Graphical Abstract

SUMMARY
Mouse oocyte maturation, fertilization, and reprogramming occur in the absence of transcription and thus changes in mRNA levels and translation rate are regulated through post-transcriptional mechanisms [1]. Surprisingly, microRNA function, which is a major form of post-transcriptional regulation, is absent during this critical period of mammalian development [2,3]. Here, we investigated the mechanisms underlying the global suppression of microRNA activity. In both mouse and frogs, microRNA function was active in growing oocytes, but then absent during oocyte maturation. RNA-Seq of mouse oocytes uncovered that the microRNA effector protein AGO2 is predominantly expressed as an alternative isoform that encodes a truncated protein lacking all of the known essential domains. Full length Ago2 as well as the related Argonautes (Ago1, Ago3, and Ago4) were lowly expressed in maturing mouse oocytes. Reintroduction of full-length AGO2 together with an exogenous microRNA in either mouse or frog oocytes restored translational repression of a target reporter. However, levels of endogenous transcripts remained unchanged. Consistent with a lack of microRNA activity, analysis of transcripts with alternative polyadenylation sites showed increased stability of transcripts with a longer 3′ UTR during oocyte maturation. Redundant mechanisms protecting endogenous transcripts and the conserved loss of microRNA activity suggest a strong selection for suppressing microRNA function in vertebrate oocytes.
RESULTS
Suppression of miRNA function in maturing oocytes is conserved
MicroRNAs (miRNAs) and endogenous siRNAs (endo-siRNAs) represent two types of small RNAs that are defined by their differential biogenesis [4]. MiRNAs and endo-siRNAs form a complex with one of four Argonaute proteins (AGO1-4); the resulting complex then binds the 3′ UTR of target mRNAs through either partial or complete complementation. Partial complementation of an AGO-small RNA complex with a target mRNA leads to the recruitment of additional proteins that inhibit translation and destabilize the transcript through exonucleolytic decay [5,6]. In contrast, perfect or near-perfect complementation of an AGO2-small RNA complex with a target mRNA, results in AGO2 endonucleolytic cleavage of the mRNA target [7]. Genetic studies show that endo-siRNAs and the endonucleolytic activity of AGO2 are essential for oocyte maturation, while miRNAs are not [2,8–11]. Furthermore, removal of miRNAs has no discernable effect on the transcriptome of mouse oocytes, in striking contrast to other mouse cell types [2,12].
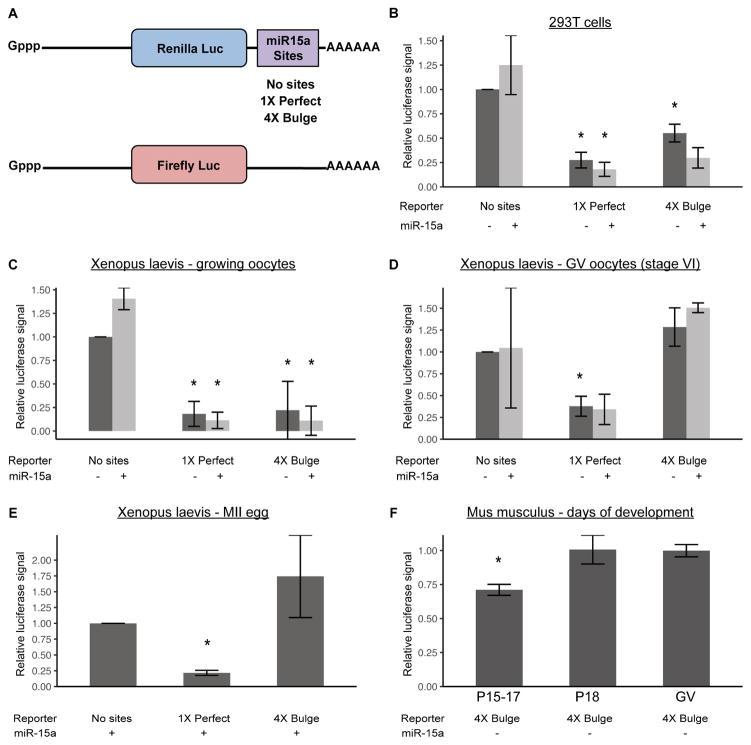
To identify potential mechanisms underlying this surprising result, we first asked whether the loss of miRNA-driven translational repression and exonucleolytic destabilization is conserved across vertebrate species. To differentiate endonucleolytic cleavage from translational repression and exonucleolytic destabilization by small RNAs, we built renilla luciferase reporters carrying either a perfect match or four bulge sites to the miRNA, miR-15a (Figures 1A, S1). MiR-15a is highly conserved across Xenopus laevis (henceforth referred to as Xenopus), mouse, and human and is one of the most highly expressed miRNAs in oocytes [3,13]. We used firefly luciferase to normalize reporter activity. The ability of the reporters to readout both types of miR-15a activity was first confirmed in 293T cells, where both perfect match and bulge reporters showed strong suppression in presence of miR-15a (Figure 1B).
Figure 1. Suppression of miRNA activity in maturing oocytes is conserved from mouse to Xenopus.
