ABSTRACT
Metastatic basal cell cancer (BCC) is an ultra-rare malignancy with no approved therapies beyond Hedgehog inhibitors. We characterized the genomics, tumor mutational burden (TMB), and anti-PD-1 therapy responses in patients with locally advanced or metastatic BCC. Overall, 2,039 diverse cancer samples that had undergone comprehensive genomic profiling (CGP) were reviewed. Eight patients with locally advanced/metastatic BCC were identified (two had two CGP analyses; total, 10 biopsies). Two tumors demonstrated PD-L1 amplification. Seven patients had >1 actionable alteration. The TMB (mutations/mb) (median (range)) was 90 (3-103) for the BCCs versus 4 (1-860) for 1637 cancers other than BCC (P < 0.0001). Median progression-free survival (PFS) for all four patients treated with PD-1 blockade was 10.7 months (range, 3.8 to 17.6+ months); three patients had an objective response. In conclusion, advanced/metastatic BCC often has biological features (high TMB; PD-L1 amplification) predictive of immunotherapy benefit, and patients frequently respond to PD-1 blockade.
KEYWORDS: Basal cell carcinoma, Immunotherapy, tumor mutational burden, PD-1, PD-L1, Checkpoint blockade, Biomarkers, Therapeutic antibodies
Introduction
Basal cell carcinoma is the most common malignancy with approximately 2.8 million cases diagnosed annually in the United States. Rarely, basal cell carcinomas can become locally destructive and/or metastasize.1 The development of metastatic basal cell carcinoma is highly dependent on the primary tumor size. Previous studies have shown that there is an approximately 2% incidence of metastasis for tumors larger than 3 cm in diameter. The incidence increases to 25% for tumors larger than 5 cm in diameter and is 50% for tumors larger than 10 cm in diameter. In addition, a greater than 90% metastasis rate has been observed for tumors that are greater than 25 cm2.2,3
Most basal cell carcinomas harbor mutations in the Hedgehog pathway.4 Currently, the Hedgehog inhibitors vismodegib and sonidegib are approved by the Food and Drug Administration (FDA) for treating locally advanced/metastatic basal cell carcinoma.5,6 However, there are no approved therapies for patients who progress on Hedgehog inhibitors. Furthermore, response rates to Hedgehog inhibitors are only 30–45%, with a median duration of response of approximately six months.5,6
Antibodies blocking programmed death receptor (PD-1)/programmed death receptor – ligand 1 (PD-L1) show efficacy in both solid tumors and lymphoma.7,8 Biomarkers for response include PD-L1 positivity by immunohistochemistry, microsatellite instability, and high tumor mutations burden (TMB).7,8 Interestingly, approximately 90% of basal cell carcinomas are positive for PD-L1 by immunohistochemistry,9 and the intensity of PD-L1 staining on basal cell carcinoma cells increases with the number of prior treatment modalities.9 Furthermore, basal cell carcinoma is one of the most mutated types of human cancer.10 Similar to melanoma, the majority of mutations identified in basal cell carcinoma have an ultraviolet (UV) signature. Therefore, advanced basal cell carcinomas have features (PD-L1 positivity and high TMB) that would predict checkpoint inhibitor response.
There are a few case reports of successful treatment of refractory basal cell carcinoma with anti-PD-1 therapy.1,11,12 Herein, we present the genomic correlates of the largest case series to date of patients with advanced/metastatic basal cell cancer treated with anti-PD-1 therapy.
Results
Patient Characteristics: Nine biopsies from eight patients with locally advanced or metastatic basal cell carcinoma and a calculated TMB were identified (Supplemental Figure 1). Patient characteristics are listed in Table 1 and Supplemental Table 1. Five patients had locally advanced disease while three patients had metastatic disease.
Table 1.
