Abstract
Noncoding markers have a particular appeal as tools for phylogenomic analysis because, at least in vertebrates, they appear less subject to strong variation in GC content among lineages. Thus far, ultraconserved elements (UCEs) and introns have been the most widely used noncoding markers. Here we analyze and study the evolutionary properties of a new type of noncoding marker, conserved nonexonic elements (CNEEs), which consists of noncoding elements that are estimated to evolve slower than the neutral rate across a set of species. Although they often include UCEs, CNEEs are distinct from UCEs because they are not ultraconserved, and, most importantly, the core region alone is analyzed, rather than both the core and its flanking regions. Using a data set of 16 birds plus an alligator outgroup, and 3600–3800 loci per marker type, we found that although CNEEs were less variable than bioinformatically derived UCEs or introns and in some cases exhibited a slower approach to branch resolution as determined by phylogenomic subsampling, the quality of CNEE alignments was superior to those of the other markers, with fewer gaps and missing species. Phylogenetic resolution using coalescent approaches was comparable among the three marker types, with most nodes being fully and congruently resolved. Comparison of phylogenetic results across the three marker types indicated that one branch, the sister group to the passerine falcon clade, was resolved differently and with moderate (70%) bootstrap support between CNEEs and UCEs or introns. Overall, CNEEs appear to be promising as phylogenomic markers, yielding phylogenetic resolution as high as for UCEs and introns but with fewer gaps, less ambiguity in alignments and with patterns of nucleotide substitution more consistent with the assumptions of commonly used methods of phylogenetic analysis.
Keywords: Biased-gene conversion, conserved element, incomplete lineage sorting, intron, multispecies coalescent
As a result of advances in DNA sequencing and phylogenetic theory, as well as broader and more aggressive taxon sampling and access to museum specimens, phylogenetics is undergoing a renaissance. “Phylogenomics,” although a term originally coined to denote the increasing need for a phylogenetic perspective when inferring genome function (Eisen et al. 1997; Eisen 1998), is now meant also to signify the expanded scale in which phylogenetics typically is executed in the era of high-throughput sequencing (Delsuc et al. 2005; Posada 2016). This scaling up has taken two principleforms: increased taxon sampling as a means of producing greater phylogenetic accuracy, and perhaps even more pointedly, increased amounts of sequence data and numbers of loci generated to test a given phylogenetic hypothesis. Many phylogenies now contain hundreds, if not thousands of taxa, although in many cases highly taxon-rich studies still employ a modest number of loci or base pairs in the phylogenetic analysis. Through a variety of next-generation sequencing technologies, systematists now also have access not only to large numbers of loci for phylogenetic analysis but also a wide diversity of genes and noncoding regions for building phylogenetic trees (Bi et al. 2012; Faircloth et al. 2012; Chen et al. 2015). This access to a diversity of loci for building trees has inevitably increased interest in functional ties between phylogeny and genome history, thereby helping recapture some of the original intent of the term “phylogenomics.” For example, comparison of coding regions generated by transcriptomes across species can reveal key events in the history of adaptation of a clade (Pease et al. 2016), and phylogenetic analyses of conserved noncoding elements and transposable elements in vertebrates have yielded insight into major phases of regulatory evolution (Lowe et al. 2011) and sources of genomic innovation, respectively (Novick et al. 2009).
Despite this progress, in many ways systematists are still constrained by technology in their choice of marker loci for building trees, and this constraint has begun to yield cracks in the vision for phylogenomics going forward (Edwards 2016). For example, transcriptomes are widely used in plant, invertebrate and vertebrate phylogenomics, and with considerable success, in part due to their ease of access in organisms without available genomes and their relative ease of alignment across broad evolutionary distances. Yet, particularly in vertebrate phylogenetics, the deficiencies of coding regions for phylogenetic analysis have long been noted, even in the PCR-era of phylogenetics. For example, Chojnowski et al. (2008) suggested that introns were superior to coding regions in the phylogenetic analysis of birds in part because of their higher variability. Although coding regions provide effective phylogenetic resolution atshallower taxonomic levels in vertebrates (Blom et al. 2017; Potter et al. 2016), it is also widely recognized that the third positions of codons can become saturated in vertebrate data sets encompassing deeper divergences, and exhibit high variance in GC content among lineages (Weber et al. 2014), consequently providing less reliable phylogenetic signal at deep nodes. This trend was previously thought to be confined to fast-evolving mitochondrial genes, but is now generally acknowledged for nuclear genes as well, in many cases necessitating removal of 3rd positions of codons orthe use of amino acids rather than nucleotides (Cummins and McInerney 2011; Pisani et al. 2015). A compelling exampleof the challenges of coding regions for phylogenomic analysis has recently been found for birds, where, among all marker types tested, coding regions showed the highest level of among-lineage variation in base composition, resulting in severe challenges for phylogenetic analysis and ultimately yielding gene and species trees with lower congruence than other types of markers (Jarvis et al. 2014). Some of these deficiencies for phylogenetic analysis can be compensated for by improved models of molecular evolution (Phillippe et al. 2011; Pisani et al. 2015), partitioning, use of amino acids instead of nucleotides or dropping sites from analysis, yet at the same time there is a clear need for additional kinds of markers that may yield signals more commensurate with the major assumptions of many tree-building algorithms, such as base compositional stationarity. Reddy et al. (2017) recently provided compelling evidence, albeit in a concatenation framework, that the marker types currently in use in avian phylogenomics influence phylogenomic results even more so than taxon sampling, implying that additional marker types may be useful going forward.
Ultraconserved elements (UCEs) have also emerged as a major type of marker for phylogenomics, particularly in vertebrates (Faircloth et al. 2012; McCormack et al. 2012; Lemmon and Lemmon 2013; McCormack et al. 2013). These markers, which consist of and whose signal is dominated by the more variable regions flanking highly conserved core regions, are found throughout vertebrate and other genomes and have a number of features making them attractive for phylogenetics. They are numerous, allowing the accumulation of thousands of markers for a given study, and most importantly, the flanking regions are characterized by high variability, much more so than the conserved regions that are used to identify them. Although this higher variability yields large numbers of informative sites for phylogenetic analysis, it comes at the cost of decreasing reliability of alignments as one moves away from the core, conserved region (Faircloth et al. 2012; McCormack et al. 2013). Perhaps the most useful aspect of UCEs is their convenience: they can be isolated, through hybrid capture or other methods, without knowing anything about the genome of the species under study. In a similar fashion, anchored hybrid enrichment, whereas not focusing specifically on UCEs, also yields loci easily comparable among genomically novel taxa (Lemmon et al. 2012). Such loci have been readily isolated from hundreds of taxa that are otherwise genomically unstudied. Although many bioinformatics pipelines specifically exclude UCEs that include coding regions, in some studies, UCEs or ‘anchored’ conserved loci include exons (e.g., Lemmon et al. 2012; Prum et al. 2015). Additionally, in several studies not explicitly focused on UCEs, the more variable introns flanking exons have also been accessed in genomically unstudied species in a way similar to the flanking regions of UCEs, using approaches such as exon-capture or anchored enrichment (Lemmon et al. 2012; Hamilton et al. 2016). The convenience of collecting transcriptome data or using sequence capture when studying organisms whose genomes are not yet sequenced is a major driving force of marker choice in phylogenomics today (Sun et al. 2014; Edwards 2016; e.g., from birds, see Hosner et al. 2016; McCormack et al. 2016). These markers open up vast areas of biodiversity whose genomes have not yet been sequenced, either due to the unavailability of financial resources, small body size (and hence low DNA yield) of the studied organisms, excessively large or complex genomes, or other factors (Blaimer et al. 2016).
Conserved Nonexonic Elements in Phylogenomics
Here we analyze a new type of marker for phylogenomics that appears a promising addition to the systematists’ toolkit. Conserved nonexonic elements (CNEEs) are noncoding regions of the genome that are designated as ‘conserved’ because they evolve slower than a putatively neutral class of sites in the focal clade of organisms (Fig. 1). They are called nonexonic to distinguish them from exons, which also usually evolve more slowly than neutral regions of the genome. CNEEs share some overlap with the core regions of UCEs and could in principle also overlap with some anchored-enrichment loci (Lemmon et al. 2012; McCormack et al. 2012). However, CNEEs differ from UCEs and anchored loci in how they are identified in genomes, how phylogenetically widespread they are, and how they are analyzed in phylogenomic pipelines (Table 1). In truth, the definition of UCEs has expanded since its original definition (Bejerano et al. 2004) and its increased use in phylogenomics. For example, Bejerano et al. (2004) did not consider flanking regions in their discussion of UCEs, whereas most phylogeneticists consider the flanking regions of UCEs as part of their definition and use as phylogenetic markers. In the following, when discussing methods of procurement and phylogenetic distribution, we refer to the original definition of UCEs, which was erected by researchers studying genome function and characteristics, as opposed to the broader definition of UCEs that has subsequently been adopted and practiced by phylogenomicists. However, when discussing their use in phylogenomics, we also consider the fact that the flanking regions of UCEs comprise an integral part of their definition.
Figure 1.
Hypothetical schematic of comparisons of evolutionary rates of different noncoding markers discussed in this article. The shading of each bar is meant to indicate the distribution of rates within the range indicated by each bar. Thus, as found in this article, introns are somewhat more variable than UCEs, and CNEEs are less conserved than classically defined core regions of UCEs but not as variable as introns or the neutral rate.
Table 1.
Differences between vertebrate UCEs and CNEEs for phylogenetic analysis
| UCEs | CNEEs |
|---|---|
| Individual elements found throughout vertebrates, from fish to mammals/birds | CNEEs arise variably in evolution, and are not necessarily found across all vertebrate clades |
| Information in flanking regions principally used for phylogenetic analysis | Core region used for phylogenetic analysis |
| Can be coding or noncoding | Only noncoding |
| Core regions are ultraconserved | Core regions evolve slower than neutral regions, and thus can evolve faster than UCE core regions |
| Discovered via arbitrary conservation and length thresholds on synteny-aware whole-genome alignments | Discovered via tuned hidden Markov model |
Note: This table focuses on the classical definition of UCEs as originally described by Bejerano et al. (2004) and Faircloth et al. (2012).
Firstly, CNEEs differ from UCEs in not being “ultra-conserved”: whereas the core regions of UCEs areoften identified on the basis of >95% or higher sequence identify between genomes, the core regions of CNEEs are designated as conserved only because they evolve more slowly than a putatively neutral rate. As a result, CNEEs often exhibit moderate levels of variability, especially when compared with the core regions of UCEs (Siepel et al. 2005). This tendency raises the possibility that CNEEs might contain sufficient variability to be useful in phylogenetics, whereas at the same time exhibiting alignments of a quality that matches or exceeds those of the flanking regions of UCEs or transcriptomes.
