Abstract
Systems-biology approaches in immunology take various forms, but here we review strategies for measuring a broad swath of immunological functions as a means of discovering previously unknown relationships and phenomena and as a powerful way of understanding the immune system as a whole. This approach has rejuvenated the field of vaccine development and has fostered hope that new ways will be found to combat infectious diseases that have proven refractory to classical approaches. Systems immunology also presents an important new strategy for understanding human immunity directly, taking advantage of the many ways the immune system of humans can be manipulated.
A systems approach to an area can be defined as a broad strategy for understanding how a complex set of interacting components works to produce certain outcomes. Long ago, prescient immunologists coined the term ‘immune system’ in recognition of just how well this label fits the field, but for much of its long history, most insights into the immune system came from painstaking examination of the parts and their largely individual properties and not so much of activities across the whole system. This is understandable, as there are more parts to the immune system than even the reputed 400 cheeses in France, with 350 CD (‘differentiation cluster’) antigens, over 100 cytokines and chemokines and at least that many cell subsets, and thousands of genes. This is daunting, to say the least, but advances in technology have now made it feasible to measure a good swath of this complexity in a relatively workable way. This has resulted in the discovery of new relationships and participants in immunology, insight into why some vaccines are more effective than others, and important connections with human diseases and mouse models of disease. These data are also now being used to model important aspects of the immune response, and this will be one of the most important goals.
Two main types of systems immunology can be seen in the literature, one of which uses this approach to elucidate signaling pathways that are important in the immune system1,2. This has resulted in a new understanding of these pathways in general and the creation of useful models of how different cells of the innate immune system function. These approaches have been very capably reviewed elsewhere3 and will not be discussed here. Instead, here we describe another type of systems approach to immunology that focuses on the cells and the molecules they use for communication and how they respond to vaccines, infections and so on. The reason for this is that the main effector activities of the immune system are mediated by specialized cells that communicate with each other and various tissues via cytokines and chemokines. These cells are relatively autonomous, and their only means of sensing what they need to do comes through their various cell-surface receptors. They can be mobilized en masse, but each and every cell must receive the correct signals. Thus, specialized cells are the principal unit of activity in the immune system. A CD4+ T cell can detect even a single complex of peptide and major histocompatibility complex (MHC) on another cell, and it serves as the detector, the signal integrator and the effector (in this case, by secreting cytokines)4,5. The more of a particular antigen that comes into the system, the more T cells with that specificity are recruited. Since white blood cells that make up the bulk of the response repertoire are largely mobile, with the exception of resident memory cells (which were once mobile), it is reasonable to think that they are all autonomous. Fortunately, the technology now exists to measure, en masse, most of the known cell types, their state and function, and their products (including cytokines, chemokines and metabolites) and the genes encoding those molecules6,7. This has led to a major change in how research is performed in many laboratories, since it is now feasible to use high-throughput and high-bandwidth methods for unbiased exploration of a variety of immune responses, which has revealed key participants that contribute to health and disease, and to generate new hypothesis and mechanistic studies (Fig. 1). This ability to generate large data sets has evolved together with technological advances in informatics and statistics that have now made it feasible to generate integrative models of the human immune response6–8. Thus, a systems approach with the objective of measuring the responses of much of the immune system comprehensively is quite feasible and is the principal subject of this Review. This is not meant to supplant traditional forms of enquiry in immunological research; indeed, as new phenomena are discovered, each must be carefully investigated down to its specific mechanism, and typically the tremendous power of the mouse system is the best way to do that, if the initial data are from studies of humans. So what do researchers hope to achieve with this type of systems immunology? First, it can be seen as a logical evolution of the field from an intense study of its parts to a broader and more integrated view of how those parts work together to produce particular results. Furthermore, it can reveal (and has revealed) unexpected examples of higher-order cooperativity between those parts, both cell types and pathways, that have provided a more complete idea of what is actually going on. Second, and perhaps at least as important, since cells and molecules and gene expression can be measured accurately in a blood sample (Fig. 1), it is providing a powerful view of the human immune system, which is much needed, especially because most of the methodologies now used for mouse immunology are not typically possible in human research. In contrast to the focus of most mouse immunology, there is a strong focus on blood in human studies, as this can be obtained easily from almost any willing human, healthy or not, and while the circulatory system is not an immunological organ, it is the conduit by which almost all important immunological traffic occurs. Being able to apply this to humans also provides the advantage of being able to assess the consequences of the thousands of immunological perturbations that humans are exposed to, meaning every vaccine, infectious disease, autoimmune disease, immunomodulatory treatment and so on. More broadly, it is helping researchers to overcome the inherent limitations of a field that has relied largely on a single experimental system, inbred mice, for which normal genetic variation has been eliminated and very few environmental influences are permitted. While such compromises were and are critical to solving key problems, they are, nonetheless, compromises, and their effects need to be considered and accounted for. This is where humans fit in perfectly. Humans are largely outbred and are exposed to myriad environmental stimuli with broad variations across the globe. That has resulted in the emergence of many important discoveries about basic immunology via human studies, such as the observation that negative selection is not nearly as efficient as thought on the basis of classic mouse experiments dating back 25 years9, and thus self-specific T cells are very abundant in healthy people, possibly because the threat of infectious disease is much greater from an evolutionary perspective than is the threat of autoimmunity.
