Abstract
Phylogenetic trees underpin reconstructions of evolutionary history and tests of evolutionary hypotheses. They are inferred from both molecular and morphological data, yet the relative value of morphology has been questioned in this context due to perceived homoplasy, developmental linkage, and nonindependence of characters. Nevertheless, fossil data are limited to incomplete subsets of preserved morphology, and different regions are treated as equivalent. Through meta-analysis of 40 data sets, we show here that the dental and osteological characters of mammals convey significantly different phylogenetic signals, and that osteological characters are significantly more compatible with molecular trees. Furthermore, the application of simplified paleontological filters (retaining only dental data) results in significantly greater loss of phylogenetic signal than random character ablation. Although the mammal fossil record is largely comprised of teeth, dental data alone are generally found to be less reliable for phylogenetic reconstruction given their incongruence with osteological and molecular data. These findings highlight the need for rigorous meta-analyses of distributions of homoplasy in morphological data. These tests, and consequent refinements to phylogenetic analyses that they permit, promise to improve the quality of all macroevolutionary studies that hinge on accurate trees. [Homoplasy; Mammalia; morphology; osteology; phylogeny; teeth
As with all data used to infer phylogeny, morphological characters should be independent, and character states homologous. For molecular sequence data, modeling approaches provide objective tests and qualification of these assumptions. Morphological data, however, are more intractable; anatomical character complexes are subject to comparatively greater levels of developmental and functional linkage, ecological convergence, and subjective interpretation (O’Keefe and Wagner 2001; Scotland 2003; DeGusta 2004; Kangas et al. 2004; Evans et al. 2007; Minelli 2007; Springer et al. 2007; Sadleir and Makovicky 2008; Harjunmaa et al. 2014). Nevertheless, morphology can be hugely informative under many circumstances. Not only can it increase resolution and reveal hidden support (Gatesy et al. 1999; Gatesy and Baker 2005), but because it is usually the only class of data available from fossils, it is the best way to address historic patterns, break up long branches, and calibrate molecular clocks (Wiens 2004; Cobbett et al. 2007; Benton et al. 2005). It is, therefore, necessary to know whether some classes of morphology are more reliable than others. A better understanding of the distribution of homoplasy across different character types would enable more sophisticated treatments of morphological data and would be especially beneficial where the decisiveness of data is poor.
Here we focus on the dental and osteological data of mammals. Dental data have been used alone and alongside other morphological data to reconstruct a wide range of evolutionary transitions from the radiation of mammals (Bi et al. 2014), to the relationships of human ancestors (Strait and Grines 2004). The focus and reliance on dental data are necessitated in large measure by the taphonomic filter particular to the group; the enhanced preservation potential of mammal teeth relative to bones means that many fossil species are known only from dental data. This makes testing for differences between readily fossilizable data (teeth) and less-fossilizable data (osteology) a priority because taphonomic biases can systematically undermine evolutionary inferences (Sansom et al. 2010, 2011, 2013, 2014; Sansom and Wills 2013). In addition to that, particular concerns about character linkage and overall levels of homoplasy in teeth have been raised on several grounds. Studies of tooth development in mice have revealed dramatic and correlated changes in the number and shape of cusps resulting from minor manipulation of developmental pathways (Kangas et al. 2004). This suggests that dental characters are nonindependent and this is supported by subsequent developmental studies focusing on phenotypes and traits (Harjunmaa et al. 2014). Furthermore, measures of phenotypic complexity indicate that the morphology of mammal teeth is strongly tied to function such that form is highly homoplastic and contingent on diet (Evans et al. 2007). Aside from developmental and functional linkage, comparison of levels of dissimilarity in molecular and dental characters of phyllostomid bats found the latter to be oversaturated and potentially nonindependent (Dávalos et al. 2014), with the inference that this problem could be more widespread. Nondental anatomy is also expected to exhibit some degree of nonindependence and linkage; for example, serial homology is observed in tetrapod limbs (e.g., Ruvinksy and Gibson-Brown 2000; Young and Hallgrímsson 2005). Nevertheless, it is mammal teeth for which developmental linkage of phylogenetic characters has been empirically demonstrated (Harjunmaa et al. 2014).
Investigations of morphological partitions of empirical data (dental, cranial, and postcranial) have found similar levels of homoplasy in some cases (e.g., Sánchez-Villagra andWilliams 1998; Mounce et al. 2016). As such, the characters partitions have been interpreted as equally informative. The same is true for isolated studies of primates (Williams 2007) and Cetartiodactylia (O’Leary et al. 2003), although an earlier analysis of the latter’s data found teeth to be “markedly different from the rest of the morphological data” (Naylor and Adams 2001, p. 451) [see O’Leary et al. (2003) and Naylor and Adams (2001) for further discussion]. This raises a number of questions: Why is there an apparent discordance between developmental studies and empirical morphological data regarding the relative phylogenetic informativeness of tooth morphology? Are there differences between the phylogenetic signals contained within dental and osteological morphology? Can dental data alone be used reliably to reconstruct the evolutionary history of mammals? To answer these questions we compiled morphological data sets comprising dental and osteological characters for a broad range of mammal clades, both extant and extinct. Rather than use a single data set as a case study, we take a meta-analytical approach to maximize statistical power, maximize taxonomic coverage, and identify broad-scale patterns. While there are alternative partitions that might be applied to partitions of morphological data [e.g., cranial vs. postcranial (Mounce et al. 2016), axial vs. appendicular], our focus on dental versus osteological characters allows us to address specific concerns raised regarding the developmental and functional linkage and oversaturation of dental characters, and the taphonomic biases particular to mammals (i.e., the enhanced preservation potential of enamel over bone).
It is impossible to know the evolutionary history of empirical taxa with certainty, but congruence between trees inferred from different sources of data offers a means for cross-validation. In this context, we used trees derived from molecular data to assess the congruence of different classes of morphological data. Sequence data provide a qualitatively different and vastly larger data source that serves as a suitable and well-validated benchmark. No single case study is compelling, but meta-analyses of combined data from different sources, clades, and authors seek broad-scale patterns and generalizations from a statistically meaningful sample of independent data (e.g., Pisani et al. 2007). We, therefore, use a meta-analytical approach to address the following three questions:
(1) Do morphological partitions of dental and osteological characters convey a homogenous phylogenetic signal? More specifically, we test the null hypothesis that the partitions do not exhibit significant partition heterogeneity according to the incongruence length difference test (ILD: Farris et al. 1995a, 1995b) and incongruence relationships difference test (IRD: Mounce et al. 2016).
(2) Are dental and osteological partitions of morphological data sets equally consistent with trees derived from independent molecular sequence data? The specific null hypothesis is that dental and osteological partitions do not retain different levels of relative homoplasy as assessed by the ensemble retention index (RI: Farris 1989).
(3) Do the generalized taphonomic filters that occur during fossilization (i.e., loss of osteological data, retention of dental data) degrade the phylogentic signal any more or less than equivalent random filters? Specifically, are a similar number of nodes recovered by matrices missing osteological characters compared with matrices missing identical amounts of random characters (as assessed by the node recovery test Sansom and Wills 2013).
