Abstract
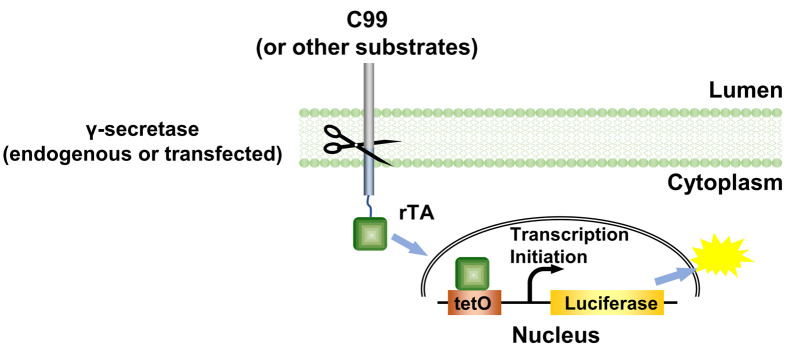
γ-Secretase epsilon-cleavage assay is derived from the cell-based Tango assay ( Kang et al., 2015 ), and is a fast and sensitive method to determine the initial cleavage of C99 by γ-secretase. In this protocol, we use HTL cells, which are HEK293 cells with a stably integrated luciferase reporter under the control of the bacterial tetO operator element, in which C99 C terminally fused to a reversed tetracyclin-inducible activator (rTA) transcriptional activator is expressed. Endogenous or transfected γ-secretase cleaves a C terminally fused rTA transcriptional activator from C99, allowing rTA to move to the nucleus to activate a luciferase reporter gene as a measurement for γ-secretase cleavage activity.
Keywords: γ-Secretase, Cleavage, Activity, Epsilon-cleavage assay, C99
Background
Alzheimer’s disease (AD) is the most prevalent chronic neurodegenerative disease. AD is closely associated with the formation of amyloid plaques. These plaques mainly consist of aggregated amyloid β, which is generated by the cleavage of the C terminal 99 amino acids fragment (C99) of the amyloid precursor protein by γ-secretase. In spite of extensive efforts, it remains unknown how γ-secretase recognizes its substrates. The conventional Tango assay was designed to monitor the activation of GPCRs ( Barnea et al., 2008 ) by engineering a TEV cleavage site and a transcription activator to the cytoplasmic C terminus to of GPCRs and a TEV protease linked to human β-arrestin2. Here we used endogenous or transfected γ-secretase to cleave the transmembrane portion of its substrates, i.e., the C terminus of C99 is fused with rTA to establish the γ-secretase Epsilon-cleavage assay (Figure 1). The assay was first established to investigate the initial cleavage of C99 by γ-secretase ( Xu et al., 2016 ).
Figure 1. The principle of the γ-secretase epsilon-cleavage assay.
. Upon cleavage of the C99 (or other γ-secretase substrates) hybrid protein by γ-secretase, the rTA is released from the membrane and enters nucleus to bind tetO DNA-binding site to stimulate luciferase reporter gene activity as measurement for total cleavage, both by endogenous and by transfected γ-secretase variants. ( Xu et al., 2016 )
Here we use the Dual-luciferase reporter assay kit. The stably integrated luciferase-Firefly reads represent the γ-secretase cleavage activity, while the transfected Renilla luciferase reads serve as a normalization standard.
Materials and Reagents
Pipette tips (VWR)
24-well plate (Corning, Costar®, catalog number: 3524)
96-well plate (Corning, catalog number: 3595)
Loading pipette tips
Cell culture flask (Corning, catalog number: 430639)
96-well OptiPlate (PerkinElmer, catalog number: 6005290)
HTL cells (a gift from G Barnea and R Axel, Brown University and Columbia University, resp.)
Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Fisher Scientific, GibcoTM, catalog number: 11965092)
Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 26140079)
Trypsin 0.25%-EDTA (Thermo Fisher Scientific, GibcoTM, catalog number: 25200056)
Opti-MEM (Thermo Fisher Scientific, GibcoTM, catalog number: 31985062)
X-tremeGENE 9 Reagent (Roche Diagnostics, catalog number: 06365787001)
Sodium phosphate dibasic (Na2HPO4) (Fisher Scientific, catalog number: S374-1)
Potassium phosphate monobasic (KH2PO4) (EMD Millipore, catalog number: PX1565-1)
Sodium chloride (NaCl) (EMD Millipore, catalog number: SX0420-5)
Potassium chloride (KCl) (Fisher Scientific, catalog number: BP366-500)
Dual-luciferase reporter assay kit (Promega, catalog number: E1960)
10x phosphate buffered saline (PBS buffer) (see Recipes)
1x passive lysis buffer (PLB) (see Recipes)
LAR2 substrate (see Recipes)
Stop & Glo® Reagent (see Recipes)
Equipment
Micro pipette (Eppendorf)
Eppendorf® Research® Pro electronic single channel pipette (20-1,000 µl)
37 °C, 5% CO2 incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: FormaTM Series II 3110 Water-Jacketed)
Cell culture microscope (Nikon Instruments, model: Eclipse TS100)
Shaker (ARMA LAB, model: Orbital Shaker 100)
Biosafety cabinet (The Baker, model: SterilGARD® e3)
EnVision Multilabel Plate Reader (PerkinElmer, model: EnVision Multilabel)
Software
GraphPad Prism 5 (https://www.graphpad.com/scientific-software/prism/)
Procedure
-
Cell culture
Grow HTL cells in DMEM supplemented with 10% (v/v) FBS at 37 °C under a humidified 5% CO2 atmosphere to about 80% confluence.
Treat the cells with 0.25% trypsin-EDTA at 37 °C for about 2 min. Dilute the cells to 0.4 x 106/ml with DMEM medium (supplemented with 10% (v/v) fetal bovine serum) and split at 50,000 per well into a 24-well plate one day prior to transfection.
-
Transfection
-
Prepare Master Mix for one sample:
To a sterile 1.5 ml Eppendorf tube, add:
10 µl Opti-MEM medium
0.195 µl X-tremeGENE 9 Transfection Reagent.
Notes:
Avoid touching the side of the tube while adding reagent.
Make a Master Mix for larger number of samples: for example, for 10 samples, multiply by 11 to prepare some extra mix. (10 x 11 = 110 µl medium + 0.195 x 11 = 2.145 µl X-tremeGENE 9 Transfection Reagent).
-
Add 10 µl Opti-MEM medium per well in a 96-well plate and then add 65 ng total DNA into each well.
Note: We usually dilute the DNA plasmid to the concentration of 100 ng/µl and store them at 4 °C.
Add 10 µl Master Mix to DNA mix per well and mix gently by pipetting.
Incubate at room temperature for about 30 min.
-
Carefully transfer 20 µl mix by pipetting into each well of cells cultured in 24-well plate.
Note: In this particular case, for coexpression with γ-secretase, 20 ng substrate-encoding DNA, 5 ng phRG-tk Renilla normalization standard and 10 ng of each of the γ-secretase subunit expression plasmids were transfected. For the substrate-only control, 20 ng substrate, 5 ng phRG-tk Renilla and 40 ng pBSK mock plasmid were transfected. As a positive control, 10 ng Rho(4M)-TEV-site-rTA, 10 ng Arr(3A)-TEV, 5 ng phRG-tk Renilla and 40 ng pBSK mock plasmid were co-transfected (Kang et al., 2015).
Culture the cells in DMEM supplemented with 10% (v/v) fetal bovine serum at 37 °C under a humidified 5% CO2 atmosphere.
-
-
Luciferase measurement
After one day growth, remove the medium from the cultured cells by vacuum pump using loading pipette tips, and gently apply a sufficient volume (i.e., 500 µl/well) of PBS (see Recipes) to rinse the bottom of each well.
-
Dispense 100 µl of 1x PLB (see Recipes) into each well and gently shake the culture plate for 15 min at room temperature (e.g., 70 rpm).
Note: We use Eppendorf® Research® Pro electronic single channel pipette (20-1,000 µl) to dispense the PLB buffer. However, any laboratory pipettes which can reach 100 µl should be fine.
-
Transfer 20 µl PLB lysate without cell debris into each well of a 96-well OptiPlate.
Note: Generally, it is unnecessary to clear lysates of residual cell debris prior to performing the assay.
Program the EnVision Multilabel Plate Reader to perform a 2-sec premeasurement delay, followed by a 10-sec measurement period using 96 Plate US Luminescence aperture (700 nm emission filter) for each reporter assay.
Dispense 50 µl LAR2 substrate (see Recipes), mix by pipetting 2 or 3 times and measure Firefly luciferase activity using EnVision Multilabel Plate Reader. The Luminescence reads represent the Firefly luciferase activity.