A) Schematic of constructs used in all subsequent luciferase assays. miR-15a sites were cloned in the 3′ UTR of Renilla luciferase (Luc) - either a 1X site with a perfect match or a 4X site with a bulged match to miR-15a. A control of no sites was also used. Firefly luciferase was used as an injection control. All injected transcripts were transcribed, capped, and polyadenylated in vitro. B) Luciferase assay in HEK-293T cells transfected with or without miR-15a mimic and psiCHECK2, a vector containing Firefly luciferase and Renilla luciferase with either no miR-15a sites, 1X Perfect, or 4X Bulge sites in the 3′ UTR of Renilla luciferase. N=2. C,D) Luciferase assays in Xenopus laevis (C) growing (stage III–V) and (D) GV (stage VI), using 1X Perfect and 4X Bulge templates, with or without miR-15a mimic. N=2. E) Luciferase assay as in (B), but in Xenopus MII eggs, with miR-15a mimic. N=2. F) Luciferase assays in oocytes at various stages of postnatal mouse development P15–23 (x-axis), with miR-15a mimic. For P15–17 N=4, P18 N=3, GV (P23) N=4. All luciferase data is a ratio of Renilla signal over Firefly signal. All error bars represent standard deviation. Asterisks (*) represent significantly different data from the corresponding ‘No sites’ control data, except in F) where they represent significantly different data from GV (Student’s T-test, p<0.05). See also Figure S1.
Next, these reporters were introduced into growing and GV/MII stage Xenopus oocytes. Since transcription is silenced in oocyte development, the reporters were in vitro transcribed, polyadenylated, and then injected. Similar to the 293T cells, growing Xenopus oocytes (stages II–IV) showed a strong repression of both reporters, which was further enhanced with co-injection of exogenous miR-15a (Figure 1C). However, once Xenopus oocytes had matured to the GV stage (stage VI), suppression of the bulge reporter was absent even when co-injected with miR-15a (Figure 1D). In contrast, knockdown of the perfect match reporter persisted. Repression of the bulge reporter was absent through oocyte maturation to MII stage eggs in Xenopus (Figure 1E). These results were similar to findings previously described for let-7 and miR-30 in early growing versus fully grown GV/MII mouse oocytes [3]. To pinpoint when translational repression is lost during oocyte development, we performed a time course analysis throughout early postnatal development in mice. Loss of translational repression began between postnatal day 17 and 18 (Figure 1F), just prior to the loss of transcription in mouse oocytes [1]. Together, these results show a conserved loss in the ability of small RNAs that imperfectly bind their targets to drive translational repression of reporters following the transition from growing to maturing oocytes.
Exogenous hAGO2 and miRNA can rescue translational suppression of a reporter
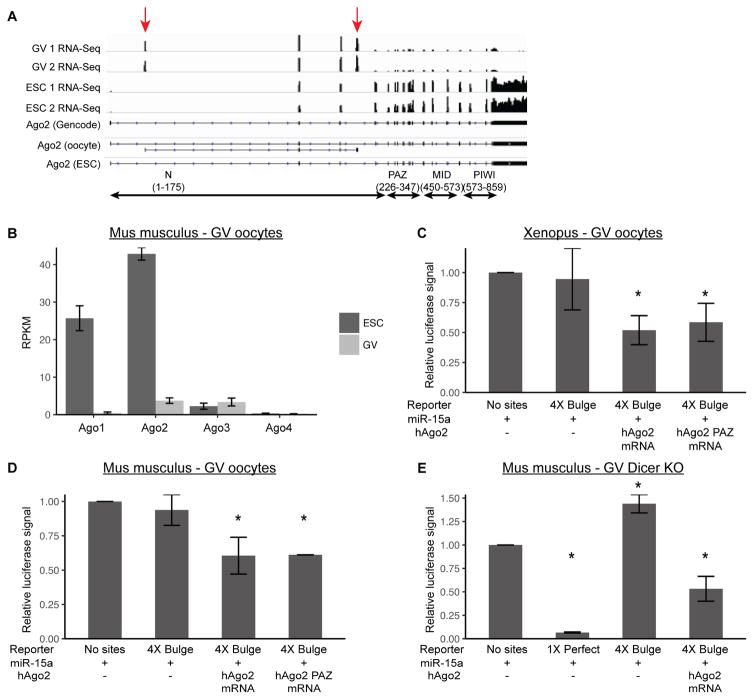
One possible explanation for the failure of GV and MII oocytes to suppress the bulge reporter is that there are rate-limiting amounts of AGO2, enough for endonucleolytic cleavage, but not translational repression and exonucleolytic destabilization. We performed RNA-sequencing on mouse GV oocytes, and saw that full length Ago2 is lowly expressed. Surprisingly, Ago2 is predominantly expressed as an unannotated truncated isoform which is not present in mouse embryonic stem cells (ESCs) (Figure 2A). RT-qPCR confirmed the presence of this isoform in oocytes and its absence in ESCs (Figure S2A). Additionally, RNA-Seq showed that Ago1, Ago3, and Ago4 were also lowly expressed in mouse GV oocytes (Figure 2B). Since the four AGO proteins are functionally equivalent for translational suppression, we decided to focus primarily on AGO2 [14]. To directly test whether AGO was rate-limiting for translational repression, we injected in vitro transcribed full-length human Ago2 (hAgo2) mRNA together with exogenous miR-15a and the bulge reporter. This approach showed a robust and reproducible repression of the reporter in both Xenopus and mouse GV oocytes (Figures 2C, D). The exogenous hAGO2 had no effect on a reporter without bulge sites (Figures S2B, C). Surprisingly, a mutant form of hAGO2 with 10 mutations in the PAZ domain [15], which has been shown not to bind miRNAs, was also able to rescue suppression of the reporter when injected together with exogenous miR-15a (Figures 2C, D). However, recent data suggests that this mutant likely retains some low-level activity, which in the context of overexpression may be enough to re-activate function [16]. These data show that exogenous hAGO2 can restore small RNA driven translational repression of an exogenously introduced reporter in maturing Xenopus and mouse oocytes.
