Abstract
Removal of toxic Cr(VI) by microbial reduction is a promising approach to reducing its ecotoxicological impact. To develop bioremediation technologies, many studies have evaluated the application of microorganisms isolated from Cr(VI)-contaminated sites. Nonetheless, little attention has been given to microbes from the environments without a history of Cr(VI) contamination. In this study, we aimed to characterize the Cr(VI) tolerance and removal abilities of a filamentous fungus strain, SL2, isolated from indoor air. Based on phenotypic characterization and rDNA sequence analysis, SL2 was identified as Penicillium oxalicum, a species that has not been extensively studied regarding Cr(VI) tolerance and reduction abilities. SL2 showed high tolerance to Cr(VI) on solid and in liquid media, facilitating its application to Cr(VI)-contaminated environments. Growth curves of SL2 in the presence of 0, 100, 400, or 1000 mg/L Cr(VI) were well simulated by the modified Gompertz model. The relative maximal colony diameter and maximal growth rate decreased as Cr(VI) concentration increased, while the lag time increased. SL2 manifested remarkable efficacy of removing Cr(VI). Mass balance analysis indicated that SL2 removed Cr(VI) by reduction, and incorporated 0.79 mg of Cr per gram of dry biomass. In electroplating wastewater, the initial rate of Cr(VI) removal was affected by the initial contaminant concentration. In conclusion, P. oxalicum SL2 represents a promising new candidate for Cr(VI) removal. Our results significantly expand the knowledge on potential application of this microorganism.
Introduction
Chromium (Cr) is a cause for considerable environmental concern because of its improper release into the environment from anthropogenic activities [1, 2]. Controlling the chemical state of Cr is vital for reducing its ecotoxicological impact. In nature, Cr mainly exists as Cr(VI) and Cr(III) compounds [3], which differ in their mobility and toxicity [4]. Cr(VI) compounds are water soluble in the full pH range and are toxic to humans [5, 6], animals [7], plants [8], and microorganisms [9], whereas Cr(III) compounds are less water soluble and serve as essential nutrients for energy metabolism [10]. Thus, reducing Cr(VI) to Cr(III) can minimize its harm to the environment and human health.
Various technologies have been developed to reduce Cr(VI) [11], including physicochemical and biological approaches [12]. Physicochemical remediation using functional materials such as polymers and nano materials is effective at Cr(VI) removal [13], however, most of them are expensive for large scale application and produce secondary environmental pollution. Alternatively, bioremediation by means of Cr(VI)-tolerant and -reductive microorganisms is considered particularly promising, and is eco-friendly and cost-effective [14]. Since Cr(VI) reduction by Pseudomonas dechromaticans was reported in the 1970s [15], many microorganisms with Cr(VI)-tolerant and -reductive properties have been isolated [16]. Nonetheless, most of these isolates are likely to be susceptible to Cr(VI) toxicity at higher concentrations, and have low efficacy of Cr(VI) removal [17], limiting their bioremediation applications. Hence, isolation of high-performance microorganisms is necessary to develop highly effective biological treatment technologies for Cr(VI) removal. Moreover, previous studies have mainly focused on Cr(VI)-tolerant and -reductive microorganisms isolated from Cr(VI)-contaminated sites [18], while little attention has been paid to microbes living without Cr(VI) selection pressure. To the best of our knowledge, no microorganisms have been isolated from indoor air for Cr(VI) removal. The isolation of Cr(VI)-tolerant and -reductive microorganisms from those environments with no history of Cr(VI) contamination may provide new candidates for Cr(VI) removal. Additionally, although fungi have certain advantages over bacteria [19], they have received less attention in studies on bioremediation of Cr(VI) contaminated environments. Hence, in the present study, we attempted to isolate a fungal strain from indoor air with the goal of characterizing its Cr(VI) tolerance and removal abilities via the modified Gompertz model and mass balance analysis. This study provides a new candidate for Cr(VI) removal, and the results significantly expand our knowledge on the utility of this microorganism.
Experimental methods
Ethics statement
No specific permits were required for the present study. Isolation of the Cr(VI)-tolerant fungus from indoor air of our work room did not cause any disturbance to the environment or involve protected species.
Isolation of a Cr-tolerant microorganism
This fungal strain, which was named SL2, was isolated by a method similar to that previously employed for isolating Cr(VI)-tolerant fungi from air contaminated with industrial vapors [20]. The potato dextrose agar (PDA) solid medium was used for microorganism enrichment, and consisted of 1 g of dextrose, 1.8 g of agar, and 100 mL of filtered soup of 20 g of potatoes boiled for 30 min. The medium was autoclaved at 115°C for 20 min, cooled to approximately 60°C, supplemented with filter-sterilized potassium dichromate (300 mg/L) as a Cr(VI) source, and poured into dishes with 12 cm diameter. The dishes were placed in the open to collect potential Cr(VI)-tolerant and -reductive filamentous fungi from the indoor air of our work room in the Nongshenghuan Building at Zhejiang University in Hangzhou, Zhejiang, China (30°17'51.3"N 120°05'27.3"E). Upon growth and sporulation of the filamentous fungi, a spore suspension (SS) was prepared by rinsing the dishes in sterile water and then serially diluting the water 10-fold. The diluted SS (0.1 mL) was used to obtain single colonies by the spread plate method.
Identification of the isolate
The isolated filamentous fungus was identified by phenotypic characterization and phylogenetic analysis. The latter was based on ribosomal DNA (rDNA; 18S rDNA, ITS, and 26S rDNA) amplification and sequence comparison. The primers for rDNA amplification are listed in Table 1. PCR was run as follows: pre-denaturation at 94°C for 10 min, then 32 cycles of denaturation at 94°C for 60 s, annealing at 55°C for 60 s, and polymerization at 72°C for 60 s (90 s for 18S rDNA). The obtained amplicon sequences were compared with published fungal sequences via BLAST (http://www.ncbi.nlm.nih.gov/blast) to identify the species of the isolate.
Table 1. Primers for the PCR amplification of rDNA.
