCouch et al. 2002, Temnykh et al. 2000), and insertion/deletion (InDel) markers, which were developed from a BLASTN alignment between the genome sequences of japonica cv. Nipponbare and indica cv. 9311. The genomic sequence was obtained from the Gramene website (http://ensembl.gramene.org/Oryza_sativa/Info/Index). Polymerase chain reaction (PCR) primers were designed using Primer Premier 5.0. A 15 μL reaction mixture was applied for PCR. The mixture was composed of 50 ng of template DNA, 0.3 μL of 10 mM each deoxynucleotide triphosphate (dNTP), 0.4 units of Taq DNA polymerase, 1.5 μL of 10× PCR buffer with Mg2+ (CWbio, Beijing, China), and 1.2 μL of 10 μmol/L forward and reverse primers. Amplification was carried out using the following reaction conditions: initial denaturation at 94°C for 5 min, followed by 35 cycles of 30 s at 94°C, 30 s at 56°C, and 30 s at 72°C, with a final extension at 72°C for 10 min. PCR products were separated on 8% polyacrylamide denaturing gels, and the bands were revealed by the silver-staining protocol (Panaud et al. 1996).
Determination of the physical location of molecular markers
A physical map of the polymorphic SSR and InDel molecular markers was constructed to verify the density and even distribution along the chromosomes. Using the online NCBI BLAST tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi), polymorphic markers were physically mapped to all twelve chromosomes of rice Nipponbare pseudomolecules, which were constructed by the International Rice Genome Sequencing Project (2005).
Data analysis and QTL mapping
The construction of graphical genotypes and calculation of the percentage of the total genome in CSSLs were performed using Graphical Genotypes (GGT) (van Berloo 1999). The underlying assumption of the GGT algorithm is that if two neighboring loci come from the same parent, the segment between them is likely to be from the same parental genome; in contrast, if the alleles at a pair of adjacent loci are inherited from different parents, a recombination event is assumed to have happened at the mid-point of the interval.
To explore the genetic basis for the four grain traits in different environments, a procedure of likelihood ratio test combined with stepwise regression (RSTEP-LRT), developed for QTL mapping in non-ideal or ideal CSSL populations was used (Wang et al. 2006). In the method of RSTEP-LRT, there are three steps. Firstly, a multiple linear model for phenotypes and adjusted phenotypic values adopted in RSTEP-LRT are summarized as follows. Assuming there were n CSS lines with m marker segments, the averaged phenotypic values for a quantitative trait in interest of the ith CSS line, yi, is represented by the following linear model,
| (1) |
where i = 0 (background parent), 1, 2, …, n, b0 is the intercept, bj (j = 1, …, m) is the partial regression coefficient of phenotype on the jth marker segment, xij is the indicator variable for the jth marker segment in the ith CSS line, and ei is the random error. Considering the number of marker segments was always larger than the population size in plant genetics, to avoid model overfitting, stepwise regression of bidirectional selection was utilized in model (1) to select important markers associated with the trait in interest, where probabilities for marker segments entering into the model (1) (PIN) was set as 0.001; and probabilities for marker segments moving out of the model (1) (POUT) was set as 0.002 for all the four traits used in this study. Secondly, to test if there is a QTL linked with the jth marker segment, the phenotypic values in model (1) can be adjusted by,
| (2) |
where Δyi contained the QTL information on the current segment, and at the same time excluded the genetic effects from the other segments. Then, likelihood ratio test is conducted for the adjusted phenotypic values to determine whether a QTL exists in the testing segment. In this study, 1000 times of permutation test was used to determine LOD thresholds to declare significant QTL (P < 0.05), and also to calculate the genome-wide type I error for each QTL. Each time we reshuffled the phenotypic data and conducted the whole process of RSTEP-LRT. We define the P-value based on the permutation test result as ‘genome-wide P-value’. For example, if the value of LOD score corresponds to the 100th largest value in the permutation test of 1000 cycles, the genome-wide P-value of the LOD score is regarded as 0.1. When the LOD score of QTL is larger than the maximum of 1000 LOD scores from permutation tests, P-values was set as 0.001, while the LOD score of QTL is smaller than the minimum of 1000 LOD scores from permutation tests, P-values was set as 1. The QTL detection by RSTEP-LRT, the calculation of phenotypic variation explained by each QTL, and the estimation of QTL additive effects, were implemented by software QTL IciMapping v4.1 (Meng et al. 2015). QTL nomenclature followed the recommendations of McCouch (2008).
A combined analysis of variance (ANOVA) over the six environments (lines × environments) was used to estimate the variance components, as described by Liu et al. (2016). The phenotypic variation of the four grain traits was estimated and a correlation analysis was performed to detect the association between grain traits based on the CSSL population data using SPSS19.0.
Results
Development of CSSLs
A total of 435 SSR markers and 175 InDel markers distributed across the 12 rice chromosomes were examined for polymorphisms between CWR274 and 9311; 119 SSR (27.3%) and 62 InDel (41.3%) polymorphic makers were used for the CSSL analysis (Supplemental Table 1). Selection of the CWR274 substitution lines in the 9311 background was achieved using these markers.
