Abstract
Pancreatic ductal adenocarcinoma (PDAC) remains a very challenging malignancy with late presentation, metastatic potential, chemoresistance, and poor prognosis. Therefore, there is an urgent need for novel diagnostic and prognostic biomarkers. miRNAs are small noncoding RNAs that regulate the expression of multitude number of genes. Aberrant expression of miRNAs has been linked to the development of various malignancies, including PDAC. A series of miRNAs have been defined as holding promise for early diagnostics, as indicators of therapy resistance, and even as markers for prognosis in PDAC patients. In this review, we summarize the current knowledge on the role of miRNAs in diagnosis, chemoresistance, and prognosis in PDAC patients.
Keywords: pancreatic ductal adenocarcinoma, miRNA, diagnosis, prognosis, chemoresistance
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of death by cancer in the USA, with 53,670 new cases expected in 2017, a number that has been steadily increasing for more than a decade.1 Due to a late diagnosis and the lack of effective treatment measures, PDAC has an extremely poor prognosis and its 5-year survival rate does not exceed 5%.2 Numerous studies have indicated that among those patients who were found occasionally through imaging modalities to have early-stage carcinoma, the improved 5-year survival rate is 30% for those with a 2 cm carcinoma, 57% for those with a 1 cm carcinoma, and 100% for patients with a ductal epithelium tumor measuring <1 cm.3 Thus, in light of the disappointing statistics in the prognosis of PDAC, there is an urgent, unmet need for development of valid, reliable biomarkers for early detection and monitoring of PDAC. The current most widely used biomarkers for PDAC are carbohydrate antigen 19-9 (CA19-9),4 carcinoembryonic antigen,5 and/or genetic markers such as K-RAS and p53.6 However, these markers are neither sensitive nor specific for screening, but are used to follow known disease if they were initially elevated, and are not recommended for screening and diagnosis of early disease.7
miRNAs are a class of short, non-coding RNAs ranging approximately from 17 to 25 nucleotides, which contain a seed sequence for binding to imperfect complementary regions in the 3′ untranslated region of the target mRNAs, inhibiting their translation or leading to their degradation.8,9 Collective evidences have demonstrated that miRNAs have a critical regulatory role in the development, differentiation, and apoptosis of normal cells, as well as in the expression of many target mRNAs simultaneously, playing a critical role in tumorigenesis, invasion, metastasis, and chemoresistance of cancer cells.10 Indeed, miRNAs exhibit tissue-specific and disease-specific expression that could provide the basis for their development as novel diagnostic, prognostic, and/or therapeutic targets, as well as chemoresistance.11,12
Since abnormal expression of miR-15 and miR-16 was reported in chronic lymphocytic leukemia,13 more and more evidences have shown that miRNA mutations or misexpression correlates with various human cancers and indicate that miRNAs can act as either tumor suppressors or oncogenes.14 Remarkably, miRNAs have been profiled in many different malignancies including breast,15 lung,16 and PDAC,17,18 and differential expression was detected with these malignancies, all of which have made miRNAs promising biomarkers. The aim of this review is to present the evidence on the utility of miRNAs in the diagnosis, chemoresistance, and prognosis of PDAC.
Role of miRNAs in the diagnosis of PDAC
miRNAs in tissues and fluids have several exceptionally appealing characteristics – they are stable as they are resistant to cleavage by ribonucleases and survive extreme pH and temperature conditions, their isolation is noninvasive, and their amplification is technically easy and inexpensive.13,19 Accumulating evidence is showing that altered levels of miRNAs in tissues, blood, and body fluids can distinguish patients with cancer from healthy individuals.20
Many researches are engaged in analysis of aberrant expression of miRNAs in normal pancreatic tissue and PDAC. For the first time in 2007, Bloomston et al showed that PDAC may have a distinct miRNA expression pattern that may differentiate it from normal pancreas (NP) and chronic pancreatitis (CP).21 Particularly, the results revealed that 21 miRNAs with increased expression and 4 with decreased expression were identified that correctly differentiated PDAC from NP in 90% of samples by cross-validation. Moreover, 15 overexpressed and 8 underexpressed miRNAs differentiated PDAC from CP with 93% accuracy.21 Similarly, Schultz et al used a diagnostic classifier including 19 miRNAs to discriminate pancreatic and ampullary adenocarcinomas from CP and NP with a sensitivity of 98.5% and a positive predictive value of 97.8% with an accuracy of 97.0%.22 For the single miRNA as a diagnostic biomarker, miR-21 has been considered a very promising biomarker for the reason that overexpression of miR-21 was persistently observed in PDAC compared to healthy tissues and/or tissues of CP in several studies.23–25 Zhang et al recently demonstrated that miR-132 was downregulated in PDAC compared to their respective benign tissues by TaqMan miRNA assays.26 Another diagnostic biomarker is miR-96, which is downregulated in pancreatic cancer as compared to normal tissues.27
With the lack of reliable approaches based on imaging techniques and routine tumor markers, the detection of miRNAs in peripheral body fluids, especially blood or serum, has currently a considerable potential for use in clinical practice.28 Compared to traditionally used markers, serum miR-1290 distinguished patients with low-stage PDAC from controls better than CA19-9 did.29 Moreover, blood samples collected from pancreatic cancer patients had higher expression levels of miR-192 with sensitivity toward cancer at 76% and specificity at 55%.30 Another potential biomarker is miR-155, which occurs at high levels in the plasma of 80% of early pancreatic lesions (stage II) in microdissected pancreatic intraepithelial neoplasias tissues.31 In addition, circulating miR-18a in the plasma of 36 PDAC patients was found to be significantly increased when compared to 30 healthy volunteers.32
Other studies are focused on using combination of several circulating miRNAs to increase the diagnostic accuracy rate. It has been reported that the combination of miR-196a and miR-217 expression patterns differentiated PDAC from healthy controls and CP cases.33 Moreover, plasma levels of miR-16 and miR-196a in combination with CA 19-9 have been shown to work very efficiently for improving the prognostic prediction of early PDAC.5 Furthermore, another group of researchers observed much higher levels of circulating miR-200a, 200b, and 210 in the plasma of PDAC patients.34
The main miRNAs isolated from the histologic samples, serum or plasma, stool, and pancreatic juices with a potential role in diagnosis of pancreatic cancer are shown in Table 1. These reports indicate the importance of miRNAs as potential biomarkers for the diagnosis of pancreatic cancer.