Patient profiles and treatment response.
| Patient | Age (years)1 | Sex | Locally advanced or metastatic disease | TMB (mutations/mb) | Number of characterized alterations | Potentially actionable alterations with either an on or off-label FDA approved drug | Genomic alterations2 | Best response/PFS (months) to a PD-1 inhibitor (agent) | Best Response/PFS (months) to a Hedgehog inhibitor (agent) | Disease status at biopsy |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 65 | M | Locally advanced (large lesion of right auricle with recurrent disease after multiple resections and radiation) | 96 | 8 | 6 | PTCH1 Q576* | PR/34.2+ (vismodegib) | Locally advanced disease | |
| ERBB4 D861N | ||||||||||
| CDKN2A p14ARF P105S | ||||||||||
| EPHA3 E930K | ||||||||||
| KEAP1 R169C | ||||||||||
| TP53 G266R | ||||||||||
| TP53 R196* | ||||||||||
| TP53 S33fs*10 | ||||||||||
| 2A3 | 56 | M | Metastatic | N/A | 10 | 5 | PTCH1 Q1366* | PR/17.6+ (nivoulmab) | PD/2.0 (vismodegib/paclitaxel) | Metastatic disease |
| PTCH1 W197* | Also received sonidegib/buparlisib with progression at 1.9 months | |||||||||
| CDKN2A P81L | ||||||||||
| TP53 P278S | ||||||||||
| CDKN1A R140Q | ||||||||||
| CTNNA1 R383H | ||||||||||
| LRP1B splice site 9121-1G>A | ||||||||||
| NOTCH1 W287* | ||||||||||
| SLIT2 K325* | ||||||||||
| SMARCA4 Q1166* | ||||||||||
| 2B3 | 103 | 19 | 11 | CD274 (PD-L1) amplification | Metastatic disease | |||||
| FLT1 E487K | ||||||||||
| JAK2 amplification | ||||||||||
| PDCD1LG2 (PD-L2) amplification | ||||||||||
| PDGFRA E459K | ||||||||||
| PIK3R2 Q412* | ||||||||||
| PTCH1 Q1366* | ||||||||||
| PTCH1 W197* | ||||||||||
| CDKN2A P81L | ||||||||||
| TP53 P278S | ||||||||||
| CDKN1A R140Q | ||||||||||
| CTNNA1 R383H | ||||||||||
| LRP1B splice site 9121-1G>A | ||||||||||
| LRP1B W2334* | ||||||||||
| MLL2 splice site 4132-1G>A | ||||||||||
| NOTCH1 W287* | ||||||||||
| SLIT2 K325* | ||||||||||
| SMARCA4 Q1166* | ||||||||||
| TERT promoter -139_-138CC>TT * | ||||||||||
| 3A3 | 53 | F | Metastatic | 90 | 6 | 2 | PTCH1 E684* | PR/3.8 (nivolumab) | PR/4.5 (vismodegib) | Locally advanced disease |
| TP53 R342* | ||||||||||
| LRP1B 2553* | ||||||||||
| WT1 S461F | ||||||||||
| KDM5A P325S | ||||||||||
| CREBBP H397fs*38 | ||||||||||
| 3B3 | 90 | 12 | 5 | PTCH1 E684* | Metastatic disease | |||||
| CD274 (PD-L1) amplification | ||||||||||
| JAK2 amplification – equivocal | ||||||||||
| PDCD1LG2 (PD-L2) amplification | ||||||||||
| TP53 R342* | ||||||||||
| CREBBP H397fs*38 | ||||||||||
| KDM5A P325S | ||||||||||
| LRP1B R2553* | ||||||||||
| STAG2 Q914* | ||||||||||
| TAF1 splice site 2119-1G>A | ||||||||||
| TERT promoter -138_-139CC>TT | ||||||||||
| WT1 S461F | ||||||||||
| 4 | 66 | M | Locally advanced (involving left auricle and left lower extremity) | 52 | 6 | 3 | PTCH1 Q889* | Locally advanced | ||
| GRM3 E49K | ||||||||||
| TP53 G245N | ||||||||||
| TP53 H179Y | ||||||||||
| NOTCH1 Q475* | ||||||||||
| NOTCH2 Q1870* | ||||||||||
| 5 | 62 | M | Locally advanced (unresectable 10 × 11 cm tumor located on back) | 53 | 12 | 6 | PTCH1 R770*-subclonal | CR/8.1+* (Nivolumab and vismodegib) | CR/8.1+* (Nivolumab and vismodegib) | Locally advanced disease |
| PTCH1 spice cite 1504-1G>T | ||||||||||
| PTEN splice cite 210–2A>T | ||||||||||
