ABSTRACT
Oral squamous cell carcinoma (OSCC) is one of the most aggressive and lethal malignancies affecting the head and neck region with a general 5-year survival rate about 50%. Long non-coding RNAs (lncRNAs) are believed to participate in diverse biological processes and are emerging as convenient and minimally invasive diagnostic/prognostic/therapeutic markers. The aim of this study was to explore CEBPA-AS1 role and mechanism in OSCC tumorigenesis. In this study, CEBPA-AS1 localized in the cytoplasm and the peri-nuclear cellular compartment functioning as a potential oncogene up-regulated in OSCC was correlated with poor differentiation, lymph node metastasis and high clinical stage, which made it considered to be a prognostic biomarker. Silence of CEBPA-AS1 inhibited OSCC cells proliferation and induced cells apoptosis, migration and invasion by targeting CEBPA and via a novel pathway CEBPA/Bcl2. Our findings provided the first evidence for the lncRNA CEBPA-AS1 regulatory network in OSCC tumorigenesis, which might be helpful to improve the effects of clinical treatment in OSCC.
KEYWORDS: Oral squamous cell carcinoma, LncRNAs, CEBPA-AS1, CEBPA
Introduction
Oral squamous cell carcinoma (OSCC) represents ∼ 3% of all malignancies, is one of the most aggressive cancer affecting the head and neck region.1 Recent years, OSCC incidence is increasing with more than 274,000 newly diagnosed cancer cases every year worldwide and accounts for 80% ∼ 90% of the oral mucosa of primary malignant tumor.2 Although significant progress in the diagnosis and clinical treatment such as surgery in combination with radiotherapy, neoadjuvant chemotherapy and targeted therapy has been made, the general survival rate of OSCC patients had a 3 year survival rate of 50% ∼ 75% and a 5-year survival rate of about 50%.3,4 Notably, this number has not been improved significantly for more than 50 years.5 Therefore, it's urgent to find suitable prognostic or metastatic biomarkers and therapeutic strategies for OSCC.
Long non-coding RNAs (lncRNAs), a class of non-coding RNA with transcripts longer than 200 nucleotides, are emerging notable molecular markers paid more attention in various types of cancer, including OSCC recent years. Accumulating evidences suggest that lncRNAs involved in regulating gene expression are validated to be associated with cancer progression by functioning as oncogene or tumor suppressor genes, thus made lncRNAs to be a novel target for cancer diagnosis and therapy.6,7 However, there are only 30 papers were published about correlation between lncRNAs and OSCC till November 16, 2017. Numerous of them were focus on the functional research on the already known lncRNA like HOTAIR, MALAT1, UCA1, FTH1P3, TUG1 and so on.8-12 Minority researches discussed the regulation mechanism mediated by microRNA. For example, MALAT1 modulate STAT3 expression by absorbing miR-125b in OSCC, LINC00668 promotes OSCC tumorigenesis via miR-297/VEGFA axis, FTH1P3 acts as a molecular sponge of miR-224-5p to modulate fizzled 5 expression.9,11,13 All these evidences made lncRNAs to be new targets for OSCC diagnosis and therapy. A clear understanding of the alterations in lncRNA expression occurring in OSCC will require larger-scale studies than those yet reported and as far as we know.
CEBPA-AS1, also known as LOC80054, located at 19q13.11 was first found enriched in human gastric cancer tissues and the plasma of patients in March 2017. Combination of CEBPA-AS1 with circulating long noncoding RNAs AK001058, INHBA-AS1, MIR4435-2HG in plasma might be used as diagnostic or prognostic markers for gastric cancer patients.14 There was on record of CEBPA-AS1 expression, function or regulation mechanisms in other diseases, including cancers. Our present results provided the first evidence for lncRNA CEBPA-AS1 regulatory network CEBPA-AS1/CEBPA/Bcl2 in OSCC, which might helpful to improve the effects of clinical treatment in OSCC.
Materials and methods
Sample collection
60 pairs of OSCC tissues and matched paraneoplastic normal tissues (NT) used in this study were selected from OSCC patients treated at Affiliated Stomatology Hospital of China Medical University between July 2011 and April 2012. Patient characteristics are shown in Table 2 and 3. Written informed consent was obtained from all patients and the study protocol was approved by the Ethics Committees of Affiliated Stomatology Hospital of China Medical University (2014016). All the samples had been collected before any kind of therapeutic measures, and fresh frozen immediately after surgery, and store at −80°C.
Table 2.
Correlation between CEBPA-AS1 expression and clinical pathological parameters of OSCC patients (n = 60).
| Relative CEBPA-AS1 expression |
|||||
|---|---|---|---|---|---|
| Parameter | Number | Low | High | P value | |
| Gender | Male | 39 | 18 | 21 | 0.4168 |
| Female | 21 | 12 | 9 | ||
| Age | ≤50 | 15 | 8 | 7 | 0.0889 |
| >50 | 45 | 22 | 23 | ||
| Smoking | Yes | 25 | 14 | 11 | 0.8953 |
| No | 35 | 19 | 16 | ||
| Drinking | Yes | 18 | 8 | 10 | 0.2820 |
| No | 42 | 25 | 17 | ||
| Position | Tongue | 30 | 14 | 16 | 0.5491 |
| Gingiva | 14 | 6 | 8 | ||
| Buccal mucosa | 8 | 5 | 3 | ||
| Lip | 3 | 1 | 2 | ||
| Palate | 5 | 4 | 1 | ||
| Differentiation | Well, moderately | 44 | 28 | 18 | 0.0134* |
| Poorly | 16 | 4 | 12 | ||
| T stage | T1 and T2 | 40 | 24 | 16 | 0.0677 |
| T3 and T4 | 20 | 7 | 13 | ||
| N stage | N0 | 33 | 8 | 25 | <0.0001* |
| N1-N3 | 27 | 22 | 5 | ||
| Clinical stage | I and II | 31 | 20 | 11 | 0.0395* |
| III and IV | 29 | 11 | 18 | ||
Chi-square test,
P < 0.05.