MATERIALS AND METHODS
Morphological data matrices of mammals were compiled from published sources (Google and Google Scholar searches for “clade phylogeny  /
/ morphology” and references and citations therein, and MorphoBank (O’Leary and Kaufman 2011) from August 2013 to April 2015. Characters were categorized as either osteological or dental on the basis of tissue type (i.e., characters relating directly and explicitly to teeth vs. osteological tissues such as the mandible). Soft tissue characters were excluded. To ensure a balance in the distribution of missing data, taxa with greater than 50% missing entries in either partition were removed, and data sets with a difference of greater than 10% missing entries between partitions were edited further by removing characters with high proportions of missing data (after Sansom and Wills 2013). All uninformative characters were removed. Thresholds were set for the minimum data set dimensions (10 taxa, 30 characters) and minimum percentage of characters in the smallest partition (20%; Sansom and Wills 2013). To ensure independence of data, data sets with appreciable taxonomic overlap were eliminated by prioritizing the more recent source. Rejected data sets, and the reasons for their rejection, are listed in the Supplementary Materials available on Dryad at http://dx.doi.org/10.5061/dryad.k23mq.
morphology” and references and citations therein, and MorphoBank (O’Leary and Kaufman 2011) from August 2013 to April 2015. Characters were categorized as either osteological or dental on the basis of tissue type (i.e., characters relating directly and explicitly to teeth vs. osteological tissues such as the mandible). Soft tissue characters were excluded. To ensure a balance in the distribution of missing data, taxa with greater than 50% missing entries in either partition were removed, and data sets with a difference of greater than 10% missing entries between partitions were edited further by removing characters with high proportions of missing data (after Sansom and Wills 2013). All uninformative characters were removed. Thresholds were set for the minimum data set dimensions (10 taxa, 30 characters) and minimum percentage of characters in the smallest partition (20%; Sansom and Wills 2013). To ensure independence of data, data sets with appreciable taxonomic overlap were eliminated by prioritizing the more recent source. Rejected data sets, and the reasons for their rejection, are listed in the Supplementary Materials available on Dryad at http://dx.doi.org/10.5061/dryad.k23mq.
We applied the ILD test (Farris et al. 1995a, 1995b; Barker and Lutzoni 2002) to our matrix partitions to test for heterogeneity of signal. The ILD test has the null of partition homogeneity and assesses this by comparing the combined length of shortest trees inferred from each partition to combined lengths of shortest trees inferred from randomly allocated partitions of the same size as the original partitions (Fig. 1a). We used scripts in TNT (Tree analysis with New Technology Goloboff et al. 2008), implementing 999 random replicates. The suitability of the ILD test has been questioned, particularly on the grounds of high type I error rate (false positives) (Dolphin et al. 2000; Hipp et al. 2004; Planet and Sarkarm 2005; Planet 2006; Ramirez 2006; Mounce et al. 2016), but it remains widely used nonetheless. In addition to the ILD test, we implemented the IRD test of Mounce et al. (2016). This is also a randomization-based test, but rather than using differences in inferred tree length, partitions are compared via the distances between the optimal trees that result from them. There are many tree-to-tree distance metrics, but here we used the symmetric difference distance (RF; Robinson and Foulds 1981; Pattengale et al. 2007) for reasons of familiarity and ease of computation. Sets of multiple optimal trees were compared by calculating the mean RF distance between each tree in one set and its nearest neighbor (the nearest neighbor distance) in the other set (Cobbett et al. 2007). As with the ILD test,  values were approximated from 999 random partitions of the data to assess whether the value from the original partitions (either combined lengths of most parsimonious trees or distance between most parsimonious trees) falls significantly outside the range of values observed in random partitions of the same data.
values were approximated from 999 random partitions of the data to assess whether the value from the original partitions (either combined lengths of most parsimonious trees or distance between most parsimonious trees) falls significantly outside the range of values observed in random partitions of the same data.
Figure 1.
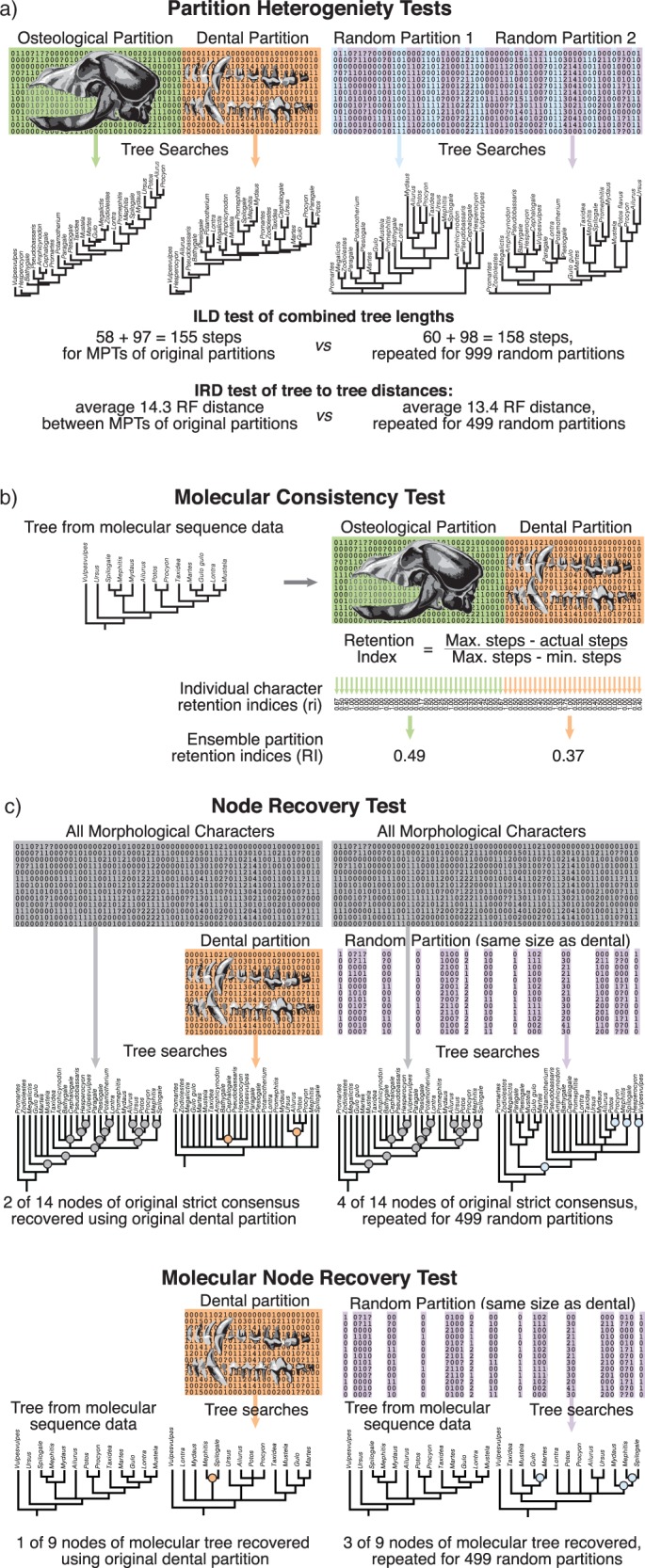
Analyses applied to morphological data. a) Partition heterogeneity tested by comparing combined tree length (ILD) or nearest neighbor tree-to-tree distances of trees (IRD) resulting from searches of each morphological character partition (osteological and dental) and random character partitions of the same size (Farris et al. 1995a, 1995b; Mounce et al. 2016). b) Molecular consistency tested by applying morphological data to a molecular tree and calculating retention indices of characters and partitions. c) Node recovery tested by comparing strict consensus trees resulting from searches using only dental characters and strict consensus trees resulting from searches using random subsets of characters in the same proportion (i.e., loss of signal with either systematic or random character removal). Signal recovery is assessed in terms of nodes shared with either the strict consensus tree using all characters or the molecular tree. Morphological data set example from Finarelli (2008) with Flynn et al. (2005) molecular tree.
