ABSTRACT
Fibroblast growth factor receptor-1 (FGFR1) over-expression was broadly found in squamous cancer, where it induced cellular proliferation, differentiation, and metastasis by activating various signaling pathway. However, the role of FGFR1 gene expression in predicting prognosis of Esophageal Squamous Cell Carcinoma (ESCC) and its regulatory function in the progression of ESCC are not well understood. Therefore, we performed an analysis of FGFR1 mRNA expression by quantitative RT-PCR in tumor tissue of 145 patients with ESCC. The relationships between FGFR1 gene expression and clinicopathological parameters, also the prognosis were further examined. Results suggested that higher FGFR1 gene expression predicted worse overall survival (HR = 1.502, 95%[CI] = 1.005–2.246, P = 0.045). Disease-free survival tends to be shorter in patients with higher FGFR1 expression but without statistical significance (HR = 1.398, 95%[CI] = 0.942–2.074, P = 0.096). FGFR1 was up regulated in multiple ESCC cell lines. Subsequent in vitro experiments demonstrated that anti-FGFR1 treatment by PD173074 inhibited TE-1 and EC9706 cell viability along with the attenuation of MEK-ERK signaling pathway. In vivo, PD173074 administration also had shown potent ESCC growth arresting effect. Overall, our study suggested that FGFR1 gene expression could be an independent prognosis predictive factor in patients with ESCC. Anti-FGFR1 inhibited ESCC growth and could be a potential strategy in ESCC targeted therapy.
KEYWORDS: esophageal squamous cell carcinoma, fibroblast growth factor receptor-1, MEK-ERK pathway, prognosis, gene expression
Abbreviations
- ESCC
esophageal squamous cell carcinoma
- FGFR1
fibroblast growth factor receptor
- DFS
disease-free survival
- OS
overall survival
Background
Esophageal cancer comprises two main histological subtypes – esophageal squamous cell carcinoma (ESCC) and adenocarcinomas (EADC). ESCC accounts for the majority of cases and are mostly prevalent in Asian populations whereas EADC represents about 70% of the total in the Western hemisphere.1 Despite a rapid development of novel therapies in EADC, quite a few progresses of therapeutical strategies have been made in ESCC, in which the primary treatments still mostly based on traditional surgery, chemotherapy, and radiotherapy. Recently, targeted therapies against thoracic cancer, including ESCC were introduced. Tyrosine kinase inhibitors (TKIs) targeting epidermal growth factor receptor and vascular endothelial growth factor receptor display clinical efficacy in ESCC.2 Increasing evidence suggests that Fibroblast Growth Factor Receptor (FGFR) could be another potential target in ESCC.
The FGFR family consists of four highly conserved members of tyrosine kinase receptors FGFR1-4, which share similar structure within a cytoplasmic tyrosine kinase domain. FGFR1, a key member of FGFR family, was reported to play a key role in diverse solid tumors. Specific ligands binding with FGFR1 to trigger a cascade of downstream signals, including RAS, MEK-ERK, and AKT1 to regulate cell proliferation, differentiation, migration, and angiogenesis.3 Several studies suggest that blocking FGFR1 promotes apoptosis or inhibit cell proliferation in various cancer.4 By now, several phase I clinical trials confirmed the preliminary antitumor activity of FGFR1 inhibitors in lung squamous cell carcinoma.5 However, it is not known whether inhibiting FGFR1 is a rational and valid approach in ESCC.
The role of aberrant FGFR1 expression has been well elucidated in squamous cancer. Previous research demonstrated that FGFR1 gene amplification was found in 10% to 20% squamous cell lung cancer patients and predicted poor prognosis.6–8 FGFR1 is also one of the most frequently amplified genes in ESCC.9 However, FGFR1 amplification seems insufficient to predict ESCC prognosis and the response to anti-FGFR therapy in clinical trials.5 Combining these and the fact that no studies reported the clinical significance of FGFR1 gene expression in ESCC, we focused on the role of FGFR1 gene expression in ESCC. Correlation analysis between FGFR1 gene expression, clinical characteristics, and survival data were conducted in an independent cohort of 145 ESCC patients. We also investigated the gene expression of FGFR1 and its function on cell apoptosis, cell proliferation in vivo and in vitro.
Results
Relationships between FGFR1 expression and patients' characteristics
One hundred and forty-five patients with surgically resected ESCC were enrolled. The majority of patients were male (91.0%), former or current smokers (71%), and with alcohol drinking history (60%). All patients received radical surgery, with evidence of pathologic stage II in 49.3% and stage III in 50.7%. Among 145 patients, 77 (53.1%) received chemotherapy and 85 (58.7%) received radiotherapy. The expression of FGFR1 was significantly correlated with alcohol drinking (P = 0.002) and radiotherapy (P = 0.038) (Table 1).
Table 1.
Association between FGFR1 m-RNA expression and clinicopathological data of patients with ESCC.
