ABSTRACT
Recent studies have shown that long non-coding RNAs (lncRNAs) are involved in a number of biological processes; however, further study is still warranted to comprehensively reveal their functions. In this study, we showed that the lncRNA in non-homologous end joining (NHEJ) pathway 1 (LINP1) was related to breast cancer cell proliferation, metastasis and chemoresistance. Loss- and gain-of function studies were used to assess the role of LINP1 in promoting breast cancer progression. LINP1 knockdown mitigated breast cancer cell growth by inducing G1-phase cell cycle arrest and apoptosis. LINP1 also promoted breast cancer cell metastasis and influenced the expression of epithelial-mesenchymal transition-related markers. We identified p53 as a regulator of LINP1, and LINP1 overexpression could restore the metastatic effects of p53. Furthermore, LINP1 was upregulated in doxorubicin- and 5-fluorouracil-resistant cells and induced chemoresistance. We also observed that LINP1 enrichment played a critical functional role in chemoresistance by inhibiting chemotherapeutics-induced apoptosis. Moreover, LINP1 in tumors was associated with lower overall survival and disease-free survival. In conclusion, LINP1 may serve as a potential oncogene and chemoresistance-related regulator of breast cancer cells, suggesting that LINP1 might be a potent therapeutic target and might reduce chemoresistance in breast cancer.
KEYWORDS: LINP1, oncogene, chemoresistance, breast cancer
Introduction
Breast cancer is the most common cancer among women and is the fifth most common cause of cancer death in the world.1 Statistical data from the ACS showed that, more than 1,676 million new cases of breast cancer and 521,900 breast cancer deaths have occurred worldwide. Breast cancer is a heterogeneous carcinoma, with both genetic and epigenetic changes contributing to breast cancer initiation, development and metastasis. Even though many researchers have struggled to clarify the mechanisms of tumor carcinogenesis and development, further understanding is warranted to provide novel key molecules for early diagnosis and prognostic predications.
Long non-coding RNAs (lncRNA) are a novel class of RNA transcripts that contain more than 200 nucleotides in length and cannot be encoded into proteins.2 Recent studies have shown that lncRNAs are involved in various biological processes, such as cell proliferation, apoptosis, metabolism, gene expression, and the regulation of epigenetic signatures.3 More importantly, aberrant lncRNA expression may be involved in inducing or inhibiting cancer development and progression, metastasis, and chemoresistance.4–7 LncRNA MEG3 functions as a tumor suppressor in a variety of cancers, including osteosarcoma,8 NSCLC,9 cervical cancer10 and breast cancer.11 LncRNAs are also known to play oncogenic roles. Sun M et al found that linc00152 was overexpressed in breast cancer and participated in controlling cell proliferation and cell-cycle gene expression.12 LncRNA NKILA could interact with the NF-kB/IkB complex and was associated with metastasis and breast cancer prognosis.13 The lncRNA Malat1 inhibits tumor growth and metastasis, and can be a treatment target for breast cancer.14 Although lncRNAs were found to be novel targets for cancer treatment, more studies are needed to clarify the lncRNA regulation mechanisms.
The current clinical treatment options for patients with breast cancer include surgery and subsequent chemotherapy, targeted therapy or radiotherapy.15 Although the combination of surgery and drug therapy can effectively prevent recurrence and metastasis and improve the prognosis of patients with breast cancer, the frequent development of drug resistance is still a major obstacle in the treatment of breast cancer.16 However, the underlying mechanism of chemoresistance remains to be fully elucidated. Therefore, there is an urgent need to develop new therapeutic strategies and targets to enhance the effectiveness of chemotherapy and reduce the rate of resistance.
In the present study, we uncovered that LINP1 mediates its oncogenic role in breast cancer by suppressing cell growth and metastasis. Additionally, LINP1 overexpression was found in 5-FU- and doxorubicin (DOX)- resistant breast cancer cell lines and was positively associated with chemoresistance. Finally, our studies indicated that upregulated LINP1 expression in breast cancer was associated with an unfavorable prognosis. Our study revealed a new role for LINP1 as an oncogene, and helped to elucidate the epigenetic mechanism of chemoresistance, which may facilitate the development of effective clinical treatments for breast cancer.
Results
LINP1 promoted proliferation via inhibiting apoptosis in breast cancer cells
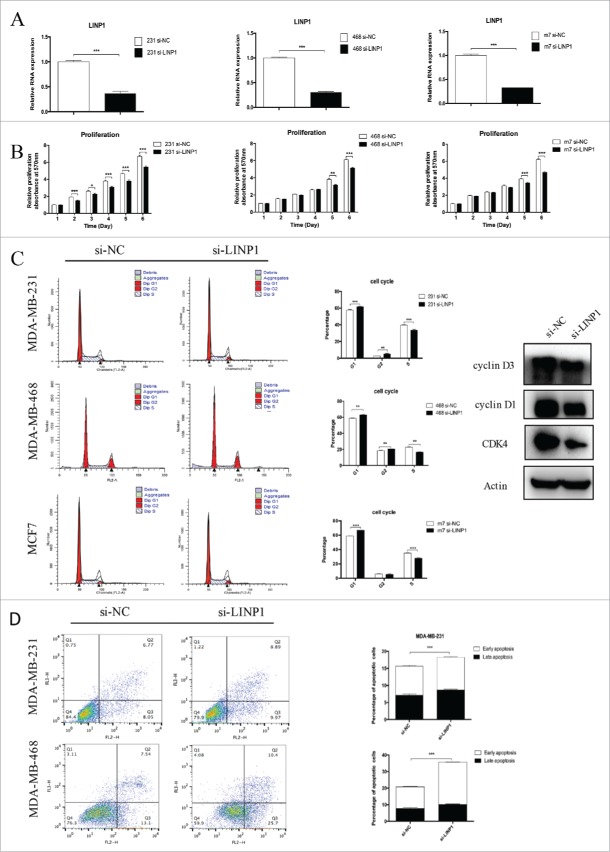
To investigate the biological functions of LINP1 in breast cancer cells, cells were transfected with si-NC or si-LINP1 and the knockdown efficiency was confirmed by qRT-PCR (Figure 1A). MTT assays showed that significant reduction of cell numbers was observed in LINP1 knockdown cells in comparison with control cells (Figure 1B). In contrast, a promotion of proliferation was observed in the LINP1-overexpressed cells (Supplementary Figure 1A). Furthermore, flow cytometry was performed to evaluate the effect of LINP1 on cell proliferation. The results showed that inhibition of LINP1 expression mainly increased the G1 phase population and decreased the S phase population in the three breast cancer cell lines (Figure 1C). Western blot was then performed to prove that LINP1 knockdown could decrease the expression of cyclin D1, cyclin D3, and CDK4 (Figure 1C). We next examined the effects of LINP1 on the apoptosis of breast cancer cells transfected with siRNAs by flow cytometry. The results showed that LINP1 knockdown increased the total apoptosis rate of MDA-MB-231 cells from 14.82% to 18.86% compared to scrambled siRNAs (Figure 1D). In the MDA-MB-468 cells, the apoptosis rate increased from 20.64% to 36.1% (Figure 1D). Consistently, LINP1 overexpression inhibited breast cancer cell apoptosis (Supplementary Figure 1B). These results suggest that LINP1 could play important roles in promoting cell proliferation as an oncogene in breast cancer cells.