Figure 2. Exogenous hAGO2 and miRNA can rescue translational suppression of a reporter.
A) RNA-Seq coverage of Ago2 isoforms in mouse GV oocytes and embryonic stem cells. Lower tracks represent Gencode Ago2 isoform and Stringtie reconstructed isoforms in oocytes and embryonic stem cells. Arrows denote exons present only in the oocyte isoform. B) RNA-Seq in mouse GV oocytes and mouse ESCs showing expression of the four Ago proteins. C, D) Luciferase assays in (C) Xenopus GV oocytes or (D) mouse GV oocytes with injection of miR-15a mimic and either wild-type hAGO2 or hAGO2-PAZ10 mutant mRNA. C, N=10; D, AGO2 N=7, PAZ10 N=2. E) Luciferase assays in DICER KO mouse GV oocytes, with miR-15a mimic and either a 1X Perfect or 4X Bulge template, with or without addition of hAGO2. N=2. Luciferase data is a ratio of Renilla signal over Firefly signal. All error bars represent standard deviation. Asterisks (*) represent significantly different data from the corresponding ‘No sites’ control data (Student’s T-test, p<0.05). See also Figure S2.
Since small RNA-induced endonucleolytic cleavage is vital in mouse oocytes, we considered the possibility that translational repression may be suppressed due to competition for AGO by the large number of endo-siRNAs [17,18]. To test this possibility, we repeated the luciferase assays in DICER knockout oocytes, which lack all endogenous siRNAs and miRNAs [2]. Even in the absence of endogenous small RNAs, exogenous miR-15a was not sufficient to knockdown the reporter (Figure 2E). Robust repression was only observed with both exogenous hAGO2 and miRNA (Figure 2E), suggesting that endo-siRNAs sequestering AGO2 is not the reason for the loss of translational repression.
MiRNAs can both repress the translation of their mRNA targets and destabilize them through exonucleolytic decay [19]. The predominant mechanism of action is believed to be context dependent [19–23]. Reduced luciferase activity could be due to either mechanism. To determine whether overexpression of hAGO2 affected Renilla mRNA levels, we performed RT-qPCR on injected Renilla and Firefly mRNA. While overexpression of hAGO2 and exogenous miR-15a reduced Renilla mRNA levels in 3 of 4 experiments (Figure S2D), the combined results did not reach significance and the effect was small, suggesting that the primary mechanism behind the reduced luciferase levels is translational repression.
Exogenous AGO2 and miRNA have a minimal impact on endogenous targets
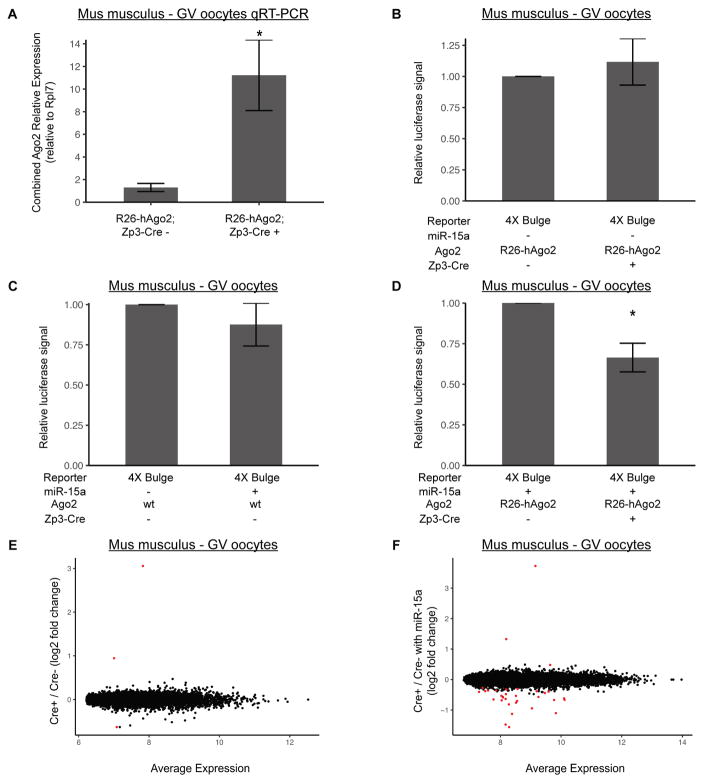
To identify the developmental and molecular consequences of re-activating translational repression in the oocyte, we turned to a genetic system in the mouse where exogenous full length hAGO2 is expressed from the Rosa26 promoter following Cre-mediated excision (R26-lox-stop-lox-GFP-myc-hAGO2) [24]. We crossed this mouse to an oocyte specific Cre (Zp3-Cre) to drive oocyte specific overexpression of full length hAGO2 [25]. RT-qPCR showed a 10-fold increase in expression of total Ago2 expression in Cre-positive GV oocytes (Figure 3A). This increase in expression alone did not repress the bulge reporter (Figure 3B). Similarly, exogenous miR-15a alone induced only a slight and inconsistent downregulation (Figure 3C). However, the two together showed robust knockdown of the bulged reporter (Figure 3D), similar to that seen with injection of the exogenous AGO2 mRNA along with miRNA.
Figure 3. Both Exogenous hAGO2 and miRNA are required to rescue miRNA activity, but have minimal impact on endogenous targets.
