Abstract
Recent data have shown that the G-protein-coupled receptor GPR54 (also known as KiSS-1 receptor) regulates GnRH release from the hypothalamus. This essential role of GPR54 in controlling the hypothalamic-pituitary-gonadal axis makes it an attractive target for therapeutic intervention in reproductive and cancer medicine. Currently, there are no small-molecule modulators of GPR54 function for experimental or clinical use. To identify small-molecule compounds that modify GPR54 signal transduction, the authors have adapted a cell-based functional assay for high-throughput screening (HTS) using a commercially available homogeneous time-resolved fluorescence assay for inositol phosphate accumulation. They generated stable Chinese hamster ovary cell transfectants that express human GPR54 for use in this assay. After optimization in an automated HTS environment, they screened a library of 110,000 small-molecule compounds using 2 protocols, one to identify agonists and one to identify antagonists. Hits obtained in the primary screen were confirmed to be active in secondary in vitro assays. Compounds identified as agonists or antagonists from HTS and secondary screening will be characterized to identify agents with the potential to be developed as novel orally active agents to treat hormone-dependent disorders such as abnormal puberty, infertility, endometriosis, and sex steroid-dependent tumors.
Keywords: GPR54, kisspeptin, agonist, antagonist, high-throughput screening
INTRODUCTION
In 2003, three independent research teams discovered that GPR54, a G-protein-coupled receptor, is critical for the attainment of reproductive function.1–3 These studies reported that humans and mice with inactivating mutations of GPR54 (also known as KiSS-1 receptor) failed to undergo puberty. GPR54-deficient animals were found to have normal hypothalamic GnRH content and to respond to exogenous GnRH administration with gonadotropin production, suggesting that GPR54 is involved in GnRH processing or release.1 Activation of GPR54 by its ligand, kisspeptin, was found to result in GnRH-dependent gonadotropin release in multiple species, including humans.4–9 Since then, an explosion of research has taken place exploring the mechanisms and sites of action of kisspeptin and GPR54 relevant to reproductive function.
Recent studies have shed light on the kisspeptin-GPR54 system as an integrator of metabolic, reproductive, and developmental cues to signal changes in GnRH output.10 In the mouse, kisspeptin administration stimulated depolarization of >90% of GnRH neurons in adult animals but only 27% of GnRH neurons in juvenile animals.11 Approximately 90% of GnRH neurons expressed GPR54 mRNA without detectable differences between prepubertal and postpubertal mice, whereas KiSS-1 mRNA increased markedly after puberty in the anteroventral periventricular nucleus (AVPV).11 These observations suggest that the activation of GnRH neurons by kisspeptin after puberty may be due to heightened responsiveness of GnRH neurons and to additional kisspeptin inputs.11
There is evidence that KiSS-1 neurons in the AVPV are instrumental in induction of the luteinizing hormone (LH) surge during the menstrual cycle. KiSS-1 neurons express estrogen receptor-α, which is required for the positive feedback of estrogen necessary to generate the LH surge.12–14 Estrogen replacement in ovariectomized mice diminished KiSS-1 mRNA in the arcuate nucleus (ARC) yet upregulated KiSS-1 mRNA in the AVPV.13,15 In the rat, KiSS-1 expression increased in the AVPV over the morning to evening transition during proestrus preceding the LH surge.14 This regulation of KiSS-1 suggests that KiSS-1 plays a role in mediating GnRH output through sex steroid feedback.
Prior to the discovery of its reproductive significance, kisspeptin was studied for its antimetastatic effects on tumors. KISS1, the gene encoding the kisspeptins, was first described in 1996 as a tumor suppressor gene.16 Overexpression of the gene in melanoma and breast cancer cells in vitro was found to inhibit metastasis.17 Hori et al.18 found that kisspeptin significantly suppressed cell motility in a chemotaxis assay and wound-healing assay at 10 to 100 nM. Microarray analysis performed on MDA-MB-435S cells expressing GPR54 and stimulated by kisspeptin-10 showed an upregulation of genes controlling cell cycle and apoptosis.19 Moreover, kisspeptin-10 and GPR54 have been found to suppress the chemotactic activity of the CXCR 4 receptor, a G-protein-coupled receptor involved in metastasis.20
Due to its central nervous system location of action and primary control of the GnRH-gonadotropic-gonadal axis, GPR54 presents itself as an ideal target for pharmacologic agents in the treatment of reproductive disorders. Given the role of GPR54 in tumor metastasis, modulators of the receptor may also be useful in the management of cancers, especially sex steroid-dependent tumors where the dual actions of metastasis suppression and sex steroid regulation would prove especially valuable. Currently, only injectable peptidic GnRH agonists and/or antagonists are available for manipulation of the reproductive axis; small-molecule modulators that are orally active have proven elusive. Here we report the development of a high-throughput screen (HTS) to identify small-molecule agonists and antagonists that could activate or inhibit GPR54 and its downstream targets in vitro. For this HTS, we created a stably transfected Chinese hamster ovary (CHO) cell line that expresses human GPR54 (hGPR54). Modulation of GPR54 activation was measured using the cell-based IP One HTRF™ assay that quantifies the accumulation of inositol monophosphate (IP1), the stable breakdown product of IP3 generated following activation of Gq/11-coupled receptors.
MATERIALS AND METHODS
Reagents
Dulbecco’s Modified Eagle’s Medium: Nutrient Mixture F-12 (DMEM:F12) was purchased from Invitrogen (Carlsbad, CA), fetal bovine serum (FBS) was from Omega Scientific (Tarzana, CA), and penicillin-streptomycin (pen-strep) and geneticin were from Invitrogen. All restriction enzymes were purchased from New England Biolabs (Beverly, MA). Mouse and human kisspeptin-10 (mkiss-10 and hkiss-10) were synthesized by the Tufts University Core Facility (Boston, MA). Human kiss-10 was best solubilized in DMSO, whereas mkiss-10 was soluble in water.
Generation of DNA construct pIRESneo3(hGPR54)
hGPR54 cDNA was subcloned into the bicistronic mammalian expression vector pIRESneo3 (Clontech, Mountain View, CA) to favor the selection of clones with high expression of the transgene. We excised the full-length hGPR54 cDNA from the pCMVsport6 hGPR54 expression vector (Invitrogen) (1) by NotI and EcoRI digestion. This fragment was then ligated to pIRESneo3 that had been digested and linearized with EcoRI and NotI. The plasmid construct was amplified in bacteria in medium containing ampicillin. Colonies were isolated and DNA extracted by the Qiagen Miniprep Kit (Qiagen, Valencia, CA). PCR was performed on miniprep DNA to verify that the hGPR54 cDNA was incorporated into the vector at the multiple cloning site and confirmed by sequence analysis.
