Abstract
Background
HER2 (ERBB2) gene amplification and its corresponding overexpression are present in 15–30% of invasive breast cancers. While HER2-targeted agents are effective treatments, resistance remains a major cause of death. The American College of Surgeons Oncology Group Z1041 trial (NCT00513292) was designed to compare the pathologic complete response (pCR) rate of distinct regimens of neoadjuvant chemotherapy and trastuzumab, but ultimately identified no difference.
Patients and methods
In supplement to tissues from 37 Z1041 cases, 11 similarly treated cases were obtained from a single institution study (NCT00353483). We have extracted genomic DNA from both pre-treatment tumor biopsies and blood of these 48 cases, and performed whole genome (WGS) and exome sequencing. Coincident with these efforts, we have generated RNA-seq profiles from 42 of the tumor biopsies. Among patients in this cohort, 24 (50%) achieved a pCR.
Results
We have characterized the genomic landscape of HER2-positive breast cancer and investigated associations between genomic features and pCR. Cases assigned to the HER2-enriched subtype by RNA-seq analysis were more likely to achieve a pCR compared to the luminal, basal-like, or normal-like subtypes (19/27 versus 3/15; P = 0.0032). Mutational events led to the generation of putatively active neoantigens, but were overall not associated with pCR. ERBB2 and GRB7 were the genes most commonly observed in fusion events, and genomic copy number analysis of the ERBB2 locus indicated that cases with either no observable or low-level ERBB2 amplification were less likely to achieve a pCR (7/8 versus 17/40; P = 0.048). Moreover, among cases that achieved a pCR, tumors consistently expressed immune signatures that may contribute to therapeutic response.
Conclusion
The identification of these features suggests that it may be possible to predict, at the time of diagnosis, those HER2-positive breast cancer patients who will not respond to treatment with chemotherapy and trastuzumab.
ClinicalTrials.gov identifiers
Keywords: breast cancer, HER2, ERBB2, trastuzumab, pathologic complete response, residual disease
Introduction
In breast cancer, ERBB2 gene amplification, and expression of its corresponding protein, HER2, is observed in ∼15–30% of invasive tumors [1, 2], and is associated with poor prognosis [3]. In these cases, the addition of HER2-targeted agents (e.g. trastuzumab, pertuzumab, and lapatinib) to neoadjuvant chemotherapy increases pathological complete response (pCR) rates [4], which in turn reduces the risk of recurrence and death [5, 6]. Nonetheless, therapeutic resistance remains a concern, with ∼40–50% of patients having residual disease (RD) after neoadjuvant treatment with chemotherapy and trastuzumab [4, 7].
The American College of Surgeons Oncology Group Z1041 trial (NCT00513292) was designed to compare the pCR rate of HER2-positive breast cancer patients that received either a sequential or concurrent regimen of chemotherapy and trastuzumab (Methods). Initial results from the trial identified no significant difference between the arms [8], suggesting that concurrent administration of trastuzumab with anthracyclines offers no additional benefit to patients.
As a monoclonal antibody, trastuzumab relies in part on the immune system to recognize and target HER2-expressing cells. However, there are conflicting reports of whether germline polymorphisms in Fc-gamma receptors, found on the surface of various immune cells and involved in the response to antibodies including trastuzumab, are associated with different outcomes in HER2-positive breast cancer cases that received adjuvant chemotherapy and trastuzumab [9–11]. Moreover, germline polymorphisms in ABCB1, an efflux pump that transports chemotherapeutic agents out of cells, have been identified as prognostic markers in HER2-positive patients [10]. Additionally, in the neoadjuvant setting it has been reported that somatic events are associated with differential response rates to chemotherapy and trastuzumab. Breast cancers assigned to the HER2-enriched subtype have been reported as having higher pCR rates than those assigned to the luminal A or luminal B subtypes [12]. Similarly, expression of the estrogen receptor alpha (ER) has been associated with higher RD rates [12]. While it has been reported that cases with PIK3CA mutations have higher rates of RD [13–15], these findings have not been universally observed [12].
Overall however, mechanisms of resistance remain poorly understood, and without a comprehensive molecular understanding of these mechanisms, therapeutic advances will be delayed. We have therefore performed massively parallel sequencing on 48 HER2-positive breast tumor biopsies and matched blood accrued from either the Z1041 trial or a local neoadjuvant tissue collection study (NCT00353483). All cases were identified as HER2-positive at diagnosis and received neoadjuvant chemotherapy and trastuzumab. We subsequently analysed the genomic landscape that defines HER2-positive breast cancer, and furthermore, identified molecular features that differentiate those cases that achieved a pCR from those that had RD.