CNEEs also differ in the means by which they are identified in genomes (Table 1). UCEs were initially identified using arbitrary thresholds of conservation and minimum length applied to synteny-aware whole genome alignments of a few exemplar taxa (Bejerano et al. 2004); they are often localized in additional genomes by blast searches using previously identified UCEs (McCormack et al. 2012). By contrast, CNEEs are delimited using statistical approaches, such as hidden Markov models (HMMs), wherein the rate of a candidate genomic region is compared with the rate at a class of putatively neutral sites (Siepel et al. 2005, Table 1). These models are usually applied to the entire set of species under analysis (although here we use a hybrid approach in which vertebrate CNEEs previously identified using a set of aligned vertebrate genomes are identified by blast in additional bird genomes). Four-fold degenerate sites of protein-coding genes are the most commonly used class of site to generate a baseline pattern of substitution. (It is reasonable to question whether 4-fold degenerate sites in coding regions are genuinely neutral (e.g., Chamary et al. 2006) and alternatives, such as ancient transposable elements that do not appear to have assumed functions, have also been used as the putatively neutral class (Siepel et al. 2005)).
CNEEs also differ from UCEs in their analysis in phylogenomic pipelines and their taxonomic distribution. Whereas it is the flanking regions of UCEs that typically provide what phylogenetic information is available for phylogenetic analysis, here we use only the CNEE itself, without any flanking regions, to generate phylogenetic hypotheses. Although CNEEs do overlap with the core regions of UCEs in genomes, and include all noncoding UCE core regions in principle, the use of only the core region of CNEEs distinguishes this class of marker and ensures that the sequences we use here for phylogenetics do not fully overlap with those of UCEs. Additionally, depending on the thresholds used to identify UCEs and tuning parameters of the HMM used to identify CNEEs, some UCE core regions will not be found in the set of CNEEs identified; this frequently happens when the core regions of UCEs are short, resulting in nonsignificant log-odds scores, which depend on length, when comparing the likelihood of CNEE sequences on trees with ‘neutral’ or ‘conserved’ branch lengths, the two states often used in the HMM. Finally, whereas UCEs were originally by definition found throughout the clade of interest (e.g., vertebrates or mammals, Bejerano et al. 2004), CNEEs are not highly phylogenetically conserved in this sense. They can appear at variable nodes in the Tree of Life; indeed, the appearance and disappearance of CNEEs within vertebrates has been studied to gain insight into the origin of phenotypic traits (e.g., Lowe et al. 2011; Lowe et al. 2014). Seki et al. (2017) recently explored the functions of conserved elements that were found only in birds (avian-specific highly conserved elements, or ASHCEs), 99% of which were found in noncoding regions. Whereas these authors focused on conserved elements found only in birds, regardless of their coding or noncoding status, here we emphasize primarily the fact that the conserved elements studied here are noncoding.
The definition and biological function of CNEEs helps further clarify their uniqueness as we define them here or their similarity to other markers. By defining CNEEs as nonexonic, we do not include any elements that overlap exons, or that overlap 3’ or 5’ untranslated regions (UTRs) or noncoding RNAs as annotated in recent chicken genome builds (Lowe et al. 2014). Otherwise, we believe our definition of CNEEs, which we borrow from Lowe et al. (2011), is similar to “CNEs,” or conserved-noncoding elements, a term that is also used in the literature, perhaps more widely than CNEEs (Lee et al. 2011; Marcovitz et al. (2016)). Sometimes transcribed noncoding regions, both for UTRs of coding mRNAs and for putatively noncoding transcripts, are called exons in the molecular biology literature (Guttman et al. 2009). Additionally, it is often unclear whether genomic regions are transcribed and made into proteins, and recent work from the ENCODE and other studies suggests that many regions previously thought not to encode proteins may in fact be transcribed and translated (Ji et al. 2015). Ultimately the details of bioinformatics pipelines will dictate which elements are included in any given analysis. What is generally agreed on, however, is that, like UCE core regions, CNEEs are known or suspected to act as regulatory enhancers, recruiting transcription factors to influence the expression of nearby or distant genes (Kvon et al. 2016; Leal and Cohn 2016).
In this Point of View, we compare the phylogenetic performance and evolutionary dynamics of three classes of noncoding genomic markers: CNEEs, UCEs, and introns. We focus on noncoding regions because they appear to be promising for vertebrate phylogenetics, and we agree with suggestions that transcriptomes may have undesirable phylogenetic properties, especially at high taxonomic levels. Our questions in analyzing CNEEs in a phylogenetic context include: do CNEEs resolve phylogenetic relationships as well as UCEs or introns? How do the substitution dynamics of CNEEs compare with those of UCEs and introns? Do CNEEs exhibit alignment and evolutionary properties that are desirable for phylogenomic analysis? And finally, how easily can CNEEs be accessed in nonmodel species, and what sorts of protocols are recommended for their large-scale deployment in phylogenomics?
Methods
Compiling CNEEs in Genomic Data
We previously explored the use of CNEEs as markers of regulatory evolution in vertebrates (Lowe et al. 2014). As part of an effort to understand the genomic basis of feather evolution using CNEEs, we first aligned 19 vertebrate genomes using BlastZ, MultiZ and chaining of local alignments (Kent et al. 2003; Schwartz et al. 2003; Blanchette et al. 2004) to determine regions of the genome (both coding and noncoding) that evolved slower than a benchmark set of 4-fold degenerate sites. The phylogeny of the 19 vertebrates used by Lowe et al. (2014) is well known and was assumed as fixed for all genes and genomic regions prior to analysis. This assumption is standard in pipelines for identifying CNEEs; although it ignores the possibility of incomplete lineage sorting (ILS), discordance due to ILS between the local genomic region and the vertebrate species tree we assumed, which has relatively long branches, is likely to be rare. Still, the potential biases incurred by assuming a fixed tree when identifying CNEEs should be explored, since ILS is known to influence parameter estimates of other macroevolutionary phenomena, such as molecular clocks, substitution rates and reconstruction of ancestral sequences (Burbrink and Pyron 2011; Groussin et al. 2015; Mendes and Hahn 2016). Branch lengths of the tree for the neutral class of sites were determined using maximum likelihood to find optimal branch lengths for the set of 4-fold degenerate sites. Conserved sites were defined as those exhibiting a better fit to a tree with branches no greater than 0.3 (30%) of the length of the 4-fold degenerate tree. The HMM had two states, “conserved” and “neutral” and the tuning parameters for the transition rate between states in the HMM were set with an expectation that CNEEs would on average have a length of 45 bp. This protocol yielded 957,409 conserved elements in total, of which 605,756 fulfilled the criteria for a CNEE. Whereas Lowe et al. (2014) used 602,539 CNEEs in their study, we retained 3207 CNEEs that were discarded in Lowe et al. (2014) because they were not assigned to chromosomes in the chicken assembly used (galGal3), making for a starting total of 605,756 CNEEs. For a detailed account of the bioinformatics pipeline by which we initially determined a working set of CNEEs, see Lowe et al. (2014). Candidate CNEEs were filtered from this vertebrate-wide set of 605,746 elements referenced on chicken to retain loci 400 bp in length (). We focused on CNEEs 400 bp long so as to use a set of loci expected to contain at least a moderate number of variable and parsimony-informative sites.
We then chose 14 exemplar species from the Avian Phylogenomics Project (Jarvis et al. 2014), including chicken, as a test case for phylogenomic analysis (see Supplementary File S1 available on Dryad at http://dx.doi.org/10.5061/dryad.25f7g). These species were chosen so as to capture major branches of the avian tree as it is now known, and in some cases pairs of species were chosen to determine if our analyses could recapitulate known or expected relationships (e.g., flamingo and grebe, penguin and loon). This group of 14 species also contains clades that are still unresolved or contentious, such as the precise order of the multiple outgroups to passerine birds (Hackett et al. 2008; Jarvis et al. 2014; Prum et al. 2015). We also included data from draft genomes of an Emu (Dromaius novaehollandiae) and Chilean Tinamou (Nothoprocta perdicaria) from Baker et al. (2014) to explore the hypothesis of ratite paraphyly (Harshman et al. 2008; Phillips et al. 2010; Smith et al. 2013), a total of 16 ingroup species. This Targeted Locus Study project (for new sequences of Chilean Tinamou and Emu) has been deposited at DDBJ/EMBL/GenBank under the accessions KBAG00000000 and KBAF00000000, respectively. The versions described in this article are the first versions, KBAG01000000 and KBAF01000000, respectively. Using an American Alligator (Alligator mississippiensis, Green et al. 2014) genome as an outgroup brought the total taxa used to 17 (Supplementary File S1 available on Dryad). Blastn searches with chicken query CNEE sequences were used against each of the 16 nonchicken target genomes at an e-value cutoff 1e. CNEEs with no missing species were retained (), and de novo aligned with default global alignment parameters in MAFFT v. 7.245 (Katoh and Standley 2013).
Intron alignments were assembled from the Avian Phylogenomics Project data of Jarvis et al. (2015); however, individual introns were used rather than alignments concatenating introns within each protein-coding gene. The SATé-MAFFT alignments provided by Jarvis et al. (2015) were reduced to the taxon subset of interest and gap-only columns removed. Loci greater than 400 bp in aligned sequence length, including the alligator outgroup sequence and with no more than three missing species were retained (). It is noteworthy that it was straightforward to compile 3700 fully populated CNEE alignments of 400 bp or greater, whereas there were only 998 (26.7%) fully populated orthologous introns from birds available; we will return to this point in the discussion. Orthologous sequences from Emu and Chilean Tinamou were identified with blastn searches against draft genome assemblies for these species with chicken, Common Ostrich, and White-throated Tinamou queries, and were profile aligned to the existing Jarvis et al. (2015) alignment with MAFFT. Because SATé-MAFFT yields relatively gappy alignments that are nonetheless “better” than MAFFT only alignments by some optimality criteria (B. Faircloth, pers. comm.), comparing alignment statistics using SATé-MAFFT and MAFFT may bias the results. We therefore applied both SATé-MAFFT and MAFFT to all three marker types to enable side-by-side comparisons. For the CNEE alignments, we recapitulated the precise SATé-MAFFT alignment protocol of Jarvis et al. (2014), including postalignment trimming with their custom python script ‘filter_alignment_fasta_v1.3B.pl’, except that we used SATé v. 2.2.7 with MAFFT v. 6.717 (Jarvis et al. 2015 used SATé 2.1.0 and MAFFT 6.860b). UCEs (, representing the full set from Jarvis et al. 2015) were compiled as described for introns. There was a higher number of fully populated alignments for UCEs of total length of 400 bp or greater (; 99.7%) than for introns.
As expected, there was overlap between our sets of CNEEs, UCEs, and introns. For example, 1497 (39.2%) CNEEs overlapped at least one UCE. The degree of overlap between introns and the other two data sets was much lower: there were six introns overlapping CNEEs and three introns overlapping UCEs (both <0.2%). Because UCE loci typically include the conserved core region in addition to the flanking regions, this overlap could lead to nonindependence of our analyses. Therefore, in addition to analyzing the full set of UCEs and CNEEs, we also analyzed nonoverlapping data sets of CNEEs and UCEs; in general, we found that our results held for overlapping and nonoverlapping subsets of data, and we suggest that even if our CNEE and UCE data sets overlapped completely, analyzing just the core or flanking regions alone would help clarify the difference in dynamics and performance between these genomic regions.