| ASXL1 Q760* | ||||||||||
| INPP4B W521* | ||||||||||
| KEL 130Q | ||||||||||
| PIK3R1 R534* | ||||||||||
| RAC1 P29S | ||||||||||
| TERT promoter-124C>T | ||||||||||
| TP53 Q100* | ||||||||||
| TP53 R196* | ||||||||||
| WT1 C350R | ||||||||||
| 6 | 69 | M | Locally advanced (lesion involving the right neck/submental area with recurrent disease following multiple surgeries and radiation) | 3 | 2 | 0 | TET2 F1287fs*76 | PR/12.2 (vismodegib) | Locally advanced | |
| GLI1 A670S4 | ||||||||||
| 7 | 61 | M | Locally advanced (large nodular lesion involving the nose with patient refusing surgery and radiation) | 20 | 8 | 3 | SMO W535L | PR/9.2 (vismodegib) | Locally advanced | |
| KDR G1145E | ||||||||||
| ARID1A Q1894* | ||||||||||
| MLL2 S3463fs*39 | ||||||||||
| PIK3R1 R534* | ||||||||||
| RUNX1 S100F | ||||||||||
| SPTA E638K | ||||||||||
| TERT promoter-146C>T | ||||||||||
| 8 | 50 | F | Metastatic | 102 | 10 | 5 | PTCH1 G854* | PD/2.5 (pembrolizumab) | PR/11.1 (vismodegib) | Metastatic |
| PTCH1 splice cite 2560+1G>A | ||||||||||
| TSC1 loss exon 9–23 | ||||||||||
| GRIN2A S929F | ||||||||||
| MAGI2 W688* | ||||||||||
| NOTCH2 R1838* | ||||||||||
| RBM10 splice cite 633+1G>A | ||||||||||
| TERT promoter-146C>T | ||||||||||
| TP53 Q136* | ||||||||||
| TP53 R213* |
Patient's age is at the time of locally advanced/metastatic disease.
Alterations in bold are considered potentially actionable by either an on- or off-label FDA approved drug.
Patients 2 and 3 each had multiple different biopsies sent for next generation sequencing. Patient 2 has been previously reported1.
The variant GLI1 p.A670S is common in healthy people from European origin (1/333 individuals – 1000 Genomes database) and is considered neutral by several algorithms (SIFT, Provean, Polyphen-2). However, these algorithms only consider the similarities between amino acids (A and S are both polar uncharged amino acids). The addition of a serine residue within Gli1 sequence creates an additional phosphorylation site, and Gli1 is exclusively regulated by phosphorylation.
The new phosphorylation site created by the A670S variant is not depending on PKA, and therefore may lead to the activation of the Hedgehog pathway. This variant might be pathogenic in the context of basal cell carcinoma.
Patient 5 received the combination of nivolumab and vismodegib.
All three patients who received immunotherapy as monotherapy received immunotherapy after treatment with a hedgehog inhibitor.
Genomics: The median (range) number of genomic alterations (characterized alterations, excludes variants of unknown significance) found per patient biopsy was 9 (2-19). The most common alterations identified were in the TP53, PTCH1, and TERT genes (Fig. 1). All patients had ≥1 genomic alterations. Amplification of CD274 (PD-L1), PDCD1LG2 (PD-L2), and JAK2 (chromosome 9p24.1 amplification) was identified in two patients. The median (range) number of potentially actionable alterations with either an on- or off -label FDA approved drugs was 5 (0-11); seven patients had ≥1 such potentially actionable alterations (Supplemental Figures 2 and 3). Excluding PTCH1 and SMO alterations, seven patients had one or more actionable alterations with an off-label approved drug (median (range) = 3 (0-9)).