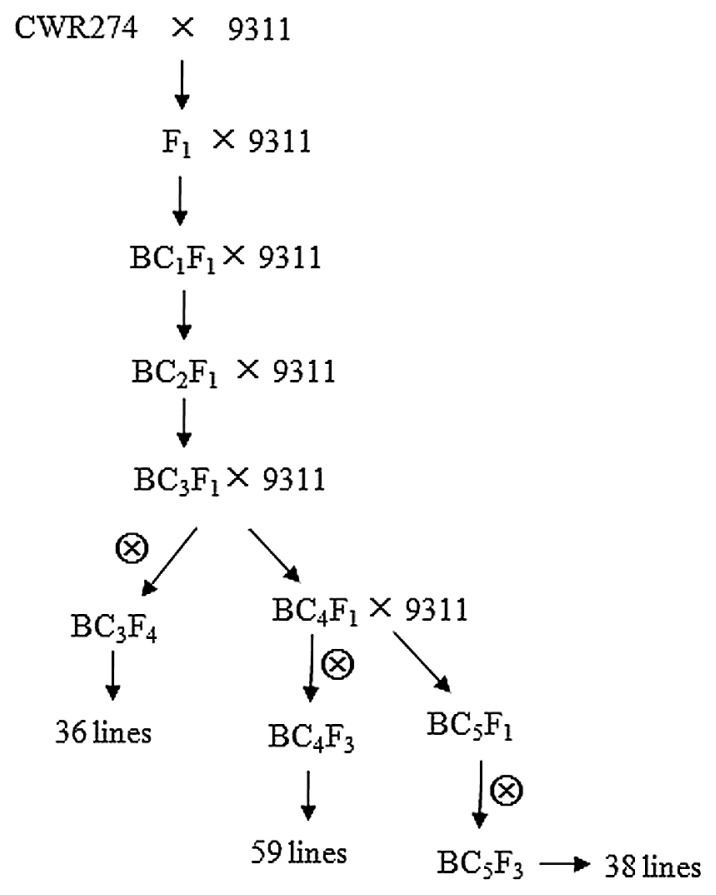
To identify the CSSLs, the F1 plant derived from a cross between CWR274 and 9311 was backcrossed to 9311 three times to produce 324 BC3F1 plants. The candidate BC3F1 individuals were selected using 181 polymorphic markers based on the following criteria: (i) a single, relatively large chromosomal segment from CWR274 substituted into the target chromosome; (ii) a high level of residual homozygosis of 9311 alleles in non-target chromosomal regions; and (iii) partially overlapping segments in the chosen lines in order to cover all 12 rice chromosomes. These plants were backcrossed continuously by MAS following the same criteria and were then self-pollinated several times. Finally, we selected 36 lines from BC3F4, 59 lines from BC4F3, and 38 lines from BC5F3, to construct the CSSL population. The procedure of CSSL development is schematically described in Fig. 1.
Fig. 1.
Breeding scheme for the development of CSSLs carrying CWR274 segments in the 9311 background.
Genotypes of the CSSLs
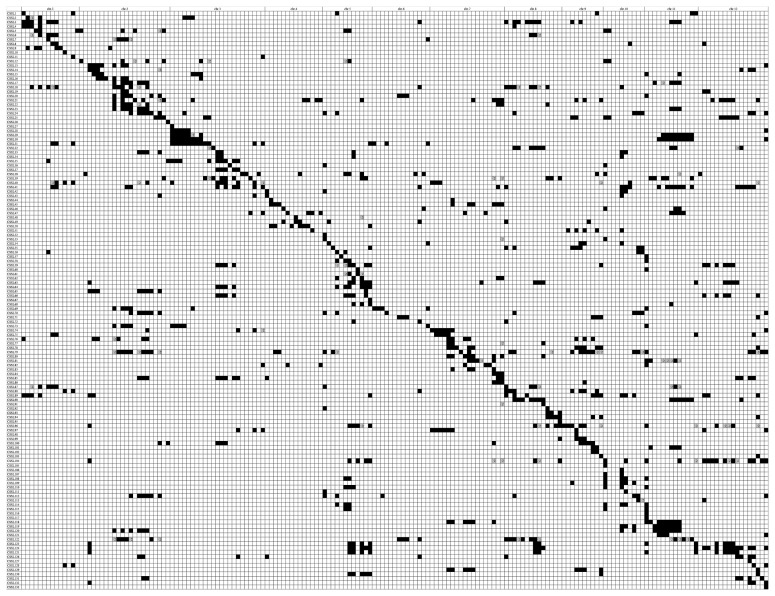
Graphical genotypes of the 133 CSSLs were generated using the linkage map of 119 SSR and 62 InDel markers (Fig. 2). The substituted chromosome segments covered most of the 12 chromosomes, except for seven gaps on Chr. 1 (defined by RM449 and RM543), Chr. 6 (defined by RM276, RM527 and RM3827), and Chr. 10 (defined by RM216 and RM239). The numbers of substituted chromosome segments among the CSSLs ranged from 1 to 14, with an average of 4.8 segments per CSSL. The roughly estimated lengths of the substituted segments ranged from 0.1 Mb to 7.9 Mb, with an average of 2.06 Mb per segment. The segment coverage was approximately 93.24% of the whole wild rice genome.
Fig. 2.
Graphical representation of the CSSLs developed in this study. Each row represents a candidate CSSL line and each column represents a marker locus. Black bars indicate homozygous regions from CWR274; white bars indicate the 9311 genetic background; gray regions indicate heterozygous regions.
Phenotypic evaluations of the four rice grain-related traits in CSSL populations across six environments
The phenotypic variation of the four grain-related traits among the six environments is summarized in Table 2. Compared with the recurrent parent 9311, high phenotypic variation was detected in four grain-related traits in the CSSL populations in six environments. The highest degree of variation was observed for TGW, with a range of 24.43 to 39.58 g in E5. The mean TGW for 9311 differed among the six environments, ranging from 31.83 to 33.66 g, and the coefficient of variation ranged from 6.15% to 7.10%. In addition, all four grain-related traits showed continuous distributions and had ultra-parental genetic types in the CSSL populations, the results show that segment from CWR274 had positive and negative effect in the four traits, suggesting that these traits were quantitatively inherited and polygenic.
Table 2.