Table 3.
Influence of CEBPA-AS1 expression and clinical characteristics on overall survival in OSCC patients (n = 60).
| Univariate analysis |
Multivariate analysis |
||||
|---|---|---|---|---|---|
| Factors | Subset | HR (95% CI) | P value | HR (95% CI) | P value |
| Gender | Male/Female | 1.76 (0.77-1.92) | 0.658 | ||
| Age | ≤50/>50 | 2.25 (1.27-3.34) | 0.152 | ||
| Smoking | Yes/No | 2.18 (0.69-2.08) | 0.314 | ||
| Drinking | Yes/No | 1.54 (1.08-2.59) | 0.784 | ||
| Position | Tongue/Gingiva/Buccal mucosa/Lip/Palate | 1.58 (0.83-2.31) | 0.468 | ||
| Differentiation | Well, moderately/Poor | 2.63 (0.96-1.94) | 0.001 | ||
| CEBPA-AS1 expression | High/Low | 5.46 (2.57-6.58) | <0.001* | 6.71 (3.61-8.73) | < 0.001 * |
| T stage | T1 and T2 / T3 and T4 | 1.49 (0.79-2.25) | 0.474 | ||
| N stage | N0 / N1-N3 | 3.52 (1.54-3.31) | 0.005 | 3.25 (1.72-5.33) | 0.001* |
| Clinical stage | I and II/III and IV | 2.16 (1.15-3.26) | 0.137 | ||
Cell culture
The OSCC cell lines (Tca8113 and Cal27) and normal oral keratinocyte hNOK were obtained from the Chinese Academy of Sciences (Shanghai, China). The cells were grown in RPMI 1640 medium and DMEM medium (Hyclone, USA), supplemented with 10% fetal bovine serum (Gibco, USA) and maintained at 37°C in a humidified incubator containing 5% CO2.
Quantitative Real-time PCR (qRT-PCR)
Total RNA was extracted with Trizol reagent (Invitrogen, USA). Then the isolated total RNA was reverse transcribed into cDNA using SuperScript® VILO™ cDNA conversing kit (Invitrogen, US). The produced cDNA was amplified by the specific primer sets listed in Table 1. One Step SYBR RT-PCR Kit (TaKaRa, Japan) was used to detect the lncRNA and mRNA expression through the 7500 Real-Time PCR System (Applied Biosystems, USA) and normalized with the GAPDH levels using 2−CT method.
Table 1.
The sequence for primers and Smart Silencer Target Sequence.
| Primers used for qRT-PCR | |
|---|---|
| GAPDH F | GGGAGCCAAAAGGGTCAT |
| GAPDH R | GAGTCCTTCCACGATACCAA |
| CEBPA-AS1 F | GCTTCGTTTTCGGTCCAGA |
| CEBPA-AS1 R | CCCTCCACAGGTGAATGCTAT |
| CEBPA F | TTTGCTCGGATACTTGCCA |
| CEBPA R | AAAGGAAAGGGAGTCTCAGACC |
| Bcl2 F | GATTGAAGACACCCCCTCGT |
| Bcl2 R | CCGGTTATCGTACCCTGTTCT |
| Ribo™ h-CEBPA-AS1 Smart Silencer Target Sequence | |
| CEBPA-AS1-1# | GGAATAAGACTTTGTCCAA |
| CEBPA-AS1-2# | CCACAGGTCCTCCTACAAT |
| CEBPA-AS1-3# | GGAGGCAGAGATCAGATTT |
| CEBPA-AS1-4# | ATGGGAATCAAGGGTGGATG |
| CEBPA-AS1-5# | ACGTGCGTCCCTCGCATTCT |
| CEBPA-AS1-6# | AGGAGTATCCCGAGGCTGCA |
Microarray analysis
Total RNA was extracted as before. The profiles of lncRNAs/mRNAs were finished by Kangcheng Bio Corporation (Shanghai, China) with Human 8 × 60K LncRNA expression array. In details, the RNA was first transcribed into cDNAs. After cleaned and purified, the labeled cDNA were hybridized with Agilent Gene Expression Hybridization Kit (Agilent, USA). The arrays were scanned and the data were extracted by using Agilent Feature Extraction Software.
Vectors construction and transfection
Smart Silencers and negative controls of CEBPA-AS1 were synthesized by RiboBio Corporation (Guangzhou, China) with its targeting sequence listed in Table 1. The pcDNA-CEBPA and blank vector, CEBPA-AS1-mut were constructed by Genescript company (Nanjing, China). Then Lipofectamine™ 3000 (Invitrogen, USA) was applied to finish the transfection according to the manufacturer's instructions, and qRT-PCR was applied to detect the transfected effects.
Cell proliferation assay
The cell proliferation ability was assayed by both EDU (5-ethynyl-2′-deoxyuridine) and MTT (3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide) methods. For the EDU assay, cells were cultured in 96-well plate and fixed in paraformaldehyde for 10 min, then incubated with EDU from EDU DNA Cell Proliferation Kit (RiboBio, China) at 37°C for 4 h. After counterstained with 4′-6′ diamidino-2-phenylindole (DAPI), the images were observed under a fluorescence microscope. For the MTT assay, 5000 cells were inoculated into 96-well plate with each group 5 wells. Then 10 μl PBS with 5% MTT was added. After incubating at 37°C for 4 h, 100 μl DMSO was added to the wells containing cells for dissolving the formazon. Absorbance at 570 nm was recorded using the Infinite F200 micro-plate reader (Tecan, Switzerland).
Flow cytometry assay
The cell cycle and cell apoptosis were finished by flow cytometry (Canto II, BD). Annexin IV-FITC apoptosis detection kit (Biosea, China) was used to detect the apoptosis 72 h after transfection according to the manufacturer's protocol. The results were analyzed using Diva 8.0 software (BD, USA). For the cell cycle assay, cells were fixed in 1 ml 70% ethanol and stained with PI (Dingguo, China) at 37°C for 40 min. All the assays were done within 1 h.