Heterologous receptor expression and functional analysis in mammalian cells
To obtain mammalian cells stably expressing pIRESneo3 (hGPR54) or the empty pIRESneo3 vector, CHO cells were transfected with 2 µg of either construct using Geneporter (Genlantis, San Diego, CA), following the manufacturer’s instructions. Transfected cells then underwent 4 rounds of selection in DMEM:F12 with 10% FBS, 1% pen-strep, and 800 µg/mL of geneticin. Individual cell colonies were selected by overlaying separate colonies with sterile cloning disks (Scienceware, Pequannock, NJ). Stable cell lines were later maintained in the same medium.
Total RNA was extracted from clonal cell lines using TRI reagent (Sigma-Aldrich, St. Louis, MO), and 10 µg of RNA from each clone was subjected to Northern blot analysis to confirm GPR54 expression. Total RNA was separated on a denaturing MOPS gel, blotted onto a nylon membrane (Schleicher & Schuell, Keene, NH), and hybridized with a GPR54 cRNA probe labeled with [α-32P]UTP (PerkinElmer, Waltham, MA) using RNA polymerase (New England Biolabs). Membranes were also probed for cyclophilin as a housekeeping gene.
3H-inositol phosphate (IP) assay
3H-IP assays were performed on pIRESneo3(hGPR54)- and empty vector-transfected stable CHO cells plated in 6-well plates, as previously described.21 After incubation of the cells for 24 h at 37°C, medium was replaced with 1 mL inositol-free DMEM for 2 h, and then 1 mL of the same medium containing 2 µCi myo-(2-3H) inositol (PerkinElmer), followed by the addition of 10 mM LiCl 15 min later. After an 18-h incubation at 37°C, cells were stimulated with increasing concentrations of hkiss-10 for 45 min. Cells were extracted twice for 30 min on ice with 20 mM formic acid, and lysates were neutralized to pH 7.5 with 7.5 mM HEPES plus 150 mM KOH. After centrifuging for 2 min at 14,000 g, protein content in the lysates was measured (Coomassie Plus protein assay reagent, Pierce Chemical Co., Rockford, IL), and the supernatants were loaded onto Ag-X8 resin anion exchange columns (Bio-Rad Laboratories, Hercules, CA) previously equilibrated with 2 mL of 1 M NaOH, 2 mL of 1 M formic acid, and 5 × 5 mL ddH2O. The columns were washed with 5 mL ddH2O and then 5 mL of 5 mM borax and 60 mM sodium formate, and IPs were eluted with 3 mL of 0.9 M ammonium formate, 0.1 M formic acid. The incorporation of radioactivity in the eluates was measured in a scintillation counter, and each sample was corrected for protein content. All assay points were performed in triplicate, and each experiment was repeated at least 3 times.
Western blot analysis of ERK-1/2 phosphorylation
The pIRESneo3(hGPR54)- and empty vector-transfected stable CHO cells were plated in 6-well plates. After incubation for 24 h at 37°C, cells were incubated an additional 24 h in serum-free DMEM:F12, followed by stimulation with increasing concentrations of hkiss-10 for 5 min. Initial time course studies indicated that hkiss-10 induced phosphorylated ERK-1/2 (pERK) maximally at 5 min. Cells were lysed on ice with RIPA buffer (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) containing 0.1 mg/mL phenylmethylsulfonyl fluoride and 1 mM sodium orthovanadate. Cell lysates were sonicated for 20 s on ice and centrifuged for 10 min at 14,000 g at 4°C. The protein content in the supernatant was measured, and 20 µg of denatured protein/well was loaded in either singlet or duplicate onto 12% polyacrylamide gels, and electrophoresis was carried out according to standard protocols. Proteins were transferred to nitrocellulose membranes and incubated overnight with a mouse anti-pERK IgG (1:5000; SC-7383, Santa Cruz), followed by incubation with goat antimouse IgG-HRP (1:5000; SC-2060, Santa Cruz). Immunoreactive bands were detected using luminol chemiluminescence reagent (PerkinElmer), and pERK bands were normalized to total ERK in the same membranes after strip washing (Restore buffer, Pierce Chemical Co.). Total ERK was determined as above after overnight incubation with a rabbit anti-ERK IgG (1:4000; SC-94, Santa Cruz), followed by incubation with donkey antirabbit IgG-HRP (1:8000; SC-2313, Santa Cruz).
Testing of HTS assays
Three screening assays amenable to adaptation to HTS automation were tested, including (1) a fluorescence polarization assay, (2) an amplified luminescent proximity homogeneous assay for phosphorylated ERK (AlphaScreen™, PerkinElmer), and (3) a fluorescence resonance energy transfer (FRET)–based IP-One homogeneous time-resolved fluorescence (HTRF™) assay (CIS-US, Bedford, MA).
The fluorescence polarization assay was designed to measure ligand binding to GPR54 by combining compounds and BODIPY-labeled hkiss-10 (Synpep, Dublin, CA) with membrane preparations from stably transfected GPR54-expressing cells in black microplates (Corning, Inc., Corning, NY). Then, 50 µL of test compound was mixed with 100 µL of varying dilutions of BODIPY-labeled hkiss-10, followed by addition of 50 µL of membrane preparation. After mixing briefly at room temperature, the plates were then read at 525-nm and 565-nm wavelengths on an instrument capable of detecting fluorescence polarization (LJL Analyst, Molecular Devices, Sunnyvale, CA).
The AlphaScreen SureFire™ ERK assay measures ERK phosphorylation in cell lysates as the result of activation of Gq-coupled receptors. Binding of phospho-ERK to antibody-coated AlphaScreen™ donor and acceptor beads generates an amplified signal that is proximity based. The day after plating cells in 384-well plates, the media over the cells were replaced with serum-free media. On the following day, the cells were stimulated with test compounds and hkiss-10 as a positive control; both 5- and 10-min time points were tested. After stimulation, media were aspirated and lysis buffer was added to each well. After the plate was shaken, 20 µL of cell lysate from each well was transferred to a white microplate, 5 µL activation buffer was added, and 6 µL of the donor/acceptor bead mixture was added in reaction buffer under dark conditions. After plates were sealed, shaken, and incubated at reader temperature for 2 h, luminescent signal was detected by a plate reader with AlphaScreen™ detection capability (EnVision™, PerkinElmer) at 680-nm and 570-nm wavelengths.
The IP-One HTRF™ assay directly measures IP1 resulting from the activation of Gq-coupled receptors in cultured cells. A monoclonal antibody specific for IP1 labeled with the inert rare earth fluorescent tracer Europium (Eu) cryptate competes for endogenous IP1 and IP1 coupled to the dye d2. FRET between the Eu cryptate donor and the d2 XL665 acceptor generates a long-lived signal that is detected at 2 different wavelengths (620 nm and 665 nm), using HTRF™ reader technology to minimize the detection of autofluorescence from unbound fluorophores. Calculation of the signal ratio at 2 different wavelengths also corrects for possible photophysical interference of the media and/or colored compounds. The specific signal is inversely proportional to the concentration of IP1 in the cell lysate. This assay was pursued further than the other two and optimized as described below.