Methods
37 cases were accessed from the American College of Surgeons Oncology Group Z1041 trial (NCT00513292) that compared the pCR rate of patients with HER2-positive breast cancer. Patients were randomly assigned to one of two arms; the first received fluorouracil 500 mg/m2, epirubicin 75 mg/m2, and cyclophosphamide 500 mg/m2 (FEC-75) on day 1 of a 21-day cycle for four cycles followed by paclitaxel 80 mg/m2 and trastuzumab 2 mg/kg (after a 4-mg/kg loading dose) once per week for 12 weeks, while the second group received paclitaxel and trastuzumab once per week for 12 weeks followed by four cycles of FEC-75 (on day 1 of each 21-day cycle) and once-weekly trastuzumab, in the same doses as the first group. In addition to the patients enrolled in the Z1041 trial, 11 HER2-positive breast cancer cases included in this study were enrolled in a local trial at Washington University School of Medicine (NCT00353483) and received neoadjuvant treatment with trastuzumab in combination with other chemotherapies. All cases were females with invasive breast cancer, not pregnant, and ≥ 18 years of age.
Institutional review boards of the participating institutions approved the study. All participants gave written informed consent.
For all cases, pre-treatment snap-frozen tumor tissue and normal blood were obtained. All cases analysed in this study were identified as HER2-positive by IHC and/or FISH.
All 48 pre-treatment tumor biopsy and blood pairs were assayed by whole genome sequencing (WGS) and whole exome sequencing (WES), and 42 of these tumor biopsies were additionally assayed by RNA sequencing (RNA-seq) (supplementary Figure S1, available at Annals of Oncology online). Alignment and variant calling were performed as previously described [16]. All sequence data have been deposited in the Database of Genotypes and Phenotypes (dbGAP) (accession: phs001291.v1.p1).
Additional details are provided in the Supplementary Methods, available at Annals of Oncology online.
Results
Patient characteristics
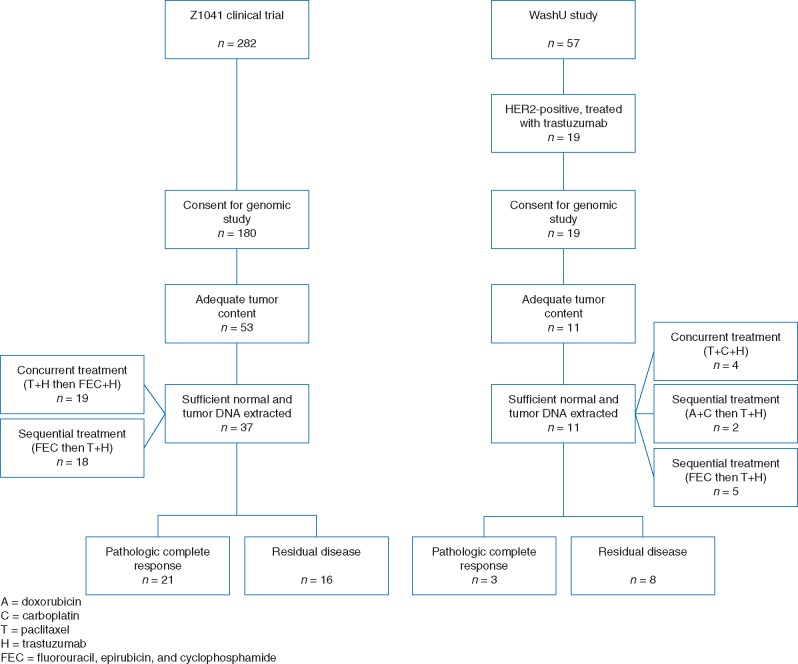
The patient cohort investigated in this study consists of 48 HER2-positive breast cancer cases treated with neoadjuvant chemotherapy and trastuzumab. Half of selected patients (n = 24) achieved a pCR after treatment, while the other half (n = 24) had RD (Figure 1 and Table 1; supplementary Table S1, available at Annals of Oncology online).
Figure 1.
REMARK diagram detailing the study cohort.
Table 1.