Cell invasion and migration assay
Cell invasion ability was assayed using Transwell chamber (Costar, USA) according to the manufacturer's protocol. In details, 5 × 104 cells with 10% serum medium were seeded to the top chambers and the bottom chambers were filed with 500 μl supernatant of human NIH3T3. After a 12 h incubation period in 37°C, cells invading to the lower surface of the chamber were stained with hematoxylin and eosin, and counted in eight random fields under microscope. For the migration assay, scratches were made by using a 100 μl pipette tip after cells confluence reached approximately 80% post-transfection. The images were observed by exhaustively washed detached cells with PBS.
RNA-Fluorescence in Situ Hybridization (Rna-FISH)
CEBPA-AS1, U6 and 18S oligonucleotide probes were bought from RiboBio Company (Guangzhou, China) and the procedures were following the manual from FISH Kit also in RiboBio. In details, 6 × 104 cells were placed on the autoclaved glass coverslips in 24 well plates. The following day, coverslips were washed in PBS and fixed in 4% formaldehyde for 10 min. After exhaustively washed with PBS containing 5% Triton X-100 at 37°C for 10 min, coverslips were pre-hybridized with the pre-binding solution for about 30 min. Then the cells were hybridized with anti-U6, anti-18S, or anti-CEBPA-AS1 probes separately for 12 h at 37°C. After counterstained with DAPI, the images were acquired by using a confocal immunofluorescence microscope.
RNA pull-down assays
CEBPA-AS1 or CEBPA probes were biotin-labeled with the Biotin RNA Labeling Mix (Roche, Switzerland) and purified with the RNeasy Mini Kit (Qiagen, USA). Then, cell extract (2 mg) was mixed with biotinylated RNA (100 pmol). Dynabeads M-280 Streptavidin (Invitrogen, USA) were added to each binding reaction and further incubated at room temperature for 10 min. Finally, beads were washed briefly three times and the co-precipitated RNAs were detected by qRT-PCR. Total RNAs and controls were also assayed to demonstrate that the detected signals were from specific RNAs. The probe sequence was 5′-Bio-CATTGCACAA-3′.
Western blot assay
Total protein was extracted using a pre-cold cell lysis buffer containing protease and phosphatase inhibitors (KEYGEN, China) and the concentration was determined using the BCA assay (KEYGEN, China) according to manufacturer's instructions. Protein lysates (30 μg) were loaded and separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to a 0.22 μm polyvinylidene fluoride (PVDF) membrane. After blocked at room temperature for 2 h with 5% powdered non-fat milk, the membranes were hybridized with primary antibodies included rabbit anti-CEBPA (1/500, Cat# YT0551, Immunoway), and mouse anti-Bcl2 (1/500, Cat# YM3041, Immunoway) overnight at 4°C. Then, the membranes were incubated with secondary antibodies at room temperature for 2 h and the immunobilized proteins were detected by Dual Color Infra-red Laser Imaging System (Gene, HK, China). The band density was quantified by Image J software (National Institutes of Health, USA). β-actin (1/1000, Cat# YM3028, Immunoway) was used as the inference gene.
Statistical analysis
All statistical analysis was performed using SPSS V22.0 (IBM, USA). The data were presented as mean ± SD of three independent experiments and data from surgical resection to overall survival time of the patients were analyzed using the Kaplan-Meier survival curve. Other compared using one-way analysis of variance (ANOVA), log-rank test, Chi-square test, and Pearson correlation analysis. A P-value of less than 0.05 was considered to be statistically significant and indicated by (* and #).
Results
CEBPA-AS1 is up-regulated in OSCC
Using the lncRNAs array assay, total 941 differentially expressed lncRNAs were identified including only 10 lncRNAs which were bidirectional relationship with their nearby coding genes in the micro-dissected OSCC tissues (Fig. 1A). In comparison with other 9 lncRNAs, CEBPA-AS1 showed a significant 7.04-fold up-regulation in OSCC tissues than that in the NT tissues (Fig. 1B, P = 0.02). Following qRT-PCR results verified that CEBPA-AS1 levels in 60 OSCC tissues were markedly higher than that in the NT (P < 0.05) (Fig. 1C). As well as CEBPA-AS1 levels in both OSCC Tca8113 and Cal27 cells were higher than that in the normal oral epithelial cells hNOK, although there were some expression level differences (Fig. 1D). These results suggested that CEBPA-AS1 participate in the process of OSCC tumorigenesis.
Figure 1.
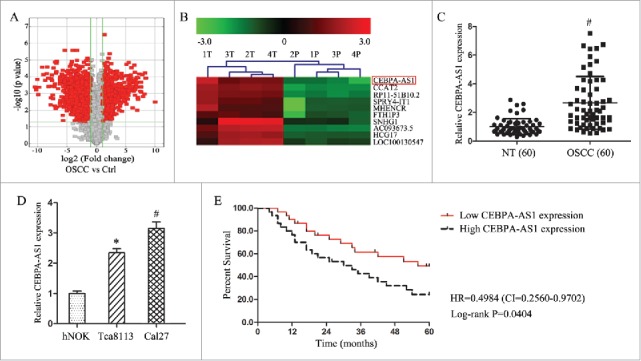
CEBPA-AS1 is an independent prognosis factors in OSCC patients. A, volcano plots of lncRNA array in 4 paired OSCC and control tissues. B, clustering map of CEBPA-AS1 in 4 paired OSCC and control tissues. 1T-4T: OSCC tissues, 1P-4P: control tissues. C, qRT-PCR assay of the CEBPA-AS1 expression levels in OSCC tissues (n = 60). # means P < 0.01 verse NT. D, qRT-PCR assay of the CEBPA-AS1 expression levels in OSCC cells. E: Kaplan-Meier overall survival curves by CEBPA-AS1 expression level. * means P < 0.05 and # means P < 0.01 verse hNOK.