| rDNA | Primer name | Sequence |
|---|---|---|
| 18S | NS1 | 5′-GTAGTCATATGCTTGTCTC-3′ |
| NS8 | 5′-TCCGCAGGTTCACCTACGGA-3′ | |
| ITS1 | ITS1 | 5′-TCCGTAGGTGAACCTGCGG-3′ |
| ITS4 | 5′-TCCTCCGCTTATTGATATGC-3′ | |
| 26S | NL1 | 5′-GCATATCAATAAGCGGAGGAAAAG-3′ |
| NL4 | 5′-GGTCCGTGTTTCAAGACGG-3′ |
Growth of the isolate under Cr(VI) stress
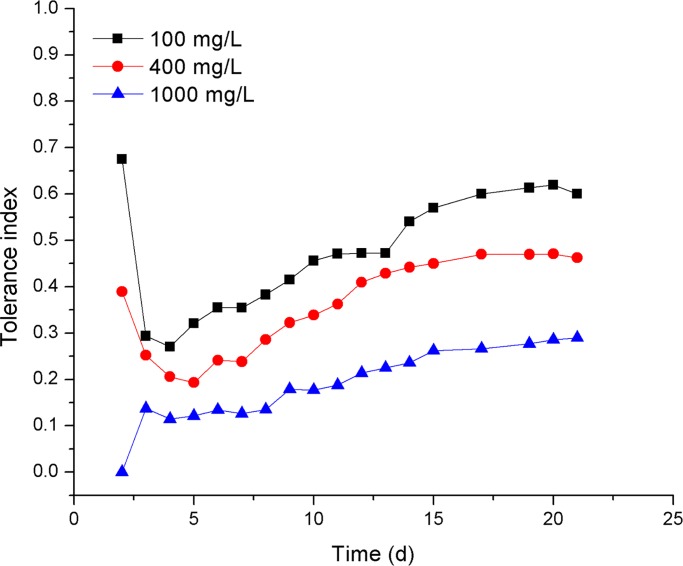
The isolate was tested for Cr(VI) tolerance in PDA supplemented with different Cr(VI) concentrations. In accordance with another study that evaluated the heavy metal tolerance of fungi [21], the diameter of colonies was measured to quantify the growth of the isolate. The effect of Cr(VI) concentration on growth was analyzed by comparing the diameters of the fungal colonies. The PDA plates were subdivided into four sets and supplemented with 0, 100, 400, or 1000 mg/L Cr(VI), inoculated with 0.5 μL of SS, and incubated at 30°C. Cultures grown in the absence of Cr(VI) served as controls. The response of the isolate to Cr(VI) was expressed as a tolerance index, which was calculated as the mean diameter of the colonies exposed to Cr(VI) divided by that of the unexposed colonies [22].
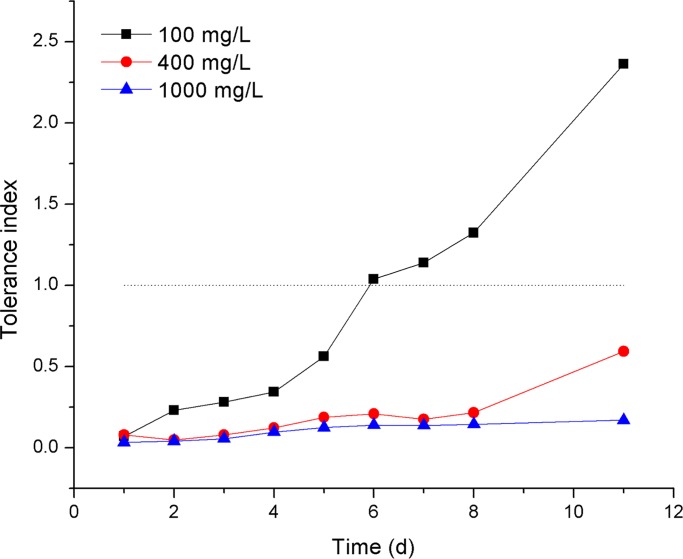
Cr(VI) has been reported to have different toxic effects on microorganisms in a liquid medium as compared to a solid medium [23]. Thus, Cr(VI) tolerance of the isolate was assessed in a potato dextrose liquid medium (PDL) supplemented with different Cr(VI) concentrations. In keeping with the composition of PDA, PDL consisted of 1 g of dextrose and 100 mL of filtered soup of potato, and was autoclaved at 115°C for 20 min before use. PDL was divided into four aliquots and supplemented with 0, 100, 400, or 1000 mg/L Cr(VI). PDL without Cr(VI) served as the control. The groups were inoculated with SS and incubated in 250-mL conical flasks at 30°C in a rotary shaker (200 rpm). For each group, 3 replicates were filtered at regular intervals to obtain the mycelia, which were dried to a constant weight at 65°C in an oven for the analysis of dry biomass. The effects of Cr(VI) on growth were analyzed by comparing the dry biomass of the isolates grown in PDL containing different Cr(VI) concentrations. The tolerance index was calculated as the dry weight of the mycelia exposed to Cr(VI) divided by that of the control.
Modeling the isolate’s growth curve under Cr(VI) stress
Growth curves of SL2 exposed to Cr(VI) were simulated by means of the modified Gompertz model, which is a three-parameter system and can describe microbial growth quantitatively [24]. The model can be expressed by the following equation [25]:
| (1) |
where y is the relative colony diameter, t is time, and the three parameters, A, μm, and λ are the relative maximal colony diameter, maximal growth rate, and lag time, respectively. A and μm indicate the growth of a microorganism, and λ means the time required for the microorganism to get to the reproductive stage [26].
The data for this analysis were obtained from the Cr(VI) tolerance test in PDA as described above. To train and validate the model, the diameters of the colonies grown at each Cr(VI) concentration were subdivided into a training data set and a validation data set. The training set consisted of diameters obtained on days 2, 3, 4, 6, 7, 8, 9, 11, 12, 13, 14, 17, 19, and 21. The validation set consisted of diameters obtained on days 5, 10, 15, and 20. The performance of the model was assessed by the coefficient of determination (R2), mean squared error (MSE), and Nash–Sutcliffe efficiency coefficient (NS). R2 measures the correlation between the experimental data and predicted values; MSE indicates the discrepancy between the measured and calculated values; and NS provides information on the predictive capability of the model. A perfect fit between the measured and predicted values would look like R2 = 1.0, MSE = 0, and NS = 1.0 [27].
Scanning electron microscopy (SEM) analysis
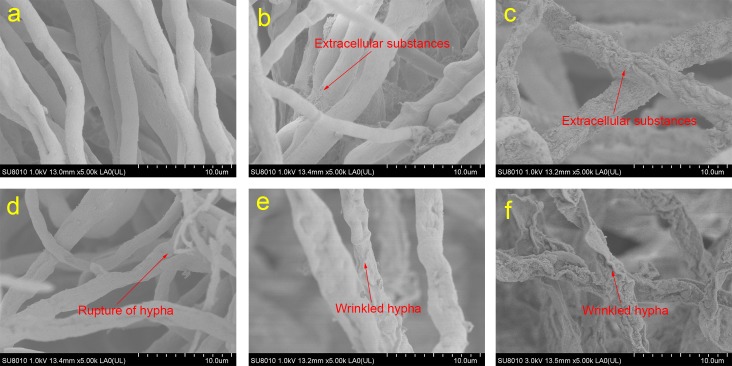
SEM is useful for characterizing materials with an application potential in pollutant removal [28, 29]. In the present study, SEM analysis was carried out to detect morphological changes of the isolate when grown in PDL supplemented with 100 or 1000 mg/L Cr(VI). PDL cultures were inoculated with SS and incubated in 250 mL conical flasks at 30°C in a rotary shaker (200 rpm). A culture without Cr(VI) served as a control. After incubation for 48 or 144 h, fungal mycelia were collected and processed for SEM examination according to another study [30]. Details of the processing method are described in Supplementary Information. The processed mycelia were imaged at 20 kV under an SU-8010 Ultra-High-Resolution scanning electron microscope (Hitachi Corp., Tokyo, Japan).
