ABSTRACT
While the importance of iron for tumor development is widely appreciated, the exact sources of tumor-supporting iron largely remain elusive. The possibility that iron might be provided by stromal cells in the tumor microenvironment was not taken into account so far. In the present study, we show that tumor-associated macrophages (TAM) acquire an iron-release phenotype upon their interaction with tumor cells, thereby increasing the availability of iron in the tumor microenvironment. Mechanistically, TAM expressed elevated levels of the high-affinity iron-binding protein lipocalin-2 (LCN-2), which appeared to be critical for the export of iron from TAM, and in turn enhanced tumor cell proliferation. Moreover, in PyMT-mouse tumors as well as in primary human breast tumors LCN-2 was predominantly expressed in the tumor stroma as compared to tumor cells. LCN-2 expression in the stroma further correlated with enhanced tumor proliferation in vivo. Our data suggest a dominant role of TAM in the tumor iron-management and identify LCN-2 as a critical iron transporter in this context. Targeting the LCN-2 iron export mechanism selectively in stromal cells might open for future iron-targeted tumor therapeutic approaches.
KEYWORDS: TAM, iron, lipocalin-2, iron-trafficking, tumor microenvironment, tumor stroma
Introduction
MΦ exhibit a remarkable heterogeneity and functional plasticity described by two extremes, i.e. the classically vs. alternatively activated phenotype, determined by the local environment.1 Tumor secreted factors, such as IL-10 or TGF-β as well as tumor hypoxia polarize infiltrating monocytes to an anti-inflammatory, alternative MΦ phenotype.2 As recently suggested, classical MΦ sequester iron, which is part of the host defense against pathogens in order to restrict iron availability for bacterial growth.3 In contrast, alternatively activated MΦ show an increased phagocytic activity, efficiently recycle iron, and release it to their microenvironment in order to promote cell proliferation, tissue repair, and regeneration.4,5 Along these lines, breast cancer cells show an increased uptake and intracellular storage of iron in order to support their enhanced metabolism,6 thereby developing a more aggressive phenotype.7 Molecularly, this is reflected by an increased expression of iron-regulated genes like the transferrin receptor (TfR)8 for iron uptake, ferritin heavy (FTH) and ferritin light (FTL)9 for iron storage, and the iron-regulatory protein 2 (IRP2).10 Expression of these genes correlated with a bad prognosis, poor outcome as well as increased tumor grade.7 In addition, reduced expression of the only characterized iron exporter ferroportin (FPN) in tumor cells and an increase of its natural ligand hepcidin (HAMP), provoking FPN degradation, significantly enhanced metastasis and increased proliferation.11 However, the sources of iron within the tumor microenvironment to meet the increased iron demand of tumor cells still remain elusive. We previously showed that TAM, following the interaction with apoptotic tumor cells, produced and secreted LCN-2,12 a well-established high affinity iron carrier protein in the bacteriostatic context.13,14 Mechanistically, the production of LCN-2 was connected to apoptotic cell released sphingosine-1-phophate (S1P) triggered STAT3 activation.15 MΦ-derived LCN-2 further promoted tumor progression and breast cancer metastasis,12,16 while depletion of LCN-2 inhibited breast tumorigenesis, angiogenesis, lymphangiogenesis,15,17,18 and metastasis16,19,20 in experimental mammary tumor models. These observations were in line with the previous finding that LCN-2 expression is associated with reduced disease-free survival.21 Based on the apparent pro-tumor function of MΦ-released LCN-2 and the established high iron-binding affinity of LCN-2, we aimed to determine if TAM-derived LCN-2 might serve as a novel, so far unappreciated iron transporter during tumor progression.
In the present study, we provide evidence that MΦ acquire an iron-release phenotype in the tumor context, thus serving as an important source of iron to meet the elevated iron demand of tumor cells and consequently supporting tumor cell proliferation. We further identify LCN-2 as critical determinant of the macrophage iron-release phenotype, which further emerged as an additional iron transporter, alternatively to FPN.
Results
Primary human MΦ acquire an alternatively activated, iron-release phenotype after co-culture with breast cancer cells
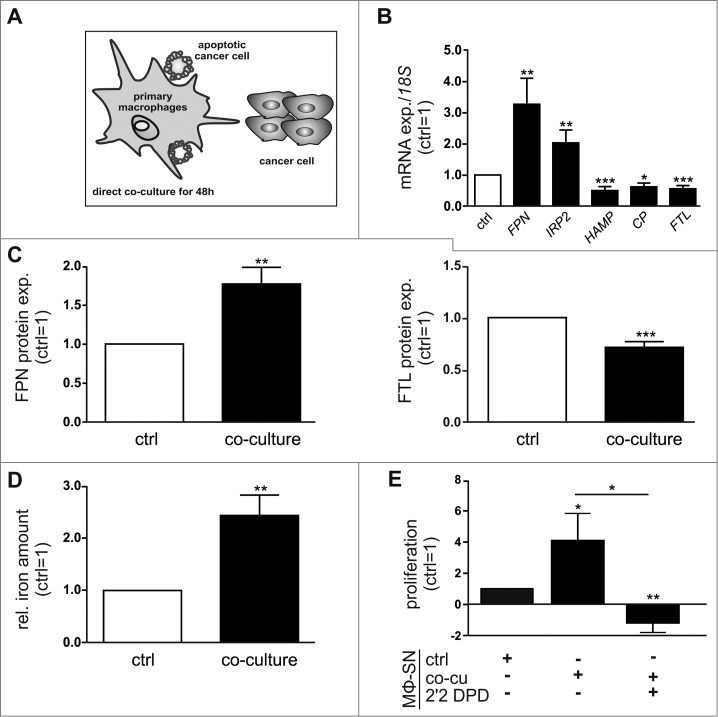
Initially, we aimed to determine how tumor cells affect the phenotype and iron-handling properties of MΦ. To this end, we co-cultured primary human MΦ with human MCF-7 breast cancer cells (Fig. 1A). To assess if the MΦ/tumor cell interplay affects the iron-handling properties of MΦ upon co-culture for 48 h, we analyzed a panel of iron-regulated genes. After 48 h of co-culture, mRNA expression of the iron exporter FPN and the cellular iron sensor IRP2 was strongly induced, while HAMP, CP, and FTL, representing iron sequestering markers, were significantly reduced in MΦ (Fig. 1B), suggesting an iron-release phenotype of MΦ. Co-culturing primary MΦ with other breast tumor cell lines, i.e. MDA-MB-231 or T47D cells, confirmed the iron-release pattern (Fig S1A). Changes at mRNA expression upon co-culture were in a similar range as previously observed after typical alternative-polarizing cytokines such as IL-105 and IL-4/Il-13 4. In line with changes at the mRNA level, FPN protein expression was significantly elevated, while expression of the iron storage protein FTL was reduced in MΦ upon co-culture with MCF-7 (Fig. 1C, Fig S1B) or other breast tumor cells (Fig S1C,D). To verify the functionality of the iron-release phenotype, we determined the iron content in the supernatants of MΦ by atomic absorption spectrometry (AAS). Supernatants of MΦ co-cultured with MCF-7 cells contained significantly elevated levels of iron compared to those of control MΦ (Fig. 1D). As tumor cells are known to require high amounts of iron to maintain their enhanced metabolic demand, we next aimed to test how changes in the iron amount of MΦ supernatants might affect tumor cell proliferation. In fact, the supernatants of MΦ co-cultured with tumor cells enhanced proliferation of MCF-7 cells determined as real-time measurements using the xCELLigence system (Fig. 1E) and by measuring the proliferation marker PCNA at mRNA level (Fig S1E). Stimulation of tumor cells with supernatants of naïve macrophages did not significantly enhance tumor cell proliferation. Adding the well described iron-chelator 2’2 DPD to the iron-release MΦ reversed the effect. Upon co-culture for 48 h, also the surface expression of the alternative MΦ activation markers MHCII, CD206, and CD163 was markedly elevated in MΦ compared to naïve control MΦ (Fig S1F). In contrast, classical-activation markers CD80, CD86, and CD14 appeared unaltered. Along these lines, mRNA expression of the alternative MΦ marker IL-10 was enhanced in the co-culture, whereas mRNA expression of the classical MΦ markers CCL-18, TNF-α, and IL-1β was reduced (Fig S1G). Taken together, we found that MΦ acquire an alternative iron-release phenotype upon interaction with tumor cells, which in turn supports tumor growth.
Figure 1.
Macrophages acquire an alternatively activated, iron-release phenotype in vitro. (A) Schematic overview of the direct co-culture model. Primary human MΦ were co-cultured with MCF-7 breast cancer cells for 48 h, unless indicated otherwise. (B) mRNA expression of iron-regulated genes (FPN, IRP2, HAMP, CP, FTL) in MΦ after co-culture (solid bars) was analyzed by qPCR and normalized to 18 S expression (n = 9-20). (C) Protein expression of FPN (surface, left panel) and FTL (intracellular, right panel) in MΦ after co-culture (solid bars) was determined by flow cytometry and normalized to the respective expression in naïve control MΦ (ctrl, open bars) (n = 9). (D) Iron amount in the supernatant of MΦ after co-culture (solid bar) was quantified by AAS and normalized to the supernatants of naïve control MΦ (ctrl, open bar) (n = 5). (E) Proliferation of MCF-7 cells upon stimulation with supernatants from MΦ (MΦ-SN) co-cultured with tumor cells (co-cu) was determined on an xCELLigence instrument as an increase in the impedance (slope 1/h) and is given relative to MCF-7 cells treated with supernatants from naïve control MΦ (ctrl) (n = 5). Data are means ± SEM from independent experiments, *p < 0.05, **p < 0.01, ***p < 0.001 vs. the respective controls unless indicated otherwise.