A) RT-qPCR of combined mouse and human Ago2 mRNA levels in LSL-GFP-myc-hAGO2/+; Zp3-Cre+ and Zp3-Cre− mouse GV oocytes. Primers recognize both mouse and human AGO2. Rpl7 is used as a reference gene. N=4. B) Luciferase assay in LSL-GFP-myc-hAGO2/+; Zp3-Cre+ and Zp3-Cre− mouse GV oocytes with no miR-15a mimic and 4x Bulge template. N=2. C) Luciferase assay in wildtype mouse GV oocytes with miR-15a mimic and 4x Bulge template. N=4. D) Luciferase assay in LSL-GFP-myc-hAGO2/+; Zp3-Cre+ and Zp3-Cre− mouse GV oocytes with miR-15a mimic and 4x Bulge template. N=3. Illumina Bead Array RNA profiling of: E) LSL-GFP-myc-hAGO2/+; Zp3-Cre+ versus Zp3-Cre− mouse GV oocytes. F) LSL-GFP-myc-hAGO2/LSL-GFP-myc-hAGO2; Zp3-Cre+ versus Zp3-Cre− mouse GV oocytes injected with miR-15a mimic. Significant differentially expressed transcripts shown in red (FDR < 0.05). E, N=3; F, N=4. Data is a ratio of Renilla signal over Firefly signal. All error bars represent standard deviation. Asterisks (*) represent significantly different data from the corresponding control data (Student’s T-test, p<0.05). See also Figure S3.
Next, we asked how this system affects the endogenous transcriptome. We performed microarray profiling on both control and Rosa26 driven hAGO2-overexpressing mouse GV oocytes. Only 3 genes were differentially expressed at a false discovery rate (FDR) < 0.05, including Ago2 itself (Figure 3E). These findings are consistent with the reporter data showing that hAGO2 overexpression alone failed to rescue suppression. Surprisingly though, hAGO2 overexpression along with exogenous miR-15a injection also had very little impact on the transcriptome (Figure 3F). 29 genes were down-regulated with a FDR < 0.05, while 3 genes were stabilized (i.e. ‘up-regulated’) including Ago2 itself. We further evaluated the 29 down-regulated genes to determine if they were enriched for miR-15a targets. Of 1121 conserved targetscan miR-15a targets, 778 are found on the array and 444 are expressed in oocytes, but only 1 was among the down-regulated genes. Biological replicates were well correlated, and thus the lack of changes is not due to noise between replicates (Figure S3). These findings show that exogenous hAGO2-miRNA complexes have little effect on the transcriptome of oocytes either through destabilization of direct miR-15a targets or through indirect targets which would likely be altered due to translational changes of miR-15a targets. This finding suggests that AGO2 levels alone are unlikely to explain the loss of miRNA activity in maturing oocytes.
Isoforms with longer 3′ UTRs are more stable in maturing oocytes
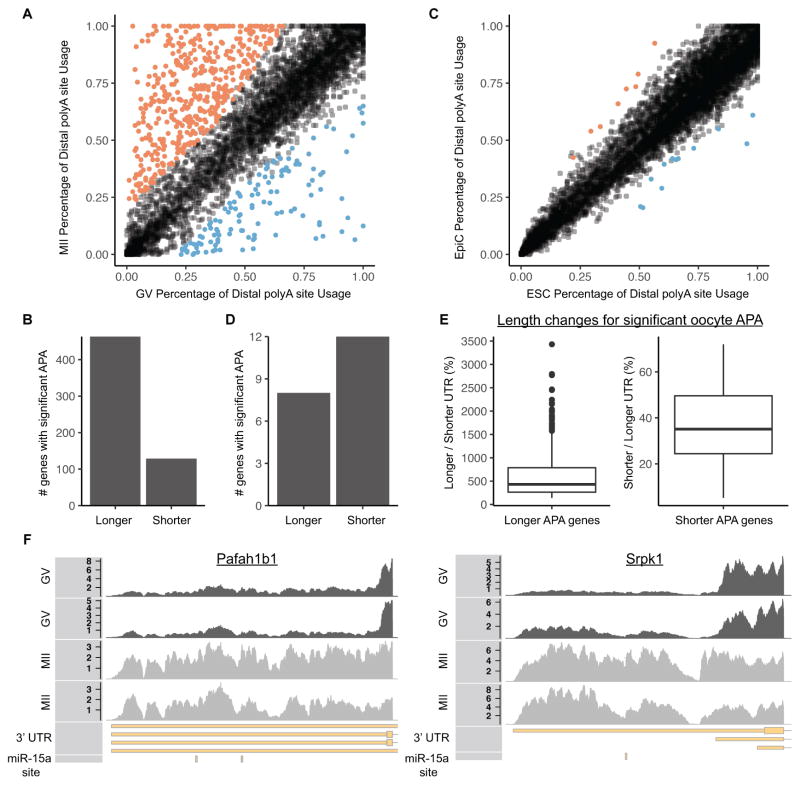
An additional mechanism of suppressing miRNA activity could occur at the level of the target transcript. Alternative polyadenylation (APA) produces 3′ UTRs of differing lengths [26]. In somatic cells, a shorter 3′ UTR is generally associated with greater mRNA stability, at least in part due to a decrease in the number of miRNA target sites [27–29]. Therefore, we evaluated alternative polyadenylation usage during the GV to MII transition. Hundreds of transcripts showed differential 3′ UTR length during the GV to MII transition (Figures 4A, B). For comparison, we looked at an alternative transition, the embryonic stem cell (ESC) to epiblast like cell (EpiC) transition, a time of abundant miRNA activity [30]. There were very few changes in 3′ UTR length during this transition (Figures 4C, D). Surprisingly, the changes in the maturing oocyte showed a strong enrichment for distal polyadenylation usage in MII versus GV oocytes (Figures 4B vs. 4D). Four hundred and sixty-three genes showed preferential stability of transcripts using the distal polyadenylation site (median 4.3 fold increase in 3′ UTR length) while one hundred and twenty-nine genes showed preferential stability of transcripts using the proximal polyadenylation site (median 2.8 fold decrease in 3′ UTR length) (Figure 4E). Individual tracks for the Pafah1b1 and Srp1k genes exemplified the striking switch between short and long forms between GV and MII oocytes (Figure 4F). Many of the 3′ UTRs gained during the GV to MII transition had target sites for miR-15a, but were not impacted by introduction of exogenous miR-15a and AGO2 (Figure 4F and Table S2). Given the absence of transcription during the GV to MII transition, these changes must reflect the preferential degradation of the short 3′ UTR isoforms, directly contrasting the previous findings in non-oocyte populations, where the long isoforms were preferentially degraded [27–29]. Thus, preferential expression of short 3′ UTR isoforms does not underlie the resistance of maturing oocytes to miRNA driven exonucleolytic decay.