To assess the role of taphonomic biases, we used the node recovery test (Sansom and Wills 2013), also implemented in TNT. The strict consensus trees resulting from maximum parsimony searches using dental characters alone were compared with the strict consensus trees resulting from searches using the entire data set in terms of number shared nodes (i.e., percentage of original signal recovered). This percentage of nodes was then compared with the distribution of percentage of nodes recovered from similar analyses of 500 equivalent matrices, each comprising the same number of characters (i.e., the number of dental characters), but drawn randomly from the entire data set (Fig. 1c).
Molecular trees were sourced in the same way as morphological data. In selecting molecular trees for each morphological data set, priority was given to the degree of taxonomic overlap, and subsequently, the underlying amount of sequence data, rather than the date of publication. Prioritizing taxonomic overlap maximized the data available for analyses because taxa not included in the molecular trees (principally extinct taxa) were removed, and consequently uninformative characters were deleted from all calculations. Both maximum parsimony and likelihood trees were used where available. In the event that multiple trees derived from the same method were presented in a single molecular study, priority was given to the tree derived from the greater amount of sequence data, and subsequently to that which showed greatest resolution. Details of all the molecular trees used are given in the Supplementary Material available on Dryad. Mesquite (Maddison and Maddison 2015) was used to construct trees from published figures and to derive retention indices of morphological characters and partitions. Retention indices are influenced by the data set dimensions and, therefore, cannot be directly compared across matrices. Differences between the partitions were, therefore, assessed using 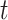 -tests of the ensemble retention index (RI i.e., for the partition as a whole) and mean character retention index (ri i.e., averaged for all characters in a partition), with values from partitions within the same data set paired.
-tests of the ensemble retention index (RI i.e., for the partition as a whole) and mean character retention index (ri i.e., averaged for all characters in a partition), with values from partitions within the same data set paired.
Molecular data were also used for a modified node recovery test (Fig. 1c). Instead of using the strict consensus tree derived from all morphological characters as the baseline for node recovery, a molecular tree was used. This enabled an assessment of whether trees derived using only dental data recovered significantly fewer molecularly compatible nodes than trees derived from a random subset of morphological characters of the same size drawn from across partitions. For this test, taxa for which molecular data were not available were removed, as were the characters subsequently rendered uninformative following the exclusion of those taxa.
RESULTS
Forty edited data sets (Supplementary Material available on Dryad) were compiled, constituting a combined total of 1234 taxa and 7403 characters and a near comprehensive sampling of the available morphological data for mammals (Table 1).
Table 1.
Data sets analyzed and partition heterogeneity test values
| Edited matrix | Test 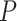 values values |
||||
|---|---|---|---|---|---|
| Data source | Ingroup | Taxa | Characters | ILD | IRD |
| Ahrens (2012) | ‡Procyonidae | 12 | 66 | 0.271 | 0.467 |
| Asher et al. (2010) |
 Chrysochloridae
Chrysochloridae |
30 | 115 | 0.004 | 0.004 |
| Asher et al. (2005) |
 Glires
Glires |
57 | 221 | 0.004 | 0.062 |
| Bi et al. (2014) |
 Mammalia
Mammalia |
37 | 402 | 0.001 | 0.114 |
| Billet (2011) | †Notoungulata | 41 | 119 | 0.072 | 0.152 |
| Boessenecker and Churchill (2013) | †Odobenidae | 17 | 85 | 0.001 | 0.090 |
| Boisserie (2005) | †Hippopotamidae | 14 | 36 | 0.104 | 0.246 |
| Bryant et al. (1993) |
 Mustelidae
Mustelidae |
23 | 36 | 0.024 | 0.042 |
| Carleton (1980) |
 Muroidea
Muroidea |
71 | 42 | 0.443 | — |
| Carstens et al. (2002) |
 Phyllostomidae
Phyllostomidae |
42 | 62 | 0.519 | 0.019 |
| Cerdeño (1995) | †Rhinocerotidae | 41 | 71 | 0.001 | — |
| Churchill et al. (2014) | ‡Otariidae | 24 | 82 | 0.064 | 0.433 |
| Domning (1994) | †Sirenia | 31 | 59 | 0.073 | 0.559 |
| Finarelli (2008) |
 Arctoidea
Arctoidea |
25 | 75 | 0.001 | 0.006 |
| Fracasso et al. (2011) |
 Chiroptera
Chiroptera |
23 | 130 | 0.114 | 0.012 |
| Froehlich (1999) | †Perissodactyls | 24 | 110 | 0.357 | 0.509 |
| Gaubert et al. (2005) |
 Feliformia
Feliformia |
44 | 261 | 0.003 | 0.293 |
| Gentry (1992) |
 Bovidae
Bovidae |
27 | 110 | 0.222 | 0.555 |
| Gheerbrant et al. (2005) | †Proboscidae | 12 | 113 | 0.424 | 0.990 |
| Giannini and Simmons (2005) |
 Pteropodidae
Pteropodidae |
49 | 165 | 0.548 | 0.782 |
| He et al. (2012) |
 Erinaceidae
Erinaceidae |
12 | 73 | 0.001 | 0.002 |
| Kielan-Jaworowska and Hurum (2001) | †Allotheria | 11 | 51 | 0.002 | 0.018 |
| Ladevèze et al. (2010) | †Condylarthra | 12 | 57 | 0.013 | 0.236 |
| Lecompte et al. (2002) | ‡Muridae | 18 | 37 | 0.204 | 0.014 |
| Ni et al. (2013) |
 Primatomorpha
Primatomorpha |
51 | 893 | 0.001 | 0.036 |
| O’Leary et al. (2003) |
 Theria
Theria |
46 | 2200 | 0.001 | 0.004 |
| Olivares and Verzi (2014) |
 Echimyidae
Echimyidae |
21 | 59 | 0.624 | 0.255 |
| Oliveira et al. (2011) |
 Didephinae
Didephinae |
51 | 80 | 0.261 | 0.225 |
| Prideaux and Warburton (2010) |
 Macropodidae
Macropodidae |
25 | 81 | 0.183 | 0.645 |
| Sánchez-Villagra et al. (2006) |
 Talpidae
Talpidae |
17 | 152 | 0.028 | 0.253 |
| Silcox et al. (2009) | †Apatemyidae | 21 | 220 | 0.001 | 0.359 |
| Spaulding et al. (2009) |
 Artiodactyla
Artiodactyla |
50 | 502 | 0.001 | 0.371 |
| Springer et al. (1997) |
 Marsupials
Marsupials |
16 | 83 | 0.001 | 0.172 |
| Steppan (1993) |
 Phyllotini
Phyllotini |
49 | 82 | 0.058 | 0.012 |
| Strait and Grines (2004) | ‡Hominoidia | 14 | 103 | 0.163 | 0.756 |
| Thorington et al. (2002) |
 Pteromyinae
Pteromyinae |
20 | 73 | 0.001 | 0.132 |
| Tomiya (2011) |
 Carnivora
Carnivora |
47 | 86 | 0.001 | 0.287 |
| Weksler (2006) |
 Oryzomyini
Oryzomyini |
52 | 64 | 0.656 | 0.333 |
| Wroe and Musser (2001) |
 Thylacinidae
Thylacinidae |
25 | 70 | 0.019 | 0.609 |
| Zrzavý and ŘIčánková (2004) |
 Canidae
Canidae |
32 | 77 | 0.007 | 0.082 |
| Totals | 1234 | 7403 |
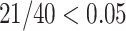
|
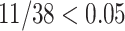
|
|
Note: Bold values,  .