TAM show an iron-release phenotype
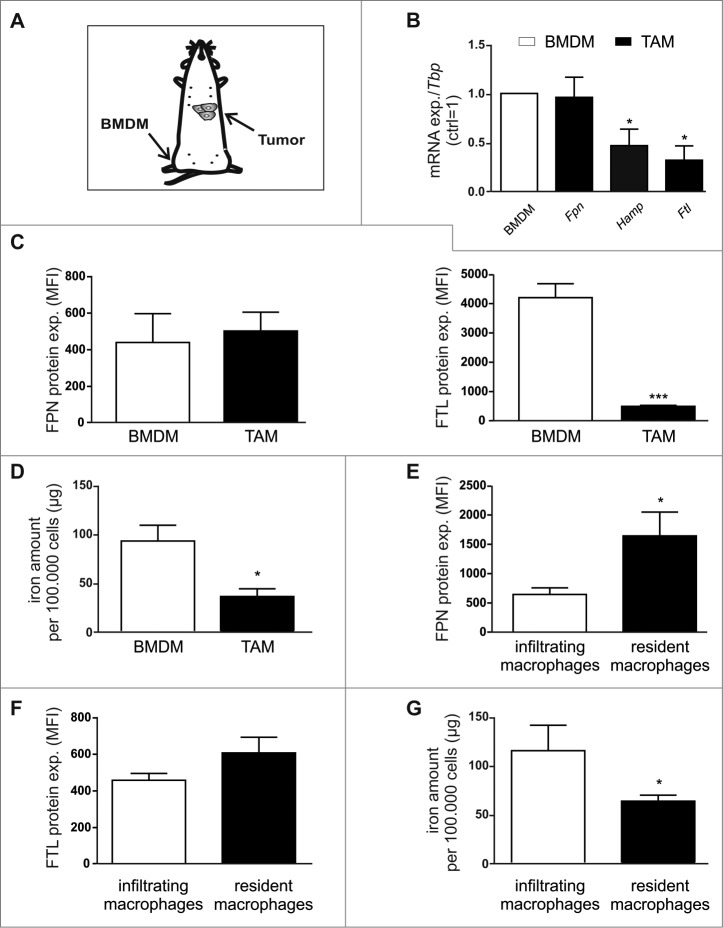
Based on our in vitro findings, we next tested if these observations hold true for TAM in vivo. Therefore, we compared the expression of Fpn, Hamp, and Ftl in F4/80+ cells isolated from spontaneously developed mammary tumors of wildtype PyMT mice, i.e. TAM, with the expression in bone marrow-derived MΦ (BMDM) isolated from the same mouse (Fig. 2A). While Fpn mRNA was expressed at similar levels in TAM and BMDM, both Hamp and Ftl mRNA expression was significantly reduced (Fig. 2B). In line, FPN protein expression remained unaltered, whereas FTL protein was massively reduced (Fig. 2C) in TAM as compared to BMDM. Despite the unaltered expression of the iron exporter FPN, the intracellular iron amount was markedly lower in TAM compared to control MΦ (Fig. 2D), suggesting enhanced iron release. Thus, similar to the in vitro observation, TAM also adopt an iron-release phenotype in vivo. To investigate the different iron-handling ability of MΦ subpopulations (Fig S2A), i.e. infiltrating and resident MΦ, we analyzed the protein expression of FPN and FTL by flow cytometry. In contrast to infiltrating MΦ, resident MΦ significantly upregulate FPN expression (Fig. 2E), but show no difference in FTL expression (Fig. 2F), indicating the iron release phenotype of this subpopulation. As a functional read-out, we quantified the iron amount in cellular lysates of sorted MΦ subpopulations. In line with previous FACS results, we observed reduced iron amounts in resident MΦ (Fig. 2G). Furthermore, we performed Multi-Epitope-Ligand Cartography stainings (MELC) to detect the localization of MΦ subpopulations within the tumor. Interestingly, we observed that resident MΦ preferentially appear close to CD31+ vessels (Fig S2B).
Figure 2.
TAM adopt an iron-release phenotype in vivo. (A) Schematic overview of sample preparation. (B) mRNA expression of iron-regulated genes (Fpn, Hamp, Ftl) in F4/80+ TAM sorted from tumors of PyMT mice was analyzed by qPCR and normalized to Tbp expression. Changes were determined relative to the respective expression in BMDM from the same animal (n = 5–8). (C) Protein expression of FPN (surface, left panel) and FTL (intracellular, right panel) in TAM (solid bars) and BMDM (open bars) was determined by flow cytometry (MFI, mean fluorescence) (n = 4–8). (D) Iron amount in the lysates of TAM (solid bar) was quantified by AAS relative to the cell number (open bar) (n = 7–13). Protein expression of (E) FPN and (F) FTL in infiltrating MΦ (open bars) and resident MΦ (solid bars) was determined by flow cytometry (MFI, mean fluorescence) (n = 11–15). (G) Iron amount in the lysates of infiltrating MΦ (open bar) and resident MΦ (solid bar)was quantified by AAS relative to the cell number (n = 8–10). Data are means ± SEM from independent experiments, *p < 0.05, **p < 0.01, ***p < 0.001 vs. the respective controls unless indicated otherwise.
FPN knockdown in primary human MΦ does not alter the iron-release phenotype
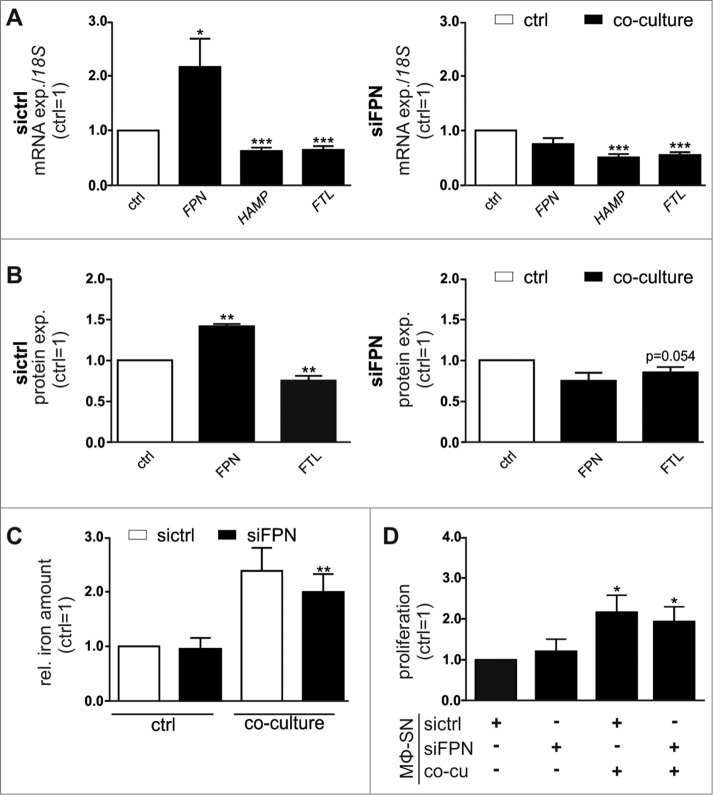
As the well-established iron exporter FPN was unaltered in TAM in vivo, yet they still released elevated levels of iron similar to MΦ in the in vitro co-culture setting, we next determined the role of FPN in the iron-release phenotype of MΦ in the tumor context. Therefore, we depleted FPN in primary MΦ using siRNAs (Fig S3A), prior to co-culturing them with MCF-7 cells. Importantly, the FPN-knockdown efficiently prevented co-culture-induced FPN expression both at mRNA (Fig. 3A, Fig S3B) and protein level (Fig. 3B). In contrast, the expression of the iron-sequestering markers HAMP and FTL were still reduced in FPN-depleted MΦ after their co-culture with MCF-7 cells. Similarly, BMDM isolated from wildtype mice reproduced the iron-release phenotype upon co-culture with syngenic E0771 murine breast tumor cells independent of the presence or absence of FPN (Fig S3C) and were still able to release iron into the supernatant, measured by AAS (Fig S3D). Despite the fact that FPN is considered as the primary iron exporter, the elevated iron amounts in the supernatants of MΦ upon co-culture with MCF-7 cells were not reduced, when FPN was depleted in the MΦ (Fig. 3C). As the iron availability is critical for tumor progression, we next tested how the tumor cell proliferation-stimulating effect of supernatants of co-cultured MΦ is affect by attenuating FPN expression. In line with the unaltered iron-release characteristics, supernatants of co-cultured wildtype (sictrl) and FPN-depleted (siFPN) MΦ similarly induced proliferation of MCF-7 cells (Fig. 3D, Fig S3E), further supporting an alternative iron-release mode.
Figure 3.