Measures of Alignment Quality and Substitution Dynamics
Alignment lengths, proportions of variable and parsimony informative sites, GC content, and the amount of missing data per alignment matrix (here defined as the number of gaps and uncalled bases per total cells in the nucleotide matrix) were calculated with AMAS (Borowiec 2016). Average pairwise nucleotide identity between species within each locus, and the proportion of gaps per base pair of aligned sequence were calculated with custom Perl scripts. Unlike the AMAS calculation of missing data, gaps per bp aligned considers only genuine gap characters (ignoring uncalled bases) and excludes leading and trailing gaps as well as gaps adjacent to uncalled bases; it is equivalent to internal gaps in the alignment per total called bases. TrimAl v. 1.2rev59 (Capella-Gutiérrez et al. 2009) was used for column-based alignment filtering, with the ‘automated1’ option to choose trimming parameters heuristically based on input alignment characteristics. We recognize that trimAl and other alignment trimmers may not necessarily improve phylogenetic analysis in some cases (Tan et al. 2015), but we use them here strictly as a standard metric for comparing alignment “quality,” without subsequent phylogenetic analysis on the trimmed alignments. Additionally, we note that in many of the analyses in Tan et al. (2015), alignment trimmers performed marginally worse only under unsustainably high levels of trimming. Model-averaged transition/transversion rate ratios (Ti/Tv), the proportion of invariant sites when appropriate and the gamma shape parameter () were estimated for each alignment with jModelTest v. 2.1.7 (Darriba et al. 2012). jModelTest runs included six substitution models (JC, F81, K80, HKY, SYM, and GTR), with invariant sites and unequal base frequencies allowed and rate variation modeled with four gamma categories.
Phylogenetic Analyses and Measures
RAxML v. 8.1.4 (Stamatakis 2014) was used to construct 200 bootstrap replicate gene trees from each unpartitioned alignment for each locus with a GTR substitution model; these were rooted with the American Alligator outgroup in DendroPy v. 3.12.0 (Sukumaran and Holder 2010; Sukumaran and Holder 2015). To measure and compare gene tree variation for each marker class, we calculated matching split distances between gene trees using TreeCmp v. 1.1-b308 (Bogdanowicz and Giaro 2012). Split distances were calculated on ML gene trees (as opposed to consensus bootstrap gene trees) using a GTR GAMMA model and 20 independent tree searches in RaxML starting from a different random starting tree. MP-EST v. 1.5 (Liu et al. 2010) was used to infer species trees for each marker type from the input set of rooted RAxML bootstrap trees. Each analysis used three full MP-EST runs starting with a different random number seed and 10 independent tree searches within each run. Highest scoring trees from each search were used to build a majority-rule extended (MRE) consensus tree for each MP-EST run using RAxML. Per-site and consistency (Kluge 1989) and retention (Farris 1989) indices (CI and RI, respectively) were calculated with PAUP v. 4a149 (Swofford 2002) using the MRE consensus gene tree of the 200 RAxML bootstrap replicates for each locus. We did not compute CI or RI on species trees because gene tree heterogeneity can distort such statistics when all gene trees are forced onto a single topology (Edwards 2009; Mendes and Hahn 2016). Average bootstrap supports are also reported for MRE consensus gene trees.
Phylogenomic Subsampling
Phylogenomic subsampling (Edwards 2016; Blom et al. 2017) was used to assess the stability of specific clades for different subsets of each of the CNEE, intron and UCE data sets. Data sets of increasing numbers of loci (, 100, 200, 300, 400, 500, 1000, 1500, 2000, 2500, 3000, and 3500 loci) were built by sampling loci with replacement from within each marker type, and repeating the process to generate 10 independent replicates of a given number of loci within each marker type. MP-EST was then run on each of the 10 replicates as described above, except that only a single MP-EST run (but with 10 independent tree searches) was performed for each replicate. Summary measures are reported by counting the frequency of splits from among the set of MP-EST output trees for each replicate rather than from a consensus tree.
Results
Alignment and Variability Metrics for Noncoding Markers in Birds
Alignment lengths and variability
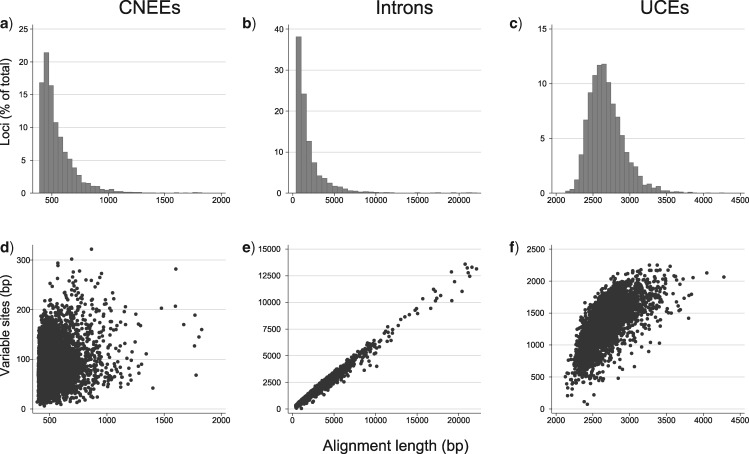
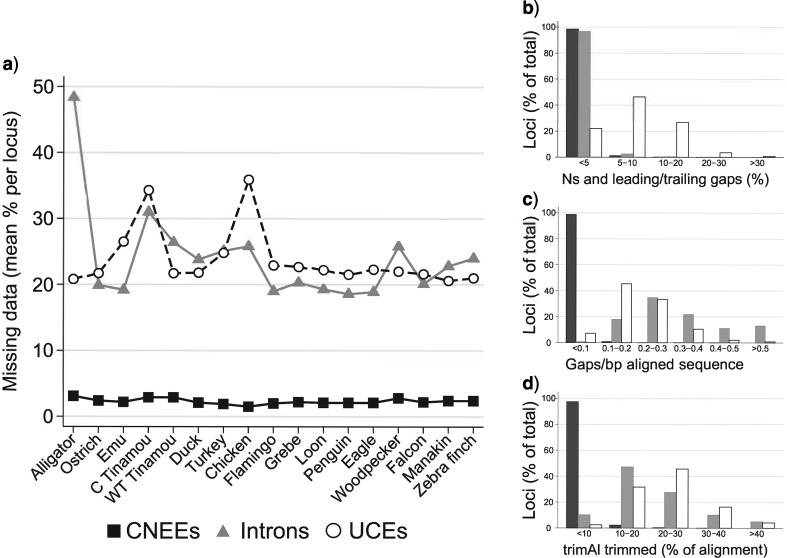
Figure 2 shows the distribution of alignment lengths among the three marker types and the percentage of variable sites within each alignment. With the constraint that each alignment must equal or exceed 400 bp, introns had longer alignments (up to 22,138 bp) than CNEEs (longest alignment, 1829 bp; Fig. 2a–c). UCE alignments based on those of Jarvis et al. (2014) varied from 2,126 – 4279 bp. CNEE alignments exhibit a higher fraction of populated bases per alignment than do introns and UCEs, with 1210 out of 3822 CNEE alignments (31.7%) possessing >99 % of populated bases (Fig. 3a,b). No intron alignments and only a single UCE alignment possessed this high a nucleotide matrix occupancy, whether considering any undetermined base or gaps between called sequence alone (Fig. 3c). CNEEs also exhibited a much lower percent of each alignment that was deemed low quality by trimAl than did introns or UCEs (Fig. 3d, Supplementary File S2 available on Dryad. Whereas 1003 out of 3822 CNEE alignments (26.2%) retained >99 % of bases after trimming, only 1 of the UCE alignments and none of the intron alignments retained this much after trimming (Fig. 3d). As expected, both introns and UCEs were more variable than CNEEs (Fig. 2d–f; Supplementary Fig. S1a available on Dryad). The number of parsimony informative sites per alignment varied among markers in a similar way, with CNEEs having the fewest and introns having the most (Supplementary Fig. S1b available on Dryad). The number of variable sites scaled more linearly with alignment length for introns () than for UCEs () or CNEEs (; Fig. 2d–f). Although the alignment and variability statistics for UCEs changed significantly when analyzed using the MAFFT-only pipeline we used for CNEEs, the magnitude of the differences was small and trends among markers did not change (Supplementary File S3 available on Dryad). Similarly, when we realigned all three marker types with the SATé-MAFFT approach used by Jarvis et al. (2014), overall trends and differences between markers were unchanged (Supplementary File S4 available on Dryad). Visual inspection of individual alignments is perhaps the best way to appreciate the differences in alignment characteristics among markers (available on Dryad).
Figure 2.
Top row: Distribution of aligned sequence lengths for a) CNEEs (3822 loci), b) introns (3579 loci), and c) UCEs (3679 loci). Bottom row: Correlations between alignment length and number of variable sites for d) CNEEs (), e) introns (), and f) UCEs ().
Figure 3.
Variation in alignment gappiness among marker types. a) Average percentage of undetermined bases per alignment for each marker type and taxon. Here, undetermined bases indicate both gaps and Ns in each alignment. Alignments that were completely missing any species were excluded before analysis. These data are separated into separate graphs for b) Ns and gaps at the very beginnings or ends of alignments (e.g., true missing data) and c) alignment gaps (e.g., gaps between ACGT bases that are introduced internal to the sequence alignment). d) Distribution of the percentage of each alignment remaining after trimming with trimAl. See Methods for further discussion.
GC content and substitution dynamics of noncoding markers
CNEEs exhibited systematically lower GC contents than did introns or UCEs (Fig. 4a,b; Supplementary File S2 available on Dryad). There was a correlation between the GC contents of different noncoding markers across species, presumably indicating a genome-wide effect on base composition that influences all three marker types (Supplementary File S5 available on Dryad). A notable outlier in GC content across all three marker types is the Downy Woodpecker (Picoides pubescens), with average GC contents of 37.52% (CNEEs), 42.44% (introns) and 40.48% (UCEs), values that deviate from the grand mean for each marker type often 10 times more than for other species (Fig. 4a,b). We found that the variance in GC content among species was lowest for CNEE markers (average variance 0.82), and higher for intron and UCE markers (average variance 5.76 and 3.91, respectively; Fig. 4c). These substitution dynamics held in nonoverlapping sets of CNEEs and UCEs (Supplementary File S6 available on Dryad).
Figure 4.
Patterns of GC content variation among markers and taxa. Alignments in which a taxon is entirely absent are omitted from all calculations. a) Per-taxon values for mean GC content for each marker type. b) Distribution of GC content among species for each marker type. c) Variance in GC content among taxa for each marker type.
