Abstract
Large quantities of immunoglobulin A (IgA) are constitutively secreted by intestinal plasma cells to coat and contain the commensal microbiota, yet the specificity of these antibodies remains elusive. Here, we profiled the reactivities of single murine IgA plasma cells by cloning and characterizing large numbers of monoclonal antibodies. IgAs were not specific to individual bacterial taxa but rather polyreactive, with broad reactivity to a diverse but defined subset of microbiota. These antibodies arose at low frequencies among naïve B cells, and were selected into the IgA repertoire upon recirculation in Peyer’s patches. This selection process occurred independent of microbiota or dietary antigens. Furthermore, while some IgAs acquired somatic mutations, these did not substantially influence their reactivity. These findings reveal an endogenous mechanism driving homeostatic production of polyreactive IgAs with innate specificity to microbiota.
Main Text
An innate barrier of mucus, antimicrobial peptides, and immunoglobulin A (IgA) serves as a first line of defense at mucosal surfaces (1–3). Of these components, IgAs uniquely derive from plasma cells (PCs) of the adaptive immune system, yet the specificity of these antibodies has long remained enigmatic. IgA responses occur constitutively under normal homeostatic conditions through both T-dependent (TD) and T-independent (TI) pathways in mucosal lymphoid tissues such as Peyer’s patches (PPs) (4–6). While some studies have suggested that IgAs may be highly specific to individual components of the commensal microbiota (7–10), continuous generation of high-affinity responses against the vast and dynamic array of exogenous antigens encountered daily could be overwhelmingly complex in practice. Instead, others have suggested that IgAs may be polyreactive, with individual antibodies able to naturally bind and neutralize multiple targets with low affinity (11–13). In support of the latter hypothesis, we recently found that T cells, germinal centers (GCs), and somatic hypermutation were largely dispensable for polyclonal IgA coating of microbiota (4). Previous studies of IgA-derived monoclonal antibodies (mAbs) have generally failed to assess reactivity to microbiota (14–16), leaving open the central question of their specificity. Here, we interrogated at the single-cell level the specificity and origins of IgA responses with an unbiased, large-scale analysis of mAbs derived from murine IgA PC populations and other B cell subsets.
Microbiota-reactivity and polyreactivity of mAbs from IgA PCs or naïve B cell subsets
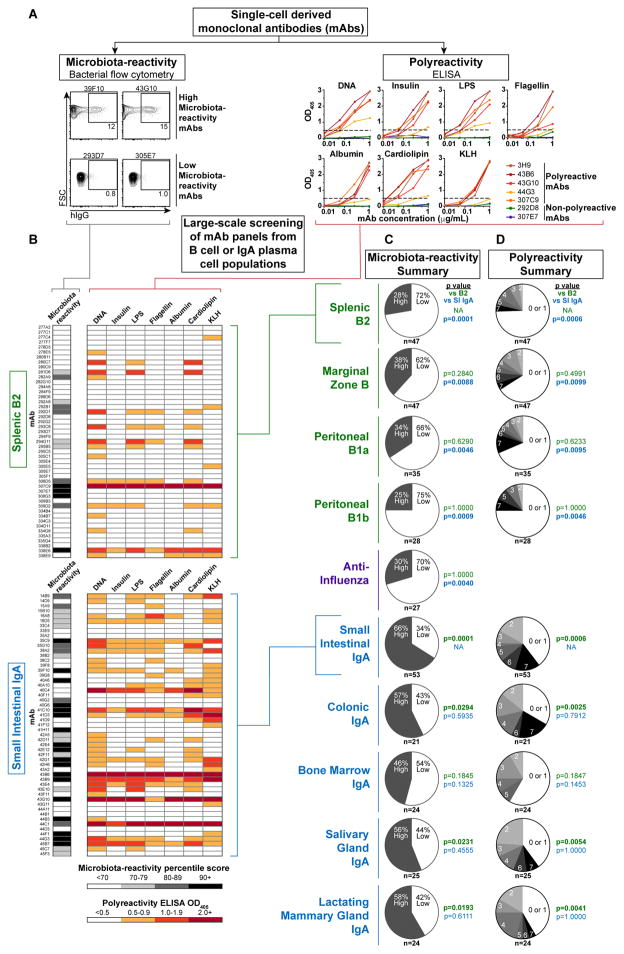
We first sought to establish the frequency of microbiota-reactive specificities within the repertoire of murine small intestinal lamina propria (SI) IgA PCs compared with naïve B cell populations that express IgM and IgD. To allow direct comparison of specificities across different isotypes, all mAbs were cloned from sorted single cells and expressed as monomeric human IgG1/Igκ chimeras (17). mAb panels were derived from pools of mice in multiple independent sorting experiments, and their microbiota-reactivity was assessed by staining and bacterial flow cytometry of SI microbiota taken directly ex vivo from Rag1−/− mice to avoid potential epitope saturation by endogenous IgA (Fig. 1A)(4). Initial experiments indicated that most IgA-derived mAbs bound microbiota, consistent with their intestinal PC origin, whereas most mAbs from naïve splenic B2 cells, peritoneal B1b, or anti-influenza human plasmablasts did not (Fig. S1A). Surprisingly, some mAbs from naïve B cells or anti-influenza plasmablasts also bound microbiota and resembled IgA mAbs, though these were found more rarely at a frequency of ~30%. Dose titration suggested that microbiota-reactive mAbs, regardless of origin, were moderate to low affinity (Fig. S1A). We did not find mAbs that bound a small fraction of microbiota brightly; instead, staining intensity correlated with percent of bacteria bound (Fig. S1B). Though absolute staining values of individual mAbs showed minor variation across independent experiments, presumably due to differences in microbiota composition, mAb rank order was well-preserved (Fig. S1C). Based on these observations, and to facilitate screening of large numbers of mAbs, we established standardized scoring and inclusion criteria for microbiota staining experiments (Materials and Methods). mAbs were assigned a microbiota-reactivity percentile score according to a control splenic B2 or anti-influenza distribution assayed side-by-side. Scores of ≥70 were considered high microbiota-reactivity, and <70 low microbiota-reactivity; details of threshold selection and scoring are described under Materials and Methods.
Figure 1. Microbiota-reactivity and polyreactivity of monoclonal antibodies from IgA plasma cell populations and naïve B cell subsets.
(A) Workflow for characterizing monoclonal antibodies (mAbs) cloned from sorted single cells. All mAbs were expressed as monomers with murine variable regions and human IgG1/Igκ constant regions except anti-influenza mAbs, which had fully human variable regions. mAbs were scored for microbiota-reactivity by bacterial flow cytometry against Rag1−/− SI microbiota or polyreactivity by ELISA against indicated antigens. (B) Microbiota-reactivity percentile scores and polyreactivity ELISA OD405 values for individual mAbs from indicated panels. (C) Summaries of indicated panels scored for microbiota-reactivity, indicating percent of mAbs that scored either high or low microbiota-reactivity; or (D) Polyreactivity summaries, indicating percent of mAbs with indicated number of positive reactivities. Numbers of mAbs analyzed in each panel are indicated below each chart. P values calculated by Fisher’s exact test against B2 (green) or SI IgA (blue) panels. Data compiled from >30 independent experiments.
We extended this analysis to 157 mAbs from all known naïve B cell subsets (Fig. 1B–C, S1D, S2A–D, Database S1). While B2 and B1b cells are the predominant precursors to IgA PCs (4, 5), peritoneal B1a and splenic marginal zone (MZ) B cells were also examined, as they have been suggested to encode natural antibacterial and polyreactive antibodies (18). We found that a similar frequency of 25–38% of mAbs showed natural, germline-encoded high microbiota-reactivity across all naïve B cell subsets, as did 30% of anti-influenza controls (Fig. 1C, S2A–D). In contrast, 66% of SI IgA mAbs (35 of 53) showed high microbiota-reactivity (Fig. 1B–C, Database S1), demonstrating that microbiota-reactive specificities occur naturally in all naïve B cell populations but are considerably enriched in the IgA repertoire.
As microbiota-reactivity could be a consequence of a broader pattern of polyreactivity, we next assessed the frequency of polyreactive specificities within the naïve B cell or IgA repertoires by screening mAbs against a panel of seven structurally diverse antigens including DNA, insulin, lipopolysaccharide (LPS), flagellin, albumin, cardiolipin, and keyhole limpet hemocyanin (KLH) by enzyme-linked immunosorbent assay (ELISA) (Fig. 1A). mAbs that bound two or more antigens with an OD405 of ≥0.5 at a concentration of ≤1 μg/mL were considered polyreactive, in accordance with prior studies (19–21). We found that 25–34% of naïve B cell mAbs were polyreactive, consistent with previous estimates (22), with no major differences between naïve B cell subsets (Fig. 1B, D, S2B–D). In contrast, 60% of IgA-derived mAbs were polyreactive (Fig. 1B, D), indicating that polyreactive specificities arise naturally in all naïve B cell subsets but are enriched in the IgA repertoire.
Flagellin and LPS are proposed antigenic targets of IgA (23–27). Although 43% of IgAs bound each of these antigens, these antibodies were consistently polyreactive and low affinity (Fig. 1B). While polyreactive mAbs lacked the anti-nuclear reactivity of disease-associated autoantibodies (Fig. S3A), they exhibited a pattern of self-reactivity in vivo, as suggested by their reduced retention in serum after injection into mice (Fig. S3B–C)(28). Furthermore, polyreactivity persisted after purification of mAbs by size exclusion chromatography, excluding potential artifacts of recombinant protein expression such as abnormal folding or aggregation (Fig. S3D).
Polyreactivity and microbiota-reactivity were significantly but not systematically associated in our assays (Fig. 1B, S2B–D), and average polyreactivity of individual mAbs correlated with their microbiota-reactivity (Fig. S4A). Neither polyreactivity nor microbiota-reactivity were significantly correlated with antibody features such as length, charge, or hydrophobicity (Fig. S4B), nor with particular variable gene usage (Fig. S4C), consistent with several previous studies (20, 22). Altogether, microbiota-reactive and polyreactive specificities accounted for 83% of SI IgAs versus only 45% of naïve B cell mAbs, underscoring the fundamental differences between these repertoires (p<0.0001, Fisher’s exact test).
