Abstract
Significance: L-ergothioneine is synthesized in actinomycetes, cyanobacteria, methylobacteria, and some fungi. In contrast to other low-molecular-weight redox buffers, glutathione and mycothiol, ergothioneine is primarily present as a thione rather than a thiol at physiological pH, which makes it resistant to autoxidation. Ergothioneine regulates microbial physiology and enables the survival of microbes under stressful conditions encountered in their natural environments. In particular, ergothioneine enables pathogenic microbes, such as Mycobacterium tuberculosis (Mtb), to withstand hostile environments within the host to establish infection.
Recent Advances: Ergothioneine has been reported to maintain bioenergetic homeostasis in Mtb and protect Mtb against oxidative stresses, thereby enhancing the virulence of Mtb in a mouse model. Furthermore, ergothioneine augments the resistance of Mtb to current frontline anti-TB drugs. Recently, an opportunistic fungus, Aspergillus fumigatus, which infects immunocompromised individuals, has been found to produce ergothioneine, which is important in conidial health and germination, and contributes to the fungal resistance against redox stresses.
Critical Issues: The molecular mechanisms of the functions of ergothioneine in microbial physiology and pathogenesis are poorly understood. It is currently not known if ergothioneine is used in detoxification or antioxidant enzymatic pathways. As ergothioneine is involved in bioenergetic and redox homeostasis and antibiotic susceptibility of Mtb, it is of utmost importance to advance our understanding of these mechanisms.
Future Directions: A clear understanding of the role of ergothioneine in microbes will advance our knowledge of how this thione enhances microbial virulence and resistance to the host's defense mechanisms to avoid complete eradication. Antioxid. Redox Signal. 28, 431–444.
Keywords: : ergothioneine, thiols, microbes, oxidative stresses, redox homeostasis
Introduction
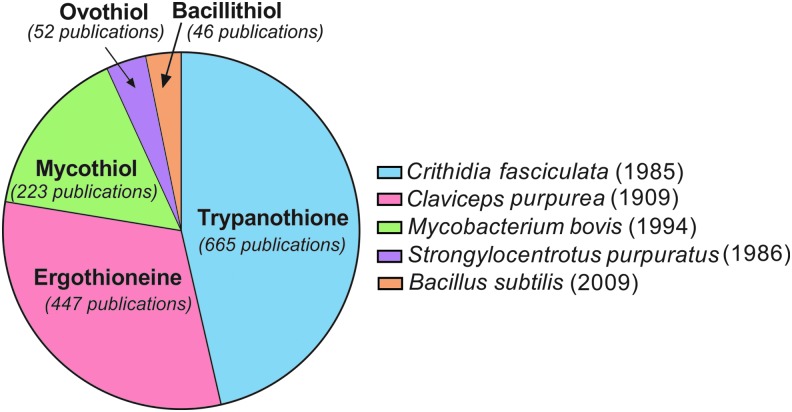
Thiols and thiones are integral parts of the prokaryotic and eukaryotic cellular machinery involved in diverse biological reactions such as intracellular redox homeostasis, reactive oxygen species (ROS) and reactive nitrogen species (RNS) detoxification, cellular signaling, transcription, and metabolic adaptations. The history of thiols and thiones in biology can be traced back as early as 1888 when Joseph de Rey-Pailhade first discovered the sulfur-containing compound, philothion (13), which was later rediscovered as glutathione (GSH) by Frederick Gowland Hopkins (30). GSH is the most abundant thiol in both eukaryotes and prokaryotes (3). In contrast, many microbes, especially gram-positive bacteria, do not produce GSH. Instead, they have developed the biosynthetic machinery to produce alternate low-molecular-weight thiols and thiones, which regulate several facets of their physiology and pathogenesis (15). These include bacillithiol (17), trypanothione (42), mycothiol (MSH) (46, 47, 65), ovothiol (10), and ergothioneine (EGT) (19–21, 34, 51, 52, 54). Of these, EGT is one of the earliest known and most extensively studied redox buffers (Fig. 1).
FIG. 1.
Pie chart indicating the percentage of publications for each naturally occurring low-molecular-weight thiol, excluding glutathione, since 1900 (PubMed search with individual thiol name). Right panel shows the species in which the thiol was first discovered and the year when it was first reported. Ergothioneine is one of the earliest discovered and most extensively studied redox buffers.
Early evidence of EGT biosynthesis was discovered in 1909 by Charles Tanret, who isolated EGT from Claviceps purpurea, an ergot fungus, hence the name ergothioneine (63). Barger and Ewins determined the structure and found that EGT is a trimethylbetaine derivative of the amino acid histidine, with a thiol group attached to the C2-position (ɛ-position) of the imidazole ring (6) (Fig. 2A). EGT has been shown to regulate microbial physiology, redox homeostasis, cellular metabolism, and pathogenesis of several microbes, including the deadliest bacterial pathogen to date, Mycobacterium tuberculosis (Mtb) (8, 50–52). In this study, we provide a concise review of the scientific advances in the field of EGT and evaluate its significance in maintaining redox homeostasis, signaling, and metabolic adaptations in different microbes of the bacterial and fungal world.
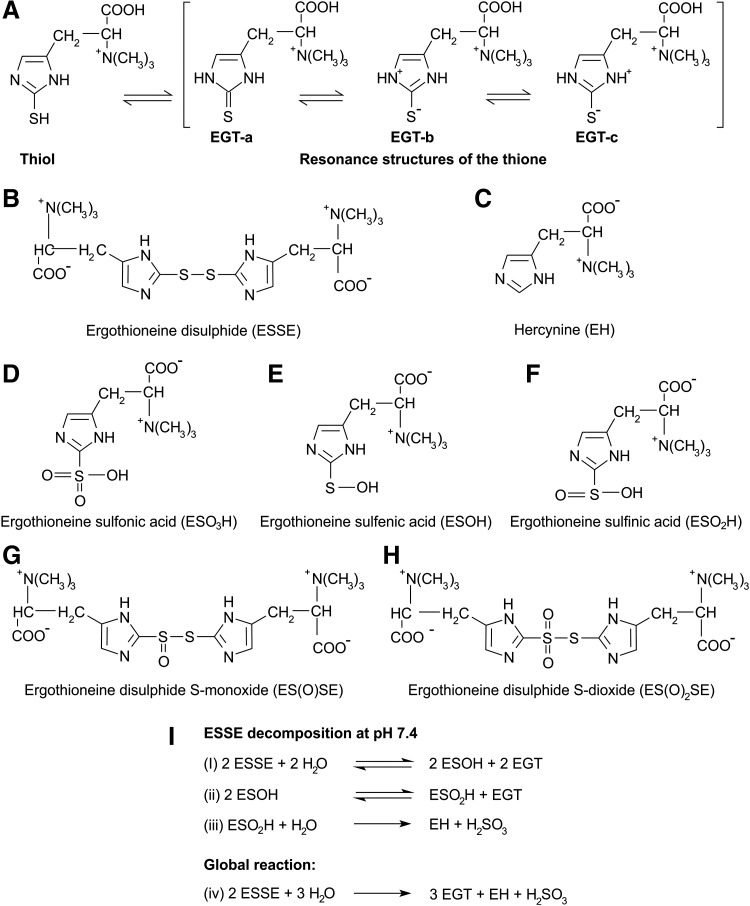
FIG. 2.
Tautomerism of EGT and chemical structures of oxidation states of EGT. (A) EGT exists as a tautomer between its thiol and thione forms. The thione has resonance structures that are thiolate in character and contribute to weak thiol-like activity of EGT. At physiological pH, EGT exists primarily in the thione form, which prevents it from undergoing autoxidation. (B) ESSE is formed when EGT is exposed to hydrogen peroxide, peroxynitrite, or hypochlorite, but due to its instability at physiological pH, it decomposes disproportionately into EGT and hercynine (C). Servillo et al. (57) proposed two oxidation patterns for EGT: (i) EGT is oxidized via the ESSE pathway and in the presence of excess oxidants, ESSE can be transformed into EGT disulfide S-monoxide (G) and EGT disulfide S-dioxide (H); (ii) EGT is first oxidized into EGT sulfenic acid (E), which is further oxidized into EGT sulfinic acid (F) that is oxidized into EGT sulfonic acid (D); Servillo et al. (57) also proposed a pathway for the decomposition of ESSE at neutral pH (I). EGT, ergothioneine; ESSE, ergothioneine disulfide.
Since the focus of this Forum edition is on microbes, we will minimize references to higher eukaryotes such as mammals. In addition, since the current Forum edition (Alternate thiol-based redox buffers) features comprehensive reviews on other low-molecular-weight thiols, readers are encouraged to peruse them as well.
Distribution of Ergothioneine
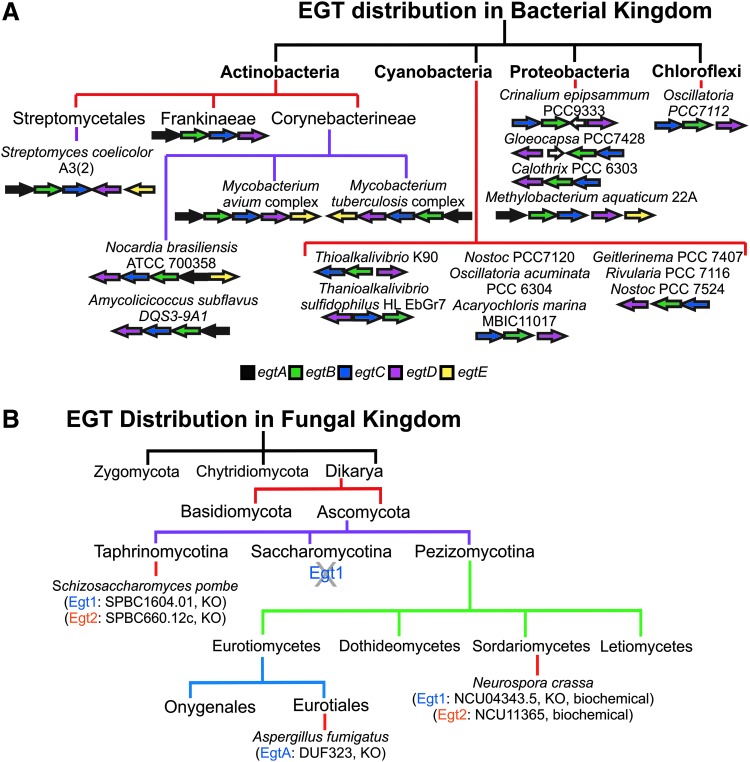
To date, EGT biosynthesis is documented in several nonyeast fungal species, including Neurospora crassa and other filamentous fungi (19, 40), Actinomycetales, including mycobacterial species such as Mtb (21, 38, 51, 52) and Mycobacterium smegmatis (20, 54), certain cyanobacteria (48), and Methylobacterium species (2) (Fig. 5).