Statistics of grain traits of 9311 and CSSLs in the six environments
| Trait and environments | 9311 | CSSLs | Trait and environments | 9311 | CSSLs | ||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| Mean ± SD | Range | CV (%) | Mean ± SD | Range | CV (%) | ||||
| TGW (g) GL (mm) | |||||||||
| E1 | 32.89 | 30.01 ± 2.11 | 22.97–36.36 | 7.04 | E1 | 9.77 | 9.74 ± 0.53 | 7.97–11.39 | 5.46 |
| E2 | 31.83 | 31.35 ± 1.93 | 24.28–36.44 | 6.15 | E2 | 9.69 | 9.68 ± 0.43 | 8.26–11.38 | 4.49 |
| E3 | 31.94 | 30.48 ± 2.11 | 24.00–36.38 | 6.94 | E3 | 9.62 | 9.74 ± 0.35 | 8.52–10.85 | 3.55 |
| E4 | 32.36 | 30.24 ± 2.02 | 23.57–36.22 | 6.67 | E4 | 9.46 | 9.47 ± 0.36 | 8.19–10.62 | 3.77 |
| E5 | 33.66 | 32.90 ± 2.34 | 24.43–39.58 | 7.10 | E5 | 9.58 | 9.66 ± 0.36 | 8.32–10.99 | 3.73 |
| E6 | 32.5 | 31.17 ± 2.01 | 23.38–36.93 | 6.45 | E6 | 9.57 | 9.64 ± 0.39 | 8.21–10.85 | 4.03 |
| LWR (mm) GW (mm) | |||||||||
| E1 | 3.40 | 3.48 ± 0.22 | 2.76–4.16 | 6.40 | E1 | 2.88 | 2.80 ± 0.09 | 2.52–3.00 | 3.33 |
| E2 | 3.44 | 3.46 ± 0.20 | 2.84–4.05 | 5.89 | E2 | 2.81 | 2.80 ± 0.09 | 2.44–3.01 | 3.29 |
| E3 | 3.42 | 3.46 ± 0.17 | 3.00–4.01 | 4.82 | E3 | 2.81 | 2.82 ± 0.10 | 2.49–3.08 | 3.43 |
| E4 | 3.38 | 3.43 ± 0.17 | 2.85–4.01 | 5.09 | E4 | 2.81 | 2.78 ± 0.10 | 2.48–3.10 | 3.66 |
| E5 | 3.41 | 3.45 ± 0.17 | 3.06–4.03 | 5.05 | E5 | 2.82 | 2.81 ± 0.10 | 2.42–3.07 | 3.64 |
| E6 | 3.35 | 3.38 ± 0.17 | 2.82–3.96 | 5.13 | E6 | 2.85 | 2.86 ± 0.09 | 2.58–3.06 | 3.23 |
TGW: thousand grain weight; GL: grain length; LWR: length-width ratio; GW: grain width; CV: coefficient of variation; SD: standard deviation. Naming of six environments is the same as Table 1.
Correlations among the rice traits for the six conditions are summarized in Table 3. The positive correlations between TGW and the other three grain-related traits were significant (P < 0.01), the correlation between GL and GW was not significant.
Table 3.
Correlations among grain-related traits in combined environments
The abbreviations for the four traits (TGW, LWR, GL and GW) are the same as Table 2. Asterisks indicate significant correlation coefficients:
P ≤ 0.05;
P ≤ 0.01.
The results of the ANOVA for TGW, GL, GW, and LWR of the CSSL populations across the six different environments are summarized in Table 4. Significant effects of lines, environments, and their interaction (lines × environments) were observed for the four traits. The analysis of variance for grain traits in the CSSL population across two years × three locations is also summarized in Supplemental Table 2. Significant effects of CSSL, Site, Year, CSSL × Site, CSSL × Year, Site × Year, and their interaction (CSSL × Site × Year) were observed for the four traits.
Table 4.
Analysis of variance for grain traits of the CSSL population across combined six environments
| SOD df | Line 133 | Environment 5 | Line × Environment 665 | Block (Environment) 6 |
|---|---|---|---|---|
| TGW | 61.46** | 506.51** | 5.37** | 1.88 |
| LWR | 101.33** | 99.82** | 5.40** | 1.1 |
| GL | 63.14** | 120.86** | 4.94** | 1.75 |
| GW | 52.03** | 104.24** | 3.39** | 46 |
SOD: sources of difference. df: degrees of freedom. The abbreviations for the four traits (TGW, LWR, GL and GW) are the same as those in Table 2. Asterisks indicate significance levels:
P ≤ 0.05;
P ≤ 0.01.
QTL analyses for grain traits
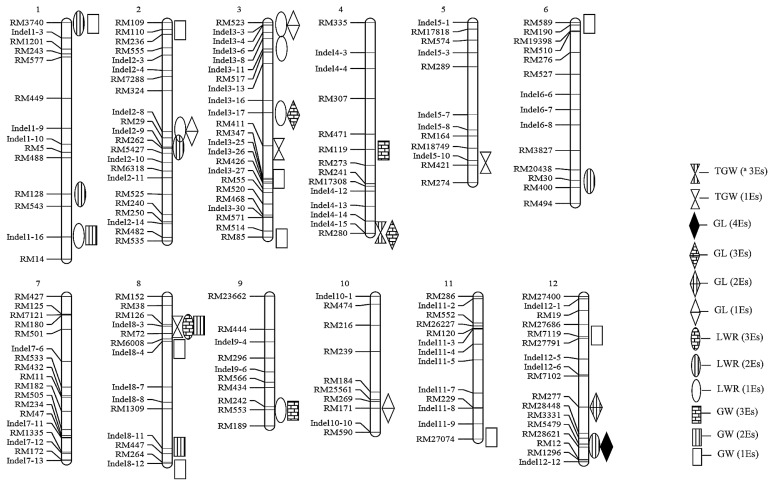
Thirty-seven QTLs for the four traits were identified in the six environments and then mapped to twelve chromosomes (Table 5, Fig. 3). For TGW, the QTL qTGW4 was consistently detected across three environments, with an average phenotypic variation explained (PVE) of 10.2%, and the wild rice allele at the qTGW4 locus decreased TGW by an average of 2.24 g. For LWR, qLWR8 were observed in three environments, with average PVE of 11.13%. qLWR1.2, qLWR1.1, qLWR2.2, qLWR6, and qLWR12 were consistently observed in two environments. For the QTLs of GL, qGL3.2, qGL4, qGL12.1, and qGL12.2 were consistently detected across two to four environments, and qGL12.2 was mapped to chromosome 12 with a PVE of 16.54%. The wild rice allele at the qGL12.2 locus increased GL by an average of 0.28 mm. For the GW QTLs, qGW8.1, qGW9, qGW1.2, qGW4, and qGW8.3 were identified in two to three environments, with a PVE of 5.33–18.16%. For all QTLs that were detected in one or two environments, the performance of 37 QTLs in six environments were listed in Supplemental Table 3.
Table 5.