Figure 4. Isoforms with longer 3′ UTRs are more stable in maturing oocytes.
(A) Percentage of distal alternative polyadenylation site usage in GV and MII oocytes. Significant increases in distal alternative polyadenylation usage are colored in orange, significant decreases are colored in blue. Each dot represents one gene. (B) Number of significant changes in alternative polyadenylation ratios resulting in longer 3′ UTRs or shorter 3′ UTRs during the GV to MII transition. (C) Same as A, except in mouse ESCs and EpiCs. (D) Same as B, except in mouse ESCs and EpiCs. (E) Boxplots showing change in 3′ UTR length for genes with a significant change in alternative polyadenylation during the GV to MII transition. Left plot shows the ratio of the longer 3′ UTR relative to the shorter 3′ UTR for genes where the distal polyadenylation site is more stable (n = 463 genes). Right plot shows the ratio of the shorter 3′ UTR relative to the longer 3′ UTR for genes where the proximal polyadenylation site is more stable (n = 129 genes). (F) RNA-Seq coverage showing stabilization of distal alternative polyadenylation site in GV to MII transition at two genes (Pafah1b1 and Srpk1). Both genes are on the negative strand (transcribed from right to left). 2 GV replicates, 2 MII replicates, Gencode annotated 3′ UTR, miR-15a TargetScan predicted target site. See also Figure S4. See also Table S2.
DISCUSSION
In this study, we sought to uncover the mechanism underlying the surprising finding that miRNA function is absent during oocyte maturation, a discovery originally made in mouse oocytes [2,3]. Here, we show that translational repression is also absent in maturing Xenopus oocytes, showing conservation across vertebrates. In both species, translational repression was suppressed around the transition from oocyte growth to maturation. Furthermore, in both species, the introduction of exogenous hAGO2 and miRNA were able to rescue repression of reporter constructs. Recent studies have shown that miRNA levels and AGO levels are intimately connected; each stabilizing the other [31,32]. This mutual dependency could explain why introducing both together is necessary for robust knockdown.
In mouse oocytes, RNA-Sequencing showed that Ago2 predominantly exists as a truncated form of AGO2 missing the PAZ, MID, and PIWI domains, making it highly unlikely that it retains activity [33]. The low levels of full length AGO2 may be enough for the observed endonucleolytic cleavage, but not translational repression. Indeed, slicing is a more efficient process than translational repression and thus likely to require significantly less AGO2 [34]. Alternatively, truncated AGO2 might act as a dominant negative, but overexpression of the truncated form in 293Ts did not negatively impact repression (Figure S4A). Therefore, the most likely explanation is that low levels of full length AGO2 contributes to the loss of miRNA induced repression.
However, this mechanism alone does not fully explain the lack of miRNA repression in oocytes. Even in the presence of exogenous AGO2 and miRNAs, there was minimal degradation of luciferase mRNA. This finding contrasts the effects seen in somatic cells where exonucleolytic decay plays a predominant role in the inhibition of target mRNAs [22]. In addition, expression profiling showed that endogenous targets of miR-15a were not destabilized at the transcript level upon introduction of exogenous AGO2 and miR-15a. Furthermore, there was a near absence of transcriptional changes globally. While it is possible that miRNAs solely repress translation in this context, the altered translation of hundreds of miR-15a targets would be expected to produce secondary mRNA changes. Therefore, additional mechanisms are likely blocking miRNA function in oocytes.
The nature of additional mechanisms that could underlie the global suppression of miRNA activity in oocytes is unclear. However, additional effectors of miRNA activity are also lowly expressed in the oocyte. Most notably, several proteins involved in exonucleolytic decay including Xrn1, the primary 5′ to 3′ exonuclease downstream of miRNA activity, are also lowly expressed in oocytes (Figure S4B). Furthermore, components of the de-adenylation and decapping complexes are lowly expressed and minimally active in GV oocytes [35,36]. The reduced exonucleolytic activity could also explain the previously described persistence of RNA fragments in oocytes [37]. Recent work has also shown that post-translational modifications of AGO2 are essential for normal function [38]. In particular, cycles of phosphorylation and dephosphorylation are essential for optimal activity. Future measurements of the activities of the kinase and phosphatase responsible for these cycles could uncover a potential role for these factors in the loss of miRNA activity in maturing oocytes.