Dispense 50 µl freshly prepared Stop & Glo® Reagent (see Recipes) in the same plate, mix by pipetting 2 or 3 times and measure the Renilla luciferase activity by EnVision Multilabel Plate Reader. The Luminescence reads represent the Renilla luciferase activity.
-
Note: Normalize the relative activity using WT activity as 100.
Data analysis
Each experiment is performed at least in triplicate (Table1), calculated activities are shown in Table 2. Data are presented as grouped bar graph type. The statistical analysis of data can be accomplished with GraphPad Prism using the two-tailed Student’s t-test versus control (Figure 2). A practical example of the current γ-secretase Epsilon-cleavage assay was recently shown in ( Yan et al., 2017a and 2017b).
Table 2. Relative activity and Normalized activity.
| Group name | Relative activity | Normalized activity | ||||
|---|---|---|---|---|---|---|
| γ-secretase+C99-T4L-rTA | 3.24901 | 3.62455 | 2.88981 | 293.415 | 327.329 | 260.976 |
| C99-T4L-rTA | 1.21520 | 1.08992 | 1.01682 | 109.743 | 98.429 | 91.828 |
| C99-T4L-FLAG | 0.20193 | 0.20993 | 0.24283 | 18.236 | 18.958 | 21.929 |
| γ-secretase+C99-T4L-FLAG | 0.23061 | 0.21134 | 0.21633 | 20.826 | 19.086 | 19.536 |
| γ-secretase | 0.07020 | 0.20301 | 0.28279 | 6.340 | 18.334 | 25.538 |
| Renilla | 0.13762 | 0.08426 | 0.12391 | 12.429 | 7.609 | 11.191 |
Note: For activity normalization, each relative activity is divided by the average of the relative activity of WT (C99-T4L-rTA) and times 100.
Figure 2. Relative reporter gene activity using C99-T4L-rTA.
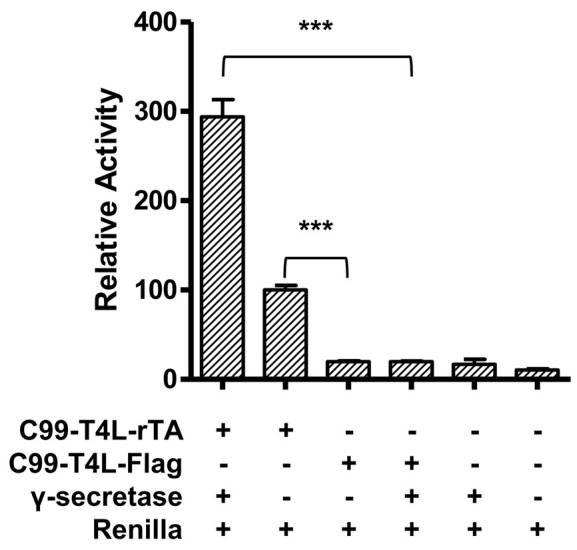
Error bars = SEM, n = 3, P-values (two-tailed Student’s t-test): *P < 0.05; **P < 0.01; ***P < 0.001).
Table 1. Original Firefly and Renilla activity reads.
| Group name | Firefly activity | Renilla activity | ||||
|---|---|---|---|---|---|---|
| γ-secretase+C99-T4L-rTA | 2514996 | 2122104 | 1842544 | 7.741E+05 | 5.855E+05 | 6.376E+05 |
| C99-T4L-rTA | 3275492 | 3228512 | 3472228 | 2.695E+06 | 2.962E+06 | 3.415E+06 |
| C99-T4L-FLAG | 119520 | 131960 | 145920 | 5.919E+05 | 6.286E+05 | 6.009E+05 |
| γ-secretase+C99-T4L-FLAG | 436543 | 345336 | 332059 | 1.893E+06 | 1.634E+06 | 1.535E+06 |
| γ-secretase | 41400 | 113200 | 92280 | 5.898E+05 | 5.576E+05 | 3.263E+05 |
| Renilla | 162120 | 101360 | 123320 | 1.178E+06 | 1.203E+06 | 9.952E+05 |
Notes
The status of the cells is very important. When performing data analysis, pay attention to the Renilla luciferase signal (Table 3). It should be around 1,000,000 photo counts.