FIG. 5.
Distribution of EGT in the (A) bacterial and (B) fungal kingdoms. (A) Distribution of EGT in the bacterial kingdom has been illustrated based on the analysis of the presence or absence of the 5-gene cluster characteristic of Actinobacteria in other bacterial phyla (modified from Ref. 33). (B) Distribution of EGT in the fungal kingdom is illustrated in the main lineages based on orthologs of NcEgt1. Documented evidence for occurrence of EGT is demonstrated in specific species by the annotation of knockout (KO) or biochemical characterization examining the synthesis of EGT in these organisms. No Egt1 homologs were identified in the Saccharomycotina species.
Various fungal species are shown to produce significant amounts of EGT, ranging from 1.7 to 46.6 mg/100 g of dry weight (19). During conidial germination stages of Colletotrichum graminicola and N. crassa, EGT production was measured at 11.6 and 25.4 nmol/mg of protein, respectively. Interestingly, during mycelium formation, these levels were reduced to 0.7 and 5.2 nmol/mg in C. graminicola and N. crassa, respectively (8). It has been reported that in Schizosaccharomyces pombe, 0.3 μM of EGT was present in its normal vegetative state, whereas during nitrogen and glucose starvation, intracellular levels of EGT were increased to 157.4 μM and 41.6 μM, respectively (50). Concentrations of EGT expressed in molarities need to be regarded with discernment as the volumes of cells have been estimated from the average dimensions of the cells, which may change during growth. The significance of these differences in EGT levels during different life stages/stresses is discussed later.
Genghof and Vandamme showed that the various mycobacterial strains isolated from human and nonhumans produce EGT levels ranging from 0 to 116 mg per 100 g of dry weight (21), with Mtb H37Rv producing up to ∼50 mg of EGT per 100 g of dry weight (21). In a more recent study by Sao Emani et al., mean EGT levels in M. smegmatis were reported as 17 pg of EGT per 105 bacterial cells (54). Similarly, Saini et al. found EGT levels in Mtb ranging from 40 to 300 ng/108 cells (52). Methylobacterial strains also produce significant levels of EGT, ranging from 0 to 100 μg/g of cells (2). However, like GSH, EGT production is not common to all bacterial species and the above studies demonstrated the absence of EGT biosynthesis in Bacillus, Clostridium, Corynebacterium, Escherichia, Lactobacillus, Propionibacterium, Proteus, Pseudomonas, Staphylococcus, Streptococcus, and Vibrio.
Plants and animals do not produce EGT; however, they acquire it from the environment and accumulate it in tissues or cells (14). EGT is concentrated in human cells that express an EGT-specific transporter, called organic cation/carnitine transporter 1 (OCTN1/ETT), a product of the SLC22A4 gene (23). As the focus of the review is on the role of EGT in microbial systems, readers are advised to follow Cheah and Halliwell for an in-depth review on the role of EGT in humans and its relevance to diseases (12), as well as Yoshida et al. for immunomodulatory functions as a Toll-like receptor (TLR) agonist (68).
Biosynthesis of Ergothioneine
Studies using radiolabeled sulfur demonstrated that the biosynthesis of EGT proceeded from the amino acid, L-histidine, through an intermediate precursor, called hercynine, which then incorporates sulfur from cysteine to form the final product (27, 29, 39). There are two main reactions in the EGT biosynthesis, namely the oxidative C-S bond formation and the subsequent C-S bond cleavage reaction, which result in the net transfer of the sulfur atom from cysteine or γ-glutamyl cysteine to the imidazole side chain of histidine (31).
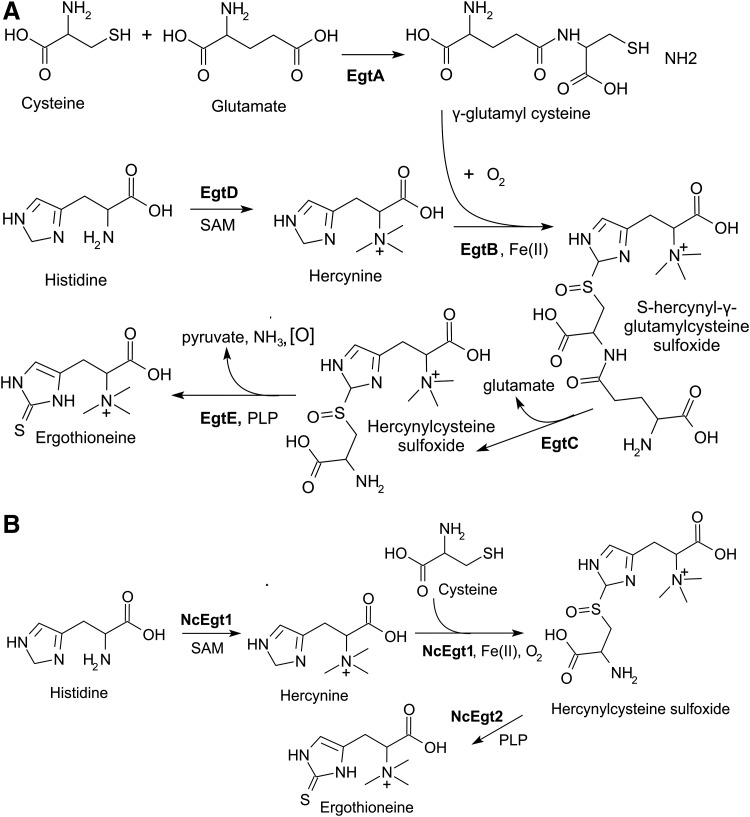
In vitro reconstitution studies using protein preparations of Erwinia tasmaniensis and M. smegmatis (56) and a genetic study in M. smegmatis (54) both provided genetic and biochemical evidence of the production of EGT in M. smegmatis. Seebeck (56) discovered a putative five-gene cluster (egtABCDE) responsible for the biosynthesis of EGT in mycobacteria (Fig. 3A). In mycobacteria, it is now clear that EgtD is a histidine methyltransferase that trimethylates the NH2 group of histidine to generate hercynine (Fig. 4A) in an S-adenosylmethionine (SAM)-dependent manner. EgtA catalyzes the synthesis of an alternate thiol source to cysteine, γ-glutamyl cysteine. In the presence of FeSO4 and oxygen, γ-glutamyl cysteine is conjugated to the imidazole side chain of hercynine by EgtB to produce S-hercynyl-γ-glutamylcysteine sulfoxide (Fig. 4A). EgtC removes the glutamate to yield hercynylcysteine sulfoxide, which in the presence of pyridoxal 5-phosphate, is converted into EGT by EgtE with the concomitant release of pyruvate and ammonia (Fig. 4A). The crystal structures of EgtB, EgtC, and EgtD have recently been determined (22, 66, 67).
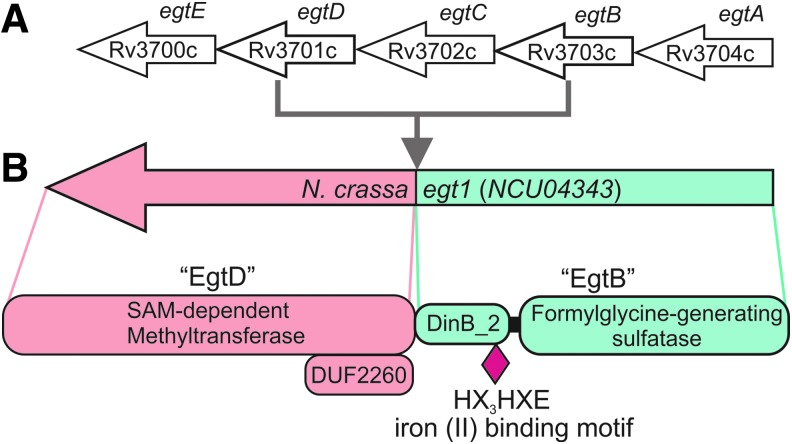
FIG. 3.
(A) Five-gene cluster in Mtb H37Rv encoding EGT biosynthetic enzymes; (B) Three protein domains common in egt1 homologs of fungi. (A) An example of the EGT five-gene cluster responsible for EGT biosynthesis that has only been found in Actinobacteria (51, 52). (B) N. crassa Egt1 is a multidomain protein, with an N-terminal SAM-dependent methyltransferase, with 27% identity to M. smegmatis EgtD and C-terminal DinB_2 and FGE-sulfatase domains that have 24% identity to M. smegmatis EgtB (modified from Ref. 9). The domains of NcEgt1 corresponding to Mtb EgtB and EgtD have been indicated. SAM, S-adenosylmethionine.
FIG. 4.
Biosynthetic pathways of EGT in (A) mycobacteria and (B) N. crassa and other fungi. (A) In mycobacteria, EGT biosynthesis occurs through five enzymatic steps where L-histidine is methylated into hercynine by methyltransferase (EgtD), followed by formation of a hercynyl γ-glutamylcysteine sulfoxide intermediate after addition of γ-glutamylcysteine by a formylglycine-generating enzyme-like protein (EgtB). Glutamate is removed from this intermediate by a glutamine amidotransferase (EgtC) to form hercynlcysteine sulfoxide that is converted into ergothioneine by a pyridoxal 5-phosphate-dependent β-lyase (EgtE). EgtA is a γ-glutamyl cysteine synthetase. (B) In N. crassa, EGT is synthesized with two enzymes: Egt1 methylates histidine and forms the C-S bond with cysteine to form hercynylcysteine sulfoxide; and Egt2 cleaves the C-S bond to form ergothioneine.