FPN knockdown in primary human MΦ does not affect the iron-release phenotype. Primary human MΦ were transfected with siRNA targeting FPN (siFPN) or a scrambled control siRNA (sictrl). 24 h post-transfection, cells were co-cultured with MCF-7 cells for 48 h. (A) mRNA expression of iron-regulated genes (FPN, HAMP, FTL) in sictrl (left panel) or FPN-depleted (right panel) MΦ after co-culture (solid bars) was analyzed by qPCR and normalized to 18 S expression. Changes were determined relative to the respective expression in naïve control MΦ (ctrl, open bar) (n = 17). (B) Protein expression of FPN (surface, left panel) and FTL (intracellular, right panel) in sictrl (open bars) or FPN-depleted (solid bars) MΦ after co-culture was determined by flow cytometry and normalized to the expression in naïve, sictrl transfected MΦ (n = 5). (C) Iron amount in the supernatant of sictrl (open bars) or FPN-depleted (solid bars) MΦ after co-culture was quantified by AAS and normalized to supernatants of naïve, sictrl transfected MΦ (n = 4). (D) Proliferation of MCF-7 cells upon stimulation with supernatants (MΦ-SN) from FPN-depleted MΦ co-culture (co-cu) was determined on an xCELLigence instrument and is given relative to MCF-7 cells treated with supernatant from sictrl transfected MΦ (n = 3–9). Data are means ± SEM from independent experiments, *p < 0.05, **p < 0.01, ***p < 0.001 vs. the respective controls unless indicated otherwise.
LCN-2 serves as an alternative iron transporter
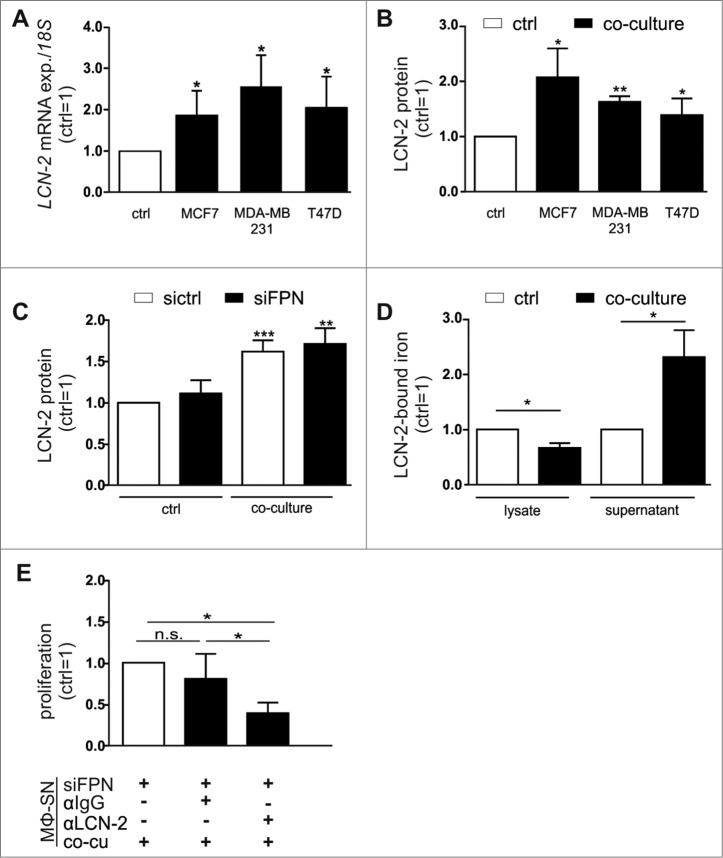
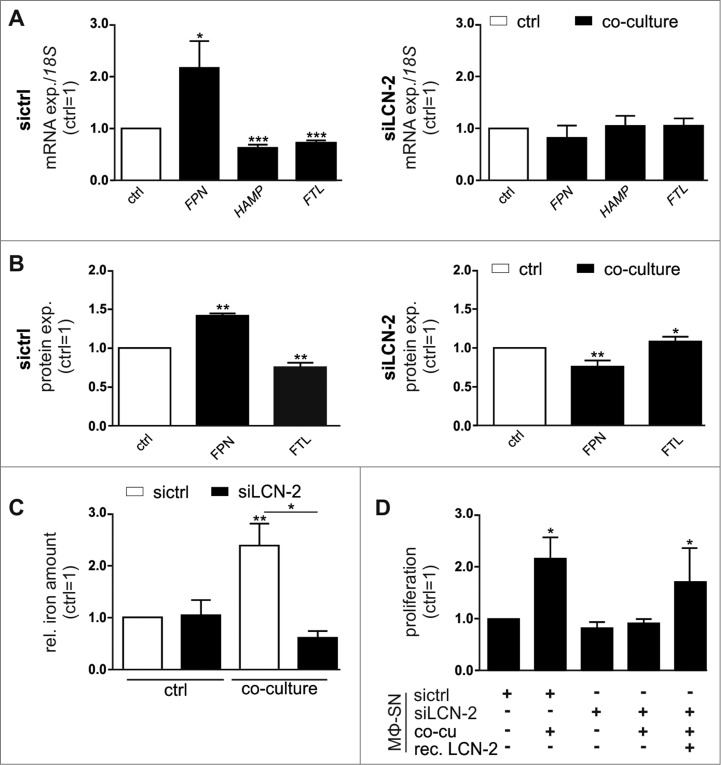
We previously found that LCN-2 is secreted from MΦ upon stimulation with tumor cell supernatants.12,15,16 Taking the high iron-binding affinity of LCN-2 into account, we questioned whether LCN-2 might serve as the alternative iron transporting protein under these conditions. Indeed, LCN-2 was significantly up-regulated in MΦ after co-culture with various breast cancer cell lines both at mRNA (Fig. 4A) and protein level (Fig. 4B). Of note, LCN-2 protein expression in MΦ after co-culture with tumor cells remained increased, even in FPN knockdown MΦ (Fig. 4C, Fig S4A). Similarly, depletion of Fpn in BMDM isolated from wildtype mice reproduced these results upon co-culture with syngenic E0771 murine breast tumor cells (Fig S4B). To assess, if elevated LCN-2 levels affect the iron-release from MΦ, we determined the iron-loading status of LCN-2. Therefore, we immunoprecipitated LCN-2 both from supernatants as well as from cell lysates of naïve or co-cultured MΦ and determined the relative iron content by AAS. LCN-2-bound iron was markedly elevated in the supernatants after co-culture, while it decreased in the cellular fraction (Fig. 4D), suggesting that iron-loaded LCN-2 is efficiently released from MΦ. To assess if MΦ-supplied iron-loaded LCN-2 might contribute to FPN-independent iron-mediated increase in tumor cell proliferation, we neutralized LCN-2 in the supernatant of FPN-depleted MΦ using a specific antibody. In fact, depleting LCN-2 in the supernatants of FPN knockdown MΦ significantly reduced tumor cell proliferation (Fig. 4E, Fig S4C).
Figure 4.
LCN-2 is up-regulated in primary human MΦ after co-culture with breast cancer cells. (A) LCN-2 mRNA expression in MΦ after co-culture with MCF-7, MDA-MB-231, or T47D breast tumor cells (solid bars) was analyzed by qPCR and normalized to 18 S expression, relative to expression in naïve control MΦ (ctrl; open bar) (n = 4–8). (B) LCN-2 protein in the supernatants of primary human MΦ after co-culture with MCF-7, MDA-MB-231, or T47D breast tumor cells (solid bars) was determined by ELISA, normalized to total protein in the supernatants and is given relative to the supernatants of respective naïve control MΦ (ctrl; open bar) (n = 5). (C) LCN-2 protein in the supernatants of sictrl (open bars) or FPN-depleted MΦ (solid bars) after the co-culture with MCF-7 cells was determined by ELISA, relative to the supernatants of naïve, sictrl transfected MΦ (n = 11). (D) Iron content of immunoprecipitated LCN-2 in the supernatant or lysates of MΦ after co-culture with MCF-7 cells was quantified by AAS and is given relative to supernatants or lysates of naïve control MΦ (n = 6). (E) Proliferation of MCF-7 cells upon stimulation with supernatants (MΦ-SN) from the FPN-depleted MΦ co-culture (co-cu) in the presence of IgG (1 µg/ml) or a LCN-2-neutralizing antibody (αLCN-2; 1 µg/ml) was determined on an xCELLigence instrument and is given relative to MCF-7 cells treated with supernatant from co-cultures with FPN-depleted MΦ (MΦ-SN) (n = 5). Data are means ± SEM from independent experiments, *p < 0.05, **p < 0.01, ***p < 0.001 vs. the respective controls unless indicated otherwise.
To provide further evidence for the importance of LCN-2 in TAM, we knocked down LCN-2 in primary human MΦ (Fig S5A,B), and found that LCN-2-depleted MΦ did not show an iron-release gene expression profile after tumor cell co-culture, i.e. neither was FPN increased nor were HAMP or FTL reduced (Fig. 5A, Fig S5C). In line, LCN-2 knockdown MΦ did not show elevated FPN or reduced FTL protein expression upon co-culture with tumor cells in contrast to sictrl-transfected MΦ (Fig. 5B). Importantly, the analysis of the iron content of the supernatants further revealed that depletion of LCN-2 in MΦ completely abolished the co-culture-induced iron-release (Fig. 5C). Consequently, supernatants of LCN-2-depleted MΦ after co-culture failed to enhance proliferation of tumor cells (Fig. 5D, Fig S5D). To further proof the importance of LCN-2 as an iron-supply system for tumor cells in this context, we supplemented supernatants of LCN-2-knockdown MΦ with recombinant iron-loaded LCN-2 (Fig S5E) and observed that the addition of recombinant iron-loaded LCN-2 to the supernatant of siLCN-2 MΦ rescued cancer cell proliferation (Fig. 5D, Fig S5D).