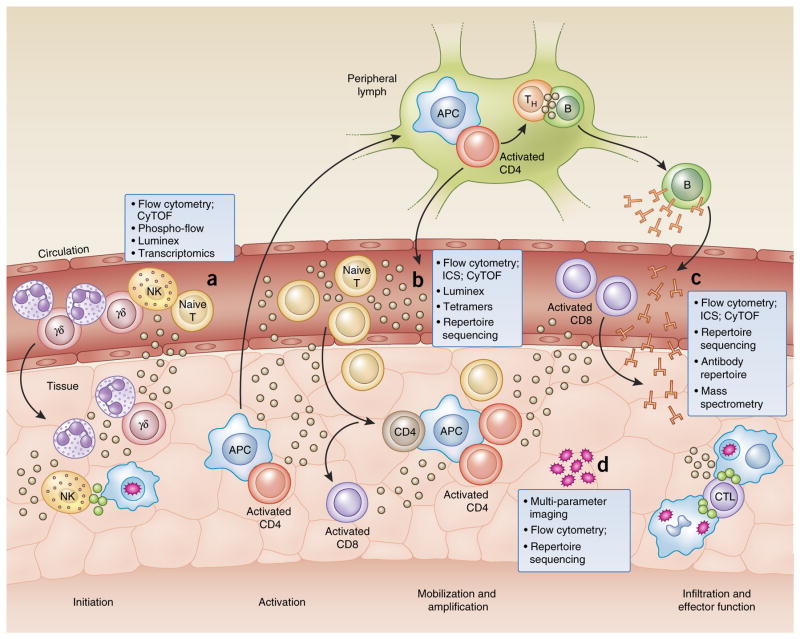
Figure 1.
Tools of the trade: a systems-immunology view. Investigating the immune system can be accomplished with a variety of techniques applied to the analysis of peripheral blood samples obtained from healthy donors, as well as those obtained at specific time points during infection, disease or after vaccination. Innate cell subsets can be characterized by RNA-seq, flow cytometry, cytometry by time-of-flight (CyTOF) and flow cytometry of phosphorylated proteins (Phospho-flow) to reveal mediators of early inflammation, and their signaling pathways (a). Cells can be classified by phenotype and functional capacity on the basis of a variety of cell-surface markers and receptor expression. Effector proteins, such as cytokines and cytolytic granules, along with other products of activated cells, can be detected intracellularly by flow cytometry or in the serum with other methodologies, such as bead-based multiplex assays (Luminex) or immunoassay kits (from Mesoscale Discovery), to rapidly survey more than 100 analytes in a single, small sample (b). Cell subsets of the adaptive immune system can be investigated for antigen specificity by tetramer technology and mass spectrometry to explore peptide presentation, as well as sequencing of the TCR and BCR repertoire for the identification of clonally expanded populations. Furthermore, antibody-repertoire sequencing can be used to characterize the extent of isotype class switching and affinity maturation (c). Many of these techniques are currently being adapted for the analysis of smaller tissue-biopsy samples to extract information about the local inflammatory environment and the infiltrating cell populations (d). s APC, antigen-presenting cell; TH, helper T cell; B, B cell; CD4, CD4+ T cell; NK, natural killer cell; γδ, γδ T cell; T, T cell; ICS, intracellular cytokine staining; CD8, CD8+ T cell; CTL, cytotoxic T lymphocyte.
The need for teamwork
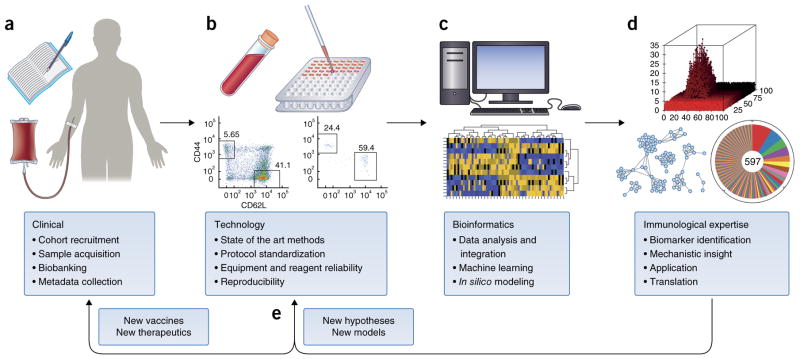
Perhaps one the greatest barriers to implementing projects in systems immunology is that a variety of high-level skill sets are needed. Just the fact that a team approach is needed is a major barrier in modern biology, in which researchers have long subscribed to the mythology of the lone hero going into the cave and slaying the dragon. Perhaps that worked in the middle ages, but today’s reality is that cooperative approaches are more and more the norm as the complexities of modern work extend beyond even the most talented people. The good news is that most medical research centers have the requisite skills, but there just must be a willingness to work together. The core skills are clinical, technical, bioinformatics and immunological (Fig. 2). Clinical skills are required for the obvious reason that this typically involves live human beings, not to mention the wealth of knowledge needed, especially if the study is about a particular disease and the collection and collation of valuable meta-data from each participant. Bioinformatics, with inputs from computer sciences and statistics, is essential because this systems approach requires the ability to manipulate large data sets to extract meaning, but standardized user-friendly analysis systems are not yet available. Crucial to the success of any systems study is the interaction of bioinformatician and immunologists. Expertise in the field of immunology is needed to direct focus on biologically relevant results obtained from computational analyses, to interpret the mechanistic implications according to immunological theory and, ultimately, to design new experimental questions and models and to introduce translational applications. Finally, technology is a must, because a systems-immunology approach requires instrumentation that can extract large amounts of information rapidly and relatively inexpensively from often fairly small samples. Ideally, this means a core facility with the most-advanced equipment, and especially valuable is one in which there are trained staff to run the assays. This is important not only for reproducibility and reliability but also to free research personnel and trainees to focus on the data. Here at Stanford University School of Medicine we have such a facility, the Human Immune Monitoring Center, which fulfills this role extremely well and has enabled hundreds of studies, mostly but not exclusively human, some large and many small (Figs. 1 and 2). The other aspect of technology that has been especially critical to advancing human sudies is the input of research groups and bioengineers who are developing new technologies or who are early adopters, keeping investigators and facilities up to date on which technology is the best to use.