Analysis of the Mtb genome has led to the prediction that the EGT biosynthetic operon is located at the locus Rv3704c-Rv3700c (52) (Fig. 3A). The lack of EGT production in transposon mutants established that these genes are indeed responsible for EGT biosynthesis in Mtb; and reverse transcriptase-PCR analysis demonstrated that egtA through egtD is indeed operonic, while egtE is not (52). Genes homologous to egtABCDE in Mycobacterium were also found in the Methylobacterium species, but egtB and egtD were encoded tandemly on the largest plasmid, and egtA, C, and E were separately encoded in distant loci in the chromosome (2) (Fig. 5A). Cyanobacterial species, which produce high levels of EGT, lack orthologs of egtA, egtC, and egtE (48). Furthermore, a glutathione mutant of cyanobacterium Synechocystis PC6803, gshB, did not produce EGT, whereas accumulation of its precursor hercynine was observed, suggesting that Synechocystis synthesizes EGT through a de novo pathway that is dependent on the synthesis of GSH (45). Orthologs for the five-gene EGT cluster were also found in the genome database of Streptomyces coelicolor A3 (2), where SC00910 to SC00913 were identified as genes corresponding to egtA to egtD in M. avium, and SCO1921, located outside the gene cluster, was suggested to be an ortholog of egtE (44) (Fig. 5A).
The first filamentous fungal biosynthetic EGT gene, egt1, was identified in N. crassa (8). This gene encodes a protein with both SAM-dependent methyltransferase and formylglycine-generating enzyme sulfatase domains that are similar to those found in M. smegmatis egtB and egtD, respectively (Fig. 3B). It was proposed that NcEgt1 catalyzes the first two steps in the biosynthesis of EGT-trimethylation of histidine to hercynine, followed by sulfoxidation of hercynine to hercynylcysteine sulfoxide (Fig. 4B) (2). EGT was not detected in the egt1 knockout strain (9). The C-S bond formation activity of Egt1 was subsequently reconstituted in vitro with purified recombinant Egt1 expressed in Escherichia coli (31). Using genome mining, NcEgt2, a pyridoxal phosphate (PLP)-binding cysteine desulfurase enzyme that converts hercynylcysteine sulfoxide to EGT, was identified and the C-S lyase activity of Egt2 (NCU11365) to generate EGT was reconstituted in vitro (31) (Fig. 4B).
In S. pombe, it has been proposed that only two genes are required for EGT biosynthesis: egt1, which has functional homology to egtB and egtD of M. smegmatis; and egt2 as a potential homolog of egtE from M. smegmatis (50). S. pombe egt1 has been found to be regulated by various transcription factors controlling sexual differentiation, such as Ste11, Mei4, atf21+, and atf31+ (50). Knockout of S. pombe egt1 results in a loss of EGT, whereas when egt2 is knocked out, small amounts of EGT are still present. This is consistent with results reported by Seebeck (56), who demonstrated that hercynylcysteine sulfoxide could spontaneously convert into EGT in the presence of PLP or the reaction could be catalyzed by an unrelated PLP-binding enzyme.
Both N. crassa and S. pombe use cysteine rather than γ-glutamyl cysteine to produce EGT because both fungi appear to lack the glutamine amidotransferase that catalyzes the hydrolysis of the γ-glutamyl amide bond from S-hercynyl-γ-glutamylcysteine sulfoxide (8, 50). Thus, Bello et al. proposed that the egt1 homologs in fungi have a unique set of three protein domains (Fig. 3B): an SAM-dependent methyltransferase domain with a domain of an unknown function 2260; a DinB_2 domain with an HX3HXE iron binding motif; and a formylglycine-generating sulfatase domain (8). Recently, Sheridan et al. (58) demonstrated that the pathogenic fungus, Aspergillus fumigatus, has a trimodular enzyme, EgtA, with a histidine-specific SAM-dependent methyltransferase at the N-terminal, a sulfatase-modifying factor enzyme 1 at the C-terminal, and a central 5-histidylcysteine sulfoxide synthase domain. Deletion of egtA abolished EGT biosynthesis in this pathogen (58).
Phylogenetic analysis of the EGT 5-gene cluster in bacterial and archaeal species by Jones et al. (34) revealed that the five clustered genes, egtABCDE, are only found in actinomycetes (Fig. 5A). However, Alamgir et al. (2) reported that five genes homologous to egtABCDE are found in Methylobacterium aquaticum strain 22A and in other Methylobacterium species, although not clustered. Furthermore, the same analysis found that egtB and egtD occurred in a number of diverse bacterial phyla, but predominantly in Actinobacterial, Proteobacterial, and Cyanobacterial species (Fig. 5A). In many cases, these two genes were genetically linked as they were often located in close proximity to each other.
Analysis of the other genes revealed that egtA is found almost solely in Actinobacteria and Proteobacteria and egtE is present mainly in Actinobacteria, although an egtE homolog has been identified in methylobacteria. This suggests that EgtB and EgtD are key enzymes in the production of ergothioneine in bacteria and functions of the enzymes, EgtA and EgtE, may be performed by as yet unknown enzymes (34). In the fungal kingdom, Egt1 from the fungus N. crassa and Egt1 from S. pombe have domains homologous to bacterial EgtB and EgtD from M. smegmatis (8, 50). Thus, the egtB and egtD genes appear to be a gene signature common to EGT biosynthesis in microbes.
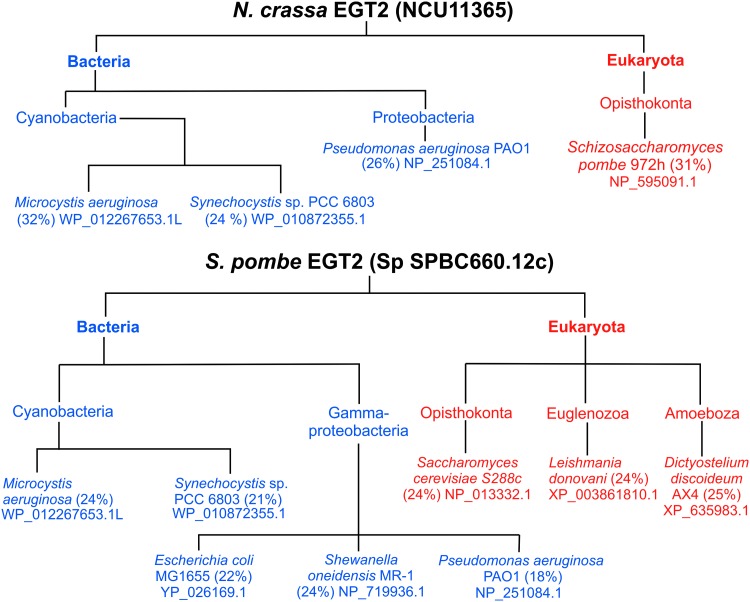
A protein database search with the Blastp tool using N. crassa Egt2 (NcEgt2) and S. pombe Egt2 (SpEgt2) as query sequences revealed bacterial homologs in cyanobacteria, including Microcystis aeruginosa and Synechocystis sp. PCC 6803, and in proteobacteria, including Pseudomonas aeruginosa (Fig. 6). Furthermore, proteobacterial homologs of SpEgt2 were found in E. coli and Shewanella oneidensis. Other eukaryotic homologs of SpEgt2 were found in Saccharomyces cerevisiae, Leishmania donovani, and Dictyostelium discoideum. This further supported that the cleavage of the C-S bond in the final stages of EGT biosynthesis in organisms outside of Actinobacteria is potentially carried out by unidentified enzymes that do not have homology with egtE, but yet have homology to enzymes with a C-S lyase activity in the formation of other metabolites in other organisms.
FIG. 6.
Homologs of NcEgt2 and SpEgt2 from Blastp searches of model organisms. The amino acid sequence of NcEgt2 and SpEgt2 was used to do a blastp search to discover egt2-like homologs in other microbes that might be responsible for the C-S lyase activity in the final stages of EGT biosynthesis as EgtE, responsible for the C-S lyase activity in mycobacteria, appears to be limited to Actinobacteria. The percentage identity of homologs with the largest coverage of the query sequence is given in brackets and the accession number of the homolog is given below the species name.
Properties of Ergothioneine
EGT (2-mercaptohisitidine trimethylbetaine) is an odorless colorless compound with a molecular weight of 229.30 g/mol and a high aqueous solubility limit of 0.9 M at 25°C (26). EGT exists in a tautomeric state between its thiol and thione forms in solution; however, at a physiological pH, it exists predominantly in its thione form (Fig. 2A) (11). The thione form has resonance forms EGT-b and EGT-c that are thiolate in character and support the weak thiolate reactivity of EGT (16). At pH 7, the thiol–disulfide couple of EGT has a more electropositive standard redox potential (−60 mV) in contrast to other naturally occurring thiols, which range typically between −200 and −320 mV (33). Both the tautomer and the high redox potential account for the greater stability of the EGT preventing it from undergoing autoxidation at physiological pH as readily as other thiols such as GSH (32, 41) and heavy metal-catalyzed autoxidation (15). In fact, EGT chelates metal ions such as Cu2+ and Fe2+, forming complexes of two moles of ergothioneine to one mole of metal (25, 43).
The high redox potential of EGT makes it an effective antioxidant through quenching singlet oxygen and scavenging of hydroxyl radicals (16, 24). However, EGT does not rapidly react with hydrogen peroxide (H2O2) or superoxide (O2•−), but it does decompose HOCl and it inhibits lipid peroxidation induced by mixtures of heme compounds and H2O2 (1, 26). Furthermore, EGT detoxifies peroxynitrite (ONOO−) that is formed by the reaction of nitric oxide with the superoxide radical, and EGT is thus able to protect α1-antiproteinase against inactivation and tyrosine against nitration by peroxynitrite (4).