Figure 1.
LINP1 knockdown inhibited proliferation and induced apoptosis in breast cancer cells. (A) MDA-MB-231 (right), MDA-MB-468 (middle) and MCF-7 (left) cells were transfected with NC (negative control) or LINP1 siRNAs to inhibit the expression of LINP1. (B) MTT assays were used to determine the cell viability of siLINP1-transfected MDA-MB-231 (right), MDA-MB-468 (middle) and MCF-7 (left) cells. Experiments were performed in triplicate. (C) Flow cytometry was performed to determine the effect of LINP1 on changes in cell cycle distribution (right). Statistical diagrams show significant differences (middle). Western blotting was used to detect cyclin D1, cyclin D3, and CDK4 expression (left). (D) LINP1 knockdown promoted apoptosis in MDA-MB-231 and MDA-MB-468 cells, *P < 0.05, **P < 0.01, and, ***P < 0.001 by the Student's t test.
LINP1 promoted migration and invasion by inducing EMT in breast cancer cells
To examine the role of LINP1 in regulating cell mobility, we performed cell migration and invasion assays in breast cancer cells transfected with siRNAs and a negative control. The wound healing assay showed that cell migration was inhibited in LINP1-knockdown breast cancer cells compared to the negative control (Figure 2A). Moreover, the transwell assay indicated a significant reduction in cell migration and invasion ability after siRNA transfection into breast cancer cells (Figure 2B-C). Consistently, LINP1 overexpression enhanced the migration ability of MDA-MB-231 and MDA-MB-468 cells (Supplementary Figure 2A). Taken together, these results suggest that LINP1 may act as a tumor promoter by promoting cell migration and invasion. The epithelial-mesenchymal transition (EMT) process is one of the important mechanisms for migration and invasion. After transfection with si-LINP1, MDA-MB-231 cells displayed a round-like morphology, which is typical of the epithelial phenotype of cells compared with the corresponding parental cells (Supplementary Figure 2B). To investigate whether LINP1 is associated with EMT, we examined the expression of epithelial and mesenchymal markers in LINP1 knockdown, overexpression, and control cells. After transfection with siRNA, the epithelial markers E-cadherin was upregulated, whereas the mesenchymal markers vimentin and N-cadherin were significantly downregulated (Figure 2D). LINP1 overexpression led to the opposite results (Supplementary Figure 2C). Thus, gene expression suggested that LINP1 may be able to drive the breast cancer cells from an epithelial to mesenchymal status.
Figure 2.
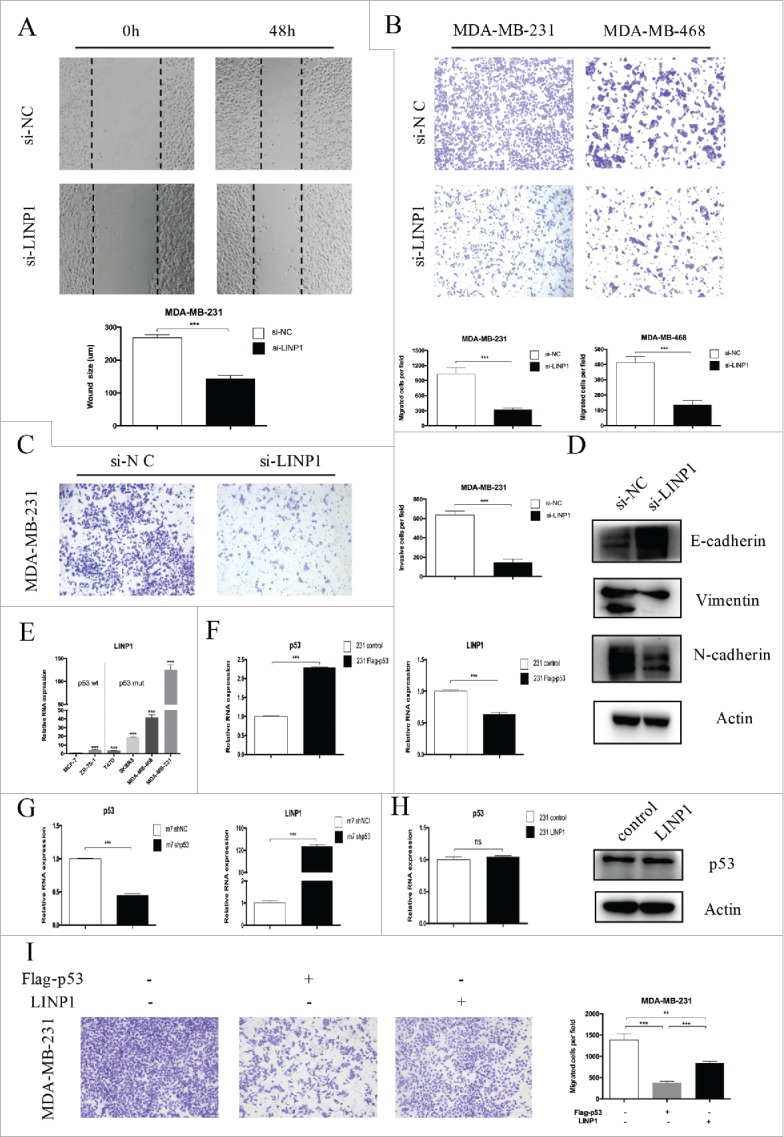
LINP1 knockdown inhibited migration and invasion in breast cancer cells. (A) Wound healing assays and (B) Transwell migration assays demonstrated that LINP1 knockdown inhibited cell migration. The columns are the average of three independent experiments. (C) Transwell invasion assays were used to measure the impaired invasion capacities of MDA-MB-231 cells. The columns are the average of three independent experiments. (D) LINP1 knockdown led to increased E-cadherin expression and decreased N-cadherin and vimentin expression. (E) LINP1 expression levels were measured in six breast cancer cell lines with quantitative real-time PCR (qPCR). Actin was used as the endogenous control. (F) MDA-MB-231 cells were transfected with flag-p53; then the efficacy (right) and effect on LINP1 expression (left) were evaluated by qPCR. (G) P53 was knockdown via transfection with shp53 in MCF-7 cells. Then, the efficacy (right) and effect on LINP1 expression (left) were evaluated by qPCR. (H) LINP1 overexpression did not influence p53 expression in MDA-MB-231 cells. (I) P53 overexpression inhibited MDA-MB-231 cell migration and LINP1 could attenuate this effect. The columns are the average of three independent experiments, *P < 0.05, **P < 0.01, and, ***P < 0.001 by the Student's t test.