QTLs affecting four grain-related traits detected using CSSL population across six environments
| Trait | QTL | Marker | Chr. | Environment | LOD Threshold | LOD | PVE (%) | Add |
|---|---|---|---|---|---|---|---|---|
| TGW | qTGW4 | RM280 | 4 | E2 | 3.03 | 3.05 | 8.17 | −1.85 |
| E5 | 3.11 | 3.67 | 11.18 | −2.62 | ||||
| E6 | 3.32 | 3.87 | 11.26 | −2.27 | ||||
| qTGW5 | Indel5-10 | 5 | E3 | 2.87 | 3.65 | 12.38 | −1.33 | |
| qTGW3 | RM411 | 3 | E1 | 3.06 | 5.82 | 16.53 | −3.53 | |
| qTGW8 | Indel8-3 | 8 | E1 | 4.72 | 12.44 | 2.50 | ||
| LWR | qLWR8 | RM126 | 8 | E3 | 3.10 | 4.54 | 7.58 | 0.08 |
| E4 | 3.16 | 6.31 | 11.14 | 0.10 | ||||
| E5 | 3.14 | 6.20 | 14.66 | 0.11 | ||||
| qLWR1.1 | RM3740 | 1 | E1 | 3.04 | 4.59 | 12.91 | 0.20 | |
| E6 | 3.11 | 3.86 | 9.19 | 0.14 | ||||
| qLWR6 | RM30 | 6 | E2 | 2.97 | 4.57 | 13.49 | 0.15 | |
| E5 | 3.14 | 6.74 | 0.09 | |||||
| qLWR1.2 | RM128 | 1 | E3 | 3.38 | 5.69 | 0.12 | ||
| E6 | 3.49 | 8.37 | 0.15 | |||||
| qLWR2.2 | Indel2-9 | 2 | E4 | 3.60 | 6.06 | −0.06 | ||
| E5 | 5.44 | 12.69 | −0.09 | |||||
| qLWR12 | RM28621 | 12 | E3 | 3.11 | 5.08 | 0.07 | ||
| E4 | 3.51 | 5.89 | 0.08 | |||||
| qLWR3.2 | Indel3-11 | 3 | E4 | 3.58 | 6.11 | −0.10 | ||
| qLWR1.3 | Indel1-16 | 1 | E4 | 6.18 | 10.89 | 0.14 | ||
| qLWR2.1 | Indel2-8 | 2 | E3 | 7.05 | 12.79 | −0.11 | ||
| qLWR3.1 | Indel3-3 | 3 | E1 | 3.97 | 11.06 | −0.17 | ||
| qLWR3.3 | Indel3-17 | 3 | E4 | 4.32 | 7.25 | −0.16 | ||
| qLWR9 | RM553 | 9 | E3 | 4.30 | 6.99 | 0.09 | ||
| GL | qGL12.2 | RM28621 | 12 | E2 | 3.35 | 6.40 | 19.87 | 0.34 |
| E4 | 3.14 | 3.24 | 7.14 | 0.17 | ||||
| E5 | 3.41 | 9.70 | 24.29 | 0.32 | ||||
| E6 | 3.17 | 5.94 | 14.85 | 0.27 | ||||
| qGL3.2 | Indel3-17 | 3 | E3 | 3.11 | 5.90 | 11.94 | −0.38 | |
| E4 | 5.41 | 12.09 | −0.41 | |||||
| E6 | 3.02 | 7.22 | −0.35 | |||||
| qGL4 | RM280 | 4 | E4 | 3.41 | 7.53 | −0.33 | ||
| E5 | 3.76 | 8.48 | −0.35 | |||||
| E6 | 3.86 | 9.30 | −0.40 | |||||
| qGL12.1 | RM277 | 12 | E3 | 5.65 | 11.39 | 0.25 | ||
| E4 | 4.19 | 9.17 | 0.23 | |||||
| qGL3.1 | Indel3-3 | 3 | E1 | 2.95 | 4.28 | 12.50 | −0.43 | |
| qGL2 | Indel2-8 | 2 | E3 | 6.71 | 14.47 | −0.23 | ||
| qGL10 | RM171 | 10 | E3 | 5.90 | 11.90 | 0.24 | ||
| GW | qGW4 | RM119 | 4 | E4 | 3.15 | 3.82 | 7.66 | −0.12 |
| E5 | 3.49 | 6.40 | 12.62 | −0.15 | ||||
| E6 | 3.14 | 3.84 | 8.41 | −0.11 | ||||
| qGW9 | RM553 | 9 | E1 | 2.84 | 3.74 | 7.86 | −0.05 | |
| E2 | 3.32 | 5.13 | 11.14 | −0.06 | ||||
| E3 | 3.08 | 3.03 | 8.60 | −0.06 | ||||
| qGW1.2 | Indel1-16 | 1 | E4 | 8.35 | 18.16 | −0.10 | ||
| E6 | 6.90 | 15.97 | −0.09 | |||||
| qGW8.1 | RM126 | 8 | E1 | 4.71 | 10.15 | −0.05 | ||
| E5 | 6.10 | 11.96 | −0.06 | |||||
| qGW8.3 | Indel8-11 | 8 | E5 | 3.35 | 6.26 | −0.06 | ||
| E6 | 4.06 | 8.94 | −0.06 | |||||
| qGW1.1 | RM3740 | 1 | E4 | 4.74 | 9.67 | −0.08 | ||
| qGW2 | RM110 | 2 | E5 | 5.94 | 11.61 | −0.20 | ||
| qGW3.1 | Indel3-26 | 3 | E6 | 3.33 | 7.23 | 0.05 | ||
| qGW3.2 | RM85 | 3 | E1 | 4.07 | 8.70 | −0.07 | ||
| qGW6 | RM589 | 6 | E4 | 3.27 | 6.48 | 0.11 | ||
| qGW8.4 | RM264 | 8 | E3 | 3.85 | 8.39 | −0.11 | ||
| qGW8.2 | RM6008 | 8 | E2 | 5.75 | 12.27 | −0.19 | ||
| qGW11 | RM27074 | 11 | E4 | 3.66 | 7.53 | 0.07 | ||
| qGW12 | RM7119 | 12 | E2 | 4.20 | 8.72 | 0.06 |
Fig. 3.
Map locations of identified QTLs for 1000-grain weight (TGW), grain length (GL), grain width (GW), and length-width ratio (LWR) of the CSSL population, detected in six environments. 4Es represents QTLs detected in four environments, and the same terminology is used for all of the following QTLs; i.e., 1Es represents one environment; 2Es, two environments, etc.