Mass balance of Cr(VI) removal
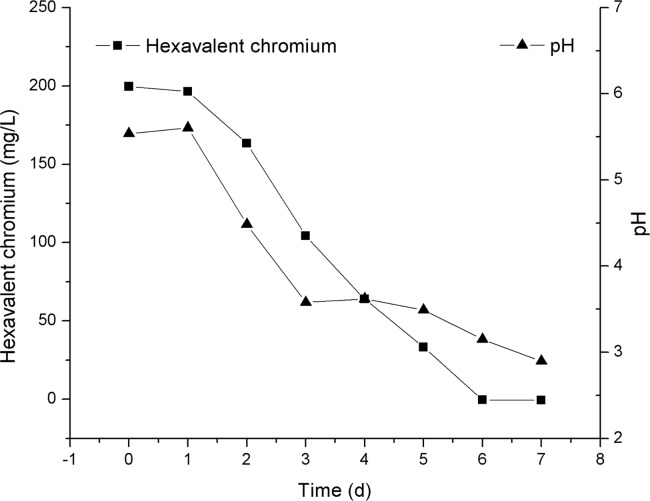
Because Cr(VI) may be removed by fungi through uptake and/or reduction [31], mass balance analysis was carried out to identify the fate of Cr(VI) after incubation with the isolate in PDL containing 199.6 mg/L Cr(VI). The medium was inoculated with SS, and a culture without inoculum served as a control. All the cultures were incubated in 250-mL conical flasks at 30°C in a rotary shaker (200 rpm). At days 0–6, samples of the culture medium were collected and filtered for analysis of pH, Cr(VI), and total Cr content. The concentration of Cr(VI) was determined by the diphenylcarbazide method, which has a detection limit of 0.2 μg/L [32]. The concentration of total Cr was determined by flame atomic absorption spectrophotometry on an MKILM6 spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
For analysis of Cr uptake by the biomass, mycelia were collected from cultures by filtration after cultivation, washed with deionized water, dried to a constant weight at 65°C in an oven, and acid digested [33]. The method for digesting mycelia was described elsewhere[34]. Triplicate dry mycelia were prepared, resuspended in concentrated HNO3, and left at 30°C for 30 min, then incubated at 60°C for 30 min and then at 120°C for 30 min on a heating panel. When the digested solutions cooled to room temperature, they were supplemented with 30% H2O2 and heated again at 120°C for 15 min. After cooling to room temperature, the digested solutions were transferred to 100-mL volumetric flasks for analysis of total Cr.
Removal of Cr(VI) from electroplating wastewater
To explore the utility of SL2 for removing Cr(VI) from actual wastewater, we tested electroplating wastewater, collected from a factory in Jinhua, Zhejiang, China. The concentrations of metals in this electroplating wastewater are shown in Supplementary Information. Given that the initial contaminant concentration may influence Cr(VI) removal [35], the electroplating wastewater was divided into 3 aliquots and diluted with deionized water at different ratios. For cultivation of mycelia, the diluted electroplating wastewater was adjusted to pH 7.0 with NaOH and supplemented with modified PDL as nutrition. The modified PDL consisted of 2 g of dextrose and 100 mL of filtered soup of 40 g of potatoes boiled for 30 min. The processed electroplating wastewater was inoculated with SS, and incubated in 250-mL conical flasks at 30°C in a rotary shaker (200 rpm). At regular intervals, samples were collected and filtered for the analysis of Cr(VI).
Data analysis
Statistical analysis was carried out in the SPSS 20.0 software (IBM Corp., Armonk, NY). The paired-sample t test was performed to evaluate the differences in colony diameter and the tolerance index of the isolate at different Cr(VI) concentrations. One-way analysis of variance was conducted to compare the initial rates of Cr(VI) removal by the isolate from electroplating wastewater at various dilutions. Differences with p < 0.05 were regarded as statistically significant.
Results and discussion
Isolation and identification of the fungal strain
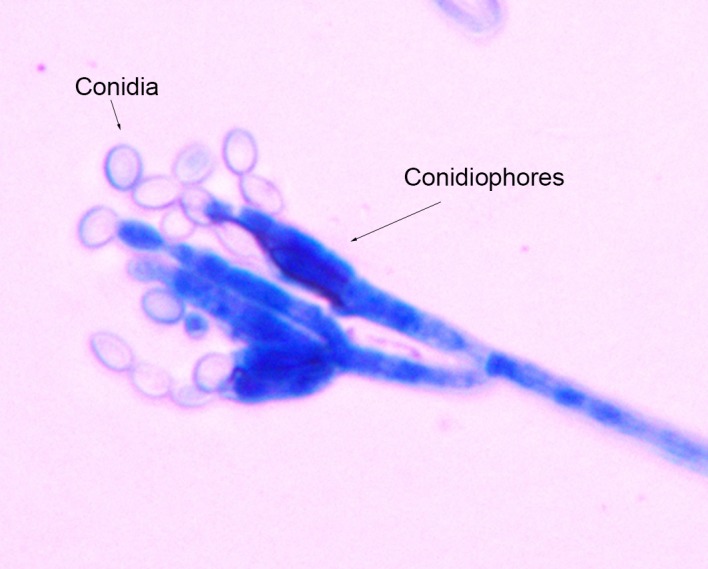
A potential Cr(VI)-tolerant filamentous fungus strain, SL2, was isolated from indoor air and then purified in PDA supplemented with 300 mg/L Cr(VI). Colonies of SL2 grew rapidly and matured within 3 days of incubation at 30°C. The texture of its colony was velvety. The initial color was white, and gradually became dark green. The reverse color was dirty white. Microscopic observation showed that SL2 had septate hyphae, branched conidiophores, swollen phialides, and unicellular conidia (Fig 1). The conidia were oval, rough, and formed long chains. Sequence comparison revealed high similarities between the rDNA sequences (18S rDNA, ITS, and 26S rDNA) of SL2 and published Penicillium oxalicum sequences. Details are presented in Supplementary Information. According to phenotypic characterization and rDNA sequence analysis, SL2 was identified as P. oxalicum, a species that has not been extensively studied regarding Cr(VI) tolerance and reduction abilities.
Fig 1. Microscopic examination of Penicillium oxalicum strain SL2, focusing on conidiophores and conidia (400× magnification).
Cr(VI) contamination is known to change the composition of microbial communities, including enrichment of microorganisms with elevated metal tolerance. Hence, most of the Cr-tolerant microorganisms reported in other studies have been isolated from Cr(VI)-contaminated sites [36, 37]. Unlike those microorganisms, P. oxalicum strain SL2 was isolated from an environment without a history of Cr(VI) contamination. The presence of Cr(VI)-tolerant microorganisms in indoor air provides evidence in support of these species being widespread in the environment. Thus, in addition to the microbiotas from sites contaminated by Cr(VI), microbial communities from sites without such contamination can harbor Cr(VI)-tolerant microorganisms holding promise for bioremediation.
Effects of Cr(VI) concentration on strain SL2 growth
SL2 showed Cr(VI) tolerance in both PDA and PDL. SL2 grew after the inoculation of 0.5 μL SS onto PDA containing 1000 mg/L Cr(VI). Nevertheless, the diameters of SL2 colonies exposed to Cr(VI) were smaller than those in the unexposed control, resulting in tolerance indices less than 1.0 (Fig 2). Furthermore, the tolerance index of SL2 decreased with an increasing Cr(VI) concentration, indicating a negative relation between SL2 growth and Cr(VI) concentration in PDA. Besides, SL2 grew in PDL containing 0 mg/L, 100 mg/L, or 1000 mg/L Cr(VI). The differences between the dry weights of biomass of SL2 grown in PDL under 100 mg/L Cr(VI) stress and those in the control were not statistically significant (p = 0.11). In addition, after incubation for 6 days, SL2 developed more biomass when grown in PDL containing 100 mg/L Cr(VI) than in the control, which resulted in a tolerance index greater than 1.0 for SL2 grown under 100 mg/L Cr(VI) stress in PDL (Fig 3). This observation could be explained by SL2’s entering the death phase earlier in the control than at 100 mg/L Cr(VI), as a result, the biomass of SL2 in the control decreased and at the measurement time point was less than that in PDL containing 100 mg/L Cr(VI), which went into the death phase later. These results suggested that the growth of SL2 had different responses to Cr(VI) in liquid and solid media.