Using the online software program PROMO, we decided to explore the role of p53 in regulating LINP1 expression. LINP1 expression was lower in p53-wt cells (such as MCF-7 and ZR-75-1), and higher in p53-mut cells (such as MDA-MD-231and MDA-MB-468) (Figure 2E). Thus, we overexpressed p53 by transfecting MDA-MB-231 cells with pcDNA3.1-p53, and LINP1 expression decreased (Figure 2F). Consistently, LINP1 expression was upregulated after the transfection of MCF-7 cells with siRNA (Figure 2G). However, neither the p53 mRNA nor protein expression levels were affected by LINP1 (Figure 2H and Supplementary Figure 2D). We then delineated the role of p53 in LINP1-regulated tumor metastasis. Transwell assay showed that p53 overexpression inhibited breast cancer cell migration and that LINP1 overexpression could partly attenuate the effects of p53 on migration (Figure 2I). Together, these findings demonstrated that p53 was involved in regulating LINP1 expression in different breast cancer cells.
LINP1 is associated with chemoresistance in breast cancer cells
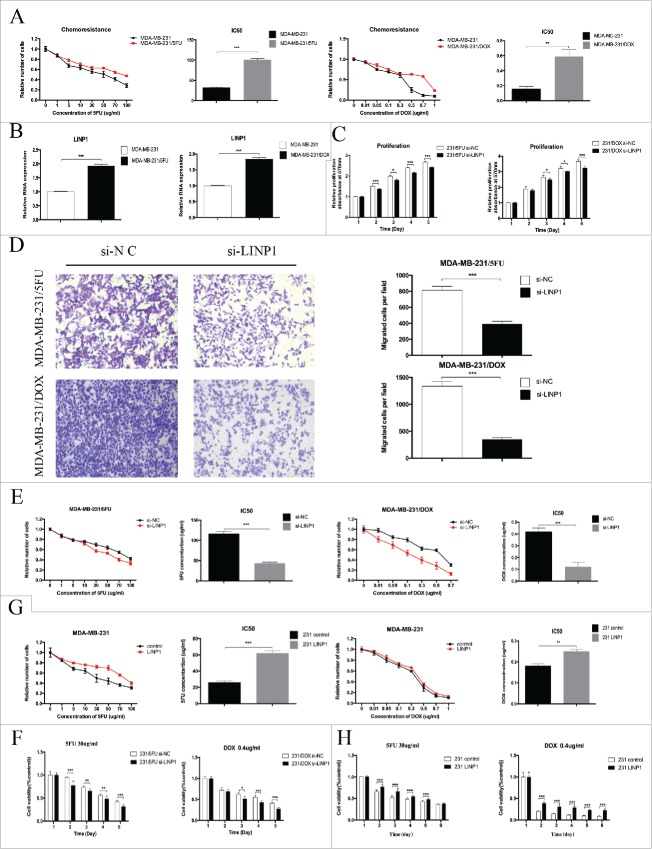
5-fluoroutacil and doxorubicin are two effective chemotherapeutics against breast cancer. However, acquired resistance to 5FU and DOX has been a major obstacle in the clinical treatment of breast cancer. To determine whether LINP1 is involved in regulating breast cancer cell resistance to 5FU and DOX, we first established 5FU- (MDA-MB-231/5FU) and DOX-resistant cell lines (MDA-MB-231/DOX). The IC50 of 5FU in the MDA-MB-231/5FU cells was 100.5 ± 2.309 µg/ml, which is significantly higher than in MDA-MB-231 cells at 31.69 ± 1.16 µg/ml (P < 0.05). The calculated resistance index (RI) was 3.17. The IC50 of DOX in the MDA-MB-231/DOX cells was 0.59 ± 0.06 μg/ml, which is also significantly higher than in MDA-MB-231 cells at 0.16 ± 0.017 μg/ml (P < 0.05) (Figure 3A). The IC50 in the drug-resistant cells was much higher, with a resistance index (RI) of 3.69. Next, we examined the expression of LINP1 in the drug-resistant cells and the corresponding parental cell lines. As shown in Figure 3B, increased LINP1 expression was observed in the two drug-resistant cell lines. These results suggest that LINP1 might be involved in the generation of the chemoresistance phenotype in breast cancer.
Figure 3.
LINP1 contributes to multidrug resistance in breast cancer cells. (A) The IC50 values of MDA-MB-231/5FU (upper) and MDA-MB-231/DOX (down) cells was higher than that of their parental cells. (B) qPCR analysis showed that LINP1 expression levels were higher in MDA-MB-231/5FU (left) and MDA-MB-231/DOX (right) cell lines compared with their parental cell lines. (C and D) LINP1 knockdown inhibited drug-resistant cell proliferation and migration. The columns are the average of three independent experiments. (E) MTT assays indicated that LINP1 knockdown in drug-resistant cells decreased cell viability under the stress of 5FU or DOX. (F) 30 μg/ml 5FU or 0.4 μg/ml DOX was used to test the inhibition. of drug-resistance caused by LINP1 knockdown in MDA-MB-231/5FU (left) and MDA-MB-231/DOX (right) cells, respectively. (G) MTT assays indicated that LINP1 overexpression in MDA-MB-231 cells increased resistance to both 5FU and DOX. (H) 30 μg/ml 5FU or 0.4 μg/ml DOX was used to test the promotion of drug-resistance caused by LINP1 overexpression in MDA-MB-231 cell lines, *P < 0.05, **P < 0.01, and, ***P < 0.001 by the Student's t test.
Given that LINP1 was overexpressed in the drug-resistant cells, we speculated that LINP1 could regulate the breast cancer cells with chemotherapy. First, we transfected 231/5FU and 231/DOX cells with control or LINP1-specific siRNAs. MTT and transwell assays showed that LINP1 knockdown led to decreased drug-resistant cell viability and motility (Figure 3C-D), indicating a potential role for LINP1 in regulating chemoresistance.
We subsequently determined the role of LINP1 in regulating the IC50 of 5FU and DOX in the 231/5FU and 231/DOX cells. The IC50 of 5FU in the MDA-MB-231/5FU control and LINP1 knockdown cells were 116 ± 3.46 and 43.28 ± 1.732 μg/ml, respectively, and the IC50 of DOX in the MDA-MB-231/DOX control and LINP1 knockdown cells were 0.42 ± 0.017 and 0.12 ± 0.02 μg/ml (Figure 3E), respectively, suggesting that LINP1 knockdown might attenuate the drug-resistant cell resistance to 5FU and DOX. Thus, we chose 5-FU at a concentration of 30 μg/ml and DOX at a concentration of 0.4 μg/ml to treat the drug-resistant cells. MTT assays revealed that LINP1 knockdown decreased the survival of cells undergoing treatment with the chemotherapeutics (Figure 3F). LINP1 knockdown in sensitive breast cancer cells further increased the sensitivity to 5FU and DOX (Supplementary Figure 3A-B).