Figure 5.
LCN-2 knockdown in primary human MΦ attenuates the iron-release phenotype upon co-culture with breast cancer cells. Primary human MΦ were transfected with siRNA targeting LCN-2 (siLCN-2) or a scrambled control siRNA (sictrl). 24 h post-transfection, cells were co-cultured with MCF-7 cells for 48 h. (A) mRNA expression of iron-regulated genes (FPN, HAMP, FTL) in sictrl (left panel) or LCN-2-depleted (right panel) MΦ after co-culture (solid bars) was analyzed by qPCR and normalized to 18 S expression (n = 10–19). Changes were determined relative to the respective expression in naïve control MΦ (ctrl, open bar). (B) Protein expression of FPN (surface, left panel) and FTL (intracellular, right panel) in sictrl (open bars) or LCN-2-depleted (solid bars) MΦ after co-culture was determined by flow cytometry, normalized to the expression in naïve, sictrl transfected MΦ (n = 11–21). (C) Iron amount in the supernatant of sictrl (open bars) or LCN-2-depleted (solid bars) MΦ after co-culture was quantified by AAS and normalized to supernatants of naïve, sictrl transfected MΦ (n = 4). (D) Proliferation of MCF-7 cells upon stimulation with supernatant (MΦ-SN) from LCN-2-depleted MΦ co-culture (co-cu) was determined on an xCELLigence instrument and is given relative to MCF-7 cells treated with supernatant from naïve, sictrl transfected MΦ (n = 4). Data are means ± SEM from independent experiments, *p < 0.05, **p < 0.01, ***p < 0.001 vs. the respective controls unless indicated otherwise.
Thus, LCN-2 emerges as an alternative iron transporter in the interplay between tumor cells and MΦ, where it critically contributes to the iron transport from MΦ to the tumor cell, thereby supporting tumor progression.
The iron-release phenotype of TAM depends on LCN-2
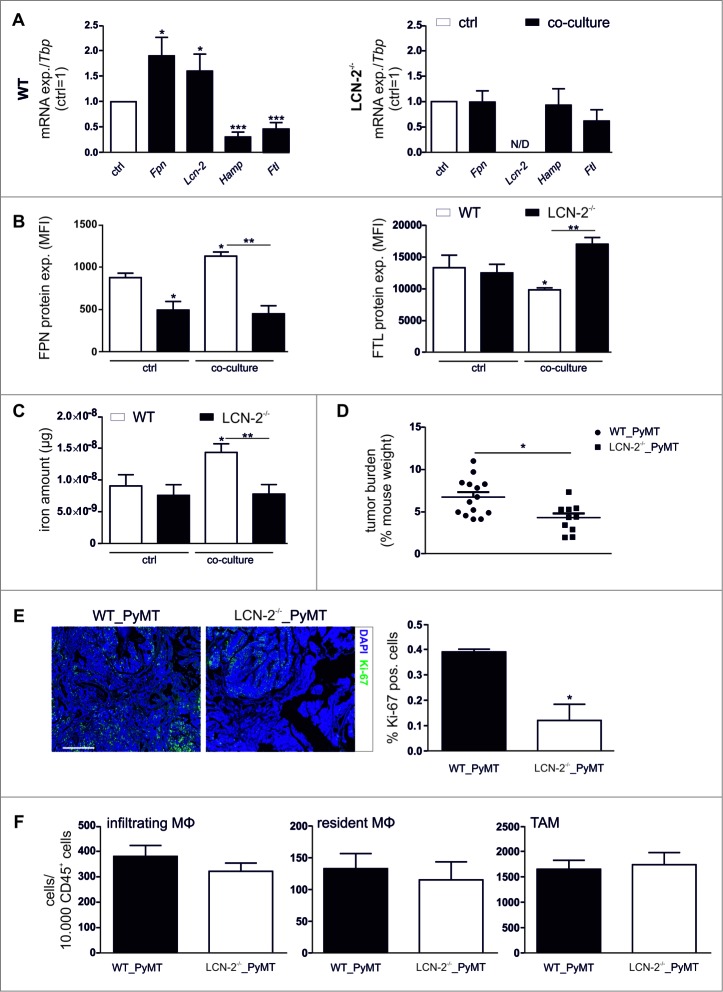
Since LCN-2 appeared to be involved in the TAM-induced pro-tumorigenic effects, we next explored the role of LCN-2 in the MΦ iron-release phenotype by using LCN-2−/− macrophages. To this end, we co-cultured BMDM isolated from wildtype or LCN-2−/− mice with syngenic E0771 mouse breast cancer cells. The iron-regulated gene expression profile obtained in wildtype BMDM after co-culture with E0771 cells pointed to an iron-release profile (Fig. 6A, left panel), closely resembling the observation in primary human MΦ (Fig. 1D). In contrast, in LCN-2-deficient MΦ the expression of Fpn, Hamp, and Ftl was not affected by the co-culture (Fig. 6A, right panel). Similarly, the enhanced FPN protein expression upon co-culture in wildtype BMDM was absent in LCN-2−/−-BMDM and the FTL protein reduction observed in wildtype BMDM upon co-culture was even reversed in LCN-2-deficient BMDM (Fig. 6B). To test, if these iron-marker profiles were functionally effective, we determined the iron release of BMDM upon co-culture. In analogy to the human system, supernatants of BMDM contained higher iron amounts after tumor cell co-culture than naïve BMDM (Fig. 6C, open bars). In contrast, LCN-2−/−-BMDM released less iron upon co-culture compared to naïve BMDM (Fig. 6C, solid bars). Thus, LCN-2-knockout MΦ did not adopt an iron-release phenotype upon contact with tumor cells in vitro. Corroborating the role of LCN-2 as high-affinity iron-binding protein delivering iron from macrophages to the tumor microenvironment. Surprisingly, the important iron-exporter FPN appeared to be dispensable in the macrophage-tumor interplay, whereas loss of LCN-2 not only attenuated iron-release from macrophages, but also the associated tumor cell proliferation.
Figure 6.
LCN-2−/− attenuates the iron-release phenotype in TAM. (A) mRNA expression of iron-regulated genes (Fpn, Lcn-2, Hamp, Ftl) in wildtype (WT) (left panel) or Lcn-2−/− (right panel) BMDM after co-culture with E0771 cells (solid bars) was analyzed by qPCR and normalized to Tbp expression, relative to the respective expression in naïve control MΦ (ctrl, open bar) (n = 9–18). (B) Protein expression of FPN (surface, left panel) and FTL (intracellular, right panel) in Lcn-2−/− (solid bars) and WT BMDM (open bars) was determined by flow cytometry (MFI, mean fluorescence) (n = 3). (C) Iron amount in the supernatant of WT (open bars) or LCN-2−/− (solid bars) BMDM after co-culture was quantified by AAS and normalized to supernatants of naïve, sictrl transfected MΦ (n = 8–14). (D) Tumor burden at the day of death of wildtype (WT) (n = 14) or LCN-2−/− (n = 11) mice. P-value was calculated using two-tailed Student's t test. (E) Sections from wildtype (WT) and LCN-2−/− PyMT tumors were stained for Ki-67 with the Opal dye technique. DAPI was used as nuclear stain. Scale bar is 100 µm. Representative pictures are shown. Quantification was done using the Inform software (n = 3). (F) Infiltration of MΦ subpopulations (infiltrating and resident MΦ) as well as total amount of F4/80+ TAM into wildtype (WT) and LCN-2−/− PyMT tumors was detected via flow cytometry (n = 15). Data are means ± SEM from independent experiments, *p < 0.05, **p < 0.01, ***p < 0.001 vs. the respective controls unless indicated otherwise.