Fig 2. The tolerance index of SL2 in PDA solid media containing different concentrations of Cr(VI).
Fig 3. The tolerance index of SL2 in PDL media containing different concentrations of Cr(VI).
It has been reported that Cr(VI) has greater toxic effects on microorganisms in a liquid medium than on a solid one, thus leading to a difference in the minimum inhibitory concentration (MIC) of Cr(VI) [23, 38]. In the present study, SL2 was tolerant to 1000 mg/L Cr(VI) in PDA and PDL, implying that the MIC of Cr(VI) for SL2 is greater than 1000 mg/L on solid and in liquid media. Some studies indicate that Cr(VI)-tolerant microorganisms can survive in a wide range of Cr(VI) concentrations, but most of those microorganisms have a MIC less than 1000 mg/L [39]. Some Cr(VI)-tolerant microorganisms grow only at Cr(VI) concentrations of less than 100 mg/L [40–43]. Therefore, SL2 can tolerate a higher Cr(VI) concentration than most microorganisms can. Compared with other strains in the genus Penicillium [34], SL2 manifested distinctly higher tolerance to Cr(VI) as well. In general, SL2 showed high tolerance to Cr(VI) on solid and in liquid media, and this ability may facilitate its application to Cr(VI)-contaminated environments.
The modified Gompertz model was successfully applied to simulate the growth of strain SL2 under Cr(VI) stress. As shown in Table 2, the training data obtained at each of the tested concentrations fitted well to the modified Gompertz model, with R20mg/L, R2100mg/L, R2400mg/L, and R21000mg/L close to 1.0. Moreover, the newly developed model was capable of predicting the growth curve of SL2 under Cr(VI) stress with good NS0mg/L, NS100mg/L, NS400mg/L, and NS1000mg/L in the validation period. The good fit of the modified Gompertz model indicated that the growth curve patterns of SL2 were not significantly different with and without Cr(VI) stress. In addition, the newly developed model revealed that the growth of SL2 was affected by the Cr(VI) concentration (Table 3). The relative maximal colony diameter (A) and the maximal growth rate (μm) decreased as Cr(VI) concentration increased from 0 to 1000 mg/L, while lag time (λ) increased, indicating that SL2 needed more time to adapt to greater Cr(VI) stress and get to the reproductive stage. As the modified Gompertz model successfully describes the growth of SL2 under Cr(VI) stress, it may be an effective tool for improving this microbe’s performance in practice.
Table 2. Performance statistics of the modified Gompertz model during training and validation periods for the modeling of growth curves of strain SL2 under Cr(VI) stress.
| Cr(VI) concentration | Training | Validation | ||||
|---|---|---|---|---|---|---|
| R2 | MSE | NS | R2 | MSE | NS | |
| 0 mg/L | 0.996 | 0.023 | 0.996 | 0.984 | 0.078 | 0.975 |
| 100 mg/L | 0.997 | 0.009 | 0.996 | 0.996 | 0.014 | 0.994 |
| 400 mg/L | 0.990 | 0.018 | 0.989 | 0.991 | 0.020 | 0.987 |
| 1000 mg/L | 0.993 | 0.004 | 0.993 | 0.986 | 0.008 | 0.986 |
R2, coefficient of determination; MSE, mean squared error; NS, Nash–Sutcliffe efficiency coefficient.
Table 3. Parameters of the modified Gompertz model for describing the growth of strain SL2 at different Cr(VI) concentrations.
| Cr(VI) concentration | A | μm | λ |
|---|---|---|---|
| 0 mg/L | 8.36 | 0.90 | 0.99 |
| 100 mg/L | 5.65 | 0.37 | 1.92 |
| 400 mg/L | 4.22 | 0.34 | 2.83 |
| 1000 mg/L | 2.68 | 0.18 | 3.00 |
A, relative maximal colony diameter; μm, maximal growth rate; λ, lag time.
SEM analysis
SEM micrographs of SL2 reveal the effects of Cr(VI) on its surface topography (Fig 4). After incubation for 48 h, strain SL2 grown without Cr(VI) stress had a regular shape, with few extracellular substances on the cell surface (Fig 4A). In contrast, strain SL2 grown under Cr(VI) stress showed increased amounts of extracellular substances on the cell surface (Fig 4B and 4C). In addition, SL2 grown under 1000 mg/L Cr(VI) stress had a relatively irregular shape. The results were in accordance with data from another study on an arbuscular mycorrhizal fungus that formed increased amounts of particles on its cell surface under Cr(VI) stress [44]. It was reported that alcohol, carboxyl, and amino groups may interact with heavy metals [45], and this interaction may result in their biosorption and flocculation [16, 46]. Hence, the production of extracellular substances containing these groups induced by Cr(VI) stress may be a strategy by which SL2 alleviates the poisonous effects of Cr(VI).
Fig 4. Micrographs of strain SL2 grown in PDL under Cr(VI) stress.
Panels a, b, and c are images of strain SL2 grown for 48 h in the PDL medium containing 0, 100, or 1000 mg/L Cr(VI), respectively. Panels d, e, and f are images of SL2 grown for 144 h in the PDL medium containing 0, 100, or 1000 mg/L Cr(VI), respectively.
After incubation for 144 h without Cr(VI) stress, SL2 went into the death phase of its growth curve, and wrinkling and rupture of hypha were detected (Fig 4D). In the same period, hypha under 100 mg/L Cr(VI) stress were also wrinkled and irregular, but no rupture of hypha was detected in the 100 and 1000 mg/L Cr(VI) stress conditions (Fig 4E and 4F). These results supported the hypothesis that SL2 entered the death phase later under Cr(VI) stress than in the control condition. Moreover, the hypha of SL2 grown under 100 and 1000 mg/L Cr(VI) stress were bigger than those of strain SL2 grown without Cr(VI) stress. These results are consistent with those of other studies, which showed increases in the microbe size of some strains when exposed to Cr(VI) [47, 48]. The change in size of microorganisms could be a Cr(VI) stress response, allowing a strain to adapt to its environment.
Mass balance of Cr(VI) removal
Cr(III) and Cr(VI) are the stable forms of Cr found in the environment, and Cr(III) is more difficult to quantify than Cr(VI) or total Cr, hence, most of the studies characterize Cr(VI) reduction ability of microorganisms by measuring the decrease in Cr(VI) concentration [49, 50], and the production of Cr(III) is indicated by the difference between total Cr and Cr(VI). In the present study, the isolated strain manifested remarkable efficacy at lowering the Cr(VI) level in PDL (Fig 5). After incubation for 7 days, Cr(VI) concentration decreased from the initial value of 199.6 mg/L to an undetectable level. Meanwhile, total Cr content in the medium did not change significantly over time, suggesting that Cr(VI) was reduced to Cr(III) and it accounted for most of the decrease in Cr(VI) concentration in PDL. In addition, the color of the liquid medium changed from the yellow color of soluble Cr(VI) to a slightly turbid brownish color, which may have been caused by the chemical conversion of chromium compounds in PDL. Our results are in agreement with other studies on Aspergillus sp., Penicillium sp., and Paecilomyces sp. [34, 37], which can quantitatively reduce Cr(VI) to Cr(III) and cause color changes in the medium. As reduction of Cr(VI) to Cr(III) can minimize the harmful effects of Cr, the Cr(VI) reduction ability should make fungal strain SL2 useful for bioremediation.