We also overexpressed LINP1 via the transfection of pcDNA3.1-LINP1 into sensitive breast cancer cells (Supplementary Figure 3C). Breast cancer cells overexpressing LINP1 displayed an increased tolerance to 5FU and DOX treatment compared with the response of the control cells, and the 5FU and DOX IC50 values were significantly increased in the LINP1 overexpression cells (Figure 3G and Supplementary Figure 3D). MTT assays also revealed that LINP1 overexpression decreased the sensitivity to cytotoxicity due to chemotherapy compared with the controls (Figure 3H and Supplementary Figure 3E-F). Taken together, these results indicated that LINP1 overexpression could enhance the resistance of breast cancer cells to 5FU and DOX.
LINP1 promotes chemoresistance by regulating apoptosis-related proteins in breast cancer cells
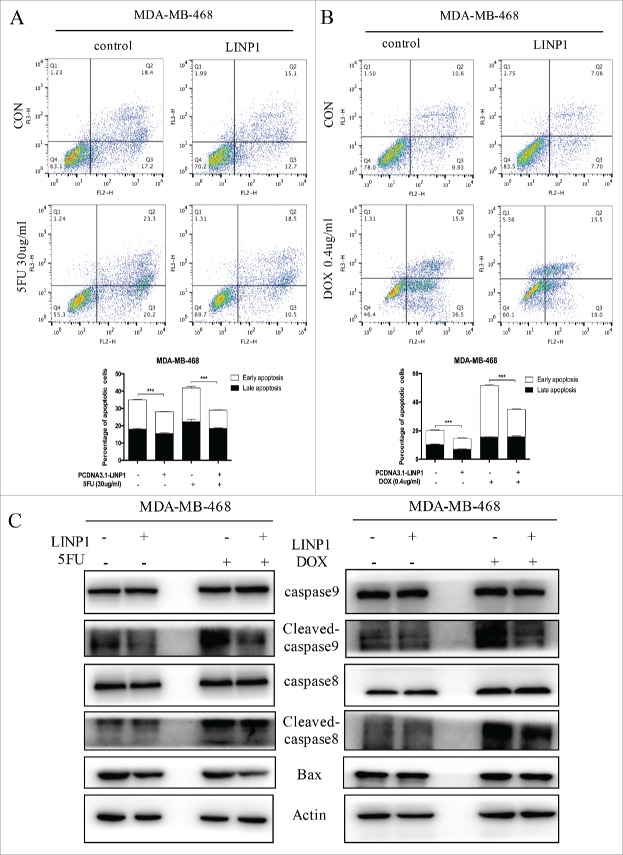
We used flow cytometry analyses to assess the effects of LINP1 on apoptosis upon exposure to chemotherapeutic drugs. MDA-MB-468 cells were first transfected with LINP1-overexpression or control vector and then treated with 5FU or DOX for 48 h before harvesting for flow cytometry assays. Compared with the negative controls, LINP1 overexpression caused a decrease in apoptosis in MDA-MB-468 cells treated with 5FU (0 μg/ml or 30 μg/ml) or DOX (0 μg/ml or 0.4 μg/ml), indicating that LINP1 overexpression significantly increase the cell's ability to resist chemotherapeutic drugs (Figure 4A-B). TUNEL assay also demonstrated the inhibiting effects of LINP1 on apoptosis (Supplementary Figure 4A-B).
Figure 4.
LINP1 protected breast cancer cells from chemotherapeutic-induced apoptosis. (A and B) MDA-MB-468 cells were transiently transfected with LINP1 overexpression plasmid or control plasmid, followed by 5FU or DOX treatment. The apoptosis rates were determined by FACS analysis. Representative results are shown, and the data are presented as the mean ± SD. (C) Western blot analysis was performed to detect the effects of LINP1 overexpression on the protein levels of apoptosis-related proteins with or without drug treatment. Actin served as a loading control for Western blots, *P<0.05, **P<0.01, and, ***P<0.001 by the Student's t test.
Next, we measured the levels of apoptosis-related proteins (cleaved caspase-8, total caspase-8, cleaved caspase-9, total caspase-9, and Bax proteins). LINP1 overexpression decreased BAX levels in MDA-MB-468 cells. LINP1 overexpression also decreased the expression level of cleaved-caspase9 induced by 5FU treatment, thought total proteins levels remained unchanged (Figure 4C and Supplementary Figure 4C). Moreover, LINP1 overexpression decreased cleaved caspase-8/9 levels in MDA-MB-468 cells induced by DOX treatment, while total proteins levels remained unchanged (Figure 4C and Supplementary Figure 4C). In supplementary experiments, we detected a role for LINP1 in apoptosis caused by Huaier, a traditional Chinese medicine proven to be important in inducing apoptosis (Supplementary Figure 4D).17 Therefore, LINP1 might inhibited the caspase-9/Bax during 5FU treatment or caspase8/9 during DOX treatment. Taken together, these data suggest that LINP1 overexpression decreased the in vitro chemosensitivity of breast cancer cells, while enhancing their proliferation and reducing apoptosis.
LINP1 expression was correlated with poor prognosis in breast cancer
Given our abovementioned results, we further investigated the role of LINP1 in predicting the prognosis of breast cancer patients. The associations between LINP1 expression and the clinicopathological characteristics of breast cancer patients are depicted in Table 1. High LINP1 expression levels were significantly correlated with distant metastasis (P = 0.011) and advanced clinical stage (P = 0.035). There was no significant correlation between LINP1 expression and age, tumor size or lymph node metastasis (all P > 0.05, Table 1). We then investigated whether increased LINP1 levels were associated with an unfavorable outcome in breast cancer patients. Kaplan–Meier assay showed that patients with high LINP1 expressions in tumors, lymph node metastases or distant metastases had significantly high risks of death (Table 2). LINP1 relative expression detected in breast cancer tissues was significantly associated with shorter overall survival and disease-free survival in breast cancer patients (P = 0.0221, 0.0085; Figure 5A-B). Consistently, we detected much higher LINP1 level in primary tumor tissues from patients who developed distant metastases during follow-up (Figure 5C), suggesting that LINP1 dysregulation might contribute to breast cancer metastasis. Multivariate analysis showed major effects of LINP1 overexpression and metastasis on the patients' prognosis (Table 3). In summary, our results showed that LINP1 overexpression was associated with unfavorable prognoses and that LINP1 may serve as a prognostic marker in breast cancer.
Table 1.