Screening for GPR54 agonists
We performed additional optimization experiments of the IP-One HTRF™ assay in 384-well white plates with or without clear bottoms (Nunc, Thermo Fisher Scientific, Rochester, NY). Cells were passaged using TrypLE™Express (Invitrogen) and added in medium to plates using the ThermoScientific Multidrop (Waltham, MA) 1 day prior to assay. Plates were sealed with gas-permeable aeraseals (Bellco Glass, Vineland, NJ) and incubated overnight in a humidified 5% CO2/95% air ThermoForma (Waltham, MA) incubator at 37°C. The following day, all medium was removed using a 384-well plate aspirator (ELx405™, Biotek, Winooski, VT) just prior to compound addition. Immediately thereafter, 10 µL of IP-One stimulation buffer containing compound for a final concentration of 1 µM in 0.1% final concentration of DMSO was added. Human kiss-10 was used as a positive control at a dose of 0.1 µM and was added at a final DMSO concentration of 1%. After incubation at 37°C for 1 h, 5 µL of IP1-d2 conjugate followed by 5 µL of Eu-cryptate-labeled anti-IP1 antibody were added. All additions were performed with the Biomek™ FX Laboratory Automation Workstation (Beckman Coulter, Fullerton, CA). After incubation at room temperature for 1 h, HTRF™ at 620 nm and 665 nm was measured on the PHERA star multimodal plate reader (BMG Labtech, Offenburg, Germany).
Screening for GPR54 antagonists
As in the agonist protocol, 1 day after plating, all medium was removed using a 384-well plate aspirator (ELx405™) immediately prior to compound addition. During the time of optimization of the antagonist screen, a newer version of the IP-One HTRF™ kit was tested and incorporated into the screen. Using the new kit, 7 µL of IP-One stimulation buffer containing compound for a final concentration of 1 µM in 0.1% final concentration of DMSO was added. After incubation at 37°C for 1 h, 7 µL of hkiss-10 at 2 × 10−7 M was added. This submaximal dose was found to approach the EC80 after we identified a shift in the EC50 after scaling the assay up to 30+ plates per run, particularly in the antagonist screen protocol that had more steps and took more time. The shift in EC50 was thought to be due to compound loss in the source plate on the robot and extended plate time outside of the incubator that was unavoidable with larger numbers of plates.
After incubation at 37°C for 45 min, 3 µL of IP1-d2 conjugate was added, followed by 3 µL of Eu-cryptate-labeled anti-IP1 antibody. All additions were performed with the Biomek FX. After incubation at room temperature for 1 h, HTRF™ at 620 nm and 665 nm was measured on the PHERA star plate reader.
Cell viability assays
Cell viability assays were performed using the tetrazolium dye MTS (CellTiter 96 Aqueous One Solution, Promega, Madison, WI) following the manufacturer’s instructions, to ensure that plating with the Multidrop was consistent. One day after cells were plated with the Multidrop, 10 µL of MTS tetrazolium compound was added to all wells and the plates incubated at 37°C for 2 h. MTS is bioreduced by cells into a formazan product that causes a color change in the medium that may be measured at 490 nm. The plates were read at this wavelength on the SpectraMax Plus 384 plate reader (Molecular Devices, Sunnyvale, CA), and the percent coefficient of variance between wells remained below 10%.
Small-molecule library used for HTS
The small-molecule library of the Laboratory for Drug Discovery in Neurodegeneration (LDDN) at Brigham and Women’s Hospital was screened. The LDDN compound library contains approximately 110,000 compounds. The collection consists of compounds purchased from 10 primary vendors. Most compounds in the library are from commercial sources with resupply readily available. All compounds in the LDDN in-house collection were plated in 384-well plates. Typically, 352 unique compounds are arrayed in a 384-well plate, leaving columns 23 and 24 vacant. Compounds were dissolved in DMSO to a 10-mM final concentration and stored desiccated at −20°C. The target average final concentration of the compound library for screening in cell-based assays was 1 µM.
Various computational filters were applied to select compounds for the LDDN library with an increased probability of oral bioavailability and blood-brain barrier penetration, including calculations of polar surface area, Lipinski’s “rule of 5,”22 and filters to reduce common toxicophores and undesirable/non-drug-like functionalities. The result has been a high-quality collection of drug-like molecules that has been tested successfully in a number of screening campaigns.
Statistical analysis
The data from dose-response experiments were subjected to nonlinear regression analysis, and the EC50 was calculated using Prism 3.0 (GraphPad Software, Inc., San Diego, CA). Results from independent experiments were analyzed by analysis of variance (ANOVA) followed by a Tukey-Kramer multiple-comparison post hoc test using Instat 3.0 (GraphPad) to determine statistical difference in pairwise comparisons. Error bars represent SEM, and p-values <0.05 were considered statistically significant in all figures.
RESULTS
Generation and characterization of stable CHO cell transfectants
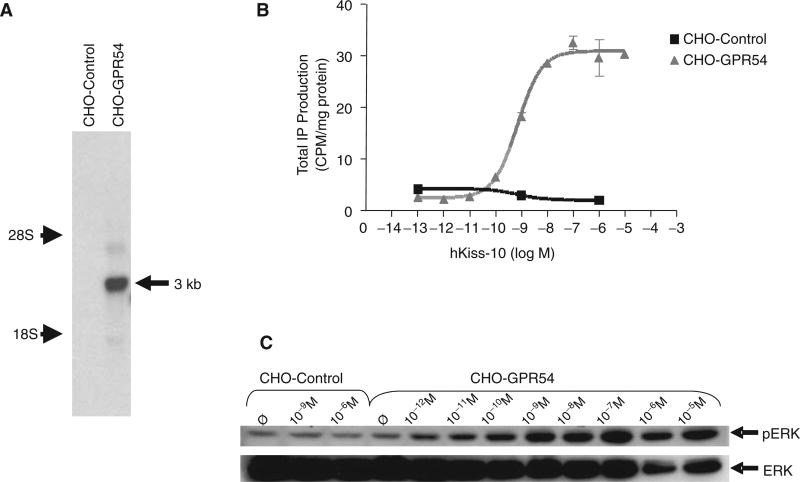
For use in our HTS assay, we created clonal cell lines stably expressing pIRESneo3(hGPR54) or the empty pIRESneo3 vector. The empty pIRESneo3 vector-expressing line (referred to as CHO-control) was created to serve as a negative control when testing compounds for GPR54 specificity. After introduction of the expression vectors into CHO cells, the transfected cells underwent 4 rounds of selection in medium containing 800 µg/mL of geneticin using cloning disks. Seventeen clones were selected and expanded, and total RNA was extracted. Clones were screened for IP induction in response to hkiss-10, and the 4 clones showing the highest response were screened by Northern blot analysis using a GPR54 riboprobe to confirm the presence or absence of GPR54 expression, as well as to compare the levels of expression. Empty pIRESneo3 vector-expressing clones were also screened to confirm the specificity of bands observed by Northern blot with the GPR54 riboprobe. A band approximately 3 kb in size, consistent with the size of hGPR54 cDNA, was present in stable CHO cell transfectants expressing hGPR54 but not in CHO-control (Fig. 1A). Northern blot analysis was also performed with a cyclophilin riboprobe to confirm the integrity of the RNA samples (not shown).