Previously, it was shown that the stability of EGT results in slow degradation and resistance to disulfide formation (36, 37) and that the disulfide form (Fig. 2B) was only formed at very low pH in the presence of copper and H2O2 (28). However, using high-performance liquid chromatography coupled on-line with flow injection analysis (FIA) electron spray ionization (ESI) acquired mass spectroscopy with a quadrupole ion trap (HPLC-FIA-ESI-MS/MS), Servillo et al. (57) detected EGT disulfide (ESSE, Fig. 2B) during the oxidative burst of neutrophils in the presence of EGT and when EGT was exposed to biologically relevant oxidants, such as hypochlorite, peroxynitrite, and hydrogen peroxide, at a physiological pH of 7.4. FIA enabled analysis of these fast reactions, and ESI enabled detection of ions of macromolecules such as ESSE, which usually fragment upon ionization. Other oxidation products that were detected were hercynine (EH, Fig. 2C) and EGT sulfonic acid (ESO3H, Fig. 2D), which had significant formation when EGT was oxidized with hypochlorite.
ESSE was reported to have an unusual behavior that was not common to other alkyl disulfides (28). Although ESSE is stable in a very acidic aqueous solution, Servillo et al. found that ESSE was unstable at physiological pH and undergoes a progressive disproportionate decomposition to three moles of EGT and 1 mole of hercynine from two moles of EGT disulfide, rather than a reductive reconversion to EGT (57). Servillo et al. proposed a decomposition pathway for ESSE (Fig. 2I): at neutral pH, ESSE is hydrolyzed to EGT and highly reactive EGT sulfenic acid (ESOH, Fig. 2E and I i), which then disproportionates into EGT sulfinic acid (ESO2H, Fig. 2F, I ii) and EGT. ESO2H then decomposes irreversibly into hercynine and H2SO3 (Fig. 2I iii), and this irreversible decomposition drives the entire process (Fig. 2I iv). Notably, EGT is partially reformed from ESSE without any reducing agents. Furthermore, sulfurous acid (H2SO3), which is also formed (Fig. 2I iv), is another effective reducing agent that could counteract cell oxidative stress by conversion into sulfate.
Thus, Servillo et al. indicate that this oxidation of EGT to EH and sulfate makes EGT a rich source of reducing equivalents involving the irreversible transfer of six electrons per EGT molecule to intracellular oxidants. This contrasts with classical oxidation of alkylthiols into their respective alkyl disulfides, which involve the transfer of one electron per thiol molecule to oxidants. In addition, cellular reducing equivalents are required to regenerate alkylthiols contrary to partial reformation of EGT upon ESSE decomposition at physiological pH (57).
Servillo et al. proposed two oxidative pathways of EGT: (i) EGT is oxidized through the formation of ESSE; and in the presence of excess oxidants, ESSE may not only be disproportionate but also be transformed into more oxidized species, such as EGT disulfide S-monoxide (ES(O)SE, Fig. 2G) and EGT disulfide S-dioxide (ES(O)2SE, Fig. 2H), which have in common the partial formation of EGT. (ii) EGT is oxidized first to ESOH (Fig. 2E), which can decompose further as depicted in Figure 2I, but in the presence of excess oxidants, ESOH is oxidized to ESO2H (Fig. 2F) that is then oxidized to ESO3H (Fig. 2D). However, the key intermediate in both oxidative pathways is ESO2H, which undergoes irreversible decomposition to EH and sulfurous acid.
Ergothioneine in Mycobacteria
Mycobacteria, like most gram-positive bacteria, do not produce GSH, rather they synthesize mycothiol and EGT (52). Similar to GSH, EGT serves as an antioxidant to protect bacteria from environmental oxidative stress (52). In Mtb, EGT production is regulated by WhiB3, which is a 4Fe-4S cluster DNA-binding protein that maintains redox homeostasis by sensing NO and O2 generated by the host. Metabolite analysis by liquid chromatography coupled with tandem mass spectrometry revealed a 7.3-fold increased production of EGT in MtbΔwhiB3 mutant. Complementation of whiB3 restored the EGT levels similar to that in wild-type Mtb, suggesting that WhiB3 modulates the production of EGT (52). WhiB3 also initiates a metabolic switchover to the preferred in vivo carbon source (fatty acids) and integrates redox signals with central carbon metabolism (59, 60). MtbΔwhiB3 has significantly increased levels of EGT when catabolizing a diverse set of fatty acids, such as stearate, myristate, and palmitate, when compared with glucose (52). This indicates that WhiB3 regulates EGT production in response to catabolism of fatty acid precursors and cholesterol.
Singh et al. reported that the transposon mutant of egtA in M. smegmatis did not produce EGT; however, there is no difference in growth compared with the wild type (61). The egtA mutant, R119, was shown to be highly susceptible to oxidative agents such as cumene hydroperoxide and diamide; alkylating agents such as N-ethylmalemide, iodoacetamide, and chlorodinitrobenzene; and metals such as Cd2+, Cu2+, SeO32−, TeO32−, and CrO42− (Fig. 7). The sensitivity to all of these agents was reversed in the complemented strain demonstrating the importance of EGT in M. smegmatis (61).
FIG. 7.
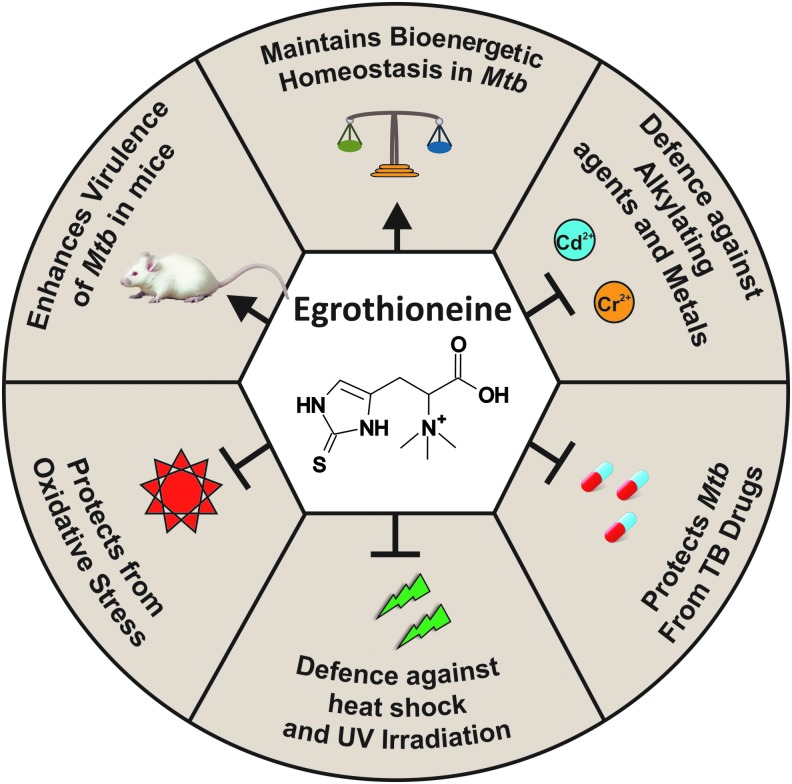
Functions of EGT in bacteria. This figure illustrates functions of ergothioneine in various bacteria, such as maintaining bioenergetic homeostasis in Mycobacterium tuberculosis, modulating susceptibility of M. tuberculosis toward TB drug, protection against diverse oxidative stressors in M. tuberculosis, Methylobacterium aquaticum, and Streptomyces coelicolor, resistance against alkylating agents and metals in M. smegmatis, and defense against heat shock and UV irradiation in M. aquaticum. EGT is essential for survival of M. tuberculosis in macrophages and virulence of M. tuberculosis in mice.
R119 had less protein carbonylation and less lipid oxidation than the wild type, whereas a transposon mutant of mshC in M. smegmatis that does not produce MSH had greater protein carbonylation and greater lipid oxidation than the wild type. The complemented strain of R119 had levels of protein carbonylation and lipid oxidation that were not significantly different from the wild type. Thus, the lack of EGT results in less damage to cellular proteins and lipids, and lack of MSH results in increased damage. This decrease in protein carbonylation and lipid peroxidation in the EGT mutant can be explained by the upregulation of Ohr (organic hydroperoxide reductase) and other enzymes involved in reduction of oxidants, such as manganese-dependent catalase, KatG, and AhpC. In addition, this suggests that MSH has a greater protective effect than EGT against protein carbonylation and lipid oxidation.
Peroxidase activity in the EGT mutant was assessed to determine if EGT acts as an electron donor to peroxidase(s) by measuring the decomposition of H2O2, cumene hydroperoxide, and tert-butyl hydroperoxide (61). The amount of peroxides in the EGT mutant were the same as that in wild type, showing that EGT is not involved directly in the reduction of peroxides in M. smegmatis or another mechanism that detoxifies the peroxides is induced in the EGT mutant, such as Ohr, KatG, or AhpC (61). However, these experiments do not rule out the presence of an EGT-dependent peroxidase activity in M. smegmatis. Further experimentation is needed to demonstrate EGT-dependent enzyme-mediated detoxification pathways of different peroxides.
Sao Emani et al. (54) showed that an M. smegmatis mutant lacking egtD was unable to synthesize EGT and they also demonstrated that EGT protects M. smegmatis against peroxides such as tert-butyl hydroperoxide and cumene hydroperoxide (54). The M. smegmatis egtD mutant was more susceptible to the lipid peroxides, cumene peroxide and tert-butyl hydroperoxide, in contrast to the M. smegmatis egtA mutant, which was only sensitive to cumene peroxide (61). Greater susceptibility to cumene hydroperoxide, hydrogen peroxide, and paraquat was also observed in an Mtb egtD transposon mutant than in an Mtb egtA transposon mutant (52). This may be due to the accumulation or loss of other intermediates in the EGT synthetic pathway, for example, possibly an accumulation of γ-glutamyl cysteine in the egtD mutant, which has differing redox buffering capacities. Furthermore, Sao Emani et al. showed that a double mutant lacking both EGT and MSH is significantly more sensitive to peroxide stress than single mutants, suggesting that MSH and EGT may be able to partly compensate for each other (54). However, as the levels of EGT are many-fold lower than MSH in M. smegmatis, EGT is less likely to be able to compensate for MSH (54, 61).