Associations between patient characteristics and LINP1 expression.
| LINP1 expression |
||||
|---|---|---|---|---|
| Variables | Cases (%) | Low (n = 34) | High (n = 33) | P-valuea |
| Age | ||||
| ≤ 50 | 31 (46.2%) | 14 | 17 | 0.396 |
| > 50 | 36 (53.7%) | 20 | 16 | |
| Tumor size (cm) | ||||
| ≤ 2 | 44 (65.7%) | 21 | 23 | 0.494 |
| > 2 | 23 (34.3%) | 13 | 10 | |
| Positive lymph nodes | ||||
| 0 | 33 (49.3%) | 17 | 16 | 0.901 |
| ≥ 1 | 34 (50.7%) | 17 | 17 | |
| Distant metastasis | ||||
| M0 | 54 (80.6%) | 32 | 22 | 0.005 |
| M1 | 13 (19.4%) | 2 | 11 | |
| Clinical stage | ||||
| I | 14 (20.9%) | 8 | 6 | 0.035 |
| II | 35 (52.2%) | 21 | 14 | |
| III | 5 (7.5%) | 3 | 2 | |
| IV | 13 (19.4%) | 2 | 11 | |
| ER | ||||
| Negative | 11 (16.4%) | 8 | 3 | 0.111 |
| Positive | 56 (83.6%) | 26 | 30 | |
| PR | ||||
| Negative | 14 (20.9%) | 8 | 6 | 0.59 |
| Positive | 53 (79.1%) | 26 | 27 | |
| HER-2 | ||||
| Negative | 64 (95.5%) | 33 | 31 | 0.537 |
| Positive | 3 (4.48%) | 1 | 2 | |
Chi-square detection.
Table 2.
Influence of LINP1 expression and different clinicopathological parameters on overall survival for breast cancer patients.
| Univariate analysis | P-valuea |
|---|---|
| Age | 0.249 |
| Tumor size | 0.259 |
| Positive lymph nodes | 0.024 |
| Distant metastasis | NAb |
| Clinical stage | NAb |
| LINP1 expression | 0.022 |
Kaplan-Meier survival analysis.
Data are not available due to low number of patients.
Figure 5.
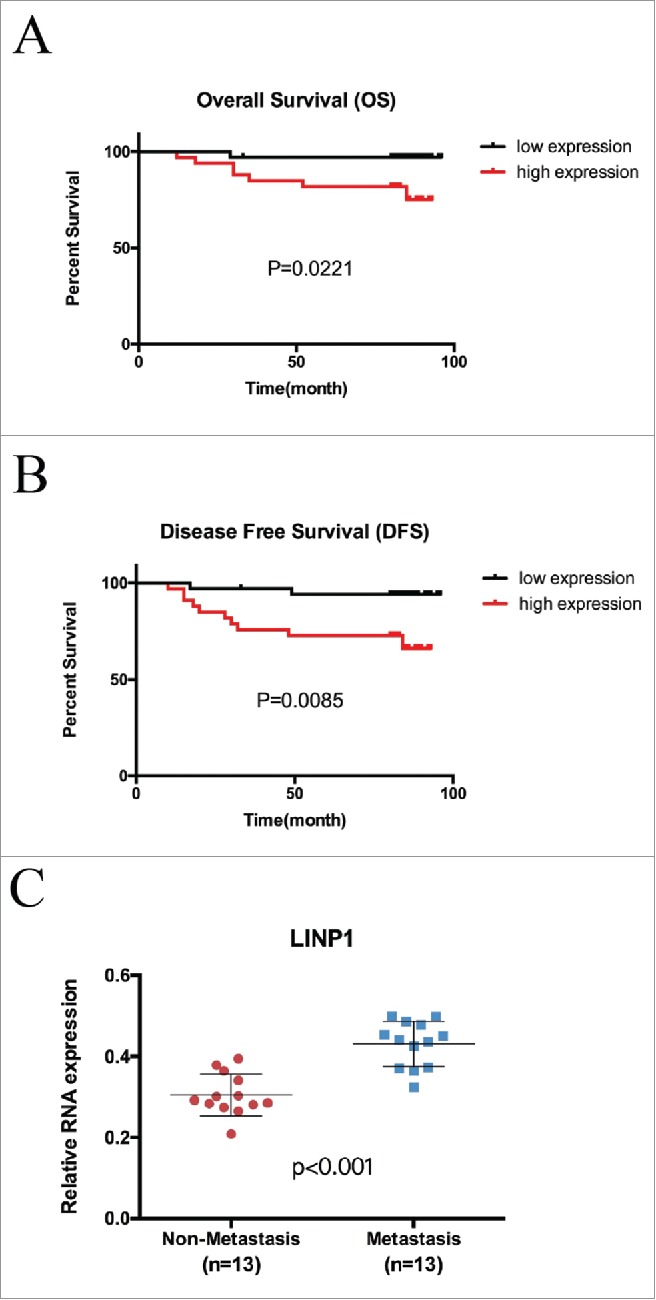
LINP1 was an unfavorable prognostic marker in breast cancer. Kaplan-Meier analysis for (A) overall survival and (B) disease-free survival in 67 breast cancer tissue donors stratified for low and high relative LINP1 expression. (C) LINP1 expression in primary breast cancers with or without distant metastasis. Actin was used as an endogenous control.
Table 3.
Cox proportional hazard multivariate analysis: Influence of HOTAIR tumor levels and positive lymph nodes on overall survival for breast cancer patients.
| Multivariate analysis | P-valuea | Hazard ratio | Confidence interval | |
|---|---|---|---|---|
| Positive lymph nodes | 0.047 | 0.120 | 0.015 | 0.975 |
| LINP1 expression | 0.045 | 0.117 | 0.014 | 0.57 |
Cox proportional hazards model multivariate analysis.
Discussion
Over the past decade, increasing numbers of long non-coding RNAs (lncRNAs) have been identified,18 and accumulating evidence has highlighted the key roles of lncRNAs in various diseases, especially cancer. Mounting lncRNAs have been found to function as potential tumor suppressor genes or oncogenes and be correlated with early diagnosis and prognosis prediction in various cancers.19–21 However, the regulatory roles of lncRNAs played in cancers remain to be fully illustrated. Interestingly, many lncRNAs are emerging as potential biomarkers for diagnosis, prediction of prognosis and drug-resistance in breast cancer.7,22–24
LINP1, which is located in chromosome 10, is abnormally expressed in breast cancer and highly expressed in p53 mutant types. A previous study showed that LINP1 enhanced the survival of breast cancer cells exposed to radiation, suggesting a potential role for LINP1 in the treatment of the disease.25 However, the function of LINP1 in tumor development and chemoresistance remains unclear. In this study, we uncovered a new role for LINP1 in promoting proliferation and mobility and inhibiting apoptosis in breast cancer cells. Mechanistically, p53, a key tumor suppressor, plays a role in repressing LINP1 expression, and LINP1 could partially reverse the inhibitory effects of p53 on proliferation and migration. Moreover, LINP1 expression was positively correlated with 5FU and DOX-resistance in breast cancer cells. Finally, the Kaplan-Meier analysis indicated that patients with high LINP1 expression had a worse prognosis, as supported by shorter disease-free and overall survival.