Fig. 1.
Characterization of pIRESneo3(hGPR54)- and pIRESneo3-transfected stable Chinese hamster ovary (CHO) cells (CHO-GPR54 and CHO-control, respectively). (A) Northern blot analysis of total RNA extracted from CHO-GPR54 and CHO-control cells, hybridized with a GPR54 riboprobe. A band was present in the CHO-GPR54 cell line but not in the CHO-control cell line. The band (indicated by the arrow) was estimated to be 3 kb in size based on the positions of 28S and 18S rRNA bands, consistent with the size of hGPR54 cDNA. (B) Stimulation of inositol phosphate (IP) production by increasing concentrations of hkiss-10 in the CHO-GPR54 cell line with an EC50 of 1.0 nM. CHO-control cells showed no response to hkiss-10. Symbols and error bars represent the mean ± SE of 3 replicates from a representative experiment. (C) Stimulation of ERK1/2 phosphorylation by increasing concentrations of kisspeptin in CHO-GPR54 and CHO-control cell lines. Cells were deprived of serum for 24 h, followed by stimulation with increasing concentrations (10−12 M to 10−5 M) of hkiss-10 for 5 min. Cells were lysed, extracts were isolated, and Western blot analysis was performed. Membranes were reprobed with ERK1/2 antibody to correct for protein loading. Samples were loaded in singlet; a representative blot is shown.
Clonal cell lines were subjected to IP assay after stimulation with increasing concentrations of hkiss-10 to assess the functional status of hGPR54 expressed by cells. The hGPR54-expressing clone with the greatest induction of IP production in response to hkiss-10 stimulation was used in all subsequent experiments and will be referred to as CHO-GPR54 (Fig. 1B). The EC50 for hkiss-10-induced IP production in these studies was between 1.2 and 3.9 nM, consistent with previously published data.23 pIRES ne03-transfected (“empty vector”) stable CHO-control cells showed no response to hkiss-10 (Fig. 1B), confirming the absence of functional GPR54 receptors in CHO cells.
Results from the IP assay were confirmed by measurement of ERK1/2 phosphorylation by Western blot analysis following stimulation with increasing doses of hkiss-10. These studies revealed a greater increase in pERK1/2 levels 5 and 10 min after hkiss-10 stimulation in this clonal line (CHO-GPR4) than in the other GPR54-expressing stable transfectants generated and studied (data not shown). The stimulation of ERK phosphorylation in this clonal line was also dependent on kisspeptin dose (Fig. 1C). This CHO-GPR54 clone, which was the most highly responsive to hkiss-10 in both the IP assay and Western blot analysis for pERK, was selected for use in high-throughput assays. pIRES ne03-transfected (“empty vector”) stable CHO cells (CHO-control) showed no increase in ERK1/2 phosphorylation in response to hkiss-10 (Fig. 1C).
HTS assay selection and optimization of IP-One HTRF™ assay
Of the 3 assays tested, ultimately we selected the IP-One HTRF™ assay for use in our HTS assay (see Materials and Methods). In the fluorescence polarization assay, the BODIPY-labeled kisspeptin tended to aggregate in solution, leading to high values in all wells. The aggregation improved only at high salt concentrations (NaCl 500 mM) that would also have interfered with compound binding to receptor. Another advantage of the IP-One HTRF™ assay over the fluorescence polarization assay was the ability to measure receptor function in cells as opposed to simply binding. The AlphaScreen SureFire™ ERK assay was not selected due to the need to serum deprive cells prior to the assay, adding an additional day to the HTS protocol, and the requirement of dark assay conditions due to light and temperature sensitivity of the beads. There was also a tendency for high background readings with the SureFire™ ERK assay, even in negative control wells.
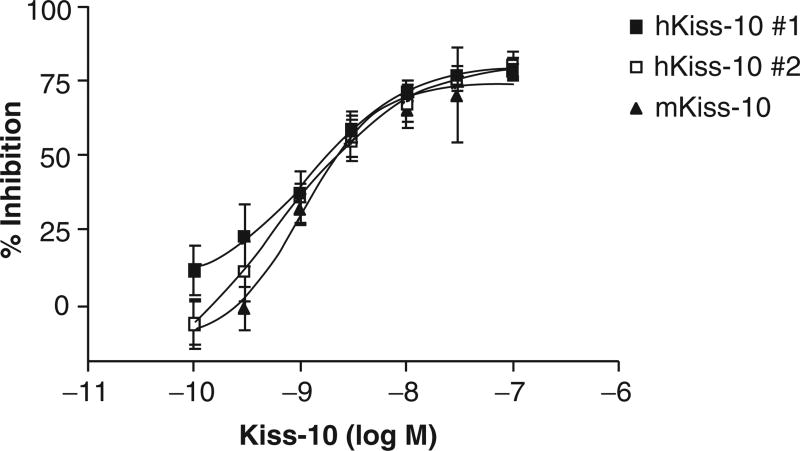
We performed optimization experiments of the IP-One HTRF™ assay in 384-well white microplates and tested plating densities of 10,000, 20,000, 30,000, 40,000, and 60,000 cells/ well and volumes of 40 and 80 µL. We found the optimal plating density to be 30,000 cells/well in a volume of 40 µL as this density yielded the most consistent hkiss-10 dose response with the lowest background readings in the smallest amount of media. Serum deprivation did not improve results. Preliminary automated IP1 dose-response experiments using both mkiss-10 and hkiss-10 performed on different days yielded inhibition curves with an EC50 of 0.6 to 1.3 nM, values consistent with published Kd values (Fig. 2).23 IP1 results comparing hkiss-10 (dissolved in DMSO) and mkiss-10 (dissolved in water) were similar, providing background information on the lack of nonspecific or toxic effects of DMSO when testing library compounds.
Fig. 2.
Accumulation of IP1 by increasing concentrations (10−10 to 10−7 M) of human and mouse kisspeptin-10 (hkiss-10 and mkiss-10, respectively) as measured by the IP-One HTRF™ assay in the CHO-GPR54 cell line as percent inhibition of the fluorescence resonance energy transfer (FRET) signal. Compounds and reagents were added by the Biomek FX robotic workstation in high-throughput screening format.