Saini et al. showed that disruption of egtA and egtD genes involved in EGT biosynthesis in Mtb abrogates EGT production (52). EGT and MSH are the major redox buffers, which are physiologically linked in Mtb. Significantly increased levels of EGT were observed in an Mtb strain deficient in MSH production. However, lack of EGT did not show a corresponding increase in MSH, which correlated with the increased endogenous ROS levels in ΔegtA mutant (52), as revealed by flow cytometry analysis showing that the ΔegtA mutant had 25-fold more ROS-producing cells than wild type; and ΔmshA mutant had eight-fold more ROS-producing cells than wild type (52). Similarly, the M. smegmatis mshA::Tn5 mutant markedly increased production of EGT by 26-fold in the exponential phase of growth and five-fold in the stationary phase (62). In addition, the mshA::Tn5 mutant displayed increased expression of the Ohr protein, while no expression was observed in WT and complemented strains. This study suggests that increased expression of Ohr provides protection to mshA::Tn5 by reducing peroxide levels in cells and confers resistance to cumene hydroperoxide (62).
Lack of either EGT or MSH was shown to alter the respiration in Mtb and is critical for maintaining bioenergetic homeostasis in Mtb (52). In addition, transcriptome analysis of respective mutants in Mtb revealed that EGT and MSH have overlapping, but distinct, functions in Mtb (52). The distinct functions between EGT and MSH may abrogate competition between the EGT and MSH biosynthetic pathways for cysteine to a certain degree, with the required redox function needed at a moment in time, based on the current redox status of the cell, determining which redox buffer will have preferential synthesis. However, the elevated levels of EGT observed in the MSH mutant are not reciprocated with compensatory elevated levels of MSH in the EGT mutant. This may be due to a predominant protective effect of MSH over EGT inducing an MSH biosynthetic competitive edge over EGT for cysteine.
Mycobacterial adaptation to environmental conditions is dependent on classical bacterial two-component systems made up of histidine kinases and response regulators in addition to Ser/Thr protein kinases (STPKs) (5). Richard-Greenblatt et al. found egtD, which is essential for EGT synthesis in Mtb, to be negatively regulated by phosphorylation with STPK, PknD. In addition, intracellular levels of EGT were shown to be increased during the late log phase and required for Mtb survival during starvation (51). This was further supported by a microarray analysis of global adaptation of Mtb to nutrient starvation, revealing that pknD was downregulated after 4 h (7). Lack of EGT alters the respiration of Mtb, suggesting that EGT plays a role in maintaining bioenergetic homeostasis (Fig. 7). Saini et al. (52) also showed that egtA and egtD mutants of Mtb are significantly more sensitive to oxidative stress agents, such as hydrogen peroxide, cumene hydroperoxide, paraquat, and menadione (Fig. 7).
Saini et al. employed egtA and egtD mutants to investigate the role of EGT in Mtb susceptibility to anti-TB drugs. Following exposure to rifampicin (inhibits bacterial RNA polymerase) and isoniazid (inhibits mycolic acid synthesis), the percentage survival of the egtA and egtD mutants was 40% and 25% less than that of the wild type, respectively. Similarly, the survival of egtA and egtD mutants was reduced to <70% and <50% when exposed to clofazimine (inhibits Complex I [NADH dehydrogenase] of the bacterial electron transport chain) and bedaquiline (inhibits Complex V [ATP synthase]), respectively. Of particular interest is clofazimine, which is involved in ROS-mediated killing of Mtb, from which EGT could protect the mycobacterium. Thus, egtA and egtD mutants lacking EGT would be more susceptible to killing by clofazimine. Genetic complementation restored the sensitivity of the mutants similar to that of the wild type. In addition, the MSH mutant, ΔmshA, was shown to be more susceptible to clofazimine and bedaquiline than Mtb and egt mutants. This study shows that lack of EGT modulates the susceptibility of Mtb to frontline anti-TB drugs (52).
In contrast, Sao Emani et al. demonstrated that the M. smegmatis egtD mutant did not display susceptibility to anti-TB drugs compared with the parental strain, suggesting that EGT deficiency does not alter susceptibility to antibiotics (54). These differences in antibiotic susceptibility are expected as M. smegmatis and Mtb differ naturally in their resistance to many drugs. Therefore, this different drug-resistant profile could be attributed to many genetic, physiological, and metabolic factors, including, but not limited to, M. smegmatis being a fast-growing nonvirulent strain (90-min doubling time) in contrast to Mtb that is a slow-growing virulent strain (24-h doubling time). Also, Saini et al. showed that EGT and MSH may have overlapping, but distinct, functions in Mtb physiology. These results provide important insights into the role of EGT in oxidative stress and drug susceptibility.
Studies to assess whether EGT can protect Mtb from the microbiocidal effects of macrophages were carried out in J774A.1 macrophages. The ΔegtD mutant demonstrated a five-fold reduction in CFU after a 120-h infection of murine J774A.1 macrophages in comparison with infection with the wild type, and a three-fold reduction in intracellular survival of egtA and egtD Mtb transposon mutants was observed at 120 h postinfection in murine RAW264.7 macrophages (52). Complementation restored the intracellular survival comparable with wild-type Mtb. These studies demonstrate that lack of EGT led to reduced survival of Mtb in macrophages and EGT may be required to a greater extent at later stages of infection (51, 52).
Saini et al. also demonstrated (for the first time) the role of EGT in the mouse model of infection. Mice infected with both egtA and egtD Mtb mutants demonstrated a four-fold reduction in bacillary burden in lungs when compared with lungs of wild-type Mtb-infected mice. In addition, reduced pathology was observed in the lungs of mice infected with the egt mutant strains. Complementation of mutant strains with egtA and egtD, respectively, restored the bacillary burden and pathology of lungs similar to that of wild-type infected mice (52). These studies provide strong evidence that EGT-deficient Mtb strains are attenuated in the mouse model of tuberculosis and establish the significance of EGT in Mtb intracellular survival and virulence (Fig. 7). Genes involved in EGT biosynthesis might be attractive drug targets for the development of new therapeutic molecules against mycobacterial infections.
Ergothioneine in Methylobacteria
Methylobacteria are facultative methylotrophic bacteria, which can use methanol and other carbon sources, such as ethanol, succinate, and glucose, as substrates. Metabolic analysis of methanol-grown M. aquaticum 22A revealed a large accumulation of EGT (2). Although ergothioneine has been reported to be secreted in mycobacteria, it was not secreted in Methylobacterium spp. The intracellular concentration of EGT in M. aquaticum 22A was estimated to be ∼183 mM based on an estimation of the cell volume from the cell dimensions.
When methylobacteria were grown on other carbon sources, such as ethanol, succinate, and glucose, although succinate did not induce EGT production, glucose and ethanol did, but with much lower yields than when the methylobacteria were grown in methanol. Compared with carbon sources, nitrogen sources such as ammonium nitrate, ammonium chloride, and ammonium sulfate were less effective for EGT production. Available carbon sources will induce switches in central carbon metabolism to produce the energy requirements for the survival of the bacterium. These changes in metabolism can alter the intracellular redox status that will drive the production of redox buffers such as EGT and MSH. In the case of strain 22A, growth in methanol alters the core intermediary metabolism with concomitant changes in the intracellular redox state such that it stimulates the production of prominent levels of EGT.
Blast analysis revealed that egt operon genes were found in methylobacterium strain 22A with 25–46% identity to Mtb (2). A deletion mutant of egtBD (Δegt) in Methylobacterium completely abolished EGT production. Complementation recovered the EGT production to levels greater than that in the wild type (2). These observations suggest that the egtBD genes are indeed involved in EGT synthesis in methylobacteria and that the pathway might be similar to that in Mycobacterium species (2). In the presence of methanol, the Δegt mutant demonstrated increased growth and cell numbers, indicating that EGT is not required for methylotrophic growth of the bacterium.
The Δegt strain was more sensitive to heat shock at 46°C than the wild type after 20 min of treatment. UV irradiation (at 253 nm) severely reduced the viability of the Δegt cells and decreased the intracellular EGT concentration in the methanol-grown 22A cells, suggesting that EGT protects the cells against sunlight to which methylobacteria are exposed to in the phyllosphere (2) (Fig. 7). Interestingly, Alamgir et al. also showed that the Δegt mutant showed higher resistance to H2O2 than the wild type, possibly due to the presence of other antioxidant molecules that could compensate for the loss of EGT. In addition, these authors showed that Δegt strain displayed slightly reduced growth (less than one-fold, p < 0.1) compared with wild type when they are independently grown on Arabidopsis thaliana leaves, suggesting that EGT is not essential for methylobacteria's growth on the leaves (2). This was further supported when the Δegt strain was coinoculated with the wild type on A. thaliana leaves and lesser growth of the wild type was observed possibly due to the biological cost of EGT synthesis (2).
Ergothioneine in Streptomyces
Deletion of the egtA gene (SCO0910) encoding γ-glutamyl-cysteine synthetase in S. coelicolor synthesized reduced levels of EGT, possibly due to utilization of L-cysteine instead of γ-glutamyl cysteine. Intracellular and secreted levels of EGT in the egtA mutant were estimated to be 20% and 85% in comparison with wild type (44). Strains with reduced EGT (egtA mutant) showed delayed growth at the late logarithmic phase, which may be due to increased levels of L-cysteine in the egtA mutant that become toxic as L-cysteine cytotoxicity was previously reported in E. coli even at low concentrations (35). Both the egtA mutant and an mshC mutant lacking MSH were more sensitive to hydrogen peroxide and cumene hydroperoxide than the parental strain. However, the egtA mutant that still produced a reduced level of EGT was more susceptible than the mshC mutant, suggesting that the levels of EGT present in the bacterium play a significant role in protection against oxidative stress (44).