Figure 1.
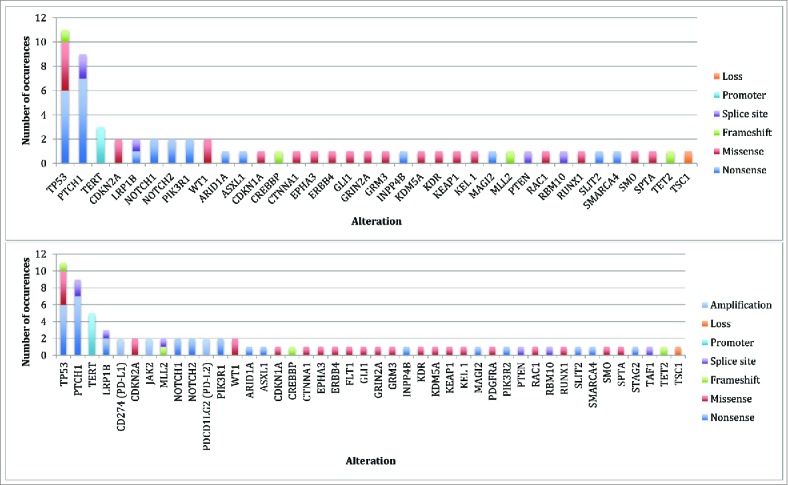
Genomic alterations identified* Top Panel: Total alterations (N = 62) identified by NGS on initial biopsy (N = 8 biopsies). Bottom Panel: Total alterations (N = 77) identified by NGS on initial and subsequent biopsies (N = 10 biopsies). *Some patients had multiple alterations in the same gene (i.e. TP53 and PTCH1).
TMB: Nine biopsies from eight patients had TMB calculated. The median TMB (mutations/mb) (range) for nine basal cell carcinoma samples was 90 (3-103) while the median TMB (range) for 1637 samples with cancers other than basal cell carcinoma was 4 (1-860) (P < 0.0001) (Supplemental Table 1 and Figure 2). Of the eight patients evaluable for TMB, seven had a high TMB (≥ 20 mutations/megabase); six of these patients had a very high TMB (≥ 50 mutations/megabase).
Figure 2.
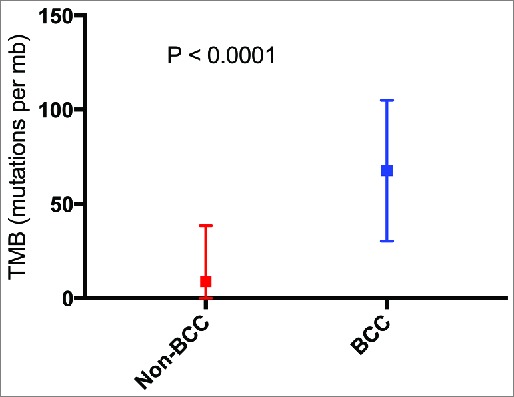
Mean tumor mutational burden for cancers other than basal cell carcinoma (N = 1,637) vs. basal cell carcinoma (N = 9 biopsies with available data). P value calculated using Mann Whitney U test. Squares represent mean TMB. Bars represent the standard deviation of the mean. Abbreviations: BCC = basal cell carcinoma; mb = megabase; TMB = tumor mutational burden.
Treatment with PD-1 blockade (Table 1): Four patients were treated with PD-1 blockade. Three of these patients had had tumor progression on ≥1 Hedgehog inhibitor while one patient received a combination of nivolumab and vismodegib.
Patient 2, with metastatic disease and a chromosome 9p24.1 amplification, has an ongoing near complete response (CR) on nivolumab (17.6+ months duration). Patient 3, also with metastatic disease and a chromosome 9p24.1 amplification, had a partial response (PR) of 4.2 months duration on nivolumab. Patient 5, with locally advanced disease (10 × 11 cm unresectable tumor), has an ongoing CR of 8.1+ months duration on the combination of nivolumab and vismodegib. Patient 8, with metastatic disease, had progressive disease on pembrolizumab after 2.5 months. The median PFS for all four patients treated with PD-1 blockade was 10.7 months. For the 7 patients treated with a Hedgehog inhibitor, the median PFS was 11.1 months (P = 0.91) (Supplemental Figure 4).