Table 1.
Selected miRNA candidates which are correlated to diagnosis in PDAC
| miRNA | Histologic samples | Serum or plasma | Stool | Pancreatic juice |
|---|---|---|---|---|
| miR-10b | ↑35 | ↑36 | ||
| miR-16 | ↑37 | |||
| miR-18a | ↑32,38 | |||
| miR-20a | ↑39 | |||
| miR-21 | ↑23–25 | ↑39,40 | ↑25 | |
| miR-24 | ↑39 | |||
| miR-25 | ↑39 | |||
| miR-27a-3p | ↑41 | |||
| miR-29c | ↓42 | |||
| miR-30a-3p | ||||
| miR-30c | ||||
| miR-31 | ||||
| miR-34a | ↑23 ↓43 | ↓43 | ||
| miR-96 | ↓27 | |||
| miR-99a | ↑39 | |||
| miR-101 | ||||
| miR-103 | ↑44 | |||
| miR-106b | ||||
| miR-107 | ↑44 | |||
| miR-130b | ||||
| miR-132 | ↑45 ↓26 | |||
| miR-135b | ||||
| miR-139-3p | ||||
| miR-141 | ↓46 | |||
| miR-143 | ↑46 | ↓41 | ||
| miR-145 | ↑46 | |||
| miR-146a | ↑46 | |||
| miR-148a | ↓46,47 | |||
| miR-148b | ↓46 | |||
| miR-150 | ↓43 | ↓43 | ||
| miR-155 | ↑24,25 | ↑40 | ↓41 | ↑25 |
| miR-181a | ↑21 | |||
| miR-181b | ↑21 | ↑48 | ||
| miR-181d | ↑21 | |||
| miR-185 | ↑39 | |||
| miR-191 | ↑39 | |||
| miR-192 | ↑30,49 | |||
| miR-194 | ↑49 | |||
| miR-196a | ↑46 | ↑37,40,50 | ↓41 | |
| miR-196b | ↑51 | ↑50 | ||
| miR-200a | ↑52 | |||
| miR-200b | ↑52 | |||
| miR-203 | ↑53 | ↑54 | ||
| miR-210 | ↑22 | ↑40,55 | ↑48 | |
| miR-212 | ↑56 | |||
| miR-216 | ↓46 | |||
| miR-216a | ↓41 | |||
| miR-217 | ↓23 ↓46 | |||
| miR-222 | ↑21,46,56 | |||
| miR-223 | ↑46 | |||
| miR-369-5p | ↑54 | |||
| miR-373 | ↓57 | |||
| miR-375 | ↓54 | |||
| miR-376a | ↑54 | |||
| miR-492 | ↓58 | |||
| miR-494 | ↓46 | |||
| miR-663a | ↓58 | |||
| miR-1246 | ↑59 | |||
| miR-1290 | ↑29 | |||
| miR-3976 | ↑59 | |||
| miR-4306 | ↑59 | |||
| miR-4644 | ↑59 |
Notes: ↑, upregulated; ↓, downregulated.
Abbreviation: PDAC, pancreatic ductal adenocarcinoma.
Role of miRNAs in the chemoresistance of PDAC
Chemotherapy (eg, gemcitabine) represents an important therapeutic strategy for most patients with PDAC.60 However, it has been shown that the limited response to current chemotherapy results in an exceptionally poor prognosis.61 Despite investigations into the mechanisms underlying chemoresistance over the past 50 years, the exact mechanism of this phenomenon is still unknown. It has been suggested cancer chemoresistance can arise from physiological barriers to drug absorption or penetration into the target tissues or from biologic mechanisms within individual tumor cells which reduce the effectiveness at their intended site of action, such as increased expression of enzymes involved in drug catabolism or antiapoptotic proteins.62
Recent studies have indicated that miRNAs appear to be critical regulators of chemoresistance in PDAC cells.63 The levels of the oncogenic miR-155 were shown to increase after pancreatic cancer cells were treated with gemcitabine.64 Moreover, downregulation of miR-200 family expression was observed in gemcitabine-resistant pancreatic cancer cells.65 Furthermore, miR-34 is reported to involve in the self-renewal of pancreatic cancer stem cells, while the loss of miR-34 in pancreatic cancer is associated with an enrichment of cancer stem cells that are insensitive to chemotherapy.66 miRNAs with a putative impact on chemoresistance are shown in Table 2.
Table 2.
miRNA candidates which are correlated to chemoresistance in PDAC
| miRNA | Gemcitabine | Cisplatin | 5-fluorouracil | Docetaxel | Irradiation |
|---|---|---|---|---|---|
| miR-let-7 | ↓70 | ||||
| miR-10b | ↑71 | ||||
| miR-21 | ↑72–76 | ↑72,77 | |||
| miR-29a | ↑78 | ||||
| miR-34 | ↓66 | ↓66 | ↓66 | ↓66 | |
| miR-99b | ↑79 | ||||
| miR-101-3p | ↓80 | ||||
| miR-125b | ↑81 | ||||
| miR-142-5p | ↓82 | ||||
| miR-155 | ↑83,84 | ||||
| miR-181b | ↑85 | ||||
| miR-200a | ↓70 | ||||
| miR-200b | ↓70 | ||||
| miR-204 | ↓82 | ||||
| miR-210 | ↓86 | ||||
| miR-214 | ↑87 | ||||
| miR-221 | ↑72 | ↑72 | |||
| miR-320a | ↑88 | ||||
| miR-320c | ↑89 | ||||
| miR-365 | ↑67 | ||||
| miR-374b | ↓90 | ||||
| miR-1246 | ↑68 |
Notes: ↑, upregulated; ↓, downregulated.