HTS Assays for GPR54 agonists and antagonists
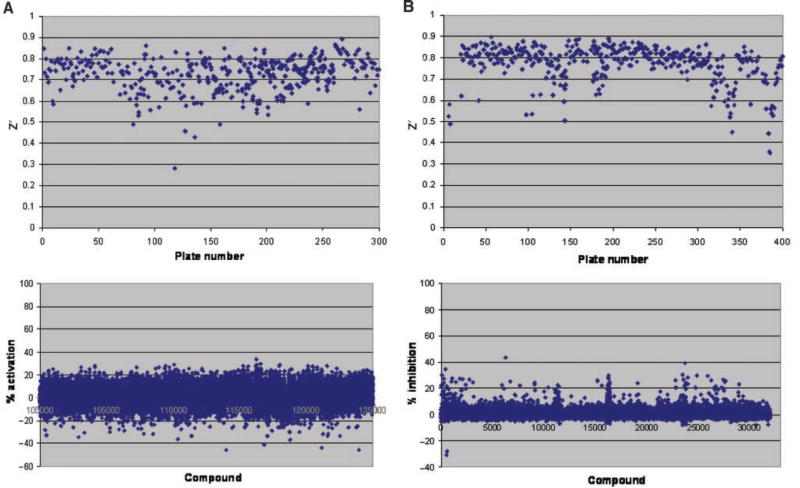
We screened the LDDN library of 110,000 small-molecule compounds to search for agonist compounds. Compounds with greater than 30% activation of IP1 at a concentration of 1 µM were designated as “hits” and subjected to further verification of activity (Fig. 3A, lower panel). Thirty percent activation was chosen arbitrarily as the cutoff for selecting activators to generate a hit rate of <0.5%. Our overall hit rate was 0.06% (72 compounds). We used the statistical calculation for the Z′ factor24 to define signal dynamic range and the data variation associated with the IP1 signal measurements. In general, Z′ factors greater than 0.5 indicate excellent quality for an assay.24 Of the 338 plates tested in our screen, most had a Z′ value between 0.65 and 0.75, indicating a robust assay (Fig. 3A, upper panel).
Fig. 3.
Scatter plots showing Z prime (Z′) factor per plate and compound IP-One assay readings from high-throughput screens. (A) Z′ factor plot for plates (upper panel) and IP1 activation by compounds at a 1-µM concentration (lower panel) in screen for GPR54 agonists. (B) Z′ factor plot for plates (upper panel) and inhibition of IP1 accumulation by 0.2 µM hkiss-10 by compounds at 10 µM concentration (lower panel) in screen for GPR54 antagonists.
We rescreened the library for antagonist compounds that inhibited IP1 induction by 0.2 µM hkiss-10 (submaximal, EC80 concentration), which was added to all wells except negative control wells 1 h after compound additions. Because there were no known antagonists available for our use at the time of the screen, readings from negative control wells lacking hkiss-10 were used to define 100% inhibition. Compounds with greater than 50% inhibition of kisspeptin-stimulated IP1 at 10 µM concentration were subjected to further verification of activity (Fig. 3B, lower panel), and of the 387 plates tested, most had a Z′ value between 0.75 and 0.85 (Fig. 3B, upper panel). The LDDN compound library had expanded during the time between the agonist screen and the antagonist screen, thus accounting for additional plates in the antagonist screen. Fifty percent inhibition was chosen arbitrarily as the cutoff for selecting antagonists to generate a hit rate of <0.5%. Our overall hit rate was 0.05% (64 compounds).
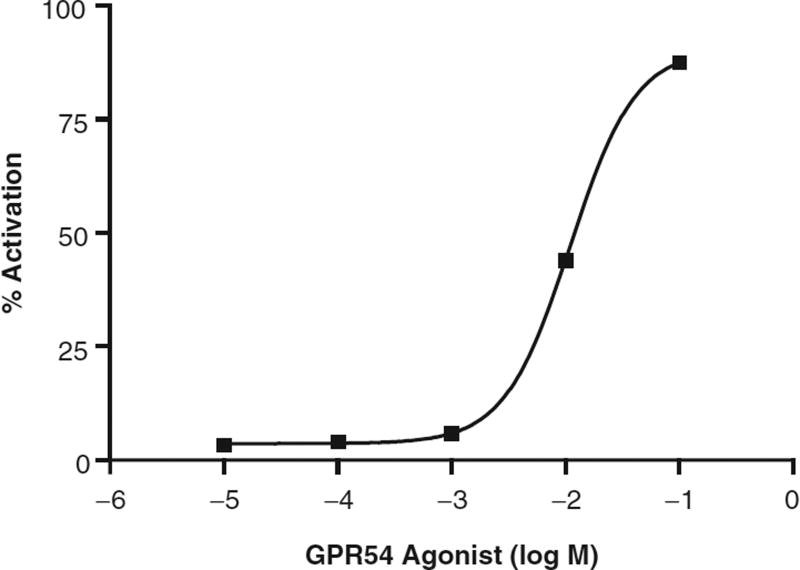
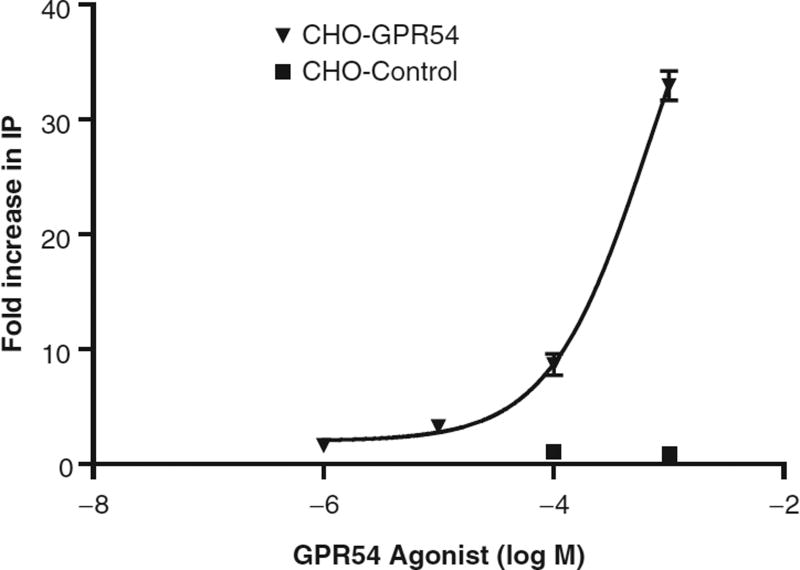
Of the 72 compounds identified in our initial agonist screen, only 1 of these hits was verified on repeat screening. This candidate compound also showed evidence of dose-dependent activation of cellular IP1 accumulation (Fig. 4) and was not cytotoxic on MTS testing, whereas the remaining 71 compounds either were cytotoxic or did not show dose-dependent activation. 3H-IP assays of the CHO-GPR54 cell line also showed a significant response to the candidate agonist in the CHO-GPR54 cell line, with a maximal induction of 33.0 ± 1.3-fold at a 10−3 M concentration of the agonist, the highest concentration tested (Fig. 5). By comparison, 10−10 M hkiss-10 resulted in a 23.7 ± 0.93-fold increase in IP production in the same assay (data not shown). In contrast, the empty vector CHO-control cell line did not respond to the candidate agonist with any increase in IP production (Fig. 5). IP induction triggered by this agonist indicates activation of the Gq/11 pathway, known to be coupled to GPR54. Also, the absence of activation in the empty vector CHO-control cell line that does not express GPR54 demonstrates that the IP response is GPR54 specific.