While the SI harbored the most abundant population of IgA PCs, smaller populations were also apparent in the colonic lamina propria, bone marrow (BM), lung, salivary gland (SG), and lactating mammary gland (LMG) (Fig. S5A). Analysis of somatic mutation distribution and T-cell-deficient Tcrb−/−d−/− mice indicated that SI, colonic, and SG IgA PCs were mixtures of TD and TI specificities, whereas BM and LMG IgAs were largely TD (Fig. S5A–C). To assess the specificity of these IgAs, we cloned 94 mAbs from colonic, BM, SG, or LMG PCs (Database S1). These mAb panels bore striking resemblance to SI IgAs and showed similar enrichment for both microbiota-reactive and polyreactive specificities, with the possible exception of BM IgAs, which displayed a somewhat lower frequency of such reactivities (Fig. 1C–D, S6A–D). Therefore, regardless of their tissue of origin, IgA PC populations predominantly expressed microbiota-reactive and polyreactive antibodies.
Microbiota-reactive antibodies bind a diverse subset of commensal bacteria
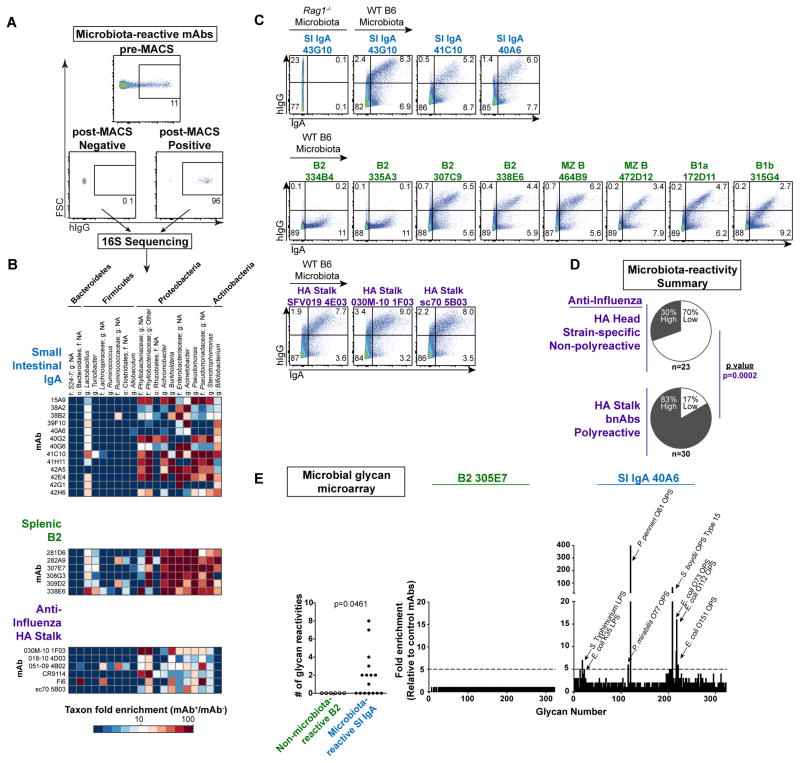
Polyclonal IgA coats a distinct subset of microbiota in vivo (4, 9, 29–33). To identify the targets of individual microbiota-reactive IgA-derived mAbs, we purified and characterized mAb+ and mAb− bacteria by 16S rRNA gene-targeted (16S) amplicon sequencing (Fig. 2A)(4). IgA-derived mAbs showed a variety of binding patterns but typically bound multiple distinct microbial taxa with frequent reactivity to Proteobacteria; however, most Bacteroidetes and Firmicutes were not bound (Fig. 2B, S7A–B). Importantly, the 16S microbiota-binding patterns of many individual mAbs closely resembled previously published patterns of binding by endogenous polyclonal IgA (4). Furthermore, co-purification of polyclonal IgA+ or mAb+ fractions from colonic microbiota of wild-type (WT) B6 mice demonstrated strongly overlapping patterns of microbial targeting (Fig. S7A). Several Proteobacteria-reactive mAbs also bound segmented filamentous bacteria (SFB), a member of the Firmicutes and known stimulator of IgA responses (Fig. S7A)(4, 30, 34–36). Additionally, in double-staining experiments on WT colonic microbiota, mAbs selectively bound to bacteria that were endogenously coated by IgA in vivo (Fig. 2C). Together, these data indicate that IgA-derived mAbs recapitulate the binding patterns of endogenous IgAs that coat the microbiota.
Figure 2. Microbiota-reactive antibodies bind a broad but defined subset of commensal bacteria.
(A) Representative plots depicting pre-MACS and post-MACS positive or negative fractions used for 16S sequencing analysis of mAb-bound or –unbound bacteria purified from Rag1−/− SI microbiota. (B) Microbial taxa bound by individual mAbs from indicated panels; enrichment in 16S sequencing data calculated by relative abundance in mAb+/mAb− fractions. All mAbs in a given panel were co-purified in the same experiment. Data compiled from three independent experiments. Taxonomy abbreviations: o= order; f=family; g=genus. (C) Flow cytometry of Rag1−/− or WT B6 colonic microbiota comparing bacteria stained by indicated mAbs with those endogenously coated by polyclonal IgA. Data compiled from three independent experiments. (D) Summary of microbiota-reactivity for panels of non-polyreactive strain-specific mAbs against the HA head or polyreactive bnAbs against the HA stalk. Numbers of mAbs analyzed in each panel are indicated below each chart. P value calculated by Fisher’s exact test. (E) Summary of microbial glycan microarray reactivities for microbiota-reactive IgA mAbs or non-microbiota-reactive B2 mAbs (left), and representative reactivities of individual mAbs (right). P value calculated by Fisher’s exact test. Data expressed as enrichment over average background of six B2 negative control mAbs. Annotated peaks showed >5-fold enrichment. Data compiled from two independent experiments.
Notably, naturally microbiota-reactive mAbs from naïve B cells showed the same patterns of microbiota-binding by 16S sequencing as those derived from IgAs, and also selectively stained endogenously IgA-coated microbiota (Fig. 2B–C, S8A) suggesting that, although less frequent, they and IgAs both belong to the same category of natural polyreactive antibodies. To test whether this property of microbiota-reactivity extended to other unrelated polyreactive antibodies, we examined a panel of 30 human broadly neutralizing antibodies (bnAbs) to influenza that recognize the conserved hemagglutinin (HA) stalk region (Database S1). These bnAbs were previously reported to show extensive polyreactivity to the same DNA, insulin, and LPS antigens used in our study (19). Strikingly, we found that a majority of bnAbs showed high microbiota-reactivity with similar binding patterns to IgA-derived mAbs, including selective staining of endogenously IgA-coated microbiota (Fig. 2C–D, S9A). In contrast, 23 non-polyreactive strain-specific mAbs against the influenza HA head typically showed little reactivity (Fig. 2D, S9A, Database S1), similar to a previous independent panel (Fig. 1C, S2A). Interestingly, bacteria naturally coated with the highest levels of endogenous IgA were usually the most brightly stained by mAbs (Fig. 2C). Together, these data show that polyreactive antibodies of various origins, whether derived from IgA PCs, naïve B cells, or anti-influenza responses, exhibit similar patterns of reactivity to a broad yet defined subset of microbiota, reminiscent of pattern recognition by innate immune receptors.
While we observed robust mAb staining of microbiota ex vivo, we did not detect binding to a series of bacterial strains cultured in vitro from the mAb+ fraction (Fig. S10A–B), suggesting that relevant surface antigen expression was dependent upon the in vivo niche. Indeed, numerous reports have documented modulation of bacterial surface antigens including capsular polysaccharides, LPS, flagella, and adhesion factors in response to different growth or environmental conditions (37–43), and additional biochemical modifications by host- or microbiota-derived factors are also possible (44–46). Moreover, ex vivo mAb staining of microbiota was unaltered by pre-treatment with nuclease or pre-washing with low pH glycine buffer (Fig. S10C), indicating that the observed reactivity was not due to bacterial adsorption of free nucleic acids nor non-covalently-associated factors from the host or other microbes.
To further understand the molecular basis for microbiota surface-reactivity, we asked whether IgAs might bind microbial glycans by screening mAbs against a microbial glycan microarray (47). Numerous glycan reactivities were observed among microbiota-reactive mAbs from IgA PCs (Fig. 2E, S11A–B), whereas non-microbiota-reactive mAbs from B2 cells showed little reactivity. Significantly, microbiota-reactive IgA mAbs typically bound multiple distinct microbial glycans, often at low to moderate affinity. For example, mAb 40A6 showed both high-binding and low-binding interactions with glycans from Proteus penneri, Proteus mirabilis, Shigella boydii, Salmonella Typhimurium, and various strains of Escherichia coli. These findings support the conclusion that microbial surface glycans are common targets of microbiota-reactive IgAs (15, 48).