We first investigated the oncogenic role of LINP1 in breast cancer by evaluating its effects on proliferation and metastasis in breast cancer cells. In the current study, we demonstrated that LINP1 knockdown could inhibit proliferation and growth through cell cycle arrest in G0/G1 stage, whereas LINP1 overexpression significantly promoted cell proliferation and growth. Moreover, our present study also found that CDK4, cyclin D1 and cyclin D3 protein levels, which are low in quiescent cells,26 were positively correlated with LINP1, indicating that LINP1 might regulate cell cycle by modulating these cyclins. The ability of cancer cells to escape apoptosis is one of the mechanism developed during tumor progression, and we demonstrated that LINP1 overexpression inhibited apoptosis. Moreover, transwell and wound healing assays demonstrated that LINP1 upregulation promoted the invasion and migration of breast cancer cells. A major mechanism of metastasis is epithelial-mesenchymal transition (EMT), which is characterized by an altered cell phenotype.27 The hallmarks of EMT include the downregulation of epithelial markers such as E-cadherin and the upregulation of mesenchymal makers such as N-cadherin and vimentin.28 In the present study, we found that LINP1 knockdown could inhibit the process of EMT by influencing its key regulators. Thus, LINP1 may play a key role in the regulation of breast cancer development and progression.
Considering that p53, a major tumor suppressor, is crucial in a number of cell processes, including cell cycle, apoptosis and metastasis,29 we further explored the interaction between p53 and LINP1. A previous study demonstrated that p53 negatively regulated LINP1 expression by increasing miR-29 expression level.25 Our results also demonstrated that p53 overexpression led to lower LINP1 levels, whereas p53 knockdown led to the opposite results. In contrast, LINP1 expression levels had no effect on p53 expression at either the protein or mRNA levels, indicating that LINP1 may be not a regulator of p53 and that there was no feedback regulation between LINP1 and p53. P53 mutations frequently occurred in breast cancer, resulting in the loss of normal ability of p53. Mutant p53 could prevent wt p53 from interacting with its downstream target genes. In contrast, mutant p53 could also activate the transcription of genes that are normally inhibited by wt p53. Our results and a previous report showed that LINP1 expression was higher in cells with mutant p53 than cells with normal p53. We also revealed that the p53 expression in T47D cells was higher than in MCF-7 cells, though there were no significant differences between ZR-75-1 and T47D cells, potentially due to other molecular biological difference between these two cell types. However, more experiments were needed to further clarify the concrete mechanism by which mutant p53 is related to higher LINP1 expression. Moreover, the promotion of LINP1 attenuated the inhibitory effects of p53 on cancer cell metastasis, further revealing a functional association between p53 and LINP1. These results indicated that p53 negatively regulated LINP1 and contributed to various biological functions and tumor suppression.
The major treatment for breast cancer patients is surgery. Postoperative adjuvant therapy also plays important roles in breast cancer treatment. However, chemoresistance remains a major obstacle in the clinical treatment of breast cancer, and its mechanism has not been fully elucidated. 5-fluoroutacil (5-FU) and doxorubicin (DOX) are two effective and important chemotherapeutics against breast cancer,30 which could interact with DNA by inhibiting macro-molecular biosynthesis,31 thereby causing DNA damage. However, the clinical use of 5-FU and DOX has been limited due to development of chemoresistance.32 Recently, several studies have revealed a role for lncRNAs in regulating chemoresistance.33 Tsang and Kwok showed that H19 induces drug resistance in liver cancer cells.34 CCAT1 was upregulated in LAD tissues and promoted the acquisition of doxorubicin-resistance. Yu Fan et al reported that UCA1 increased the cisplatin resistance of bladder cancer cells by regulating Wnt signaling.35 Therefore, it is very important to understand the role of lncRNAs in chemoresistance and identify predictive markers of the therapeutic response. Given that DNA repair capacity is critical for the survival of cancer cells upon therapeutic DNA damage, we further explored the role of LINP1 in drug-resistant breast cancer cells. We demonstrated that drug-resistant cells had higher LINP1 expression levels compared with parental cells. We first found that LINP1 overexpression could decrease the sensitivity of breast cancer cells to 5FU and DOX. Recent studies have emphasized the importance of disrupting apoptosis during the acquisition of chemoresistance.36 Our findings indicated that LINP1 overexpression may reduce the sensitivity of breast cancer cells to chemotherapeutics through the inhibition of apoptosis. The Bax and caspase families are key regulators of cell apoptosis. We identified that LINP1 mainly inhibited the expression of caspase9/Bax induced by 5FU or the expression of caspase8/9 induced by DOX, leading to subsequent drug resistance. In contrast, LINP1 inhibition increased breast cancer cell sensitivity to these chemotherapy drugs. These results indicated that LINP1 induced breast cancer chemoresistance through the regulation of key apoptosis-related genes and that targeting LINP1 may be a potential strategy for reversing 5FU and DOX chemoresistance in breast cancer.
A number of lncRNAs have been found to be related with cancer prognosis, such as HOTAIR,37 LIMT,38 and ANCR.39 However, more specific predictors are needed for breast cancer. Our findings suggested that LINP1 may be used as a predictor of therapeutic effect and prognosis in breast cancer. To verify this hypothesis, we first evaluated the correlation between breast cancer clinicopathological features and LINP1 expression. We demonstrated that LINP1 expression levels were significantly associated with distant metastasis and advanced clinic stage. Consistently, LINP1 was highly upregulated in the patients with distant metastases. We also analyzed the overall and disease-free survival of patients with breast cancer according to LINP1 expression levels. The results from the log rank test showed that high LINP1 levels in breast cancer tissues were associated with an unfavorable prognosis, specifically being related with a high risk of death and shorter survival. Of note, though an obvious trend was observed between distant metastases and poor prognosis according to the Kaplan-Meier analysis, we failed to evaluate the influence of metastases on overall survival using multivariate analysis. A plausible explanation is that the follow-up time was too short to obtain significant prognostic results in the LINP1-downregulated group. Additionally, the small size of the cohort may also account for these findings. Thus, a larger sample size and longer follow-up time needs to be further investigated. In summary, these reports suggest that LINP1 plays certain roles in breast cancer carcinogenesis and highlights LINP1 as a promising therapeutic target in breast cancer. However, more tissue samples are needed to further analyze the correlation between LINP1 expression and patient clinicopathological features to evaluate the potential of LINP1 as an independent biomarker in breast cancer. Moreover, the universality of LINP1 expression in various cancers deserves further study.
Materials and methods
Cell culture and reagents
Human breast cancer cells (MDA-MB-231, MDA-MB-468 and MCF7) were purchased from the American Type Culture Collection (Manassas, VA, USA). The cell lines have been tested and authenticated. Cells were routinely maintained in Dulbecco's modified Eagle's medium (Gibco-BRL, Rockville, IN, USA) supplemented with 10% fetal bovine serum (Haoyang Biological Manufacture Co. Ltd., Tianjin, China), 100 U/ml penicillin and 100 μg/ml streptomycin. A 5FU-resistant and DOX-resistant breast cancer cell lines, MDA-MB-231/5FU and MDA-MB-231/DOX, respectively, were derived from the MDA-MB-231 cell line by continuous exposure to 5FU (final concentration of 10 μg/ml) (Sigma-Aldrich, Shanghai, China) or DOX (final concentration of 0.2 μg/ml) (Sigma-Aldrich, Shanghai, China) over a period of 12 months. The drug-resistant cell lines are capable of proliferation in the presence of chemotherapeutics. At least two weeks before further experiments, normal culture medium replaced those with the drugs. Cells were cultured at 37°C with 5% CO2 in a humidified incubator.