Abbreviation: PDAC, pancreatic ductal adenocarcinoma.
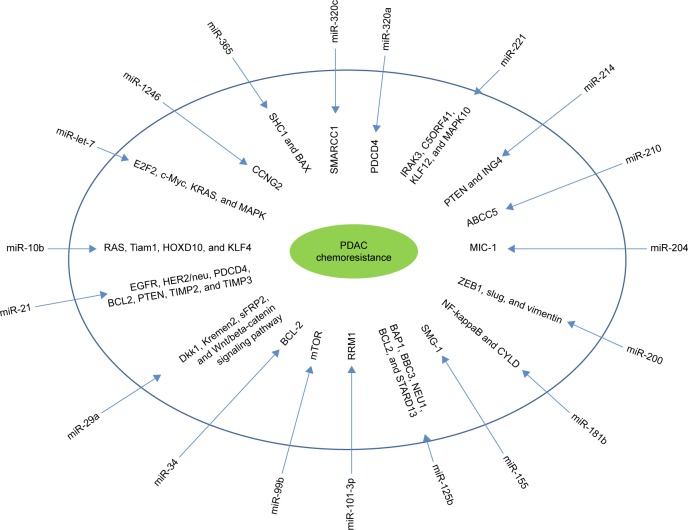
The mechanisms through which miRNAs induce chemoresistance have been clarified in several studies. It has been shown that miR-365 induced chemoresistance through directly targeting the adaptor protein Src Homology 2 Domain Containing 1 and apoptosis-promoting protein BAX.67 Another study showed that miR-1246 expression induced chemoresistance through downregulating CCNG2, a family of cyclins.68 In addition, a recent study also indicates that miRNAs might regulate the epithelial–mesenchymal transition (EMT) through the regulation of cadherin1 and other molecules,69 which mediate various types of cellular drug resistance mechanisms. Many members of the let-7 family are downregulated in EMT-type cells that are resistant to gemcitabine. In an investigation of the expression levels of miR-200 and let-7 in EMT-phenotype pancreatic cancer cells that are resistant to gemcitabine, re-expression of the downregulated miR-200 family upregulates cadherin1 and downregulates ZeB1 and vimentin (EMT inducers).70 Table 3 and Figure 1 show the mechanisms by which miRNAs cause chemoresistance. These results clearly suggest the potential role of miRNAs as novel targets to improve chemoresistance.
Table 3.
The mechanisms through which miRNAs induce chemoresistance
| miRNA | Mechanisms | Reference |
|---|---|---|
| miR-let-7 | E2F2, c-Myc, KRAS, and MAPK | 70 |
| miR-10b | RAS, Tiam1, HOXD10, and KLF4 | 71 |
| miR-21 | EGFR, HER2/neu, PDCD4, BCL2, PTEN, TIMP2, and TIMP3 | 73, 74 |
| miR-29a | Dkk1, Kremen2, sFRP2, and Wnt/beta-catenin signaling pathway | 78 |
| miR-34 | BCL-2 | 66 |
| miR-99b | mTOR | 80 |
| miR-101-3p | RRM1 | 80 |
| miR-125b | BAP1, BBC3, NEU1, BCL2, and STARD13 | 81 |
| miR-155 | SMG-1 | 83 |
| miR-181b | NF-kappaB and CYLD | 85 |
| miR-200 | ZEB1, slug, and vimentin | 70 |
| miR-204 | MIC-1 | 82 |
| miR-210 | ABCC5 | 86 |
| miR-214 | PTEN and ING4 | 87 |
| miR-221 | IRAK3, C5ORF41, KLF12, and MAPK10 | 72 |
| miR-320a | PDCD4 | 88 |
| miR-320c | SMARCC1 | 89 |
| miR-365 | SHC1 and BAX | 67 |
| miR-1246 | CCNG2 | 68 |
Figure 1.
The mechanisms through which miRNAs induce chemoresistance.
Abbreviation: PDAC, pancreatic ductal adenocarcinoma.
Role of miRNAs in the prognosis of PDAC
As one of the most lethal human cancers, PDAC is known for its very poor overall prognosis.91 Thus, finding prognostic markers to assess probable course of the disease prior to treatment is highly desirable. A number of literature reports are devoted to the use of miRNAs as prognostic markers in PDAC.
In particular, many studies have been carried out on RNA extraction of histologic tissue. A meta-analysis involving 1,525 patients has shown that overall and/or disease-free survivals were significantly shorter in patients with high tumoral miR-21.92 Multivariate analyses have confirmed that a low level of miR-218 expression was an independent predictor of poor prognosis in PDAC patients.93 Similarly, low expression of miR-497 was also an independent adverse prognostic factor of PDAC.94 Recently, Zou et al showed that miR-29c expression was significantly lower in the PDAC tissue of 109 patients compared with pair-matched adjacent paracancerous tissues, suggesting that a lower level of miR-29c is associated with a poor prognosis.95
miRNAs as prognostic biomarkers have also been evaluated in the serum or plasma of PDAC patients. It was reported that serum miR-196a expression level had a potential value in predicting the median survival time of PDAC patients (high-level miR-196a, 6.1 months versus low-level miR-196a, 12.00 months; p=0.007), indicating that serum miR-196a could be a potential noninvasive marker for PDAC prognosis.96 More recently, Hua et al demonstrated that serum miR-373 expression was greatly downregulated in a total of 103 PDAC patients who had shorter 5-year overall survival.57 Consistent with the predictive role of miR-21 isolated from the histologic samples in poor prognosis, it has been shown that serum miR-21 levels in PDAC patients were also significantly associated with overall survival.39
The main miRNAs isolated from the histologic samples, serum or plasma, and pancreatic juices with a potential role in poor prognosis of pancreatic cancer are shown in Table 4. These data demonstrate that miRNA-based biomarker can serve as an effective approach for PDAC prognosis.