.
 Data sets for which molecular trees were available.
Data sets for which molecular trees were available.
†, ‡Data sets for which molecular data were unavailable: either because the ingroup were largely fossil taxa (b) or failed to meet minimum criteria following editing (c), for example, number of taxa, ratio of osteological to dental characters.
Homogeneity of Signal
Application of the ILD test (Farris et al. 1995a, 1995b) found significant (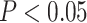 ) heterogeneity between the osteological and dental character partitions in 21 out of 40 data sets, and a highly significant difference overall (Fisher’s combined probability
) heterogeneity between the osteological and dental character partitions in 21 out of 40 data sets, and a highly significant difference overall (Fisher’s combined probability 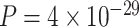 , Table 1). Applying more stringent thresholds for significance still found widespread heterogeneity between osteological and dental partitions (17 out of 40 data sets had
, Table 1). Applying more stringent thresholds for significance still found widespread heterogeneity between osteological and dental partitions (17 out of 40 data sets had 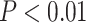 , and 12 out of 40 had
, and 12 out of 40 had 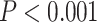 ). Using the tree-to-tree distance based test (IRD; Mounce et al. 2016) also identified widespread heterogeneity between dental and osteological partitions (11 out of 38 data sets have
). Using the tree-to-tree distance based test (IRD; Mounce et al. 2016) also identified widespread heterogeneity between dental and osteological partitions (11 out of 38 data sets have 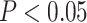 , Fisher’s combined probability
, Fisher’s combined probability 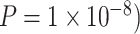 .
.
Relative Molecular Consistency
Molecular data are unavailable for almost all extinct taxa [an exception here being Thylacinus from Wroe and Musser (2001)]. Following the removal of fossil taxa, and the characters subsequently rendered uninformative, 14 morphological data sets were omitted from molecular comparisons (of which, 10 were composed largely of fossil taxa and 4 failed to meet the minimum dimensions for inclusion following taxon and character removal). This resulted in 26 data sets (Table 1) for which molecular data were available (seven of which molecular data were drawn directly from the same study as the morphological data). The subset of morphological matrices with molecular data available comprised a combined total of 698 taxa and 5589 morphological characters, and like the total data set, it also showed significant partition heterogeneity (Fisher’s combined probabilities of 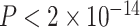 for ILD tests and
for ILD tests and 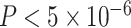 for IRD tests). More importantly, osteological characters were found to have significantly higher retention indices than dental characters when optimized onto molecular trees (Fig. 2). This difference was significant both for average retention indices of individual characters (paired
for IRD tests). More importantly, osteological characters were found to have significantly higher retention indices than dental characters when optimized onto molecular trees (Fig. 2). This difference was significant both for average retention indices of individual characters (paired 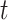 -test,
-test, 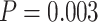 ) and for ensemble retention indices for whole partitions (paired
) and for ensemble retention indices for whole partitions (paired 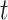 -test,
-test, 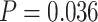 ). Furthermore, the difference was significant irrespective of the tree building methods that were used to infer molecular trees (i.e., maximum likelihood vs. maximum parsimony; see Supplementary Material available on Dryad). We note that it was not necessary to control for differences in numbers of characters in the dental and osteological partitions, using, for example, a jackknifing approach. While the Retention Index of most parsimonious trees is sensitive to differences in data set dimensions, our approach here was to optimize characters onto an existing tree (the molecular) in each case. The retention index (ri) for a given character is then the same regardless of whether this is calculated as part of the original partition, as part of a jackknifed subsample of this partition, or as a singly optimized character.
). Furthermore, the difference was significant irrespective of the tree building methods that were used to infer molecular trees (i.e., maximum likelihood vs. maximum parsimony; see Supplementary Material available on Dryad). We note that it was not necessary to control for differences in numbers of characters in the dental and osteological partitions, using, for example, a jackknifing approach. While the Retention Index of most parsimonious trees is sensitive to differences in data set dimensions, our approach here was to optimize characters onto an existing tree (the molecular) in each case. The retention index (ri) for a given character is then the same regardless of whether this is calculated as part of the original partition, as part of a jackknifed subsample of this partition, or as a singly optimized character.
Figure 2.
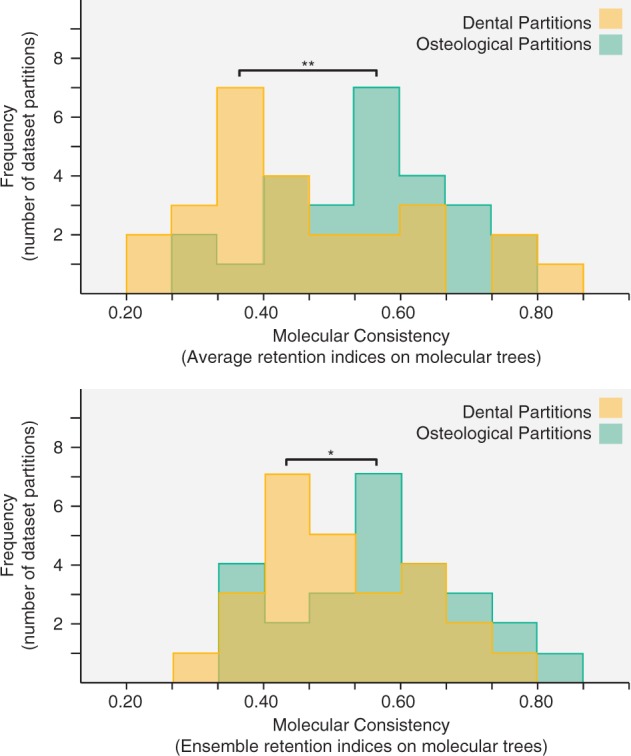
Tests for molecular consistency of morphological characters found significant differences between osteological and dental partitions (paired 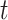 -test,
-test, 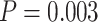 for average retention indices, ri; paired
for average retention indices, ri; paired 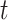 -test,
-test, 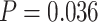 for ensemble retention indices, RI).
for ensemble retention indices, RI).