Discussion
We demonstrate that the majority of patients with advanced and/or metastatic basal cell carcinoma have numerous genomic alterations that are potentially targetable by both on- and off-label FDA approved drugs. Importantly, seven of eight patients (84%) had a high TMB, which can be a marker of response to immunotherapy.13 Further, two of the patients had amplification of PD-L1, which is the hallmark of Hodgkin lymphoma, known to be exquisitely responsive to checkpoint inhibitors in Hodgkin lymphoma.14 Finally, we present four patients with locally advanced/metastatic basal cell carcinoma treated with PD-1 blockade (seven of whom had previously failed ≥1 Hedgehog inhibitors, while one patient with an ongoing CR received concomitant vismodegib and nivolumab). The response rate in our cohort was 75% (3 of 4 patients), with a median PFS of 10.7 months (range, 3.8 to 17.6+ months).
TMB, measured by CGP has been shown to correlate with response to PD-1/PD-L1 blockade in patients with melanoma15 and urothelial carcinoma.16 Herein, TMB reveals that advanced basal cell carcinoma has an extremely high mutational burden. Indeed, six patients had a TMB of ≥50 mutations/megabase (≥20 mutations/megabase is considered high). Prior studies have also demonstrated a high TMB and ultraviolet (UV) signature in basal cell carcinoma (Supplemental Table 3).10,17 This observation suggests that, like melanoma, basal cell carcinoma has a marker associated with a high likelihood of responding to checkpoint blockade.
Interestingly, two patients had amplification of chromosome 9p24.1, the region containing PD-L1, PD-L2, and JAK2. Chromosome 9p24.1 copy number alterations are a disease-defining feature in Hodgkin lymphoma, with 97% of patients having 9p24.1 copy number alterations.18 Heavily-pretreated Hodgkin lymphoma demonstrates response rates as high as 87% to PD-1 blockade.14
Patients with advanced/metastatic basal cell carcinoma have a poor prognosis after progressing on Hedgehog inhibitor therapy, with limited therapeutic options. Our data presents a biologic rational and clinical data that support the use of checkpoint blockade with PD-1 inhibitors in these patients. Larger clinical trials are needed to confirm these findings.
Methods
Patient selection: We evaluated 2,039 cancer samples that underwent comprehensive genomic profiling (CGP). Only patients with a diagnosis of locally advanced/metastatic basal cell carcinoma confirmed by a dermatopathologist (PRC) are presented. This study was performed in accordance with UCSD Institutional Review Board guidelines for the PREDICT study (NCT02478931) and for any investigational treatments for which patients gave consent in accordance with the Declaration of Helsinki
Next Generation Sequencing and Assessment of Tumor Mutational Burden (TMB): Formalin-fixed paraffin embedded tumor samples were submitted for CGP to Foundation Medicine (N = 2039 patients) (clinical laboratory improvement amendments (CLIA)-certified lab): the FoundationOne (hybrid-capture-based CGP; 182, 236 or 315 genes, depending on the time period). (http://www.foundationone.com/).19
For TMB (mutations per megabase (mb)), the number of somatic mutations detected on NGS (interrogating 1.2 mb of the genome) were quantified and that value extrapolated to the whole exome using a validated algorithm.16 Alterations likely or known to be bona fide oncogenic drivers and germline polymorphisms were excluded.
Definition of actionable alteration: An alteration was defined as potentially actionable if its protein product is a component of a molecularly defined pathway for which there is at least one available FDA-approved drug that may impact the function of the protein product of the alteration or the immediate downstream effectors of the protein product.