Another potential mechanism to avoid miRNA function would be preferential expression of transcript isoforms with short 3′ UTRs reducing the number of potential miRNA targeting sites. However, in contrast to previous work on non-oocytes [27–29], we found that longer 3′ UTRs are associated with increased rather than decreased stability of the cognate transcript during oocyte maturation. This finding suggests inhibition of miRNA function upstream of the target transcript. How longer 3′ UTRs stabilize transcripts in the oocyte is unclear. It is possible that the binding of stabilizing RNA binding proteins (RBPs) to the 3′ UTRs in the absence of miRNA activity underlies this stabilization and might even serve as a redundant mechanism to prevent miRNA activity. Indeed, oocytes express many RBPs that bind the 3′ UTRs of transcripts to promote cytoplasmic polyadenylation, which in turn promotes stabilization, or that directly antagonize miRNA function [19,39–41]. Thus, the longer 3′ UTRs could provide docking sites for these RBPs, which in turn suppresses degradation of the cognate mRNA.
In summary, we have identified an alternative truncated isoform of AGO2, which is the dominant AGO species in maturing oocytes. Re-introduction of full-length hAGO2 together with an exogenous miRNA represses reporters in a setting where endogenous miRNA function is absent. However, endogenous transcripts are still resistant to miRNA driven exonucleolytic activity. Therefore, there appear to be multiple mechanisms of globally suppressing miRNA activity in the maturing vertebrate oocyte, one of which is a dramatic reduction in the levels of functional AGO-miRNA complexes.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Robert Blelloch (robert.blelloch@ucsf.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
R26-hAGO2 mice
Rosa26 lox-stop-lox-myc-GFP-hAGO2 (R26-hAGO2) mice [24] were obtained from the Jackson Laboratory (JAX stock #017626). These mice were crossed to C57/B6 Zp3-Cre mice [25] to generate R26-hAGO2; Zp3-Cre mice. For all experiments, R26-hAGO2; Zp3-Cre− females were bred to R26-hAGO2; Zp3-Cre+ males. Oocytes were collected from 3–4 week old female Cre− and Cre+ littermates. Mice were housed per UCSF and IACUC guidelines.
Wildtype mice
C57/B6 mice were ordered from the Jackson Laboratory (JAX stock #00064) and Simonsen Laboratory (Stock #C57BL/6NCrSim). Oocytes were collected from 3–4 week old females.
Xenopus laevis
Mature female Xenopus laevis were ordered from Nasco (Stock #LM00535MX) and cared for per supplier instructions and IACUC guidelines. Mature females were staged by length (9+ cm) according to Nasco instructions.
Cell lines and cell culture
293Ts were grown in DMEM supplemented with 10% Fetal Bovine Serum. ESCs were grown in Knockout DMEM (Invitrogen) supplemented with 15% Fetal Bovine Serum, LIF and 2i (PD0325901 and CHIR99021). To generate Epi cells, ESCs were plated at 400,000 cells in a 15 plate, 24 hours later LIF and 2i were removed and Epi Cells were collected ~56 hours later [42]. ESCs are male. 293Ts are female. ESC lines were authenticated via RNA-Seq.
METHOD DETAILS
Constructs
miRNA binding sites were ordered as oligos from IDT. Oligos were phosphorylated, annealed, and cloned into the 3′ UTR of psiCHECK2 (Promega) using the restriction enzymes NotI and XhoI. Truncated AGO2 was amplified from oocyte cDNA and cloned into pGAMA. Full length hAGO2 was amplified from an existing plasmid and cloned into pGAMA (see Table S1 for oligonucleotide sequences).
In vitro transcription
Linearized psiCHECK2 vector (Promega), pCDNA3-myc-hAGO2 [7], or pCDNA3-RFP was in vitro transcribed and capped (Ambion). mRNA was then polyadenylated by Poly(A) tailing kit (Ambion).
Mouse oocyte collection and injection
48 hours prior to injection mice were primed with 5 units of PMSG. GV oocytes were collected in MEM medium supplemented with 2uM milrinone to prevent resumption of meiosis. In vitro transcribed mRNA and 2uM miR-15a mimic (Dharmacon) were injected on a Leica DMI3000 B with an Eppendorf FemtoJet. Oocytes were cultured in MEM medium supplemented with 2uM milrinone overnight at 37° and 5% CO2 for luciferase assays.
Luciferase assay
Luciferase assays were performed using the Dual Luciferase Reporter Assay kit (Promega) and measured on a SpectraMaxL Luminometer (Molecular Devices). Renilla signal was normalized to firefly signal for all experiments.
Microarray
RNA was extracted from GV oocytes with the PicoPure RNA Isolation Kit (ARCTURUS). RNA was sent to the UCLA Neuroscience Genomics Core where cDNA was amplified (Nugen) and then hybridized to Illumina BeadChips. Data was pre-processed and normalized in R version 3.3.2 using bead array version 2.24.0 [43]. Differential expression was performed in R using limma version 3.30.4 [44].
AGO2 RT-qPCR
RNA was extracted with TRIzol (Invitrogen). cDNA was made using Superscript III First-strand Synthesis kit (Invitrogen) following the standard protocol. qPCR was performed using Power SYBR Green mix (Applied Biosystems) on an ABI 7900HT 384-well PCR machine.
Luciferase RT-qPCR
RNA was extracted with PicoPure RNA Isolation Kit (ARCTURUS). cDNA was made using Maxima RT cDNA kit (Thermo Scientific) following the standard protocol. qPCR was performed using Roche SYBR Green mix (Roche) on an ABI 7900HT 384-well PCR machine.