Recovery of Nodes by Dental and Osteological Character Partitions
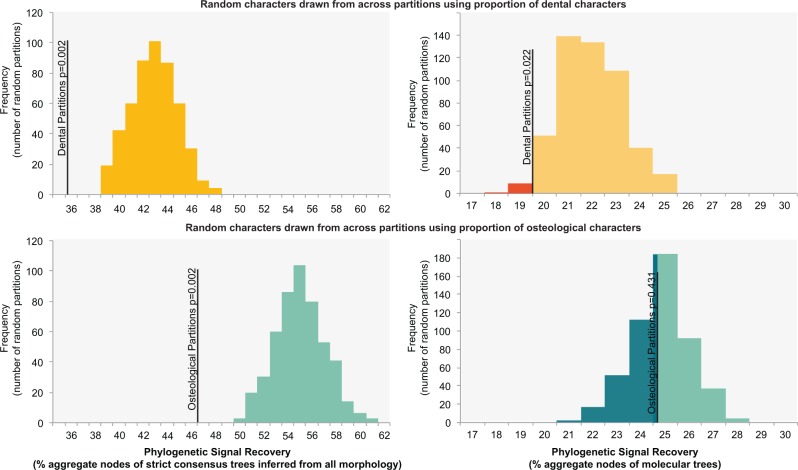
The node recovery test (Sansom andWills 2013) found that strict consensus trees inferred from exclusively dental characters recovered only 36% of the nodes of strict consensus trees inferred using all morphological characters as aggregated across data sets. This is a significantly lower percentage than strict consensus trees inferred using the same number of characters as dental characters, but drawn at random from across both partitions (median of 43% of aggregated nodes across all studies) (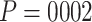 , Fig. 3, see Supplementary Material available on Dryad). Strict consensus trees recovered using only osteological characters also recovered significantly fewer of the original strict consensus nodes compared to the searches using the same number of characters drawn at random from across partitions (47% of aggregated nodes for osteological only searches vs. median of 56% from random partitions of the same size;
, Fig. 3, see Supplementary Material available on Dryad). Strict consensus trees recovered using only osteological characters also recovered significantly fewer of the original strict consensus nodes compared to the searches using the same number of characters drawn at random from across partitions (47% of aggregated nodes for osteological only searches vs. median of 56% from random partitions of the same size; 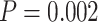 , see Supplementary Material available on Dryad). The modified node recovery test using molecular trees as a baseline for signal recovery yielded slightly different results. Strict consensus trees from searches using both osteological and dental characters of taxa for which molecular data were available recovered 29% of the nodes of molecular trees (as aggregated across data sets). Strict consensus trees from searches using only dental characters of these same taxa recovered just 19.8% of the molecular trees nodes; this is a significantly lower percentage of molecular tree nodes than strict consensus trees inferred using the same number of characters drawn at random from across both partitions (median of 22.2% of aggregated molecular nodes;
, see Supplementary Material available on Dryad). The modified node recovery test using molecular trees as a baseline for signal recovery yielded slightly different results. Strict consensus trees from searches using both osteological and dental characters of taxa for which molecular data were available recovered 29% of the nodes of molecular trees (as aggregated across data sets). Strict consensus trees from searches using only dental characters of these same taxa recovered just 19.8% of the molecular trees nodes; this is a significantly lower percentage of molecular tree nodes than strict consensus trees inferred using the same number of characters drawn at random from across both partitions (median of 22.2% of aggregated molecular nodes; 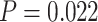 ; Fig. 3; Supplementary Material available on Dryad). However, strict consensus trees recovered using only osteological characters fell within the distribution of molecular node recovery seen in equivalent searches using the same number of characters drawn at random from across partitions (25.0% of aggregated molecular nodes for osteological searches vs. median of 25.2% from random partitions of the same size;
; Fig. 3; Supplementary Material available on Dryad). However, strict consensus trees recovered using only osteological characters fell within the distribution of molecular node recovery seen in equivalent searches using the same number of characters drawn at random from across partitions (25.0% of aggregated molecular nodes for osteological searches vs. median of 25.2% from random partitions of the same size; 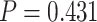 ).
).
Figure 3.
Results of node recovery tests showing distribution of phylogenetic signal recovery using either total morphological signal as a benchmark (left, percentage aggregate nodes of strict consensus trees inferred using both osteological and dental characters recovered) or molecular data as a benchmark (right, percentage aggregate nodes of molecular trees recovered). Characters were drawn from across both partitions using either the proportion of dental characters (above) or the proportion of osteological characters (below). The node recovery of the original partitions (i.e., dental or osteological, black lines) is mapped onto the distributions of equivalent randomly sampled partitions with estimated 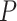 values (using proportion of random replicates recovering less signal recovery).
values (using proportion of random replicates recovering less signal recovery).
DISCUSSION
Within morphological data sets, osteological and dental character partitions exhibit significant heterogeneity as evidenced from both length-based tests (ILD) and tree-to-tree distance based tests (IRD). This indicates that a different phylogenetic signal is contained within each morphological partition; however, these tests alone do not indicate whether partitions differ in their ability to capture evolutionary history. Since it is impossible to know relationships with certainty, we used congruence with a qualitatively different and operationally distinct data source—namely molecules—as a benchmark of phylogenetic fidelity. Of the two morphological partitions, dental characters are a significantly poorer fit with independent molecular trees (Fig. 2). We interpret this to indicate that teeth are relatively less phylogenetically informative, which bears out predictions from developmental studies (Kangas et al. 2004; Harjunmaa et al. 2014) but contradicts findings from smaller scale empirical studies (Sánchez-Villagra andWilliams 1998; O’Leary et al. 2003;Williams 2007). That dental characters are less congruent with molecular data is unfortunate from a palaeontological perspective given that teeth constitute so much of the mammalian fossil record. Here we demonstrate that the application of simplified paleontological filters (the removal of osteology in node recovery tests) yields a loss of signal far in excess of the amount predicted from random incompleteness (Fig. 3). A similar pattern was found in an analysis of a single primate data set by Pattinson et al. (2015) whereby “artificial fossil templates” using largely dental characters were found to result in appreciably less signal recovery than “random templates”. Indeed, they state that “for most fossils in our data set known for fewer than 180 sampled characters (i.e., >50% missing data), taphonomically induced bias decreases phylogenetic accuracy” (Pattinson et al. 2015, p. 179). Their “random templates” are akin to the random missing data analysis used in our study (unlike their “random states” that we interpret as providing a baseline of node recovery for phylogenetically random data with the same general properties of state frequencies). Pattinson et al. (2015) found that sampling across morphological partitions performed better than sampling predominately from a single partition and interpreted this as supporting the principle that characters should be sampled from as many sources as possible (Kluge 1989; O’Leary et al. 2003; Mounce et al. 2016). However, the empirical distribution of characters was such that artificial templates dominated by one partition were always dominated by dental characters. As such, insufficiency of a single partition equates to insufficiency of dental characters in this case. In our meta-analyses of 40 data sets (26 with molecular trees), we have explicitly tested both dental only partitions and osteology only partitions. Both dental only and osteology only partitions recover significantly less signal (percentage of original strict consensus nodes when using all morphology as benchmark) than partitions of equivalent size that sample characters randomly from across partitions. This in itself would imply that characters should be sampled widely from across both partitions. When molecular trees are used as a benchmark of signal recovery, however, dental only searches are found to perform significantly worse than random partitions of equivalent size, whereas osteological only searches do not (Fig. 3). Application of molecular data, therefore, indicates that dental data are generally less-reliable indicators of phylogenetic history than osteological data. Not only do dental characters exhibit elevated levels of relative homoplasy, but they are also unlikely to be sufficient for accurate phylogeny reconstruction on their own.