Statistical Analysis and Outcome Evaluation: Mann Whitney U test was used to assess continuous variables. Responses were assessed based on physician notation; physicians used RECIST criteria. Progression-free survival (PFS) was calculated by the method of Kaplan and Meier (P values by log-rank (Mantel-Cox) test). For patients who received multiple treatment regimens, the treatment with the longest PFS was chosen for analysis. Patients were censored at date of last follow up for PFS if they had not progressed. Statistical analyses were performed using Graph-Pad Prism version 7.0 (San Diego, CA, USA).
Supplementary Material
Funding Statement
Funded in part by National Cancer Institute grant P30 CA016672 and the Joan and Irwin Jacobs Fund philanthropic fund.
Abbreviations
- CR
complete response
- FDA
Food and Drug Administration
- f
female
- m
male
- mb
megabase
- N/A
not available
- PD
progressive disease
- PFS
progression free survival
- PR
partial response
- TMB
tumor mutational burden
Conflict of interest
Dr. Frampton, Dr. Stephens, and Dr. Miller are employees and equity holders of Foundation Medicine. Dr. Kurzrock receives research funding from Genentech, Merck, Serono, Pfizer, Sequenom, Foundation Medicine, and Guardant, as well as consultant fees from X Biotech, and Actuate Therapeutics and has an ownership interest in Curematch Inc.
References
- 1.Ikeda S, Goodman AM, Cohen PR, Jensen TJ, Ellison CK, Frampton G, Miller V, Patel SP, Kurzrock R. Metastatic basal cell carcinoma with amplification of PD-L1: exceptional response to anti-PD1 therapy. Npj Genomic Med. 2016;1:16037. doi: 10.1038/npjgenmed.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uzquiano MC, Prieto VG, Nash JW, Ivan DS, Gong Y, Lazar AJ, Diwan AH. Metastatic basal cell carcinoma exhibits reduced actin expression. Mod Pathol. 2008;21(5):540–3. doi: 10.1038/modpathol.3801051. PMID:18223552. [DOI] [PubMed] [Google Scholar]
- 3.Vu A, Laub D. Metastatic Basal Cell Carcinoma: A Case Report and Review of the Literature. Eplasty. 2011;11,5(Suppl 1): S26–S29. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3088016/. Accessed November4, 2017. PMID:21559224 [PMC free article] [PubMed] [Google Scholar]
- 4.Epstein EH. Basal cell carcinomas: attack of the hedgehog. Nat Rev Cancer. 2008;8(10):743–54. doi: 10.1038/nrc2503. PMID:18813320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sekulic A, Migden MR, Oro AE, Dirix L, Lewis KD, Hainsworth JD, Solomon JA, Yoo S, Arron ST, Friedlander PA, et al.. Efficacy and Safety of Vismodegib in Advanced Basal-Cell Carcinoma. N Engl J Med. 2012;366(23):2171–9. doi: 10.1056/NEJMoa1113713. PMID:22670903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Migden MR, Guminski A, Gutzmer R, Dirix L, Lewis KD, Combemale P, Herd RM, Kudchadkar R, Trefzer U, Gogov S, et al.. Treatment with two different doses of sonidegib in patients with locally advanced or metastatic basal cell carcinoma (BOLT): a multicentre, randomised, double-blind phase 2 trial. Lancet Oncol. 2015;16(6):716–28. doi: 10.1016/S1470-2045(15)70100-2. PMID:25981810. [DOI] [PubMed] [Google Scholar]
- 7.Goodman A, Patel SP, Kurzrock R. PD-1-PD-L1 immune-checkpoint blockade in B-cell lymphomas. Nat Rev Clin Oncol. 2017;14:203–20. doi: 10.1038/nrclinonc.2016.168. PMID:27805626 [DOI] [PubMed] [Google Scholar]
- 8.Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther. 2015;14(4):847–56. doi: 10.1158/1535-7163.MCT-14-0983. PMID:25695955. [DOI] [PubMed] [Google Scholar]