The egtA mutant had a five-fold increase in MSH levels in comparison with the wild type, even though the egtA mutant still produced low levels of EGT, whereas equivalent levels of EGT were found in the mshC mutant and the parental strain. This study shows that EGT plays a more important role than MSH in S. coelicolor (44). Zhao et al. demonstrated that disruption of egtD in Streptomyces lincolnensis resulted in reduced levels of EGT (16.8%) compared with wild type. In addition, ΔegtD cells have reduced lincomycin A levels compared with wild type, presumably due to reduced EGT levels. Authors showed that in S. lincolnesis, EGT serves as a carrier in the first S-glycosylation step of the biosynthesis of lincomycin A to channel the lincosamine unit and mediate its condensation with propyl-L-proline in the biosynthesis of lincomycin A, a C-8 sugar antibiotic (69).
Ergothioneine in Neurospora
N. crassa mycelia have an equal content of EGT and GSH (5.2 nmol and 5.5 nmol/mg protein, respectively). However, during conidial stages, EGT levels were significantly higher than the GSH (25.4 nmol compared with 3.0 nmol/mg protein) and were shown to protect N. crassa during conidial germination (8). Deletion of the gene NCU04343, which encodes NcEgt-1 (NcΔegt-1) in N. crassa, completely abolished the EGT biosynthesis, showing that NcEgt-1 is the primary gene involved in EGT biosynthesis in N. crassa (8). The mutant strain deficient in EGT biosynthesis had no significant growth defects compared with wild-type N. crassa, either in nitrogen-rich or -deficient medium. However, the EGT-deficient mutant had significantly reduced longevity, coupled with ≈20% reduction in conidial life span, regardless of whether the conidia were stored at low relative humidity (52%) or high relative humidity (97%) (8, 9). This clearly suggests that EGT plays a crucial role in the life cycle between conidiogenesis and conidial germination in N. crassa, even under environmental conditions inducing desiccation (Fig. 8).
FIG. 8.
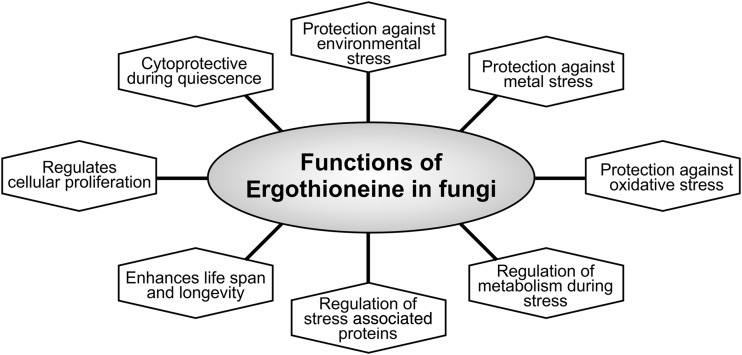
Functions of EGT in fungi. Schematic overview of the wide physiological functions played by ergothioneine in different fungal species. EGT has been shown to protect conidial germination, thereby enhancing the longevity and overall life span of N. crassa as well as in A. fumigatus. In N. crassa, along with its roles in longevity, EGT is also involved in maintaining cellular redox homeostasis. Similarly, in A. fumigatus, EGT is protective against oxidative stress contributed by free radicals and metal ions such as iron, cobalt, and copper. Furthermore, lack of EGT is also suggested to be responsible for the disruption of iron metabolism in A. fumigatus. In fission yeast, S. pombe, EGT protects the cells during quiescent stages and against environmental stresses, including nitrogen and glucose starvation, and heavy metal stress such as contributed by cadmium (Cd2+).
Using a bathocuproinedisulfonic acid-based copper reduction assay, Bello et al. showed that EGT accounts for one-third of the antioxidant capacity in conidia (9). Upon treatment with tertbutyl hydroperoxide, the germination of NcΔegt-1 was severely compromised compared with WT demonstrating that EGT protects N. crassa against peroxide damage during germination, in vivo, by quenching peroxide (8) (Fig. 8). Conversely, abrogation of EGT biosynthesis had no significant effect on growth or germination under menadione or Cu2+-stressed conditions (8). This proposes an interesting dynamic of the antioxidant properties of EGT where it handles superoxide and peroxide stresses differently, suggesting selective antioxidant properties of EGT against different free radicals and peroxides. Bioinformatic-coupled biochemical characterization of EGT biosynthesis by Hu et al. showed that N. crassa has a regulated EGT biosynthetic pathway because of its nonheme iron enzyme, Egt1, which instead of using γ-glutamyl cysteine as the substrate, uses cysteine (31).
Ergothioneine in Aspergillus
EGT was first identified in Aspergillus species in 1956 when it was isolated from hot water extracts of Aspergillus niger mycelium (40). Later in 2012, Gallagher et al. detected EGT in A. fumigatus, a saprotroph and opportunistic pathogen, which is widespread in nature and causes severe allergic or invasive diseases in immunocompromised humans (18). Early evidence for a role of EGT in A. fumigatus life cycle was demonstrated during disruption of gliotoxin biosynthesis by deletion of the gliK gene (γ-glutamyl cyclotransferase), a key gene of gli cluster of A. fumigatus, which resulted in substantial oxidative stress and significantly increased intracellular EGT levels (18).
In a more recent study, Sheridan et al. demonstrated that the egtA gene of A. fumigatus is essential for EGT biosynthesis (58). Deletion of egtA blocked EGT production, attenuated gliotoxin production, and significantly increased GSH levels in the A. fumigatus strain AfS77 (58). However, it is still unclear that this compensatory mechanism protects A. fumigatus from redox stress. Furthermore, EGT deficiency in A. fumigatus led to an increased susceptibility to oxidative stress agents, hydrogen peroxide (H2O2) and menadione, clearly demonstrating that EGT is crucial for A. fumigatus resistance against high concentrations of superoxide radicals that probably form a product that interacts with EGT given the slow reaction rate of EGT with superoxide radicals.
In addition, the deletion of EGT biosynthesis in a redox-sensitive strain of A. fumigatus, Δyap1 (Yap1 is a oxidative stress response regulator), resulted in significantly increased susceptibility to oxidative stressors such as H2O2, paraquat, menadione, and tert-butylhydroperoxide, as well as heavy metals such as iron, zinc, copper, and cobalt (58) (Fig. 8). Although EGT deficiency decreased resistance of ΔegtA to H2O2 levels at 3 μM, radial growth of ΔegtA was unaffected at 1 and 2 μM H2O2, suggesting that EGT functions as an auxiliary antioxidant that is required for growth at elevated oxidative stress conditions (58).
Global proteome analysis revealed that that the loss of EGT in A. fumigatus (ΔegtA) also resulted in impairment of several proteins involved in maintaining redox such as reductases, oxidases, stress response proteins, and enzymes with oxidizing products (58). This further highlights the role of EGT as a key redox regulator in A. fumigatus. Of note, HapC, the redox-sensitive subunit of the CCAAT-binding complex, which is involved in the oxidative stress response (64), is absent in ΔegtA indicating a defective oxidative system in A. fumigatus (58). In addition, the proteome profiles indicated that cystathionine metabolism was dysregulated in ΔegtA.
Under basal conditions, cystathionine γ-synthase that catalyzes the formation of cystathionine from cysteine and homoserine was absent in ΔegtA compared with wild type, proposing that cystathionine production is reduced when EGT biosynthesis is dysfunctional, possibly to provide more cysteine for GSH production. However, after addition of H2O2, there was a significant increase in cystathionine β-lyase (CBL), which catalyzes the conversion of cystathionine to homocysteine, ammonia, and pyruvate. The resulting increased homocysteine formation can be converted into methionine, which is required for SAM biosynthesis that is needed for the trimethylation step catalyzed by EgtA in EGT biosynthesis. Thus, increased CBL in egtA in response to H2O2 could lead to a shift toward increasing SAM availability, via methionine, in an attempt to synthesize EGT. The proteomic data also show an increased abundance of HisF involved in the biosynthesis of histidine and sulfite reductase, indicating an increased need for sulfur, both further supporting the attempts of ΔegtA to synthesize EGT (58).
During iron starvation, A. fumigatus produces an intracellular siderophore, ferricrocin (Fc), and two extracellular siderophores, fusarinine-C (FsC) or triacetylfusarinine-C (TAFC). Lack of siderophores, leading to a lower iron uptake, resulted in attenuated virulence in A. fumigatus (55). Lack of EGT in A. fumigatus increased levels of intracellular Fc, and combined with Yap1 deficiency, EGT deficiency increased production of extracellular FsC and decreased TAFC, although total extracellular siderophore production was not significantly altered. Northern blots revealed transcriptional upregulation of egtA during iron starvation compared with iron sufficiency (58). SidG, an acetyltransferase that mediates the conversion of FsC to TAFC, is transcriptionally upregulated during iron starvation and downregulated in ΔegtA consistent with increased FsC and decreased TAFC production. As iron homeostasis is also important for maintaining a stable oxidative environment, this clearly implicates another facet of EGT-mediated control of redox homeostasis in A. fumigatus (Fig. 8).
Similar to N. crassa, EGT deficiency results in paler conidia and lower levels of conidiation, indicating that EGT is also important during conidial germination in A. fumigatus (9, 58) (Fig. 8). Taken together, these findings establish EGT as a central redox regulator in A. fumigatus and it underlies various mechanisms by which it maintains intracellular redox homeostasis under various intra- and extracellular stresses.
Ergothioneine in S. pombe
EGT was identified in S. pombe by Pluskal et al. as one of the metabolites in their extensive metabolite screening of S. pombe using liquid chromatography–mass spectrometry (49). In a later study, Pluskal et al. showed that EGT levels in the vegetative state of S. pombe were minimal (0.3 μM), whereas the levels were increased significantly under low-glucose starvation (41.5 μM) and nitrogen starvation (157.4 μM). In addition to the elevated EGT levels, its precursor, trimethyl histidine, and trehalose were also significantly elevated compared with their levels present under normal culture conditions, thereby implicating EGT as a necessity to control oxidative stress under low-glucose or nitrogen starvation conditions (50).
Similarly, in an environmental stress experiment, the mRNA levels of the egt1+ gene increased by ∼10 and ∼4-fold in the presence of 0.5 mM H2O2 and cadmium ions (Cd2+), respectively. This clearly implicates EGT as a stress-responsive element in S. pombe and is important against oxidant-mediated damage during environmental and heavy metal stress conditions (59) (Fig. 8). Similarly, under nitrogen starvation, the levels of EGT, its S-methyl derivative (S-methyl-EGT), and hercynylcysteine sulfoxide, an EGT precursor, were significantly elevated in S. pombe (53). Taken together, EGT appears to be highly inducible under stressful conditions and its intracellular levels play important roles in maintaining redox homeostasis.