When results from different studies are compared, a coherent picture emerges. Dental morphology is found to convey a phylogenetic signal that is different to that derived from osteology and is comparatively less consistent with molecular data. That dental characters are more homoplastic accords with the observation that they are oversaturated (Dávalos et al. 2014), as well as with developmental evidence for the relative ease and correlation of shape changes in teeth (Kangas et al. 2004; Harjunmaa et al. 2014). Furthermore, loss of osteology (the common palaeontological case) causes significant and disproportionate degradation of phylogenetic signal, such that analyses of dental morphology alone are less likely to reconstruct relationships accurately. Notwithstanding all of the above considerations, we do not advocate the wholesale exclusion of dental morphology from phylogenetic and evolutionary studies. Not only would that exclude many taxa known only from teeth, but also neglect the reliable phylogenetic signal that must exist in at least some dental characters, as well as the hidden support that may exist between dental and other characters (Gatesy et al. 1999); the inclusion of dental characters alongside osteological recovers 29% of molecular tree nodes compared to 25% when using osteology only. Instead, it is necessary to identify subsets of morphology that are less subject to convergence and to directly address the issue of oversaturation and nonindependence (Dávalos et al. 2014) [e.g., by identifying suites and cliques of convergent or correlated characters (O’Keefe and Wagner 2001; Holland et al. 2010)]. The phenomenon appears to be widespread for all mammals; it is possible that it could extend to other vertebrates, although the condition of heterodonty in mammals may have compounded the issue, potentially through failure to properly account for serial homology. In all instances, we strongly advocate meta-analyses of cladistics data sets; it is only when data from a wide diversity of clades are comprehensively sampled and compared that large-scale patterns such as these become apparent (Sansom andWills 2013; Mounce et al. 2016).
Acknowledgments
We are grateful to all the authors who created the data that were included in the meta-analysis. Feedback was kindly provided by Manchester Evolution research group, Rob Asher (Cambridge), and three anonymous reviewers. The authors declare no conflict of interest.
SUPPLEMENTARY MATERIAL
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.k23mq.
FUNDING
This work was supported by Biotechnology and Biological Sciences Research Council [BBSRC Manchester DTP Summer research experience placement to T.W. and R.S.S., and grants BB/N015827/1 to R.S.S., BB/K015702/1 and BB/K006754/1 to M.A.W.]; Natural Environment Research Council [NERC Fellowship NE/I020253/1 to R. S.S.]; Leverhulme Trust [grant F/00351/Z to M. A. W.]; and John Templeton Foundation [grant 43915 to Mark Wilkinson and M.A.W.].
References
- Ahrens H.E. 2012. Craniodental characters and the relationships of Procyonidae (Mammalia: Carnivora). Zool. J. Linn. Soc.164:669–713. [Google Scholar]
- Asher R.J.,, Maree S.,, Bronner G.,, Bennett N.C.,, Bloomer P.,, Czechowski P.,, Hofreiter M. 2010. A phylogenetic estimate for golden moles (Mammalia, Afrotheria, Chrysochloridae). BMC Evol. Biol.10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher R.J.,, Meng J.,, Wible J.R.,, McKenna M.C.,, Rougier G.W.,, Dashzeveg D.,, Novacek M.J. 2005. Stem Lagomorpha and the antiquity of Glires. Science307:1091–1094. [DOI] [PubMed] [Google Scholar]
- Barker F.K.,, Lutzoni F.M. 2002. The utility of the incongruence length difference test. Syst. Biol. 51:625–637. [DOI] [PubMed] [Google Scholar]
- Benton M.J., Donoghue P.C.J.,, Asher R.J,, Friedman M.,, Near T.J.,, Vinther J. 2005. Constraints on the timescale of animal evolutionary history Paleontol. Electron. 18.1.1FC:1–106. [Google Scholar]
- Bi S.,, Wang Y.,, Guan J.,, Meng J. 2014. Three new Jurassic euharamiyidan species reinforce early divergence of mammals. Nature514:579–584. [DOI] [PubMed] [Google Scholar]
- Billet G. 2011. Phylogeny of the Notoungulata (Mammalia) based on cranial and dental characters. J. Syst. Palaeontol.9:481–497. [Google Scholar]
- Boessenecker R.W.,, Churchill M. 2013. A reevaluation of the morphology, paleoecology, and phylogenetic relationships of the enigmatic walrus Pelagiarctos. PLoS One 8:e54311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisserie J.-R. 2005. The phylogeny and taxonomy of Hippopotamidae (Mammalia: Artiodactyla): a review based on morphology and cladistic analysis. Zool. J. Linn. Soc.143:1–26. [Google Scholar]
- Brunet M. et al. 2002. A new hominid from the Upper Miocene of Chad, Central Africa. Nature418:145–151. [DOI] [PubMed] [Google Scholar]
- Bryant H.N.,, Russell A.P.,, Fitch W.D. 1993. Phylogenetic relationships within the extant Mustelidae (Carnivora): appraisal of the cladistic status of the Simpsonian subfamilies. Zool. J. Linn. Soc.108:301–334. [Google Scholar]
- Carleton M.D. 1980. Phylogenetic relationships in neotomine-peromyscine rodents (Muroidea) and a reappraisal of the dichotomy within New World Cricetinae. Misc. Pub. Mus. Zool. Univ. Michigan157:1–140. [Google Scholar]
- Carstens B.C.,, Lundrigan B.L.,, Myers P. 2002. A phylogeny of the neotropical nectar-feeding bats (Chiroptera: Phyllostomidae) based on morphological and molecular Data. J. Mamm. Evol.9:23–53. [Google Scholar]
- Cerdeño E. 1995. Cladistic analysis of the family Rhinocerotidae (Perissodactyla). Am. Mus. Noviates3143:1–25 [Google Scholar]
- Churchill M.,, Boessenecker R.W.,, Clementz M.T. 2014. Colonization of the Southern Hemisphere by fur seals and sea lions (Carnivora: Otariidae) revealed by combined evidence phylogenetic and Bayesian biogeographical analysis. Zool. J. Linn. Soc.172:200–225. [Google Scholar]