- 9.Chang J, Zhu GA, Cheung C, Li S, Kim J, Chang ALS. Association Between Programmed Death Ligand 1 Expression in Patients With Basal Cell Carcinomas and the Number of Treatment Modalities. JAMA Dermatol. 2017;153(4):285–90. doi: 10.1001/jamadermatol.2016.5062. [DOI] [PubMed] [Google Scholar]
- 10.Jayaraman SS, Rayhan DJ, Hazany S, Kolodney MS. Mutational landscape of basal cell carcinomas by whole-exome sequencing. J Invest Dermatol. 2014;134(1):213–20. doi: 10.1038/jid.2013.276. PMID:23774526. [DOI] [PubMed] [Google Scholar]
- 11.Lipson EJ, Lilo MT, Ogurtsova A, Esandrio J, Xu H, Brothers P, Schollenberger M, Sharfman WH, Taube JM. Basal cell carcinoma: PD-L1/PD-1 checkpoint expression and tumor regression after PD-1 blockade. J Immunother Cancer. 2017;176(2):498–502. doi: 10.1186/s40425-017-0228-3. PMID:28344809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winkler JK, Schneiderbauer R, Bender C, Sedlaczek O, Fröhling S, Penzel R, Enk A, Hassel JC. Anti-programmed cell death-1 therapy in nonmelanoma skin cancer. Br J Dermatol. 2017;176(2):498–502. doi: 10.1111/bjd.14664. PMID:27061826. [DOI] [PubMed] [Google Scholar]
- 13.Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, Stephens PJ, Daniels GA, Kurzrock R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther. 2017;16(11):2598–608. doi: 10.1158/1535-7163.MCT-17-0386. PMID:28835386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, Schuster SJ, Millenson MM, Cattry D, Freeman GJ, et al.. PD-1 Blockade with Nivolumab in Relapsed or Refractory Hodgkin's Lymphoma. N Engl J Med. 2015;372(4):311–9. doi: 10.1056/NEJMoa1411087. PMID:25482239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson DB, Frampton GM, Rioth MJ, Yusko E, Xu Y, Guo X, Ennis RC, Fabrizio D, Chalmers ZR, Greenbowe J, et al.. Targeted Next Generation Sequencing Identifies Markers of Response to PD-1 Blockade. Cancer Immunol Res. 2016;4(11):959–67. doi: 10.1158/2326-6066.CIR-16-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, Dawson N, O'Donnell PH, Balmanoukian A, Loriot Y, et al.. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet Lond Engl. 2016;387(10031):1909–20. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonilla X, Parmentier L, King B, Bezrukov F, Kaya G, Zoete V, Seplyarskiy VB, HJ1 Sharpe, McKee T, Letourneau A, et al.. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat Genet. 2016;48(4):398–406. doi: 10.1038/ng.3525. PMID:26950094. [DOI] [PubMed] [Google Scholar]
- 18.Roemer MGM, Advani RH, Ligon AH, Natkunam Y, Redd RA, Homer H, Connelly CF, Sun HH, Daadi SE, Freeman GJ, et al.. PD-L1 and PD-L2 Genetic Alterations Define Classical Hodgkin Lymphoma and Predict Outcome. J Clin Oncol. 2016;34(23):2690–97. doi: 10.1200/JCO.2016.66.4482. PMID:27069084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, Schnall-Levin M, White J, Sanford EM, An P, et al.. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023–31. doi: 10.1038/nbt.2696. PMID:24142049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armand P, Shipp M, Ribrag V, et al.. PD-1 Blockade with Pembrolizumab in Patients with Classical Hodgkin Lymphoma after Brentuximab Vedotin Failure: Safety, Efficacy, and Biomarker Assessment (KEYNOTE-013) [abstract]. American Society of Hematology Annual Meeting 2016;34(31):3733–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: From Discovery to Therapy. Cancer Discov. 