Plasmid construction and transfection
For LINP1 overexpression, the LINP1 cDNA was cloned into the multiple cloning sites in the pcDNA3.1 vector (Invitrogen, Carlsbad, CA, USA). The expression plasmid vector and the empty vector were used to transfect breast cancer cells using Lipofectamine 2000 (Invitrogen, MA, USA) to establish the LINP1 overexpression and control cell lines. For LINP1 knockdown, si-LINP1 and si-control were purchased from Applied Biological Materials (ABM, Canada). Transfections were performed using the Lipofectamine 2000 to establish the si-NC and si-LINP1 cell lines.
Quantitative real-time reverse transcriptase (RT)-PCR
Total RNA was prepared from breast cancer cells using the Trizol reagent (Invitrogen, CA, USA) according to the manufacturer's instructions. Reverse transcription was performed with the PrimeScript RT Reagent Kit (Takara, Japan) according to the manufacturer's protocol. qRT-PCR was performed using a SYBR Green PCR kit (Takara, Japan). The cDNA reaction products were amplified with the following primers: (1) LINP1 forward, 5′- TGCCACTGCCATTAGAAGAAC-3′, and LINP1 reverse, 5′- GCTCACAGAGGAGCTACCCA-3′; (2) p53 forward, 5′- CCTCA GCATCTTATCC GAGTGG -3′, and p53 reverse, 5′- TGGATGGTGGTACAGTCA GAGC -3′; and (3) β-actin forward, 5′- CATGTACGTTGCTATCCAGGC -3′, and β-actin reverse, 5′- CTCCTTAATGTCACGCACGAT -3′. The results are represented as the log10 (2−ΔΔCT). β-actin was used as an internal control for calculating lncRNAs and mRNA expression.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and in vitro chemosensitivity assay
Cells were plated into 96-well plates and grown overnight at 37°C under 5% CO2. After treatment with or without appropriate concentrations of 5FU or DOX for the indicated times, 20 μl of MTT solution (5 mg/ml) was added to each well and incubated for 4–6 h at 37°C. Then, the media was removed and 100 µl DMSO was added to each well. The optical density (OD) was measured with a Microplate Reader (Bio-Rad, Hercules, CA, USA) at 570 nm to assess the relative number of surviving cells. All assays were performed in triplicate.
Cell-cycle analysis
Cells were seeded in 6-well plates and starved in serum-free medium at 37°C under 5% CO2. Forty-eight hours after transfection, the cells were trypsinized and washed with cold PBS twice. Then, the cell plates were suspended with 300 µl PBS, followed by the addition of 500 µl cell cycle staining buffer (MultiSciences (Lianke) Biotech Co., Ltd.). After a 30 min incubation at room temperature, a flow cytometer was used to analyze the DNA contents of the cells, and the data were analyzed with the ModFitLT V2.0 software (Becton Dickinson).
Western blot analysis
Cells were collected and lysed with lysis buffer (1 × PBS, 1% NP40, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate, 5 mM ethylenediaminetetraacetic acid, and 1 mM sodium orthovanadate) containing protease inhibitors. Subsequently, equal amounts of proteins were assayed by SDS-PAGE and then transferred to a polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA, USA). After blocking with 5% non-fat milk, the membrane was incubated with primary antibody (Immuno-Way, Newark, DE, USA) overnight at 4°C followed by labeling with a horseradish peroxidase-coupled secondary antibody. The protein bands were detected with enhanced chemiluminescence. β-actin expression was used as the endogenous control.
PE Annexin V apoptosis assay
Cells transfected with the indicated siRNAs or plasmids were collected after 48 h. For detecting chemotherapeutic-induced apoptosis, cells were divided into two aliquots 24 h after transfection. After adherence, 30 µg/ml 5FU or 0.4 µg/ml DOX were used to induced apoptosis for 24 h to 48 h. Apoptosis was measured by flow cytometry with a PF/Annexin V Apoptosis Detection Kit (BD Biosciences, USA) according to the manufacturer's instructions. All samples were assayed in triplicate.
Wound healing assay
Cells were seeded onto 6-well plates and transfected with si-NC or si-LINP1 for 48 h. Then, the cells were serum-starved for 12 h and replenished with 10% fetal bovine serum-DMEM medium. When the cells reached 80% confluence, an artificial wound was carefully created by scratching the confluent cell monolayer with a 200 µl pipette tip. The wound was imaged immediately and at 24 h and 48 h.
Cell invasion and migration assay
Invasion and migration assays were performed using the Transwell system (8-µm pore, Corning Costar, Lowell, MA, USA). In total, 1 × 105 cells were suspended in 100 µl serum-free medium and added to the upper chamber that was pre-coated with or without Matrigel (50 µl/well; Becton Dickinson, Bedford, USA). The lower chamber was filled with 700 µl medium with 20% FBS. After 24–48 h of incubation, the cells adhering to the lower surface of the membrane were fixed with methanol and stained with 0.1% crystal violet; the remaining invaded cells were counted in 5 different fields with each filter.
Terminal-deoxynucleotidyl transferase-mediated nick end labeling (TUNEL) assay
Cells were transfected with si-NC or si-LINP1 for 24 h and then divided into two aliquots. The cells were treated with 30 µg/ml 5FU or 0.4 µg/ml DOX. The TUNEL apoptosis assay was performed using the One Step TUNEL Apoptosis Assay Kit (Beyotime, Jiangsu, China) according to the guidelines. Apoptotic cells (red fluorescence) were imaged under a fluorescent microscope.
Patient samples and ethics statement
Studies using human tissues were reviewed and approved by the Ethical Committee of Shandong University. Tumor samples were obtained from breast cancer patients admitted to Qilu Hospital from January 2004 to December 2011. The follow-up period for all the patients was 1–96 months, with a median of 43 months. All the participants in this study provided written informed consent for the use of the clinical materials obtained in the study.
Statistical analysis
The SPSS software (version 18.0) was used for the statistical analysis. Two group comparisons were performed with the student's t test. Survival rates were calculated by Kaplan–Meier survival analysis. Cox proportional hazards model multivariate analyses were used to evaluate the influence of LINP1 expression and the clinicopathological features on overall survival. All the performed tests were two-sided and error bars represent the standard error of the mean (SEM) of three experiments. Differences with p < 0.05 were considered to be statistically significant.
Supplementary Material
Funding Statement
This work was supported by the National Natural Science Foundation of China (No.81272903; No.81672613), the Key Research and Development Program of Shandong Province (No.2016GGE2775), the Shandong Science and Technology Development Plan (2016CYJS01A02) and a Special Support Plan for National High-Level Talents (“Ten Thousand Talents Program”) to Qifeng Yang.