| All patients | High FGFR1 expression | Low FGFR1 expression | ||
|---|---|---|---|---|
| Characteristics | No. (%) | No. (%) | No. (%) | p-value |
| Age | 0.369 | |||
| <70 | 132(9.0) | 68(93.2) | 64(88.8) | |
| ≥70 | 13(91.0) | 5(6.8) | 8(11.2) | |
| Gender | 0.751 | |||
| Male | 132 (91.0) | 67 (91.8) | 65(90.3) | |
| Female | 13(9.0) | 6(9.2) | 7(9.3) | |
| BMI | 0.197 | |||
| Low | 20(17.4) | 7(11.7) | 13(26.0) | |
| Mid | 82(71.3) | 47(78.3) | 35(64.8) | |
| High | 13(11.4) | 6(10.0) | 6(9.2) | |
| Smoking | 0.675 | |||
| No | 34(29.0) | 19 (26.0) | 15 (20.8) | |
| Yes | 111 (71.0) | 54(74.0) | 57(79.2) | |
| Drinking | 0.002* | |||
| Constantly | 87(60.0) | 53(72.6) | 33(45.8) | |
| Never or seldom | 58(40.0) | 20(27.4) | 39(55.2) | |
| TNM stage | 0.455 | |||
| II | 72(49.3) | 34(46.6) | 38(52.7) | |
| III | 73(50.7) | 39(53.4) | 34(47.3) | |
| T stage | 0.637 | |||
| 2 | 31(21.4) | 17(23.2) | 14(19.4) | |
| 3 | 96(66.2) | 46(63.0) | 50(69.4) | |
| 4 | 17(11.7) | 10(13.7) | 8(11.2) | |
| N stage | 0.589 | |||
| 0 | 61(42.1) | 29(39.7) | 32(44.4) | |
| 1 | 44(30.3) | 25(34.2) | 19(26.4) | |
| 2 | 40(27.6) | 19(26.1) | 21(29.0) | |
| WHO classification | 0.381 | |||
| G3 | 42(29.0) | 22(30.1) | 20(27.7) | |
| G2 | 98(67.6) | 50(68.5) | 48(66.7) | |
| G1 | 5(3.4) | 1(1.4) | 4(5.6) | |
| Pathological type | 0.856 | |||
| Ulcerated | 90(63.8) | 43(58.9) | 47(65.3) | |
| Medullary | 28(20.0) | 15(20.5) | 13(18.1) | |
| Protruded | 17(12.1) | 9(12.3) | 8(11.1) | |
| Other | 6(4.1) | 6(8.2) | 4(5.5) | |
| Location | 0.937 | |||
| Upper | 8(5.5) | 3(4.3) | 5(6.8) | |
| Middle | 64(44.1) | 31(44.9) | 33(44.6) | |
| Under | 71(50.4) | 35(48.6) | 36(48.7) | |
| Radiotherapy | 0.038* | |||
| No | 60(41.3) | 31(42.4) | 43(59.7) | |
| Yes | 85(58.7) | 42(57.6) | 29(40.2) | |
| Chemotherapy | 0.450 | |||
| No | 68(46.9) | 37(50.6) | 31(43.0) | |
| Yes | 77(53.1) | 36(49.3) | 41(57.0) |
χ2-test: P < 0.05. BMI, body mass index; FGFR1, fibroblast growth factor receptor-1
Higher expression of FGFR1 predicts poor prognosis
Univariate analysis showed that only TNM stage (HR = 1.898, 95% [CI]: 1.271–2.836, P = 0.002), N stage (HR = 1.308, 95% [CI]: 1.049–1.632, p = 0.017), and FGFR1 (HR = 1.512, 95% [CI]: 1.014–2.256, P = 0.043) had statistically significant effects on OS (Table 2). Multivariate analysis indicated that FGFR1 and the TNM stage could be independent factors in predicting prognosis of ESCC patients in OS. Patients with higher FGFR1 expression had a significantly greater risk of death than those with lower FGFR1 gene expression after adjusting for the pathologic stage (OS: HR = 1.502, 95%[CI] = 1.005–2.246, P = 0.045). Advanced TNM stage (HR = 1.667, 95%[CI] = 1.005–2.767, P = 0.048) also predicted poor prognosis (Table 3). Consistent with OS analysis result, patients with advanced TNM stage (P = 0.002), advanced N stage (P = 0.026), or without radiotherapy (P = 0.017) had a shorter DFS (Table 2). DFS tend to be shorter in patients with higher FGFR1 expression but without statistical significant difference comparing to whom with lower FGFR1 expression (HR = 1.398, 95%[CI] = 0.942–2.074, P = 0.096) (Table 2).
Table 2.
Univariate analyses of OS and DFS in ESCC patients.
| OS |
DFS |
|||||
|---|---|---|---|---|---|---|
| Univariate analysis | HR | 95%CI | P | HR | 95%CI | P |
| Age (<70 / ≥70) | 1.36 | 0.71–2.26 | 0.35 | 1.297 | 0.675–2.493 | 0.435 |
| Gender (Male/Female) | 1.634 | 0.757–3.526 | 0.211 | 0.598 | 0.277–1.29 | 0.19 |
| BMI (Low, Middle/High) | 1.072 | 0.775–1.483 | 0.673 | 1.038 | 0.752–1.427 | 0.828 |
| Smoking (No/Yes) | 1.021 | 0.678–1.538 | 0.921 | 0.973 | 0.794–1.194 | 0.795 |
| Drinking (Constantly/Never, Seldom) | 1.299 | 0.820–2.059 | 0.265 | 1.354 | 0.855–2.142 | 0.196 |
| WHO Classification (G3/G2, G1) | 0.879 | 0.534–1.320 | 0.534 | 0.899 | 0.704–1.123 | 0.322 |
| TNM stage (III/II) | 1.898 | 1.271–2.836 | 0.002* | 1.884 | 1.267–2.801 | 0.002* |
| T stage (4/2,3) | 1.342 | 0.966–1.866 | 0.08 | 1.37 | 0.985–1.907 | 0.062 |
| N stage (2/1,0) | 1.308 | 1.049–1.632 | 0.017* | 1.288 | 1.031–1.609 | 0.026* |
| Chemotherapy (No/Yes) | 0.79 | 0.533–1.172 | 0.241 | 0.83 | 0.562–1.227 | 0.35 |
| Radiotherapy (No/Yes) | 1.454 | 0.979–2.159 | 0.064 | 0.621 | 0.42–0.919 | 0.017* |
| FGFR1 (High/Low) | 1.512 | 1.014–2.256 | 0.043* | 1.398 | 0.942–2.074 | 0.096 |
Statistical analysis was evaluated by a proportional hazard model (Cox).
P < 0.05. OS overall survival; DFS disease free survival; FGFR1 fibroblast growth factor receptor 1; BMI Body Mass Index
Table 3.
Multivariate analyses of OS and DFS in ESCC patients.
| OS |
DFS |
||||||
|---|---|---|---|---|---|---|---|
| Multivariate analysis | HR | 95%CI | P | HR | 95%CI | P | |
| TNM stage (III/II) | 1.667 | 1.005–2.767 | 0.048* | TNM stage (III/II) | 1.68 | 1.006–2.806 | 0.047* |
| N stage (2/1,0) | 0.451 | 0.836–1.496 | 0.451 | N stage (2/1,0) | 1.105 | 0.824–1.481 | 0.506 |
| FGFR1 (High/Low) | 1.502 | 1.005–2.246 | 0.045* | Radiotherapy | 0.614 | 0.413–0.913 | 0.016* |
Statistical analysis was evaluated by a proportional hazard model (Cox).
P < 0.05. OS overall survival; DFS disease free survival; FGFR1 fibroblast growth factor receptor 1
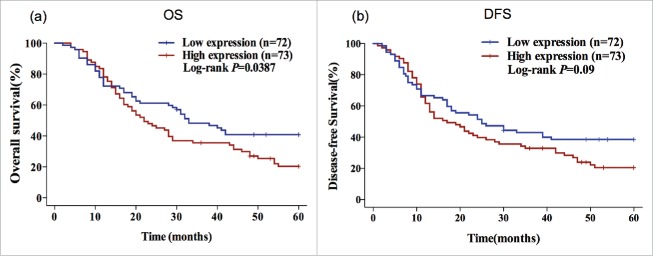
Kaplan-Meier survival curves also suggested that patients with higher FGFR1 gene expression had significantly shorter OS (OS: 22.00 vs 33.00 months; P = 0.0387) than those with lower FGFR1 gene expression. However, the analysis of DFS indicated there was no significant association between FGFR1 expression and DFS (DFS: 17.00 vs 25.00 months, P = 0.09) (Fig. 1).
Figure 1.