Table 4.
miRNA candidates which are correlated to poor prognosis in PDAC
| miRNA | Histologic samples | Serum or plasma | Pancreatic juice |
|---|---|---|---|
| miR-let-7g | ↓97 | ||
| miR-7 | ↓98 | ||
| miR-10b | ↑99 | ||
| miR-17-5p | ↑100 | ||
| miR-21 | ↑23,24,101 | ↑39 | |
| miR-29c | ↓95 | ||
| miR-30a-3p | ↓21 | ||
| miR-30d | ↓101 | ||
| miR-31 | ↑102 | ||
| miR-34a | ↓101 | ↓43 | |
| miR-105 | ↓21 | ||
| miR-127 | ↓21 | ||
| miR-130b | ↓103 | ||
| miR-143 | ↓104 | ||
| miR-143 | ↓105 | ||
| miR-148a | ↓97 | ||
| miR-155 | ↑106 | ||
| miR-186 | ↑107 | ||
| miR-187 | ↓21,97 | ||
| miR-196a | ↑96,108 | ||
| miR-196a-2 | ↑21 | ||
| miR-198 | ↑23 | ||
| miR-203 | ↑53,106 | ||
| miR-205 | ↑20 | ||
| miR-210 | ↑106 | ↑20 | |
| miR-212 | ↑97 | ||
| miR-218 | ↓93 | ||
| miR-219 | ↑109 | ||
| miR-222 | ↑106,110 | ||
| miR-223 | ↑111 | ↑111 | |
| miR-326 | ↑107 | ||
| miR-373 | ↓57 | ||
| miR-452 | ↓21 | ||
| miR-492 | ↑20 | ||
| miR-497 | ↓94 | ||
| miR-518a-2 | ↓21 | ||
| miR-675 | ↑97 | ||
| miR-1207-3p | ↑102 | ||
| miR-1247 | ↓112 | ↑20 | |
| miR-1249 | ↑102 | ||
| miR-1274a | ↑102 | ||
| miR-1290 | ↑102 | ||
| miR-1914 | ↑102 | ||
| miR-4281 | ↑102 |
Notes: ↑, upregulated; ↓, downregulated.
Abbreviation: PDAC, pancreatic ductal adenocarcinoma.
Conclusion
Taken together, accumulating evidence supports the view that miRNAs have proven effective for PDAC diagnosis, chemoresistance, and prognosis. Despite the many efforts that have been taken, a practical application to be used in the clinic is still lacking. Additional studies in larger homogeneous populations with validated methodology using the emerging miRNAs as markers within prospective trials, to see if they can aid clinical decision making are needed.
Acknowledgments
This project was supported by the External Science and Technology Cooperation Planning Projects of Anhui Province of China (No. 1604b0602021).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Ganepola GA, Rutledge JR, Suman P, Yiengpruksawan A, Chang DH. Novel blood-based microRNA biomarker panel for early diagnosis of pancreatic cancer. World J Gastrointest Oncol. 2014;6(1):22–33. doi: 10.4251/wjgo.v6.i1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J, Bamlet WR, Oberg AL, et al. Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19-9 blood markers. Sci Transl Med. 2017;9(398):pii:eaah5583. doi: 10.1126/scitranslmed.aah5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou G, Liu X, Wang X, et al. Combination of preoperative CEA and CA19-9 improves prediction outcomes in patients with resectable pancreatic adenocarcinoma: results from a large follow-up cohort. Onco Targets Ther. 2017;10:1199–1206. doi: 10.2147/OTT.S116136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt C. Early detection tools for pancreatic cancer. J Natl Cancer Inst. 2012;104(15):1117–1118. doi: 10.1093/jnci/djs348. [DOI] [PubMed] [Google Scholar]
- 7.Poruk KE, Gay DZ, Brown K, et al. The clinical utility of CA 19-9 in pancreatic adenocarcinoma: diagnostic and prognostic updates. Curr Mol Med. 2013;13(3):340–351. doi: 10.2174/1566524011313030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galasso M, Sandhu SK, Volinia S. MicroRNA expression signatures in solid malignancies. Cancer J. 2012;18(3):238–243. doi: 10.1097/PPO.0b013e318258b5f4. [DOI] [PubMed] [Google Scholar]
- 9.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27(34):5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M, Marin-Muller C, Bharadwaj U, Chow KH, Yao Q, Chen C. MicroRNAs: control and loss of control in human physiology and disease. World J Surg. 2009;33(4):667–684. doi: 10.1007/s00268-008-9836-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12(11):847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brunetti O, Russo A, Scarpa A, et al. MicroRNA in pancreatic adenocarcinoma: predictive/prognostic biomarkers or therapeutic targets? Oncotarget. 2015;6(27):23323–23341. doi: 10.18632/oncotarget.4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25(46):6163–6169. doi: 10.1038/sj.onc.1209909. [DOI] [PubMed] [Google Scholar]
- 15.Zare M, Bastami M, Solali S, Alivand MR. Aberrantly miRNA promoter methylation and EMT-involving miRNAs in breast cancer metastasis: diagnosis and therapeutic implications. J Cell Physiol. 2017 Aug;:3. doi: 10.1002/jcp.26116. Epub. [DOI] [PubMed] [Google Scholar]
- 16.Switlik WZ, Szemraj J. Circulating miRNAs as non-invasive biomarkers for non-small cell lung cancer diagnosis, prognosis and prediction of treatment response. Postepy Hig Med Dosw (Online) 2017;71(0):649–662. doi: 10.5604/01.3001.0010.3845. [DOI] [PubMed] [Google Scholar]
- 17.Bimonte S, Barbieri A, Leongito M, et al. The role of miRNAs in the regulation of pancreatic cancer stem cells. Stem Cells Int. 2016;2016:8352684. doi: 10.1155/2016/8352684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawa Z, Haque I, Ghosh A, Banerjee S, Harris L, Banerjee SK. The miRacle in pancreatic cancer by miRNAs: tiny angels or devils in disease progression. Int J Mol Sci. 2016;17(6):pii:E809. doi: 10.3390/ijms17060809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kishikawa T, Otsuka M, Ohno M, Yoshikawa T, Takata A, Koike K. Circulating RNAs as new biomarkers for detecting pancreatic cancer. World J Gastroenterol. 2015;21(28):8527–8540. doi: 10.3748/wjg.v21.i28.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Raimondo M, Guha S, et al. Circulating microRNAs in pancreatic juice as candidate biomarkers of pancreatic cancer. J Cancer. 2014;5(8):696–705. doi: 10.7150/jca.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297(17):1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 22.Schultz NA, Werner J, Willenbrock H, et al. MicroRNA expression profiles associated with pancreatic adenocarcinoma and ampullary adenocarcinoma. Mod Pathol. 2012;25(12):1609–1622. doi: 10.1038/modpathol.2012.122. [DOI] [PubMed] [Google Scholar]