Naturally microbiota-reactive recirculating naïve B cells are selected into the IgA repertoire in PPs
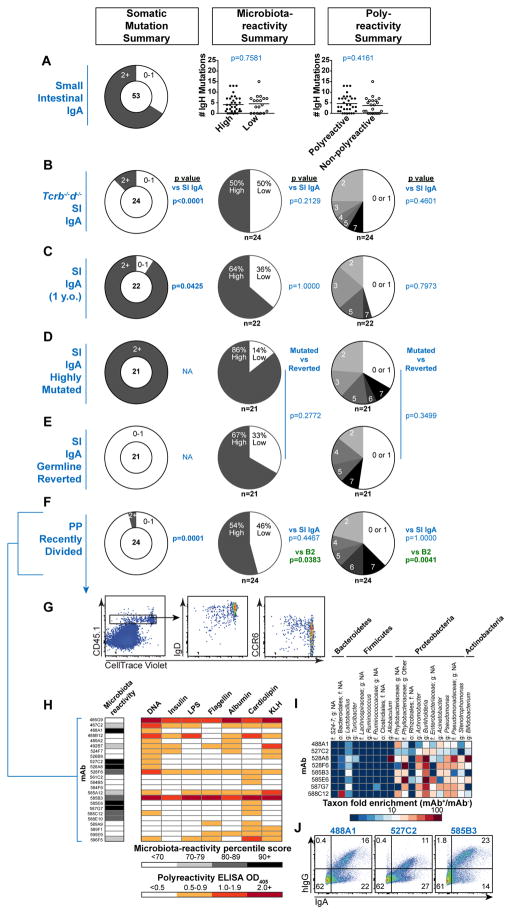
To determine the mechanisms by which microbiota-reactive and polyreactive specificities are selected into the IgA repertoire, we first considered whether selection might preferentially occur through either TD or TI pathways. Some hypotheses suggest TI IgA responses may favor generation of polyreactive specificities (8, 49), whereas others suggest polyreactivity may be acquired in GCs (20, 50). In our SI IgA mAb panel, which derived from 8-to-15-week-old mice, about two-thirds of sequences harbored somatic mutations (Fig. 3A, S12A). However, there was no association between microbiota-reactivity or polyreactivity and the presence or extent of somatic mutation (Fig. 3A). Furthermore, a panel of 24 TI mAbs cloned from Tcrb−/−d−/− SI IgAs showed similar frequencies and patterns of microbiota-reactivity and polyreactivity to WT SI IgAs, despite a near absence of mutations (Fig. 3B, S12A–C, Database S1). Conversely, a panel of 22 mAbs cloned from SI IgA PCs of one-year-old WT mice displayed an even greater frequency of mutated sequences than 8-to-15-week-old mice, as reported (51), but showed comparable microbiota-reactivity and polyreactivity to younger mice (Fig. 3C, S12A, D–E, Database S1). To directly test the contribution of somatic mutations to mAb reactivity, we reverted 21 highly mutated SI IgAs to germline configuration (Fig. S13A, Database S1). Reverted mAbs showed no significant differences in overall microbiota-reactivity or polyreactivity (Fig. 3D–E), although there were a few examples of either gain or loss of polyreactivity and/or microbiota-reactivity (Fig. S13B–C). These results are consistent with the observation that only ~70% of IgA mutations were nonsynonymous, a rate that does not support affinity selection (Fig. S13D), and with a recent report suggesting that PP GCs lack evidence of antigen-driven selection (52). While T cells did not substantially modulate the specificity of IgA, they did promote an increase in IgA PC numbers and were largely required for the generation of extraintestinal IgA PC populations (Fig. S5A). We conclude that selection of microbiota-reactive and polyreactive IgAs occurs independent of GCs and that these specificities can differentiate via either TI or TD pathways.
Figure 3. Mechanisms of IgA selection.
Summary of somatic mutations, microbiota-reactivity, and polyreactivity of mAb panels derived from (A) SI IgA PCs from 8–15 week old WT B6 mice, (B) Tcrb−/−d−/− SI IgA PCs, or (C) SI IgA PCs from one-year-old WT B6 mice. Data compiled from >5 independent experiments. (D–E) Summary of 21 highly mutated SI IgAs and their engineered germline revertants, or (F) recently divided CD45.1+ donor-derived cells in PPs 7d after transfer to MD4Tg recipients, isolated as indicated in (G). (H) Microbiota-reactivity percentile scores and polyreactivity ELISA OD405 values of individual mAbs from recently divided cells in PPs, compiled from three independent experiments, (I) Microbial taxa binding patterns of individual mAbs as determined by 16S sequencing, or (J) double staining of colonic microbiota comparing individual mAbs with endogenously IgA-coated bacteria. Numbers of mAbs analyzed in each panel are indicated below each chart. P values calculated by unpaired t test in panel A and Fisher’s exact test in panels B–F.
Searching for an early post-selection progenitor to intestinal IgA, we asked whether the recently identified IgM+IgD+CCR6+ dividing B cell population in PPs, which originates from recirculating naïve B cells (6), might exhibit signs of selection for microbiota-reactivity. We transferred fluorescently-labeled CD45.1+ polyclonal splenic B cells into CD45.2+ MD4 Ig-transgenic (MD4Tg) recipients that have little endogenous PP B cell activation (6), and isolated 24 mAbs from recently-divided CD45.1+ IgD+/lo CCR6+ cells in PPs (Fig. 3F–G, Database S1). While these mAbs were largely unmutated, as expected, they exhibited a striking enrichment in microbiota-reactivity and polyreactivity (Fig. 3F–H), and displayed 16S microbiota-binding patterns that resembled SI IgAs (Fig. 3I, 2B) as well as selective binding to bacteria endogenously coated with polyclonal IgA (Fig. 3J). Such specificities accounted for 88% of mAbs and were significantly enriched relative to naïve B cell panels (p=0.0001, Fisher’s exact test). These data indicate that naturally microbiota-reactive and polyreactive naïve B cells are extracted from circulation to become intestinal IgA precursors in PPs. Expression of CCR6 then guides their migration to the PP subepithelial dome, where they receive signals that direct class-switch recombination to the IgA isotype and imprinting for gut homing (6, 53–55).
An endogenous mechanism driving natural IgA selection
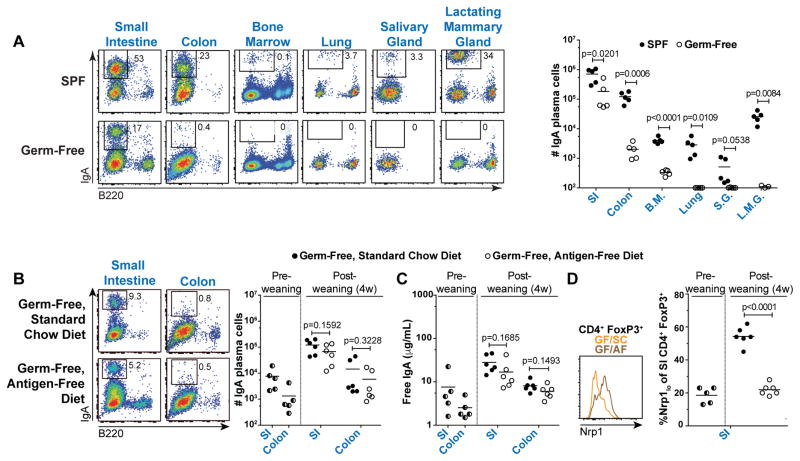
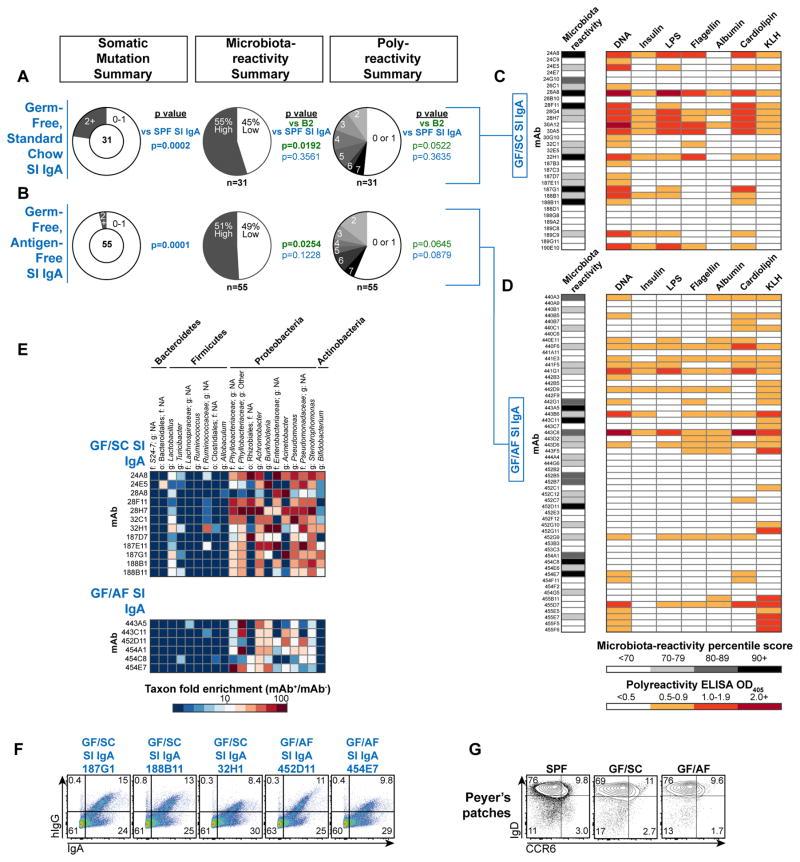
These observations prompted us to examine the role of exogenous microbiota or dietary antigens in IgA selection. IgA production is thought to be microbiota-dependent, as titers are diminished in germ-free (GF) relative to specific-pathogen-free (SPF) mice (56). Consistent with this notion, we found that IgA PCs were severely reduced or absent from most tissues in GF mice, including the colon, BM, lung, SG, and LMG (Fig. 4A). However, in the SI, the predominant site of IgA production, IgA PCs were largely preserved and showed only a minor decrease (Fig. 4A). Differentiation of SI IgAs in GF mice might represent a response to dietary antigens. To explore this possibility, we weaned three-week-old GF mice onto either a standard chow (SC) or chemically-defined antigen-free (AF) diet (Table S1)(57, 58). Remarkably, a dramatic expansion of SI IgA PCs and luminal free IgA titers was apparent after weaning of GF mice fed both SC and AF diets, indicating that neither microbiota nor dietary antigens were required for the post-weaning IgA response (Fig. 4B–C). Notably, as previously reported, SI Nrp1lo T regulatory cells failed to accumulate in GF/AF compared to GF/SC mice, confirming the major impact of the AF diet on other aspects of intestinal immunity (Fig. 4D)(57).
Figure 4. Small intestinal IgA plasma cell differentiation does not require exogenous microbiota or dietary antigens.