High gene expression of FGFR1 predicts poor OS in ESCC patients. Kaplan–Meier plots of the association of FGFR1 mRNA expression with OS (a) and DFS (b) in ESCC patients. P-values are verified by the log-rank test.
FGFR1 is over-expressed in ESCC cell lines
Comparative analysis of FGFR1 mRNA and protein expression in 8 ESCC cell lines and a normal human esophageal mucosa cell line-Het1A indicated that all ESCC cell lines over-expressed FGFR1, but at varying degrees (P<0.05) (Fig. 2a), corroborating the FGFR1 protein expression which was significantly higher in ESCC cells (Fig. 2b). Among the eight ESCC cell lines, TE-1 and EC9706 showed 4 to 5 fold-increasing expression of FGFR1 compared to Het1A and were therefore selected for further studies.
Figure 2.
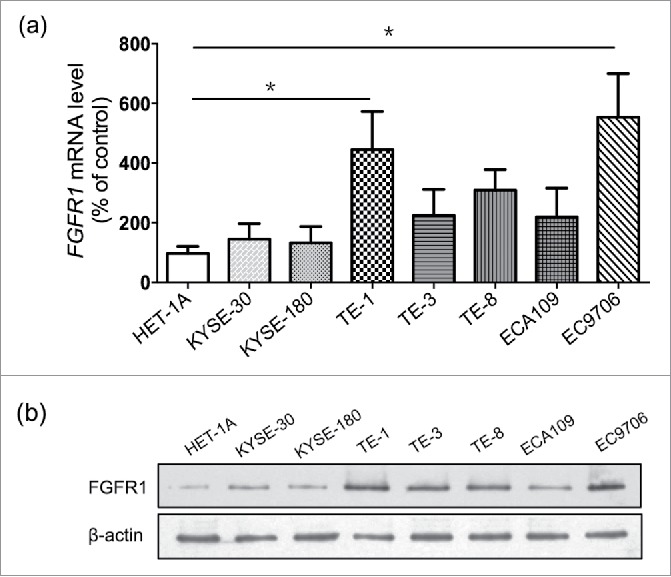
FGFR1 is over-expressed in ESCC cell lines. mRNA expression (a) and protein expression (b) of normal human esophageal mucosa cell line (Het-1A) and eight human esophageal carcinoma cell lines including KYSE-30, KYSE-180, ECA109, EC706 and TE series (TE-1, 3, 8). Data are presented as the mean ± SD; *P < 0.05 significantly different compared to each other.
Inhibition of FGFR1 effects the proliferation but not apoptosis of ESCC
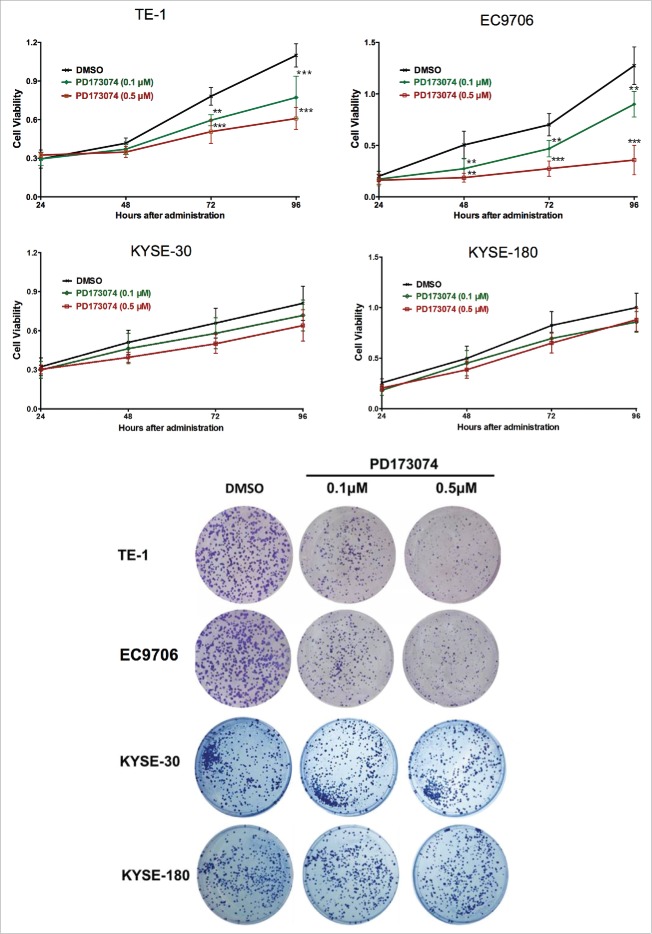
Observations made with clinical data prompted the exploration of the biological function of FGFR1 in the pathogenesis of ESCC. This was determined by inhibiting FGFR1 with PD173074, a selective inhibitor of FGFR1. As shown in Fig. 3a, measurement of cell viability indicated a significant difference in number of viable cells between PD173074 and DMSO treatment. PD173074 treatment decreased TE1 and EC9706 cell proliferation in a dose- and time-dependent manner with the strongest inhibitory effect on proliferation exhibited at 0.5 μM concentration and administrated for 72h (P<0.001). In regarding of the KYSE-30 and KYSE-180, the cells with lower FGFR1 expression, a tendency of cell growth inhibition were also observed but without significance.
Figure 3.
Anti-FGFR1 affects the proliferation of ESCC cells expressing high FGFR1. (a) Cell viability of TE-1, EC9706, KYSE-30 and KYSE-180 with treatment of PD173074 (0.1 μM, 0.5 μM) or DMSO detecting by MTT at 24, 48, 72, and 96 hours; (b) Representative images of colony formation assays of TE-1, EC9706, KYSE-30 and KYSE-180 with treatment of PD173074 (0.1 μM, 0.5 μM) or DMSO for 14 days. (Data are presented as the mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001 significantly different compared to control group.
To further explore the role of FGFR1 in ESCC cells viability, colony formation assay was performed. Compared to the vehicle treatment, inhibition of FGFR1 by PD173074 significantly decreased the number of colonies in a dose-dependent manner in TE-1 and EC9706 but not in KYSE-30 or KYSE-180 (Fig. 3b). Flow cytometry analysis showed that PD173074 had no significant effect on induction of ESCC apoptosis (Data not shown).