Fig 5. Cr(VI) removal and pH change during the incubation of SL2 in the PDL medium.
Cr(III) can be oxidized to Cr(VI) by strong oxidants [51], hence, the generated Cr(III) needs to be further removed from this reduction system. As a culture medium is an organic-matter-rich environment, Cr(III) may not be fully immobilized by strain SL2 owing to the coordination caused by soluble small organic molecules such as amino acids and organic acids in the culture medium [52]. Organo-Cr(III) species resulting from the coordination of Cr(III) and organic molecules are intermediates in the Cr biogeochemical cycle, and may convert into a Cr(OH)3 precipitate over time [53], and then could be removed by filtration.
Cr uptake by the biomass of strain SL2 was found to be 0.79 mg/g dry mass, and it accounted for ~0.95% of total Cr initially present in PDL. Compared with Pleurotus sp. [54], magnetic carbon-iron nanoadsorbents [55], polyaniline/ethyl cellulose [56], polyaniline/carbon fabrics [57], magnetic carbons [58], magnetic carbon nanoadsorbents [59], polyethylenimine/ethyl cellulose [60], and extracellular polymeric substances-modified polyaniline nanocomposites [61], strain SL2 incorporated less Cr into its biomass. Nevertheless, strain SL2 incorporated more Cr than did Aspergillus sp. and Penicillium sp. grown in the presence of 50 mg/L Cr(VI) [34]. It is important to note that Cr uptake by microorganisms may vary among different environments [34, 62]. Consequently, Cr uptake by the biomass of strain SL2 may change in different environmental conditions.
pH influences the chemical speciation, solubility, and bioavailability of Cr(VI). Therefore, the change of pH in PDL during Cr(VI) removal was studied next. Cr(VI) reduction has been reported to be a proton consuming process [63]. Accordingly, pH should theoretically increase as Cr(VI) is reduced. In contrast, as shown in Fig 5, the removal of Cr(VI) was accompanied by a decrease in pH in this study. This effect could be explained as follows: strain SL2 produced acidic metabolites [22], which provided more protons than those consumed during Cr(VI) removal. In addition to providing proton for Cr(VI) reduction, the decreased pH provided a high protonation level of the biomass with lots of positive charges, which thereby attracted the negative Cr(VI) ion. During the incubation of SL2, pH in PDL decreased from 5.54 to 2.90, and this change might have influenced the growth and Cr uptake by the biomass. Nevertheless, to gain a comprehensive understanding of the influence of pH, further study is needed.
Removal of Cr(VI) from electroplating wastewater
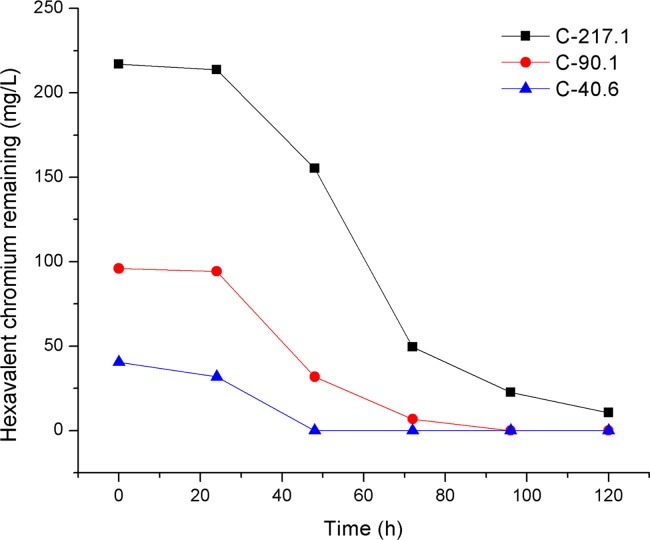
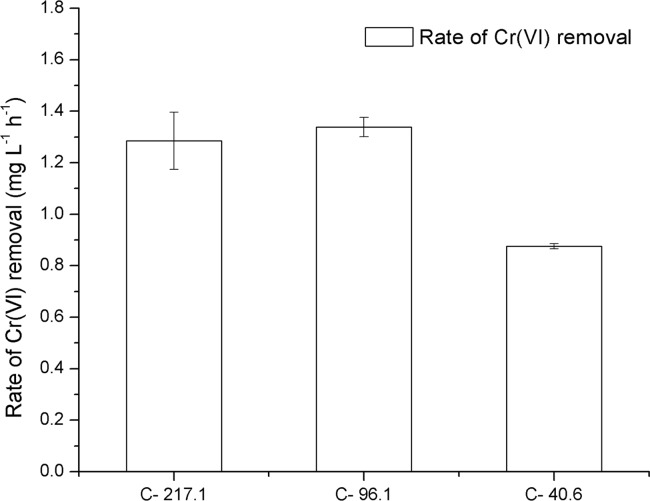
Cr(VI) in electroplating wastewater was effectively removed by fungal strain SL2. As shown in Fig 6, after inoculation of spores of SL2, Cr(VI) in diluted electroplating wastewater at 40.6 and 96.1 mg/L was completely removed within 48 and 96 h, respectively. Furthermore, 89.6% of Cr(VI) in diluted electroplating wastewater at an initial concentration of 217.1 mg/L was removed in 96 h. The initial rate of Cr(VI) removal by SL2 was affected by the initial contaminant concentration. As depicted in Fig 7, Cr(VI) removal by SL2 was faster at 217.1 and 96.1 mg/L Cr(VI) in electroplating wastewater samples than in the sample with 40.6 mg/L Cr(VI), but there was no difference in the rate of Cr(VI) removal between the concentrations 217.1 and 96.1 mg/L. This phenomenon may be explained by the rate of collision of Cr(VI) with active sites on the cell surface [39]. The collision rate should increase as the initial contaminant concentration increases when the contaminant concentration was insufficient to inhibit the growth of the microorganism, and, as a result, the rate of Cr(VI) removal increased. In contrast, after the contaminant concentration exceeded the level that inhibits microbial growth, an increase in contaminant concentration decreased the number of active sites on the cell surface, as a result, the collision rate and Cr(VI) removal rate should not increase with an increase in contaminant concentration. Therefore, it is important to control the initial contaminant concentration when using SL2 to remove Cr(VI) from wastewater.
Fig 6. Cr(VI) removal by SL2 at different dilutions of electroplating wastewater.
C-217.1, C-96.1, and C-40.6 represent treatments with initial Cr(VI) concentrations of 217.1, 96.1, and 40.6 mg/L, respectively.
Fig 7. The initial rate of Cr(VI) removal by SL2 at different dilutions of electroplating wastewater.
C-217.1, C-96.1, and C-40.6 represent treatments with initial Cr(VI) concentrations of 217.1, 96.1, and 40.6 mg/L, respectively. Error bars represent standard deviation. Treatments marked with the same letter are not significantly different.