CEBPA-AS1 is an independent prognosis factors in OSCC patients
To assess the clinical significance of CEBPA-AS1, all 60 patients were divided equally into higher and lower CEBPA-AS1 expression group from the median value of relative expression levels. Then we evaluated the correction between its level and clinic-pathological parameters. Results revealed that CEBPA-AS1 levels were remarkably corrected with differentiation grade, lymph node metastasis and clinical stage in OSCC. Nevertheless, CEBPA-AS1 levels were not associated with other clinical characteristics, such as gender, age and position in OSCC (Table 2). Additionally, univariate and multivariate Cox regression analysis revealed that high CEBPA-AS1 expression was an independent predictor of over-all survival (OS) in OSCC patients (Table 3). Kaplan-Meier analysis indicated that high CEBPA-AS1 expression was related to a poorer OS (Fig. 1E). These results confirmed that high CEBPA-AS1 might be crucial in OSCC, and CEBPA-AS1 was an independent prognostic biomarker for OSCC patients.
CEBPA-AS1 functions as an oncogene in OSCC cells
To investigate the functional role of CEBPA-AS1, we used smart silencers to depress CEBPA-AS1 expression in both Tca8113 and Cal-27 cells. Cell morphology observation and qRT-PCR were used to test the knockdown efficiency. The results showed that cells transfected with ss-CEBPA-AS1 had a significant cell numbers decreased, and the cells became round (data not shown). qRT-PCR results showed that cells transfected with ss-CEBPA-AS1 presented a significantly decreased CEBPA-AS1 expression level compared with the NC group in both cells, while there were no statistic difference in blank control and NC group (P < 0.05, Fig. 2A). Therefore, subsequent cell biology studies were performed with those smart silencers.
Figure 2.
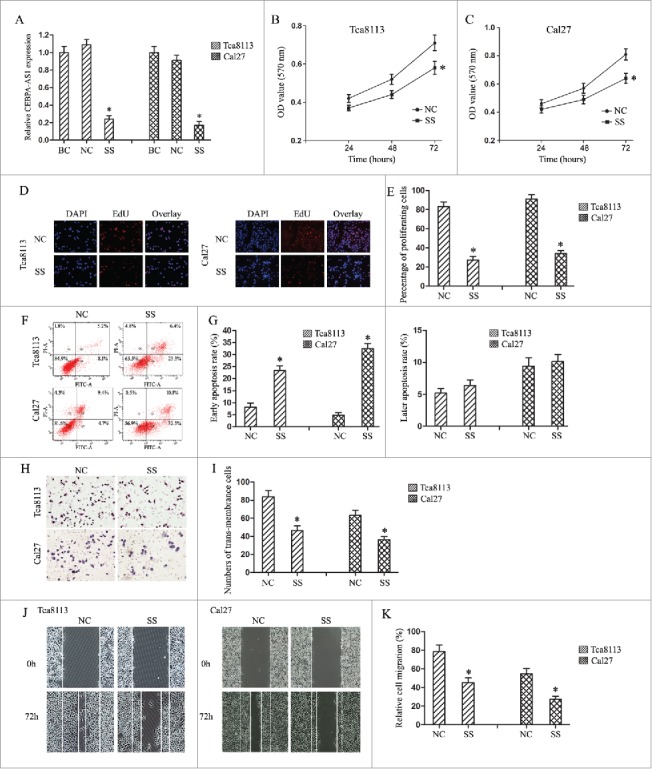
Modulation of CEBPA-AS1 expression effects cell proliferation, cell apoptosis and cell invasion in OSCC cells. BC, blank control group; NC, negative control smart silencers transfection group; SS, CEBPA-AS1 smart silencers transfection group. Each bar represents triplicate analyses of mean ± SD and * means the significant difference from the negative control (P < 0.05). A, qRT-PCR detects the CEBPA-AS1 smart silencers transfection efficiency. B and C, cells proliferation ability was determined by MTT assay. D and E, cells proliferation ability was determined by EDU assay. F and G, cells apoptosis were stained for Annexin V/PI and analyzed by flow cytometry. The percentage of cells positive for Annexin V and negative for PI staining are considered as early apoptosis. The percentage of cells positive for Annexin V and PI staining are considered as late apoptosis. Each of them was presented in quadrant. H and I, the invasive potential of CEBPA-AS1 in OSCC cells was examined by transwell invasion assay (visual field representative of 1 experiment). J and K, the migration potential of CEBPA-AS1 in OSCC cells was examined by a wound healing assay. The percent surface area between the wounds was determined using NIS elements software.
Next, the cell proliferation viability was assessed using MTT method as well as EdU nucleic acid labeling technology in Tca8113 and Cal-27 cells. Depress of CEBPA-AS1 remarkably inhibited cell proliferation at 72 h (Fig. 2B and 2C). These results were confirmed by EdU nucleic acid labeling assays (Fig. 2D and 2E). Further, flow cytometry was conducted to analyze the role of CEBPA-AS1 on cell cycle in OSCC cells and the results showed no statistic significance (data not shown). These results indicated that depression of CEBPA-AS1 might repress cell growth in both Tca8113 and Cal-27 cells.
We also explored the efficiency of CEBPA-AS1 on cell apoptosis using flow cytometry. As shown in Fig. 2F and 2G, depressed CEBPA-AS1 led to the fraction of Annexin positive cells a ∼3.2- folds and ∼5.9-folds increase in Tca8113 and Cal-27 cells separately in comparison to that of each control groups. Interestingly, no significant differences were observed in propidium iodide-positive cells, indicating that depression of CEBPA-AS1 facilitated cell apoptosis primarily through early apoptosis, but not necrosis in Tca8113 and Cal-27 cells.
Finally, we examined the effect of CEBPA-AS1 on invasion and migration in Tca8113 and Cal-27 cells using Transwell chamber and wound healing assay. The results of the invasion assay shows that depressed expression of CEBPA-AS1 inhibits OSCC cells invasion in comparison to control groups. In detail, depressed CEBPA-AS1 nearly suppressed 37% and 54% of the cells' invasion activity in Tca8113 and Cal-27 cells, respectively (Fig. 2H and 2I). Wound and healing assay showed that depressed expression of CEBPA-AS1 inhibits OSCC cells occurring wound closure in comparison to control groups (Fig. 2J and 2K), these results were consistent with the transwell chamber assay, which indicated that depression of CEBPA-AS1 retarded cell invasion and migration ability in Tca8113 and Cal-27 cells.