Figure 2.
The ideal team. Cross-disciplinary efforts have allowed considerable advances in human medical research. The integration of clinical research (a) was made possible by the invention of new technologies (b) for comprehensive analysis of precious blood and tissue samples. The development of new algorithms for the visualization and analysis of such large data sets (c) has enabled novel insights and the generation of new hypotheses (d). Such collaborations should help to bridge the gap between bench and bedside by accelerating the translational pipeline (e).
The need for innovative technologies and analyses
Since many of the standard manipulations used for the study of mouse immunology cannot be used in ethical work studying humans, there has been a strong need for technologies that can extract as much information as possible from often very limited clinical specimens, particularly blood. A mainstay here has been gene-expression arrays, which, while not a new technology, when applied to whole-blood analysis, are very ‘noisy’ because of the wide variation in subsets of human white blood cells. This results in a gene-expression pattern that is more reflective of particular person’s cell types rather than differences in the content of mRNA per cell. Another problem is that of ‘false discovery’, in which the sheer number of genes being surveyed increases the probability of finding a difference that is an artifact. Fortunately, in the latter case, grouping co-expressed genes into modules whose products act in concert provides way to find true changes that is both simplified and statistically valid10,11, and for the former problem, methods have been developed to deconvolute gene-expression data on the basis of minimal information on the frequency of immune-cell subsets from healthy people and in patients with disease as well12–16. Particularly promising in this context is the CIBERSORT program, which can predict cell frequencies from gene-expression data and has thus opened up the large amount of gene-expression data in the public domain for analysis17. A third challenge in data science is to deal with so-called ‘overfitting’, a statistical concept that refers to the propensity of a model to capture ‘noise’ in the data rather than a genuine relationship, which is often the result of models that are too complex. In such cases, the model learned from a given data set and by a given statistical method will probably fail to generalize to additional data sets. Various methods have been developed to avoid this problem. For example, in regression analysis, regularization methods impose mathematical constraints on the regression coefficients, which enables ‘simpler’ solutions—in other words, the selection of models with fewer immunological components that are found to be relevant for a given mathematical task, which thus avoids overfitting18–20. Finally, another major analytical effort has focused on deriving key gene ‘signatures’ from data obtained by microarray or RNA-based next-generation sequencing (RNA-seq) to elucidate key events in human immunity, such as the progression to active disease in tuberculosis21,22 or the differences in the immune response to infection with influenza virus versus the response to other viral or bacterial infections23. Methodologies such as these have great potential to affect clinical diagnostic abilities, as well as the ability to monitor and predict responses to treatment and recovery from disease.
Technological advances have been equally important, and while most are discussed in the context of the sections below, those involving ‘deep’ cell phenotyping and repertoire analysis have been the most important. In particular, the invention of mass cytometry and its subsequent development24–26 was a watershed moment during which research went from 12 colors, with flow cytometry, to 25 and 45 or more labels on single cells; this essentially redefined all the major subsets of white blood cells, given the wealth of data now possible27–29. It can also work on such small numbers of cells (fewer than 1 × 106 peripheral blood mononuclear cells) that studies of samples obtained from pediatric subjects are more easily performed30,31. Additional technological advances have been extremely useful in the functional genomics arena. For example, methods using the CRISP-Cas9 system, such as Perturb-Seq32, CROP-Seq33 and CRISP-Seq34, have enabled the investigation of the effect of hundreds or thousands of genetic changes in parallel. By taking advantage of RNA-seq as a ‘readout’, these approaches overcome the limitations of previous ones and allow the analysis of perturbations and the phenotype of each cell simultaneously.