(A) Representative flow cytometry plots and cell number summary of IgA PCs in SPF or GF B6 mice. Plots gated on lineage− lymphocytes. L.M.G. were obtained from females 3–7d post-partum; all other tissues were from males. P values calculated by unpaired t test. (B) Representative flow cytometry plots and cell number summary of IgA PCs or (C) Luminal free IgA titers determined by ELISA in indicated mice, and (D) Representative flow cytometry of SI CD4+ FoxP3+ cells and summary of percent Nrp1lo among CD4+ FoxP3+ cells in indicated mice. P values calculated by unpaired t test. Data compiled from four independent experiments.
We assessed the specificities of 31 GF/SC and 55 GF/AF SI IgA-derived mAbs (Database S1). Despite their selection in the absence of exogenous antigens, both panels showed frequent microbiota-reactivity and polyreactivity (Fig. 5A–D) with similar 16S binding profiles to SPF SI IgAs (Fig. 5E, 2B), and selectively bound bacteria endogenously coated with IgA in vivo (Fig. 5F). Together, microbiota-reactive and polyreactive specificities accounted for 69% of GF/SC and GF/AF SI IgA mAbs, a significant enrichment relative to naïve B cell panels (p=0.0005, Fisher’s exact test). GF/SC and GF/AF SI IgAs were largely unmutated (Fig. 5A–B) and resembled IgAs from SPF Tcrb−/−d−/− mice in mutation frequency, mAb reactivity, and intestinal/extraintestinal PC distribution (Fig. S5A, 3B, S12A–C), consistent with a requirement for T cell-intrinsic sensing of microbiota through Toll-like receptors for T cell help (29), and with the observation that individual IgA clones in GF mice persist and progressively acquire somatic mutations after colonization with microbiota (51). As IgD+CCR6+ cells were readily observed in GF/SC and GF/AF PPs (Fig. 5G), natural IgA selection presumably occurred through the same pathway described in SPF mice (Fig. 3). We conclude that a natural IgA response occurs in PPs that is largely independent of exogenous antigens or T cell help, and may instead result from autoreactive stimulation by endogenous antigens. This innate response preferentially recruits naturally polyreactive and microbiota-reactive recirculating naïve B cells to enter the intestinal IgA PC repertoire and secrete the IgA that coats the commensal microbiota.
Figure 5. Natural IgA selection.
Summary of somatic mutations, microbiota-reactivity, and polyreactivity of mAb panels derived from (A) GF/SC SI IgA PCs or (B) GF/AF SI IgA PCs. Numbers of mAbs analyzed in each panel are indicated below each chart. P values calculated by Fisher’s exact test. (C–D) Microbiota-reactivity percentile scores and polyreactivity ELISA OD405 values of individual mAbs from GF/SC and GF/AF SI IgA panels. Data compiled from >6 independent experiments. (E) Microbial taxa binding patterns of individual mAbs as determined by 16S sequencing, or (F) double staining of colonic microbiota comparing individual mAbs with endogenously IgA-coated bacteria. (G) Flow cytometry of CD19+IgD+CCR6+ cells in PPs of SPF, GF/SC, or GF/AF mice. Representative of two independent experiments.
Discussion
Our studies have elucidated the specificity of homeostatic intestinal IgA and demonstrate its natural origins. They also define an endogenous mechanism of selection in PPs that enriches microbiota-reactive and polyreactive specificities into the IgA repertoire (Fig. S14). Of note, this response appears distinct from natural serum IgM responses that derive from peritoneal B1a cells (59) as canonical B1a rearrangements were not observed in the SI IgA PC repertoire (4), and these specificities were not conspicuously enriched in microbiota-reactivity.
We found that IgA PCs from a diverse array of intestinal and extraintestinal tissues typically expressed microbiota-reactive and polyreactive specificities. These data suggest a common mechanism by which IgAs found in serum, intestinal, salivary, and mammary secretions promote homeostasis in various tissue contexts and in nursing offspring (60–62). Notably, IgA PCs in tissues other than the SI were severely reduced in the absence of microbiota and T cells; this may reflect defects in cell trafficking, proliferation, or maintenance in the absence of the TD response, which requires T cell-intrinsic innate immune signals from the microbiota (29). In contrast to these sites, SI IgA PCs were readily detected in GF mice and showed a four-fold reduction, comparable to previous reports (5, 34, 35, 63). Two-thirds of SI IgA PCs were somatically mutated in WT mice, and we previously reported a three-fold reduction in in CD4-cre Bcl-6fl/fl mice, which lack T follicular helper cells (Tfh) and germinal centers but otherwise retain an intact T cell compartment (4). As T cell-deficient Tcrb−/−d−/− mice showed a more significant ten-fold reduction, T cells appear to have additional effects on IgA PCs outside the canonical Tfh pathway, potentially by regulating plasma cell maintenance, trafficking, or proliferation, cytokine production, and/or mucosal barrier integrity.
We identified a broad but distinct subset of microbiota that was bound by IgA antibodies in vivo. Yet, it remains unclear whether this binding has functional effects on commensal fitness or physiology; it is possible that IgA binding either enhances or diminishes fitness. These effects could include alterations in microbial gene expression, motility, and/or spatial localization and may be mediated by mechanisms such as agglutination, enchained growth, neutralization, and immune exclusion (15, 24, 64–66). While we observed numerous IgA reactivities to bacterially-derived antigens including flagellin, LPS, and various glycans, the relevant targets of these natural antibodies in vivo remain unclear as relevant culture conditions, biochemistry, and genetics have not been established for the targeted commensal organisms. However, we hypothesize that recognition may involve low-affinity interactions with numerous antigens on the bacterial surface. It is also possible that polyreactivity enhances binding to entropically unfavorable targets such as carbohydrates, reminiscent of its role in increasing the apparent affinity of broadly neutralizing antibodies against the HIV envelope spike (20). The structural basis for antibody polyreactivity remains enigmatic; in particular, it is unclear whether similar or distinct binding mechanisms are utilized for different antigens. As we did not discern clear repertoire features associated with polyreactivity or microbiota-reactivity, it is probable that individual antibodies can achieve this type of recognition via a variety of structural mechanisms. One previous study identified the heavy-chain complementarity determining region 3 (CDR3) as a key determinant of polyreactivity, though this analysis was limited to a single antibody (67), and silencing of heavy chain polyreactivity by light chains has been described (68). Detailed structural studies including crystallography of these antibodies in complex with various ligands are warranted.
Our analysis of GF/AF mice demonstrated that IgA plasma cells with microbiota-reactive specificities can arise in the absence of exogenous microbiota or dietary antigens. While exogenous antigen exposure was dramatically reduced in these mice, as exemplified by a lack of SI regulatory T cell induction (57), it is possible that they encountered trace quantities of dietary antigen during the first few weeks of life, potentially transmitted via lactation. Long-term maintenance on the AF diet results in emergence of micronutrient insufficiencies – notably deficiency of vitamin A, a known regulator of IgA class-switch recombination and gut trafficking, and thus new caveats emerge in mice fed this diet for several generations (53, 54, 57). Nonetheless, our ontogenetic analysis revealed a dramatic expansion of IgA plasma cells and luminal titers after weaning of mice onto the AF diet, comparable to mice weaned onto SC, supporting our conclusion that these plasma cells can differentiate in the absence of exogenous antigen. While our studies focused on the C57BL/6 strain, GF BALB/c mice were reported to produce significantly higher titers of intestinal IgA than GF C57BL/6 and these antibodies appeared to be polyreactive, suggesting that the polyreactive and microbiota-reactive nature of IgAs extends to different strain backgrounds (12, 16).
Natural IgA secretion may represent an innate-like mechanism for mucosal defense and may be evolutionarily ancient; indeed, specialized mucosal antibodies of TI origin have been described in a wide variety of jawed vertebrates (69, 70), suggesting evolutionary benefit to antibody coating of microbiota. Curiously, natural IgAs bind a broad but defined subset of microbiota; thus, evolutionary pressure may have led certain bacterial taxa to express a cell surface that either attracts or evades these antibodies. IgA responses are amplified in inflammatory bowel diseases (30) and enteric infections (71), yet the extent to which these responses differ from homeostatic mechanisms remains unclear. Notably, recruitment of new, antigen-specific clones into the SI IgA repertoire was observed after infection with S. Typhimurium (51), suggesting that more specific responses may be induced in response to particularly strong inflammatory stimuli. B cells expressing polyreactive specificities have been implicated in lymphomas of mucosal origin (72), are counterselected during central tolerance (21, 73), and are elevated in systemic autoimmune disease (74), which had suggested a detrimental role for these antibodies. However, recent reports that inherently polyreactive antibodies constitute the bulk of broadly neutralizing antibody responses to influenza virus and HIV (19, 20, 75, 76), and our finding that these bnAbs are of the same type that fuels the homeostatic intestinal IgA response, provide concrete examples demonstrating the protective value of polyreactive antibodies in a variety of homeostatic as well as pathological contexts; exploiting the naturally polyreactive IgA response may provide novel opportunities to elicit bnAbs by mucosal vaccination.
Materials and Methods
Mice
C57BL/6J, one-year-old C57BL/6J, Tcrb−/−d−/− (B6.129P2-Tcrbtm1Mom Tcrdtm1Mom/J), CD45.1+ (B6.SJL-Ptprca Pepcb/BoyJ), Rag1−/− (B6.129S7-Rag1tm1Mom/J), and MD4Tg (C57BL/6-Tg(IghelMD4)4Ccg/J) mice were purchased from Jackson Laboratories. For most experiments, mice were analyzed at 6–12 weeks of age and compared to littermate controls unless otherwise indicated. Mice were maintained in a specific pathogen-free environment at the University of Chicago. Germ free C57BL/6 mice were housed in isolators at the University of Chicago gnotobiotic facility under carefully monitored germ-free conditions; germ free cages contained sterile pine shavings as bedding material. Experimental guidelines were approved by the Institutional Animal Care and Use Committee (IACUC).
Single-cell cloning and production of monoclonal antibodies
Single cells were sorted from the following populations using the indicated markers in parentheses: Splenic B2 (CD19+ B220+ CD21+ CD23+); Splenic MZ B (CD19+ B220+ CD21hi CD23−); Peritoneal B1a (CD19+ CD23− CD5+); Peritoneal B1b (CD19+ CD23− CD5−); dividing donor-derived cells in MD4Tg recipients (CD45.1+ CD19+ CellTrace Violetdiluted); or IgA plasma cells (Lineage− B220− IgA+).