Inhibition of FGFR1 arrestes ESCC tumor growth in vivo
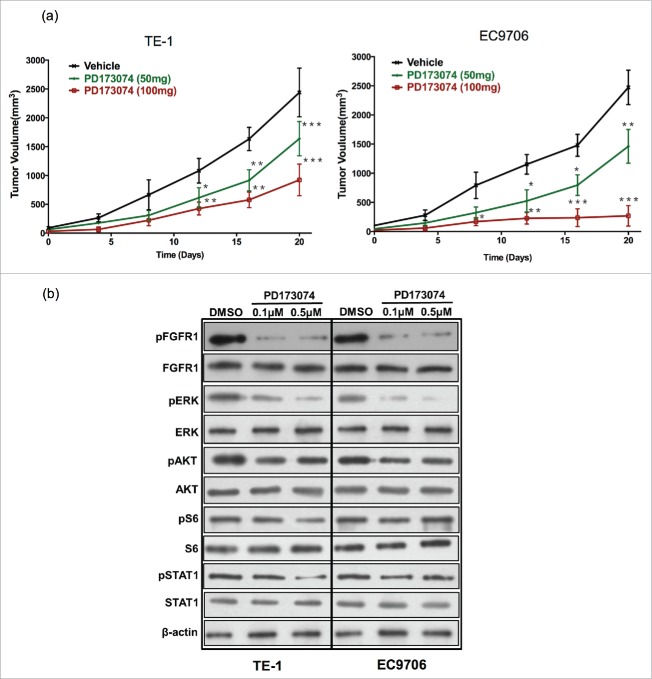
To examine the effect of inhibiting FGFR1 on ESCC growth in vivo, TE-1 and EC9706 tumor-bearing mice were treated with PD173074. Blocking FGFR1 signaling with PD173074 resulted in a significant inhibition of TE-1 and EC9706 tumor growth in a dose-dependent manner. A potent anti-tumor growth was observed both in 100mg/kg and 50mg/kg groups. TE-1 tumor volume decreased by approximately 80% and EC9706 tumor volume reduced by 90% at 20 days after PD173074 (100mg/kg) administration (P<0.001) (Fig. 4a).
Figure 4.
Anti-FGFR1 arrests ESCC tumor growth in vivo and attenuates the MAPK/ERK signaling pathway. (a) Antitumor effect of anti-FGFR1 treatment. Nude mice model established with TE-1 or EC9706 cells were treated orally with PD173074 at a dose of 50 or 100 mg/kg or with vehicle alone twice daily for 20 days. Data are presented as the mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001 significantly different compared to vehicle group. (b) The effect of anti-FGFR1 therapy on FGFR1, ERK, AKT, S6 and STAT1 phosphorylation detecting by western blotting.
Inhibition of FGFR1 blocks the MEK-ERK signaling pathway
As expected, PD173074 decreased the phosphorylation of FGFR1 in a dose-dependent manner in TE-1 and EC9706 cell lines (Fig. 4b). We then explored the effect of RAS, MEK-ERK, and PI3K-Akt pathway after inhibition of FGFR1. Western-blotting result showed that the phosphorylation of ERK was almost completely suppressed by inhibition of FGFR1 with PD173074. In TE-1, the phosphorylation of STAT1 and S6 were significant decreased with treatment of PD173074 in a higher dose. Similarly, we observed a lower expression of phosphate-STAT1 after inhibition of FGFR1 in EC9706. PD173074 also slightly suppressed the activation of AKT both in TE-1 and EC9706 without a dose-dependent manner (Fig. 4b).
Discussion
In this study, we investigated the prognostic role of FGFR1 gene expression in resected ESCC. Compared to FGFR1 gene over-expression, FGFR1 gene amplification was broadly reported in lung, head and neck, and breast cancers8,10,11 and as well in 6%–9.4% ESCC patients.12 However, the prognosis value of FGFR1 amplification in ESCC remains controversial.9,12 Though gene amplification seems would increases its expression, results have been inconclusive.13 One study suggested no correlation of FGFR1 amplification and mRNA expression in ESCC.14 Compared to gene amplification, there have been relatively fewer studies exploring the role of FGFR1 over-expression in the prognosis of cancer patients. FGFR1 protein expression was correlated with significantly worse OS in breast cancer.11,15,16 In lung cancer, FGFR1 protein expression was associated with increasing average micro-vessel density and predicted poor survival.1,18
Our data suggested that higher gene expression of FGFR1 could be an independent marker to predict poor prognosis of OS in resected ESCC. To our knowledge, this is the first report on the prognostic role of FGFR1 over-expression in the largest cohort of resected ESCC patients. Another study with detection of FGFR1 by IHC in 79 ESCC patients suggested a combined effect of FGF and FGFR1 but solely FGFR1 was not found to be an independent predictor.19 These contradictions might be due to smaller sample size or the difference between protein and mRNA expression. Given the fact that alcohol consumption is a common risk factor for ESCC development, it was not surprising that FGFR1 was higher in patients with history of alcohol use in our study. There was no significant effect of smoking on FGFR1 expression in our study, which contradicted previous studies showing that smoking increases FGFR1 gene expression in other SCC, like lung cancer.20 This might partly due to the heterogeneity of SCC type.
In vitro data demonstrated that FGFR1 was over-expressed in ESCC cell lines compared to a normal human esophageal mucosa cell line suggesting it key role in ESCC. Previous studies demonstrated that it was involved in the progression of various cancers.21 Upon binding to FGFR1, FGF triggers several signaling pathways. As predominant pathways, MEK-ERK and PI3K–Akt are central to regulate cell growth and proliferation. Activation of FGFR1 induces cellular proliferation and survival in acute myeloid leukemia22 and bladder cancer.23 FGFR1 activation stimulates the STAT pathway directly or indirectly through JAKs.3 We report that blocking FGFR1 inhibited the proliferation of ESCC cells both in vivo and in vitro and partially maybe due to the inhibition of the MEK-ERK pathways, which were inhibited mostly comparing to PI3K–Akt or STAT1 signaling network. This suggesting MEK-ERK might comprise the major signaling pathway mediated by FGFR1 in ESCC progression (Fig. 5). Published studies also revealed the role of FGF1-FGFR1 axis in modulating the epithelial-mesenchymal transition (EMT) pathway to induce metastasis of various carcinomas.24–26 Moreover, FGFR1 affect the maintenance of cancer ‘stemness’ through the regulation of GLI2 expression via the ERK pathway, to promote the stem cell-like phenotype of lung SCC.27 In breast cancer, FGF/FGFR also plays a role in the paracrine signaling pathway activated by estrogen to expand stem-like cells.28
Figure 5.