QTL clusters on chromosomes
Of 37 additive effect QTLs affecting four grain traits, 16 were detected under at least two environments and two QTL clusters were observed on Chr. 4 and Chr. 8 (Fig. 3). Near the RM280 marker on Chr. 4, one cluster harboring two QTLs (qTGW4 and qGL4) controlled TGW and GL, near the closely linked markers RM126 (qGW8.1 and qLWR8) and InDel8-3 (TGW8) on Chr. 8 has two QTLs affected GW and TGW.
Comparison of TGW between CSSLs and 9311 to select an elite resource
Based on the mean improvement in TGW across six environments, to select several high TGW lines for rice breeding, and detect QTLs in these lines that confirm the results of the QTL analysis, three highest-TGW CSSLs, CSSL85, CSSL126, and CSSL132, were selected (Table 6). Compared to 9311, these CSSLs exhibited increases in TGW by 8.11–10.44%, on average, in the six environments. In addition, four lowest-TGW CSSLs, CSSL21, CSSL31, CSSL35, and CSSL82, showing the most decreases in TGW compared to 9311 in six environments were selected. Compared to 9311, these CSSLs showed apparent decreases in TGW by 17.79–23.85%. Both high-TGW and low-TGW CSSLs carried positive and negative QTLs, respectively. Due to the positive correlation index between TGW and GL, LWR, and GW, the high-TGW CSSLs were selected as elite resources for rice breeding.
Table 6.
Number of high-TGW and low-TGW CSSLs and QTLs included in the substituted segments
| Numbers of CSSLs | Thousand grain weight (TGW) (g) | Positive QTL | Negative QTL | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| E1 | E2 | E3 | E4 | E5 | E6 | |||
| High-TGW | ||||||||
| 85 | 35.47 | 36.44 | 34.07 | 33.44 | 37.83 | 33.98 | qGL12.1, qGL12.2, qLWR12 | |
| 126 | 33.33 | 35.7 | 33.87 | 36.22 | 39.58 | 36.93 | qLWR6, qLWR12, qGL12.2, qGL12.1 | |
| 132 | 33.78 | 35.55 | 34.96 | 34.09 | 37.42 | 35.21 | qGL12.2, qLWR12 | |
| Low-TGW | ||||||||
| 21 | 28.37 | 27.15 | 24 | 26.9 | 27.68 | 26.37 | qLWR3.2, qGL4, qTGW4, qTGW5, qLWR2.2, qLWR2.1, qGL2 | |
| 31 | 25.6 | 27.03 | 27.59 | 25.4 | 28.13 | 26.33 | qLWR3.2, qGL3.1, qGL3.2 | |
| 35 | 25.42 | 26.97 | 25.22 | 26.38 | 26.33 | 25.66 | qLWR3.1, qLWR3.3, qGL3.2, qGL3.1 | |
| 82 | 27.37 | 24.28 | 25.57 | 23.57 | 24.43 | 23.38 | qTGW4, qGL4 | |
Naming of six environments (E1–E6) is the same as Table 1.
Discussion
Previous studies have shown that O. rufipogon from southern China is the ancestor of current rice cultivars; more than 20% of alleles in the wild species have been lost during the domestication of rice (Huang et al. 2012, Sun et al 2001). The exploitation and utilization of favorable alleles from wild rice that were lost in current cultivars have become increasingly important. Some CSSLs have been utilized to identify new QTLs that may improve existing rice cultivars (Adachi et al. 2010, Marzougui et al. 2012), and many positive QTLs related to yield have been identified from common wild rice collected from various locations (Fu et al. 2010, Furuta et al. 2014, Septiningsih et al. 2003, Tan et al. 2007). In this study, we developed 133 CSSLs harboring chromosomal segments from the donor parent CWR274 collected from Hainan island of China, which has unique morphological and ecological characteristics that are primitive and distinguishable from those of cultivated rice. The common wild rice from that area is rarely used for genomics research, but is an important and unique genetic resource for rice genome and domestication studies (Qi et al. 2013).
Graphical genotypes of the 133 CSSLs were generated using a linkage map of 181 molecular markers. There were seven gaps in the population, possibly because MAS was performed from the BC3 generation, other biological factors, such as genetic hybrid sterility and both the complexity and instability expression of photo-thermo-sensitive genes, offer an explanation for this observation.
Grain size and weight play important roles in the evolution and domestication of cultivated rice. The grains of wild relatives are usually small and round in shape; these traits are often favored under natural selection because they are associated with high fertility and are advantageous for dispersal by natural vectors. Using data for the CSSL population from six environments, a total of 37 QTLs for TGW, GL, GW, and LWR located on 12 chromosomes were identified in this study (Table 5).
A combined analysis of variance (ANOVA) of grain traits across years and locations showed that these traits were significantly influenced by genotype and environment, as well as genotype × environment interactions (Table 4). The interaction between environmental factors, year and location, also significantly influenced the performance of grain traits (Supplemental Table 2). The data indicated that minor QTL with low PVE effects are affected by environmental conditions.
Sixteen major QTLs were detected across more than two environments, among them, seven positive QTLs (qLWR1.2, qLWR8, qLWR1.1, qLWR6, qLWR12, qGL12.2, and qGL12.1) for grain size were identified from common wild rice accessions. Most of these QTLs appear to coincide to loci that were described previously (www.gramene.org). Two QTLs for TGW (qTGW3 and qTGW5) were identified and located in the vicinity of QTLs detected in previous reports (Bian et al. 2010, Tian et al. 2005). Four loci associated with GL and LWR (qGL3.2, qGL4, qLWR8, and qLWR2.2) were probably the same locus described by Fan and other research teams (Fan et al. 2006, Jing et al. 2011, Wan et al. 2006). Three GW QTLs (qGW8.1, qGW8.3, and qGW9) may share the same location as those detected in other populations (Aluko et al. 2004, Jing et al. 2011, Nagata et al. 2015, Redona and Mackill 1998). As suggested by Tanksley (1993) and Zhuang et al. (1997), QTLs with major effects are more likely to be stable across multiple environments and various genetic backgrounds. For example, qGL3.2 in the local InDel3-17 on chromosome 3 (Fig. 3), which was associated with GL, shared the same location as GS3, which has been identified in other interspecific crosses (Fan et al. 2006, Zhang et al. 2012). These results suggested that our CSSLs are an efficient population for grain-related QTL identification.