- 23.Vychytilova-Faltejskova P, Kiss I, Klusova S, et al. MiR-21, miR-34a, miR-198 and miR-217 as diagnostic and prognostic biomarkers for chronic pancreatitis and pancreatic ductal adenocarcinoma. Diagn Pathol. 2015;10:38. doi: 10.1186/s13000-015-0272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caponi S, Funel N, Frampton AE, et al. The good, the bad and the ugly: a tale of miR-101, miR-21 and miR-155 in pancreatic intraductal papillary mucinous neoplasms. Ann Oncol. 2013;24(3):734–741. doi: 10.1093/annonc/mds513. [DOI] [PubMed] [Google Scholar]
- 25.Sadakari Y, Ohtsuka T, Ohuchida K, et al. MicroRNA expression analyses in preoperative pancreatic juice samples of pancreatic ductal adenocarcinoma. JOP. 2010;11(6):587–592. [PubMed] [Google Scholar]
- 26.Zhang S, Hao J, Xie F, et al. Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis. 2011;32(8):1183–1189. doi: 10.1093/carcin/bgr105. [DOI] [PubMed] [Google Scholar]
- 27.Yu S, Lu Z, Liu C, et al. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer Res. 2010;70(14):6015–6025. doi: 10.1158/0008-5472.CAN-09-4531. [DOI] [PubMed] [Google Scholar]
- 28.Halkova T, Cuperkova R, Minarik M, et al. MicroRNAs in pancreatic cancer: involvement in carcinogenesis and potential use for diagnosis and prognosis. Gastroenterol Res Pract. 2015;2015:892903. doi: 10.1155/2015/892903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li A, Yu J, Kim H, et al. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin Cancer Res. 2013;19(13):3600–3610. doi: 10.1158/1078-0432.CCR-12-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao C, Zhang J, Zhang S, et al. Diagnostic and biological significance of microRNA-192 in pancreatic ductal adenocarcinoma. Oncol Rep. 2013;30(1):276–284. doi: 10.3892/or.2013.2420. [DOI] [PubMed] [Google Scholar]
- 31.Ryu JK, Hong SM, Karikari CA, et al. Aberrant MicroRNA-155 expression is an early event in the multistep progression of pancreatic adenocarcinoma. Pancreatology. 2010;10(1):66–73. doi: 10.1159/000231984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morimura R, Komatsu S, Ichikawa D, et al. Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br J Cancer. 2011;105(11):1733–1740. doi: 10.1038/bjc.2011.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park JY, Helm J, Coppola D, et al. MicroRNAs in pancreatic ductal adenocarcinoma. World J Gastroenterol. 2011;17(7):817–827. doi: 10.3748/wjg.v17.i7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56(11):1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Preis M, Gardner TB, Gordon SR, et al. MicroRNA-10b expression correlates with response to neoadjuvant therapy and survival in pancreatic ductal adenocarcinoma. Clin Cancer Res. 2011;17(17):5812–5821. doi: 10.1158/1078-0432.CCR-11-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cote GA, Gore AJ, McElyea SD, et al. A pilot study to develop a diagnostic test for pancreatic ductal adenocarcinoma based on differential expression of select miRNA in plasma and bile. Am J Gastroenterol. 2014;109(12):1942–1952. doi: 10.1038/ajg.2014.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Gao J, Du Y, et al. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int J Cancer. 2012;131(3):683–691. doi: 10.1002/ijc.26422. [DOI] [PubMed] [Google Scholar]
- 38.Komatsu S, Ichikawa D, Takeshita H, et al. Circulating miR-18a: a sensitive cancer screening biomarker in human cancer. In Vivo. 2014;28(3):293–297. [PubMed] [Google Scholar]
- 39.Liu R, Chen X, Du Y, et al. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58(3):610–618. doi: 10.1373/clinchem.2011.172767. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Chen J, Chang P, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila) 2009;2(9):807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang WS, Liu LX, Li GP, et al. Combined serum CA19-9 and miR-27-a-3p in peripheral blood mononuclear cells to diagnose pancreatic cancer. Cancer Prev Res (Phila) 2013;6(4):331–338. doi: 10.1158/1940-6207.CAPR-12-0307. [DOI] [PubMed] [Google Scholar]
- 42.Lu Y, Hu J, Sun W, et al. MiR-29c inhibits cell growth, invasion, and migration of pancreatic cancer by targeting ITGB1. Onco Targets Ther. 2016;9:99–109. doi: 10.2147/OTT.S92758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Long LM, Zhan JK, Wang HQ, et al. The clinical significance of miR-34a in pancreatic ductal carcinoma and associated molecular and cellular mechanisms. Pathobiology. 2017;84(1):38–48. doi: 10.1159/000447302. [DOI] [PubMed] [Google Scholar]