Fig. 4.
Dose-dependent increase in IP1 accumulation induced by a candidate GPR54 agonist as measured in the IP-One HTRF™ assay in the CHO-GPR54 cell line. Values and error bars represent the mean ± SE of quadruplicate samples, and results are expressed as percent activation of IP1.
Fig. 5.
Secondary screening of a candidate GPR54 agonist using [3H]-inositol phosphate assays shows specificity for GPR54. In the CHO-GPR54 cell line, the GPR54 agonist stimulates IP production in a dose-dependent fashion. The CHO-control cell line shows no response to the agonist or to hkiss-10. Points represent the mean ± SE of 3 replicates.
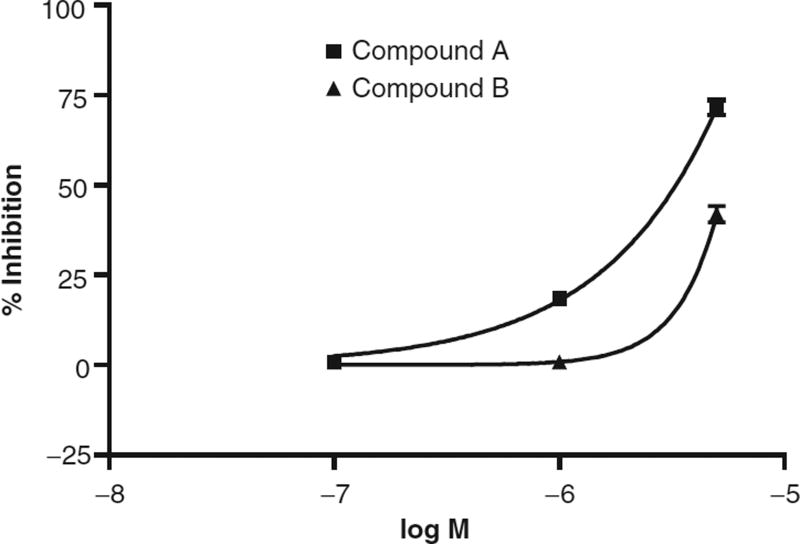
Of the 64 compounds identified in the antagonist screen, 7 of these hits showed both dose-dependent inhibition and were not cytotoxic on MTS testing, whereas the remaining compounds either were cytotoxic or did not show dose-dependent inhibition. Dose dependency was determined by testing the ability of 0.1-, 1.0-, and 5.0-µM concentrations of the compounds to inhibit IP1 activation by 0.2 µM hkiss-10 (Fig. 6).
Fig. 6.
Dose-dependent inhibition of hkiss-10-stimulated IP1 activation by candidate small-molecule GPR54 antagonists in the IP One HTRF™ assay. Dose dependency was determined by testing 0.1-, 1.0-, and 5.0-µM concentrations of the compounds against 0.2 µM hkiss-10. Shown are representative dose responses for 2 antagonist compounds, compound A and compound B.
DISCUSSION
Kisspeptins are a family of peptides encoded by the KISS1 gene and expressed in the placenta and in the central nervous system, particularly in the AVPV and ARC nuclei of the hypothalamus.13,23 They activate the G-protein-coupled receptor, GPR54, to mediate the release of GnRH from hypothalamic GnRH neurons. GPR54, a member of the rhodopsin subclass, is widely expressed in human tissues but its highest concentration is in placenta, brain, and pituitary.23 The kisspeptin/GPR54 dyad coordinates signals conveying the pubertal, hormonal, and metabolic status of the organism to the brain,10 modulating GnRH and hence gonadotropin release accordingly. As discussed earlier, this system has also been shown to inhibit tumor metastasis in melanoma and breast cancer cells by suppressing cell motility and chemotaxis.17,18
We present herein an HTS strategy for small-molecule agonists and antagonists of GPR54 using a pIRESneo3(hGPR54)-transfected CHO cell line (CHO-GPR54), with IP1 as the primary signal as detected by the IP One HTRF™ assay. We have compared several screening assays in this study and selected this functional cellular signaling assay as the preferred assay for this Gq-coupled receptor over others tested for several reasons. In the fluorescence polarization assay, the BODIPY-labeled hkiss-10 aggregated in solution, leading to high nonspecific background values that made it difficult to detect true positives over the background signal. High salt concentrations decreased compound aggregation but also interfered with compound binding to the receptor, raising the concern that we would increase our false-negative rate. Another disadvantage of the fluorescent membrane binding assay compared to the functional assays was its inability to distinguish agonists from antagonists in the HTS. Yet another potential disadvantage of the fluorescence polarization assay was that it would only detect compounds that interfered with BODIPY-hkiss-10 binding to GPR54, so that compounds that could interact with GPR54 through allosteric mechanisms would not be detected. For these reasons, we preferred a functional assay. The IP-One HTRF™ assay was selected over the AlphaScreen SureFire™ ERK assay because of the more complex nature of the AlphaScreen SureFire™ ERK assay. The need to serum deprive cells prior to the assay, adding an additional day to the HTS protocol, and the requirement for dark assay conditions due to light and temperature sensitivity of the beads both made the ERK assay more difficult for HTS assays. The IP-One HTRF™ assay may be a preferred HTS strategy for identifying small-molecule agonists and antagonists of Gq-coupled receptors in general.
In the IP-One HTRF™ assay, the Z′ factor indicated a robust assay. In the agonist screen, for which we chose a 30% IP1 activation threshold, we identified 72 compounds (hit rate of 0.06%). On further verification, one of these hits showed dose-dependent activation and was not cytotoxic on MTS testing. On the antagonist screen, we set a threshold of 50% inhibition of IP1 and identified 64 compounds for a hit rate of 0.05%. Seven of these hits showed dose-dependent inhibition and were not cytotoxic on MTS testing. The false-positive rate with the IP-One kit was high, and the source of the false positives is unknown but could be due to occasional pipetting errors, although our Z′ rate would argue against this. There may be something inherent to the components of the kit or HTRF™ technology that produces a higher false-positive rate. The use of the IP One HTRF™ assay has been described previously for this target as part of a triple assay screening campaign.25 Unlike fluorescent membrane binding assays, the advantage of this particular assay in screening activators or inhibitors of Gq/11-coupled receptors is that receptor function in the cell is measured directly. The drawback is that the assay is cell based and as such is more cumbersome for high-throughput application.