When analyzing GC content of only variable sites (Supplementary File S2, Fig. S2 available on Dryad), we found that GC content was actually slightly higher in CNEEs (43.40%) than for introns (42.61%) or UCEs (42.45%). In this case, both tinamous and the Downy Woodpecker exhibited higher than expected GC content, suggesting genome-wide shifts in substitution dynamics or recombination across all markers (Meunier and Duret 2004). Additionally, for polymorphic sites, the variance in GC content among lineages was actually highest in CNEEs ([, 1 standard deviation [SD]), as compared with introns () or UCEs (), although this was likely a consequence of the small number of variable sites at CNEE loci (Supplementary File S2, Fig. S2 available on Dryad; see Discussion).
Using jModelTest, we evaluated the substitution dynamics and optimal substitution model for each alignment. On average, CNEEs exhibited higher transition/transversion rate ratios (average 2.44) than did introns (1.90) or UCEs (1.79; Supplementary Fig. S1d available on Dryad). CNEEs also exhibited intermediate estimates of the gamma shape parameter (average 1.46) compared with introns (7.81) or UCEs (0.92; Supplementary Fig. S1 available on Dryad). Overall, although all three markers displayed a similar range of nucleotide substitution models, the most complex models (GTRI and GTR) were least prevalent as the best-fitting model for CNEEs (7.2 and 29.2% of loci, respectively) than for introns (13. 7 and 73.2%) or UCEs (74.7 and 23.6%; Supplementary File S7 available on Dryad). CNEEs displayed significantly higher CI (mean 0.92 for full and nonoverlapping set) than UCEs (mean 0.82; <0.00001) or introns (mean 0.82; <0.00001); Supplementary Fig. S1, File S2 available on Dryad). A similar pattern was found for RI (Supplementary Fig. S1, File S2, available on Dryad).
Phylogenomic Signal and Consistency of Noncoding Sequences
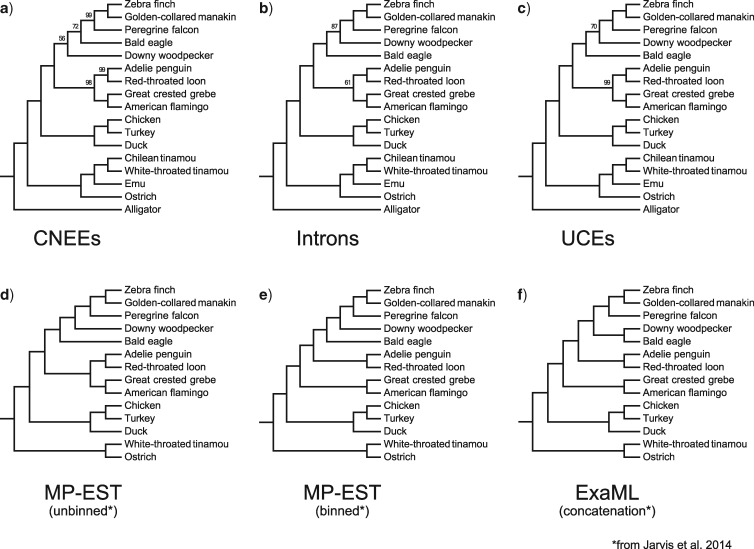
As expected from the rank order of variability of each of the three marker types, gene trees made from CNEE alignments exhibited the lowest average bootstrap support, with introns and UCEs having progressively higher support (Supplementary Fig. S1c available on Dryad). Consequently, split distances among gene trees were higher for CNEEs (Mean SD 0.61 0.16) than for introns (Mean SD 0.38 0.13) or UCEs (Mean SD 0.31 0.11; Supplementary Fig. S3 available on Dryad). However, the estimates of overall phylogenetic relationships and clade support as judged by species tree analyses were generally concordant among marker types and with previous analyses using larger data sets (Jarvis et al. 2014). All markers recovered ratite paraphyly, with the Emu clustering with the two tinamous to the exclusion of the Common Ostrich at 100% bootstrap support (Fig. 5a–c). In all three trees, the Neognathae are monophyletic and the three taxa representing Galloanseres (chicken, turkey, and domestic duck) were monophyletic at 100%, appearing as expected as sister to all the remaining taxa (Neoaves). All branches in the MP-EST species trees in this study achieved 95% for all marker types, except for two branches in the total CNEE tree, two branches in the total intron tree, and one branch in the total UCE tree. The branches in question invariably involved relationships among the outgroups to passerine birds and falcons, a clade termed Australaves (Yuri et al. 2013, Fig. 5d–f). Whereas the total CNEE tree suggests that the Bald Eagle is closer to this clade than the Downy Woodpecker (albeit with only 72% and 56% bootstrap support, respectively, for these two branchings), both the total intron and UCE trees support the reverse branching order, with first Downy Woodpecker (at 87% and 70% bootstrap support for introns and UCEs, respectively), then Bald Eagle (with 100% support in both cases) forming successive sister groups to the Australaves. Depending on how one likes to draw bootstrap support cutoffs in phylogenomics analyses, there is no case among the total marker trees of strongly supported conflict in overall species tree estimates among the three marker types for any cutoff greater than 87%. This trend largely held for phylogenetic analysis of the nonoverlapping subsets of CNEEs and UCEs (Supplementary Fig. S4 available on Dryad): support values increase for CNEEs (72% to 89% for eagle falcon/passerines and 56% to 85% for woodpecker other ‘land birds’), and decrease for UCEs (70% to 62% for woodpecker falcon/passerines). When we confine phylogenetic analysis to the 1000 loci with the highest variability or most highly supported gene trees, the results are largely similar (Supplementary Fig. S5 available on Dryad).
Figure 5.
Species tree topologies discussed in this study. Top row: MP-EST species trees for a) CNEEs (3822 loci), b) introns (3579 loci), and c) UCEs (3679 loci), with support values <100% indicated. Bottom row: TENT trees, each built from 2516 introns, 3769 UCEs, and 8251 protein-coding genes, from Jarvis et al. (2014), pruned to the taxon set used in the current study. d) MP-EST unbinned analysis, e) MP-EST binned analysis, and f) concatenated analysis. Support values are omitted from the pruned Jarvis et al. trees. The main tree presented in Prum et al. (2015) is identical to the tree depicted in panel d, assuming that Chilean Tinamou and Emu would fall where found in other studies.
When all loci were analyzed together, the resulting trees differed in nonsignificant ways from trees made from a single marker type or from each other. A total evidence noncoding marker tree (9739 loci), including UCEs (2232 loci) and introns (3685 loci) that did not overlap other markers yielded the CNEE-only tree (Fig. 5a), albeit with all branches resolved at 100% except the branch leading to eagle/falcon/passerines at 94% (Supplementary Fig. S6a available on Dryad). Total evidence (9682 loci) analysis of all UCEs (3679 loci), and CNEEs (2318 loci) and introns (3685 loci) that did not overlap other markers yielded a similar tree with 51% support for the same branch in two of three MP-EST runs, and a tree resembling those in Figure 5b,c, with the branch leading to woodpecker/falcon/passerines with 52% support (Supplementary Fig. S6b available on Dryad).
The relationships obtained for the three marker types are also similar to those found in a more taxon-rich tree for birds produced with fewer (259) loci, most of which were derived from coding sequences (Prum et al. 2015). A source of disagreement for the taxa that we have sampled involved the sister group to Australaves (Prum et al. 2015). Although both Prum et al. (2015) and some trees in Jarvis et al. (2014) placed woodpeckers closer to Australaves than eagles, neither paper produced this result unambiguously; whereas the Jarvis et al. (2014) achieved 100% support for a sister clade to Australaves that included both woodpeckers and eagles in their total evidence concatenation tree (TENT) using ExaML, other analyses from Jarvis et al. (2014), as well as the results of Prum et al. (2015), placed woodpeckers as sister to Australaves, with eagles falling outside this clade, albeit with highly varying levels of support. The relationships among waterbirds (penguin, loon, flamingo, and grebe), although consistent across analyses and markers in this study, constitute another region of disagreement with studies employing more taxa. Whereas this study and Prum et al. (2015) suggest monophyly of the four water birds sampled here (Aequorlitornithes), many of the Jarvis analyses, including their TENT analysis, suggested paraphyly of this clade. Although overall our results appear to be more congruent with the results of Prum et al. (2015), because the two studies with more extensive taxon sampling differ in several key areas relevant to this article, it is difficult to assess which topology for the two clades of interest are better corroborated by our analyses.
We conducted phylogenomic subsampling to study the accumulation of signal as the number of loci increases for two expected clades that ultimately achieve high certainty for all data sets as well as for the two uncertain clades described above. The two high-confidence relationships we examined were the paraphyly of ratites and the sister group to passerines (i.e., falcons; Fig. 6a,b). We found that all three marker types established high confidence in the paraphyly of ratites by 200 genes, with introns accumulating signal somewhat faster than CNEEs and UCEs (Fig. 6a). By contrast, the falcon passerine clade achieved consistent 100% support at 1000 loci for introns and UCEs, whereas CNEEs did not achieve an average of 100% support for the number of loci analyzed here, peaking at 98% support at 3500 loci and 99% with the full data set (Fig. 6b). For the monophyly of the waterbird clade (Fig. 6c), we found that the accumulation of signal was more rapid for CNEEs and UCEs, and less rapid for introns. Introns achieved an average bootstrap support of only 70% for subsamples of 3500 loci (only 61% for the full data set), whereas average support of similarly sized subsamples of CNEEs and UCEs approached 100% (98 and 99%, respectively, for the full data set). For this clade, no marker type exhibited monotonically increasing average support with larger subsamples of loci, although the lack of monotonic increase was much more pronounced for introns than for the other two markers (Fig. 6c). The subsampling results for the sister to Australaves are more interesting, in so far as they begin to suggest genuine conflicts between the marker types. Whereas both introns and UCEs accumulate stronger signal favoring a woodpecker Australaves clade (87 and 70%, respectively; Figs. 5b,c and 6d), the CNEEs instead accumulate stronger signal favoring an eagle Australaves clade, approaching 72% (Figs. 5a and 6d). Whereas CNEEs exhibit a threshold of sorts for the accumulation of signal for the waterbird clade, increasing in average support and number of replicates achieving >70% support at 500 loci (in part an artifact of the particular intervals chosen for subsampling), introns suggest a threshold at 1500 loci for the woodpecker/Australaves clade (Supplementary Fig. S7 available on Dryad).
Figure 6.
Phylogenomic subsampling, with MP-EST species trees inferred for 12 data sets of increasing numbers of randomly chosen loci, and with 10 replicates per data set. In each row, left panels plot the mean bootstrap support among the 10 MP-EST replicates for each marker type and data set size. At right are the two branches, indicated by stars, whose support is investigated by subsampling, with open and solid markers indicating support for one or the other branch. a) Trends in support with increasing numbers of loci for paraphyly of ratites. b) Trends in support for the branch uniting falcons as the sister group to passerine birds. c) Trends in support for competing hypotheses placing either core landbirds (songbirds falcon eagle woodpecker) or (flamingo grebe) as the sister group to (penguin loon). d) Trends in support for either Bald Eagle or Downy Woodpecker as the sister to (songbirds falcon).