- 44.Roldo C, Missiaglia E, Hagan JP, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24(29):4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 45.Park JK, Henry JC, Jiang J, et al. miR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor. Biochem Biophys Res Commun. 2011;406(4):518–523. doi: 10.1016/j.bbrc.2011.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szafranska AE, Davison TS, John J, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26(30):4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 47.Hanoun N, Delpu Y, Suriawinata AA, et al. The silencing of microRNA 148a production by DNA hypermethylation is an early event in pancreatic carcinogenesis. Clin Chem. 2010;56(7):1107–1118. doi: 10.1373/clinchem.2010.144709. [DOI] [PubMed] [Google Scholar]
- 48.Ren Y, Gao J, Liu JQ, et al. Differential signature of fecal microRNAs in patients with pancreatic cancer. Mol Med Rep. 2012;6(1):201–209. doi: 10.3892/mmr.2012.862. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J, Zhao CY, Zhang SH, et al. Upregulation of miR-194 contributes to tumor growth and progression in pancreatic ductal adenocarcinoma. Oncol Rep. 2014;31(3):1157–1164. doi: 10.3892/or.2013.2960. [DOI] [PubMed] [Google Scholar]
- 50.Slater EP, Strauch K, Rospleszcz S, et al. MicroRNA-196a and -196b as potential biomarkers for the early detection of familial pancreatic cancer. Transl Oncol. 2014;7(4):464–471. doi: 10.1016/j.tranon.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu J, Li A, Hong SM, et al. MicroRNA alterations of pancreatic intraepithelial neoplasias. Clin Cancer Res. 2012;18(4):981–992. doi: 10.1158/1078-0432.CCR-11-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li A, Omura N, Hong SM, et al. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. 2010;70(13):5226–5237. doi: 10.1158/0008-5472.CAN-09-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ikenaga N, Ohuchida K, Mizumoto K, et al. MicroRNA-203 expression as a new prognostic marker of pancreatic adenocarcinoma. Ann Surg Oncol. 2010;17(12):3120–3128. doi: 10.1245/s10434-010-1188-8. [DOI] [PubMed] [Google Scholar]
- 54.Yabushita S, Fukamachi K, Tanaka H, et al. Circulating microRNAs in serum of human K-ras oncogene transgenic rats with pancreatic ductal adenocarcinomas. Pancreas. 2012;41(7):1013–1018. doi: 10.1097/MPA.0b013e31824ac3a5. [DOI] [PubMed] [Google Scholar]
- 55.Ho AS, Huang X, Cao H, et al. Circulating miR-210 as a novel hypoxia marker in pancreatic cancer. Transl Oncol. 2010;3(2):109–113. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120(5):1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hua Y, Chen H, Wang L, et al. Low serum miR-373 predicts poor prognosis in patients with pancreatic cancer. Cancer Biomark. 2017;20(1):95–100. doi: 10.3233/CBM-170231. [DOI] [PubMed] [Google Scholar]
- 58.Lin MS, Chen WC, Huang JX, et al. Aberrant expression of microRNAs in serum may identify individuals with pancreatic cancer. Int J Clin Exp Med. 2014;7(12):5226–5234. [PMC free article] [PubMed] [Google Scholar]
- 59.Madhavan B, Yue S, Galli U, et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer. 2015;136(11):2616–2627. doi: 10.1002/ijc.29324. [DOI] [PubMed] [Google Scholar]
- 60.Gong J, Munoz AR, Pingali S, et al. Downregulation of STAT3/NF-kappaB potentiates gemcitabine activity in pancreatic cancer cells. Mol Carcinog. 2017;56(2):402–411. doi: 10.1002/mc.22503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Costello E, Greenhalf W, Neoptolemos JP. New biomarkers and targets in pancreatic cancer and their application to treatment. Nat Rev Gastroenterol Hepatol. 2012;9(8):435–444. doi: 10.1038/nrgastro.2012.119. [DOI] [PubMed] [Google Scholar]
- 62.Giovannetti E, Erozenci A, Smit J, et al. Molecular mechanisms underlying the role of microRNAs (miRNAs) in anticancer drug resistance and implications for clinical practice. Crit Rev Oncol Hematol. 2012;81(2):103–122. doi: 10.1016/j.critrevonc.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 63.Wang Z, Li Y, Ahmad A, et al. Pancreatic cancer: understanding and overcoming chemoresistance. Nat Rev Gastroenterol Hepatol. 2011;8(1):27–33. doi: 10.1038/nrgastro.2010.188. [DOI] [PubMed] [Google Scholar]
- 64.Xia QS, Ishigaki Y, Sun L, et al. Effect of anti-cancer drugs on the expression of BIC/miR-155 in human pancreatic cancer PANC-1 cells. Zhonghua Yi Xue Za Zhi. 2010;90(2):123–127. [PubMed] [Google Scholar]
- 65.Park JK, Lee EJ, Esau C, et al. Antisense inhibition of microRNA-21 or -221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas. 2009;38(7):e190–e199. doi: 10.1097/MPA.0b013e3181ba82e1. [DOI] [PubMed] [Google Scholar]
- 66.Ji Q, Hao X, Zhang M, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4(8):e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hamada S, Masamune A, Miura S, et al. MiR-365 induces gemcitabine resistance in pancreatic cancer cells by targeting the adaptor protein SHC1 and pro-apoptotic regulator BAX. Cell Signal. 2014;26(2):179–185. doi: 10.1016/j.cellsig.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 68.Hasegawa S, Eguchi H, Nagano H, et al. MicroRNA-1246 expression associated with CCNG2-mediated chemoresistance and stemness in pancreatic cancer. Br J Cancer. 2014;111(8):1572–1580. doi: 10.1038/bjc.2014.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wellner U, Schubert J, Burk UC, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11(12):1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 70.Li Y, VandenBoom TG, 2nd, Kong D, et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69(16):6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Setoyama T, Zhang X, Natsugoe S, et al. microRNA-10b: a new marker or the marker of pancreatic ductal adenocarcinoma? Clin Cancer Res. 2011;17(17):5527–5529. doi: 10.1158/1078-0432.CCR-11-1477. [DOI] [PubMed] [Google Scholar]