Detailed protocols for single-cell cloning and mAb production have been published separately (17, 77, 78). Briefly, single cells were sorted into 96-well plates (Biorad) using a BD AriaII cell sorter. In all plates, one row was left empty as a negative control. For most plasma cell populations, cells were sorted into 10 μL/well nuclease-free 10 mM Tris-HCl with 1 U/μL SUPERase-In RNase inhibitor (Ambion) and immediately frozen on dry ice. Single-cell cDNA synthesis and paired amplification of rearranged immunoglobulin VH and Vκ genes was then performed using a cocktail of primers as described (17). Non-plasma cell populations amplified inefficiently with this method and were instead sorted into 5 μL/well TCL buffer (Qiagen) with 1% beta-mercaptoethanol (v/v). RNA was purified using RNase-free SPRI beads (Beckman Coulter) as described (17), and cDNA synthesis and paired amplification were performed as described above. PCR products from single cells were sequenced and cloned using gene-specific primers into human IgG1/Igκ expression vectors as described (17); plasmid midipreps were performed with a Qiagen HiSpeed Midi Kit. All sequences were identified and processed using IMGT’s default settings (www.imgt.org) (79, 80). Somatic mutations were identified and quantified using IMGT’s default settings. In some cases, mutations introduced at the beginning or end of the sequence by degenerate primers were discarded from analysis. For germline reverted mAbs, heavy and light chain sequences were reverted to inferred germline as determined using IMGT, synthesized, and cloned into expression vectors using the same methods described above. If identical mAbs were obtained from multiple cells, only a single copy was retained. mAbs were expressed by transient PEI transfection of 293T cells as described (17). Culture supernatants were harvested 6–7d after transfection, cleared of cells by centrifugation at 900g for 10 mins, and mAbs purified with Protein A agarose (Thermo) as described (17). mAbs were stored at 4 °C in TBS pH 8.0 with 0.5% BSA, 150 mM NaCl, and 0.05% sodium azide.
Protein expression and concentration were determined by ELISA performed as follows: ELISA plates (Thermo) were coated with 100 μL/well goat anti-human kappa capture antibodies (Southern Biotech) at 10 μg/mL in carbonate buffer (Bethyl) for 1 hour, washed 4x with TBS-T, and blocked with 200 μL/well TBS 1% BSA (Sigma) for 1 hour. Plates were washed 4X with TBS-T, incubated for 1 hour with 100 μL/well mAbs or purified human IgG1-kappa standard (Sigma) each run in duplicate and serially diluted, washed 4X with TBS-T, incubated for 1 hour with goat anti-human IgG HRP detection antibodies (Southern Biotech, 1:20,000 in TBS-T 1% BSA), washed 4X with TBS-T, revealed with TMB HRP substrate (Bethyl), and quenched with 0.18M H2SO4. OD450 was read using an ELISA plate reader (BioTek) and concentration was determined by comparison to standards fit to a four-parameter logistic curve.
Lymphocyte flow cytometry and antibodies
Cells were incubated with anti-CD16/32 (Fc block) prior to staining with the following fluorophore or biotin conjugated monoclonal antibodies purchased from Biolegend, eBioscience, or BD unless otherwise indicated (clone in parentheses): B220 (RA3-6B2), CCR6 (29-2L17), CD4 (GK1.5 or RM4-5), CD5 (53-7.3), CD8 (53-6.7), CD11c (HL3), CD19 (6D5), CD21 (7G6), CD23 (B3B4), CD45.1 (A20), CD45.2 (104), F4/80 (BM8), FoxP3 (FJK-16s), Gl7 (GL7), GR1 (RB6-8C5), IgD (11-26c.2a), IgM (Il/41), NK1.1 (PK136), TER-119 (TER-119), Tcrb (H57-597), goat anti-mouse IgA(Southern Biotech), and goat anti-mouse Nrp1 (RD Systems). Lineage cocktail for plasma cell staining included CD3, CD4, CD11c, F4/80, NK1.1, Tcrb, and TER-119. Doublet exclusion was used when possible. FoxP3 intracellular staining was performed using a FoxP3 staining kit (eBioscience). Cell numbers were calculated by flow cytometry with counting beads (Spherotech).
Lymphocyte isolation from tissues
Small intestinal or colonic (excluding the cecum) lamina propria isolation was performed as follows: tissues were excised and fat, Peyer’s patches, and intestinal contents removed. Tissues were opened longitudinally, cut into ~1 cm pieces, and placed into a 50 mL tube with 10 mL RPMI 1% FCS 1 mM EDTA. Small intestines were processed as two halves in separate tubes and combined after lamina propria lymphocyte isolation as noted below; colons were processed in a single tube. Samples were incubated at 37 °C with shaking for 15 mins. Pieces were collected on a 100 μm cell strainer (Fisher) and placed in fresh RPMI 1% FCS 1mM EDTA for another 15 mins with shaking at 37 °C. Pieces were again collected on a 100 μm cell strainer and flow-through was discarded as it predominantly contained epithelial cells and intraepithelial lymphocytes. Intestinal pieces were then placed into 10 mL RPMI 20% FCS with 0.5 mg/mL collagenase A (Roche) and 0.1 mg/mL DNase I (Roche) at 37 °C with shaking for 30 mins. Supernatant was collected by filtering through a fresh 100 μm cell strainer and intestinal pieces were placed in fresh RPMI 20% FCS with collagenase and DNase for another 30 mins with shaking at 37 °C. Supernatant was again collected, remaining tissues was mashed, and the cell strainer was washed with 30 mL RPMI. The two lamina propria lymphocyte fractions were combined after centrifugation and further enriched by centrifugation in 40% percoll (Sigma) to remove epithelial cells and debris, washed in flow cytometry buffer, and resuspended for cell staining.
Salivary gland, lactating mammary gland, and lung isolations were performed as follows: after excision, fat was removed and tissues were chopped into small pieces and placed into a 50 mL tube containing 5 mL RPMI 4% BSA (Fisher), 2 mg/mL collagenase type I (Gibco), and 0.1 mg/mL DNase (Roche). Samples were incubated for 45 mins at 37 °C with shaking, then filtered through a 100 μm cell strainer. Remaining tissue was mashed and the strainer was washed with 30 mL RPMI. After centrifugation, lymphocytes were enriched by centrifugation in 40% percoll (Sigma), washed in flow cytometry buffer, and resuspended for cell staining.
Bacterial flow cytometry and antibodies
Small intestinal contents from two or more 6-to-20-week-old Rag1−/− mice were isolated by running forceps along the length of the intestinal tissue, pooled, and placed into a 2 mL Eppendorf Biopur tube on ice. 1 mL PBS (Corning) was added and samples were homogenized by taping the Eppendorf tube horizontally on a vortex and vortexing vigorously for 5 mins. Samples were centrifuged for 5 min at 400g to pellet large debris, and supernatant was filtered through a sterile 70 μm cell strainer (Fisher) and transferred to a new 2 mL tube. Samples were centrifuged at 8000g for 5 min and supernatant was removed by aspiration. The bacterial pellet was resuspended in ~3 mL per mouse PBS 0.25% BSA with SYTO BC (1:7500, Life Technologies) and incubated ~30 mins on ice. 50 μL/well of bacterial suspension was added to a 96-well V bottom plate (Fisher) and cells pelleted by centrifugation at 4700g for 15 mins. Supernatant was removed and bacteria were resuspended in 100 μL mAb pre-diluted to 10 μg/mL (or desired concentration) in PBS 0.25% BSA and incubated for 20 mins on ice. Suspensions were washed once with 100 uL PBS 0.25% BSA and cells pelleted by centrifugation at 4700g for 15 mins. Supernatant was removed and pellets were resuspended in 100 μL PBS 0.25% BSA 5% normal mouse serum (Jackson Immunoresearch) with mouse anti-human IgG-APC (1:800; HP6017, Biolegend) and incubated for 20 mins on ice. Suspensions were washed once with 100 uL PBS 0.25% BSA and cells pelleted by centrifugation at 4700g for 15 mins, then resuspended in PBS 0.25% BSA with DAPI (Life Technologies) prior to flow cytometry. Staining of live bacteria was visualized by gating on FSC+SSC+SYTO BC+DAPI− cells. In most experiments, samples were run using an LSRII flow cytometer (BD) equipped with a high throughput sampler (HTS).
For pre-treatment of microbiota with nuclease, SI microbiota were isolated as described above and incubated with 1 U/mL nuclease (Benzonase, Sigma) for 10 mins at room temperature in PBS 0.25% BSA. Cells were washed twice with 1 mL PBS 0.25% BSA, and resuspended in PBS 0.25% BSA with SYTO BC for 15 mins on ice and stained as described above. For pre-washing of microbiota with low pH buffer, SI microbiota were resuspended in 0.1M glycine-HCl pH 3.0 (Boston Bioproducts) and incubated for 1 min at room temperature. Cells were centrifuged for 1 min at 8000g, and supernatant was aspirated. Cells were washed once with 1 mL PBS 0.25% BSA, centrifuged 5 mins at 8000g, and resuspended in PBS 0.25% BSA with SYTO BC for 15 mins on ice and stained as described above. Untreated controls were processed side by side and washed twice with PBS 0.25% BSA prior to resuspension in PBS 0.25% BSA with SYTO BC and staining as described above.