Disclosure of potential conflicts of interest
The authors declare no conflict of interest. All authors approved the final version of the manuscript for submission.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al.. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Wang J, Ye C, Xiong H, Shen Y, Lu Y, Zhou J, et al.. Dysregulation of long non-coding RNA in breast cancer: an overview of mechanism and clinical implication. Oncotarget. 2017;8:5508–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–87. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kunej T, Obsteter J, Pogacar Z, Horvat S, Calin GA. The decalog of long non-coding RNA involvement in cancer diagnosis and monitoring. Critical reviews in clinical laboratory sciences 2014;51:344–57. doi: 10.3109/10408363.2014.944299. [DOI] [PubMed] [Google Scholar]
- 6.Chen QN, Wei CC, Wang ZX, Sun M. Long non-coding RNAs in anti-cancer drug resistance. Oncotarget. 2017;8:1925–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majidinia M, Yousefi B. Long non-coding RNAs in cancer drug resistance development. DNA repair. 2016;45:25–33. doi: 10.1016/j.dnarep.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Tian ZZ, Guo XJ, Zhao YM, Fang Y. Decreased expression of long non-coding RNA MEG3 acts as a potential predictor biomarker in progression and poor prognosis of osteosarcoma. International journal of clinical and experimental pathology. 2015;8:15138–42. [PMC free article] [PubMed] [Google Scholar]
- 9.Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, et al.. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC cancer. 2013;13:461. doi: 10.1186/1471-2407-13-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Lin Z, Gao Y, Yao T. Downregulation of long noncoding RNA MEG3 is associated with poor prognosis and promoter hypermethylation in cervical cancer. Journal of experimental & clinical cancer research: CR. 2017;36:5. doi: 10.1186/s13046-016-0472-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang JJ, Guo SH, Jia BQ. Down-regulation of long non-coding RNA MEG3 serves as an unfavorable risk factor for survival of patients with breast cancer. European review for medical and pharmacological sciences. 2016;20:5143–7. [PubMed] [Google Scholar]
- 12.Sun M, Gadad SS, Kim DS, Kraus WL. Discovery, Annotation, and Functional Analysis of Long Noncoding RNAs Controlling Cell-Cycle Gene Expression and Proliferation in Breast Cancer Cells. Molecular cell. 2015;59:698–711. doi: 10.1016/j.molcel.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, et al.. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer cell. 2015;27:370–81. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, et al.. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes & development. 2016;30:34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Senkus E, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rutgers E, et al.. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annals of oncology: official journal of the European Society for Medical Oncology. 2015;26 Suppl 5:v8–30. doi: 10.1093/annonc/mdv298. [DOI] [PubMed] [Google Scholar]
- 16.Osako T, Horii R, Matsuura M, Ogiya A, Domoto K, Miyagi Y, et al.. Common and discriminative clinicopathological features between breast cancers with pathological complete response or progressive disease in response to neoadjuvant chemotherapy. Journal of cancer research and clinical oncology. 2010;136:233–41. doi: 10.1007/s00432-009-0654-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song X, Li Y, Zhang H, Yang Q. The anticancer effect of Huaier (Review). Oncology reports. 2015;34:12–21. doi: 10.3892/or.2015.3950. [DOI] [PubMed] [Google Scholar]
- 18.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al.. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer biology & medicine. 2015;12:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin K, Luo G, Xiao Z, Liu Z, Liu C, Ji S, et al.. Noncoding RNAs as potential biomarkers to predict the outcome in pancreatic cancer. Drug design, development and therapy. 2015;9:1247–55. doi: 10.2147/DDDT.S77597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin CY, Xu HM. Novel perspectives of long non-coding RNAs in esophageal carcinoma. Carcinogenesis. 2015;36:1255–62. doi: 10.1093/carcin/bgv136. [DOI] [PubMed] [Google Scholar]
- 22.Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, et al.. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35:2746–55. doi: 10.1038/onc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Sharma S, Watabe K. Roles of lncRNA in breast cancer. Frontiers in bioscience. 2015;7:94–108. doi: 10.2741/s427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang M, Wu WB, Wang ZW, Wang XH. lncRNA NEAT1 is closely related with progression of breast cancer via promoting proliferation and EMT. European review for medical and pharmacological sciences. 2017;21:1020–6. [PubMed] [Google Scholar]
- 25.Zhang Y, He Q, Hu Z, Feng Y, Fan L, Tang Z, et al.. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nature structural & molecular biology. 2016;23:522–30. doi: 10.1038/nsmb.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–8. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 27.Diepenbruck M, Christofori G. Epithelial-mesenchymal transition (EMT) and metastasis: yes, no, maybe? Current opinion in cell biology. 2016;43:7–13. doi: 10.1016/j.ceb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Peinado H, Cano A. New potential therapeutic targets to combat epithelial tumor invasion. Clinical & translational oncology: official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2006;8:851–7. doi: 10.1007/s12094-006-0148-z. [DOI] [PubMed] [Google Scholar]
- 29.Stracquadanio G, Wang X, Wallace MD, Grawenda AM, Zhang P, Hewitt J, et al.. The importance of p53 pathway genetics in inherited and somatic cancer genomes. Nature reviews Cancer. 2016;16:251–65. doi: 10.1038/nrc.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nature reviews Cancer. 2003;3:330–8. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 31.Tacar O, Sriamornsak P, Dass CR. Doxorubicin: an update on anticancer molecular action, toxicity and novel drug delivery systems. The Journal of pharmacy and pharmacology. 2013;65:157–70. doi: 10.1111/j.2042-7158.2012.01567.x. [DOI] [PubMed] [Google Scholar]
- 32.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al.. Cancer statistics, 2008. CA: a cancer journal for clinicians. 2008;58:71–96. [DOI] [PubMed] [Google Scholar]
- 33.Liu Z, Sun M, Lu K, Liu J, Zhang M, Wu W, et al.. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PloS one. 2013;8:e77293. doi: 10.1371/journal.pone.0077293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsang WP, Kwok TT. Riboregulator H19 induction of MDR1-associated drug resistance in human hepatocellular carcinoma cells. Oncogene. 2007;26:4877–81. doi: 10.1038/sj.onc.1210266. [DOI] [PubMed] [Google Scholar]
- 35.Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, et al.. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. The FEBS journal. 2014;281:1750–8. doi: 10.1111/febs.12737. [DOI] [PubMed] [Google Scholar]
- 36.Si X, Zang R, Zhang E, Liu Y, Shi X, Zhang E, et al.. LncRNA H19 confers chemoresistance in ERalpha-positive breast cancer through epigenetic silencing of the pro-apoptotic gene BIK. Oncotarget. 2016;7:81452–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, et al.. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer research. 2011;71:6320–6. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 38.Sas-Chen A, Aure MR, Leibovich L, Carvalho S, Enuka Y, Korner C, et al.. LIMT is a novel metastasis inhibiting lncRNA suppressed by EGF and downregulated in aggressive breast cancer. EMBO molecular medicine. 2016;8:1052–64. doi: 10.15252/emmm.201606198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z, Hou P, Fan D, Dong M, Ma M, Li H, et al.. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell death and differentiation. 2017;24:59–71. doi: 10.1038/cdd.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.