Discussion
In this study, we explored the evolution of CNEEs, a class of noncoding marker that has not received attention in terms of its utility for phylogenomics, and compared them to the performance of two other classes of noncoding markers, introns and UCEs. Overall, the full data set of CNEEs performed well compared with introns and UCEs, with a similar number (1–2) of branches in our 17-taxon tree achieving less than 95% support. The utility of CNEEs for phylogenomic analysis will depend somewhat on the values held by different researchers. If a researcher values high support for branches achieved quickly as numbers of loci are increased, at the expense of more uncertain and gappy alignments with missing species, then introns and UCEs clearly outperform CNEEs. However, if a researcher favors higher certainty and quality of alignment, and a better fit of alignments to the equilibrium assumptions of most phylogenetic models of nucleotide substitution, then CNEEs may offer advantages. The major advantages of CNEEs are the ease of obtaining large numbers of high-quality alignments without missing species, their low homoplasy and their low variance in GC across species. Despite their low variability, and the correspondingly weak support in gene trees, CNEEs produced a species tree that rivaled those produced by a similar number of intron and UCE alignments (Fig. 6). Indeed, given that the CNEE alignments were the shortest of the three marker types, one could argue for the overall efficiency of CNEEs in terms of phylogenetic resolution per base pair sequenced as compared with the other two markers. CNEEs identified using more liberal tuning parameters of the HMM, such as using a conserved tree with branch lengths greater than 30% of the neutral tree such as we used here, might also yield a broader set of markers with higher resolving power.
The means by which markers are collected for phylogenomic studies will influence the relative utility of CNEEs versus UCEs. Our study in some ways gave UCEs the best chance for optimal performance compared with CNEEs, because we used long UCEs generated bioinformatically from sequenced genomes. By contrast, the shorter UCE loci (core plus flanking regions) generated by sequence capture methods often exhibit very limited variation per locus—for example, <2% of the UCE sites in the recent study by Meiklejohn et al. (2016) were parsimony informative and only 54 of the 1289 UCEs with at least one parsimony informative site had >25 parsimony informative sites per locus. This level of information content led to very noisy gene trees and poor performance of species tree methods, as expected (Xi et al. 2015), even if this effect was ameliorated by using the most informative loci to generate gene trees for input into two-step coalescent methods (Meiklejohn et al. 2016, Liu et al. 2015). CNEEs may well yield higher quality gene trees than the short UCE loci that are often harvested by hybrid capture. Additionally, because the phylogenetic information in CNEEs is in the same core regions that would be used as baits to capture loci, we predict that the phylogenetic informativeness of CNEEs would not be compromised as much as for UCEs when using hybrid capture. We verified that CNEEs might also be collected by hybrid capture by comparing the sequence divergence between Emu and a passerine for anchored loci collected by hybrid capture in the Prum et al. (2015) study with that found in this study for CNEEs (using the Yellow-wattled Bulbul and the Zebra Finch, respectively, as the passerine). We found that CNEEs for this comparison averaged 0.041 substitutions per site, whereas loci for this comparison in Prum et al. (2015) averaged 0.057. (The anchored loci in Prum et al. (2015) were not UCEs, and so we cannot conclude from these values that hybrid-captured UCEs have higher variability than CNEEs.) We conclude that CNEEs are likely amenable to hybrid capture and may prove even more informative than UCEs when both markers are generated by hybrid capture or other laboratory methods.
Number of Markers for Phylogenomic Analysis
Critical factors in phylogenetics are the number of markers of reasonable length available for analysis, and the ease of producing fully populated alignments, since these factors could place a limit on phylogenomic resolution of a particular marker type, especially when using coalescent methods (Edwards 2009; McCormack et al. 2012; McCormack et al. 2013). Introns are numerous in vertebrate genomes, on the order of several times the number of genes, which usually number about 15,000–20,000. However, orthologous introns often vary substantially in length among taxa (Vinogradov 2002; Waltari and Edwards 2002; Pozzoli et al. 2007; Zhang and Edwards 2012); due to their high variability and length differences, gaps will be frequent, with many alignments >400 bp having large numbers of unfilled (missing) bases. Conserved elements are very numerous in vertebrate genomes, with as many as 3.6 million elements detected in mammals, over 80% of which are noncoding (Lindblad-Toh et al. 2011). However, the average length of these elements is often > 50 bp. Faircloth et al. (2012) were able to assemble 5599 unique UCEs, which need to achieve a certain minimum length of the core region to be detectable by hybrid capture methods. Bejerano et al. (2004) found only 481 fully conserved UCEs longer than 200 bp in vertebrate genomes, and the total number of UCEs >100 bp in vertebrates is estimated to be 14,000 (Stephen et al. 2008). Today, markers designated as “UCEs” often contain loci that are not strictly UCEs, but rather mildly conserved (coding or noncoding) elements, whose core is often more variable than the original definition of UCEs (Bejerano et al. 2004). In this respect, the number of “UCE” loci has increased in recent studies and can overlap even more with loci designated here as CNEEs. The more restricted phylogenetic distribution of some CNEEs is probably not a severe issue, since many phylogenomic studies focus on similarly restricted set of taxa; in the same way, some studies using UCEs have been able to augment the number of loci by using loci with restricted phylogenetic distribution (Faircloth et al. 2012). This phylogenetic flexibility of UCEs and CNEEs may ultimately be an advantage, and in some ways captures the spirit of the original definition of UCE, which explored conservation at various levels in phylogeny (Bejerano et al. 2004). CNEEs are also prevalent in plants and invertebrates (Siepel et al. 2005; Kritsas et al. 2012; Ryu et al. 2012; Haudry et al. 2013; Burgess and Freeling 2014; Villar et al. 2014), suggesting that they may have wide phylogenomic utility, although it is unclear whether the challenges of orthology in some plant UCEs also apply to CNEEs (Reneker et al. 2012).
It was straightforward to compile a data set of several thousand CNEE markers >400 bp which contained all species, and most of which contained <2 alignment indels. By contrast, because of their intrinsic variability or reliance on variable flanking regions, both introns and UCEs had between 20–40% missing data (gaps) per species per alignment, a consequence of their high-indel rate, and it was challenging to find intron alignments that contained all 17 species in our study. These trends were evident despite the fact that all three marker types were harvested from whole genomes, as opposed to being generated using molecular methods such as hybrid capture. The reasons for the lower incidence of fully populated alignments for introns does not seem to lie in the lower coverage of some of the genomes used since the CNEEs were harvested from the same source data. Rather, it seems to lie in the greater challenges of detecting introns via blast, or by challenges with genome annotations, or the great length of many introns, which undercuts search algorithms.
GC content and Patterns of Nucleotide Substitution
The three marker types exhibited differing patterns of nucleotide substitution, which could influence their phylogenetic performance, and which appear to be driven in part by overall levels of variation. For example, across all sites, CNEEs had the lowest level of among-lineage variation in GC content, a trait that conforms well with the equilibrium assumptions of most models of nucleotide substitution. High variance in GC content can complicate phylogenomic analyses, since most phylogenetic models assume that all species in the analysis share a similar equilibrium base composition (Lockhart et al. 1994; Foster and Hickey 1999; Mooers and Holmes 2000). However, we also found that, at variable sites only, the GC content and the variance in GC content among lineages were both highest for CNEEs, a result that undermine their utility in phylogenomics more so than other markers. Here, we reasoned that because the number of variable sites in CNEEs is small, small numbers of substitutions could drastically change the GC content of variable sites. Consistent with this hypothesis, we found that the correlation between the percentage of variable sites per CNEE and the variance in GC among lineages was higher (0.40, <2.2e-16) than the correlation between total CNEE length and variance in GC (-0.07 <6.22e-06), although both were significant (but both without any correction for phylogeny). Additionally, the HMM to identify CNEEs may partly drive the lower GC content of CNEEs versus other markers: if the 4D sites on which the neutral branch lengths are estimated have a high-GC content, then it may be easier to reject a neutral tree if you have a lower GC content. Regardless, we view the high variance in GC content at variable sites as an interesting phenomenon worthy of study, but not damaging to our case on the utility of CNEEs in phylogenomics. Weber et al. (2014) have discussed the interplay of forces influencing GC content variation among bird lineages, which include life history traits and the extent of local recombination and GC-biased gene conversion.
To our knowledge we are the first to report the anomalous GC content of the Downy Woodpeker genome. It is likely that the higher fraction of transposable elements (TEs) in this genome (22%) compared with other birds investigated thus far, as reported by (Zhang et al. 2014), is linked to the outlier status of the Downy Woodpecker in terms of GC content, although we have not verified the prediction that TEs in this genome are higher in GC than other genomic regions. As expected due to their inclusion of the slowly evolving core as well as more rapidly evolving flanking sequences, UCEs exhibited high levels of among-site rate variation (low compared with introns and CNEEs. Although not necessarily detrimental to phylogenetic analysis, it is widely acknowledged that high levels of among site rate variation are more difficult to model than low levels (Vogler et al. 2005; Marshall et al. 2006; Holland et al. 2013). On the other hand, CNEEs exhibited the highest transition/transversion (ts/tv) ratio among the markers; although high ts/tv ratios, like among-site rate variation, often lead to homoplasy, the higher consistency index among CNEEs appears here to be driven more by their low substitution rate than ts/tv ratio. CNEEs were markedly more AT-rich than the other two classes of markers, which, as a group, tend to be more AT-rich than coding regions. Although AT- versus GC-rich markers may not appear to present any obvious advantages, Romiguier et al. (2013) recently suggested that, in mammals, GC-rich markers result in higher gene tree heterogeneity than AT-rich markers, possibly due to biased gene conversion, making phylogenetic analysis more challenging.
Information Content of CNEEs for Phylogenetic Analysis
Overall, we found that CNEEs delivered an estimate of phylogenetic relationships that was as strong as that for UCEs and introns. For some expected phylogenetic results, such as the paraphyly of the ratites, the approach to phylogentic “certainty” (100% bootstrap support) was as fast as that for the other two markers. However, for other questions that appear to be gaining consistent support among phylogenomic data sets, such as the falconid sister group of the passerine birds, the approach to phylogenomic resolution was markedly slower than for UCEs or introns. And yet for other clades, such as the sister relationship between penguin/loon and flamingo/grebe, it was introns that failed to achieve high resolution compared with CNEEs and UCEs. Finally, CNEEs suggested a different sister group to falconids and passerines, namely eagles, at fairly high (72%) and increasing support as more loci were accumulated, as compared with introns and UCEs, which favored woodpeckers as the sister group, again with high support. This result was the only case of moderately strong conflict among markers in our data set, and in our view, either result is plausible, given that this node was not resolved with certainty among larger data sets (Jarvis et al. 2014). The fact that among the taxa we studied the woodpecker is a base compositional outlier more strongly for introns and UCEs than for CNEEs could be driving this difference in result. We were able to achieve high and consistent confidence for nearly all branches in our analysis without binning (Mirarab et al. 2014), suggesting that large numbers of loci, rather than concatenation of loci, remains a plausible way forward for phylogenomics (Liu and Edwards 2015). Although our results point to possible differences in performance and interesting trends among these markers, because we only sampled 16 ingroup bird species as exemplars, the generality of these trends requires further investigation.