Summary of the clinical and molecular features of the cohort in this study
| Unstratified | pCR achieved | Residual disease | |
|---|---|---|---|
| Number of cases | 48 | 24 | 24 |
| Age | |||
| Median | 50 | 50 | 50 |
| Range | 36–70 | 39–70 | 36–67 |
| Unknown | 0 (0%) | 0 (0%) | 0 (0%) |
| Clinical primary tumor stage (T) | |||
| T1 | 2 (4.17%) | 1 (4.17%) | 1 (4.17%) |
| T2 | 24 (50%) | 14 (58.3%) | 10 (41.7%) |
| T3 | 16 (33.3%) | 6 (25.0%) | 10 (41.7%) |
| T4 | 6 (12.5%) | 3 (12.5%) | 3 (12.5%) |
| Clinical lymph node stage (N) | |||
| N0 | 16 (33.3%) | 7 (29.2%) | 9 (37.5%) |
| N1 | 25 (52.1%) | 14 (58.3%) | 11 (45.8%) |
| N2 | 6 (12.5%) | 2 (8.33%) | 4 (16.7%) |
| N3 | 1 (2.08%) | 1 (4.17%) | 0 (0%) |
| Unknown | 0 (0%) | 0 (0%) | 0 (0%) |
| ER status prior to neoadjuvant treatment | |||
| Positive | 28 (58.3%) | 11 (45.8%) | 17 (70.8%) |
| Negative | 20 (41.7%) | 13 (54.2%) | 7 (29.2%) |
| Unknown | 0 (0%) | 0 (0%) | 0 (0%) |
| PR status prior to neoadjuvant treatment | |||
| Positive | 21 (43.8%) | 7 (29.2%) | 14 (58.3%) |
| Negative | 26 (54.2%) | 16 (66.7%) | 10 (41.7%) |
| Unknown | 1 (2.08%) | 1 (4.17%) | 0 (0%) |
| HER2 status prior to neoadjuvant treatment | |||
| Positive | 48 (100%) | 24 (100%) | 24 (100%) |
| Negative | 0 (0%) | 0 (0%) | 0 (0%) |
| Unknown | 0 (0%) | 0 (0%) | 0 (0%) |
| Grade | |||
| 1 | 1 (2.08%) | 0 (0%) | 1 (4.17%) |
| 2 | 12 (25%) | 7 (29.2%) | 5 (20.8%) |
| 3 | 34 (70.8%) | 17 (70.8%) | 17 (70.8%) |
| Unknown | 1 (2.08%) | 0 (0%) | 1 (4.17%) |
| PAM50 subtype | |||
| Luminal A | 12 (25%) | 2 (8.33%) | 10 (41.7%) |
| Luminal B | 11 (22.9%) | 8 (33.3%) | 3 (12.5%) |
| HER2-enriched | 14 (29.2%) | 11 (45.8%) | 3 (12.5%) |
| Basal-like | 5 (10.4%) | 1 (4.17%) | 4 (16.7%) |
| Normal-like | 0 (0%) | 0 (0%) | 0 (0%) |
| Unknown | 6 (12.5%) | 2 (8.33%) | 4 (16.7%) |
| AIMS subtype | |||
| Luminal A | 2 (4.17%) | 0 (0%) | 2 (8.33%) |
| Luminal B | 8 (16.7%) | 3 (12.5%) | 5 (20.8%) |
| HER2-enriched | 27 (56.2%) | 19 (79.2%) | 8 (33.3%) |
| Basal-like | 4 (8.33%) | 0 (0%) | 4 (16.7%) |
| Normal-like | 1 (2.08%) | 0 (0%) | 1 (4.17%) |
| Unknown | 6 (12.5%) | 2 (8.33%) | 4 (16.7%) |
| Recurrence | |||
| Recurrence event | 9 (18.8%) | 1 (4.17%) | 8 (33.3%) |
| No recurrence event | 39 (81.2%) | 23 (95.8%) | 16 (66.7%) |
| Unknown | 0 (0%) | 0 (0%) | 0 (0%) |
| Source of specimens | |||
| Z1041 trial (NCT00513292) | 37 (77.1%) | 21 (87.5%) | 16 (66.7%) |
| WashU study (NCT00353483) | 11 (22.9%) | 3 (12.5%) | 8 (33.3%) |
In our study cohort, patients that achieved a pCR tended to have smaller tumors, clinical node negative disease, and be assigned to the HER2-enriched subtype. Although ER status, PR status, and grade were not associated with pCR, PAM50 subtype, AIMS subtype, and disease recurrence were significantly associated (supplementary Table S2, available at Annals of Oncology online). This trend was reduced after stratification on ER status, although the association between PAM50 and pCR as well as AIMS and pCR was significant within the ER-negative set, while only the association between PAM50 and pCR was significant in the ER-positive set. Among all cases, those assigned to the HER2-enriched subtype by RNA-seq analysis were significantly more likely to achieve a pCR compared to tumors assigned to luminal A, luminal B, basal-like, and normal-like subtypes (PAM50: 11/14 versus 11/28, P = 0.023; AIMS: 19/27 versus 3/15, P = 0.0032). About 57% of all cases were assigned to the same subtype in both the PAM50 and AIMS schemes, which is lower than the 77% overlap previously reported for these schemes [17].
Germline polymorphisms
In order to investigate the landscape of germline variants in HER2-positive breast cancer, we analysed the normal genomes of each patient for known pathogenic variants. We identified 196 pathogenic germline variants (482 total events) as well as 19 likely pathogenic germline variants (27 total events), with a median of 10 and 0 variants per patient, respectively (supplementary Table S3, available at Annals of Oncology online). Affected genes include those involved in DNA repair (ATM, BRCA2, MUTYH), cell cycle (FANCA), the Wnt pathway (LRP5, WNT10A), the PI3K pathway, (PIK3R5), and telomere maintenance (TERT). For each variant, the proportion of cases with that variant did not differ between pCR and RD groups (FDR > 0.2). Similarly, no significant differences were observed in the number of variants in a given gene between the two patient groups (FDR > 0.2).