- 72.Zhao Y, Zhao L, Ischenko I, et al. Antisense inhibition of microRNA-21 and microRNA-221 in tumor-initiating stem-like cells modulates tumorigenesis, metastasis, and chemotherapy resistance in pancreatic cancer. Target Oncol. 2015;10(4):535–548. doi: 10.1007/s11523-015-0360-2. [DOI] [PubMed] [Google Scholar]
- 73.Hwang JH, Voortman J, Giovannetti E, et al. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS One. 2010;5(5):e10630. doi: 10.1371/journal.pone.0010630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moriyama T, Ohuchida K, Mizumoto K, et al. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol Cancer Ther. 2009;8(5):1067–1074. doi: 10.1158/1535-7163.MCT-08-0592. [DOI] [PubMed] [Google Scholar]
- 75.Zhang L, Yao J, Li W, et al. Micro-RNA-21 regulates cancer-associated fibroblast-mediated frug resistance in pancreatic cancer. Oncol Res. 2017 May;:5. doi: 10.3727/096504017X14934840662335. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang P, Zhuang L, Zhang J, et al. The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer, and miR-21 expression confers chemoresistance by targeting FasL. Mol Oncol. 2013;7(3):334–345. doi: 10.1016/j.molonc.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wei X, Wang W, Wang L, et al. MicroRNA-21 induces 5-fluorouracil resistance in human pancreatic cancer cells by regulating PTEN and PDCD4. Cancer Med. 2016;5(4):693–702. doi: 10.1002/cam4.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nagano H, Tomimaru Y, Eguchi H, et al. MicroRNA-29a induces resistance to gemcitabine through the Wnt/beta-catenin signaling pathway in pancreatic cancer cells. Int J Oncol. 2013;43(4):1066–1072. doi: 10.3892/ijo.2013.2037. [DOI] [PubMed] [Google Scholar]
- 79.Wei F, Liu Y, Guo Y, et al. miR-99b-targeted mTOR induction contributes to irradiation resistance in pancreatic cancer. Mol Cancer. 2013;12:81. doi: 10.1186/1476-4598-12-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fan P, Liu L, Yin Y, et al. MicroRNA-101-3p reverses gemcitabine resistance by inhibition of ribonucleotide reductase M1 in pancreatic cancer. Cancer Lett. 2016;373(1):130–137. doi: 10.1016/j.canlet.2016.01.038. [DOI] [PubMed] [Google Scholar]
- 81.Bera A, VenkataSubbaRao K, Manoharan MS, et al. A miRNA signature of chemoresistant mesenchymal phenotype identifies novel molecular targets associated with advanced pancreatic cancer. PLoS One. 2014;9(9):e106343. doi: 10.1371/journal.pone.0106343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ohuchida K, Mizumoto K, Kayashima T, et al. MicroRNA expression as a predictive marker for gemcitabine response after surgical resection of pancreatic cancer. Ann Surg Oncol. 2011;18(8):2381–2387. doi: 10.1245/s10434-011-1602-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xia QS, Ishigaki Y, Zhao X, et al. Human SMG-1 is involved in gemcitabine-induced primary microRNA-155/BIC up-regulation in human pancreatic cancer PANC-1 cells. Pancreas. 2011;40(1):55–60. doi: 10.1097/MPA.0b013e3181e89f74. [DOI] [PubMed] [Google Scholar]
- 84.Mikamori M, Yamada D, Eguchi H, et al. MicroRNA-155 Controls exosome synthesis and promotes gemcitabine resistance in pancreatic ductal adenocarcinoma. Sci Rep. 2017;7:42339. doi: 10.1038/srep42339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Takiuchi D, Eguchi H, Nagano H, et al. Involvement of microRNA-181b in the gemcitabine resistance of pancreatic cancer cells. Pancreatology. 2013;13(5):517–523. doi: 10.1016/j.pan.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 86.Amponsah PS, Fan P, Bauer N, et al. microRNA-210 overexpression inhibits tumor growth and potentially reverses gemcitabine resistance in pancreatic cancer. Cancer Lett. 2017;388:107–117. doi: 10.1016/j.canlet.2016.11.035. [DOI] [PubMed] [Google Scholar]
- 87.Zhang XJ, Ye H, Zeng CW, et al. Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J Hematol Oncol. 2010;3:46. doi: 10.1186/1756-8722-3-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang W, Zhao L, Wei X, et al. MicroRNA-320a promotes 5-FU resistance in human pancreatic cancer cells. Sci Rep. 2016;6:27641. doi: 10.1038/srep27641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Iwagami Y, Eguchi H, Nagano H, et al. miR-320c regulates gemcitabine-resistance in pancreatic cancer via SMARCC1. Br J Cancer. 2013;109(2):502–511. doi: 10.1038/bjc.2013.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schreiber R, Mezencev R, Matyunina LV, et al. Evidence for the role of microRNA 374b in acquired cisplatin resistance in pancreatic cancer cells. Cancer Gene Ther. 2016;23(8):241–245. doi: 10.1038/cgt.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cao F, Li J, Sun H, et al. HES 1 is essential for chemoresistance induced by stellate cells and is associated with poor prognosis in pancreatic cancer. Oncol Rep. 2015;33(4):1883–1889. doi: 10.3892/or.2015.3789. [DOI] [PubMed] [Google Scholar]