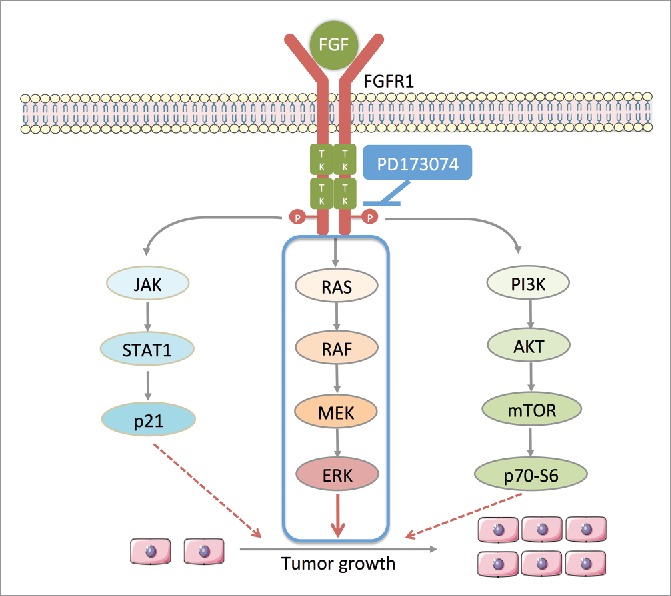
PD173074 inhibits the signal pathway downstream of FGFR1 in ESCC. Ligand (eg. FGF) binds to FGFR1 and trigger the phosphorylation of FGFR1 tyrosine kinase domains leading the activation of three main downstream pathways: 1) RAS-RAF-MAPK-ERK; 2) PI3K-AKT-mTOR-p70-S6; 3) JAK-STAT-p21; FGFR1 selective inhibitor PD173034 inhibits the phosphorylation of its tyrosine kinase domains thus inhibiting downstream signaling pathway.
PD173074 is a potent ATP-competitive inhibitor of FGFR1 with high affinity and selectivity. This inhibitor evokes anti-tumor immune response and impairs the progression of breast cancer by decreasing lung metastasis.29 Consistent with the finding that FGFR1 induces EMT, PD173074 induces the reverse process of EMT, Mesenchymal–epithelial transition (MET), by inhibiting the MAPK pathway.30 Moreover, in ESCC, PD173074 also eliminated the rescue effect induced by FGFR pathway against lapatinib treatment.31 PD173074 also reduced the expression of p-ERK and Bcl-xl to increase the anti-proliferative and apoptosis-inducing effects of 5-fluorouracil in gastric cancer.32 Our results demonstrated that FGFR1 inhibition had a strong cytotoxic effect on the ESCC cells with higher FGFR1 gene expression but not in the cells with relative lower gene expression. This suggesting the ESCC growth inhibition effect of anti-FGFR1 treatment partly relies on the FGFR1 gene expression. Moreover, we did not find significant pro-apoptotic role of PD173074 in this study, which might be attributed to the variation in cell lines characteristics or that the anti-proliferation of PD173074 might rely on other mechanisms but not the induction of apoptosis, as seen in other studies.
In view of its key role in cancer, anti-FGFR therapy has been considered as a potential treatment strategy. Although no clinical trial about PD123074 have been conducted yet. Several other small molecule inhibitors that target FGFR have been successfully applied in clinical trials with significant anti-tumor efficacy (Table 4).33–37 AZD4547, an inhibitor of FGFR1, 2, and 3, reported with high anti-tumor activity in FGFR2 amplified gastric cancer and lower activity in FGFR1 amplified breast cancer which suggested that efficacy of anti-FGFR varies in different types of cancer and genomic alternation. By now, most trials recruit patients with FGFR genetic alternation, including amplification, mutation and translocation. However, it remains a challenge to identify biomarkers that serve as good predictors of FGFR-targeted therapy response. Growing evidence suggests the emerging role of FGFR1 RNA expression in predicting treatment outcomes. One study showed that FGFR1 mRNA expression, but not gene copy number, predicts FGFR TKI sensitivity in all lung cancer cell lines.38 Similarly, in head and neck SCC patients, compared to copy-number gain, FGFR1 mRNA expression was better at predicting the response to FGFR inhibitors.39 Also, one ongoing clinical trial enrolled advanced solid tumors patients with high FGFR expression but not other genomic alternation (NCT02592785) (Table 4).
Table 4.
Summary of Clinical trials of FGFR inhibitors.
| Agent | Target | Phase | Enrolled patients | Treatment arms | Results | NCT number | Reference |
|---|---|---|---|---|---|---|---|
| AZD4547 | FGFR1/2/3 | I | Solid tumor with FGFR1 or FGFR2 gene amplification | Monotherapy/Single group | Safe and safe and with objective clinical response | NCT00979134 | 37 |
| II | Gastric, Esophageal, Breast, and lung SCC with FGFR1/2 amplification | Monotherapy/Single group | High anti-tumor activity in FGFR2 amplified gastric cancer and lower activity in FGFR1 amplified breast cancer | NCT01795768 | 36 | ||
| I/II | Advanced NSCLC | Docetaxel alone VS Docetaxel with concomitant AZD4547 | N/A | NCT01824901 | |||
| I/II | ER+ Breast Cancer patients progressed after NSAIs | AZD4547 with Anastrozole or Letrozole | N/A | NCT01791985 | |||
| II | ER+ Breast Cancer with FGFR1 polysomy or amplification | AZD4547 with Fulvestrant vs. Fulvestrant Alone | N/A | NCT01202591 | |||
| II/III | Lung SCC positive for FGFR1/2/3 | AZD4547 vs Docetaxel | N/A | NCT02154490* | |||
| II | Advanced cancer with FGFR1/2/3 mutation or translocation | Monotherapy/Single group | N/A | NCT02465060* | |||
| BGJ398 | FGFR1/2/3 | I | Advanced solid tumors with alterations of FGFR1/2/3 | Monotherapy/Single group | Safe and with objective clinical response | NCT01004224 | 33 |
| II | Recurrent Glioblastoma with amplification or translocation of FGFR1-TACC1, FGFR3-TACC-3 fusion and/or activating mutation in FGFR1/2/3 | Monotherapy/Single group | N/A | NCT01975701 | |||
| II | Head and Neck Cancer Patients with FGFR1/2/3 translocation, mutation, or amplification | Monotherapy/Single group | N/A | NCT02706691 | |||
| ASP5878 | FGFR1/2/3/4 | I | Advanced solid Tumors | Monotherapy/Single group | N/A | NCT02038673 | |
| ARQ087 | FGFR1/2/3 | I/II | Advanced solid Tumors with FGFR genetic alterations | Monotherapy/Single group | Safe and with objective clinical response | NCT01752920 | 35 |
| BAY1163877 | FGFR1/2/3/4 | I | Advanced solid tumors with high FGFR expression | Monotherapy/Single group | N/A | NCT02592785 | |
| Debio1347 | FGFR1/2/3 | I/II | Advanced solid tumor with FGFR1/2/3 alteration | Monotherapy/Single group | N/A | NCT01948297 | |
| LY2874455 | FGFR1/2/3/4 | I | Advanced cancers | Monotherapy/Single group | N/A | NCT01212107 | |
| INCB054828 | FGFR1/2/3 | II | Myeloid/lymphoid neoplasms with FGFR1 rearrangement | Monotherapy/Single group | N/A | NCT03011372 | |
| JNJ-42756493 | Pan-FGFR | I/II/III | Advanced solid tumors with FGFR alterations | Monotherapy/Single group | Phase I: Safe and with objective clinical response | NCT01703481* | 34 |
| PRN1371 | FGFR1/2/3/4 | I | Advanced solid tumors, expansion cohort in patients with FGFR 1/2/3/4 genetic alterations | Monotherapy/Single group | N/A | NCT02608125 | |
| TAS-120 | FGFR1/2/3/4 | I/II | Advanced solid tumors | Monotherapy/Single group | N/A | NCT02052778 |
Multiple arms include anti-FGFR treatment cohort; NSCLC, non-small cell lung cancer; SCC, squamous cell carcinoma; FGFR, fibroblast growth factor receptor
In summary, we have determined a role of FGFR1 in ESCC. We found that anti-FGFR1 potentially inhibits the MEK-ERK downstream pathways, thereby decreasing the proliferation of ESCC both in vitro and in vivo. These findings have implications for designing therapeutic strategies and for understanding the pathogenesis of ESCC mediated by FGFR1.