Previous studies have reported germline polymorphisms as predictive of prognostic outcome in HER2-positive breast cancer patients treated with trastuzumab and chemotherapy [10, 11]. These polymorphisms are found in genes including P-glycoprotein (ABCB1) and Fc gamma receptors (FCGR2A, FCGR2B, FCGR3A). In our study, however, the rates of alternate alleles at these polymorphisms did not differ between those patients that achieved a pCR and those with RD (P > 0.05, supplementary Table S4, available at Annals of Oncology online).
The somatic variant landscape of HER2-positive breast cancer
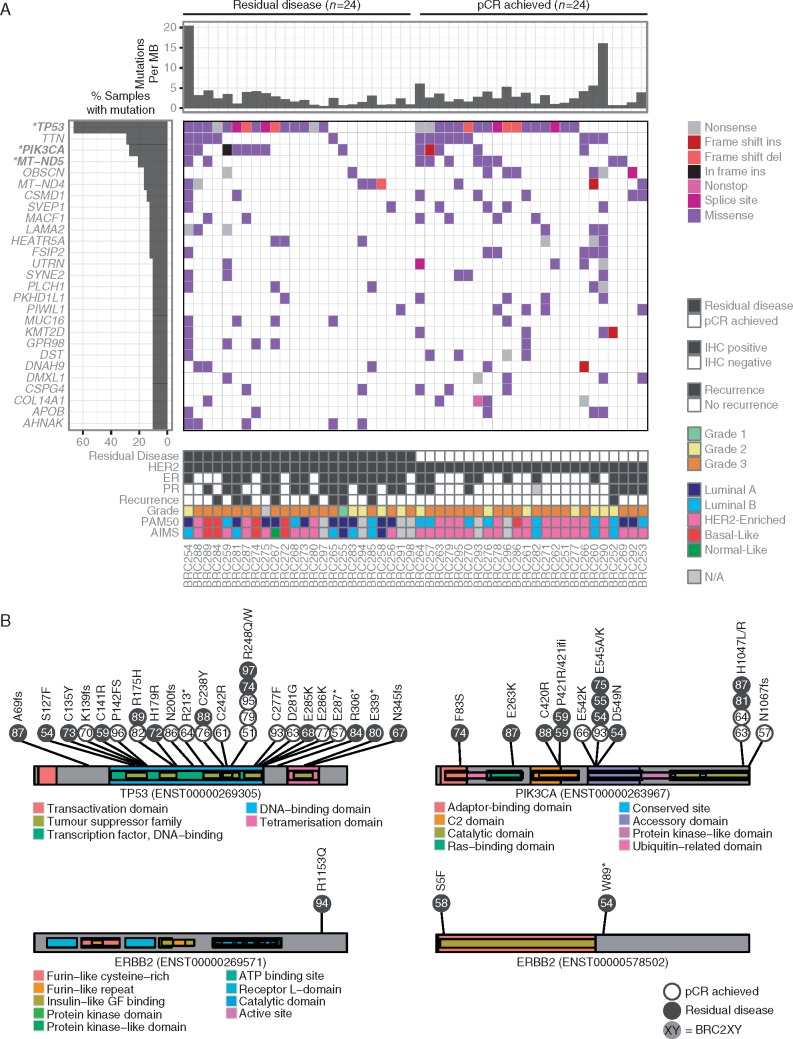
Across all 48 cases, 6685 somatic events including single nucleotide variants (SNVs) and small insertions/deletions (indels) were identified in coding regions, including 5751 missense, 531 nonsense, 20 nonstop, and 165 frameshift insertions or deletions (Figure 2A and supplementary Table S5, available at Annals of Oncology online). TP53, PIK3CA, and MT-ND2 were the only genes mutated at significantly higher rates than expected by chance (supplementary Table S6, available at Annals of Oncology online, FDR < 0.2). No significant differences were observed in the mutation rates for any gene between cases that achieved a pCR versus those that had RD (P > 0.05).
Figure 2.
(A) Somatic variants and small indels are visually represented across the most recurrently mutated genes. Only TP53, PIK3CA, and MT-ND5 were mutated at rates significantly higher than random across the entire cohort (FDR < 0.2, marked in bold with an asterisk). No genes were mutated at significantly different rates between cases that achieved a pCR versus those that had residual disease. (B) Amino acid locations affected by somatic mutations are shown for TP53, PIK3CA, and ERBB2. Individual mutations are colored by whether they achieved a pCR or had residual disease, and are labeled according to their case identifier. Mutations in TP53 were spread across the protein, but PIK3CA mutations in the adaptor-binding, C2, and Ras-binding domains were only observed among cases that had residual disease. Two of the three mutations in ERBB2 were only predicted to be found in a short isoform.