2016;6(4):353–67. doi: 10.1158/2159-8290.CD-15-0894. PMID:26658964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samadder N, Neklason DW, Boucher KM, Byrne KR, Kanth P, Samowitz W, Jones D, Tavtigian SV, Done MW, Berry T, et al.. Effect of sulindac and erlotinib vs placebo on duodenal neoplasia in familial adenomatous polyposis: A randomized clinical trial. JAMA. 2016;315(12):1266–75. doi: 10.1001/jama.2016.2522. PMID:27002448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marquez-Medina D, Popat S. Afatinib: a second-generation EGF receptor and ErbB tyrosine kinase inhibitor for the treatment of advanced non-small-cell lung cancer. Future Oncol Lond Engl. 2015;11(18):2525–40. doi: 10.2217/fon.15.183.. [DOI] [PubMed] [Google Scholar]
- 24.Furumoto Y, Gadina M. The arrival of JAK inhibitors: advancing the treatment of immune and hematologic disorders. BioDrugs Clin Immunother Biopharm Gene Ther. 2013;27(5):431–8. doi: 10.1007/s40259-013-0040-7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Infante JR, Fecher LA, Falchook GS, Nallapareddy S, Gordon MS, Becerra C, DeMarini DJ, Cox DS, Xu Y, Morris SR, et al.. Safety, pharmacokinetic, pharmacodynamic, and efficacy data for the oral MEK inhibitor trametinib: a phase 1 dose-escalation trial. Lancet Oncol. 2012;13(8):773–81. doi: 10.1016/S1470-2045(12)70270-X. PMID. [DOI] [PubMed] [Google Scholar]
- 26.Loaiza-Bonilla A, Jensen CE, Shroff S, Furth E, Bonilla-Reyes PA, Deik AF, Morrissette J. KDR Mutation as a Novel Predictive Biomarker of Exceptional Response to Regorafenib in Metastatic Colorectal Cancer. Cureus. 8(2). doi: 10.7759/cureus.478.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lasota J, Miettinen M. KIT and PDGFRA mutations in gastrointestinal stromal tumors (GISTs). Semin Diagn Pathol. 2006;23(2):91–102. doi: 10.1053/j.semdp.2006.08.006. PMID:17193822. [DOI] [PubMed] [Google Scholar]
- 28.Weigelt B, Warne PH, Lambros MB, Reis-Filho JS, Downward J. PI3 K pathway dependencies in endometrioid endometrial cancer cell lines. Clin Cancer Res Off J Am Assoc Cancer Res. 2013;19(13):3533–44. doi: 10.1158/1078-0432.CCR-12-3815.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janku F, Hong DS, Fu S, Piha-Paul SA, Naing A, Falchook GS, Tsimberidou AM, Stepanek VM, Moulder SL, Lee JJ, et al.. Assessing PIK3CA and PTEN in early-phase trials with PI3 K/AKT/mTOR inhibitors. Cell Rep. 2014;6(2):377–87. doi: 10.1016/j.celrep.2013.12.035. PMID:24440717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Said R, Hong DS, Warneke CL, Lee JJ, Wheler JJ, Janku F, Naing A, Falchook GS, Fu S, Piha-Paul S, et al.. P53 Mutations in Advanced Cancers: Clinical Characteristics, Outcomes, and Correlation between Progression-Free Survival and Bevacizumab-Containing Therapy. Oncotarget. 2013;4(5):705–14. doi: 10.18632/oncotarget.974. PMID:23670029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwaederle M, Vladimir L, Validire P, Hansson J, Lacroix L, Soria JC, Pawitan Y, Kurzrock R. VEGF-A Expression Correlates with TP53 Mutations in Non-Small Cell Lung Cancer: Implications for Anti-Angiogenesis Therapy. Cancer Res. 2015;75(7):1187–90. doi: 10.1158/0008-5472.CAN-14-2305. [DOI] [PubMed] [Google Scholar]
- 32.Koehler K, Liebner D, Chen JL. TP53 mutational status is predictive of pazopanib response in advanced sarcomas. Ann Oncol. 2016;27(3):539–43. doi: 10.1093/annonc/mdv598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iyer G, Hanrahan AJ, Milowsky MI, Al-Ahmadie H, Scott SN, Janakiraman M, Pirun M, Sander C, Socci ND, Ostrovnaya I, et al.. Genome Sequencing Identifies a Basis for Everolimus Sensitivity. Science. 2012;338(6104):221–221. doi: 10.1126/science.1226344. PMID. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