In summary, CNEEs appear to be a promising tool for phylogenomic research. Their low variability compared with introns and long UCEs captured from sequenced genomes are offset by the larger numbers of moderately long- and high-quality alignments that can be gathered from whole-genome data sets. In the future, as whole genomes become more readily available, phylogenomic data sets will increasingly be generated via statistical tools or extraction of large sets of alignments from aligned or unaligned genomes (Costa et al. 2016), rather than directly by wet lab bench work. Until that time arrives, wet-lab approaches to gathering loci, such as hybrid capture, will continue to be used. In either scenario, CNEEs should fare well, because they are readily identified by statistical means from whole genomes, and yet they would also be amenable to hybrid capture approaches. We expect that mixtures of noncoding phylogenomic markers, including CNEEs, will be helpful in understanding the dynamics of currently popular markers such as UCEs and introns and will contribute to resolving the Tree of Life.
Supplementary Material
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.25f7g.
Funding
This research was supported by NSF grant DEB 1355343 (EAR 1355292) to S.V.E. and Julia Clarke.
Acknowledgments
We thank Brant Faircloth for help in assembling the UCE data set, and Brant Faircloth, Liang Liu, Xuhua Xia, Matt Fujita, Craig Lowe, Ed Braun, Robb Brumfield, and an anonymous reviewer for helpful discussion and comments on the article. This research was generously supported by Harvard University FAS Research Computing and the Odyssey Cluster.
References
- Baker A.J.,, Haddrath O.,, McPherson J.D.,, Cloutier A. 2014. Genomic support for a moa-tinamou clade and adaptive morphological convergence in flightless ratites. Mol. Biol. Evol. 31:1686–1696. [DOI] [PubMed] [Google Scholar]
- Bejerano G.,, Pheasant M.,, Makunin I.,, Stephen S.,, Kent W.J.,, Mattick J.S.,, Haussler D. 2014. Ultraconserved elements in the human genome. Science 304:1321–1325. [DOI] [PubMed] [Google Scholar]
- Bi K.,, Vanderpool D.,, Singhal S.,, Linderoth T.,, Moritz C.,, Good J.M. 2014. Transcriptome-based exon capture enables highly cost-effective comparative genomic data collection at moderate evolutionary scales. BMC Genom. 13:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaimer B.B.,, Lloyd M.W.,, Guillory W.X.,, Brady S.G. 2014. Sequence capture and phylogenetic utility of genomic ultraconserved elements obtained from pinned insect specimens. PLoS One 11:e0161531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M.,, Kent W.J.,, Riemer C.,, Elnitski L.,, Smit A.F.A.,, Roskin K.M.,, Baertsch R.,, Rosenbloom K.,, Clawson H.,, Green E.D.,, Haussler D.,, Miller W. 2014. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14:708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom M.P.K.,, Bragg J.G.,, Potter S.,, Moritz C. 2014. Accounting foruncertainty in gene tree estimation: summary-coalescent species treeinference in a challenging radiation of Australian lizards. Syst.Biol. 66:352–366. [DOI] [PubMed] [Google Scholar]
- Bogdanowicz D.,, Giaro K. 2014. Matching split distance for unrooted binary phylogenetic trees. IEEE/ACM Trans. Comput. Biol. Bioinform. 9:150–160. [DOI] [PubMed] [Google Scholar]
- Borowiec M.L. 2014. AMAS: a fast tool for alignment manipulation and computing of summary statistics. Peer J. 4:e1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbrink F.T.,, Pyron R.A. 2014. The impact of gene-tree/species-tree discordance on diversification-rate estimation. Evolution 65:1851–1861. [DOI] [PubMed] [Google Scholar]
- Burgess D.,, Freeling M. 2014. The most deeply conserved noncoding sequences in plants serve similar functions to those in vertebrates despite large differences in evolutionary rates. Plant Cell 26:946–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S.,, Silla-Martínez J.M.,, Gabaldón T. 2014. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary J.V.,, Parmley J.L.,, Hurst L.D. 2014. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 7:98–108. [DOI] [PubMed] [Google Scholar]
- Chen M.Y.,, Liang D.,, Zhang P. 2014. Selecting question-specific genes to reduce incongruence in phylogenomics: a case study of jawed vertebrate backbone phylogeny. Syst. Biol. 64:1104–1120. [DOI] [PubMed] [Google Scholar]
- Chojnowski J.L.,, Kimball R.T.,, Braun E.L. 2014. Introns outperform exons in analyses of basal avian phylogeny using clathrin heavy chain genes. Gene 410:89–96. [DOI] [PubMed] [Google Scholar]
- Costa I.R.,, Prosdocimi F.,, Jennings W.B. 2014. In silico phylogenomics using complete genomes: a case study on the evolution of hominoids. Genome Res. 26:1257–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins C.A.,, McInerney J.O. 2014. A method for inferring the rate of evolution of homologous characters that can potentially improve phylogenetic inference, resolve deep divergence and correct systematic biases. Syst. Biol. 60:833–844. [DOI] [PubMed] [Google Scholar]
- Darriba D.,, Taboada G.L.,, Doallo R.,, Posada D. 2014. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9:772–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F.,, Brinkmann H.,, Philippe H. 2014. Phylogenomics and the reconstruction of the tree of life. Nat. Rev. Genet. 6:361–375. [DOI] [PubMed] [Google Scholar]
- Edwards S.V. 2014. Is a new and general theory of molecular systematics emerging? Evolution 63:1–19. [DOI] [PubMed] [Google Scholar]
- Edwards S.V. 2014. Phylogenomic subsampling: a brief review. Zoolog. Scripta 45:63–74. [Google Scholar]
- Edwards S.V.,, Potter S.,, Schmitt C.J.,, Bragg J.G.,, Moritz C. 2014. Reticulation, divergence, and the phylogeography–phylogenetics continuum. Proc. Natl. Acad. Sci. U.S.A. 113:8025–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J.A. 2014. Phylogenomics: improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 8:163–167. [DOI] [PubMed] [Google Scholar]
- Eisen J.A.,, Kaiser D.,, Myers R.M. 2014. Gastrogenomic delights: a movable feast. Nat. Med. 3:1076–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faircloth B.C., McCormack J.E., Crawford N.G., Harvey M.G.,, Brumfield R.T., Glenn T.C.. 2014. Ultraconserved elements anchorthousands of genetic markers spanning multiple evolutionarytimescales. Syst. Biol. 61:717–726. [DOI] [PubMed] [Google Scholar]
- Farris J.S. 2014. The retention index and the rescaled consistency index. Cladistics 5:417–419. [DOI] [PubMed] [Google Scholar]
- Foster P.G.,, Hickey D.A. 2014. Compositional bias may affect both DNA-based and protein-based phylogenetic reconstructions. J. Mol. Evol. 48:284–290. [DOI] [PubMed] [Google Scholar]
- Green R.E.,, Braun E.L.,, Armstrong J.,, Earl D.,, Nguyen N.,, Hickey G.,, Vandewege M.W.,, St John J.A.,, Capella-Gutierrez S.,, Castoe T.A.,, Kern C.,, Fujita M.K.,, Opazo J.C.,, Jurka J.,, Kojima K.K.,, Caballero J.,, Hubley R.M.,, Smit A.F.,, Platt R.N.,, Lavoie C.A.,, Ramakodi M.P.,, Finger J.W.,, Suh A.,, Isberg S.R.,, Miles L.,, Chong A.Y.,, Jaratlerdsiri W.,, Gongora J.,, Moran C.,, Iriarte A.,, McCormack J.,, Burgess S.C.,, Edwards S.V.,, Lyons E.,, Williams C.,, Breen M.,, Howard J.T.,, Gresham C.R.,, Peterson D.G.,, Schmitz J.,, Pollock D.D.,, Haussler D.,, Triplett E.W.,, Zhang G.,, Irie N.,, Jarvis E.D.,, Brochu C.A.,, Schmidt C.J.,, McCarthy F.M.,, Faircloth B.C.,, Hoffmann F.G.,, Glenn T.C.,, Gabaldoon T.,, Paten B.,, Ray D.A. 2014. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 346:1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groussin M.,, Hobbs J.K.,, Szöllősi G.J.,, Gribaldo S.,, Arcus V.L.,, Gouy M. 2014. Toward more accurate ancestral protein genotype–phenotype reconstructions with the use of species tree-aware gene trees. Mol. Biol. Evol. 32:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M.,, Amit I.,, Garber M.,, French C.,, Lin M.F.,, Feldser D.,, Huarte M.,, Zuk O.,, Carey B.W.,, Cassady J.P.,, Cabili M.N.,, Jaenisch R.,, Mikkelsen T.S.,, Jacks T.,, Hacohen N.,, Bernstein B.E.,, Kellis M.,, Regev A.,, Rinn J. L.,, Lander E.S. 2014. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458:223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett S.J.,, Kimball R.T.,, Reddy S.,, Bowie R.C.K.,, Braun E.L.,, Braun M.J.,, Chojnowski J.L.,, Cox W.A.,, Han K.L.,, Harshman J.,, Huddleston C.J.,, Marks B.D.,, Miglia K.J.,, Moore W.S.,, Sheldon F.H.,, Steadman D.W.,, Witt C.C.,, Yuri T. 2014. A phylogenomic study of birds reveals their evolutionary history. Science 320:1763–1768. [DOI] [PubMed] [Google Scholar]