More importantly, seven QTLs for TGW, LWR, GL, and GW that were detected across more than two environments have not previously been identified, indicating potentially novel alleles from common wild rice. Two of the alleles related to GL showed positive effects. One QTL associated with GL (qGL12.2) was stably detected with positive effects in four environments, actually it can be detected in five environments while the threshold was LOD > 2.5 (Supplemental Table 3), qGL12.2 with PVE of 16.54% was near RM28621 on Chr. 12. The wild rice allele at locus qGL12.2 increased GL by an average of 0.28 mm. These data showed that qGL12.2 is a novel QTL for modification of rice grain shape.
The traits of GW and GL have high heritability and are stably expressed; however, some QTLs were detected in one or two environments, and minor QTLs providing a small contribution rate were easily affected by environmental conditions. Significant interaction effects of lines and environments were observed throughout this study. Several QTLs in this study were inconsistent between different environments. Since the CSSL contained more than one introgression segment, the genetic background was also an important factor to influence the stability of QTLs and similar multi-environmental results have previously been reported by Wang et al. 2006.
We performed a comparative analysis of high-TGW and low-TGW CSSLs. Compared to 9311, most of the highest-TGW CSSLs carried positive QTL and lowest-TGW CSSLs generally carried negative QTL (Table 6); three high-TGW CSSLs carried the same positive QTL, qGL12.2. In future studies, the CSSLs harboring the positive QTLs (qGL12.2), such as CSSL132, will be backcrossed to 9311, and secondary F2 populations will be exploited for fine mapping and positional cloning of the QTL. Given that cultivar 9311 is an elite variety and has been planted on a large scale in China, some elite lines in CSSL population could be used for rice breeding.
Many QTLs affecting related traits were mapped to similar genomic regions (Fig. 3); the high trait correlations likely reflected pleiotropic genetic basis. There was no significant correlation between GL and GW, and both were positively correlated with TGW, accordingly, the QTLs associated with GL or GW might affect TGW in the same direction. The correlation was consistent with previous results (Lee et al. 2005, Wan et al. 2005). Given that LWR was positively affected by GL and negatively affected by GW, three QTL clusters, RM3740 (qLWR1 and qGW1.1) on Chr.1, RM126 (qLWR8 and qGW8.1) on Chr. 8 and RM28621 (qLWR12 and qGL12.2) on Chr. 12, might actually control a single trait, GW or GL.
Although many studies have already reported the development of CSSLs for O. rufipogon (Fu et al. 2010, Furuta et al. 2014, Marri et al. 2005), the use of different accessions of O. rufipogon may lead to the discovery of novel QTLs. In this study, 37 grain-related QTLs on 12 chromosomes were detected, among which 16 QTLs were detected in at least 2 environments. Seven novel QTLs were identified and qGL12.2 was stably detected with positive effects in four environments. These novel QTLs identified in wild rice will facilitate fine mapping and functional studies. Our CSSL is not perfect as a single segment substitution line or as NIL; improvements are still required as well as backcrossing with the recipient parent to eliminate genetic noise. However, the advantage of the CSSL in this study was also clear; we have detected several QTLs using this population, and more importantly, this population provides a germplasm resource for rice breeding and a platform for wild rice genomic research.
Supplementary Information
Acknowledgments
We thank Dr. Huihui Li and Dr. Jiankang Wang (Institute of Crop Science, Chinese Academy of Agricultural Sciences), for their critical reading and comments on the manuscript. This research was supported by The Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Science and by a grant from the National Natural Science Foundation of China (No. 31471471) to Weihua Qiao. We acknowledge funding from the Technology Development Research Project of Hainan Province (KYYS-2015-03) to Xiaoning Wang.
Literature Cited
- Adachi, S., Tsuru, Y., Kondo, M., Yamamoto, T., Arai-Sanoh, Y., Ando, T., Ookawa, T., Yano, M. and Hirasawa, T. (2010) Characterization of a rice variety with high hydraulic conductance and identification of the chromosome region responsible using chromosome segment substitution lines. Ann. Bot. 106: 803–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandratos, N. and Bruinsma, J. (2012) World agriculture towards 2030/2050: the 2012 revision, FAO, Rome, ESA Working Paper No. 12-03. [Google Scholar]
- Aluko, G., Martinez, C., Tohme, J., Castano, C., Bergman, C. and Oard, J.H. (2004) QTL mapping of grain quality traits from the interspecific cross Oryza sativa × O. glaberrima. Theor. Appl. Genet. 109: 630–639. [DOI] [PubMed] [Google Scholar]
- Ando, T., Yamamoto, T., Shimizu, T., Ma, X.F., Shomura, A., Takeuchi, Y., Lin, S.Y. and Yano, M. (2008) Genetic dissection and pyramiding of quantitative traits for panicle architecture by using chromosomal segment substitution lines in rice. Theor. Appl. Genet. 116: 881–890. [DOI] [PubMed] [Google Scholar]
- Bai, X.F., Luo, L.J., Yan, W.H., Kovi, M.R., Zhan, W. and Xing, Y.Z. (2010) Genetic dissection of rice grain shape using a recombinant inbred line population derived from two contrasting parents and fine mapping a pleiotropic quantitative trait locus qGL7. BMC Genet. 11: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian, J.M., Jiang, L., Liu, L.L., Wei, X.J., Xiao, Y.H., Zhang, L.J., Zhao, Z.G., Zhai, H.Q. and Wan, J.M. (2010) Construction of a new set of rice chromosome segment substitution lines and identification of grain weight and related traits QTLs. Breed. Sci. 60: 305–313. [Google Scholar]