In all, we got a conclusion that modulating CEBPA-AS1 expression could effect cell proliferation and apoptosis, cell invasion and migration, thus promote tumorigenesis by functioning as an oncogene in OSCC.
CEBPA-AS1 localization in OSCC cells
RNA-FISH was used to characterize CEBPA-AS1 expression in both Tca8113 and Cal27 cells, the results showed that CEBPA-AS1 is predominantly localized in the cytoplasm and the perinuclear cellular compartment of OSCC cells, little is distributed to the nucleus as a punctate pattern (Fig. 3A and 3B). Consistent with the quantitative CEBPA-AS1 expression results (Fig. 1D), RNA-FISH also revealed that CEBPA-AS1 was expressed in OSCC cells. These results suggest that CEBPA-AS1 might play a role in nuclear membrane and cytoplasm trafficking.
Figure 3.
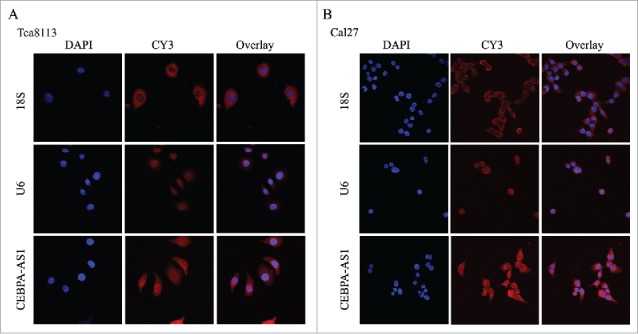
Localization of CEBPA-AS1 by RNA-FISH in OSCC cells. Nuclei are stained blue (DAPI), 18S (Cytoplasm positive), U6 (Nuclei positive) and CEBPA-AS1 labeled with CY3 are stained red. Magnification is 60 × with 4.5 × zoom.
CEBPA is a target of CEBPA-AS1 in OSCC cells
To investigate the mechanism of CEBPA-AS1 in OSCC, we next predicted the lncRNA nearby coding gene by Gene Ontology (Go) analysis (http://geneontology.org), and found that there was a bidirectional relationship between CEBPA-AS1 and CEBPA. Further enrichment analysis showed that the cell apoptosis and adhesion were the most common processes associated with CEBPA-AS1 and CEBPA. Then we tried to clarify the regulatory relationship of them by using the online server ChIPBase, an open database for decoding the transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data and found that there was a CEBPA binding site located at -2468bp of CEBPA-AS1 gene in sample HUMHG00993 (Motif locus: -2510bp). Together with the localization of CEBPA-AS1 in Tca8113 and Cal27 cells, we speculated that CEBPA might be a target of CEBPA-AS1 to influence the tumorigenesis of OSCC.
Our previous mRNA array assay showed that the expression of CEBPA had a 7.49-fold up-regulation in OSCC tissues than that in the matched paraneoplastic normal tissues. Following qRT-PCR results verified that a 3.25-fold mRNA expression level of CEBPA was observed in OSCC tissues when compared with the NT tissues (P < 0.001) (Fig. 4A, P < 0.05). The co-expression patterns (log2-scale) between CEBPA and CEBPA-AS1 in TCGA Pan-Cancer (PANCAN) showed that the expression of CEBPA-AS1 was positive correlated to the CEBPA's level, with a Pearson coefficient r = 0.4454 and p = 7.65E-29 (two tail, t-test) in head and neck squamous cell carcinoma (Fig. 4B).
Figure 4.
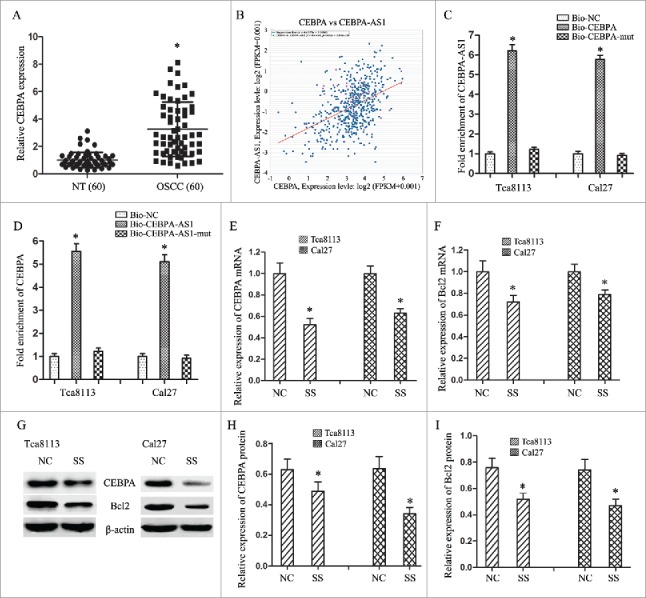
CEBPA-AS1 represses CEBPA/Bcl2 expression via targeting CEBPA in OSCC cells. All data were represented as the mean ± S.D. from three independent experiments, *P < 0.05. A, relative CEBPA levels in OSCC are detected by qPCR. B, the co-expression patterns between CEBPA and CEBPA-AS1 in head and neck squamous cell carcinoma is searched using the online server ChIPBase. C, CEBPA-AS1 RNA levels was detected in the substrate of pull-down assay by qRT-PCR. CEBPA-mut probe was used as negative control. D, CEBPA mRNA level was detected in the substrate of pull-down assay by qRT-PCR. CEBPA-AS1-mut probe was used as negative control. E and F, the level of CEBPA and Bcl2 mRNA are determined by qRT-PCR when CEBPA-AS1 down-regulated in OSCC cells. G-I, the level of CEBPA and Bcl2 protein are determined by western blot when CEBPA-AS1 down-regulated in OSCC cells. β-actin was used as reference control.