Materials and methods
Patients and samples
One hundred and forty-five patients with ESCC who underwent surgical resection at West China Hospital from Jan. 2009 to Dec. 2010 were enrolled. The patient inclusion criteria were: 1) availability of tumor tissue from primary cancer, clinical characteristics and survival data; 2) patients with stage II and III disease; 3) patients who had not received neo-adjuvant chemotherapy or radiotherapy. This study was approved by West China Hospital and informed consent was obtained from patients.
Histopathological classification and tumor stage were defined according to 2004 WHO classification system and UICC/AJCC (7th edition) for esophageal carcinoma. Clinicopathological data were obtained from patient medical records. Disease-free survival (DFS) and overall survival (OS) were calculated as the number of months from the date of surgery to the date of the initial tumor relapse and the date of death, respectively.
mRNA analysis of FGFR-1 expression
mRNA extraction procedures from cells or formalin-fixed paraffin-embedded tissue samples were conducted according to standard manufacturer's protocols (TaKaRa, Liaoning, China). mRNA expression of FGFR-1 was detected and quantified using SYBR-Green assay (Biorad, CA, USA). FGFR1 primers: sense, 5′-CTTCGTTTCTTGTTGGTATGC-3′, antisense, 5′-GGACAGGATGGAGTTTGGAC-3′, GAPDH primers: sense, 5′-ACTCCTCCACCTTTGACGCTG-3′, antisense, 5′-CTCTCTTCCTCTTGTGCTCTTGC-3′. The median mRNA expression was used as the threshold to divide these patients into high and low FGFR1 expression groups.
Cell lines and reagent
Normal human esophageal mucosa cell line (Het-1A) and eight human esophageal carcinoma cell lines including KYSE-30, KYSE-180, ECA109, EC706 and TE series (TE1, 3, 8) were selected. All cell lines were obtained as a gift from State Key Laboratory of Biotherapy (Chengdu, China); Cells were cultured in RPMI-1640 or DMEM medium (10% fetal calf serum and 1% antibiotics (penicillin and streptomycin) at 37°C with 5% CO2 and 95% humidity.
PD173074 (Selleck Chemicals, Houston, TX, USA) was dissolved in dimethyl sulfoxide (DMSO) and stored at −80°C until use. DMSO (0.1%) served as a vehicle control.
Western blotting
Total protein from cells were extracted using RIPA solution (Beyotime Biotech, Hangzhou, China) with Protease and Phosphatase Inhibitor Cocktails (Sigma, MO, USA) and resolved on 10–15% SDS-PAGE gel, followed by transfer to polyvinylidene fluoride membrane (Millipore, Massachusetts, USA). After blocking, membranes were incubated overnight at 4°C with primary antibodies specific for anti-pFGFR1 (1:500), anti-FGFR1 (1:1000), anti-pERK (1:500), anti-ERK (1:1000), anti-AKT (1:1000), anti-pAKT (1:500), anti-pS6 (1:500), anti-S6 (1:100), anti-pSTAT1 (1:500), anti-STAT1 (1:1000), and anti-β Actin (1:5000). All antibodies were purchased from (Santa Cruz, Dallas, USA). Immunoreactivity was detected with an enhanced chemiluminescence kit (Millipore, Massachusetts, USA).
Colony formation assay
Cells were seeded in triplicate 6-well plates at approximately 800 cells/well. After 24 h of incubation, cells were treated with 0.1, 0.5 μM PD173074 or DMSO and then cultured for 14 days. The colonies were fixed by 4% paraformaldehyde solution and stained with crystal violet. Colonies with >50 cells were scored.
Methylthiazoletetrazolium (MTT) assay
Cell viability was determined using an MTT assay according to the manufacturer's protocol. ESCC cells were seeded in 96-well plates at a density of 8,000 cells/well and treated with 0.1, 0.5 μM PD173074 or DMSO. After 48 h, 72h and 96h of administration, MTT at a concentration of 5 mg/ml was added to each well and incubated for an additional 4 h at 37°C. Absorbance was measured at 490 nm after adding 150 μl DMSO to each well.
Analysis of apoptosis by flow cytometry
After incubation with 0.1, 0.5 μM PD173074 or DMSO for 48 h, 72h and 96h, cells were harvested and washed with PBS. 106 cells/ml cells were double-labeled with Annexin V–fluorescein isothiocyanate (FITC) and Propidium iodide (PI). Fluorescence was analyzed using flow cytometry (FACS Calibur, BD Biosciences, CA).
Mouse model
Four-week-old female nude mice purchased from the Experimental Animal Center, Chinese Academy of Medical Science were used. The Animal Care and Use Committee of Sichuan University approved this study. Mice were randomly divided into three groups with 6 mice per group. Tumor burden was established with 5 × 106 TE-1 or EC9706 cells that injected subcutaneously in the right flank. Then the mice were treated with PD173074 (50mg/kg or 100mg/kg) or vehicle oral gavage, twice daily for 20 days. Tumor volumes were assessed every 4 days and calculated according to the formula: Tumor volume (mm3) = 0.52 × a × b2 (a represents the longer diameter and b represents the shorter diameter).
Statistical analyses
Fisher's exact test and the Chi-square test were applied to compare the clinical and demographic characteristics of patients in high or lower FGFR1 expression groups. DFS and OS were estimated using the Kaplan–Meier method. Survival data were censored at the time of the last visit for patients who were still alive at the final analysis. The log-rank test was applied to compare DFS and OS in two groups. Multivariate analysis was performed using the Cox proportional hazard regression model. Statistical tests were based on a two-sided significance level of 0.05. All statistical analyses were conducted using SPSS 17.0.0 for Windows (IBM Corp, NY, USA).
Ethics approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All procedures performed in studies involving animals were in accordance with the ethical standards of the West China Hospital, Sichuan University.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Funding Statement
National science foundation of China. [grant number. 81472808].
Conflict of interests
The authors declare no conflicts of interest.