27 somatic mutations were observed in TP53, the most frequently mutated gene (Figure 2B). Mutations were highly variable across p53 protein domains, and we did not observe apparent differences in the domain-specific mutation rates or predicted effect of mutations between cases that achieved a pCR versus those that had RD. In contrast, somatic mutations in the adaptor-binding, C2, and Ras-binding domains of PIK3CA were only observed among cases that had RD, indicating that there may be functional differences in the activity and regulation of the protein (Figure 2B). Somatic mutations in ERBB2 were identified in three cases, all of which failed to achieve pCR (Figure 2B). BRC254 was observed to have a nonsense mutation (p.W89*) that is expected to result in early truncation of the gene. However, this truncation was observed to affect a short isoform of the gene (ENST00000578502) that only contains part of the corresponding extracellular protein domains. Similarly, BRC258 was observed to have a missense mutation (p.S5F) that affected the same short isoform, causing an amino acid substitution within the insulin-like growth factor binding domain. Finally, BRC294 was observed to have a missense mutation (p.R1153Q) causing an amino acid substitution within the terminus of the intracellular portion of the protein, but not targeting known protein domains. To our knowledge, these mutations have not been previously reported, and none of the mutations were found in mutational ‘hot spots’.
We further investigated whether molecular pathways were enriched for somatic SNVs and indels. Various pathways were observed to be significantly enriched in all 48 cases, including those related to cell cycle, PI3K-Akt signaling, p53 signaling, Wnt signaling, focal adhesion, TGF-beta signaling, ErbB signaling, and immune function (supplementary Table S7, available at Annals of Oncology online). Moreover, those pathways found to be significant among cases that achieved a pCR tended to also be significant among cases that had RD. This observation is in line with our previous finding that no gene was mutated at significantly different rates between these two groups.
More broadly, using genome-wide somatic mutations we predicted the clonal architecture of individual cases. The number of subclones ranged from two to seven across all cases, but the subclone rates were not significantly different between cases that achieved a pCR versus those with RD (P = 0.356). Genome-wide mutations were dominated by C > T and C > G changes (or equivalently, G > A and G > C changes). These mutations were characterized as being predominantly attributed to mutational signature 1 (associated with age), and to a lesser extent signatures 2 and 13 (associated with the APOBEC family of cytidine deaminases), as well as signature 3 (associated with dysfunctional BRCA repair) [18, 19] (supplementary Table S8, available at Annals of Oncology online). These observations were mostly consistent with previous findings, although we did not observe contributions from signature 5 (aetiology unknown), which has been reported to be a prominent signature among HER2-positive cases [19]. Interestingly, we identified examples where the contribution of mutational signatures to the somatic landscape of tumors was significantly different between cases that achieved a pCR versus those that had RD. Signature 13 contributed significantly more to the landscape of cases with RD (P = 0.0401), and signature 3 contributed more to the landscape of cases that achieved a pCR (P = 0.0540).
Copy number changes and RNA expression associate with therapeutic response
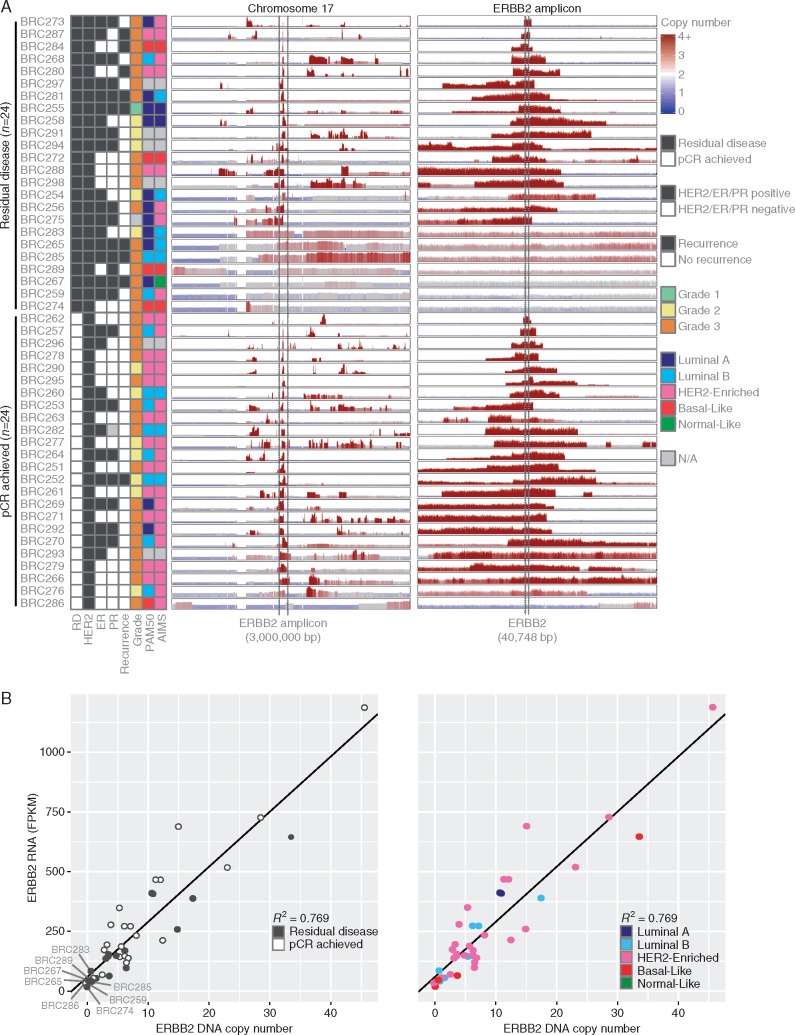
Copy number aberrations previously characterized in breast cancer were commonly observed in our cohort (supplementary Figures S2 and S3, available at Annals of Oncology online). In particular, we observed focal amplifications in 17q, whole arm gains in 1q, 8q, and 20q, and whole arm deletions in 1p, 8p, and 17p. Similar patterns were observed between cases that achieved a pCR and those with RD; in a genome-wide analysis, no regions were found to have copy number gains or losses at significantly different rates (FDR > 0.2).