Then we conducted RNA pull-down assay to explore whether CEBPA-AS1 functioned via interaction with CEBPA. The results showed that CEBPA-AS1 could be pulled down by biotinylated CEBPA, the biotinylated CEBPA-mut probe was used as negative control (Fig. 4C). We also demonstrated that CEBPA was preferentially enriched in CEBPA-AS1-containing beads compared with the beads harboring biotinylated CEBPA-AS1-mut (Fig. 4D). These results confirmed that CEBPA is a target of CEBPA-AS1 in OSCC cells.
Depressed CEBPA-AS1 expression reduces CEBPA and Bcl2 expression
To further investigate the regulation mechanism of CEBPA-AS1 on the tumorigenesis of OSCC cells, we transfected smart silencers to depress the expression of CEBPA-AS1 and observe the targeted gene CEBPA and the downstream gene Bcl2 expression. The results showed that decreased of CEBPA-AS1 inhibited CEBPA expression in both Tca8113 and Cal-27 cells. Bcl2, a target gene of CEBPA, also an important anti-apoptotic gene in many cancers, was decreased expression with the depression of CEBPA-AS1 (Fig. 4E-4I). These results suggest that CEBPA-AS1 and CEBPA might have a synergistic action on the tumorgenesis of OSCC by reducing CEBPA and Bcl2 expression.
Increased CEBPA expression promotes the CEBPA-AS1 associated tumorigenesis
To testify the oncogene role of CEBPA-AS1 was associated with the increase of CEBPA, the pcDNA-CEBPA, or pcDNA empty vector was transfected into OSCC cell lines Tca8113 and Cal27. qRT- PCR confirmed that transfection of pcDNA-CEBPA could significantly increase the CEBPA expression level in both cell lines when compared with the pcDNA empty vector transfection groups (P < 0.05) (Fig. 5A). Then we used co-transfection technology to alteration the expression of CEBPA and CEBPA-AS1. Flow cytometry and transwell chamber assays showed that cells transfected with pcDNA-CEBPA had reduced apoptosis rate and increased invasion ability compared with other groups (P < 0.05) (Fig. 5B-5E). These results pointed that pcDNA-CEBPA rescued the inhibition effect of CEBPA-AS1 silence in both Tca8113 and Cal27 cells. In consistent with the result of biological behaviour analysis, the suppression of CEBPA and anti-apoptotic genes Bcl2 were also alleviated (P < 0.05) (Fig. 5F-5H). Therefore, CEBPA-AS1 promotes the tumorigenesis via CEBPA/Bcl2 in OSCC.
Figure 5.
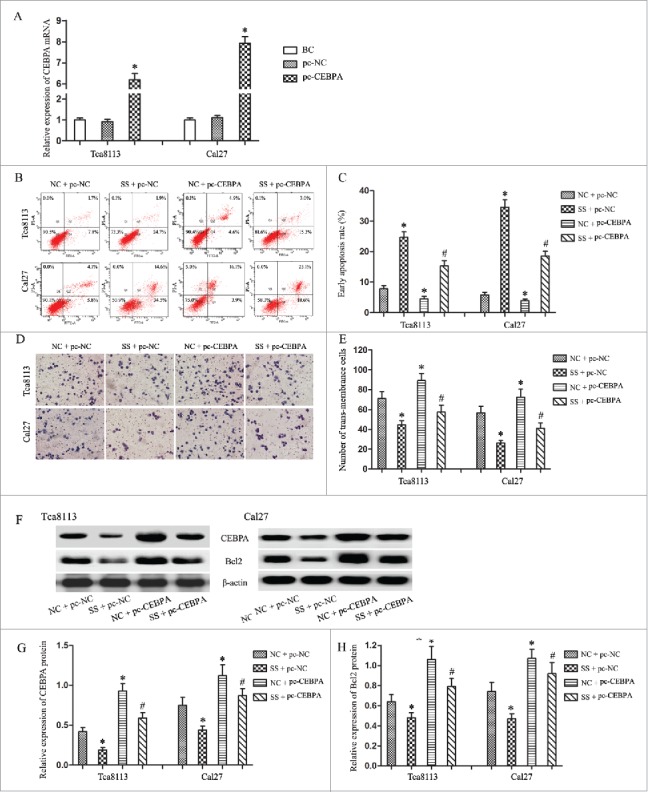
CEBPA promotes the CEBPA-AS1 associated tumorgenesis in OSCC cells by alleviated CEBPA and Bcl2 expression. Each bar represents triplicate analyses of mean ± SD, * means the significant difference from the NC + pc-NC control (P < 0.05) and # means the significant difference from the NC + pc-CEBPA group (P < 0.05). A, qRT-PCR detects the CEBPA vectors or the empty vectors transfection efficiency. B and C, cells apoptosis were analyzed by flow cytometry. D and E, the invasive ability was examined by transwell invasion assay. F-H, the level of CEBPA and Bcl2 protein are determined by western blot. β-actin was used as reference control.
Discussion
Recent years, lncRNAs are believed to participate in diverse biological processes and are emerging as convenient and minimally invasive diagnostic/prognostic/therapeutic markers.15 Some lncRNAs such as PCA3 is now routinely used in the clinical diagnosis of prostate cancer.16 SOX21-AS1 detected in the advanced OSCC patients might be a potential prognostic biomarker for OSCC.17 LncRNA FOXCUT co-amplified with FOXC1 may serve as novel biomarkers and therapeutic targets in OSCC patients who overexpress this “lncRNA-mRNA pair”.18 In the present study, we found lncRNA CEBPA-AS1 could also be an independent prognostic biomarker for OSCC patients. As for the important clinical significance of CEBPA-AS1, we next explored the potential function of CEBPA-AS1 in OSCC by using a serials assays and showed that suppressing the expression of CEBPA-AS1 could inhibit the malignant biological behaviors of OSCC cells, which revealed that CEBPA-AS1 functions as an oncogene in the process of OSCC.