Vaccination responses and infectious diseases
Responses to vaccination have received the lion’s share of attention in these early days of systems approaches because of both the need for more-effective vaccines and the relative ease with which approved vaccines can be administered to human volunteers to probe their ability to elicit an immune response. Vaccines also tend to ‘push many buttons’ in the immune system, so it has been fruitful to compare different vaccine responses in various cohorts. It even has its own subdivision as ‘systems vaccinology’35. The first broad analysis of the effect of a vaccine was research that focused on the vaccine against yellow fever36,37, believed to be one of the most successful in terms of its nearly universal response rate. Those early papers focused largely on transcriptional responses, but subsequent work incorporated more of the breadth of the immune response, including serum cytokines, cell phenotyping and functional immunological assays to study the mechanisms of vaccination against that and other diseases, such as pneumonia, measles and influenza38–45. The vaccine against influenza is particularly interesting as a standard vaccine to study, as it has a markedly variable response rate, with ~80% of younger people and less than 30% of older people judged to be stimulated enough for a protective response46. That is a dismal success rate, and the reasons for it have been a chief target of many papers in this nascent area. For the RTS,S candidate vaccine against malaria, which confers substantial protection against infection with Plasmodium falciparum in humans, research has demonstrated the utility of systems approaches in identifying molecular and cellular signatures associated with protection and immunogenicity. In particular, both the titers of immunoglobulin G antibodies to circumsporozoite protein and the population expansion of circumsporozoite-protein-specific polyfunctional CD4+ T cells producing a combination of the B cell–stimulatory molecule CD40L and the cytokines IL-2, TNF and IFN-γ after vaccination are predictive of protection against challenge with the pathogen47. Mechanisms reported to contribute to vaccine responses include a failure in the induction of cell death42, the metabolism of lipids and endocrine factors that suppress immune-system function44, prior exposure to infection48, and nutrient-sensing pathways, including the sensor GCN2, which mediates T cell responses to vaccination49. There is also a particularly interesting connection to the microbiota, in that stimulation of the receptor TLR5 on B cells in mice by microbiota-derived flagellin is crucial to the maturation of those cells into antibody-secreting cells50. Those are among some of the mechanistic insights revealed by systems studies that are helping researchers to consider and weigh the effect of factors that contribute to the variability of individual human responses to vaccination and to holistically incorporate variables derived from the host, pathogen, environment and vaccine (formulation and route of delivery). Furthermore, such systems approaches to studying vaccination responses have great potential to elucidate ways in which existing vaccines can be improved through stimulation of parts of the immune system that are not being stimulated by the current vaccines. There is also the hope that stimuli can be identified that could even generate protective responses to pathogens and diseases that have so far proven refractory to standard approaches, such as human immunodeficiency virus, malaria, dengue virus or tuberculosis in adults. This has given new life to the vaccine field, which has been facing major challenges with such diseases for which traditional approaches have failed to produce efficacious vaccines.
In addition, longitudinal studies are particularly well suited for inferring the mechanisms of action of vaccines. For example, in a study monitoring the immune system, samples were obtained every 4 weeks for a period of 8 months from patients infected with human immunodeficiency virus who were receiving a therapeutic vaccine; these showed a substantial increase in the number of polyfunctional T cells specific for human immunodeficiency virus, associated with control of viral load51. In another longitudinal study, samples were obtained periodically from young adults who received the seasonal vaccine against influenza virus, the 13-strain pneumococcal vaccine or a placebo. Gene-expression analysis showed unique and distinct signatures elicited by those vaccines52.
Aging
Vaccines in human studies have another use as a general stimulant of multiple components of the immune system for the study of the immunological consequences of various conditions, such as the influence of genetics, neoplasia and so on. This has particular relevance to aging, as there is clearly a sharp drop in the effectiveness of the immune system as most people age, especially over the 65- to 70-year mark. Early investigations noted a sharp increase in the number of CD28-CD8+ T cells as people age, which correlates with loss of function53, but with the broad approaches now possible, many more aspects of immunity can be surveyed. In particular, there seems to be a loss of the T cell repertoire54 that might relate to thymic involution and the subsequent slowing of the production of new T cells to a trickle. However, involution happens in a person’s 20s, and repertoire depletion happens many decades later55, so is that the real reason? Work on aging has demonstrated strong links between chronic inflammation in the immune system and cardiovascular disease. A longitudinal study of aging people has shown a widespread elevation of phosphorylated transcription factors of the STAT family, especially in lymphocytes from subjects in their 70s and 80s, which suggests that these lymphocytes had been exposed to inflammatory cytokines56. That takes advantage of the fact that lymphocytes are continually circulating throughout the body and thus potentially ‘report on’ pathologies that might be physically distant. The presence of those hyper-phosphorylated STAT molecules often correlated with an elevation in inflammatory cytokines in the blood and diminished ability of cells from these subjects to respond to cytokine stimulation in vitro. That cytokine unresponsiveness was then linked to atherosclerosis and other measures of cardiovascular disease. Another study of that same longitudinal cohort noted, from transcriptional data in over 5 years, that certain gene modules showing enrichment for genes encoding components inflammasomes correlated with older subjects with persistent hypertension and a lack of longevity in their family history57. That led to a metabolomics analysis that discovered aberrant nucleotide metabolism in these subjects and two previously unknown danger-associated molecular patterns that stimulated the expression of inflammasomes in vitro. In addition, those compounds caused hypertension and infiltration of lymphocytes into the kidneys of mice57. This shows how casting a broad net with a systems approach can yield important insight into a common human disease (hypertension) about which very little is known mechanistically. It also adds important specifics about the nature of chronic inflammation that is often seen in the elderly and is linked to many diseases of the elderly, especially cardiovascular disease.
Cancer
While neoplasia takes many forms, the relationship between a particular cancer type and the immune system is clearly a complex and little-understood topic that could benefit from a systems approach. The clinical relevance of having a better understanding is clear, since while new immunotherapeutic approaches involving either checkpoint inhibitors or chimeric antigen receptor (CAR) T cells have beneficial effects, the former has been effective in only a fraction of patients and the latter has not yet been effective with solid tumors. Additionally, better definition of the parameters of anti-tumor immunity will support the more-informed design of vaccines for the induction of clinical immunity to tumor antigens, which has so far fallen short of experimental expectations58. The basis of any of these deficiencies is not understood, so better understanding of the immune-system dynamics is clearly needed and could provide the basis for a new generation of anti-cancer vaccines coupled with tolerance-inhibiting agents and/or chemotherapeutics and radiation. Studies aimed at understanding the effects of checkpoint inhibitors, in which a combination of genomic and systems immunology were applied along with bioinformatics analysis, have found that this immunotherapy is associated with T cell responses to mutant tumor antigens that are often reactivated upon treatment with immune-checkpoint inhibitors59–61. Furthermore, systems analysis of a mouse model of cancer has illustrated that multiple components of the immune system are involved and has provided a template and rationale for human studies along these lines62.