Again, the carbon substrates available in the external environment regulate switches in the central carbon metabolism of S. pombe to supply the energy requirements of the organism. These changes can potentially alter the intracellular redox poise, which in turn will stimulate the production of redox buffers to maintain redox homeostasis required for survival of the organism. In this regard, the EGT overexpressing systems developed in S. pombe by Pluskal et al. may provide novel and interesting avenues to further explore the underlying molecular mechanisms of EGT-mediated redox regulation (50).
Role of EGT in Pathogenicity
Mtb EGT has been demonstrated to be essential for virulence of Mtb during the infection of both macrophages and in mice. As discussed in the section on EGT in mycobacteria, disruption of two different genes (egtA or egtD) in the Mtb EGT biosynthetic pathway significantly reduced the intracellular survival of Mtb in macrophages after 5 days, demonstrating that EGT is essential for intracellular survival in macrophages (52). Infection of mice with these same EGT mutants reduced the bacillary load in lungs and the number of lesions in the mouse lungs at 5 weeks postinfection. Furthermore, histopathological analysis revealed reduced pathology in the mouse lungs infected with the EGT mutants, that is, visible alveolar spaces with partial lung parenchyma were observed in the mouse lungs infected with the mutants in contrast to large areas of alveolar consolidation due to lymphocyte-rich granulomatous infiltration in the lungs of mice infected with the wild-type Mtb and complemented strains (52). This provides evidence for the role of EGT in establishing virulent infection of Mtb in the mouse lungs and for pathological progression of disease.
Furthermore, increased intracellular levels of EGT in Mtb were observed during late logarithmic growth and stationary phases when Mtb is under more hostile conditions of limited nutrition and low pH. This requirement of EGT for Mtb survival during the stationary phase and during starvation (51) and the reduced anti-TB drug susceptibility (52) suggest that EGT plays a role in the Mtb persistence under hostile conditions present in the host. The roles of EGT in pathogenesis of fungal infections have not yet been investigated.
EGT could also possibly enhance pathogenicity of microbes due to the proinflammatory immunomodulatory effects of EGT that have been reported by Yoshida et al. (68). EGT was found to enhance TLR signaling in bone marrow-derived macrophages as pretreatment of macrophages with EGT augmented cytokine production, in particular IL-6, IL-12p40, and IL-1β, which are characteristic cytokine responses to TLR signaling (68). These cytokines are indicative of a proinflammatory polarization of macrophages that were found to promote Th17 differentiation of naïve splenic CD4+ T cells in coculture experiments (68).
Conclusions
EGT is a unique redox buffer, in that it exists in the thione form at physiological pH that is resistant to autoxidation and exhibits potent antioxidant activities through its resonance forms. EGT exhibits peculiar stability and reactivity with a more positive redox potential that is different from other naturally occurring alkylthiols such as glutathione and mycothiol (26). Although EGT disulfide is formed when EGT is oxidized, the disulfide is unstable at physiological pH and rapidly decomposes to partially reform EGT and hercynine. This may account for the lack of evidence of dedicated enzymatic pathways to channel the reducing power of ergothioneine to different targets or the association of ergothioneine with enzymatic redox-based systems. However, the fate of the stable products of EGT oxidation, such as ESO3H, in vivo is unknown.
Biosynthesis of EGT in both pathogenic and nonpathogenic microbes is required for protection against a variety of environmental stresses contributed by peroxides, superoxides, and heavy metals, as well as nutritional limitations that microbes encounter in their natural environments; however, it is not always required for logarithmic growth of bacteria (51).
EGT is also involved in maintaining microbial redox homeostasis and in secondary metabolic adaptations of bacterial and fungal physiology to environmental changes. However, microbes often produce a second redox buffer such as GSH or MSH or have alternative antioxidant molecules that assist with maintaining redox homeostasis and protecting the microbe from its natural environment, as illustrated in Mtb, where MSH was elevated in the absence of EGT (52), and in A. fumigatus, where GSH levels were increased in EGT deficiency and EGT was shown to have a more auxiliary function of protection under very high oxidative stress conditions (58). Thus, the antioxidant, cytoprotective, and metabolic properties of EGT (Figs. 7 and 8) appear to be more evident in microbes under hostile conditions.
In Mtb, EGT plays a vital role in the protection of the microbe against the defense strategies of the host and subversion of the immune response, hence augmenting virulence of the pathogen to establish infection. Furthermore, the environmental conditions in the host often result in intracellular redox imbalance that will affect antimycobacterial drug efficacy as anti-TB drugs such as isoniazid, ethionamide, and PA-824 require bioreductive activation for their antimycobacterial action. However, the mechanisms of EGT in Mtb to maintain redox homeostasis in the host and modulate drug susceptibility remain unknown.
The identification of EGT-dependent enzymatic pathways and the mechanisms of EGT in these pathways for detoxification of different peroxides, in addition to their interactions with other intracellular redox thiols, are still unknown. Identifying these pathways in human pathogens, Mtb and A. fumigatus, would further enhance our understanding of the role of EGT in the pathogens' survival strategies during disease progression and provide new paradigms to explore for the elimination of infection and disease.
Abbreviations Used
- CBL
cystathionine β-lyase
- CFU
colony-forming units
- DUF
domain of unknown function
- EGT
ergothioneine
- EH
hercynine
- ES(O)SE
ergothioneine disulfide S-monoxide
- ES(O2)SE
ergothioneine disulfide S-dioxide
- ESI
electron spray ionization
- ESO2H
ergothioneine sulfinic acid
- ESO3H
ergothioneine sulfonic acid
- ESOH
ergothioneine sulfenic acid
- ESSE
ergothioneine disulfide
- Fc
ferricrocin
- FGE
formylglycine-generating enzyme
- FIA
flow injection analysis
- FsC
fusarinine C
- GSH
glutathione
- MSH
mycothiol
- Mtb
Mycobacterium tuberculosis
- Ohr
organic hydroperoxide reductase
- PLP
pyridoxal phosphate
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- SAM
S-adenosylmethionine
- TAFC
triacetylfusarinine C
- TLR
Toll-like receptor
Acknowledgments
This work was supported by NIH grants R01Al111940 and R21A127182 and by pilot funds from the UAB CFAR, CFRB, and Infectious Diseases and Global Health and Vaccines Initiative to A.J.C.S. A.J.C.S is a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease. The research from which this publication emanated was cofunded by the South African Medical Research Council.
References
- 1.Akanmu D, Cecchini R, Aruoma OI, and Halliwell B. The antioxidant action of ergothioneine. Arch Biochem Biophys 288: 10–16, 1991 [DOI] [PubMed] [Google Scholar]
- 2.Alamgir KM, Masuda S, Fujitani Y, Fukuda F, and Tani A. Production of ergothioneine by Methylobacterium species. Front Microbiol 6: 1185, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antelmann H. and Helmann JD. Thiol-Based Redox Switches and Gene Regulation. Antioxid Redox Signal 14: 1049–1063, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aruoma OI, Whiteman M, England TG, and Halliwell B. Antioxidant action of ergothioneine: assessment of its ability to scavenge peroxynitrite. Biochem Biophys Res Commun 231: 389–391, 1997 [DOI] [PubMed] [Google Scholar]
- 5.Av-Gay Y. and Everett M. The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol 8: 238–244, 2000 [DOI] [PubMed] [Google Scholar]
- 6.Barger G. and Ewins AJ. CCLVII.—The constitution of ergothioneine: a betaine related to histidine. J Chem Soc Trans 99: 2336–2341, 1911 [Google Scholar]
- 7.Be NA, Bishai WR, and Jain SK. Role of Mycobacterium tuberculosis pknD in the pathogenesis of central nervous system tuberculosis. BMC Microbiol 12: 7, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bello MH, Barrera-Perez V, Morin D, and Epstein L. The Neurospora crassa mutant NcDeltaEgt-1 identifies an ergothioneine biosynthetic gene and demonstrates that ergothioneine enhances conidial survival and protects against peroxide toxicity during conidial germination. Fungal Genet Biol 49: 160–172, 2012 [DOI] [PubMed] [Google Scholar]
- 9.Bello MH, Mogannam JC, Morin D, and Epstein L. Endogenous ergothioneine is required for wild type levels of conidiogenesis and conidial survival but does not protect against 254 nm UV-induced mutagenesis or kill. Fungal Genet Biol 73: 120–127, 2014 [DOI] [PubMed] [Google Scholar]
- 10.Braunshausen A. and Seebeck FP. Identification and characterization of the first ovothiol biosynthetic enzyme. J Am Chem Soc 133: 1757–1759, 2011 [DOI] [PubMed] [Google Scholar]
- 11.Carlsson J, Kierstan MP, and Brocklehurst K. Reactions of L-ergothioneine and some other aminothiones with2,2′-and 4,4′-dipyridyl disulphides and of L-ergothioneine with iodoacetamide. 2-Mercaptoimidazoles, 2- and 4-thiopyridones, thiourea and thioacetamide as highly reactive neutral sulphur nucleophils. Biochem J 139: 221–235, 1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheah IK. and Halliwell B. Ergothioneine; antioxidant potential, physiological function and role in disease. Biochim Biophys Acta 1822: 784–793, 2012 [DOI] [PubMed] [Google Scholar]