RNA-Seq
RNA from GV oocytes and mouse embryonic stem cells was extracted in TRIzol (Invitrogen) and sequencing libraries were made using the Illumina stranded paired-end Truseq kit (Illumina). Reads were mapped to the mm10 mouse genome using STAR version 2.5.1b [45]. Reads were counted with featureCounts [46] version 1.5.0-p1 using the Gencode m10 genome annotation. De novo isoform discovery was performed in stringtie version 1.2.4 [47].
Transfections
All transfections were done in 96 well plates with 200 ng of DNA following the standard Fugene 6 (Promega) protocol.
Xenopus oocyte collection and injection
To prepare Xenopus laevis for surgery, animals were anaesthetized using a 2 grams/liter solution of tricaine methane sulfonate (MS-222) buffered to a neutral pH of 7.0–7.5 using sodium bicarbonate. Autoclave-sterilized tools were used to perform live surgery, and oocytes were removed by biopsying the ovary (no more than 50%). The frogs were then re-sutured using nylon monofilament suture, and monitored in a shallow tank until they were fully awake and mobile. They were then returned to their housing tank, and observed daily for 3 days. Frogs were used for surgery only once every 3 months (or longer if necessary).
Once harvested, oocytes were dissociated in 2 mg/ml collagenase in modified Barth’s saline (1X MBS) at room temperature for 1–3 hours, being constantly monitored to avoid over digestion. The oocytes were then transferred to culture media (10% fetal calf serum, 1X penicillin/streptomycin, 100 μg/mL gentamycin, 50% L-15 medium (Leibovitz) + glutamine) for subsequent procedures and incubations.
Xenopus oocytes were injected using a microINJECTOR system (Tritech research). Concentrations of materials injected were the same as for mouse oocytes (see above). For luciferase assays, oocytes were incubated overnight at 16C, and lysed (single oocytes in individual wells of a 96 well plate) using as directed in the Dual Luciferase Reporter Assay kit (see above).
For maturation of oocytes into eggs (MII), stage VI oocytes were incubated with 3uM progesterone in culture media overnight.
APA analysis
DaPars [48] versin 0.9.1 was used to identify Percentage of Distal polyA site Usage Index (PDUI). DaPars was run with the default settings from the original paper describing the software: Adjusted P Val < 0.05, minimum ΔPDUI of 0.2, and minimum 1.5 fold change in PDUI. For genes with more than two transcripts and more than 2 ΔPDUI changes, the greatest ΔPDUI and lowest adjusted P value was used. Conserved miRNA target sites were downloaded from TargetScan 7.1.
Experimental Design
For all experiments n indicating the number of biological replicates are given in figure legends. The study was not done blinded. No data was excluded. Based on previous literature we assume luciferase data follows a t-distribution and use a t-test for calculating significance. Different litters were used for different biological replicates.
QUANTIFICATION AND STATISTICAL ANALYSIS
For all experiments n indicating the number of biological replicates and additional details are listed in the figure legend. For oocyte experiments, biological replicates refers to pools of oocytes collected on different days. For cell lines, biological replicates refers to separate culture dishes of cells. All luciferase data is a ratio of Renilla signal over Firefly signal. All error bars represent standard deviation. Asterisks (*) represent significant differences with student’s T-test, p < 0.05. For array and sequencing analysis differences were defined with an adjusted P value < 0.05. Statistical significance was calculated in R version 3.3. To the extent possible, we checked if data met assumptions of statistical methods used.
DATA AND SOFTWARE AVAILABILITY
All array and sequencing data has been submitted to GEO under accession number: GSE92768
Supplementary Material
Output of DaPars analysis for each transcript with sufficient coverage in the 3′ UTR between GV and MII oocytes. Columns show the predicted proximal APA site, long and short APA usage in GV (A1/A2) and MII (B1/B2) samples, Percentage of Distal polyA site Usage Index (PDUI) in GV (A1/A2) and MII (B1/B2) samples, the change in PDUI, an adjusted P Value for the significance of the change in PDUI, whether the PDUI change passed the significance filter (Adjusted P Val < 0.05, minimum ΔPDUI of 0.2, and minimum 1.5 fold change in PDUI), gene symbol, and whether the gene is predicted to be a TargetScan miR-15a target.
Acknowledgments
We thank the following people for critical reading of the manuscript: Amy Chen, Brian DeVeale, and Archana Shenoy. We also thank Marco Conti and the Conti lab (UCSF), and James Ferrell and the Ferrell lab, specifically Tony Tsai (Stanford University) for technical assistance with Xenopus experiments. We also acknowledge Indiana University for access to their Mason cluster of computers, supported by the National Science Foundation (DBI #1458641). This project was funded by the National Institute of Child Health and Disease (P50 HD055764), a Genentech Predoctoral Research Fellowship to J.W.F, and an AP Gianinni Postdoctoral Award to R.K.
Footnotes
AUTHOR CONTRIBUTIONS
J.W.F., R.K., and R.B. conceived the study, designed the study, and wrote the manuscript.
J.W.F., R.K., and M.S.C performed experiments.