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 81472808), International Visiting Program for Excellent Young Scholars of Sichuan University and the Fundamental Research Funds for the Central Universities (No.2017SCU11039).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. PMID:25651787 [DOI] [PubMed] [Google Scholar]
- 2.Digklia A, Voutsadakis IA. Targeted treatments for metastatic esophageal squamous cell cancer. World J Gastrointest Oncol. 2013;5:88–96. doi: 10.4251/wjgo.v5.i5.88. PMID:23799158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–29. doi: 10.1038/nrc2780. PMID:20094046 [DOI] [PubMed] [Google Scholar]
- 4.Grose R, Dickson C. Fibroblast growth factor signaling in tumorigenesis. Cytokine Growth Factor Rev. 2005;16:179–86. doi: 10.1016/j.cytogfr.2005.01.003. PMID:15863033 [DOI] [PubMed] [Google Scholar]
- 5.Weeden CE, Solomon B, Asselin-Labat ML. FGFR1 inhibition in lung squamous cell carcinoma: questions and controversies. Cell Death Discov. 2015;1:15049. doi: 10.1038/cddiscovery.2015.49. PMID:27551478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie FJ, Lu HY, Zheng QQ, Qin J, Gao Y, Zhang YP, Hu X, Mao WM. The clinical pathological characteristics and prognosis of FGFR1 gene amplification in non-small-cell lung cancer: a meta-analysis. Onco Targets Ther. 2016;9:171–81. doi: 10.2147/OTT.S91848. PMID:26793001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Gao W, Xu J, Chen X, Yang Y, Zhu Y, Yin Y, Guo R, Liu P, Shu Y, et al.. The role of FGFR1 gene amplification as a poor prognostic factor in squamous cell lung cancer: a meta-analysis of published data. Biomed Res Int. 2015;2015:763080. doi: 10.1155/2015/763080. PMID:26788508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heist RS, Mino-Kenudson M, Sequist LV, Tammireddy S, Morrissey L, Christiani DC, Engelman JA, Iafrate AJ. FGFR1 amplification in squamous cell carcinoma of the lung. J Thorac Oncol. 2012;7:1775–80. doi: 10.1097/JTO.0b013e31826aed28. PMID:23154548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon D, Yun JY, Keam B, Kim YT, Jeon YK. Prognostic implications of FGFR1 and MYC status in esophageal squamous cell carcinoma. World J Gastroenterol. 2016;22:9803–12. doi: 10.3748/wjg.v22.i44.9803. PMID:27956804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ach T, Schwarz-Furlan S, Ach S, Agaimy A, Gerken M, Rohrmeier C, Zenk J, Iro H, Brockhoff G, Ettl T. Genomic aberrations of MDM2, MDM4, FGFR1 and FGFR3 are associated with poor outcome in patients with salivary gland cancer. J Oral Pathol Med. 2016;45:500–9. doi: 10.1111/jop.12394. PMID:26661925 [DOI] [PubMed] [Google Scholar]
- 11.Cheng CL, Thike AA, Tan SY, Chua PJ, Bay BH, Tan PH. Expression of FGFR1 is an independent prognostic factor in triple-negative breast cancer. Breast Cancer Res Treat. 2015;151:99–111. doi: 10.1007/s10549-015-3371-x. PMID:25868865 [DOI] [PubMed] [Google Scholar]
- 12.Kim HS, Lee SE, Bae YS, Kim DJ, Lee CG, Hur J, Chung H, Park JC, Jung DH, Shin SK, et al.. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival in patients with resected esophageal squamous cell carcinoma. Oncotarget. 2015;6:2562–72. doi: 10.18632/oncotarget.2944. PMID:25537505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong AJ, Bigner SH, Bigner DD, Kinzler KW, Hamilton SR, Vogelstein B. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci U S A. 1987;84:6899–903. doi: 10.1073/pnas.84.19.6899. PMID:3477813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Loga K, Kohlhaussen J, Burkhardt L, Simon R, Steurer S, Burdak-Rothkamm S, Jacobsen F, Sauter G, Krech T. FGFR1 amplification is often homogeneous and strongly linked to the squamous cell carcinoma subtype in esophageal carcinoma. PLoS One. 2015;10:e0141867. doi: 10.1371/journal.pone.0141867. PMID:26555375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi YJ, Tsang JY, Ni YB, Chan SK, Chan KF, Tse GM. FGFR1 is an adverse outcome indicator for luminal A breast cancers. Oncotarget. 2016;7:5063–73. doi: 10.18632/oncotarget.6563. PMID:26673008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomiguchi M, Yamamoto Y, Yamamoto-Ibusuki M, Goto-Yamaguchi L, Fujiki Y, Fujiwara S, Sueta A, Hayashi M, Takeshita T, Inao T, et al.. Fibroblast growth factor receptor-1 protein expression is associated with prognosis in estrogen receptor-positive/human epidermal growth factor receptor-2-negative primary breast cancer. Cancer Sci. 2016;107:491–8. doi: 10.1111/cas.12897. PMID:26801869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pu D, Liu J, Li Z, Zhu J, Hou M. Fibroblast Growth Factor Receptor 1 (FGFR1), partly related to Vascular Endothelial Growth Factor Receptor 2 (VEGFR2) and microvessel density, is an independent prognostic factor for non-small cell lung cancer. Med Sci Monit. 2017;23:247–57. doi: 10.12659/MSM.899005. PMID:28088809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang F, Gao Y, Geng J, Qu D, Han Q, Qi J, Chen G. Elevated expression of SOX2 and FGFR1 in correlation with poor prognosis in patients with small cell lung cancer. Int J Clin Exp Pathol. 2013;6:2846–54. PMID:24294370 [PMC free article] [PubMed] [Google Scholar]
- 19.Sugiura K, Ozawa S, Kitagawa Y, Ueda M, Kitajima M. Co-expression of aFGF and FGFR-1 is predictive of a poor prognosis in patients with esophageal squamous cell carcinoma. Oncol Rep. 2007;17:557–64. PMID:17273733 [PubMed] [Google Scholar]
- 20.Kim HR, Kim DJ, Kang DR, Lee JG, Lim SM, Lee CY, Rha SY, Bae MK, Lee YJ, Kim SH, et al.. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival and cigarette smoking dosage in patients with resected squamous cell lung cancer. J Clin Oncol. 2013;31:731–7. doi: 10.1200/JCO.2012.43.8622. PMID:23182986 [DOI] [PubMed] [Google Scholar]
- 21.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–49. doi: 10.1016/j.cytogfr.2005.01.001. PMID:15863030 [DOI] [PubMed] [Google Scholar]