- Ebitani, T., Takeuchi, Y., Nonoue, Y., Yamamoto, T., Takeuchi, K. and Yano, M. (2005) Construction and evaluation of chromosome segment substitution lines carrying overlapping chromosome segments of indica rice cultivar ‘Kasalath’ in a genetic background of japonica elite cultivar ‘Koshihikari’. Breed. Sci. 55: 65–73. [Google Scholar]
- Fan, C., Xing, Y., Mao, H., Lu, T., Han, B., Xu, C., Li, X. and Zhang, Q. (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112: 1164–1671. [DOI] [PubMed] [Google Scholar]
- Fu, Q., Zhang, P.J., Tan, L.B., Zhu, Z.F., Ma, D., Fu, Y.C., Zhan, X.C., Cai, H.W. and Sun, C.Q. (2010) Analysis of QTLs for yield-related traits in Yuanjiang common wild rice (Oryza rufipogon Griff.). J. Genet. Genomics 37: 147–157. [DOI] [PubMed] [Google Scholar]
- Furuta, T., Uehara, K., Angeles-Shim, R.B., Shim, J., Ashikari, M. and Takashi, T. (2014) Development and evaluation of chromosome segment substitution lines (CSSLs) carrying chromosome segments derived from Oryza rufipogon in the genetic background of Oryza sativa L. Breed. Sci. 63: 468–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, R.Y., Jiang, L.R., Zheng, J.S., Wang, T.S., Wang, H.C., Huang, Y.M. and Hong, Z.L. (2013) Genetic bases of rice grain shape: so many genes, so little known. Trends Plant Sci. 18: 218–226. [DOI] [PubMed] [Google Scholar]
- Huang, X.H., Kurata, N., Wei, X.H., Wang, Z.X., Wang, A.H., Zhao, Q., Zhao, Y., Liu, K.Y., Lu, H.Y., Li, W.J.et al. (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800. [DOI] [PubMed] [Google Scholar]
- Jing, Z.B., Chen, Y., Pan, D.J., Qu, Y.Y., Fan, Z.L., Chen, J.Y. and Li, C. (2011) Development and evaluation of primary introgression lines of rice by advanced backcross QTL strategy for Gaozhou wild rice (O. rufipogon). Rice Genomics Genet. 2: 1–11. [Google Scholar]
- Kubo, T., Aida, Y., Nakamura, K., Tsunematsu, H., Doi, K. and Yoshimura, A. (2002) Reciprocal chromosome segment substitution series derived from japonica and indica cross of rice (Oryza sativa L.). Breed. Sci. 52: 319–325. [Google Scholar]
- Lee, S.J., Oh, C.S., Suh, J.P., McCouch, S.R. and Ahn, S.N. (2005) Identification of QTLs for domestication-related and agronomic traits in an Oryza sativa × O. rufipogon BC1F7 population. Plant Breed. 124: 209–219. [Google Scholar]
- Li, Y.B., Fan, C.C., Xing, Y.Z., Jiang, Y.H., Luo, L.J., Sun, L., Shao, D., Xu, C.J., Li, X.H., Xiao, J.et al. (2011) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43: 1266–1269. [DOI] [PubMed] [Google Scholar]
- Liu, X., Zhao, Z.G., Liu, L.L., Xiao, Y.H., Tian, Y.L., Liu, S.J., Chen, L.M., Wang, Y., Liu, Y.Q., Chen, S.H.et al. (2016) Construction of chromosomal segment substitution lines and genetic dissection of introgressed segments associated with yield determination in the parents of a super-hybrid rice. Plant Breed. 135: 63–72. [Google Scholar]
- Marri, P.R., Sarla, N., Reddy, L.V. and Siddiq, E.A. (2005) Identification and mapping of yield and yield related QTLs from an Indian accession of Oryza rufipogon. BMC Genet. 6: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzougui, S., Sugimoto, K., Yamanouchi, U., Shimono, M., Hoshino, T., Hori, K., Kobayashi, M., Ishiyama, K. and Yano, M. (2012) Mapping and characterization of seed dormancy QTLs using chromosome segment substitution lines in rice. Theor. Appl. Genet. 124: 893–902. [DOI] [PubMed] [Google Scholar]
- McCouch, S.R., Teytelman, L., Xu, Y.B., Lobos, K.B., Clare, K., Walton, M., Fu, B.Y., Maghirang, R., Li, Z.K., Xing, Y.Z.et al. (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9: 199–207. [DOI] [PubMed] [Google Scholar]
- McCouch, S.R., Sweeney, M., Li, J.M., Jiang, H., Thomson, M., Septiningsih, E., Edwards, J., Moncada, P., Xiao, J.H., Garris, A.et al. (2007) Through the genetic bottleneck: O. rufipogon as a source of trait-enhancing alleles for O. sativa. Euphytica 154: 317–339. [Google Scholar]
- McCouch, S.R. (2008) Gene nomenclature system for rice. Rice 1: 72–84. [Google Scholar]
- Meng, L., Li, H.H., Zhang, L.Y. and Wang, J.K. (2015) QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 3: 269–283. [Google Scholar]
- Onishi, K., Horiuchi, Y., Ishigoh-Oka, N., Takagi, K., Ichikawa, N., Maruoka, M. and Sano, Y. (2007) A QTL cluster for plant architecture and its ecological significance in Asian wild rice. Breed. Sci. 57: 7–16. [Google Scholar]
- Panaud, O., Chen, X. and McCouch, S.R. (1996) Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol. Gen. Genet. 252: 597–607. [DOI] [PubMed] [Google Scholar]
- Qi, L., Wang, X.N., Zhang, J.Z., Tang, Q.J., Meng, W.D. and Yan, X.W. (2013) Study on genetic diversity and differentiation of three wild rice species in Hainan using SRAP markers. J. Plant Genet. Resour. 14: 402–406. [Google Scholar]
- Qi, P., Lin, Y.S., Song, X.J., Shen, J.B., Huang, W., Shan, J.X., Zhu, M.Z., Jiang, L., Gao, J.P. and Lin, H.X. (2012) The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 22: 1666–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redona, E.D. and Mackill, D.J. (1998) Quantitative trait locus analysis for rice panicle and grain characteristics. Theor. Appl. Genet. 96: 957–963. [Google Scholar]
- Septiningsih, E.M., Prasetiyono, J., Lubis, E., Tai, T.H., Tjubaryat, T., Moeljopawiro, S. and McCouch, S.R. (2003) Identification of quantitative trait loci for yield and yield components in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor. Appl. Genet. 107: 1419–1432. [DOI] [PubMed] [Google Scholar]