Further characterization of the stimulatory and inhibitory compounds is currently under way, correlating their EC50 values with their capacity to upregulate or downregulate Gq/11-coupled second messengers such as inositol phosphates and phosphorylated ERK. Competitive binding assays with I125-kisspeptin are being performed to determine if the modulators are able to displace kisspeptin at functionally relevant doses or if an allosteric mode of action is responsible. In vivo testing in rodents is also being performed to determine if the compounds are able to stimulate LH secretion or inhibit kisspeptin-induced LH secretion.
The discovery of GPR54 agonists and antagonists may provide novel therapies to harness control of the neuroendocrine reproductive axis, as well as supply a means to prevent metastatic spread of malignancies. Like GnRH agonists, GPR54 agonists may serve to stimulate gonadotropin release in the short term and downregulate the gonadotropic axis when given long term. In fact, continuous intravenous administration of hkiss-10 to juvenile male rhesus monkeys for 4 days was shown to desensitize GPR54-mediated GnRH release after an initial increase in GnRH (quantified by LH response) during the first 3 h of infusion.26 Antagonists of GPR54 likely would primarily be used to suppress the gonadotropic axis without an initial stimulatory effect, or flare. The lack of flare effect may be desirable in certain clinical situations such the treatment of sex steroid-dependent cancers, where a transient spike in sex steroid production may promote tumor growth and/or invasion.
Therapeutic use of the known ligand of GPR54, kisspeptin, possesses limitations as it must be given parenterally, and stability of peptides may be problematic. HTS of a small-molecule library for GPR54 modulators provides the advantages of testing readily available and, in some cases, clinically approved drugs that may be brought to the clinical arena sooner than kisspeptin peptide analog design and synthesis, currently being undertaken by several other groups.27–30 These groups have employed alanine and D-amino acid scanning on the biologically active kisspeptin fragment to identify residues critical for receptor activation. A modified pentapeptide was generated with the most potent GPR54 agonistic activity reported to date.28 Key modifications in the carboxy-terminal 5 amino acids of the decapeptide led to the development of a peptide GPR54 antagonist.27 Peptidic approaches represent a feasible alternative method of identifying analogs for G-protein-coupled receptors (GPCR s) with known peptide ligands as it is often difficult to identify small molecules that activate these GPCR s. The only other HTS for small-molecule GPR54 agonists reported also identified only a single positive hit, in this case out of 365 primary hits.25 To our knowledge, our study is unique in that it represents only the second report of an HTS of a small-molecule library for GPR54 agonists and the first HTS for antagonists of GPR54.
Interestingly, this previous report of an HTS for kisspeptin analogs was based on a concomitant HTS of a 50,000-compound library using 3 different assays (AequoScreen™ technology, IP One HTRF™, and a multiplex assay composed of both Fluo-3 and Fura-2 loaded cells) in addition to a fluorescence-based homogeneous calcium dye assay. This screening strategy confirmed 1 selective active hit,25 similar to our agonist screen. Further characterization of this hit has not yet been published. This very low hit rate reported by Cassutt et al.25 is consistent with our findings. If the small-molecule modulator identified in the screen does act at a site separate from the kisspeptin binding site, presumably also the site of action for peptidic analogs, novel therapeutic combinations, and modes of administration may be possible.
In summary, the identification of modulators of GPR54 may permit more precise control of the hypothalamic-pituitary-gonadal axis. Developing orally active drug therapies to achieve control of the hormonal axis may facilitate the treatment of a variety of reproductive disorders and cancers. These small-molecule compounds may also be used to probe the mechanisms of activation and inhibition of this GPCR through testing of wild-type and mutant receptor isoforms.
Acknowledgments
This work was supported by National Institutes of Health SCCPRR Grant U54-HD28138 (to WFC and UBK), the ASRM/Ortho-McNeil Research Grant, Reproductive Scientist Development Program (ASRM-NICHD 5K12-HD00849 Phase I, ACOG Phase II), NICHD Grant 1K08HD053706 (to WK), and the Partners Center for Drug Discovery.
References
- 1.Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Jr, Shagoury JK, et al. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 2.de Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci USA. 2003;100:10972–10976. doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Funes S, Hedrick JA, Vassileva G, Markowitz L, Abbondanzo S, Golovko A, et al. The KiSS-1 receptor GPR54 is essential for the development of the murine reproductive system. Biochem Biophys Res Commun. 2003;312:1357–1363. doi: 10.1016/j.bbrc.2003.11.066. [DOI] [PubMed] [Google Scholar]
- 4.Gottsch ML, Cunningham MJ, Smith JT, Popa SM, Acohido BV, Crowley WF, et al. A role for kisspeptins in the regulation of gonadotropin secretion in the mouse. Endocrinology. 2004;145:4073–4077. doi: 10.1210/en.2004-0431. [DOI] [PubMed] [Google Scholar]
- 5.Dhillo WS, Chaudhri OB, Patterson M, Thompson EL, Murphy KG, Badman MK, et al. Kisspeptin-54 stimulates the hypothalamic-pituitary-gonadal axis in human males. J Clin Endocrinol Metab. 2005;90:6613. doi: 10.1210/jc.2005-1468. [DOI] [PubMed] [Google Scholar]
- 6.Matsui H, Takatsu Y, Kumano S, Matsumoto H, Ohtaki T. Peripheral administration of metastin induces marked gonadotropin release and ovulation in the rat. Biochem Biophys Res Commun. 2004;320:383–388. doi: 10.1016/j.bbrc.2004.05.185. [DOI] [PubMed] [Google Scholar]
- 7.Navarro VM, Castellano JM, Fernandez-Fernandez R, Barreiro ML, Roa J, Sanchez-Criado JE, et al. Developmental and hormonally regulated messenger ribonucleic acid expression of KiSS-1 and its putative receptor, GPR54, in rat hypothalamus and potent luteinizing hormone-releasing activity of KiSS-1 peptide. Endocrinology. 2004;145:4565–4574. doi: 10.1210/en.2004-0413. [DOI] [PubMed] [Google Scholar]