Most tumors in our cohort had observable copy number gain of the genomic region surrounding ERBB2 (Figure 3A). Consistent with what has been reported previously, nearly all these gains were focal amplifications rather than whole arm, although the copy number and size of the amplified region varied within both patient groups. There were no cases where we observed copy number gains of both alleles, indicating that possible germline variant heterozygosity of the HER2 protein was not a determinant of therapeutic response. However, we observed eight cases with either no observable (BRC259, BRC267, BRC274, BRC286) or low-level (BRC265, BRC283, BRC285, BRC289) ERBB2 copy number gain. These cases were significantly less likely to achieve pCR, with seven out of the eight cases having RD (P = 0.048). Interestingly, all four cases with no observable gain were confirmed to have a score of 3+ for HER2 IHC, and case BRC259 was additionally observed to have an average of >20 copies of ERBB2 per nucleus assessed by Fluorescent In Situ Hybridization (FISH). All eight cases had excellent WGS tumor coverage (median mean coverage = 53.5×; mean coverage range = 42×–59×), and high maximum WGS variant allele frequencies (median 67%, range 46–95%), indicating that these samples all had high tumor purity.
Figure 3.
(A) DNA copy number aberrations are shown for all cases across chromosome 17 and the ERBB2 amplicon. Predicted copy number gains are shown in red and losses in blue at 10 000 bp windows across the genome. The height of each panel was separately scaled for each case according to the maximum copy number value observed. Cases are ordered by whether they had residual disease or achieved a pCR. (B) ERBB2 RNA FPKM expression values and DNA copy number values are plotted against each other for the 42 cases where RNA sequencing was performed. The values were highly correlated (Spearman R2 = 0.769). Cases with low ERBB2 levels (bottom left corner of each plot) tended to have a residual disease and to not be classified as HER2-enriched.
Across the cohort, ERBB2 RNA-seq (FPKM) and copy number values were highly correlated (Spearman R2 = 0.769, Figure 3B). Although BRC286 achieved a pCR, the tumor from this case was observed to have copy number loss of ERBB2 and a relatively lower level of ERBB2 expression within the bulk profile (FPKM = 30.066).
More broadly within the RNA-seq data, no genes or distinct transcript isoforms were found to be univariately differentially expressed between cases that achieved pCR versus those with RD (FDR > 0.2). At the multivariate level, however, we did observe significant differences in the expression of gene signatures. About 1531 out of 10 959 signatures tested were enriched in cases that achieved a pCR (supplementary Table S9, available at Annals of Oncology online, P < 0.05 and FDR < 0.2). These signatures were predominantly related to immune categories (P = 8.69×10 − 42), with T-Cell (P = 5.49×10 − 21), B-Cell (P = 1.17×10 − 14), and inflammatory (P = 0.0203) signatures all significantly enriched. In contrast, no signatures were enriched in the RD group, indicating that these cases have greater heterogeneity than those that achieved a pCR.
A total of 311 gene fusion events were observed in the cohort, with a median of 6.5 and range of 1–27 events (supplementary Table S10, available at Annals of Oncology online). No recurrent fusion involving the same two genes was observed, however 62 genes were involved in a fusion in at least two cases. ERBB2 was the gene most frequently involved in fusion events, with five events observed in four cases (BRC260, BRC272, BRC287, BRC295), followed by GRB7 with four events in three cases (BRC251, BRC262, BRC295). Two of the four cases with ERBB2 gene fusions achieved a pCR, while all of the three cases with GRB7 gene fusions achieved a pCR. These results suggest that genomic instability in the ERBB2 amplicon is the predominant source for gene fusions in HER2-positive breast cancer, although such fusions do not lead to therapeutic resistance.