- 92.Frampton AE, Krell J, Jamieson NB, et al. microRNAs with prognostic significance in pancreatic ductal adenocarcinoma: a meta-analysis. Eur J Cancer. 2015;51(11):1389–1404. doi: 10.1016/j.ejca.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 93.Li BS, Liu H, Yang WL. Reduced miRNA-218 expression in pancreatic cancer patients as a predictor of poor prognosis. Genet Mol Res. 2015;14(4):16372–16378. doi: 10.4238/2015.December.9.5. [DOI] [PubMed] [Google Scholar]
- 94.Xu J, Wang T, Cao Z, et al. MiR-497 downregulation contributes to the malignancy of pancreatic cancer and associates with a poor prognosis. Oncotarget. 2014;5(16):6983–6993. doi: 10.18632/oncotarget.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zou Y, Li J, Chen Z, et al. miR-29c suppresses pancreatic cancer liver metastasis in an orthotopic implantation model in nude mice and affects survival in pancreatic cancer patients. Carcinogenesis. 2015;36(6):676–684. doi: 10.1093/carcin/bgv027. [DOI] [PubMed] [Google Scholar]
- 96.Kong X, Du Y, Wang G, et al. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR-196a could be a potential marker for poor prognosis. Dig Dis Sci. 2011;56(2):602–609. doi: 10.1007/s10620-010-1285-3. [DOI] [PubMed] [Google Scholar]
- 97.Schultz NA, Andersen KK, Roslind A, Willenbrock H, Wøjdemann M, Johansen JS. Prognostic microRNAs in cancer tissue from patients operated for pancreatic cancer–five microRNAs in a prognostic index. World J Surg. 2012;36(11):2699–2707. doi: 10.1007/s00268-012-1705-y. [DOI] [PubMed] [Google Scholar]
- 98.Singh S, Chitkara D, Kumar V, Behrman SW, Mahato RI. miRNA profiling in pancreatic cancer and restoration of chemosensitivity. Cancer Lett. 2013;334(2):211–220. doi: 10.1016/j.canlet.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 99.Nakata K, Ohuchida K, Mizumoto K, et al. MicroRNA-10b is overexpressed in pancreatic cancer, promotes its invasiveness, and correlates with a poor prognosis. Surgery. 2011;150(5):916–922. doi: 10.1016/j.surg.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 100.Yu J, Ohuchida K, Mizumoto K, Fujita H, Nakata K, Tanaka M. MicroRNA miR-17-5p is overexpressed in pancreatic cancer, associated with a poor prognosis, and involved in cancer cell proliferation and invasion. Cancer Biol Ther. 2010;10(8):748–757. doi: 10.4161/cbt.10.8.13083. [DOI] [PubMed] [Google Scholar]
- 101.Jamieson NB, Morran DC, Morton JP, et al. MicroRNA molecular profiles associated with diagnosis, clinicopathologic criteria, and overall survival in patients with resectable pancreatic ductal adenocarcinoma. Clin Cancer Res. 2012;18(2):534–545. doi: 10.1158/1078-0432.CCR-11-0679. [DOI] [PubMed] [Google Scholar]
- 102.Wang S, Zhao Y, Li D, Zhu L, Shen Z. Identification of biomarkers for the prognosis of pancreatic ductal adenocarcinoma with miRNA microarray data. Int J Biol Markers. 2015;30(2):e226–e233. doi: 10.5301/jbm.5000134. [DOI] [PubMed] [Google Scholar]
- 103.Zhao G, Zhang JG, Shi Y, et al. MiR-130b is a prognostic marker and inhibits cell proliferation and invasion in pancreatic cancer through targeting STAT3. PLoS One. 2013;8(9):e73803. doi: 10.1371/journal.pone.0073803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev. 2013;23(1):3–11. doi: 10.1016/j.gde.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tavano F, di Mola FF, Piepoli A, et al. Changes in miR-143 and miR-21 expression and clinicopathological correlations in pancreatic cancers. Pancreas. 2012;41(8):1280–1284. doi: 10.1097/MPA.0b013e31824c11f4. [DOI] [PubMed] [Google Scholar]
- 106.Greither T, Grochola LF, Udelnow A, et al. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126(1):73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 107.Zhang ZL, Bai ZH, Wang XB, et al. miR-186 and 326 predict the prognosis of pancreatic ductal adenocarcinoma and affect the proliferation and migration of cancer cells. PLoS One. 2015;10(3):e0118814. doi: 10.1371/journal.pone.0118814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Frampton AE, Krell J, Jacob J, et al. microRNAs as markers of survival and chemoresistance in pancreatic ductal adenocarcinoma. Expert Rev Anticancer Ther. 2011;11(12):1837–1842. doi: 10.1586/era.11.184. [DOI] [PubMed] [Google Scholar]
- 109.Papaconstantinou IG, Manta A, Gazouli M, et al. Expression of microRNAs in patients with pancreatic cancer and its prognostic significance. Pancreas. 2013;42(1):67–71. doi: 10.1097/MPA.0b013e3182592ba7. [DOI] [PubMed] [Google Scholar]
- 110.Lee C, He H, Jiang Y, et al. Elevated expression of tumor miR-222 in pancreatic cancer is associated with Ki67 and poor prognosis. Med Oncol. 2013;30(4):700. doi: 10.1007/s12032-013-0700-y. [DOI] [PubMed] [Google Scholar]
- 111.Komatsu S, Ichikawa D, Miyamae M, et al. Malignant potential in pancreatic neoplasm; new insights provided by circulating miR-223 in plasma. Expert Opin Biol Ther. 2015;15(6):773–785. doi: 10.1517/14712598.2015.1029914. [DOI] [PubMed] [Google Scholar]
- 112.Shi S, Lu Y, Qin Y, et al. miR-1247 is correlated with prognosis of pancreatic cancer and inhibits cell proliferation by targeting neuropilins. Curr Mol Med. 2014;14(3):316–327. doi: 10.2174/1566524014666140228120014. [DOI] [PubMed] [Google Scholar]