- Shim, R.A., Angeles, E.R., Ashikari, M. and Takashi, T. (2010) Development and evaluation of Oryza glaberrima steud. chromosome segment substitution lines (CSSLs) in the background of O. sativa L. cv. Koshihikari. Breed. Sci. 60: 613–619. [Google Scholar]
- Song, X.J., Huang, W., Shi, M., Zhu, M.Z. and Lin, H.X. (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39: 623–630. [DOI] [PubMed] [Google Scholar]
- Song, X.J., Kuroha, T., Ayano, M., Furuta, T., Nagai, K., Komeda, N., Segami, S., Miura, K., Ogawa, D., Kamura, T.et al. (2015) Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc. Nalt. Acad. Sci. USA 112: 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, C.Q., Wang, X.K., Li, Z.C., Yoshimura, A. and Iwata, N. (2001) Comparison of the genetic diversity of common wild rice (Oryza rufipogon Griff.) and cultivated rice (O. sativa L.) using RFLP markers. Theor. Appl. Genet. 102: 157–162. [Google Scholar]
- Tan, L.B., Liu, F.X., Xue, W., Wang, G.J., Ye, S., Zhu, Z.F., Fu, Y.C., Wang, X.K. and Sun, C.Q. (2007) Development of Oryza rufipogon and O. sativa introgression lines and assessment for yield-related quantitative trait loci. J. Integr. Plant Biol. 49: 871–884. [Google Scholar]
- Tan, L.B., Zhang, P.J., Liu, F. X., Wang, G.J., Ye, S., Zhu, Z.F., Fu, Y. C., Cai, H.W. and Sun, C.Q. (2008) Quantitative trait loci underlying domestication- and yield-related traits in an Oryza sativa × Oryza rufipogon advanced backcross population. Genome 51: 692–704. [DOI] [PubMed] [Google Scholar]
- Tanksley, S.D. (1993) Mapping polygenes. Annu. Rev. Genet. 27: 205–233. [DOI] [PubMed] [Google Scholar]
- Temnykh, S., Park, W.D., Ayres, N., Cartinhour, S., Hauck, N., Lipovich, L., Cho, Y.G., Ishii, T. and McCouch, S.R. (2000) Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor. Appl. Genet. 100: 697–712. [Google Scholar]
- Tian, F., Li, D.J., Fu, Q., Zhu, Z.F., Fu, Y.C., Wang, X.K. and Sun, C.Q. (2005) Construction of introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice (Oryza sativa L.) background and characterization of introgressed segments associated with yield-related traits. Theor. Appl. Genet. 112: 570–580. [DOI] [PubMed] [Google Scholar]
- van Berloo, R. (1999) Computer note.GGT.software for the display of graphical genotypes. J. Hered. 90: 328–329. [Google Scholar]
- Wan, X.Y., Wan, J.M., Weng, J.F., Jiang, L., Bi, J.C., Wang, C.M. and Zhai, H.Q. (2005) Stability of QTLs for rice grain dimension and endosperm chalkiness characteristics across eight environments. Theor. Appl. Genet. 110: 1334–1346. [DOI] [PubMed] [Google Scholar]
- Wan, X.Y., Wan, J.M., Jiang, L., Wang, J.K., Zhai, H.Q., Weng, J.F., Wang, H.L., Lei, C.L., Wang, J.L., Zhang, X.et al. (2006) QTL analysis for rice grain length and fine mapping of an identified QTL with stable and major effects. Theor. Appl. Genet. 112: 1258–1270. [DOI] [PubMed] [Google Scholar]
- Wang, J.K., Wan, X.Y., Crossa, J., Crouch, J., Weng, J. F., Zhai, H.Q. and Wan, J.M. (2006) QTL mapping of grain length in rice (Oryza sativa L.) using chromosome segment substitution lines. Genet. Res. 88: 93–104. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., Second, G. and Tanksley, S.D. (1992) Polymorphism and phylogenetic relationships among species in the genus Oryza as determined by analysis of nuclear RFLPs. Theor. Appl. Genet. 83: 565–581. [DOI] [PubMed] [Google Scholar]
- Wei, X., Qiao, W.H., Chen, Y.T., Wang, R.S., Cao, L.R., Zhang, W.X., Yuan, N.N., Li, Z.C., Zeng, H.L. and Yang, Q.W. (2012) Domestication and geographic origin of Oryza sativa in China: insights from multilocus analysis of nucleotide variation of O. sativa and O. rufipogon. Mol. Ecol. 21: 5073–5087. [DOI] [PubMed] [Google Scholar]
- Xiao, J.H., Grandllo, S., Ahn, S.N., McCouch, S.R., Tanksley, S.D., Li, J.M. and Yuan, L.P. (1996) Genes from wild rice improve yield. Nature 384: 223–224. [Google Scholar]
- Xie, X.B., Jin, F.X., Song, M.H., Suh, J.P., Hwang, H.G., Kim, Y.G., McCouch, S.R. and Ahn, S.N. (2008) Fine mapping of a yield-enhancing QTL cluster associated with transgressive variation in an Oryza sativa × O. rufipogon cross. Theor. Appl. Genet. 116: 613–622. [DOI] [PubMed] [Google Scholar]
- Yamamoto, T., Yonemaru, J. and Yano, M. (2009) Towards the understanding of complex traits in rice: substantially or superficially? DNA Res. 16: 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, M. and Sasaki, T. (1997) Genetic and molecular dissection of quantitative traits in rice. Plant Mol. Biol. 35: 145–153. [PubMed] [Google Scholar]
- Yoshida, S., Ikegami, M., Kuze, J., Sawada, K., Hashimoto, Z., Ishii, T., Nakamura, C. and Kamijima, O. (2002) QTL analysis for plant and grain characters of sake-brewing rice using a doubled haploid population. Breed. Sci. 52: 309–317. [Google Scholar]
- Yuan, P.R., Kim, H.J., Chen, Q.H., Ju, H.G., Lee, S.J., Ji, S.D. and Ahn, S.N. (2009) QTL dissection of agronomic and domestication traits using introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice (O. sativa L.) background. J. Crop Sci. Biotechnol. 12: 245–252. [Google Scholar]
- Zhang, X.J., Wang, J.F., Huanga, J., Lan, H.X., Wang, C.L., Yin, C.F., Wu, Y.Y., Tang, H.J., Qian, Q., Li, J.Y.et al. (2012) Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA 109: 21534–21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, K.L., Huang, N., Bennett, J. and Khush, G.S. (1995) PCR-based marker assisted selection in rice breeding. IRRI discussion paper series No. 12. [Google Scholar]
- Zhuang, J.Y., Lin, H.X., Lu, J., Qian, H.R., Hittalmani, S., Huang, N. and Zheng, K.L. (1997) Analysis of QTL × environment interaction for yield components and plant height in rice. Theor. Appl. Genet. 95: 799–808. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.