Table 3. Examples of bad cell status .
| Name | Firefly activity | Renilla activity | Relative activity | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ctrl1 | 83840 | 30640 | 53480 | 2.240E+03 | 1.800E+03 | 3.560E+03 | 37.42857 | 17.02222 | 15.02247 |
| ctrl2 | 48372 | 63752 | 65404 | 7.132E+03 | 7.476E+03 | 1.048E+04 | 6.78239 | 8.52755 | 6.24084 |
Three wells of Group ctrl1 and three wells of Group ctrl2 should be the negative controls, the low Renilla activity, which to some degree reflects the cell status and transfection efficiency, makes the relative activity much higher than normal.
Recipes
-
10x phosphate buffered saline (PBS buffer) (1 L)
11.5 g Na2HPO4
2 g KH2PO4
80 g NaCl
2 g KCl
Dissolve in 1 L of sterile, deionized water
The pH of 1x PBS should be 7.4
-
1x passive lysis buffer (PLB)
Add 1 volume of 5x PLB from Dual-luciferase reporter assay kit to 4 volumes of distilled water and mix well
The 1x PLB can be stored at 4 °C no more than one month
-
LAR2 substrate (from Dual-luciferase reporter assay kit)
Resuspend the lyophilized Luciferase Assay Substrate in Luciferase Assay Buffer 2 (10 ml for one bottle) and store at -20 °C less than 1 month or -70 °C less than 1 year
-
Stop & Glo® Reagent (from Dual-luciferase reporter assay kit)
Add 1 volume Stop & Glo® Substrate to 49 volumes Stop & Glo® Buffer and vortex for 10 sec
Acknowledgments
This work was supported by the Van Andel Research Institute, the National Natural Science Foundation of China (31300607, 31300245 and 91217311), Ministry of Science and Technology grants 2012ZX09301001, 2012CB910403, and 2013CB910600, XDB08020303, 2013ZX09507001, Shanghai Science and Technology Committee (13ZR1447600), Shanghai Rising-Star Program (14QA1404300), and the National Institute of Health grants DK071662 (H.E.X.), GM102545 and GM104212 (K. M.). The authors declare no conflict of interest.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Barnea G., Strapps W., Herrada G., Berman Y., Ong J., Kloss B., Axel R. and Lee K. F.(2008). The genetic design of signaling cascades to record receptor activation. Proc the Natl Acad Sci U S A 105(1): 64-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kang Y., Zhou X. E., Gao X., He Y., Liu W., Ishchenko A., Barty A., White T. A., Yefanov O., Han G. W., Xu Q., de Waal P. W., Ke J., Tan M. H., Zhang C., Moeller A., West G. M., Pascal B. D., Van Eps N., Caro L. N., Vishnivetskiy S. A., Lee R. J., Suino-Powell K. M., Gu X., Pal K., Ma J., Zhi X., Boutet S., Williams G. J., Messerschmidt M., Gati C., Zatsepin N. A., Wang D., James D., Basu S., Roy-Chowdhury S., Conrad C. E., Coe J., Liu H., Lisova S., Kupitz C., Grotjohann I., Fromme R., Jiang Y., Tan M., Yang H., Li J., Wang M., Zheng Z., Li D., Howe N., Zhao Y., Standfuss J., Diederichs K., Dong Y., Potter C. S., Carragher B., Caffrey M., Jiang H., Chapman H. N., Spence J. C., Fromme P., Weierstall U., Ernst O. P., Katritch V., Gurevich V. V., Griffin P. R., Hubbell W. L., Stevens R. C., Cherezov V., Melcher K. and Xu H. E.(2015). Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 523(7562): 561-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu T. H., Yan Y., Kang Y., Jiang Y., Melcher K. and Xu H. E.(2016). Alzheimer's disease-associated mutations increase amyloid precursor protein resistance to γ-secretase cleavage and the Aβ42/Aβ40 ratio. Cell Discov 2: 16026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan Y., Xu T. H., Harikumar K. G., Miller L. J., Melcher K. and Xu H. E.(2017). Dimerization of the transmembrane domain of amyloid precursor protein is determined by residues around the gamma-secretase cleavage sites. J Biol Chem 292(38): 15826-15837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan Y., Xu T. H., Melcher K. and Xu H. E.(2017). Defining the minimum substrate and charge recognition model of gamma-secretase. Acta Pharmacol Sin 38(10): 1412-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]