For co-staining of mAbs and endogenous polyclonal IgA, intestinal contents from WT B6 mice were processed as described above and resuspended in PBS 0.25% BSA with SYTO BC for 30 mins on ice. 50 μL bacterial suspension was stained with 50 μL 2X mAb master mix at a final concentration of 10 μg/mL for 20 mins on ice. Cells were washed once with 1 mL PBS 0.25% BSA, centrifuged 5 mins at 8000g, and resuspended in 50 μL PBS 0.25% BSA with 10% normal goat serum for 10 mins on ice. 50 μL 2X goat anti-human IgG biotin (1:400 final concentration, Southern Biotech) and goat anti-mouse IgA PE (1:800 final concentration, Southern Biotech) were added and incubated 20 mins on ice. Cells were washed once with 1 mL PBS 0.25% BSA, centrifuged 5 mins at 8000g, and resuspended in 100 μL PBS 0.25% BSA with streptavidin-APC (1:800, Biolegend) for 20 mins on ice. Cells were washed once with 1 mL 0.25% BSA, centrifuged 5 mins at 8000g, and resuspended in PBS 0.25% BSA with DAPI prior to flow cytometry on an LSRII flow cytometer (BD).
Microbiota-reactivity scoring and inclusion criteria
Standardized scoring and inclusion criteria for microbiota-reactivity were used to score all mAbs in this study unless otherwise noted; these were as follows: 1) All experiments were performed using Rag1−/− small intestinal microbiota. 2) Scores were assigned based on the percent hIgG+ bacteria at a mAb staining concentration of 10 μg/mL. 3) mAbs were assigned a microbiota-reactivity percentile score based on a control distribution of the 47 splenic B2 or 27 anti-influenza mAbs shown in Fig. 1, assayed side-by-side in the same experiment. Percentile scores were assigned using the Microsoft Excel PERCENTRANK.INC function. 4) Experiments were considered invalid and repeated if high positive SI IgA mAb 39F10 stained less than 6% of microbiota, and this mAb was included in all experiments. This cutoff was selected arbitrarily to avert incorrect scoring of low-positive clones if staining fell to low levels, thus impairing signal detection. 5) mAbs were classified categorically as high microbiota-reactivity if their percentile score was ≥70 and low microbiota-reactivity if their percentile score was <70. This cutoff was selected based on the observation that mAbs with scores of ≥70 showed reactivity that was reproducibly above background.
Purification of mAb-bound or unbound microbiota
Small intestinal contents from two or more 6-to-20-week-old Rag1−/− mice were isolated by running forceps along the length of the intestinal tissue, pooled, and placed into a 2 mL Eppendorf Biopur tube on ice. 1 mL sterile PBS (Corning) was added and samples were homogenized by taping the Eppendorf tube horizontally on a vortex and vortexing vigorously for 5 mins. Samples were centrifuged for 5 min at 400g to pellet large debris, and supernatant was filtered through a sterile 70 μm cell strainer (Fisher) and transferred to a new 2 mL tube. Samples were centrifuged at 8000g for 5 min and supernatant was removed by aspiration. The bacterial pellet was resuspended in 3 mL per mouse sterile PBS 0.25% BSA with SYTO BC (1:7500, Life Technologies) and incubated ~30 mins on ice. An unfractionated control sample was archived for all experiments. 250 μL bacterial suspension was added to a 1.5 mL Eppendor tube and 250 μL 2X mAb master mix diluted in PBS 0.25% BSA was added for a final staining concentration of 10 μg/mL. Samples were incubated 20 mins on ice then washed once with 1 mL PBS 0.25% BSA and pelleted by centrifugation at 8000g for 5 min. Pellets were resuspended in 500 μL PBS 0.25% BSA 5% normal goat serum (Jackson Immunoresearch) 1:400 goat anti-human IgG-biotin (Southern Biotech) and incubated for 20 mins on ice. Samples were washed once with 1 mL PBS 0.25% BSA and centrifuged 5 mins at 8000g. Pellets were then resuspended in 500 μL PBS 0.25% BSA 1:800 streptavidin-APC (Biolegend) and incubated 20 mins on ice. Samples were washed once with 1 mL PBS 0.25% BSA and centrifuged 5 mins at 8000g. Pellets were resuspended in in 500 μL PBS 0.25% BSA 1:50 anti-APC magnetic beads (Miltenyi) and incubated 20 mins on ice. Samples were washed once with 1 mL PBS 0.25% BSA and centrifuged 5 mins at 8000g then resuspended in 500 μL PBS 0.25% BSA. Samples were separated using an autoMACS separator (Miltenyi) and the posselds program. Negative samples were saved, and positive samples were further purified by running again using the posseld2 program. Samples were centrifuged at 8000g and frozen at −70 °C until DNA extraction for 16S sequencing. Aliquots of pre- and post-MACS fractions from all samples were analyzed to verify separation and purity, and negative controls were included in all experiments. In some experiments, colonic contents excluding cecal contents were removed from WT C57BL/6 mice as described above and resuspended in 6 mL PBS 0.25% BSA with SYTO BC (1:7500, Life Technologies) and incubated ~30 mins on ice, then stained with mAbs and separated as described above. For analysis of endogenous polyclonal IgA coating, bacteria were resuspended in 250 μL PBS 0.25% BSA with SYTO BC and 5% normal goat serum and incubated 20 mins on ice. 250 μL of PBS 0.25% BSA 2X goat anti-mouse IgA-biotin (1:1600 final concentration, Southern Biotech) was added and cells incubated 20 mins on ice. Samples were then processed and separated as described above using streptavidin-APC, anti-APC beads, and separation by autoMACS.
For bacterial culture after purification, the mAb+ fraction was prepared as described above, serially diluted, and plated on Schaedler’s Blood Agar (BD) or EMB Agar (BD) under either aerobic or anaerobic conditions and grown for two days at 37 °C. In a second experiment, bacteria were plated on either blood agar or MacConkey agar (Thermo) and grown for two days at 37 °C. Individual colonies were restreaked and DNA was extracted using the PureLink Microbiome DNA Extraction Kit (Invitrogen). 16S sequences were amplified using forward primer ACTCCTACGGGAGGCAGCAGT and reverse primer ATTACCGCGGCTGCTGGC using Q5 Hot Start Polymerase Master Mix (New England Biolabs) and the following PCR program: Initial denaturation at 98 °C for 30s, followed by 35 cycles of 98 °C for 10s and 72 °C for 20s, with a final cycle of 72 °C for 2 mins. PCR products were cleaned up using ExoSAP-IT (Thermo) according to the manufacturer’s protocol and subjected to Sanger sequencing. Bacterial taxonomy was assigned from sequences using the Michigan State University Ribosomal Database Project Classifer (81). For flow cytometry staining, bacteria were washed from agar plates with 5 mL sterile PBS 0.25% BSA and centrifuged for 5 min at 400g to pellet debris. Supernatant was filtered through a 70 um cell strainer, transferred to a new tube, and pelleted at 8000g for 5 mins. Bacterial pellets were resuspended in PBS 0.25% BSA with SYTO BC (1:7500) and incubated for 15 mins on ice, then stained as described above.
16S rRNA gene sequencing and analysis
DNA was extracted using the PowerSoil®-htp 96 Well Soil DNA Isolation Kit (MO BIO Laboratories, Inc.) according to the manufacturer’s instructions. Amplicons of the V4 region of 16S rRNA genes were generated using the polymerase chain reaction (PCR) using the modified 515F-806R primer set (82). The primers were Golay-barcoded to allow for multiplexing on the Illumina MiSeq platform (83). PCR reactions were carried out in triplicate for each sample using sterile, DNase-free 96 well plates with appropriate (DNA template-free) negative controls using the 5 PRIME MasterMix (Gaithersburg, MD). No significant amplification was observed in the template-free control reactions, and negative control reactions processed through the sequencing pipeline did not produce any valid sequences. PCR reactions began with an initial denaturation step of 95 °C for 3 minutes, followed by 35 cycles of the following: 95 °C for 30 seconds, 55 °C for 45 seconds, then 72 °C for 1.5 minutes. The PCR reaction was finalized using a single extension step at 72 °C for 10 minutes.
Triplicate PCR reactions were then pooled together and total DNA was quantified using the PicoGreen® dsDNA Assay (Life Technologies). Primer dimers were removed from the pooled product using the UltraClean® PCR Clean-Up Kit (MoBio Laboratories, Inc.) and the amount of amplicon in each pooled sample was normalized to a final concentration of 2 ng μL−1. Amplicons were sequenced on an Illumina MiSeq using 151×151 base pair paired-end sequencing at the Environmental Sample Preparation and Sequencing Facility at Argonne National Laboratory.
Raw sequence data was processed using the QIIME analysis pipeline (84) and other bioinformatics toolkits. Paired-end reads were joined together using PEAR (85), and any reads of insufficient quality that prevented joining were discarded (< 5% of total reads). From a total of 270 samples, 1.24 × 107 paired-end reads were generated (46,065±24,747 reads per sample). Sequences were then aligned to a database of reference sequences using PyNAST (86) and clustered into operational taxonomic units (OTUs) using UCLUST at 97% similarity (87). A consensus taxonomy from the Greengenes reference database (88) was assigned to each sequence using the UCLUST taxonomy assigner. Sequencing data are publicly available to download via MG-RAST (89).
Polyreactivity ELISAs
Polyreactivity assays were performed as described (19–21). ELISA plates (Thermo) were coated overnight with 50 μL/well antigen diluted in carbonate buffer (Bethyl) with the exception of cardiolipin, which was coated overnight in 100% ethanol left uncovered to allow evaporation. The following antigens and concentrations were used for coating: calf thymus DNA (Life Technologies), 10 μg/mL; human insulin (Fitzgerald), 5 μg/mL; LPS from E. coli (Sigma), 10 μg/mL; flagellin from S. typhimurium (InvivoGen), 2 μg/mL; cardiolipin (Sigma), 10 μg/mL; albumin from human serum, low endotoxin (Sigma), 10 μg/mL; and KLH, endotoxin free (Millipore), 10 μg/mL. Plates were washed 4x with nanopure H2O with an ELISA plate washer (BioTek) and blocked with 150 μL/well blocking buffer [1X TBS-T (Thermo), 1mM EDTA (Boston BioProducts)] for 1 hour at 37 °C. Plates were washed 4x with H2O using an ELISA plate washer (BioTek) and 50 μL/well of mAbs pre-diluted in TBS pH 7.4 were added. mAbs were assayed at 1 μg/mL and three additional 1:4 dilutions. Strongly polyreactive positive control mAb 3H9 and four TBS-only wells were included on each plate. Plates were incubated at 37 °C for one hour then washed 4x with H2O. 75 μL/well of goat anti-human IgG HRP (Southern Biotech) diluted in blocking buffer were added and plates were incubated one hour at 37 °C then washed 4x with H2O. 150 μL/well of blocking buffer was added and plates incubated 5 mins at room temperature. Plates were washed 4x with H2O and 100 μL/well of developing reagent was added (Super AquaBlue ELISA Substrate, eBioscience). Plates were monitored at OD405 using an ELISA plate reader (BioTek) and reading was stopped when the 3H9 positive control reached an OD405 of ~3.0. Average OD405 of TBS-only wells was subtracted from mAb-containing wells for analysis. mAbs were considered polyreactive if they bound two or more antigens with an OD405 of ≥0.5 at a concentration of ≤ 1 μg/mL.