Autoimmunity
The field of autoimmunity has great potential for systems studies, as clearly in autoimmunity there has been some disturbance to the normally tight regulation of self tolerance, but surprisingly few studies have been done that could be considered systems immunology. One such study has shown that administration of gluten to patients with celiac disease not only induces the expected surge in gliadin-specific CD4+ T cells 6 days after challenge but also induces oligoclonal CD8+ and γδ T cells that appear and disappear from the blood with similar kinetics63. This suggests that there is a complexity to the T cell response in patients with celiac disease that had not been appreciated previously and that this might be a factor in other diseases as well. Particularly relevant in the field of autoimmune diseases, since most of these are sex biased, are the differences in the activity of immune cells in males versus that in females. Applications of systems immunology to this area have just begun, and initial data show gross sex differences for healthy young adults, but for not older adults, in particular subsets of cells, including blood neutrophils, in their maturation status and responses to sera from patients with systemic lupus erythematosus64 that were previously unappreciated. In the domain of systemic lupus erythematosus, work has shown that the application of systems approaches to these patients enables accurate identification of molecular networks that stratify disease activity65.
Genetics
The widespread use of powerful genetic analyses has resulted in many large studies aimed at finding genes or polymorphisms that underlie various immunological diseases, particularly autoimmunity. While such studies have identified many interesting relevant loci, their contribution to disease risk is typically very small. Also, the role of environmental influences is typically not studied directly. For this reason, we embarked on an analysis of twins, both monozygotic and dizygotic, which concluded that for over 200 immunological traits, including responses to the vaccine against influenza virus, in most cases non-heritable influences were dominant, and that infection with cytomegalovirus had a substantial effect on most of these variables66. That supports the proposal that the immune system is (and must be) a highly adaptable system, and not just in the specific T lymphocytes or B lymphocytes that it produces but also in its overall makeup. It is also worth noting that because the immune system utilizes hundreds, if not thousands, of genes, the genetic load is considerable, which means that most people will have some non-functional or poorly functional genes. While this will in most cases be ameliorated by heterozygosity, it could still be deleterious, were it not for the redundancy that is continually seen in the system. This could explain the finding that despite the genetic load, susceptibility to serious infectious disease is very rare, although susceptibility to less-serious versions is not, consistent with published information on this67–69.
BCR and TCR repertoire analysis
One important aspect of an immune response that is just starting to be captured adequately for the purposes of systems immunology is the response repertoire of T lymphocytes and B lymphocytes. This is being made possible by both the rapid progress of DNA-sequencing technology and the development or application of software that is able to capture the diversity or other aspects of B cell or T cell responses. Studies of antibody responses first used high-throughput sequencing to analyze B cell responses70,71. This work was advanced further by the pioneering of single-cell sequencing of tens of thousands of B cells simultaneously72. Specific, common immunoglobulin sequence motifs have also been noted that correlate with particular specificities, which indicates that it might be possible to infer the specificity and potential efficacies of a response from sequence alone73.
In the T cell area, the ability of mass cytometry to analyze many labeled molecules simultaneously at the single-cell level led to the description of hundreds of different functional variants of human CD8+ T cells, which showed a nearly stochastic array of cytokine combinations28. Combinatorial peptide–MHC staining also has allowed over 100 different T cell specificities to be surveyed in a standard human blood sample, which has greatly expanded the ability to track T cell responses in any immune response74. Using such methods to study human responses has been a necessity, but the fact that these natural infections and vaccine responses are so diverse calls into question the current practice in mouse immunology of monitoring single-cell MHC class I– or class II–restricted epitopes of ovalbumin in a given disease model. Also, just as with analysis of the repertoire of the B cell antigen receptor (BCR) response, the explosive growth and lower cost of DNA sequencing has greatly empowered the application of that technology to analysis of the TCR repertoire. Thus, there is widescale use of both sequencing of the TCRs of different populations of T cells and high-throughput single-cell methods. Particularly noteworthy in the application of bulk sequencing of TCRs of specific T cell populations is the demonstration that the initial T cell responses to specific pathogen types result in a broad array of CD4+ T cell functionality75, but later analysis finds a much narrower, frequently monomorphic range76. In single-cell analyses, substantial clonal expansion of most CD4+ T cells in a colon carcinoma tumor from a human patient has been shown, as well as a robust method for obtaining both α-chain and β-chain TCR sequences and phenotypic information77. Single-cell RNA-seq methods have also been applied to T lymphocytes, but here the relatively small amount of mRNA in T cells has limited the ability of this method to derive TCR sequences, although some have reported success78. For analysis strategies, while BCR sequences can often be grouped according to specificity by the observation of a cluster of somatic hypermutated immunoglobulin sequences, this mechanism does not exist in TCRs, so a major question becomes whether similar, but not identical, TCR sequences are really ‘seeing’ the same peptide–MHC ligand. In this context, computational methods hold the promise of being able to group TCR sequences in accordance to their specificities79,80 (Glanville, J. et al. personal communication), which would greatly aid the ability to interpret the data.