LCN-2 is predominantly expressed in TAM and contributes to enhanced tumor growth
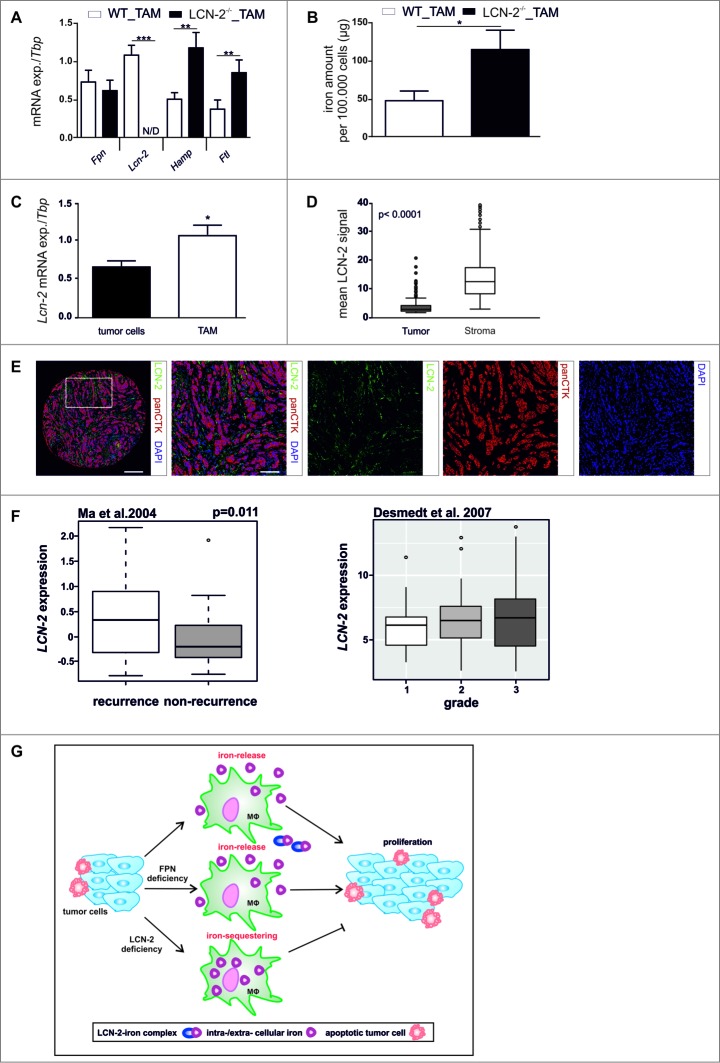
To assess, the in vivo relevance of our observations, we determined tumor cell proliferation in wildtype and LCN-2-deficient PyMT mice. Corroborating in vitro findings, the tumor burden (Fig. 6D) and number of proliferating, i.e. Ki67-positive cells (Fig. 6E), was strongly reduced in tumors of LCN-2 knockout mice as compared to wildtype mice. To rule out that changes in tumor cell proliferation were due to an altered composition of the immune infiltrate rather than differences in MΦ function, we analyzed various myeloid cell types within PyMT tumors by flow cytometry. The infiltration of monocytes, resident MΦ, and TAM was unaltered in LCN-2-deficient as compared to wildtype mice (Fig. 6F). To test if LCN-2 is similarly important for the iron-release phenotype of TAM in vivo, we sorted TAM from tumor-bearing wildtype and LCN-2−/− PyMT mice. LCN-2-deficient TAM expressed similar levels of Fpn mRNA, while the mRNA expression of iron sequestration markers Hamp and Ftl was significantly elevated compared to TAM isolated from wildtype tumors (Fig. 7A). Accordingly, the intracellular iron amount was significantly increased in LCN-2−/−-TAM (Fig. 7B, solid bars) compared to wildtype TAM (Fig. 7B, open bars), suggesting that more iron was retained in LCN-2-deficient TAM.
Figure 7.
LCN-2 is predominantly expressed in TAM and enhances tumor progression. (A) mRNA expression of iron-regulated genes (Fpn, Lcn-2, Hamp, Ftl) in WT (open bars) or LCN-2−/− (solid bars) TAM isolated from tumors of PyMT mice was analyzed by qPCR and normalized to Tbp expression (n = 5–9). (B) Iron amount in the lysates of WT (open bar) and LCN-2−/− (solid bar) TAM was quantified by AAS relative to the cell number (n = 5–7). (C) mRNA expression of Lcn-2 in tumor cells (solid bars) and TAM (open bars) isolated from tumors of WT PyMT mice was analyzed by qPCR and normalized to Tbp expression. Data are means ± SEM from independent experiments, *p < 0.05 vs. controls unless indicated otherwise (n = 6–11). (D + E) Primary human breast tumor samples were stained with antibodies against panCTK and LCN-2 with the Opal dye technique. DAPI was used as nuclear stain. (D) Quantitative comparison of the expression of LCN-2 in the tumor (solid box) and the stromal compartment (open box) (n = 198). (E) Representative picture of the costainings (scale bar = 200 µm for the core overview or 100 µm for close ups). (F) Meta-analysis of the correlation of recurrence and tumor grade with LCN-2 expression in breast cancer patients. Data are presented as box- and –whisker plots of LCN-2 gene expression in breast tumors. White and gray rectangles represent interquartile range, line in the middle of each rectangle represents the median value. Lines extending from the interquartile range mark the 5th and 95th percentile values, and the individual open circles represent values that are above the 95th percentile for each distribution. (G) Schematic representation of the impact of LCN-2 on the crosstalk of tumor cells and MΦ in terms of iron. Data are means ± SEM from independent experiments, *p < 0.05, **p < 0.01, ***p < 0.001 vs. the respective controls unless indicated otherwise.
As LCN-2 was depleted in all cells in this animal model, we next analyzed LCN-2 expression in sorted tumor cells and TAM from tumors of wildtype PyMT mice to determine the prime source of LCN-2 in this context. In line with previous observations,15,16,22 TAM expressed higher levels of Lcn-2 mRNA than the tumor cells (Fig. 7C). To gain further evidence for the differential distribution of LCN-2 within the tumor context and to further translate our findings to the human situation, we determined LCN-2 protein expression using immunohistochemistry in human breast tumor samples employing the Vectra system. In fact, LCN-2 protein was predominantly expressed in the stromal compartment of the tumors (Fig. 7D,E), supporting the concept that stroma-derived LCN-2 might serve as an iron source for tumors in vivo. We next aimed to determine if LCN-2 expression holds predictive power for breast cancer patients. To this end, we performed meta analyses of available data sets23,24 LCN-2 expression positively correlated both with the rate of recurrence in breast cancer patients as well as with the histological grade (Fig. 7F). Considering the skewed distribution of LCN-2 expression towards TAM in the tumor, these findings underline the potential of LCN-2 expression in the stroma as a novel prognostic marker for breast cancer patients.
In summary, we propose that TAM support tumor growth by supplying the latter with iron via an LCN-2-mediated, FPN-independent transport (Fig. 7G). LCN-2 in the tumor stroma therefore might be a promising target for novel therapeutic approaches aiming at restricting the iron-availability of tumors.
Discussion
In the present study, we present evidence for an iron-release phenotype of TAM contributing to tumor cell proliferation. As LCN-2 was primarily found in the stroma of primary human breast cancer samples as well, and LCN-2-deficiency limits tumor cell proliferation in vivo, LCN-2 in TAM might be a promising novel tumor therapeutic target.
While the importance of MΦ for tumor development and progression is widely appreciated, and the elevated iron-demand of tumors is also well established, a potential connection between TAM and the tumor iron supply remains largely elusive. Our observation that TAM release iron into the tumor microenvironment via an LCN-2 mediated mechanism therefore provides first insights into the role of TAM in the tumor iron management. Previously, LCN-2 expression in tumors was correlated with poor prognosis and suggested to support tumor development and metastatic spread,25,26 but the effect of MΦ-secreted LCN-2 and its iron-load during tumor progression was not investigated so far. First studies suggested that LCN-2 positive staining increases in advanced breast cancer stages.21,26 Along these lines, we previously showed that MΦ-derived LCN-2 supports metastasis.16 Our present data indicate that TAM are an important source of iron in the tumor microenvironment. Moreover, we found that TAM release iron bound to the iron transporting protein LCN-2 into the tumor microenvironment. The observation that MΦ acquire an iron-release phenotype corroborates previous reports suggesting an iron-release phenotype in alternatively activated MΦ.4,5 Interestingly, depletion of the established iron exporter FPN in TAM neither affected the iron-release from TAM nor did it attenuate enhanced proliferation of MCF-7 tumor cells upon stimulation with MΦ supernatants. In contrast, LCN-2 knockdown abolished iron release and proliferation. Thus, LCN-2 emerges as a major iron transporter in this context. In line with our findings, it was previously suggested that iron sequestration in LCN-2−/− MΦ ensures host defense against salmonella typhimurium infection by controlling MΦ iron homeostasis.27 Of note, depletion of LCN-2 also markedly reduced FPN expression, suggesting that LCN-2 and FPN might act in concert to export iron from TAM. We speculate that the depletion of LCN-2 alters iron-dependent gene expression and/or the labile iron pool, which might result in a negative regulation of FPN and/or its degradation. However, further studies are needed to determine the exact mechanistic links and/or interplay between these iron export pathways in TAM.
Tumors demand an excess of iron, both during early steps of tumor development, e.g. for enhanced survival28 and proliferation of transformed cells,29 but also during metastasis to facilitate remodeling of the extracellular matrix, and to increase motility and invasion of cancer cells.30 Our findings that MΦ actively release iron in the tumor context suggest a central role of MΦ in regulating tumor iron homeostasis. Along the same lines, high expression of a variety of iron-regulated genes such as HAMP and FTL correlated with poor prognosis, a higher tumor grade, and increased chemoresistance.31 Furthermore, clinical studies inversely correlated the expression of FPN in human breast tumors with patient survival and disease outcome.11 Interestingly, in our setting FPN depletion did not affect the iron-release from MΦ and consequently did not alter the pro-tumorigenic properties of MΦ supernatants. Similarly, it was recently suggested that MΦ release ferritin,32 thereby supporting tumor development. Moreover, enhanced ferritin expression was found in the tumor stroma.9 Since we did not observe elevated ferritin levels in TAM and taking into account that LCN-2 in tumor patients correlated with bad prognosis, we suggest that the high-affinity iron-binding protein LCN-233 serves as an alternative iron transporter, supplying tumor cells with iron. However, in the present study we observed reduced ferritin levels only at mRNA level, whereas secretion was not checked. Furthermore, the iron-handling properties of different MΦ subpopulations, i.e. infiltrating vs. resident MΦ indicated that ferritin expression was not elevated in the PyMT breast cancer model. However, it might well be that ferritin secretion accounts for enhanced iron release in a particular subpopulation, i.e. CD163+ MΦ as described previously for human tumors.9 The iron-loading status of LCN-2 has previously been shown to be decisive for its biological activity, i.e., iron-loaded LCN-2 favors cell survival and proliferation by enhancing the intracellular iron concentration and the induction of protective Bcl-2. However, internalization of iron-free LCN-2 promotes cell death by LCN-2-dependent intracellular iron deprivation and the concomitant increase of the pro-apoptotic Bim.34
In summary, we propose that TAM-derived LCN-2 serves as a crucial iron transporter in breast cancer, thereby providing an additional iron source for tumor cells. So far, most iron-targeting therapeutic approaches focused on modulating the aberrant iron homeostasis directly in tumor cells, largely ignoring the impact of infiltrating stromal cells. Our study underscores the significance of stroma-derived iron for tumor progression. Taking the previously proposed negative prognostic properties of LCN-2 for breast cancer patients and the herein characterized iron-supply function of LCN-2 in the tumor context into account, targeting LCN-2 in TAM might open a novel therapeutic approach to limit iron availability, specifically for tumors, without affecting classical iron transport routes.