Conclusion
Cr-tolerant microorganisms are widespread in the environment. Thus, in addition to microbiotas of Cr(VI)-contaminated sites, microbial communities in environments without Cr(VI) contamination can harbor Cr-tolerant microorganisms useful for Cr(VI) removal. P. oxalicum SL2, isolated from indoor air, was found to tolerate a high Cr(VI) concentration on solid and in liquid media, and this property may facilitate its application to Cr(VI)-contaminated environments. Simulated growth curves of SL2 exposed to Cr(VI) by means of the modified Gompertz model suggest that Cr(VI) decreases the relative maximal colony diameter and the maximal growth rate, and increases the period required for SL2 to adapt to Cr(VI) stress and get to the reproductive stage. SL2 showed remarkable efficiency to remove Cr(VI), and the mass balance suggests that this ion was reduced to Cr(III), but this process was not confirmed in this study. Our findings provide a new candidate for Cr(VI) removal, and will significantly expand our knowledge about the utility of this microorganism.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We thank Peihua Wang, Yige Zhao, and Chidong Zhou for their work on the analysis of Cr(VI) and total Cr in PDL. We also thank Haomin Huang for her suggestions on the draft of the manuscript.
Data Availability
The rDNA sequences (18S rDNA, ITS, and 26S rDNA) of strain SL2 are deposited in GenBank. The accession number for ITS is MG585100. The accession number for 18S rDNA is MG585101. The accession number for 26S rDNA is MG585103. Other data underlying the findings are shown in the manuscript and Supplementary Information.
Funding Statement
This work was supported by the National Natural Science Foundation of China (U1532103, 41422107), National Key Research and Development Program of China (2016YFD0800401). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cheng HG, Zhou T, Li Q, Lu L, Lin CY. Anthropogenic Chromium Emissions in China from 1990 to 2009. Plos One. 2014;9(2). doi: 10.1371/journal.pone.0087753 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H, Teng Y, Lu S, Wang Y, Wang J. Contamination features and health risk of soil heavy metals in China. Sci Total Environ. 2015;512–513:143–53. doi: 10.1016/j.scitotenv.2015.01.025 . [DOI] [PubMed] [Google Scholar]
- 3.McClain CN, Maher K. Chromium fluxes and speciation in ultramafic catchments and global rivers. Chemical Geology. 2016;426:135–57. doi: 10.1016/j.chemgeo.2016.01.021 [Google Scholar]
- 4.Nowicka AM, Stojek Z, Hepel M. Chromium(VI) but Not Chromium(III) Species Decrease Mitoxantrone Affinity to DNA. J Phys Chem B. 2013;117(4):1021–30. doi: 10.1021/jp3109094 . [DOI] [PubMed] [Google Scholar]
- 5.Yang LQ, Xia B, Yang XQ, Ding H, Wu DS, Zhang HM, et al. Mitochondrial DNA hypomethylation in chrome plating workers. Toxicol Lett. 2016;243:1–6. doi: 10.1016/j.toxlet.2015.11.031 . [DOI] [PubMed] [Google Scholar]
- 6.Wise SS, Holmes AL, Liou L, Adam RM, Wise JP, Sr. Hexavalent chromium induces chromosome instability in human urothelial cells. Toxicol Appl Pharmacol. 2016;296:54–60. doi: 10.1016/j.taap.2016.02.015 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wise SS, Wise C, Xie H, Guillette LJ, Zhu CR, Wise JP Jr, et al. Hexavalent chromium is cytotoxic and genotoxic to American alligator cells. Aquat Toxicol. 2016;171:30–6. doi: 10.1016/j.aquatox.2015.12.004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu SQ, Gu HR, Cui CY, Ji R. Toxicity of combined chromium(VI) and phenanthrene pollution on the seed germination, stem lengths, and fresh weights of higher plants. Environmental Science and Pollution Research. 2016;23(15):15227–35. doi: 10.1007/s11356-016-6701-6 . [DOI] [PubMed] [Google Scholar]
- 9.Kilic NK, Stensballe A, Otzen DE, Donmez G. Proteomic changes in response to chromium(VI) toxicity in Pseudomonas aeruginosa. Bioresource Techn. 2010;101(7):2134–40. doi: 10.1016/j.biortech.2009.11.008 . [DOI] [PubMed] [Google Scholar]
- 10.Pechova A, Pavlata L. Chromium as an essential nutrient: a review. Veterinarni Medicina. 2007;52(1):1–18. [Google Scholar]
- 11.Guemiza K, Coudert L, Metahni S, Mercier G, Besner S, Blais J-F. Treatment technologies used for the removal of As, Cr, Cu, PCP and/or PCDD/F from contaminated soil: A review. J Hazard Mater. 2017;333:194–214. doi: 10.1016/j.jhazmat.2017.03.021 . [DOI] [PubMed] [Google Scholar]
- 12.Malaviya P, Singh A. Bioremediation of chromium solutions and chromium containing wastewaters. Crit Rev Microbiol. 2016;42(4):607–33. doi: 10.3109/1040841X.2014.974501 . [DOI] [PubMed] [Google Scholar]
- 13.Yu G, Lu Y, Guo J, Patel M, Bafana A, Wang X, et al. Carbon nanotubes, graphene, and their derivatives for heavy metal removal. Advanced Composites and Hybrid Materials. 2017. doi: 10.3390/ma10060679 [Google Scholar]
- 14.Malaviya P, Singh A. Bioremediation of Chromium Solutions and Chromium Containing Wastewaters. Crit Rev Microbiol. 2014:1–27. doi: 10.3109/1040841X.2014.974501 . [DOI] [PubMed] [Google Scholar]
- 15.Romanenko VI, Koren'kov VN. Pure culture of bacteria using chromates and bichromates as hydrogen acceptors during development under anaerobic conditions. Mikrobiologiia. 1977;46(3):414–7. . [PubMed] [Google Scholar]
- 16.Karthik C, Barathi S, Pugazhendhi A, Ramkumar VS, Thi NB, Arulselvi PI. Evaluation of Cr(VI) reduction mechanism and removal by Cellulosimicrobium funkei strain AR8, a novel haloalkaliphilic bacterium. J Hazard Mater. 2017;333:42–53. doi: 10.1016/j.jhazmat.2017.03.037 . [DOI] [PubMed] [Google Scholar]
- 17.Gutiérrez AM, Cabriales JJP, Vega MM. Isolation and characterization of hexavalent chromium-reducing rhizospheric bacteria from a wetland. Int J Phytoremediat. 2010;12(4):317–34. doi: 10.1080/15226510902968118 . [DOI] [PubMed] [Google Scholar]
- 18.Dhal B, Thatoi HN, Das NN, Pandey BD. Chemical and microbial remediation of hexavalent chromium from contaminated soil and mining/metallurgical solid waste: A review. J Hazard Mater. 2013;250:272–91. doi: 10.1016/j.jhazmat.2013.01.048 . [DOI] [PubMed] [Google Scholar]