Overall, it seems likely that databases of BCRs and TCRs organized around the responses to specific pathogens will become an increasingly valuable resource with which to interpret adaptive immune responses. Questions about whether the diversity of the BCR or TCR response matters in terms of protection become very important in vaccine development, for example. Alternatively, is there a specific BCR or TCR specificity that is critical to protection? In the case of antibody responses, this is clearly the case, as many antibody specificities are clearly not effective in inhibiting viral entry, for example. In T cell responses, it remains unknown whether a diverse response is the most critical factor or whether particular specificities are important. Thus, it is the hope that methods such as these will enlighten understanding not only of the evolution of the immune response within a host but also the features of protection at the molecular level.
Summary and future directions
While a good beginning has been made in the systems-immunology approach described here, much more is ahead, as immunologists have barely scratched the surface of many aspects. Also, there are major challenges in defining mechanistic and causal relationships. Right now research in this field is largely dependent on mice, but there will doubtless be situations for which inbred mice will not mimic a human pathway. In this context, humanized mice are one alternative but are still a work in progress. An alternative that should be developed more widely would be in vitro cell and organ systems that are entirely human and functional, which would allow mechanistic studies. Particularly important in this are the implications of systems immunology for medicine81, in which this approach could open many doors for the exploration of both human health and the myriad diseases that affect humans (Fig. 3). While it is known that many diseases involve inflammation, for example, this has not been explored with systems approaches in more than a few cases, so many specifics have yet to be discovered. Furthermore, while it is currently more difficult to discern biological meaning in the adaptive immune responses to an infection as compared to innate responses, the newer methods for analyzing the TCR or BCR repertoire are making it much easier to track this in people, so we think that there are many surprises in store here, such as a study showing the ability of lymphocyte physiology to be predictive of recovery following surgery82,83. Currently, general medical practice and diagnosis do not make much use of immunology or appreciate how much it has advanced in the past decades. A case in point is that the main immunological metrics used widely in medicine are white blood cell counts and the complete blood cell count; the former was developed in 1915 and the latter was developed in 1959, it is time for an ‘upgrade’!
Figure 3.
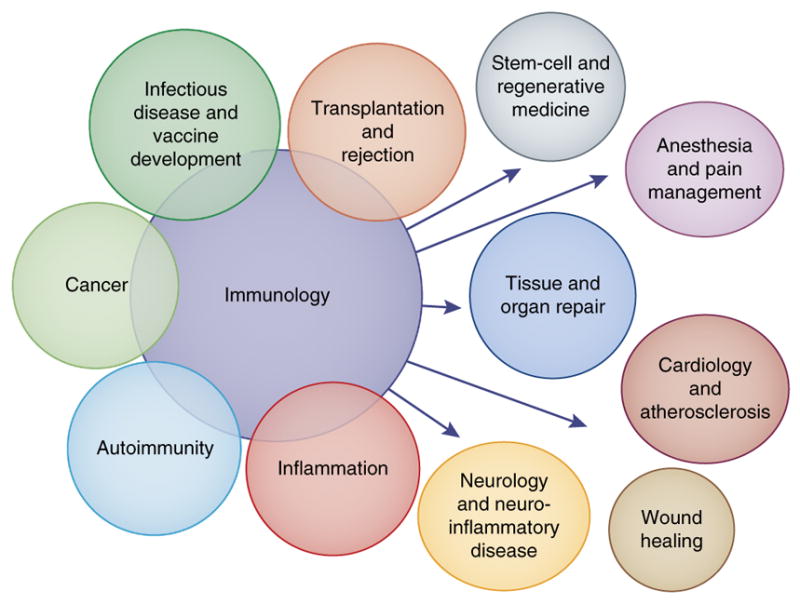
A comprehensive picture of health. The immune system has a fundamental role in a variety of diseases and disorders, including the capacity for resistance, recovery and maintenance of the health of a person.
Acknowledgments
Supported by the Howard Hughes Medical Institute, The Parker Institute for Cancer Immunotherapy and the US National Institutes of Health (U19 AI 057229).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Morrissette NS, et al. Isolation and characterization of monoclonal antibodies directed against novel components of macrophage phagosomes. J Cell Sci. 1999;112:4705–4713. doi: 10.1242/jcs.112.24.4705. [DOI] [PubMed] [Google Scholar]
- 2.Aderem A. Phagocytosis and the inflammatory response. J Infect Dis. 2003;187:S340–S345. doi: 10.1086/374747. [DOI] [PubMed] [Google Scholar]
- 3.Lienert F, Lohmueller JJ, Garg A, Silver PA. Synthetic biology in mammalian cells: next generation research tools and therapeutics. Nat Rev Mol Cell Biol. 2014;15:95–107. doi: 10.1038/nrm3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. Direct observation of ligand recognition by T cells. Nature. 2002;419:845–849. doi: 10.1038/nature01076. [DOI] [PubMed] [Google Scholar]
- 5.Huang J, et al. A single peptide-major histocompatibility complex ligand triggers digital cytokine secretion in CD4(+) T cells. Immunity. 2013;39:846–857. doi: 10.1016/j.immuni.2013.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hood L, Heath JR, Phelps ME, Lin B. Systems biology and new technologies enable predictive and preventative medicine. Science. 2004;306:640–643. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- 7.Furman D, Davis MM. New approaches to understanding the immune response to vaccination and infection. Vaccine. 2015;33:5271–5281. doi: 10.1016/j.vaccine.2015.06.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kidd BA, Peters LA, Schadt EE, Dudley JT. Unifying immunology with informatics and multiscale biology. Nat Immunol. 2014;15:118–127. doi: 10.1038/ni.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu W, et al. Clonal deletion prunes but does not eliminate self-specific αβ CD8+ T lymphocytes. Immunity. 2015;42:929–941. doi: 10.1016/j.immuni.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segal E, et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 11.Chaussabel D, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen-Orr SS, et al. Cell type-specific gene expression differences in complex tissues. Nat Methods. 2010;7:287–289. doi: 10.1038/nmeth.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y, Simon R. Gene expression deconvolution in clinical samples. Genome Med. 2010;2:93. doi: 10.1186/gm214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong Y, Liu Z. Gene expression deconvolution in linear space. Nat Methods. 2011;9:8–9. doi: 10.1038/nmeth.1830. [DOI] [PubMed] [Google Scholar]