HEp2 ANA ELISAs
mAbs were assayed at 10 μg/mL and 3 additional 1:4 dilutions using the QUANTAlite ANA ELISA kit (INOVA Diagnostics) using the manufacturer’s protocol and provided reagents. High positive control mAb 3H9 was included on each plate, as were low positive and negative control serum provided by the manufacturer.
Free IgA ELISAs
Intestinal contents were resuspended at 0.1 mg/μL in PBS (Corning) with protease inhibitors (SIGMAFAST Protease Inhibitor Tablets, Sigma) and homogenized by taping the Eppendorf tube horizontally on a vortex and vortexing vigorously for 5 mins. Samples were centrifuged 5 mins at 400g and supernatants were filtered through a 70 μm sterile strainer (Fisher) and transferred to a new tube. This tube was centrifuged at 8000g for 5 mins. Supernatant was collected and assayed for free IgA by ELISA using the Bethyl Mouse IgA Quantitation Set according to the manufacturer’s protocol. Plates were read using an ELISA plate reader (BioTek) and samples values quantified by comparison to mouse IgA standards fit to a four-parameter logistic curve.
mAb in vivo serum retention experiments
mAbs were buffer exchanged into sterile HBSS using an Amicon 30K centrifugal filter and 2 μg was injected intravenously into Rag1−/− recipients. Blood was collected 5d after injection and serum isolated using serum separator tubes (BD). mAb concentration was measured by ELISA using goat anti-human IgK capture antibodies (Southern Biotech) and goat anti-human IgG HRP detection antibodies (Southern Biotech) and concentration determined by comparison to a standard curve as described above under ‘Single cell cloning and production of monoclonal antibodies.’ Polyreactive and non-polyreactive mAbs were randomly distributed among mice of similar age and gender.
Size exclusion chromatography
mAbs were purified from several pooled 293T culture supernatants with protein A agarose, eluted in Gly-HCl pH 3.0, and neutralized with 1M Tris-HCl pH 8.0 as described above. No BSA or additional preservatives were added. Samples were analyzed and monomers purified by fast protein liquid chromatography (FPLC) using a Superdex 200 column (GE). Purified monomers were immediately used in assays as indicated.
Microbial glycan microarrays
mAbs were diluted to 10 μg/mL or 1 μg/mL in 20 mM Tris-HCl pH 7.4 150mm NaCl 2mM CaCl 2mM MgCl 0.05% Tween 20 1% BSA and assayed against the microbial glycan microarray by the Consortium for Functional Glycomics at the Protein-Glycan Interaction Core (CFG) and National Center for Functional Glycomics at Beth Israel Deaconess Medical Center in Boston, MA. Raw data are publicly available under accession numbers CFG_3246, CFG_3329, and CFG_3402 (www.functionalglycomics.org).
MD4Tg cell transfers
Spleens from multiple CD45.1+ donors were pooled and depleted of non-B cells with biotinylated anti-CD4, CD8, CD3, NK1.1, CD11c, GR1, and TER119 antibodies and streptavidin magnetic beads (Miltenyi) using an autoMACS separator (Miltenyi). Depleted samples were counted and resuspended in PBS at 20 million cells per mL. 0.0312 μL of 5mM CellTrace Violet stock solution (Life Technologies) was added per mL and cells were incubated at 37 °C for 20 mins. Reaction was quenched by adding PBS 10% FCS and incubating at 37 °C for 5 mins. Cells were centrifuged 5 min at 500g, washed with HBSS 0.25% BSA, centrifuged again at 500g, washed with PBS, and resuspended in sterile HBSS. Small aliquots were taken for counting and staining to verify purity and fluorescent labeling (anti-CD19 and CD45.1). 20–30 million B cells were injected intravenously into 6-to-10-week-old MD4Tg recipients and analyzed 7d later. For single cell cloning, Peyer’s patches from multiple recipients were pooled and MACS-enriched for CD45.1+ cells and single CD19+CD45.1+ cells that had diluted CellTrace Violet were sorted.
Antigen-free diet experiments
Three-week-old germ-free mice were weaned onto either standard chow or antigen-free diets. The antigen-free diet consisted of an aqueous solution of vitamins and minerals administered ad libitum as well as a lipid supplement administered by daily oral gavage (0.25 mL/d). Aqueous diet was refilled as needed and fresh stock solutions were introduced into the isolator every two weeks. Both components were sterilized by filtration through a 0.22 μm sterile filter (Millipore). Mice were kept on the diet for four weeks and then analyzed. Diet was prepared as described (57, 58) according to the recipe in Table S1.
Statistical analysis
Paired or unpaired t test, chi-square test, or 2×2 Fisher’s exact test were performed as indicated in figure legends using GraphPad Prism.
Supplementary Material
Acknowledgments
We thank the University of Chicago Flow Cytometry Core for assistance with cell sorting, S. Owens and S. Greenwald at the Argonne National Laboratory Environmental Sample Preparation and Sequencing Facility for assistance with 16S sequencing, the University of Chicago DNA Sequencing Core for assistance with plasmid minipreps and sequencing, B. Theriault and the University of Chicago Gnotobiotic Facility for assistance with germ-free and antigen-free experiments, the Consortium for Functional Glycomics for microbial glycan microarray screening, G. Salzman for assistance with FPLC, and S. Canavan for assistance with figure preparation. This work was supported by NIH grants T32GM007281 and F30AI124476 (to J.J.B), grants U01AI125250, R01AI038339, R01AI108643, R01GM106173, and R01HL118092 (to A.B.), grant P41GM103694 to the National Center for Functional Glycomics at Beth Israel Deaconess Medical Center, and grant P30DK042086 to the University of Chicago Digestive Diseases Research Core Center (to A.B., B.J., and D.A.A.).
Footnotes
All data to understand and assess the conclusions of this research are available in the main text, supplementary materials, and via the following repositories: antibody sequences are available via GenBank accession numbers MF466210-MF467203 and 16S rRNA sequencing data are available via MG-RAST accession number MGP80334.
Author Contributions: J.J.B. and A.B. conceived the study; J.J.B., S.A.E., and A.B. designed research; A.B. supervised research; J.J.B. and S.A.E. performed experiments and analyzed data; C.H. and P.C.W. contributed anti-influenza mAbs and advice regarding mAb production and characterization; T.M.F., J.C.K., and D.A.A. processed samples for 16S sequencing and analyzed associated data; M.M. and B.J. provided mice and assistance with gnotobiotic experiments; J.J.B. and A.B. wrote the paper and all authors approved the final manuscript.