- Cobbett A.,, Wilkinson M.,, Wills M.A. 2007. Fossils impact as hard as living taxa in parsimony analyses of morphology. Syst. Biol. 56:753–766. [DOI] [PubMed] [Google Scholar]
- Dávalos L.M.,, Velazco P.M.,, Warsi O.M.,, Smits P.D.,, Simmons N.B. 2014. Integrating incomplete fossils by isolating conflicting signal in saturated and non-independent morphological characters. Syst. Biol. 63:582–600. [DOI] [PubMed] [Google Scholar]
- DeGusta D. 2004. A method for estimating the relative importance of characters in cladistic analyses. Syst. Biol. 53:529–532. [DOI] [PubMed] [Google Scholar]
- Dolphin K.,, Belshaw R.,, Orme C.D.L.,, Quicke D.L.J. 2000. Noise and incongruence: interpreting results of the incongruence length difference test. Mol. Phylogenet. Evol.17:401–406. [DOI] [PubMed] [Google Scholar]
- Domning D.P. 1994. A phylogenetic analysis of the Sirenia. Proc. San Diego Soc. Nat. Hist.29:177–189. [Google Scholar]
- Evans A.R.,, Wilson G.P.,, Forelius M.,, Jernvall J. 2007. High-level similarity of dentitions in carnivorans and rodents. Nature445:78–81. [DOI] [PubMed] [Google Scholar]
- Farris J.S. 1989. The retention index and homoplasy excess. Syst. Biol. 38:406–407. [Google Scholar]
- Farris J.S.,, Källersjö M.,, Kluge A.G.,, Bult C. 1995a. Constructing a significance test for incongruence. Syst. Biol. 44:570–572. [Google Scholar]
- Farris J.S.,, Kallersjo M.,, Kluge A.G.,, Bult C. 1995b. Testing significance of incongruence. Cladistics10:315–319. [Google Scholar]
- Finarelli J.A. 2008. A total evidence phylogeny of the Arctoidea (Carnivora, Mammalia). J. Mamm. Evol.15:231–259. [Google Scholar]
- Flynn J.J.,, Finarelli J.A.,, Zehr S.,, Hsu J.,, Nedball M.A. 2005. Molecular phylogeny of the Carnivora (Mammalia): assessing the impact of increasing sampling on resolving enigmatic relationships. Syst. Biol. 54:317–447. [DOI] [PubMed] [Google Scholar]
- Fracasso M.P.A.,, de Oliveira Salles L.,, Perini F.A. 2011. Upper molar morphology and relationships among higher taxa in bats. J. Mammal.92:421–432. [Google Scholar]
- Froehlich D.J. 1999. Phylogenetic systematics of basal perissodactyls. J. Vertebr. Paleontol.19:140–159. [Google Scholar]
- Gatesy J.,, Baker R. 2005. Hidden likelihood support in genomic data: can forty-five wrongs make a right? Syst. Biol. 54:483–492. [DOI] [PubMed] [Google Scholar]
- Gatesy J.,, O’Grady P.,, Baker R. 1999. Corroboration among data sets in simultaneous analysis: hidden support for phylogenetic relationships among higher level artiodactyl taxa. Cladistics15:271–313. [DOI] [PubMed] [Google Scholar]
- Gaubert P.,, Wozencraft W.C.,, Cordeiro-Estrela P.,, Veron G. 2005. Mosaics of convergences and noise in morphological phylogenies: what’s in a viverrid-like carnivoran? Syst. Biol. 54:865–894. [DOI] [PubMed] [Google Scholar]
- Gentry A.W. 1992. The subfamilies and tribes of the family Bovidae. Mammal Rev.22:1–32. [Google Scholar]
- Gheerbrant E.,, Sudre J.,, Tassy P.,, Amaghzaz M.,, Bouya B.,, Iarochene M. 2005. Nouvelles données sur Phosphatherium escuilliei (Mammalia, Proboscidea) de l’Eocène inférieur du Maroc, apports à la phylogénie des Proboscidea et des ongulés lophodontes. Geodiversitas27:239–333. [Google Scholar]
- Giannini N.P.,, Simmons N.B. 2005. Conflict and congruence in a combined DNA–morphology analysis of megachiropteran bat relationships (Mammalia: Chiroptera: Pteropodidae). Cladistics21:411–437. [DOI] [PubMed] [Google Scholar]
- Gingerich P.D.,, Franzen J.L.,, Habersetzer J.,, Hurum J.H.,, Smith B.H. 2010. Darwinius masillae is a haplorhine – reply to Williams et al. (2010). J. Hum. Evol.59:574–579. [Google Scholar]
- Goloboff P.A.,, Farris J.S.,, Nixon K.C. 2008. TNT, a free program for phylogenetic analysis. Cladistics24:774–786. [Google Scholar]
- Harjunmaa E. et al. 2014. Replaying evolutionary transitions from the dental fossil record. Nature512:44–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K.,, Chen J.-H.,, Gould G.C.,, Yamaguchi N.,, Ai H.-S.,, Wang Y.-X.,, Jiang X.-L. 2012. An estimation of erinaceidae phylogeny: a combined analysis approach. PLoS One 7:e39304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp A.L.,, Hall J.C.,, Sytsma K.J. 2004. Congruence versus phylogenetic accuracy: revisiting the incongruence length difference test. Syst. Biol. 53:81–89. [DOI] [PubMed] [Google Scholar]
- Holland B.R.,, Spencer H.G.,, Worthy T.H.,, Kennedy M. 2010. Identifying cliques of convergent characters: concerted evolution in the cormorants and shags. Syst. Biol. 59:443–445. [DOI] [PubMed] [Google Scholar]
- Huson D.H.,, Scornavacca C. 2012. Dendroscope 3, an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61:1061–1067. [DOI] [PubMed] [Google Scholar]
- Kangas A.T.,, Evans A.R.,, Thesleff I.,, Jernvall J. 2004. Nonindependence of mammalian dental characters. Nature432:211–214. [DOI] [PubMed] [Google Scholar]
- Kielan-Jaworowska Z.,, Hurum J.H. 2001. Phylogeny and systematics of multituberculate mammals. Palaeontology44:389–429. [Google Scholar]
- Kluge A.G. 1989. A concern for evidence and a phylogenetic hypothesis of relationships among Epicrates (Boidae, Serpentes). Syst. Zool.38:7–25. [Google Scholar]
- Ladevèze S.,, Missiaen P.,, Smith T. 2010. First skull of Orthaspidotherium edwardsi (Mammalia “Condylarthra”) from the late Paleocene of Berru (France) and phylogenetic affinities of the enigmatic European family Pleuraspidotheriidae. J. Vertebr. Paleontol. 30:1559–1578. [Google Scholar]
- Lecompte E.,, Granjon L.,, Denys C. 2002. The phylogeny of the Praomys complex (Rodentia: Muridae) and its phylogeographic implications. J. Zool. Syst. Evol. Res. 40:8–25. [Google Scholar]
- Maddison W.P.,, Maddison D.R. 2015. Mesquite, a modular system for evolutionary analysis. Version 3.03. Available from: URLhttp://mesquiteproject.org.