- 15.Shen-Orr SS, Gaujoux R. Computational deconvolution: extracting cell type-specific information from heterogeneous samples. Curr Opin Immunol. 2013;25:571–578. doi: 10.1016/j.coi.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Oh WK, Zhu J. Disease-specific classification using deconvoluted whole blood gene expression. Sci Rep. 2016;6:32976. doi: 10.1038/srep32976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tikhonov AN. Solution of incorrectly formulated problems and the regularization method. Soviet Mathematics Doklady. 1963;4:1035–1038. [Google Scholar]
- 19.Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc B. 1996;58:267–288. [Google Scholar]
- 20.Zou HaH T. Regularization and variable selection via the elastic net. J R Stat Soc B. 2005;67:301–320. [Google Scholar]
- 21.Zak DE, et al. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet. 2016;387:2312–2322. doi: 10.1016/S0140-6736(15)01316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sweeney TE, Braviak L, Tato CM, Khatri P. Genome-wide expression for diagnosis of pulmonary tuberculosis: a multicohort analysis. Lancet Respir Med. 2016;4:213–224. doi: 10.1016/S2213-2600(16)00048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andres-Terre M, et al. Integrated, multi-cohort analysis identifies conserved transcriptional signatures across multiple respiratory viruses. Immunity. 2015;43:1199–1211. doi: 10.1016/j.immuni.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu P, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bendall SC, Nolan GP. From single cells to deep phenotypes in cancer. Nat Biotechnol. 2012;30:639–647. doi: 10.1038/nbt.2283. [DOI] [PubMed] [Google Scholar]
- 27.Horowitz A, et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med. 2013;5:208ra145. doi: 10.1126/scitranslmed.3006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36:142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simoni Y, et al. Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity. 2017;46:148–161. doi: 10.1016/j.immuni.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kay AW, Strauss-Albee DM, Blish CA. Application of mass cytometry (CyTOF) for functional and phenotypic analysis of natural killer cells. Methods Mol Biol. 2016;1441:13–26. doi: 10.1007/978-1-4939-3684-7_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao Y, et al. CyTOF supports efficient detection of immune cell subsets from small samples. J Immunol Methods. 2014;415:1–5. doi: 10.1016/j.jim.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dixit A, et al. Perturb-Seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell. 2016;167:1853–1866. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Datlinger P, et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods. 2017;14:297–301. doi: 10.1038/nmeth.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaitin DA, et al. Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-Seq. Cell. 2016;167:1883–1896. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 35.Pulendran B. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nat Rev Immunol. 2009;9:741–747. doi: 10.1038/nri2629. [DOI] [PubMed] [Google Scholar]
- 36.Gaucher D, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205:3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Querec TD, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haralambieva IH, et al. Transcriptional signatures of influenza A/H1N1-specific IgG memory-like B cell response in older individuals. Vaccine. 2016;34:3993–4002. doi: 10.1016/j.vaccine.2016.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haralambieva IH, et al. Whole transcriptome profiling identifies CD93 and other plasma cell survival factor genes associated with measles-specific antibody response after vaccination. PLoS One. 2016;11:e0160970. doi: 10.1371/journal.pone.0160970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ovsyannikova IG, et al. Gene signatures associated with adaptive humoral immunity following seasonal influenza A/H1N1 vaccination. Genes Immun. 2016;17:371–379. doi: 10.1038/gene.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakaya HI, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furman D, et al. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Mol Syst Biol. 2013;9:659. doi: 10.1038/msb.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price JV, et al. Characterization of influenza vaccine immunogenicity using influenza antigen microarrays. PLoS One. 2013;8:e64555. doi: 10.1371/journal.pone.0064555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furman D, et al. Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination. Proc Natl Acad Sci USA. 2014;111:869–874. doi: 10.1073/pnas.1321060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li S, et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol. 2014;15:195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goodwin K, Viboud C, Simonsen L. Antibody response to influenza vaccination in the elderly: a quantitative review. Vaccine. 2006;24:1159–1169. doi: 10.1016/j.vaccine.2005.08.105. [DOI] [PubMed] [Google Scholar]
- 47.Kazmin D, et al. Systems analysis of protective immune responses to RTS,S malaria vaccination in humans. Proc Natl Acad Sci USA. 2017;114:2425–2430. doi: 10.1073/pnas.1621489114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furman D. Sexual dimorphism in immunity: improving our understanding of vaccine immune responses in men. Expert Rev Vaccines. 2015;14:461–471. doi: 10.1586/14760584.2015.966694. [DOI] [PubMed] [Google Scholar]
- 49.Ravindran R, et al. Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science. 2014;343:313–317. doi: 10.1126/science.1246829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oh JZ, et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014;41:478–492. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lévy Y, et al. Dendritic cell-based therapeutic vaccine elicits polyfunctional HIV-specific T-cell immunity associated with control of viral load. Eur J Immunol. 2014;44:2802–2810. doi: 10.1002/eji.201344433. [DOI] [PubMed] [Google Scholar]