Materials and methods
Materials
If not indicated otherwise, all chemicals were purchased from Sigma Aldrich.
Primary MΦ generation
Human monocytes were isolated from commercially available buffy coats (DRK-Blutspendedienst Baden-Württemberg-Hessen, Frankfurt, Germany) using Ficoll-Hypaque gradients (PAA Laboratories). Monocytes were differentiated into primary human MΦ with RPMI 1640 containing 5% AB-positive human serum (DRK-Blutspendedienst) for 7 days and achieved approximately 80% confluence. 24 h prior to stimulation, cells were serum-starved. Conditioned media of polarized MΦ were collected, centrifuged at 1000 x g for 5 min at 4°C, and aliquots were stored at -80°C until further use. Supernatant of unstimulated MΦ served as control.
Generation of murine BMDM
Murine BMDM were generated by isolating the bone marrow of wildtype and LCN-2−/− mice. Cells were differentiated directly in 6-well plates (6 × 106 cells/well) in the presence of 10 ng/ml MΦ colony-stimulating factor (GM-CSF) (Peprotech) for up to 7 days. At day 3, fresh GM-CSF was added.
Tumor cell culture
The human breast cancer cell lines MCF-7, MDA-MB-231, and T47D as well as the murine breast cancer cell line E0771 were cultured in Dulbecco's modified Eagle medium (DMEM) with high glucose (Life Technologies), supplemented with 100 U/ml penicillin (PAA), 100 µg/ml streptomycin (PAA), and 10% fetal calf serum (FCS; PAA). Tumor cells were cultivated in a humidified atmosphere with 5% CO2 at 37°C and passaged 3 times per week. 24 h prior to stimulation, cells were serum starved. Each cell line was routinely tested for mycoplasma. 2´2 DPD was used in a 100 mM concentration.
Co-culture of primary MΦ with breast cancer cells
MΦ were isolated and differentiated as described above. For the co-culture, tumor cells were harvested by trypsin, washed once with PBS and suspended in MΦ medium. MΦ and cancer cells were cultured in a 1:1 ratio for 48 h in MΦ medium in a humidified atmosphere of 5% CO2 at 37°C. At the end of the co-culture, conditioned media was harvested and stored for further use at -80°C. The remaining tumor cells were removed by applying accutase and MΦ were processed for further analysis.
Small interfering RNA (siRNA) transfections
GenMute® transfection reagent (SignaGen) was used according to the manufacturer's instructions. 50 nM LCN-2 siRNA or 50 nM FPN siRNA (Qiagen) was transfected into primary MΦ. A non-targeting, scrambled siRNA (sictrl) was used as control.
Proliferation assay
Proliferation of human MCF-7 breast cancer cells was measured using the RTCA DP xCELLigence instrument (OLS) as described previously.15 Data are presented as the slope per hour (slope 1/h) of the normalized cell index as a measure for the time-dependent changes in impedance. To determine the proliferation marker proliferating-cell-nuclear-antigen (PCNA), MCF-7 cells were treated for 24 h with MΦ-supernatant (MΦ-SN) and gene expression was measured by qPCR.
RNA extraction and quantitative real-time PCR (qPCR)
RNA was extracted using peqGold RNAPure reagent (Peqlab). Total RNA (1 µg) was transcribed using the Maxima first-strand cDNA synthesis kit (Fermentas). qPCR was performed using the MyIQ real-time PCR system (BioRad) and Absolute Blue qPCR SYBR green fluorescein mix (Thermo Fisher). qPCR results were quantified using the Bio-Rad CFX Manager (version 3.1) software program from Bio-Rad, with 18 S mRNA expression as an internal housekeeping gene control for human samples and Tbp for murine samples. All primers were bought from Biomers, except the human LCN-2 primer, which was bought from Qiagen. Changes were determined relative to the respective expression in naïve control MΦ (ctrl).
Atomic absorption spectrometry
The iron content bound to proteins in the supernatant and the whole iron amount in the lysates was determined by graphite furnace atomic-absorption-spectrometry. Therefore, supernatants were concentrated using Amicon® Ultra-4 centrifugal filter devices (Merck Millipore). Samples were measured as triplicate with a PinAAcle™ 900 T atomic-absorption-spectrometer (PerkinElmer). A wavelength of 248.33 nm and a slit width of 0.2 mm were used as spectrometer parameters. A hollow cathode iron lamp (30 mA maximum operating current) was run at 100% maximum current. The calibration solutions (10 µg/l to 90 µg/l) were prepared by adequate dilution of iron standard for AAS (Sigma-Aldrich) stock solution. A pyrolysis temperature of 1400°C and an atomization temperature of 2100°C were used. The iron amount was quantified relative to the protein content and normalized to naïve control MΦ (ctrl), if not other indicated.
Generation of recombinant LCN-2
Recombinant human LCN-2 was produced by transformation of E.coli with a pGEX-4 T-3-NGAL plasmid as already described.16 In order to test efficient LCN-2-catechol-iron complex formation, UV-visible spectroscopy (UV-vis) was used. Therefore, 10 µM LCN-2 were incubated with 10 µM catechol (Sigma) and 10 µM iron (Sigma).
ELISA
Detection of secreted LCN-2 was performed as previously described.16 LCN-2 content was calculated by the amount per mg of total protein (ng/mg protein). The anti-LCN-2 antibody was bought from R&D (MAB1857, MAB1757) and the secondary anti-rat IgG antibody was purchased from DAKO. Both Streptavidin-HRP and color reagent were from R&D.
LCN-2 immunoprecipitation
For immunoprecipitation (IP), primary human MΦ were co-cultured for 48 h with MCF-7 tumor cells. After removing the remaining tumor cells, MΦ were lysed on ice for 30 min in IP buffer (50 mM Tris-HCl pH 7.4, 300 mM NaCl, 5 mM EDTA, 1% NP-40, 1 mM PMSF, 1x protease and phosphatase inhibitor mix, 1 mM Na3VO4). Dynabeads (Thermo Fisher) were added and 1 mg protein was incubated overnight at 4°C in the presence of a specific antibody against LCN-2 (R&D, MAB1757). Beads were precipitated using the DynaMag-2 magnet (Thermo Fisher) and washed three times with IP buffer. Protein was eluted by addition of 2x loading buffer and incubated at 95°C for 5 min.
Animal studies
MMTV-PyMT breast cancer model
Wildtype and LCN-2−/− mice with or without crossing into a Polyoma-middle-T (PyMT) background were used. For genotyping, tail-tips were lysed with KAPA Genotyping lysis buffer (Peqlab) and the resulting DNA solutions were analyzed by PCR amplification using KAPA Hotstart Genotyping reaction mix (Peqlab). Tumor growth was monitored using sliding calipers until mice developed a maximal tumor diameter of 1.5 cm. After sacrificing mice, both the tumor and BMDM were isolated.
Flow cytometric analyses
For analysis of in vitro samples, MΦ were detached using accutase (PAA). Tumors from PyMT mice were dissociated using the Tumor Dissociation Kit (Miltenyi) and the GentleMACS system (Miltenyi). Samples were acquired with a LSRII/Fortessa flow cytometer (BD) expressed as mean fluorescence intensity (MFI). All antibodies and secondary reagents were titrated to determine optimal concentrations. CompBeads (BD) were used for single color compensation to create multi-color compensation matrices. For gating, fluorescence minus one (FMO) controls were used. The instrument calibration was controlled daily using Cytometer Setup and Tracking beads (BD).
For extracellular staining of iron regulated proteins in murine tumor lysates an antibody cocktail containing CD45 Vioblue (Miltenyi, 130–102–430), F4/80 PE-Cy7 (Biolegend, 123114), CD11b BV605 (Biolegend, 101257), CD11c AlexaFluor 700 (BD, 560583), CD326 BV711 (BD, 563134), and FPN PE (Novus, NBP1-21502PE) was used. Subsequently, cells were fixed and permeabilized with Cytofix/Cytoperm (BD) for intracellular staining with an antibody against FTL (Abcam, 109373) in combination with an APC-labeled secondary antibody (ThermoFisher, A-10931). To analyze the immune cell infiltration in murine tumors, cells were stained with an antibody cocktail containing CD45 Vioblue (Miltenyi, 130–102–430), F4/80 PE-Cy7 (Biolegend, 123114), CD11b BV605 (Biolegend, 101257), CD11c AlexaFluor 700 (BD, 560583), MHCII APC (Miltenyi, 130–102–139), Ly6C PerCP-Cy5.5 (BD, 560525), and Ly6G APC-Cy7 (BD, 127624) was used.