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abe KI, Inoue A, Suzuki MG, Aoki F. Global gene silencing is caused by the dissociation of RNA polymerase II from DNA in mouse oocytes. J Reprod Dev. 2010;56:502–507. doi: 10.1262/jrd.10-068a. [DOI] [PubMed] [Google Scholar]
- 2.Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr Biol. 2010;20:271–277. doi: 10.1016/j.cub.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma J, Flemr M, Stein P, Berninger P, Malik R, Zavolan M, Svoboda P, Schultz RM. MicroRNA activity is suppressed in mouse oocytes. Curr Biol. 2010;20:265–270. doi: 10.1016/j.cub.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babiarz, Blelloch Small RNAs – their biogenesis, regulation and function in embryonic stem cells. StemBook. 2009 Available at: http://dx.doi.org/10.3824/stembook.1.47.1. [PubMed]
- 5.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 8.Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for Dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang F, Kaneda M, O’Carroll D, Hajkova P, Barton SC, Sun YA, Lee C, Tarakhovsky A, Lao K, Surani MA. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stein P, Rozhkov NV, Li F, Cárdenas FL, Davydenko O, Davydenk O, Vandivier LE, Gregory BD, Hannon GJ, Schultz RM. Essential Role for endogenous siRNAs during meiosis in mouse oocytes. PLoS Genet. 2015;11:e1005013. doi: 10.1371/journal.pgen.1005013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaneda M, Tang F, O’Carroll D, Lao K, Surani MA. Essential role for Argonaute2 protein in mouse oogenesis. Epigenetics Chromatin. 2009;2:9. doi: 10.1186/1756-8935-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shenoy A, Blelloch RH. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat Rev Mol Cell Biol. 2014;15:565–576. doi: 10.1038/nrm3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu S, Jin L, Huang Y, Zhang F, Kay MA. Slicing-independent RISC activation requires the argonaute PAZ domain. Curr Biol. 2012;22:1536–1542. doi: 10.1016/j.cub.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 19.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 20.Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D, Hsu SH, Ghoshal K, Villén J, Bartel DP. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol Cell. 2014;56:104–115. doi: 10.1016/j.molcel.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alemán LM, Doench J, Sharp PA. Comparison of siRNA-induced off-target RNA and protein effects. RNA. 2007;13:385–395. doi: 10.1261/rna.352507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, Huang ZJ. Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron. 2012;73:35–48. doi: 10.1016/j.neuron.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Vries WN, Binns LT, Fancher KS, Dean J, Moore R, Kemler R, Knowles BB. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis. 2000;26:110–112. [PubMed] [Google Scholar]
- 26.Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14:496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 27.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spies N, Burge CB, Bartel DP. 3′ UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res. 2013;23:2078–2090. doi: 10.1101/gr.156919.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu KL, Zhang Q, Yan Y, Li TT, Duan FF, Hao J, Wang XW, Shi M, Wu DR, Guo WT, et al. Pluripotency-associated miR-290/302 family of microRNAs promote the dismantling of naive pluripotency. Cell Res. 2016;26:350–366. doi: 10.1038/cr.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zamudio JR, Kelly TJ, Sharp PA. Argonaute-bound small RNAs from promoter-proximal RNA polymerase II. Cell. 2014;156:920–934. doi: 10.1016/j.cell.2014.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez NJ, Gregory RI. Argonaute2 expression is post-transcriptionally coupled to microRNA abundance. RNA. 2013;19:605–612. doi: 10.1261/rna.036434.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 34.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma J, Fukuda Y, Schultz RM. Mobilization of Dormant Cnot7 mRNA Promotes Deadenylation of Maternal Transcripts During Mouse Oocyte Maturation. Biol Reprod. 2015;93:48. doi: 10.1095/biolreprod.115.130344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma J, Flemr M, Strnad H, Svoboda P, Schultz RM. Maternally recruited DCP1A and DCP2 contribute to messenger RNA degradation during oocyte maturation and genome activation in mouse. Biol Reprod. 2013;88:11. doi: 10.1095/biolreprod.112.105312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Svoboda P, Franke V, Schultz RM. Sculpting the Transcriptome During the Oocyte-to-Embryo Transition in Mouse. Curr Top Dev Biol. 2015;113:305–349. doi: 10.1016/bs.ctdb.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 38.Golden RJ, Chen B, Li T, Braun J, Manjunath H, Chen X, Wu J, Schmid V, Chang TC, Kopp F, et al. An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature. 2017;542:197–202. doi: 10.1038/nature21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Racki WJ, Richter JD. CPEB controls oocyte growth and follicle development in the mouse. Development. 2006;133:4527–4537. doi: 10.1242/dev.02651. [DOI] [PubMed] [Google Scholar]
- 40.Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JAF, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 41.Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JAF, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Ørom UA, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 42.Krishnakumar R, Chen AF, Pantovich MG, Danial M, Parchem RJ, Labosky PA, Blelloch R. FOXD3 Regulates Pluripotent Stem Cell Potential by Simultaneously Initiating and Repressing Enhancer Activity. Cell Stem Cell. 2016;18:104–117. doi: 10.1016/j.stem.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunning MJ, Smith ML, Ritchie ME, Tavaré S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23:2183–2184. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- 44.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 47.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, Li W. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat Commun. 2014;5:5274. doi: 10.1038/ncomms6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Output of DaPars analysis for each transcript with sufficient coverage in the 3′ UTR between GV and MII oocytes. Columns show the predicted proximal APA site, long and short APA usage in GV (A1/A2) and MII (B1/B2) samples, Percentage of Distal polyA site Usage Index (PDUI) in GV (A1/A2) and MII (B1/B2) samples, the change in PDUI, an adjusted P Value for the significance of the change in PDUI, whether the PDUI change passed the significance filter (Adjusted P Val < 0.05, minimum ΔPDUI of 0.2, and minimum 1.5 fold change in PDUI), gene symbol, and whether the gene is predicted to be a TargetScan miR-15a target.