As we all known, the regulatory mechanisms of lncRNAs are diverse and complex. Some lncRNAs could function by promoting or repressing transcription, or by acting as modulators of mRNA translation. Some lncRNAs regulate the transcription of nearby genes in cis, while others act in trans.19-21 In this study, we first detected the localization of CEBPA-AS1 by RNA-FISH and found that CEBPA-AS1 is predominantly localized in the cytoplasm and the perinuclear cellular compartment of OSCC cells, which suggest that CEBPA-AS1 might play a role in nuclear membrane and cytoplasm trafficking, further function by promoting or repressing transcription. Through Go and ChIPBase analysis, we found that there was a bidirectional/positive relationship between CEBPA-AS1 and CEBPA, together with the characterization of CEBPA-AS1, we speculated that CEBPA might be a target of its nearby gene CEBPA-AS1 to influence the tumorigenesis of OSCC. As for the regulation manner was in cis or in trans, we used RNA pull down to confirmed the targeting relationship and used smart silencers to depress the CEBPA-AS1 expression and the results showed a significantly reduced CEBPA levels, which revealed that CEBPA-AS1 regulates CEBPA expression in a cis manner. Recent studies also revealed that a large proportion of lncRNAs cooperate with adjacent protein-coding genes and form “lncRNA-mRNA pairs” that impact their function.18,22 Our present study depressing CEBPA-AS1 expression markedly reduced its target gene CEBPA expression level, was coincidence with the “lncRNA-mRNA” pair, which is now regarded as a new form in the very complex gene expression modulation network.
CCAAT enhancer-binding protein alpha (CEBPA), a type of transcription factor, is an important member of the C/EBP family (including C/EBPα, β, γ, δ, ε and ζ) localized in chromosome 19q13.1.23 Normally, CEBPA plays an important role in the modulation of cell proliferation, differentiation or apoptosis in various tissues.24 Recently there was some controversy about the expression of CEBPA in solid cancers. Several studies have shown that CEBPA functions as a tumor suppressor with reduced expression and growth inhibitory effect in primary mammary carcinomas, such as in lung cancer, cervical squamous cell carcinoma and even in chronic myeloid leukemia.25-27 Some studies have showed CEBPA overexpression was significantly correlated with poor prognosis and prediction of ovarian cancer and breast patients, likely due to an oncogenic role that CEBPA apparently acquires in these neoplasms.28,29 Herein, we reported that CEBPA as a target of CEBPA-AS1 in OSCC Tca8113 and Cal27 cell lines. CEBPA-AS1 down-regulated inhibited CEBPA expression, resulting in the inhibition of proliferation, promoting of apoptosis, cell migration and invasion of OSCC cells, which revealed an oncogenic role of CEBPA in OSCC.
Since CEBPA exerts a great role in CEBPA-AS1 associated OSCC tumorigenesis, regulation of its expression was also an import part of its function exploration. Recent studies showed that CEBPA mediated its activity through direct interaction with NF-κB p50 bound to the Bcl2, which is central to the intrinsic apoptotic pathway.28,30 Our study showed that CEBPA-AS1 depressed expression inhibited CEBPA expression, causing suppression of apoptosis protective genes Bcl2, and resulted in the modulation of OSCC cells tumorigenesis. Moreover, pcDNA-CEBPA rescued the tumorigenesis inhibition of OSCC cells and the suppression of anti-apoptotic genes Bcl2 caused by CEBPA-AS1 depressed expression. Collectively, CEBPA-AS1 targeting CEBPA could some part, but not in whole inhibit tumorigenesis in OSCC cells via a novel pathway CEBPA/Bcl2.
Conclusions
Our results clarify that CEBPA-AS1 up-regulated in OSCC, correlated with poor differentiation, lymph node metastasis and high clinical stage, could be a prognostic biomarker of OSCC patients. “CEBPA-AS1-CEBPA” pair regulated in cis functions as an oncogene in OSCC by inducing proliferation, inhibiting apoptosis, cell migration and invasion. CEBPA-AS1 predominantly localized in the cytoplasm and the perinuclear cellular compartment promotes OSCC cells tumorigenesis via a novel pathway CEBPA/Bcl2. Our findings elucidate a potential mechanism of lncRNA CEBPA-AS1 in OSCC tumorigenesis, and indicate that CEBPA-AS1 could be a potential diagnostic and therapeutic target in OSCC. However, the exact mechanisms still need further exploring.
Funding Statement
National Natural Science Foundation of China [Grant number 81301834].
Conflict of interest
The authors declare that they have no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81301834).