Neoantigen profiles of HER2-positive breast cancer
Given the observed role for T-Cell immune response in patients that achieved a pCR, we predicted in silico the tumor neoantigens and binding affinity scores for each case (supplementary Figure S4, available at Annals of Oncology online). Among neoantigens with binding affinity scores low enough to putatively elicit an immune response, loads ranged from 0 to 2584 epitopes across all cases. Cases that achieved a pCR had nonsignificantly higher loads compared to those with RD (P > 0.05). The genes that most frequently produced putatively active neoantigens were similar to those that were most frequently mutated in our cohort, including TP53 and PIK3CA (supplementary Table S11, available at Annals of Oncology online). Most peptides predicted to elicit an immune response were only observed in a single case, and none were observed in more than two cases (supplementary Table S12, available at Annals of Oncology online).
Prediction of therapeutic response using combined genomic data levels
In order to identify whether interactions between different data levels of genomic features are informative of response to chemotherapy and trastuzumab, we computationally generated predictors of pCR using copy number, gene mutation, and gene isoform expression data. Using these data, we were able to achieve an area under the curve (AUC) of 0.720 for predicting which cases achieved a pCR or had RD (supplementary Figure S5, available at Annals of Oncology online). Transcriptional features were consistently selected as the most influential in making these predictions (supplementary Table S13, available at Annals of Oncology online), and included isoforms of genes such as NOP56, SCARB1, LAD1, ETFA, and LTBP3. This result is consistent with our previous observations that RNA features are able to differentiate these two classes, while copy number and mutational data have a limited ability to do so. Similar results were observed when we constructed predictors of therapeutic response using only the transcriptional, copy number, or mutational data. A successful predictor was trained using the transcriptional data (AUC = 0.725) and to a lesser extent using the mutational data (AUC = 0.663), but the copy number data was not informative (AUC = 0.550).
Discussion
Here we have characterized the genomic landscape of HER2-positive breast cancer, and further identified a set of recurrent somatic events in HER2-positive breast cancer that may aid in the identification of patients, at the time of diagnosis, who will not have a pCR after treatment with chemotherapy and trastuzumab.
It is interesting to note that while all 48 cases were clinically classified as HER2-positive, we identified four cases with no observable ERBB2 copy number gains, and an additional four with very low level, whole chromosome arm gains. Although all eight cases were scored as 3+ by IHC, seven of them had RD, together indicating that massively parallel sequencing technologies provide predictive information in complement to clinical IHC/FISH. Moreover, we observed three cases with somatic mutations in ERBB2; all three cases had RD. Although the mutations did not directly confer obvious resistance by alteration of domains within the full-length protein, it is possible that these somatic events led to the generation of neoantigens, thereby allowing the host immune system to target the cancer cells harboring these mutations.
In addition to somatic events that directly alter ERBB2, we identified broad patterns at the RNA level that were associated with response to chemotherapy and trastuzumab. This is consistent with a previous report that transcriptional features are more predictive than other data types of drug response in breast cancer cell lines [20]. In particular, cases that achieved a pCR had evidence of an activated immune response, including T-Cell, B-Cell, and inflammatory signatures. In contrast, cases with RD displayed evidence of greater heterogeneity as no RNA signatures were enriched among this group. This hypothesis is further reinforced by the observation that cases classified as HER2-enriched achieved significantly higher rates of pCR compared to those classified as luminal A, luminal B, basal-like, or normal-like.
Supplementary Material
Acknowledgements
We are grateful to all of the patients that participated in this study. We also thank the McDonnell Genome Institute’s LIMS, Analysis Pipeline, and Systems groups for developing and maintaining the automated sequence analysis pipelines used in this work.
Funding
This work was supported by grants from the National Institutes of Health (NIH) [U10CA180821, U10CA180882, U10CA76001], with support for analysis from NIH National Human Genome Research Institute (NHGRI) [U54HG003079]. OLG was supported by a NIH National Cancer Institute grant [K22CA188163]. MG was supported by a NIH NHGRI grant [K99HG007940]. The funding bodies had no role in the design of the study, in collection, analysis, and interpretation of data, or in writing the article.
Disclosure
FM-B was supported by research grants with Genentech and Puma Biotechnology, is on an advisory committee with Genentech, and receives honoraria from Roche. All remaining authors have declared no conflicts of interest.