- 8.Navarro VM, Castellano JM, Fernandez-Fernandez R, Tovar S, Roa J, Mayen A, et al. Characterization of the potent LH releasing activity of KiSS-1 peptide, the natural ligand of GPR54. Endocrinology. 2004;146:156–163. doi: 10.1210/en.2004-0836. [DOI] [PubMed] [Google Scholar]
- 9.Shahab M, Mastronardi C, Seminara SB, Crowley WF, Ojeda SR, Plant TM. Increased hypothalamic GPR54 signaling: a potential mechanism for initiation of puberty in primates. Proc Natl Acad Sci USA. 2005;102:2129–2134. doi: 10.1073/pnas.0409822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Popa SM, Clifton DK, Steiner RA. The role of kisspeptins and GPR54 in the neuroendocrine regulation of reproduction. Annu Rev Physiol. 2008;70:213–238. doi: 10.1146/annurev.physiol.70.113006.100540. [DOI] [PubMed] [Google Scholar]
- 11.Han SK, Gottsch ML, Lee KJ, Popa SM, Smith JT, Jakawich SK, et al. Activation of gonadotropin-releasing hormone neurons by kisspeptin as a neuroendocrine switch for the onset of puberty. J Neurosci. 2005;25:11349–11356. doi: 10.1523/JNEUROSCI.3328-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franceschini I, Lornet D, Cateau M, Delsol G, Tillet Y, Caraty A. Kisspeptin immunoreactive cells of the ovine preoptic area and arcuate nucleus co-express estrogen receptor alpha. Neurosci Lett. 2006;401:225–230. doi: 10.1016/j.neulet.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 13.Smith JT, Cunningham MJ, Rissman EF, Clifton DK, Steiner RA. Regulation of Kiss1 gene expression in the brain of the female mouse. Endocrinology. 2005;146:3686–3692. doi: 10.1210/en.2005-0488. [DOI] [PubMed] [Google Scholar]
- 14.Smith JT, Popa SM, Clifton DK, Hoffman GE, Steiner RA. Kiss1 neurons in the forebrain as central processors for generating the preovulatory luteinizing hormone surge. J Neurosci. 2006;26:6687–6694. doi: 10.1523/JNEUROSCI.1618-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith JT, Dungan HM, Stoll EA, Gottsch ML, Braun RE, Eacker SM, et al. Differential regulation of KiSS-1 mRNA expression by sex steroids in the brain of the male mouse. Endocrinology. 2005;146:2976–2984. doi: 10.1210/en.2005-0323. [DOI] [PubMed] [Google Scholar]
- 16.Lee JH, Miele ME, Hicks DJ, Phillips KK, Trent JM, Weissman BE, et al. KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. J Natl Cancer Inst. 1996;88:1731–1737. doi: 10.1093/jnci/88.23.1731. [DOI] [PubMed] [Google Scholar]
- 17.Lee JH, Welch DR. Identification of highly expressed genes in metastasis-suppressed chromosome 6/human malignant melanoma hybrid cells using subtractive hybridization and differential display. Int J Cancer. 1997;71:1035–1044. doi: 10.1002/(sici)1097-0215(19970611)71:6<1035::aid-ijc20>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 18.Hori A, Honda S, Asada M, Ohtaki T, Oda K, Watanabe T, et al. Metastin suppresses the motility and growth of CHO cells transfected with its receptor. Biochem Biophys Res Commun. 2001;286:958–963. doi: 10.1006/bbrc.2001.5470. [DOI] [PubMed] [Google Scholar]
- 19.Becker JA, Mirjolet JF, Bernard J, Burgeon E, Simons MJ, Vassart G, et al. Activation of GPR 54 promotes cell cycle arrest and apoptosis of human tumor cells through a specific transcriptional program not shared by other Gq-coupled receptors. Biochem Biophys Res Commun. 2005;326:677–686. doi: 10.1016/j.bbrc.2004.11.094. [DOI] [PubMed] [Google Scholar]
- 20.Navenot JM, Wang Z, Chopin M, Fujii N, Peiper SC. Kisspeptin-10-induced signaling of GPR54 negatively regulates chemotactic responses mediated by CXCR4: a potential mechanism for the metastasis suppressor activity of kisspeptins. Cancer Res. 2005;65:10450–10456. doi: 10.1158/0008-5472.CAN-05-1757. [DOI] [PubMed] [Google Scholar]
- 21.Bedecarrats GY, Lindher KD, Kaiser UB. Two common naturally occurring mutations in the human gonadotropin-releasing hormone (GnRH) receptor have differential effects on gonadotropin gene expression and on GnRH-mediated signal transduction. J Clin Endocrinol Metab. 2003;88:834–843. doi: 10.1210/jc.2002-020806. [DOI] [PubMed] [Google Scholar]
- 22.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Del Rev. 2001;46(1–3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 23.Ohtaki T, Shintani Y, Honda S, Matsumoto H, Hori A, Kanehashi K, et al. Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature. 2001;411:613–617. doi: 10.1038/35079135. [DOI] [PubMed] [Google Scholar]
- 24.Zhang JH, Chung TDY, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 25.Cassutt KJ, Orsini MJ, Abousleiman M, Colone D, Tang W. Identifying nonselective hits from a homogeneous calcium assay screen. J Biomol Screen. 2007;12:285–287. doi: 10.1177/1087057106298538. [DOI] [PubMed] [Google Scholar]
- 26.Seminara SB, DiPietro MJ, Ramaswamy S, Crowley WF, Jr, Plant TM. Continuous human metastin 45–54 infusion desensitizes GPR54-induced GnRH release monitored indirectly in the juvenile male rhesus monkey (Macaca mulatta): a finding with therapeutic implications. Endocrinology. 2006;147:2122–2126. doi: 10.1210/en.2005-1550. [DOI] [PubMed] [Google Scholar]
- 27.Roseweir AK, Kauffman AS, Smith JT, Guerriero KA, Morgan K, Pielecka-Fortuna J, et al. Discovery of potent kisspeptin antagonists delineate physiological mechanisms of gonadotropin regulation. J Neurosci. 2009;29:3920–3929. doi: 10.1523/JNEUROSCI.5740-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niida A, Wang Z, Tomita K, Oishi S, Tamamura H, Otaka A, et al. Design and synthesis of downsized metastin (45–54) analogs with maintenance of high GPR54 agonistic activity. Bioorg Med Chem Lett. 2006;16:134–137. doi: 10.1016/j.bmcl.2005.09.054. [DOI] [PubMed] [Google Scholar]
- 29.Orsini MJ, Klein MA, Beavers MP, Connolly PJ, Middleton SA, Mayo KH. Metastin (KiSS-1) mimetics identified from peptide structure-activity relationship-derived pharmacophores and directed small molecule database screening. J Med Chem. 2007;50:462–471. doi: 10.1021/jm0609824. [DOI] [PubMed] [Google Scholar]
- 30.Gutierrez-Pascual E, Leprince J, Martinez-Fuentes AJ, Segalas-Milazzo I, Pineda R, Roa J, et al. In vivo and in vitro structure-activity relationships and structural conformation of kisspeptin-10-related peptides. Mol Pharmacol. 2009;76:58–67. doi: 10.1124/mol.108.053751. [DOI] [PubMed] [Google Scholar]