References
- 1.Global burden of oral Jin LJ, Lamster IB, Greenspan JS, Pitts NB, Scully C, Warnakulasuriya S. diseases: emerging concepts, management and interplay with systemic health. Oral Dis. 2016;22(7):609–19. doi: 10.1111/odi.12428. PMID:26704694 Review. [DOI] [PubMed] [Google Scholar]
- 2.de Camargo Cancela M Voti L, Guerra-Yi M, Chapuis F, Mazuir M, Curado MP. Oral cavity cancer in developed and in developing countries: population-based incidence. Head Neck. 2010;32(3):357–67. PMID:19644932. [DOI] [PubMed] [Google Scholar]
- 3.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 4.Kessler P, Grabenbauer G, Leher A, Bloch-Birkholz A, Vairaktaris E, Neukam FW. Neoadjuvant and adjuvant therapy in patients with oral squamous cell carcinoma Long-term survival in a prospective, non-randomized study. Br J Oral Maxillofac Surg. 2008;46(1):1–5. doi: 10.1016/j.bjoms.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Ribeiro KC1, Kowalski LP, Latorre MR. Impact of comorbidity, symptoms, and patients' characteristics on the prognosis of oral carcinomas. Arch Otolaryngol Head Neck Surg. 2000;126(9):1079–85. doi: 10.1001/archotol.126.9.1079. PMID:10979120. [DOI] [PubMed] [Google Scholar]
- 6.Tano K1, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. 2012;3:219. doi: 10.3389/fgene.2012.00219 eCollection 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutschner T1, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–19. doi: 10.4161/rna.20481. doi: 10.4161/rna.20481. PMID:22664915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H, Li Z, Wang C, Feng L, Huang H, Liu C, Li F. Expression of long non-coding RNA-HOTAIR in oral squamous cell carcinoma Tca8113 cells and its associated biological behavior. Am J Transl Res. 2016;8(11):4726–4734. [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren Y, Wu Y, Mei M, Zhang L, Wang X. Long Non Coding RNA MALAT1 Promotes Tumor Growth and Metastasis by inducing Epithelial-Mesenchymal Transition in Oral Squamous Cell Carcinoma. Sci Rep. 2015;5:15972. doi: 10.1038/srep15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang YT, Wang YF, Lai JY, Shen SY, Wang F, Kong J, Zhang W, Yang HY. Long non-coding RNA UCA1 contributes to the progression of oral squamous cell carcinoma by regulating the WNT/β-catenin signaling pathway. Cancer Sci. 2016;107(11):1581–1589. doi: 10.1111/cas.13058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang CZ. Long non-coding RNA FTH1P3 facilitates oral squamous cell carcinoma progression by acting as a molecular sponge of miR-224-5p to modulate fizzled 5 expression. Gene. 2017;607:47–55. doi: 10.1016/j.gene.2017.01.009. doi: 10.1016/j.gene.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 12.Liang S, Zhang S, Wang P, Yang C, Shang C, Yang J, Wang J. LncRNA, TUG1 regulates the oral squamous cell carcinoma progression possibly via interacting with Wnt/β-catenin signaling. Gene. 2017;608:49–57. doi: 10.1016/j.gene.2017.01.024. doi: 10.1016/j.gene.2017.01.024. [DOI] [PubMed] [Google Scholar]
- 13.Zhang CZ. Long intergenic non-coding RNA 668 regulates VEGFA signaling through inhibition of miR-297 in oral squamous cell carcinoma. Biochem Biophys Res Commun. 2017;489(4):404–412. doi: 10.1016/j.bbrc.2017.05.155. PMID:28564590. [DOI] [PubMed] [Google Scholar]
- 14.Ke D, Li H, Zhang Y, An Y, Fu H, Fang X, Zheng X. The combination of circulating long noncoding RNAs AK001058, INHBA-AS1, MIR4435-2HG, and CEBPA-AS1 fragments in plasma serve as diagnostic markers for gastric cancer. Oncotarget. 2017;8(13):21516–21525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandra Gupta S, Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets. Int J Cancer. 2017;140(9):1955–1967. doi: 10.1002/ijc.30546. [DOI] [PubMed] [Google Scholar]
- 17.Yang CM, Wang TH, Chen HC, Li SC, Lee MC, Liou HH, Liu PF, Tseng YK, Shiue YL, Ger LP, Tsai KW. Aberrant DNA hypermethylation-silenced SOX21-AS1 gene expression and its clinical importance in oral cancer. Clin Epigenetics. 2016;8:129. eCollection 2016. doi: 10.1186/s13148-016-0291-5. PMID:27933111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong XP, Yao J, Luo W, Feng FK, Ma JT, Ren YP, Wang DL, Bu RF. The expression and functional role of a FOXC1 related mRNA-lncRNA pair in oral squamous cell carcinoma. Mol Cell Biochem. 2014;394(1-2):177–86. doi: 10.1007/s11010-014-2093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–81. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 20.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47(4):648–55. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–46. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, Young RA. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A. 2013;110(8):2876–81. doi: 10.1073/pnas.1221904110. PMID:23382218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.J1 Tsukada, Y Yoshida, Kominato Y, Auron PE. The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation. Cytokine. 2011;54(1):6–19. doi: 10.1016/j.cyto.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 24.RG1 Lopez, S Garcia-Silva, Moore SJ, Bereshchenko O, Martinez-Cruz AB, Ermakova O, Kurz E, Paramio JM, Nerlov C. C/EBPalpha and beta couple interfollicular keratinocyte proliferation arrest to commitment and terminal differentiation. Nat Cell Biol. 2009;11(10):1181–90. doi: 10.1038/ncb1960. [DOI] [PubMed] [Google Scholar]
- 25.B1 Halmos, CS Huettner, Kocher O, Ferenczi K, Karp DD, Tenen DG. Down-regulation and antiproliferative role of C/EBPalpha in lung cancer. Cancer Res. 2002;62(2):528–34. [PubMed] [Google Scholar]
- 26.Pan Z, Zheng W, Zhang J, Gao R, Li D, Guo X, Han H, Li F, Qu S, Shao R. Down-regulation of the expression of CCAAT/enhancer binding protein α gene in cervical squamous cell carcinoma. BMC Cancer. 2014;14:417. doi: 10.1186/1471-2407-14-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.S1 Kagita, S1 Uppalapati, S1 Gundeti, R2 Digumarti. Correlation of C/EBPα expression with response and resistance to imatinib in chronic myeloid leukaemia. Jpn J Clin Oncol. 2015;45(8):749–54. doi: 10.1093/jjco/hyv064. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Ma Y, Wang S, Chen F, Gu Y. C/EBPα inhibits proliferation of breast cancer cells via a novel pathway of miR-134/CREB. Int J Clin Exp Pathol. 2015;8(11):14472–8. eCollection 2015. [PMC free article] [PubMed] [Google Scholar]
- 29.Pabst T, Mueller BU. Complexity of CEBPA dysregulation in human acute myeloid leukemia. Clin Cancer Res. 2009;15(17):5303–7. doi: 10.1158/1078-0432.CCR-08-2941. [DOI] [PubMed] [Google Scholar]
- 30.Paz-Priel I, Ghosal AK, Kowalski J, Friedman AD. C/EBPalpha or C/EBPalpha oncoproteins regulate the intrinsic and extrinsic apoptotic pathways by direct interaction with NF-kappaB p50 bound to the bcl-2 and FLIP gene promoters. Leukemia. 2009;23(2):365–74. doi: 10.1038/leu.2008.297. [DOI] [PMC free article] [PubMed] [Google Scholar]