- 19.Leitao AL. Potential of penicillium species in the bioremediation field. Int J Environ Res Public Health. 2009;6(4):1393–417. doi: 10.3390/ijerph6041393 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arevalo-Rangel DL, Cardenas-Gonzalez JF, Martinez-Juarez VM, Acosta-Rodriguez I. Hexavalent chromate reductase activity in cell free extracts of Penicillium sp. Bioinorg Chem Appl. 2013;2013:909412 doi: 10.1155/2013/909412 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anahid S, Yaghmaei S, Ghobadinejad Z. Heavy metal tolerance of fungi. Sci Iran. 2011;18(3):502–8. doi: 10.1016/j.scient.2011.05.015 [Google Scholar]
- 22.Deng XH, Chai LY, Yang ZH, Tang CJ, Wang YY, Shi Y. Bioleaching mechanism of heavy metals in the mixture of contaminated soil and slag by using indigenous Penicillium chrysogenum strain F1. J Hazard Mater. 2013;248:107–14. doi: 10.1016/j.jhazmat.2012.12.051 . [DOI] [PubMed] [Google Scholar]
- 23.Long DY, Tang XJ, Cai K, Chen GC, Shen CF, Shi JY, et al. Cr(VI) resistance and removal by indigenous bacteria isolated from chromium-contaminated soil. Journal of Microbiology and Biotechnology. 2013;23(8):1123–32. doi: 10.4014/jmb.1301.01004 . [DOI] [PubMed] [Google Scholar]
- 24.Liu H, Chen N, Feng C, Tong S, Li R. Impact of electro-stimulation on denitrifying bacterial growth and analysis of bacterial growth kinetics using a modified Gompertz model in a bio-electrochemical denitrification reactor. Bioresource Techn. 2017;232:344–53. doi: 10.1016/j.biortech.2017.02.064 . [DOI] [PubMed] [Google Scholar]
- 25.Zwietering MH, Jongenburger I, Rombouts FM, Vantriet K. Modeling of the bacterial-growth curve. Appl Environ Microb. 1990;56(6):1875–81. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chatterjee T, Chatterjee BK, Majumdar D, Chakrabarti P. Antibacterial effect of silver nanoparticles and the modeling of bacterial growth kinetics using a modified Gompertz model. Bba-Gen Subjects. 2015;1850(2):299–306. doi: 10.1016/j.bbagen.2014.10.022 . [DOI] [PubMed] [Google Scholar]
- 27.Shoaib M, Shamseldin AY, Melville BW. Comparative study of different wavelet based neural network models for rainfall-runoff modeling. J Hydrol. 2014;515:47–58. doi: 10.1016/j.jhydrol.2014.04.055 [Google Scholar]
- 28.Wang C, Zhao M, Li J, Yu J, Sun S, Ge S, et al. Silver nanoparticles/graphene oxide decorated carbon fiber synergistic reinforcement in epoxy-based composites. Polymer. 2017;131(Supplement C):263–71. https://doi.org/10.1016/j.polymer.2017.10.049 [Google Scholar]
- 29.Zhang L, Yu W, Han C, Guo J, Zhang Q, Xie H, et al. Large Scaled Synthesis of Heterostructured Electrospun TiO 2 /SnO 2 Nanofibers with an Enhanced Photocatalytic Activity. Journal of the Electrochemical Society. 2017;164(9):H651–H6. [Google Scholar]
- 30.Chen XC, Hu SP, Shen CF, Dou CM, Shi JY, Chen YX. Interaction of Pseudomonas putida CZ1 with clays and ability of the composite to immobilize copper and zinc from solution. Bioresource Techn. 2009;100(1):330–7. doi: 10.1016/j.biortech.2008.04.051 . [DOI] [PubMed] [Google Scholar]
- 31.Poljsak B, Pocsi I, Raspor P, Pesti M. Interference of chromium with biological systems in yeasts and fungi: a review. J Basic Microb. 2010;50(1):21–36. doi: 10.1002/jobm.200900170 . [DOI] [PubMed] [Google Scholar]
- 32.Urone PF. Stability of colorimetric reagent for chromium, s-diphenylcarbazide, in various solvents. Anal Chem. 1955;27(8):1354–5. doi: 10.1021/ac60104a048 [Google Scholar]
- 33.Munoz AH, Corona FG, Wrobel K, Soto GM, Wrobel K. Subcellular distribution of aluminum, bismuth, cadmium, chromium, copper, iron, manganese, nickel, and lead in cultivated mushrooms (Agaricus bisporus and Pleurotus ostreatus). Biological Trace Element Research. 2005;106(3):265–77. doi: 10.1385/BTER:106:3:265 . [DOI] [PubMed] [Google Scholar]
- 34.Acevedo-Aguilar FJ, Espino-Saldana AE, Leon-Rodriguez IL, Rivera-Cano ME, Avila-Rodriguez M, Wrobel K, et al. Hexavalent chromium removal in vitro and from industrial wastes, using chromate-resistant strains of filamentous fungi indigenous to contaminated wastes. Canadian Journal of Microbiology. 2006;52(9):809–15. doi: 10.1139/w06-037 . [DOI] [PubMed] [Google Scholar]
- 35.Coreno-Alonso A, Acevedo-Aguilar FJ, Reyna-Lopez GE, Tomasini A, Fernandez FJ, Wrobel K, et al. Cr(VI) reduction by an Aspergillus tubingensis strain: Role of carboxylic acids and implications for natural attenuation and biotreatment of Cr(VI) contamination. Chemosphere. 2009;76(1):43–7. doi: 10.1016/j.chemosphere.2009.02.031 . [DOI] [PubMed] [Google Scholar]
- 36.Mabrouk MEM, Arayes MA, Sabry SA. Hexavalent chromium reduction by chromate-resistant haloalkaliphilic Halomonas sp. M-Cr newly isolated from tannery effluent. Biotechnology & Biotechnological Equipment. 2014;28(4):659–67. doi: 10.1080/13102818.2014.937092 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cardenas-Gonzalez JF, Acosta-Rodriguez I. Hexavalent chromium removal by a Paecilomyces sp. fungal strain isolated from environment. Bioinorg Chem Appl. 2010:6 doi: 10.1155/2010/676243 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Megharaj M, Avudainayagam S, Naidu R. Toxicity of hexavalent chromium and its reduction by bacteria isolated from soil contaminated with tannery waste. Curr Microbiol. 2003;47(1):51–4. doi: 10.1007/s00284-002-3889-0 . [DOI] [PubMed] [Google Scholar]
- 39.Narayani M, Shetty KV. Chromium-resistant bacteria and their environmental condition for hexavalent chromium removal: a review. Critical Reviews in Environmental Science and Technology. 2013;43(9):955–1009. doi: 10.1080/10643389.2011.627022 [Google Scholar]
- 40.Sarangi A, Krishnan C. Comparison of in vitro Cr(VI) reduction by CFEs of chromate resistant bacteria isolated from chromate contaminated soil. Bioresource Techn. 2008;99(10):4130–7. doi: 10.1016/j.biortech.2007.08.059 . [DOI] [PubMed] [Google Scholar]