- 52.Obermoser G, et al. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013;38:831–844. doi: 10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weng NP, Akbar AN, Goronzy J. CD28- T cells: their role in the age-associated decline of immune function. Trends Immunol. 2009;30:306–312. doi: 10.1016/j.it.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qi Q, et al. Diversity and clonal selection in the human T-cell repertoire. Proc Natl Acad Sci USA. 2014;111:13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goronzy JJ, Fang F, Cavanagh MM, Qi Q, Weyand CM. Naive T cell maintenance and function in human aging. J Immunol. 2015;194:4073–4080. doi: 10.4049/jimmunol.1500046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen-Orr SS, et al. Defective signaling in the JAK-STAT pathway tracks with chronic inflammation and cardiovascular risk in aging humans. Cell Systems. 2016;3:374–384. doi: 10.1016/j.cels.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Furman D, et al. Expression of specific inflammasome gene modules stratifies older individuals into two extreme clinical and immunological states. Nat Med. 2017;23:174–184. doi: 10.1038/nm.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Romero P, et al. The Human Vaccines Project: A roadmap for cancer vaccine development. Sci Transl Med. 2016;8:334ps9. doi: 10.1126/scitranslmed.aaf0685. [DOI] [PubMed] [Google Scholar]
- 59.Gubin MM, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Rooij N, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31:e439–e442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spitzer MH, et al. Systemic immunity is required for effective cancer immunotherapy. Cell. 2017;168:487–502. doi: 10.1016/j.cell.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han A, et al. Dietary gluten triggers concomitant activation of CD4+ and CD8+ αβ T cells and γδ T cells in celiac disease. Proc Natl Acad Sci USA. 2013;110:13073–13078. doi: 10.1073/pnas.1311861110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blazkova J, et al. Multicenter Systems analysis of human blood reveals immature neutrophils in males and during pregnancy. J Immunol. 2017;198:2479–2488. doi: 10.4049/jimmunol.1601855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Banchereau R, et al. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell. 2016;165:551–565. doi: 10.1016/j.cell.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brodin P, et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Casanova JL. Severe infectious diseases of childhood as monogenic inborn errors of immunity. Proc Natl Acad Sci USA. 2015;112:E7128–E7137. doi: 10.1073/pnas.1521651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Casanova JL. Human genetic basis of interindividual variability in the course of infection. Proc Natl Acad Sci USA. 2015;112:E7118–E7127. doi: 10.1073/pnas.1521644112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Conley ME, Casanova JL. Discovery of single-gene inborn errors of immunity by next generation sequencing. Curr Opin Immunol. 2014;30:17–23. doi: 10.1016/j.coi.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boyd SD, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med. 2009;1:12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weinstein JA, Jiang N, White RA, III, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324:807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Robinson WH. Sequencing the functional antibody repertoire--diagnostic and therapeutic discovery. Nat Rev Rheumatol. 2015;11:171–182. doi: 10.1038/nrrheum.2014.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jackson KJ, et al. Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell Host Microbe. 2014;16:105–114. doi: 10.1016/j.chom.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Newell EW, et al. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nat Biotechnol. 2013;31:623–629. doi: 10.1038/nbt.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Warren RL, et al. Exhaustive T-cell repertoire sequencing of human peripheral blood samples reveals signatures of antigen selection and a directly measured repertoire size of at least 1 million clonotypes. Genome Res. 2011;21:790–797. doi: 10.1101/gr.115428.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Becattini S, et al. T cell immunity. Functional heterogeneity of human memory CD4+ T cell clones primed by pathogens or vaccines. Science. 2015;347:400–406. doi: 10.1126/science.1260668. [DOI] [PubMed] [Google Scholar]
- 77.Han A, Glanville J, Hansmann L, Davis MM. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol. 2014;32:684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stubbington MJ, et al. T cell fate and clonality inference from single-cell transcriptomes. Nat Methods. 2016;13:329–332. doi: 10.1038/nmeth.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heather JM, Ismail M, Oakes T, Chain B. High-throughput sequencing of the T-cell receptor repertoire: pitfalls and opportunities. Brief Bioinform. 2017 doi: 10.1093/bib/bbw138. https://dx.doi.org/10.1093/bib/bbw138. [DOI] [PMC free article] [PubMed]
- 80.Cinelli M, et al. Feature selection using a one dimensional naïve Bayes’ classifier increases the accuracy of support vector machine classification of CDR3 repertoires. Bioinformatics. 2017;33:951–955. doi: 10.1093/bioinformatics/btw771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davis MM. A prescription for human immunology. Immunity. 2008;29:835–838. doi: 10.1016/j.immuni.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gaudillière B, et al. Clinical recovery from surgery correlates with single-cell immune signatures. Sci Transl Med. 2014;6:255ra131. doi: 10.1126/scitranslmed.3009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fragiadakis GK, et al. Patient-specific immune states before surgery are strong correlates of surgical recovery. Anesthesiology. 2015;123:1241–1255. doi: 10.1097/ALN.0000000000000887. [DOI] [PMC free article] [PubMed] [Google Scholar]