FACS staining of primary human MΦ
For extracellular detection of FPN, a PE-labeled anti-FPN (Novus) antibody was used. For intracellular staining of FTL (Abcam), MΦ were fixed with Cytofix/Cytoperm buffer (BD) for a maximum of 10 min on ice, followed by washing and permeabilization with Perm/Wash buffer (BD). For detection of FTL, a secondary AlexaFluor 488-labeled antibody against rabbit (Immunotools) was used. To determine the polarization profile of primary human MΦ, an antibody cocktail containing CD11 c V450 (BD, 560369), CD14 APC-H (BD, 560180), HLA-DR PE-Cy7 (BD, 560651), CD80 APC (BioLegend, 305220), CD86 FITC (BD, 555657), CD206 PE-Cy5 (BioLegend, 321108), and CD45 PE (BD, 555483) was used.
FACS sorting
Tumor single-cell suspensions were stained with an antibody cocktail containing CD45 Vioblue (Miltenyi, 130–102–430), CD326 PE (Miltenyi, 130–102–265), F4/80 PE-Cy7 (BD, 123114), CD11b Alexa Fluor 700 (BD, 101257), and 7-AAD (BD, 559925). CD45−/CD326+ living tumor cells and F4/80+/CD326− TAM were sorted using a FACS Aria (BD). Cells were harvested for AAS or directly used for RNA isolation applying the RNeasy microKit (Qiagen). For conversion of mRNA to cDNA, the Sensiscript RT kit (Qiagen) was used.
Multiplex immunohistochemistry
Tumor sections from PyMT mice were subjected to antigen retrieval and stained with an antibody against Ki-67 (Abcam, ab15580) according to manufacturer´s instructions using the Opal™ 4-Color Fluorescent IHC Kit (Perkin Elmer). DAPI was used for nuclear visualization. As secondary antibodies, linked to horseradish peroxidase (HRP) an anti-rabbit (GE Healthcare, NA934-1 ML) antibody was used. Images were acquired using the Vectra automated imaging system and analysis was performed using Inform software.
MELC
MELC robot technology involved validated distinct hardware and software components as described earlier.35 Murine primary tumors were immediately frozen and used for frozen sections (4 µM). Before staining, sections were fixed wit 4% paraformaldehyde/PBS for 10 min. Cells were permeabilized with 0.1% Triton X-100/PBS for 10 min and incubated with 3% BSA/PBS for 60 min. The coverslip was placed onto a customized sample holder (2-mm thickness, inner diameter of 16 mm) and fixed with an adapted silicon ring (outer diameter 2 cm, inner diameter 1 cm) that also served as antibody incubation chamber. The sample was positioned onto an inverted fluorescence microscope (Leica DM IRE2; 63 oil objective lens). By a robotic pipetting process, the specimen was incubated with fluorophore-labeled antibodies and wash solutions. Phase contrast and fluorescence signals were recorded, followed by a bleaching step (488 nm for FITC and 546 nm for phycoerythrin). A post-bleaching image was taken and subtracted from the image taken from the following epitope. Signals were validated using conventional immunehistochemistry. Recording and processing of all image data and the coordination of all system components were fully automatically controlled by software developed by MelTec GmbH (Magdeburg, Germany).
Primary human tumor samples
4-µm sections of the recipient block were mounted onto slides, subjected to antigen retrieval, and stained with an antibody combination consisting of LCN-2 (R&D, MAB1757) and pan-cytokeratin (panCTK) (Abcam, ab7753) according to manufacturer´s instructions using the Opal™ 4-Color Fluorescent IHC Kit (Perkin-Elmer). DAPI was used for nuclear visualization. Horseradish peroxidase (HRP) coupled anti-mouse (GE Healthcare, NA931.1 ML) or anti-rat (GE Healthcare, NA935-1 ML) secondary antibodies was used for detection. Images were acquired using the Vectra automated imaging system and analysis was performed using ImageJ. Specifically, the background was substracted using the rolling circle function and the channels were demixed during this process (the rolling circle radius was set to at least the size of the largest object that is not part of the background which was determined a priori). Thresholds were set semi-automatically for the pre-processed images. Under- or overexposed images were discarded from further analysis. For quantitative analysis, the images were then converted to binary images and the signal distribution was determined for each color individually.
Study approval
Investigations were conducted in accordance with the ethical standards according to the Declaration of Helsinki and to the national and international guidelines and have been approved by the authors' institutional review board. All procedures involving mice followed the guidelines of the Hessian animal care and use committee. Primary human tumor sections were prepared from representative areas of 198 invasive, paraffin-embedded tumors from premenopausal patients with lymph-node negative breast cancer, with the approval of the research ethics committee of Lund University (Regionala etikprövningsnämnden, LU 240–01 and Drn 2016/789).
Meta analyses of LCN-2 expression with breast cancer grade and recurrence
Correlation between LCN-2 expression in primary human breast tumors and given data were assessed with gene expression profiles from accessible microarray data sets. To this end, we used the following studies from Ma et al. (GSE1379),36 and Desmedt et al. (GSE7390).24 Statistical analysis were performed with GNU R version 3.3.2.37 The significance levels were assessed with the Welch two-sample t-test.
Statistical analysis
Statistical analysis was performed using Prism software (GraphPadSoftware Inc.). Values for all measurements were expressed as means ± SEM. Each experiment was performed at least three times (independent experiments using three technical replicates). Data were analyzed using One-way ANOVA. Patient samples were statistical analyzed using the Mann-Whitney-U test. P values were considered significant at *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary Material
Funding Statement
This work was supported by the Fritz Thyssen Stiftung under Grant Az.10.12.2.156, awarded to MJ, Goethe-University (Faculty of Medicine young researcher award, to MJ), Doktor Robert Pfleger Stiftung (awarded to MJ), and the Deutsche Krebshilfe under Grant 111578 to BB.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Ute Bahr (Institute of Pharmaceutical Chemistry, Goethe-University Frankfurt) for the technical assistance using AAS and Christine von Hayn for technical assistance with qPCR analysis. We also thank Dr. Klaus Zwicker for assisting in performing UV-visible spectra measurement and Jack B. Cowland (National University Hospital, University of Copenhagen, Denmark) for kindly providing the Lcn2−/− mice.
Funding details
This work was supported by the Fritz Thyssen Stiftung under Grant Az.10.12.2.156, awarded to MJ, Goethe-University (Faculty of Medicine young researcher award, to MJ), Doktor Robert Pfleger Stiftung (awarded to MJ), and the Deutsche Krebshilfe under Grant 111578 to BB.
Author contributions
The authors contributed in the following way: CM: data acquisition, analysis, and interpretation, and writing of the manuscript; JM: data analysis and interpretation; BÖ: data acquisition; SG: data analysis and interpretation; AW, SW: data interpretation; TS, BB: data interpretation and writing of the manuscript; MJ: study design and supervision, data acquisition and analysis, and writing of the manuscript. All authors read and agreed on the final manuscript.