- 22.Karajannis MA, Vincent L, Direnzo R, Shmelkov SV, Zhang F, Feldman EJ, Bohlen P, Zhu Z, Sun H, Kussie P, et al.. Activation of FGFR1beta signaling pathway promotes survival, migration and resistance to chemotherapy in acute myeloid leukemia cells. Leukemia. 2006;20:979–86. doi: 10.1038/sj.leu.2404203. PMID:16598308 [DOI] [PubMed] [Google Scholar]
- 23.Tomlinson DC, Lamont FR, Shnyder SD, Knowles MA. Fibroblast growth factor receptor 1 promotes proliferation and survival via activation of the mitogen-activated protein kinase pathway in bladder cancer. Cancer Res. 2009;69:4613–20. doi: 10.1158/0008-5472.CAN-08-2816. PMID:19458078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiao J, Zhao X, Liang Y, Tang D, Pan C. FGF1-FGFR1 axis promotes tongue squamous cell carcinoma (TSCC) metastasis through epithelial-mesenchymal transition (EMT). Biochem Biophys Res Commun. 2015;466:327–32. doi: 10.1016/j.bbrc.2015.09.021. PMID:26362179 [DOI] [PubMed] [Google Scholar]
- 25.Tomlinson DC, Baxter EW, Loadman PM, Hull MA, Knowles MA. FGFR1-induced epithelial to mesenchymal transition through MAPK/PLCgamma/COX-2-mediated mechanisms. PLoS One. 2012;7:e38972. doi: 10.1371/journal.pone.0038972. PMID:22701738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Acevedo VD, Gangula RD, Freeman KW, Li R, Zhang Y, Wang F, Ayala GE, Peterson LE, Ittmann M, Spencer DM. Inducible FGFR-1 activation leads to irreversible prostate adenocarcinoma and an epithelial-to-mesenchymal transition. Cancer Cell. 2007;12:559–71. doi: 10.1016/j.ccr.2007.11.004. PMID:18068632 [DOI] [PubMed] [Google Scholar]
- 27.Ji W, Yu Y, Li Z, Wang G, Li F, Xia W, Lu S. FGFR1 promotes the stem cell-like phenotype of FGFR1-amplified non-small cell lung cancer cells through the Hedgehog pathway. Oncotarget. 2016;7:15118–34. doi: 10.18632/oncotarget.7701. PMID:26936993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fillmore CM, Gupta PB, Rudnick JA, Caballero S, Keller PJ, Lander ES, Kuperwasser C. Estrogen expands breast cancer stem-like cells through paracrine FGF/Tbx3 signaling. Proc Natl Acad Sci U S A. 2010;107:21737–42. PMID:21098263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye T, Wei X, Yin T, Xia Y, Li D, Shao B, Song X, He S, Luo M, Gao X, et al.. Inhibition of FGFR signaling by PD173074 improves antitumor immunity and impairs breast cancer metastasis. Breast Cancer Res Treat. 2014;143:435–46. PMID:24398778 [DOI] [PubMed] [Google Scholar]
- 30.Nguyen PT, Tsunematsu T, Yanagisawa S, Kudo Y, Miyauchi M, Kamata N, Takata T. The FGFR1 inhibitor PD173074 induces mesenchymal-epithelial transition through the transcription factor AP-1. Br J Cancer. 2013;109:2248–58. PMID:24045665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saito S, Morishima K, Ui T, Hoshino H, Matsubara D, Ishikawa S, Aburatani H, Fukayama M, Hosoya Y, Sata N, et al.. The role of HGF/MET and FGF/FGFR in fibroblast-derived growth stimulation and lapatinib-resistance of esophageal squamous cell carcinoma. BMC Cancer. 2015;15:82. PMID:25884729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye YW, Hu S, Shi YQ, Zhang XF, Zhou Y, Zhao CL, Wang GJ, Wen JG, Zong H. Combination of the FGFR4 inhibitor PD173074 and 5-fluorouracil reduces proliferation and promotes apoptosis in gastric cancer. Oncol Rep. 2013;30:2777–84. PMID:24126887 [DOI] [PubMed] [Google Scholar]
- 33.Nogova L, Sequist LV, Perez Garcia JM, Andre F, Delord JP, Hidalgo M, Schellens JH, Cassier PA, Camidge DR, Schuler M, et al.. Evaluation of BGJ398, a fibroblast growth factor receptor 1–3 Kinase inhibitor, in patients with advanced solid tumors harboring genetic alterations in fibroblast growth factor receptors: results of a global Phase I, dose-escalation and dose-expansion study. J Clin Oncol. 2017;35:157–65. PMID:27870574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tabernero J, Bahleda R, Dienstmann R, Infante JR, Mita A, Italiano A, Calvo E, Moreno V, Adamo B, Gazzah A, et al.. Phase I Dose-escalation study of JNJ-42756493, an oral pan-fibroblast growth factor receptor inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2015;33:3401–8. PMID:26324363 [DOI] [PubMed] [Google Scholar]
- 35.Papadopoulos KP, Tolcher AW, Patnaik A, Rasco DW, Chambers G, Beeram M, et al.. Phase 1, first-in-human study of ARQ 087, an oral pan-Fibroblast Growth Factor Receptor (FGFR) inhibitor, in patients (pts) with advanced solid tumors. J Clin Oncol. 2017;35:4017–4017. doi: 10.1200/JCO.2017.35.15_suppl.4017. [DOI] [Google Scholar]
- 36.Smyth EC, Turner NC, Peckitt C, Pearson A, Brown G, Chua S, et al.. Phase II multicenter proof of concept study of AZD4547 in FGFR amplified tumours. J Clin Oncol. 2015;33:2508–2508. doi: 10.1200/jco.2015.33.15_suppl.2508. [DOI] [Google Scholar]
- 37.Andre F, Ranson M, Dean E, Varga A, Van der Noll R, Stockman PK, et al.. Abstract LB-145: Results of a phase I study of AZD4547, an inhibitor of fibroblast growth factor receptor (FGFR), in patients with advanced solid tumors. Cancer Res. 2013;73(8 Suppl):Abstract nr LB-145. doi: 10.1158/1538-7445.AM2013-LB-145. [DOI] [Google Scholar]
- 38.Wynes MW, Hinz TK, Gao D, Martini M, Marek LA, Ware KE, Edwards MG, Böhm D, Perner S, Helfrich BA, et al.. FGFR1 mRNA and protein expression, not gene copy number, predict FGFR TKI sensitivity across all lung cancer histologies. Clin Cancer Res. 2014;20:3299–309. doi: 10.1158/1078-0432.CCR-13-3060. PMID:24771645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goke F, Franzen A, Hinz TK, Marek LA, Yoon P, Sharma R, Bode M, von Maessenhausen A, Lankat-Buttgereit B, Göke A, et al.. FGFR1 expression levels predict BGJ398 sensitivity of FGFR1-dependent head and neck squamous cell cancers. Clin Cancer Res. 2015;21:4356–64. doi: 10.1158/1078-0432.CCR-14-3357. PMID:26015511 [DOI] [PMC free article] [PubMed] [Google Scholar]