- Minelli A. et al. 2007. Reconstructing animal phylogeny in the light of evolutionary developmental biology. Boca Raton (FL): CRC Press–Taylor & Francis Group. [Google Scholar]
- Mounce R.,, Sansom R.S.,, Wills M.A.W. 2016. Craniodental and postcranial characters of vertebrates often imply different trees. Why characters should be sampled holistically.Evolution 70:666–686. [DOI] [PubMed] [Google Scholar]
- Naylor G.J.P.,, Adams D.C. 2001. Are the fossil data really at odds with the molecular data? Morphological evidence for Cetartiodactyla phylogeny re-examined. Syst. Biol. 50:444–453. [PubMed] [Google Scholar]
- Ni X.,, Gebo D.L.,, Dagosto M.,, Meng J.,, Tafforeau P.,, Flynn J.J.,, Beard K.C. 2013. The oldest known primate skeleton and early haplorhine evolution. Nature 498:60–64. [DOI] [PubMed] [Google Scholar]
- O’Keefe F.R.,, Wagner P.K. 2001. Inferring and testing hypotheses of cladistic character dependence by using character compatibility. Syst. Biol. 50:657–675. [DOI] [PubMed] [Google Scholar]
- O’Leary M.A.,, Gatesy J.,, Novacek M.J. 2003. Are the dental data really at odds with the molecular data? Morphological evidence for whale phylogeny (re)re-examined. Syst. Biol. 52:853–864. [PubMed] [Google Scholar]
- O’Leary M.A.,, Kaufman S.G. 2011. MorphoBank: Phylophenomics in the “cloud”. Cladistics 27:529–537. [DOI] [PubMed] [Google Scholar]
- Olivares A.I.,, Verzi D.H. 2014. Systematics, phylogeny and evolutionary pattern of the hystricognath rodent Eumysops (Echimyidae) from the Plio–Pleistocene of southern South America.Hist. Biol. 27:1042–1061. [Google Scholar]
- Oliveira E.V.,, Villa Nova P.,, Goin F.J.,, Dos Santos Avilla L. 2011. A new hyladelphine marsupial (Didelphimorphia, Didelphidae) from cave deposits of northern Brazil. Zootaxa 3041:51–62. [Google Scholar]
- Pattengale N.D.,, Gottlieb E.J.,, Moret B.M.E. 2007. Efficiently computing the Robinson-Foulds metric. J. Comput. Biol. 14:724–735. [DOI] [PubMed] [Google Scholar]
- Pattinson D.J.,, Thompson R.S.,, Piotrowski A.K.,, Asher R.J. 2015. Phylogeny, paleontology, and primates, do incomplete fossils bias the tree of life? Syst. Biol. 64:169–186. [DOI] [PubMed] [Google Scholar]
- Pisani D.,, Benton M.J.,, Wilkinson M. 2007. Congruence of morpho logical and molecular phylogenies. Acta Biotheor. 55:269–281. [DOI] [PubMed] [Google Scholar]
- Planet P. 2006. Tree disagreement: measuring and testing incongruence in phylogenies. J. Biomed. Informat. 39: 86–102. [DOI] [PubMed] [Google Scholar]
- Planet P.J.,, Sarkarm I.N. 2005. mILD: a tool for constructing and analyzing matrices of pairwise phylogenetic character incongruence tests. Bioinformatics 21:4423–4424. [DOI] [PubMed] [Google Scholar]
- Prideaux G.J.,, Warburton N.M. 2010. An osteology-based appraisal of the phylogeny and evolution of kangaroos and wallabies (Macropodidae: Marsupialia). Zool. J. Linn. Soc. 159:954–987. [Google Scholar]
- Ramirez M.J. 2006. Further problems with the incongruence length difference test: “Hypercongruence” effect and multiple comparisons. Cladistics 22:289–295. [Google Scholar]
- Robinson D.R.,, Foulds L.R. 1981. Comparison of phylogenetic trees. Math. Biosci. 53:131–147. [Google Scholar]
- Ruvinksy I.,, Gibson-Brown J.J. 2000. Genetic and developmental bases of serial homology in vertebrate limb evolution. Development 127:5233–5244. [DOI] [PubMed] [Google Scholar]
- Sadleir R.W.,, Makovicky P.J. 2008. Cranial shape and correlated characters in crocodilian evolution. J. Evol. Biol. 21:1578–1596. [DOI] [PubMed] [Google Scholar]
- Sanchez-Villagra M.R.,, Horovitz I.,, Motokawa M. 2006. A comprehensive morphological analysis if talpid moles (Mammalia) phylogenetic relationships. Cladistics 22:59–88. [DOI] [PubMed] [Google Scholar]
- Sánchez-Villagra M.R.,, Williams B.A. 1998. Levels of homoplasy in the evolution of the mammalian skeleton. J. Mamm. Evol. 5:113–126. [Google Scholar]
- Sansom R.S.,, Gabbott S.E.,, Purnell M.A. 2010. Non random decay causes bias in fossil interpretation. Nature 463:797–800. [DOI] [PubMed] [Google Scholar]
- Sansom R.S.,, Gabbott S.E.,, Purnell M.A. 2011. Decay of vertebrate characters in hagfish and lamprey (Cyclostomata) and the implications for the vertebrate fossil record. Proc. R. Soc. B 278:1150–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom R.S.,, Gabbott S.E.,, Purnell M.A. 2013. Atlas of vertebrate decay: a visual and taphonomic guide to fossil interpretation. Palaeontology 56:457–474. [Google Scholar]
- Sansom R.S.,, Wills M.A. 2013. Fossilization causes organisms to appear erroneously primitive by distorting evolution trees. Sci. Rep. 3:2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom R.S. 2014. Bias and sensitivity in the placement of fossil taxa resulting from interpretations of missing data. Syst. Biol. 64:256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotland R.W.,, Olmstead R.G.,, Bennett J.R. 2003. Phylogeny reconstruction, the role of morphology. Syst. Biol. 52:529–548. [DOI] [PubMed] [Google Scholar]
- Silcox M.T.,, Bloch J. I.,, Boyer D.M.,, Houde P. 2009. Cranial anatomy of Paleocene and Eocene Labidolemur kayi (Mammalia: Apatotheria), and the relationships of the Apatemyidae to other mammals. Zool. J. Linnean Soc. 160:773–825. [Google Scholar]
- Spaulding M.,, O’Leary M.A.,, Gatesy J. 2009. Relationships of Cetacea (Artiodactyla) among mammals: increased taxon sampling alters interpretations of key fossils and character evolution. PLoS One 4:e7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M.S.,, Kirsch J.A.W.,, Chase J.A. 1997. In: Givnish T.J.,, Sytsma K.J., editors. The chronicle of marsupial evolution. Molecular evolution and adaptive radiation. Cambridge: Cambridge University Press. p. 129–161. [Google Scholar]
- Springer M.S. et al. 2007. The adequacy of morphology for reconstructing the early history of placental mammals. Syst. Biol. 56:673–684. [DOI] [PubMed] [Google Scholar]
- Steppan S. 1993. Phylogenetic relationships among the Phyllotini (Rodentia: Sigmodontinae) using morphological characters. J. Mamm. Evol. 1:187–213. [Google Scholar]
- Strait D.S.,, Grine F.E. 2004. Inferring hominoid and early hominid phylogeny using craniodental characters, the role of fossil taxa. J. Hum. Evol. 47:399–452. [DOI] [PubMed] [Google Scholar]
- Thorington R.W.,, Pitassy D.,, Jansa S.A. 2002. Phylogenies of flying squirrels (Pteromyinae). J. Mamm. Evol. 9:99–135. [Google Scholar]
- Tomiya S. 2011. A new basal caniform (Mammalia: Carnivora) from the Middle Eocene of North America and remarks on the phylogeny of early carnivorans. PLoS One 6:e24146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weksler M. 2006. Phylogenetic relationships of Oryzomine rodents (Muroidea, Sigmodontinae): separate and combined analyses of morphological and molecular data. Bull. Am. Mus. Nat. Hist. 296:1–48 [Google Scholar]
- Wiens J.J. 2004. The role of morphological data in phylogeny reconstruction. Syst. Biol. 53:653–661. [DOI] [PubMed] [Google Scholar]
- Williams B.A. 2007. Comparing levels of homoplasy in the primate skeleton. J. Hum. Evol. 52:480–489. [DOI] [PubMed] [Google Scholar]
- Wolpoff M.H.,, Senut B.,, Pickford M.,, Hawks J. 2002. Sahelanthropus or ‘Sahelpithecus’? Nature 419:581–582. [DOI] [PubMed] [Google Scholar]
- Wroe S.,, Musser A. 2001. The skull of Nimbacinus dicksoni (Thylacinidae: Marsupialia). Aust. J. Zool. 49:487–514. [Google Scholar]
- Young N.,, Hallgrímsson B. 2005. Serial homology and the evolution of mammalian limb covariation structure. Evolution 59:2691–2704. [PubMed] [Google Scholar]
- Zrzavý J.,, ŘIčánková V. 2004. Phylogeny of Recent Canidae (Mammalia, Carnivora): relative reliability and utility of morphological and molecular datasets. Zoologica Scripta, 33:311–333. 10.1111/j.0300-3256.2004.00152.x [DOI] [Google Scholar]