References
- 1.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nature reviews Immunology. 2008;8:958–69. doi: 10.1038/nri2448. PMID:19029990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends in immunology. 2002;23:549–55. doi: 10.1016/S1471-4906(02)02302-5. PMID:12401408. [DOI] [PubMed] [Google Scholar]
- 3.Cairo G, Recalcati S, Mantovani A, Locati M. Iron trafficking and metabolism in macrophages: contribution to the polarized phenotype. Trends in immunology. 2011;32:241–7. doi: 10.1016/j.it.2011.03.007. PMID:21514223. [DOI] [PubMed] [Google Scholar]
- 4.Recalcati S, Locati M, Marini A, Santambrogio P, Zaninotto F, De Pizzol M, Zammataro L, Girelli D, Cairo G. Differential regulation of iron homeostasis during human macrophage polarized activation. Eur J Immunol. 2010;40:824–35. doi: 10.1002/eji.200939889. PMID:20039303. [DOI] [PubMed] [Google Scholar]
- 5.Mertens C, Akam EA, Rehwald C, Brune B, Tomat E, Jung M. Intracellular Iron Chelation Modulates the Macrophage Iron Phenotype with Consequences on Tumor Progression. Plos One. 2016;11:e0166164. doi: 10.1371/journal.pone.0166164. PMID:27806101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torti SV, Torti FM. Iron and cancer: more ore to be mined. Nature reviews Cancer. 2013;13:342–55. doi: 10.1038/nrc3495. PMID:23594855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller LD, Coffman LG, Chou JW, Black MA, Bergh J, D'Agostino R Jr., Torti SV, Torti FM. An iron regulatory gene signature predicts outcome in breast cancer. Cancer Res. 2011;71:6728–37. doi: 10.1158/0008-5472.CAN-11-1870. PMID:21875943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Habashy HO, Powe DG, Staka CM, Rakha EA, Ball G, Green AR, Aleskandarany M, Paish EC, Douglas Macmillan R, Nicholson RI, et al.. Transferrin receptor (CD71) is a marker of poor prognosis in breast cancer and can predict response to tamoxifen. Breast cancer research and treatment. 2010;119:283–93. doi: 10.1007/s10549-009-0345-x. PMID:19238537. [DOI] [PubMed] [Google Scholar]
- 9.Jezequel P, Campion L, Spyratos F, Loussouarn D, Campone M, Guerin-Charbonnel C, Joalland MP, Andre J, Descotes F, Grenot C, et al.. Validation of tumor-associated macrophage ferritin light chain as a prognostic biomarker in node-negative breast cancer tumors: A multicentric 2004 national PHRC study. International journal of cancer. 2012;131:426–37. doi: 10.1002/ijc.26397. PMID:21898387. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, Deng Z, Hatcher H, Miller LD, Di X, Tesfay L, Sui G, D'Agostino RB Jr., Torti FM, Torti SV. IRP2 regulates breast tumor growth. Cancer Res. 2014;74:497–507. doi: 10.1158/0008-5472.CAN-13-1224. PMID:24285726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinnix ZK, Miller LD, Wang W, D'Agostino R Jr., Kute T, Willingham MC, Hatcher H, Tesfay L, Sui G, Di X, et al.. Ferroportin and iron regulation in breast cancer progression and prognosis. Sci Transl Med. 2010;2:43ra56. doi: 10.1126/scitranslmed.3001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung M, Weigert A, Tausendschon M, Mora J, Oren B, Sola A, Hotter G, Muta T, Brune B. Interleukin-10-induced neutrophil gelatinase-associated lipocalin production in macrophages with consequences for tumor growth. Mol Cell Biol. 2012;32:3938–48. doi: 10.1128/MCB.00413-12. PMID:22851691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell. 2002;10:1033–43. doi: 10.1016/S1097-2765(02)00708-6. PMID:12453412. [DOI] [PubMed] [Google Scholar]
- 14.Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–21. doi: 10.1038/nature03104. PMID:15531878. [DOI] [PubMed] [Google Scholar]
- 15.Jung M, Oren B, Mora J, Mertens C, Dziumbla S, Popp R, Weigert A, Grossmann N, Fleming I, Brune B. Lipocalin 2 from macrophages stimulated by tumor cell-derived sphingosine 1-phosphate promotes lymphangiogenesis and tumor metastasis. Sci Signal. 2016;9:ra64. doi: 10.1126/scisignal.aaf3241. PMID:27353364. [DOI] [PubMed] [Google Scholar]
- 16.Oren B, Urosevic J, Mertens C, Mora J, Guiu M, Gomis RR, Weigert A, Schmid T, Grein S, Brune B, et al.. Tumour stroma-derived lipocalin-2 promotes breast cancer metastasis. The Journal of pathology. 2016;239:274–85. doi: 10.1002/path.4724. PMID:27038000. [DOI] [PubMed] [Google Scholar]
- 17.Yang J, McNeish B, Butterfield C, Moses MA. Lipocalin 2 is a novel regulator of angiogenesis in human breast cancer. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2013;27:45–50. doi: 10.1096/fj.12-211730. PMID:22982376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berger T, Cheung CC, Elia AJ, Mak TW. Disruption of the Lcn2 gene in mice suppresses primary mammary tumor formation but does not decrease lung metastasis. P Natl Acad Sci USA. 2010;107:2995–3000. doi: 10.1073/pnas.1000101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leng X, Ding T, Lin H, Wang Y, Hu L, Hu J, Feig B, Zhang W, Pusztai L, Symmans WF, et al.. Inhibition of lipocalin 2 impairs breast tumorigenesis and metastasis. Cancer Res. 2009;69:8579–84. doi: 10.1158/0008-5472.CAN-09-1934. PMID:19887608. [DOI] [PubMed] [Google Scholar]
- 20.Lin CW, Yang WE, Lee WJ, Hua KT, Hsieh FK, Hsiao M, Chen CC, Chow JM, Chen MK, Yang SF, et al.. Lipocalin 2 prevents oral cancer metastasis through carbonic anhydrase IX inhibition and is associated with favourable prognosis. Carcinogenesis. 2016;37:712–22. doi: 10.1093/carcin/bgw050. PMID:27207653. [DOI] [PubMed] [Google Scholar]
- 21.Bauer M, Eickhoff JC, Gould MN, Mundhenke C, Maass N, Friedl A. Neutrophil gelatinase-associated lipocalin (NGAL) is a predictor of poor prognosis in human primary breast cancer. Breast cancer research and treatment. 2008;108:389–97. doi: 10.1007/s10549-007-9619-3. PMID:17554627. [DOI] [PubMed] [Google Scholar]
- 22.Jung M, Sola A, Hughes J, Kluth DC, Vinuesa E, Vinas JL, Perez-Ladaga A, Hotter G. Infusion of IL-10-expressing cells protects against renal ischemia through induction of lipocalin-2. Kidney Int. 2012;81:969–82. doi: 10.1038/ki.2011.446. PMID:22278021. [DOI] [PubMed] [Google Scholar]
- 23.Ma CX, Luo J, Ellis MJ. Molecular profiling of triple negative breast cancer. Breast Disease. 2010;32:73–84. doi: 10.3233/BD-2010-0309. PMID:21778575. [DOI] [PubMed] [Google Scholar]
- 24.Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, Viale G, Delorenzi M, Zhang Y, d'Assignies MS, et al.. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:3207–14. doi: 10.1158/1078-0432.CCR-06-2765. PMID:17545524. [DOI] [PubMed] [Google Scholar]
- 25.Mannelqvist M, Stefansson IM, Wik E, Kusonmano K, Raeder MB, Oyan AM, Kalland KH, Moses MA, Salvesen HB, Akslen LA. Lipocalin 2 expression is associated with aggressive features of endometrial cancer. BMC Cancer. 2012;12:169. doi: 10.1186/1471-2407-12-169. PMID:22559235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Bielenberg DR, Rodig SJ, Doiron R, Clifton MC, Kung AL, Strong RK, Zurakowski D, Moses MA. Lipocalin 2 promotes breast cancer progression. P Natl Acad Sci USA. 2009;106:3913–8. doi: 10.1073/pnas.0810617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nairz M, Schroll A, Haschka D, Dichtl S, Sonnweber T, Theurl I, Theurl M, Lindner E, Demetz E, Asshoff M, et al.. Lipocalin-2 ensures host defense against Salmonella Typhimurium by controlling macrophage iron homeostasis and immune response. Eur J Immunol. 2015;45:3073–86. doi: 10.1002/eji.201545569. PMID:26332507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bauckman K, Haller E, Taran N, Rockfield S, Ruiz-Rivera A, Nanjundan M. Iron alters cell survival in a mitochondria-dependent pathway in ovarian cancer cells. The Biochemical journal. 2015;466:401–13. doi: 10.1042/BJ20140878. PMID:25697096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steegmann-Olmedillas JL. The role of iron in tumour cell proliferation. Clinical & translational oncology: official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2011;13:71–6. doi: 10.1007/s12094-011-0621-1. [DOI] [PubMed] [Google Scholar]
- 30.Fischer-Fodor E, Miklasova N, Berindan-Neagoe I, Saha B. Iron, inflammation and invasion of cancer cells. Clujul medical. 2015;88:272–7. doi: 10.15386/cjmed-492. PMID:26609256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, He K, Liu P, Xu LX. Iron participated in breast cancer chemoresistance by reinforcing IL-6 paracrine loop. Biochem Bioph Res Co. 2016;475:154–60. doi: 10.1016/j.bbrc.2016.05.064. [DOI] [PubMed] [Google Scholar]
- 32.Alkhateeb AA, Han B, Connor JR. Ferritin stimulates breast cancer cells through an iron-independent mechanism and is localized within tumor-associated macrophages. Breast cancer research and treatment. 2013;137:733–44. doi: 10.1007/s10549-012-2405-x. PMID:23306463. [DOI] [PubMed] [Google Scholar]
- 33.Bao G, Clifton M, Hoette TM, Mori K, Deng SX, Qiu A, Viltard M, Williams D, Paragas N, Leete T, et al.. Iron traffics in circulation bound to a siderocalin (Ngal)-catechol complex. Nature chemical biology. 2010;6:602–9. doi: 10.1038/nchembio.402. PMID:20581821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Devireddy LR, Gazin C, Zhu X, Green MR. A cell-surface receptor for lipocalin 24p3 selectively mediates apoptosis and iron uptake. Cell. 2005;123:1293–305. doi: 10.1016/j.cell.2005.10.027. PMID:16377569. [DOI] [PubMed] [Google Scholar]
- 35.Pierre S, Maeurer C, Coste O, Becker W, Schmidtko A, Holland S, Wittpoth C, Geisslinger G, Scholich K. Toponomics analysis of functional interactions of the ubiquitin ligase PAM (Protein Associated with Myc) during spinal nociceptive processing. Molecular & cellular proteomics: MCP. 2008;7:2475–85. doi: 10.1074/mcp.M800201-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, Muir B, Mohapatra G, Salunga R, Tuggle JT, et al.. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–16. doi: 10.1016/j.ccr.2004.05.015. PMID:15193263. [DOI] [PubMed] [Google Scholar]
- 37.Team RC. R: A language and environment for statistical compuing R foundation for statistical computing. Vienna, Austria; 2014. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.