- 41.Xu WH, Liu YG, Zeng GM, Li X, Song HX, Peng QQ. Characterization of Cr(VI) resistance and reduction by Pseudomonas aeruginosa. T Nonferr Metal Soc. 2009;19(5):1336–41. doi: 10.1016/S1003-6326(08)60446-X [Google Scholar]
- 42.Badar U, Ahmed N, Beswick AJ, Pattanapipitpaisal P, Macaskie LE. Reduction of chromate by microorganisms isolated from metal contaminated sites of Karachi, Pakistan. Biotechnol Lett. 2000;22(10):829–36. doi: 10.1023/A:1005649113190 [Google Scholar]
- 43.Pei QH, Shahir S, Raj ASS, Zakaria ZA, Wan AA. Chromium(VI) resistance and removal by Acinetobacter haemolyticus. World J Microb Biot. 2009;25(6):1085–93. doi: 10.1007/s11274-009-9989-2 [Google Scholar]
- 44.Wu S, Zhang X, Sun Y, Wu Z, Li T, Hu Y, et al. Chromium immobilization by extra- and intraradical fungal structures of arbuscular mycorrhizal symbioses. J Hazard Mater. 2016. doi: 10.1016/j.jhazmat.2016.05.017 . [DOI] [PubMed] [Google Scholar]
- 45.Ma Y, Ling L, Guo Y, Fu Y, Shao Q, Wu T, et al. Porous lignin based poly (acrylic acid)/organo-montmorillonite nanocomposites: Swelling behaviors and rapid removal of Pb (II) ions. Polymer. 2017. [Google Scholar]
- 46.Sun XF, Wang SG, Zhang XM, Chen JP, Li XM, Gao BY, et al. Spectroscopic study of Zn2+ and Co2+ binding to extracellular polymeric substances (EPS) from aerobic granules. J Colloid Interf Sci. 2009;335(1):11–7. doi: 10.1016/j.jcis.2009.03.088 . [DOI] [PubMed] [Google Scholar]
- 47.Zakaria ZA, Zakaria Z, Surif S, Ahmad WA. Hexavalent chromium reduction by Acinetobacter haemolyticus isolated from heavy-metal contaminated wastewater. J Hazard Mater. 2007;146(1–2):30–8. doi: 10.1016/j.jhazmat.2006.11.052 . [DOI] [PubMed] [Google Scholar]
- 48.Samuel J, Paul ML, Ravishankar H, Mathur A, Saha DP, Natarajan C, et al. The differential stress response of adapted chromite mine isolates Bacillus subtilis and Escherichia coli and its impact on bioremediation potential. Biodegradation. 2013;24(6):829–42. doi: 10.1007/s10532-013-9631-8 . [DOI] [PubMed] [Google Scholar]
- 49.Long DY, Tang XJ, Cai K, Chen GC, Chen LG, Duan DC, et al. Cr(VI) reduction by a potent novel alkaliphilic halotolerant strain Pseudochrobactrum saccharolyticum LY10. J Hazard Mater. 2013;256:24–32. doi: 10.1016/j.jhazmat.2013.04.020 . [DOI] [PubMed] [Google Scholar]
- 50.Zhong L, Lai C-Y, Shi L-D, Wang K-D, Dai Y-J, Liu Y-W, et al. Nitrate effects on chromate reduction in a methane-based biofilm. Water Research. 2017;115:130–7. doi: 10.1016/j.watres.2017.03.003 . [DOI] [PubMed] [Google Scholar]
- 51.Varadharajan C, Beller HR, Bill M, Brodie EL, Conrad ME, Han R, et al. Re-oxidation of chromium(III) products formed under different biogeochemical regimes. Environmental science & technology. 2017. doi: 10.1021/acs.est.6b06044 . [DOI] [PubMed] [Google Scholar]
- 52.Cheng YJ, Yan FB, Huang F, Chu WS, Pan DM, Chen Z, et al. Bioremediation of Cr(VI) and Immobilization as Cr(III) by Ochrobactrum anthropi. Environmental Science & Technology. 2010;44(16):6357–63. doi: 10.1021/Es100198v . [DOI] [PubMed] [Google Scholar]
- 53.Cheng Y, Holman HY, Li Z. Remediation of Chromium and Uranium Contamination by Microbial Activity. Elements. 2012;8(2):107–12. doi: 10.2113/gselements.8.2.107 [Google Scholar]
- 54.Li X, Wang Y, Pan Y, Yu H, Zhang X, Shen Y, et al. Mechanisms of Cd and Cr removal and tolerance by macrofungus Pleurotus ostreatus HAU-2. J Hazard Mater. 2017;330:1–8. doi: 10.1016/j.jhazmat.2017.01.047 . [DOI] [PubMed] [Google Scholar]
- 55.Qiu B, Gu H, Yan X, Guo J, Wang Y, Sun D, et al. Cellulose derived magnetic mesoporous carbon nanocomposites with enhanced hexavalent chromium removal. J Mater Chem A. 2014;2(41):17454–62. doi: 10.1039/C4TA04040F [Google Scholar]
- 56.Qiu B, Xu C, Sun D, Yi H, Guo J, Zhang X, et al. Polyaniline Coated Ethyl Cellulose with Improved Hexavalent Chromium Removal. ACS Sustainable Chemistry & Engineering. 2014;2(8):2070–80. doi: 10.1021/sc5003209 [Google Scholar]
- 57.Qiu B, Xu C, Sun D, Wei H, Zhang X, Guo J, et al. Polyaniline coating on carbon fiber fabrics for improved hexavalent chromium removal. Rsc Advances. 2014;4(56):29855–65. doi: 10.1039/C4RA01700E [Google Scholar]
- 58.Huang J, Cao Y, Shao Q, Peng X, Guo Z. Magnetic Nanocarbon Adsorbents with Enhanced Hexavalent Chromium Removal: Morphology Dependence of Fibrillar vs Particulate Structures. Ind Eng Chem Res. 2017;56(38):10689–701. doi: 10.1021/acs.iecr.7b02835 [Google Scholar]
- 59.Qiu B, Wang Y, Sun D, Wang Q, Zhang X, Weeks BL, et al. Cr(vi) removal by magnetic carbon nanocomposites derived from cellulose at different carbonization temperatures. J Mater Chem A. 2015;3(18):9817–25. doi: 10.1039/C5TA01227A [Google Scholar]
- 60.Qiu B, Guo J, Zhang X, Sun D, Gu H, Wang Q, et al. Polyethylenimine Facilitated Ethyl Cellulose for Hexavalent Chromium Removal with a Wide pH Range. ACS Applied Materials & Interfaces. 2014;6(22):19816–24. doi: 10.1021/am505170j . [DOI] [PubMed] [Google Scholar]
- 61.Hu Q, Guo C, Sun D, Ma Y, Qiu B, Guo Z. Extracellular Polymeric Substances Induced Porous Polyaniline for Enhanced Cr(VI) Removal from Wastewater. Acs Sustainable Chemistry & Engineering. 2017. [Google Scholar]
- 62.Coreno-Alonso A, Sole A, Diestra E, Esteve I, Gutierrez-Corona JF, Lopez GER, et al. Mechanisms of interaction of chromium with Aspergillus niger var tubingensis strain Ed8. Bioresource Techn. 2014;158:188–92. doi: 10.1016/j.biortech.2014.02.036 . [DOI] [PubMed] [Google Scholar]
- 63.Chirwa EN, Wang YT. Simultaneous chromium(VI) reduction and phenol degradation in an anaerobic consortium of bacteria. Water Research. 2000;34(8):2376–84. doi: 10.1016/S0043-1354(99)00363-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
The rDNA sequences (18S rDNA, ITS, and 26S rDNA) of strain SL2 are deposited in GenBank. The accession number for ITS is MG585100. The accession number for 18S rDNA is MG585101. The accession number for 26S rDNA is MG585103. Other data underlying the findings are shown in the manuscript and Supplementary Information.