References
- 1. Press MF, Bernstein L, Thomas PA. et al. HER-2/neu gene amplification characterized by fluorescence in situ hybridization: poor prognosis in node-negative breast carcinomas. J Clin Oncol 1997; 15: 2894–2904. [DOI] [PubMed] [Google Scholar]
- 2. Slamon DJ, Clark GM, Wong SG. et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987; 235: 177–182. [DOI] [PubMed] [Google Scholar]
- 3. Ravdin PM, Chamness GC.. The c-erbB-2 proto-oncogene as a prognostic and predictive marker in breast cancer: a paradigm for the development of other macromolecular markers – a review. Gene 1995; 159: 19–27. [DOI] [PubMed] [Google Scholar]
- 4. Hamy-Petit AS, Belin L, Bonsang-Kitzis H. et al. Pathological complete response and prognosis after neoadjuvant chemotherapy for HER2-positive breast cancers before and after trastuzumab era: results from a real-life cohort. Br J Cancer 2016; 114: 44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim MM, Allen P, Gonzalez-Angulo AM. et al. Pathologic complete response to neoadjuvant chemotherapy with trastuzumab predicts for improved survival in women with HER2-overexpressing breast cancer. Ann Oncol 2013; 24: 1999–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Untch M, Fasching PA, Konecny GE. et al. Pathologic complete response after neoadjuvant chemotherapy plus trastuzumab predicts favorable survival in human epidermal growth factor receptor 2-overexpressing breast cancer: results from the TECHNO trial of the AGO and GBG study groups. J Clin Oncol 2011; 29: 3351–3357. [DOI] [PubMed] [Google Scholar]
- 7. Buzdar AU, Valero V, Ibrahim NK. et al. Neoadjuvant therapy with paclitaxel followed by 5-fluorouracil, epirubicin, and cyclophosphamide chemotherapy and concurrent trastuzumab in human epidermal growth factor receptor 2-positive operable breast cancer: an update of the initial randomized study population and data of additional patients treated with the same regimen. Clin Cancer Res 2007; 13: 228–233. [DOI] [PubMed] [Google Scholar]
- 8. Buzdar AU, Suman VJ, Meric-Bernstam F. et al. Fluorouracil, epirubicin, and cyclophosphamide (FEC-75) followed by paclitaxel plus trastuzumab versus paclitaxel plus trastuzumab followed by FEC-75 plus trastuzumab as neoadjuvant treatment for patients with HER2-positive breast cancer (Z1041): a randomised, controlled, phase 3 trial. Lancet Oncol 2013; 14: 1317–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hurvitz SA, Betting DJ, Stern HM. et al. Analysis of Fcgamma receptor IIIa and IIa polymorphisms: lack of correlation with outcome in trastuzumab-treated breast cancer patients. Clin Cancer Res 2012; 18: 3478–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim JW, Kim JH, Im SA. et al. ABCB1, FCGR2A, and FCGR3A polymorphisms in patients with HER2-positive metastatic breast cancer who were treated with first-line taxane plus trastuzumab chemotherapy. Oncology 2012; 83: 218–227. [DOI] [PubMed] [Google Scholar]
- 11. Norton N, Olson RM, Pegram M. et al. Association studies of Fcgamma receptor polymorphisms with outcome in HER2+ breast cancer patients treated with trastuzumab in NCCTG (Alliance) Trial N9831. Cancer Immunol Res 2014; 2: 962–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carey LA, Berry DA, Cirrincione CT. et al. Molecular heterogeneity and response to neoadjuvant human epidermal growth factor receptor 2 targeting in CALGB 40601, a randomized phase III trial of paclitaxel plus trastuzumab with or without lapatinib. J Clin Oncol 2016; 34: 542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Loibl S, Majewski I, Guarneri V. et al. PIK3CA mutations are associated with reduced pathological complete response rates in primary HER2-positive breast cancer: pooled analysis of 967 patients from five prospective trials investigating lapatinib and trastuzumab. Ann Oncol 2016; 27: 1519–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Loibl S, von Minckwitz G, Schneeweiss A. et al. PIK3CA mutations are associated with lower rates of pathologic complete response to anti-human epidermal growth factor receptor 2 (her2) therapy in primary HER2-overexpressing breast cancer. J Clin Oncol 2014; 32: 3212–3220. [DOI] [PubMed] [Google Scholar]
- 15. Shi W, Jiang T, Nuciforo P. et al. Pathway level alterations rather than mutations in single genes predict response to HER2-targeted therapies in the neo-ALTTO trial. Ann Oncol 2017; 28: 128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Griffith M, Griffith OL, Smith SM. et al. Genome modeling system: a knowledge management platform for genomics. PLoS Comput Biol 2015; 11: e1004274.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paquet ER, Hallett MT.. Absolute assignment of breast cancer intrinsic molecular subtype. J Natl Cancer Inst 2015; 107: 357.. [DOI] [PubMed] [Google Scholar]
- 18. Alexandrov LB, Nik-Zainal S, Wedge DC. et al. Signatures of mutational processes in human cancer. Nature 2013; 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nik-Zainal S, Davies H, Staaf J. et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016; 534: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Daemen A, Griffith OL, Heiser LM. et al. Modeling precision treatment of breast cancer. Genome Biol 2013; 14: R110.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.