- Hamilton C.A.,, Lemmon A.R.,, Lemmon E.M.,, Bond J.E. 2014. Expanding anchored hybrid enrichment to resolve both deep and shallow relationships within the spider tree of life. BMC Evol. Biol. 16:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshman J.,, Braun E.L.,, Braun M.J.,, Huddleston C.J.,, Bowie R.C.,, Chojnowski J.L.,, Hackett S.J.,, Han K.L.,, Kimball R.T.,, Marks B.D.,, Miglia K.J.,, Moore W.S.,, Reddy S.,, Sheldon F.H.,, Steadman D.W.,, Steppan S.J.,, Witt C.C.,, Yuri T. 2014. Phylogenomic evidence for multiple losses of flight in ratite birds. Proc. Natl. Acad. Sci. U.S.A. 105:13462–13467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudry A.,, Platts A.E.,, Vello E.,, Hoen D.R.,, Leclercq M.,, Williamson R.J.,, Forczek E.,, Joly-Lopez Z.,, Steffen J.G.,, Hazzouri K.M.,, Dewar K.,, Stinchcombe J.R.,, Schoen D.J.,, Wang X.,, Schmutz J.,, Town C.D.,, Edger P.P.,, Pires J.C.,, Schumaker K.S.,, Jarvis D.E.,, Mandakova T.,, Lysak M.A.,, Van Den Bergh E.,, Schranz M.E.,, Harrison P.M.,, Moses A.M.,, Bureau T.E.,, Wright S.I.,, Blanchette M. 2014. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat. Genet. 45:891–898. [DOI] [PubMed] [Google Scholar]
- Holland B.R.,, Jarvis P.D.,, Sumner J.G. 2014. Low-parameter phylogenetic inference under the general Markov model. Syst. Biol. 62:78–92. [DOI] [PubMed] [Google Scholar]
- Hosner P.A.,, Faircloth B.C.,, Glenn T.C.,, Braun E.L.,, Kimball R.T. 2014. Avoiding missing data biases in phylogenomic inference: an empirical study in the landfowl (Aves: Galliformes). Mol. Biol. Evol. 33:1110–1125. [DOI] [PubMed] [Google Scholar]
- Jarvis E.D., Mirarab S., Aberer A.J., Li B., Houde P., Li C., Ho S.Y., Faircloth B.C., Nabholz B., Howard J.T., Suh A., Weber C.C., Da Fonseca R.R., Alfaro-Nunez A., Narula N., Liu L., Burt D., Ellegren H., Edwards S.V., Stamatakis A., Mindell D.P., Cracraft J., Braun E.L., Warnow T., Jun W., Gilbert M.T., Zhang G.; Avian Phylogenomics C.. 2014. Phylogenomic analyses data of the avian phylogenomics project. Gigascience 4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis E.D.,, Mirarab S.,, Aberer A.J.,, Li B.,, Houde P.,, Li C.,, Ho S.Y.W.,, Faircloth B.C.,, Nabholz B.,, Howard J.T.,, Suh A.,, Weber C.C.,, Da Fonseca R.R.,, Li J.W.,, Zhang F.,, Li H.,, Zhou L.,, Narula N.,, Liu L.,, Ganapathy G.,, Boussau B.,, Bayzid M.S.,, Zavidovych V.,, Subramanian S.,, Gabaldon T.,, Capella-Gutierrez S.,, Huerta-Cepas J.,, Rekepalli B.,, Munch K.,, Schierup M.,, Lindow B.,, Warren W.C.,, Ray D.,, Green R.E.,, Bruford M.W.,, Zhan X.J.,, Dixon A.,, Li S. B.,, Li N.,, Huang Y.H.,, Derryberry E.P.,, Bertelsen M.F.,, Sheldon F.H.,, Brumfield R.T.,, Mello C.V.,, Lovell P.V.,, Wirthlin M.,, Schneider M.P.C.,, Prosdocimi F.,, Samaniego J.A.,, Velazquez A.M.V.,, Alfaro-Nunez A.,, Campos P.F.,, Petersen B.,, Sicheritz-Ponten T.,, Pas A.,, Bailey T.,, Scofield P.,, Bunce M.,, Lambert D.M.,, Zhou Q.,, Perelman P.,, Driskell A.C.,, Shapiro B.,, Xiong Z.J.,, Zeng Y.L.,, Liu S.P.,, Li Z.Y.,, Liu B.H.,, Wu K.,, Xiao J.,, Yinqi X.,, Zheng Q.M.,, Zhang Y.,, Yang H.M.,, Wang J.,, Smeds L.,, Rheindt F.E.,, Braun M.,, Fjeldsa J.,, Orlando L.,, Barker F.K.,, Jonsson K.A.,, Johnson W.,, Koepfli K.P.,, O’brien S.,, Haussler D.,, Ryder O.A.,, Rahbek C.,, Willerslev E.,, Graves G.R.,, Glenn T.C.,, McCormack J.,, Burt D.,, Ellegren H.,, Alstrom P.,, Edwards S.V.,, Stamatakis A.,, Mindell D.P.,, Cracraft J.,, Braun E.L.,, Warnow T.,, Jun W.,, Gilbert M.T.P.,, Zhang G.J. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346:1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, Z.,, Song R.,, Regev A.,, Struhl K. 2014. Many lncRNAs, 5’UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife 4:e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K.,, Standley D.M. 2014. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J.,, Baertsch R.,, Hinrichs A.,, Miller W.,, Haussler D. 2014. Evolution’s cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. U.S.A. 100:11484–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluge A.G. 2014. A concern for evidence and a phylogenetic hypothesis of relationships among epicrates (Boidae, Serpentes). Syst. Zool. 38:7–25. [Google Scholar]
- Kritsas K.,, Wuest S.E.,, Hupalo D.,, Kern A.D.,, Wicker T.,, Grossniklaus U. 2014. Computational analysis and characterization of UCE-like elements (ULEs) in plant genomes. Genome Res. 22:2455–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon E.Z.,, Kamneva O.K.,, Melo S.,, Dickel D.E.,, Melo S.,, Barozzi I.,, Osterwalder M.,, Mannion B.J.,, Kvon E.Z.,, Kamneva O.K.,, Pickle C.S.,, Plajzer-Frick I.,, Lee E.A.,, Kato M.,, Garvin T.H.,, Akiyama J.A.,, Afzal V.,, Lopez-Rios J.,, Rubin E.M.,, Dickel D.E.,, Pennacchio L.A. 2014. Progressive loss of function in a limb enhancer during snake evolution article. Cell 167:633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal F.,, Cohn M.J. 2014. Loss and re-emergence of legs in snakes bymodular evolution of sonic hedgehog and HOXD enhancers. Curr. Biol. 26:2966–2973. [DOI] [PubMed] [Google Scholar]
- Lee A.P.,, Kerk S.Y.,, Tan Y.Y.,, Brenner S.,, Venkatesh B. 2014. Ancient vertebrate conserved noncoding elements have been evolving rapidly in teleost fishes. Mol. Biol. Evol. 28:1205–1215. [DOI] [PubMed] [Google Scholar]
- Lemmon A.R., Emme S.A., and Lemmon E.M.. 2014. Anchored hybrid enrichment for massively high-throughput phylogenomics. Syst. Biol. 61:727–744. [DOI] [PubMed] [Google Scholar]
- Lemmon E.M.,, Lemmon A.R. 2014. High-throughput genomic data in systematics and phylogenetics. Ann. Rev. Ecol. Evol. Syst. 44:99–121. [Google Scholar]
- Lindblad-Toh K.,, Garber M.,, Zuk O.,, Lin M.F.,, Parker B.J.,, Washietl S.,, Kheradpour P.,, Ernst J.,, Jordan G.,, Mauceli E.,, Ward L.D.,, Lowe C.B.,, Holloway A.K.,, Clamp M.,, Gnerre S.,, Alfoldi J.,, Beal K.,, Chang J.,, Clawson H.,, Cuff J.,, Di Palma F.,, Fitzgerald S.,, Flicek P.,, Guttman M.,, Hubisz M.J.,, Jaffe D.B.,, Jungreis I.,, Kent W.J.,, Kostka D.,, Lara M.,, Martins A.L.,, Massingham T.,, Moltke I.,, Raney B.J.,, Rasmussen M.D.,, Robinson J.,, Stark A.,, Vilella A.J.,, Wen J.Y.,, Xie X.H.,, Zody M.C.,, Worley K.C.,, Kovar C.L.,, Muzny D.M.,, Gibbs R.A.,, Warren W.C.,, Mardis E.R.,, Weinstock G.M.,, Wilson R.K.,, Birney E.,, Margulies E.H.,, Herrero J.,, Green E.D.,, Haussler D.,, Siepel A.,, Goldman N.,, Pollard K. S.,, Pedersen J.S.,, Lander E.S.,, Kellis M.,, Inst B.,, Med B.C.,, Univ W. 2014. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478:476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.,, Edwards S.V. 2014. Comment on “Statistical binning enables an accurate coalescent-based estimation of the avian tree. Science 350:171. [DOI] [PubMed] [Google Scholar]
- Liu L.,, Yu L.,, Edwards S. 2014. A maximum pseudo-likelihood approach for estimating species trees under the coalescent model. BMC Evol. Biol. 10:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Xi Z., Wu S., Davis C. C., and Edwards S. V.. 2015. Estimating phylogenetic trees from genome-scale data. Ann. N.Y.Acad. Sci. 1360:36–53. [DOI] [PubMed] [Google Scholar]
- Lockhart P.J.,, Steel M.A.,, Hendy M.D.,, Penny D. 2014. Recovering evolutionary trees under a more realistic model of sequence evolution. Mol. Biol. Evol. 11:605–612. [DOI] [PubMed] [Google Scholar]
- Lowe C.B.,, Clarke J.A.,, Baker A.J.,, Haussler D.,, Edwards S.V. 2014. Feather development genes and associated regulatory innovation predate the origin of Dinosauria. Mol. Biol. Evol. 32:23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe C.B.,, Kellis M.,, Siepel A.,, Raney B.J.,, Clamp, Michele,, Salama S.R.,, Kingsley D.M.,, Lindblad-Toh K.,, Haussler D. 2014. Three periods of regulatory innovation during vertebrate evolution. Science 333:1019–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcovitz A.,, Jia R.,, Bejerano G. 2014. “Reverse Genomics” predicts function of human conserved noncoding elements. Mol. Biol. Evol. 33:1358–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall D.C.,, Simon C.,, Buckley T.R. 2014. Accurate branch length estimation in partitioned Bayesian analyses requires accommodation of among-partition rate variation and attention to branch length priors. Syst. Biol. 55:993–1003. [DOI] [PubMed] [Google Scholar]
- McCormack J.E.,, Faircloth B.C.,, Crawford N.G.,, Gowaty P.A.,, Brumfield R.T.,, Glenn T.C. 2014. Ultraconserved elements are novel phylogenomic markers that resolve placental mammal phylogeny when combined with species tree analysis. Genome Res. 22:746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack J.E.,, Hird S.M.,, Zellmer A.J.,, Carstens B.C.,, Brumfield R.T. 2014. Applications of next-generation sequencing to phylogeography and phylogenetics. Mol. Phylogenet. Evol. 66:526–538. [DOI] [PubMed] [Google Scholar]
- McCormack J.E.,, Tsai W.L.,, Faircloth B.C. 2014. Sequence capture of ultraconserved elements from bird museum specimens. Mol. Ecol. Resour. 16:1189–1203. [DOI] [PubMed] [Google Scholar]
- Meiklejohn K.A.,, Faircloth B.C.,, Glenn T.C.,, Kimball R.T.,, Braun E.L. 2014. Analysis of a rapid evolutionary radiation using ultraconserved elements: evidence for a bias in some multispecies coalescent methods. Syst. Biol. 65:612–627. [DOI] [PubMed] [Google Scholar]
- Mendes F.K.,, Hahn M.W. 2014. Gene tree discordance causes apparent substitution rate variation. Syst. Biol. 65:711–721. [DOI] [PubMed] [Google Scholar]
- Meunier J.,, Duret L. 2014. Recombination drives the evolution of GC-content in the human genome. Mol. Biol. Evol. 21:984–990. [DOI] [PubMed] [Google Scholar]