References and Notes
- 1.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 2.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 3.Slack E, et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. 2009;325:617–620. doi: 10.1126/science.1172747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bunker JJ, et al. Innate and Adaptive Humoral Responses Coat Distinct Commensal Bacteria with Immunoglobulin A. Immunity. 2015;43:541–553. doi: 10.1016/j.immuni.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macpherson AJ, et al. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 6.Reboldi A, et al. IgA production requires B cell interaction with subepithelial dendritic cells in Peyer’s patches. Science. 2016;352:aaf4822-1–10. doi: 10.1126/science.aaf4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fagarasan S, et al. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 2002;298:1424–1427. doi: 10.1126/science.1077336. [DOI] [PubMed] [Google Scholar]
- 8.Kubinak JL, Round JL. Do antibodies select a healthy microbiota? Nat Rev Immunol. 2016;16:767–774. doi: 10.1038/nri.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawamoto S, et al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity. 2014;41:152–165. doi: 10.1016/j.immuni.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 10.Kawamoto S, et al. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science. 2012;336:485–489. doi: 10.1126/science.1217718. [DOI] [PubMed] [Google Scholar]
- 11.Quan CP, Berneman A, Pires R, Avrameas S, Bouvet JP. Natural polyreactive secretory immunoglobulin A autoantibodies as a possible barrier to infection in humans. Infect Immun. 1997;65:3997–4004. doi: 10.1128/iai.65.10.3997-4004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fransen F, et al. BALB/c and C57BL/6 Mice Differ in Polyreactive IgA Abundance, which Impacts the Generation of Antigen-Specific IgA and Microbiota Diversity. Immunity. 2015;43:527–540. doi: 10.1016/j.immuni.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 13.Wijburg OL, et al. Innate secretory antibodies protect against natural Salmonella typhimurium infection. J Exp Med. 2006;203:21–26. doi: 10.1084/jem.20052093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benckert J, et al. The majority of intestinal IgA+ and IgG+ plasmablasts in the human gut are antigen-specific. J Clin Invest. 2011;121:1946–1955. doi: 10.1172/JCI44447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Shimoda M, Inoue Y, Azuma N, Kanno C. Natural polyreactive immunoglobulin A antibodies produced in mouse Peyer’s patches. Immunology. 1999;97:9–17. doi: 10.1046/j.1365-2567.1999.00755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho IY, et al. Refined protocol for generating monoclonal antibodies from single human and murine B cells. J Immunol Methods. 2016;438:67–70. doi: 10.1016/j.jim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mouquet H, Nussenzweig MC. Polyreactive antibodies in adaptive immune responses to viruses. Cell Mol Life Sci. 2012;69:1435–1445. doi: 10.1007/s00018-011-0872-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrews SF, et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med. 2015;7:316ra192. doi: 10.1126/scitranslmed.aad0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mouquet H, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wardemann H, et al. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 22.Notkins AL. Polyreactivity of antibody molecules. Trends Immunol. 2004;25:174–179. doi: 10.1016/j.it.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci U S A. 2009;106:19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cullender TC, et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14:571–581. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lodes MJ, et al. Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest. 2004;113:1296–1306. doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sitaraman SV, et al. Elevated flagellin-specific immunoglobulins in Crohn’s disease. Am J Physiol Gastrointest Liver Physiol. 2005;288:G403–406. doi: 10.1152/ajpgi.00357.2004. [DOI] [PubMed] [Google Scholar]
- 27.Lullau E, et al. Antigen binding properties of purified immunoglobulin A and reconstituted secretory immunoglobulin A antibodies. J Biol Chem. 1996;271:16300–16309. doi: 10.1074/jbc.271.27.16300. [DOI] [PubMed] [Google Scholar]
- 28.Sigounas G, Harindranath N, Donadel G, Notkins AL. Half-life of polyreactive antibodies. J Clin Immunol. 1994;14:134–140. doi: 10.1007/BF01541346. [DOI] [PubMed] [Google Scholar]
- 29.Kubinak JL, et al. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe. 2015;17:153–163. doi: 10.1016/j.chom.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palm NW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kau AL, et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med. 2015;7:276ra224. doi: 10.1126/scitranslmed.aaa4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Planer JD, et al. Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature. 2016;534:263–266. doi: 10.1038/nature17940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirpuri J, et al. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes. 2014;5:28–39. doi: 10.4161/gmic.26489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lecuyer E, et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40:608–620. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 35.Klaasen HL, et al. Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune system of mice. Infect Immun. 1993;61:303–306. doi: 10.1128/iai.61.1.303-306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atarashi K, et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krinos CM, et al. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature. 2001;414:555–558. doi: 10.1038/35107092. [DOI] [PubMed] [Google Scholar]
- 38.Lee JC, Takeda S, Livolsi PJ, Paoletti LC. Effects of in vitro and in vivo growth conditions on expression of type 8 capsular polysaccharide by Staphylococcus aureus. Infect Immun. 1993;61:1853–1858. doi: 10.1128/iai.61.5.1853-1858.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lebeer S, et al. Identification of a Gene Cluster for the Biosynthesis of a Long, Galactose-Rich Exopolysaccharide in Lactobacillus rhamnosus GG and Functional Analysis of the Priming Glycosyltransferase. Appl Environ Microbiol. 2009;75:3554–3563. doi: 10.1128/AEM.02919-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Logan SM. Flagellar glycosylation - a new component of the motility repertoire? Microbiology. 2006;152:1249–1262. doi: 10.1099/mic.0.28735-0. [DOI] [PubMed] [Google Scholar]
- 41.Gryllos I, et al. Induction of group A Streptococcus virulence by a human antimicrobial peptide. Proc Natl Acad Sci U S A. 2008;105:16755–16760. doi: 10.1073/pnas.0803815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamada N, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Needham BD, Trent MS. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol. 2013;11:467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coyne MJ, Reinap B, Lee MM, Comstock LE. Human symbionts use a host-like pathway for surface fucosylation. Science. 2005;307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- 45.van Houte S, Buckling A, Westra ER. Evolutionary Ecology of Prokaryotic Immune Mechanisms. Microbiol Mol Biol Rev. 2016;80:745–763. doi: 10.1128/MMBR.00011-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lebeer S, Vanderleyden J, De Keersmaecker SC. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat Rev Microbiol. 2010;8:171–184. doi: 10.1038/nrmicro2297. [DOI] [PubMed] [Google Scholar]
- 47.Stowell SR, et al. Microbial glycan microarrays define key features of host-microbial interactions. Nat Chem Biol. 2014;10:470–476. doi: 10.1038/nchembio.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peterson DA, et al. Characterizing the interactions between a naturally primed immunoglobulin A and its conserved Bacteroides thetaiotaomicron species-specific epitope in gnotobiotic mice. J Biol Chem. 2015;290:12630–12649. doi: 10.1074/jbc.M114.633800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol. 2012;12:821–832. doi: 10.1038/nri3322. [DOI] [PubMed] [Google Scholar]
- 50.Tiller T, et al. Autoreactivity in human IgG+ memory B cells. Immunity. 2007;26:205–213. doi: 10.1016/j.immuni.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindner C, et al. Diversification of memory B cells drives the continuous adaptation of secretory antibodies to gut microbiota. Nat Immunol. 2015;16:880–888. doi: 10.1038/ni.3213. [DOI] [PubMed] [Google Scholar]
- 52.Yeap LS, et al. Sequence-Intrinsic Mechanisms that Target AID Mutational Outcomes on Antibody Genes. Cell. 2015;163:1124–1137. doi: 10.1016/j.cell.2015.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cerutti A. The regulation of IgA class switching. Nat Rev Immunol. 2008;8:421–434. doi: 10.1038/nri2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mora JR, et al. Generation of gut-homing IgA-secreting B cells by intestinal dendritic cells. Science. 2006;314:1157–1160. doi: 10.1126/science.1132742. [DOI] [PubMed] [Google Scholar]
- 55.Litinskiy MB, et al. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat Immunol. 2002;3:822–829. doi: 10.1038/ni829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mestecky J. Mucosal immunology. 4. Elsevier/AP, Academic Press is an imprint of Elsevier; Amsterdam: 2015. [Google Scholar]
- 57.Kim KS, et al. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science. 2016;351:858–863. doi: 10.1126/science.aac5560. [DOI] [PubMed] [Google Scholar]
- 58.Pleasants JR, Johnson MH, Wostmann BS. Adequacy of chemically defined, water-soluble diet for germfree BALB/c mice through successive generations and litters. J Nutr. 1986;116:1949–1964. doi: 10.1093/jn/116.10.1949. [DOI] [PubMed] [Google Scholar]
- 59.Reynolds AE, Kuraoka M, Kelsoe G. Natural IgM is produced by CD5- plasma cells that occupy a distinct survival niche in bone marrow. J Immunol. 2015;194:231–242. doi: 10.4049/jimmunol.1401203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pasquier B, et al. Identification of FcalphaRI as an inhibitory receptor that controls inflammation: dual role of FcRgamma ITAM. Immunity. 2005;22:31–42. doi: 10.1016/j.immuni.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 61.Gomez de Aguero M, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351:1296–1302. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- 62.Koch MA, et al. Maternal IgG and IgA Antibodies Dampen Mucosal T Helper Cell Responses in Early Life. Cell. 2016;165:827–841. doi: 10.1016/j.cell.2016.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hapfelmeier S, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–1709. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moor K, et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature. 2017;544:498–502. doi: 10.1038/nature22058. [DOI] [PubMed] [Google Scholar]
- 65.Stokes CR, Soothill JF, Turner MW. Immune exclusion is a function of IgA. Nature. 1975;255:745–746. doi: 10.1038/255745a0. [DOI] [PubMed] [Google Scholar]
- 66.Williams RC, Gibbons RJ. Inhibition of bacterial adherence by secretory immunoglobulin A: a mechanism of antigen disposal. Science. 1972;177:697–699. doi: 10.1126/science.177.4050.697. [DOI] [PubMed] [Google Scholar]
- 67.Ichiyoshi Y, Casali P. Analysis of the structural correlates for antibody polyreactivity by multiple reassortments of chimeric human immunoglobulin heavy and light chain V segments. J Exp Med. 1994;180:885–895. doi: 10.1084/jem.180.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wardemann H, Hammersen J, Nussenzweig MC. Human autoantibody silencing by immunoglobulin light chains. J Exp Med. 2004;200:191–199. doi: 10.1084/jem.20040818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mussmann R, Du Pasquier L, Hsu E. Is Xenopus IgX an analog of IgA? Eur J Immunol. 1996;26:2823–2830. doi: 10.1002/eji.1830261205. [DOI] [PubMed] [Google Scholar]
- 70.Zhang YA, et al. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat Immunol. 2010;11:827–835. doi: 10.1038/ni.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martinoli C, Chiavelli A, Rescigno M. Entry route of Salmonella typhimurium directs the type of induced immune response. Immunity. 2007;27:975–984. doi: 10.1016/j.immuni.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 72.Craig VJ, et al. Gastric MALT lymphoma B cells express polyreactive, somatically mutated immunoglobulins. Blood. 2010;115:581–591. doi: 10.1182/blood-2009-06-228015. [DOI] [PubMed] [Google Scholar]
- 73.Isnardi I, et al. IRAK-4- and MyD88-dependent pathways are essential for the removal of developing autoreactive B cells in humans. Immunity. 2008;29:746–757. doi: 10.1016/j.immuni.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yurasov S, et al. Defective B cell tolerance checkpoints in systemic lupus erythematosus. J Exp Med. 2005;201:703–711. doi: 10.1084/jem.20042251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haynes BF, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 77.Tiller T, Busse CE, Wardemann H. Cloning and expression of murine Ig genes from single B cells. J Immunol Methods. 2009;350:183–193. doi: 10.1016/j.jim.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 78.Smith K, et al. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc. 2009;4:372–384. doi: 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Giudicelli V, Brochet X, Lefranc MP. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb Protoc. 2011;2011:695–715. doi: 10.1101/pdb.prot5633. [DOI] [PubMed] [Google Scholar]
- 81.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Walters W, et al. Improved Bacterial 16S rRNA Gene (V4 and V4–5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems. 2016;1:e00009–15. doi: 10.1128/mSystems.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Caporaso JG, et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 88.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meyer F, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Henry Dunand CJ, et al. Both Neutralizing and Non-Neutralizing Human H7N9 Influenza Vaccine-Induced Monoclonal Antibodies Confer Protection. Cell Host Microbe. 2016;19:800–813. doi: 10.1016/j.chom.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Henry Dunand CJ, et al. Preexisting human antibodies neutralize recently emerged H7N9 influenza strains. J Clin Invest. 2015;125:1255–1268. doi: 10.1172/JCI74374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dreyfus C, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.