- Mirarab S.,, Bayzid M.S.,, Boussau B.,, Warnow T. 2014. Statistical binning enables an accurate coalescent-based estimation of the avian tree. Science 1250463. [DOI] [PubMed] [Google Scholar]
- Mooers A.O.,, Holmes E.C. 2014. The evolution of base composition and phylogenetic inference. Trends Ecol. Evol. 15:365–369. [DOI] [PubMed] [Google Scholar]
- Novick P.A.,, Basta H.,, Floumanhaft M.,, McClure M.A.,, Boissinot S. 2014. The Evolutionary dynamics of autonomous non-LTR retrotransposons in the lizard Anolis carolinensis shows more similarity to fish than mammals. Mol. Biol. Evol. 26:1811–1822. [DOI] [PubMed] [Google Scholar]
- Pease J.B.,, Haak D.C.,, Hahn M.W.,, Moyle L.C. 2014. Phylogenomicsreveals three sources of adaptive variation during a rapidradiation. PloS Biol. 14: e1002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillippe H.,, Brinkmann H.,, Lavrov D.V.,, Littlewood D.T.J.,, Manuel M.,, Worheide G.,, Baurain D. 2014. Resolving difficult phylogenetic questions: why more sequences are not enough. Plos Biol. 9:e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M.J.,, Gibb G.C.,, Crimp E.A.,, Penny D. 2014. Tinamous and moa flock together: mitochondrial genome sequence analysis reveals independent losses of flight among ratites. Syst. Biol. 59:90–107. [DOI] [PubMed] [Google Scholar]
- Pisani D.,, Pett W.,, Dohrmann M.,, Feuda R.,, Rota-Stabelli O.,, Philippe H.,, Lartillot N.,, Worheide G. 2014. Genomic data do not support comb jellies as the sister group to all other animals. Proc. Natl. Acad. Sci. U.S.A. 112:15402–15407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. 2014. Phylogenomics for systematic biology. Syst. Biol. 65:353–356. [DOI] [PubMed] [Google Scholar]
- Potter S.,, Bragg J.,, Peter B.M.,, Bi K.,, Moritz C. 2014. Phylogenomics at the tips: inferring lineages and their demographic history in a tropical lizard, Carlia amax. Mol. Ecol. 25:1367–1380. [DOI] [PubMed] [Google Scholar]
- Pozzoli U.,, Menozzi G.,, Comi G.P.,, Cagliani R.,, Bresolin N.,, Sironi M. 2014. Intron size in mammals: complexity comes to terms with economy. Trends Genet. 23:20–24. [DOI] [PubMed] [Google Scholar]
- Prum R.O.,, Berv J.S.,, Dornburg A.,, Field D.J.,, Townsend J.P.,, Lemmon E.M.,, Lemmon A.R. 2014. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 526:569–573. [DOI] [PubMed] [Google Scholar]
- Reddy S., Kimball R.T., Pandey A., Hosner P.A., Braun M.J., Hackett S.J., Han K.-L., Harshman J., Huddleston C.J., Kingston S., Marks B.D., Miglia K.J., Moore W.S., Sheldon F.H., Witt C.C., Yuri T.,, Braun E.L.. 2014. Why do phylogenomic data sets yield conflictingtrees? Data type influences the avian tree of life more than taxonsampling. Syst. Biol. 10.1093/sysbio/syx041. [DOI] [PubMed] [Google Scholar]
- Reneker J.,, Lyons E.,, Conant G.C.,, Pires J.C.,, Freeling M.,, Shyu C.R.,, Korkin D. 2014. Long identical multispecies elements in plant and animal genomes. Proc. Natl. Acad. Sci. U.S.A. 109:E1183–E1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romiguier J.,, Ranwez V.,, Delsuc F.,, Galtier N.,, Douzery E.J.P. 2014. Less is more in mammalian phylogenomics: AT-Rich genes minimize tree conflicts and unravel the root of placental mammals. Mol. Biol. Evol. 30:2134–2144. [DOI] [PubMed] [Google Scholar]
- Ryu T.,, Seridi L.,, Ravasi T. 2014. The evolution of ultraconserved elements with different phylogenetic origins. BMC Evol. Biol. 12:236–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S.,, Kent W.J.,, Smit A.,, Zhang Z.,, Baertsch R.,, Hardison R.C.,, Haussler D.,, Miller W. 2014. Human-mouse alignments with BLASTZ. Genome Res. 13:103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki R.,, Li C.,, Fang Q.,, Hayashi S.,, Egawa S.,, Hu J.,, Xu L.,, Pan H.,, Kondo M.,, Sato T.,, Matsubara H.,, Kamiyama N.,, Kitajima K.,, Saito D.,, Liu Y.,, Gilbert M.T.,, Zhou Q.,, Xu X.,, Shiroishi T.,, Irie N.,, Tamura K.,, Zhang G. 2014. Functional roles of Aves class-specific cis-regulatory elements on macroevolution of bird-specific features. Nat. Commun. 8:14229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A.,, Bejerano G.,, Pedersen J.S.,, Hinrichs A.S.,, Hou M.,, Rosenbloom K.,, Clawson H.,, Spieth J.,, Hillier L.W.,, Richards S.,, Weinstock G.M.,, Wilson R.K.,, Gibbs R.A.,, Kent W.J.,, Miller W.,, Haussler D. 2014. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15:1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A.,, Haussler D. 2014. Phylogenetic estimation of context-dependent substitution rates by maximum likelihood. Mol. Biol. Evol. 21:468–488. [DOI] [PubMed] [Google Scholar]
- Smith J.V.,, Braun E.L.,, Kimball R.T. 2014. Ratite nonmonophyly: independent evidence from 40 novel Loci. Syst. Biol. 62:35–49. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen S.,, Pheasant M.,, Makunin I.V.,, Mattick J.S. 2014. Large-scale appearance of ultraconserved elements in tetrapod genomes and slowdown of the molecular clock. Mol. Biol. Evol. 25:402–408. [DOI] [PubMed] [Google Scholar]
- Sukumaran J.,, Holder M. 2014. DendroPy: a Python library for phylogenetic computing. Bioinformatics 26:1569–1571. [DOI] [PubMed] [Google Scholar]
- Sukumaran J.,, Holder M. 2014. SumTrees: Phylogenetic TreeSummarization. 4.0.0 (January312015). Available athttps://github.com/jeetsukumaran/DendroPy.
- Sun K., Meiklejohn K.A., Faircloth B.C., Glenn T.C., Braun E.L.,, Kimball R.T.. 2014. The evolution of peafowl and other taxa withocelli (eyespots): a phylogenomic approach. Proc. Biol. Sci. 281:20140823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D.L. 2014. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Tan G.,, Muffato M.,, Ledergerber C.,, Herrero J.,, Goldman N.,, Gil M.,, Dessimoz C. 2014. Current methods for automated filtering of multiple sequence alignments frequently worsen single-gene phylogenetic inference. Syst. Biol. 64:778–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D.,, Flicek P.,, Odom D.T. 2014. Evolution of transcription factor binding in metazoans [mdash] mechanisms and functional implications. Nat. Rev. Genet. 15:221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov A.E. 2014. Growth and decline of introns. Trends Genet. 18:232–236. [DOI] [PubMed] [Google Scholar]
- Vogler A.P.,, Cardoso A.,, Barraclough T.G. 2014. Exploring rate variation among and within sites in a densely sampled tree: species level phylogenetics of North American tiger beetles (Genus Cicindela). Syst. Biol. 54:4–20. [DOI] [PubMed] [Google Scholar]
- Waltari E.,, Edwards S.V. 2014. Evolutionary dynamics of intron size, genome size, and physiological correlates in archosaurs. Am. Nat. 160:539–552. [DOI] [PubMed] [Google Scholar]
- Weber C.C.,, Boussau B.,, Romiguier J.,, Jarvis E.D.,, Ellegren H. 2014. Evidence for GC-biased gene conversion as a driver of between-lineage differences in avian base composition. Genome Biol. 15:549–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z.,, Liu L.,, Davis C.C. 2014. Genes with minimal phylogenetic information are problematic for coalescent analyses when gene tree estimation is biased. Mol. Phylogenet. Evol. 92:63–71. [DOI] [PubMed] [Google Scholar]
- Yuri T.,, Kimball R.T.,, Harshman J.,, Bowie R.C.,, Braun M.J.,, Chojnowski J.L.,, Han K.L.,, Hackett S.J.,, Huddleston C.J.,, Moore W.S.,, Reddy S.,, Sheldon F.H.,, Steadman D.W.,, Witt C.C.,, Braun E.L. 2014. Parsimony and model-based analyses of indels in avian nuclear genes reveal congruent and incongruent phylogenetic signals. Biology (Basel) 2:419–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.J.,, Li C.,, Li Q.Y.,, Li B.,, Larkin D.M.,, Lee C.,, Storz J.F.,, Antunes A.,, Greenwold M.J.,, Meredith R.W.,, Odeen A.,, Cui J.,, Zhou Q.,, Xu L.H.,, Pan H.L.,, Wang Z.J.,, Jin L.J.,, Zhang P.,, Hu H.F.,, Yang W.,, Hu J.,, Xiao J.,, Yang Z.K.,, Liu Y.,, Xie Q.L.,, Yu H.,, Lian J.M.,, Wen P.,, Zhang F.,, Li H.,, Zeng Y.L.,, Xiong Z.J.,, Liu S.P.,, Zhou L.,, Huang Z.Y.,, An N.,, Wang J.,, Zheng Q.M.,, Xiong Y.Q.,, Wang G.B.,, Wang B.,, Wang J.J.,, Fan Y.,, Da Fonseca R.R.,, Alfaro-Nunez A.,, Schubert M.,, Orlando L.,, Mourier T.,, Howard J.T.,, Ganapathy G.,, Pfenning A.,, Whitney O.,, Rivas M.V.,, Hara E.,, Smith J.,, Farre M.,, Narayan J.,, Slavov G.,, Romanov M.N.,, Borges R.,, Machado J.P.,, Khan I.,, Springer M.S.,, Gatesy J.,, Hoffmann F.G.,, Opazo J.C.,, Hastad O.,, Sawyer R.H.,, Kim H.,, Kim K.W.,, Kim H.J.,, Cho S.,, Li N.,, Huang Y.H.,, Bruford M.W.,, Zhan X.J.,, Dixon A.,, Bertelsen M.F.,, Derryberry E.,, Warren W.,, Wilson R.K.,, Li S.B.,, Ray D.A.,, Green R.E.,, O’brien S.J.,, Griffin D.,, Johnson W.E.,, Haussler D.,, Ryder O.A.,, Willerslev E.,, Graves G.R.,, Alstrom P.,, Fjeldsa J.,, Mindell D.P.,, Edwards S.V.,, Braun E.L.,, Rahbek C.,, Burt D.W.,, Houde P.,, Zhang Y.,, Yang H.M.,, Wang J.,, Jarvis E.D.,, Gilbert M.T.P.,, Wang J.,, Consortium A.G. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346:1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.,, Edwards S.V. 2014. The evolution of intron size in amniotes: a role for powered flight? Genome Biol. Evol. 4:1033–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]