- 13.de Rey-Pailhade J. Sur un corps d'origine organique hydrogénant le soufre á froid. CR Acad Sci 106: 1683–1684, 1888 [Google Scholar]
- 14.Ey J, Schomig E and Taubert D. Dietary sources and antioxidant effects of ergothioneine. J Agric Food Chem 55: 6466–6474, 2007 [DOI] [PubMed] [Google Scholar]
- 15.Fahey RC. Novel thiols of prokaryotes. Annu Rev Microbiol 55: 333–356, 2001 [DOI] [PubMed] [Google Scholar]
- 16.Fahey RC. Glutathione analogs in prokaryotes. Biochim Biophys Acta 1830: 3182–3198, 2013 [DOI] [PubMed] [Google Scholar]
- 17.Gaballa A, Newton GL, Antelmann H, Parsonage D, Upton H, Rawat M, Claiborne A, Fahey RC, and Helmann JD. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc Natl Acad Sci U S A 107: 6482–6486, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallagher L, Owens RA, Dolan SK, O'Keeffe G, Schrettl M, Kavanagh K, Jones GW, and Doyle S. The Aspergillus fumigatus protein GliK protects against oxidative stress and is essential for gliotoxin biosynthesis. Eukaryot Cell 11: 1226–1238, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genghof DS. Biosynthesis of ergothioneine and hercynine by fungi and Actinomycetales. J Bacteriol 103: 475–478, 1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genghof DS. and Van Damme O. Biosynthesis of ergothioneine from endogenous hercynine in Mycobacterium smegmatis. J Bacteriol 95: 340–344, 1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genghof DS. and Vandamme O. Biosynthesis of ergothioneine and hercynine by mycobacteria. J Bacteriol 87: 852–862, 1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goncharenko KV, Vit A, Blankenfeldt W, and Seebeck FP. Structure of the sulfoxide synthase EgtB from the ergothioneine biosynthetic pathway. Angew Chem Int Ed Engl 54: 2821–2824, 2015 [DOI] [PubMed] [Google Scholar]
- 23.Grundemann D, Harlfinger S, Golz S, Geerts A, Lazar A, Berkels R, Jung N, Rubbert A, and Schomig E. Discovery of the ergothioneine transporter. Proc Natl Acad Sci U S A 102: 5256–5261, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hand CE. and Honek JF. Biological chemistry of naturally occurring thiols of microbial and marine origin. J Nat Prod 68: 293–308, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Hanlon DP. Interaction of ergothioneine with metal ions and metalloenzymes. J Med Chem 14: 1084–1087, 1971 [DOI] [PubMed] [Google Scholar]
- 26.Hartman PE. Ergothioneine as antioxidant. Methods Enzymol 186: 310–318, 1990 [DOI] [PubMed] [Google Scholar]
- 27.Heath H, Rimington C, Glover T, Mann T, and Leone E. Studies using radioactive sulphur on ergothioneine formation in the pig. Biochem J 54: 606–611, 1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heath H. and Toennies G. The preparation and properties of ergothioneine disulphide. Biochem J 68: 204–210, 1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heath H. and Wildy J. The biosynthesis of ergothioneine and histidine by Claviceps purpurea. I. The incorporation of [2-14C]acetate. Biochem J 64: 612–620, 1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hopkins FG. On glutathione: a reinvestigation. J Biol Chem 84: 269–320, 1929 [Google Scholar]
- 31.Hu W, Song H, Sae Her A, Bak DW, Naowarojna N, Elliott SJ, Qin L, Chen X, and Liu P. Bioinformatic and biochemical characterizations of C–S bond formation and cleavage enzymes in the fungus Neurospora crassa ergothioneine biosynthetic pathway. Org Lett 16: 5382–5385, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hua Long L. and Halliwell B. Oxidation and generation of hydrogen peroxide by thiol compounds in commonly used cell culture media. Biochem Biophys Res Commun 286: 991–994, 2001 [DOI] [PubMed] [Google Scholar]
- 33.Jocelyn PC. Biochemistry of the SH Group: The Occurrence, Chemical Properties, Metabolism and Biological Function of Thiols and Disulphides. London: Academic Press, 1972 [Google Scholar]
- 34.Jones GW, Doyle S, and Fitzpatrick DA. The evolutionary history of the genes involved in the biosynthesis of the antioxidant ergothioneine. Gene 549: 161–170, 2014 [DOI] [PubMed] [Google Scholar]
- 35.Kari C, Nagy Z, Kovacs P, and Hernadi F. Mechanism of the growth inhibitory effect of cysteine on Escherichia coli. J Gen Microbiol 68: 349–356, 1971 [DOI] [PubMed] [Google Scholar]
- 36.Mayumi T, Kawano H, Sakamoto Y, Suehisa E, Kawai Y, and Hama T. Studies on Ergothioneine. V. Determination by high performance liquid chromatography and application to metabolic research. Chem Pharm Bull 26: 3772–3778, 1978 [DOI] [PubMed] [Google Scholar]
- 37.Melville DB. Ergothioneine. Vitam Horm 17: 155–204, 1959 [Google Scholar]
- 38.Melville DB. and Eich S. The occurrence of ergothioneine in plant material. J Biol Chem 218: 647–651, 1956 [PubMed] [Google Scholar]
- 39.Melville DB, Eich S, and Ludwig ML. The biosynthesis of ergothioneine. J Biol Chem 224: 871–877, 1957 [PubMed] [Google Scholar]
- 40.Melville DB, Genghof DS, Inamine E, and Kovalenko V. Ergothioneine in microorganisms. J Biol Chem 223: 9–17, 1956 [PubMed] [Google Scholar]
- 41.Misra HP. Generation of superoxide free radical during the autoxidation of thiols. J Biol Chem 249: 2151–2155, 1974 [PubMed] [Google Scholar]
- 42.Mukhopadhyay R, Dey S, Xu N, Gage D, Lightbody J, Ouellette M, and Rosen BP. Trypanothione overproduction and resistance to antimonials and arsenicals in Leishmania. Proc Natl Acad Sci USA 93: 10383–10387, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Motohashi N, Mori I, Sugiura Y, and Tanaka H. Metal complexes of ergothioneine. Chem Pharm Bull 22: 654–657, 1974 [Google Scholar]
- 44.Nakajima S, Satoh Y, Yanashima K, Matsui T, and Dairi T. Ergothioneine protects Streptomyces coelicolor A3(2) from oxidative stresses. J Biosci Bioeng 120: 294–298, 2015 [DOI] [PubMed] [Google Scholar]
- 45.Narainsamy K, Farci S, Braun E, Junot C, Cassier-Chauvat C, and Chauvat F. Oxidative-stress detoxification and signalling in cyanobacteria: the crucial glutathione synthesis pathway supports the production of ergothioneine and ophthalmate. Mol Microbiol 100: 15–24, 2016 [DOI] [PubMed] [Google Scholar]
- 46.Newton GL, Buchmeier N, and Fahey RC. Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol Mol Biol Rev 72: 471–494, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newton GL, Ta P, Bzymek KP, and Fahey RC. Biochemistry of the initial steps of mycothiol biosynthesis. J Biol Chem 281: 33910–33920, 2006 [DOI] [PubMed] [Google Scholar]
- 48.Pfeiffer C, Bauer T, Surek B, Schömig E, and Gründemann D. Cyanobacteria produce high levels of ergothioneine. Food Chem 129: 1766–1769, 2011 [Google Scholar]
- 49.Pluskal T, Nakamura T, Villar-Briones A, and Yanagida M. Metabolic profiling of the fission yeast S. pombe: quantification of compounds under different temperatures and genetic perturbation. Mol Biosyst 6: 182–198, 2010 [DOI] [PubMed] [Google Scholar]
- 50.Pluskal T, Ueno M, and Yanagida M. Genetic and metabolomic dissection of the ergothioneine and selenoneine biosynthetic pathway in the fission yeast, S. pombe, and construction of an overproduction system. PLoS One 9: e97774, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richard-Greenblatt M, Bach H, Adamson J, Pena-Diaz S, Li W, Steyn AJ, and Av-Gay Y. Regulation of ergothioneine biosynthesis and its effect on Mycobacterium tuberculosis growth and infectivity. J Biol Chem 290: 23064–23076, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saini V, Cumming BM, Guidry L, Lamprecht DA, Adamson JH, Reddy VP, Chinta KC, Mazorodze JH, Glasgow JN, Richard-Greenblatt M, Gomez-Velasco A, Bach H, Av-Gay Y, Eoh H, Rhee K, and Steyn AJ. Ergothioneine maintains redox and bioenergetic homeostasis essential for drug susceptibility and virulence of Mycobacterium tuberculosis. Cell Rep 14: 572–585, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sajiki K, Pluskal T, Shimanuki M, and Yanagida M. Metabolomic analysis of fission yeast at the onset of nitrogen starvation. Metabolites 3: 1118–1129, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sao Emani C, Williams MJ, Wiid IJ, Hiten NF, Viljoen AJ, Pietersen RD, van Helden PD, and Baker B. Ergothioneine is a secreted antioxidant in Mycobacterium smegmatis. Antimicrob Agents Chemother 57: 3202–3207, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schrettl M, Bignell E, Kragl C, Sabiha Y, Loss O, Eisendle M, Wallner A, Arst HN, Jr., Haynes K, and Haas H. Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog 3: 1195–1207, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seebeck FP. In vitro reconstitution of Mycobacterial ergothioneine biosynthesis. J Am Chem Soc 132: 6632–6633, 2010 [DOI] [PubMed] [Google Scholar]
- 57.Servillo L, Castaldo D, Casale R, D'Onofrio N, Giovane A, Cautela D, and Balestrieri ML. An uncommon redox behavior sheds light on the cellular antioxidant properties of ergothioneine. Free Radic Biol Med 79: 228–236, 2015 [DOI] [PubMed] [Google Scholar]
- 58.Sheridan KJ, Lechner BE, Keeffe GO, Keller MA, Werner ER, Lindner H, Jones GW, Haas H, and Doyle S. Ergothioneine biosynthesis and functionality in the opportunistic fungal pathogen, Aspergillus fumigatus. Sci Rep 6: 35306, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh A, Crossman DK, Mai D, Guidry L, Voskuil MI, Renfrow MB, and Steyn AJ. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog 5: e1000545, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh A, Guidry L, Narasimhulu KV, Mai D, Trombley J, Redding KE, Giles GI, Lancaster JR, Jr., and Steyn AJ. Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival. Proc Natl Acad Sci U S A 104: 11562–11567, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh AR, Strankman A, Orkusyan R, Purwantini E, and Rawat M. Lack of mycothiol and ergothioneine induces different protective mechanisms in Mycobacterium smegmatis. Biochem Biophys Rep 8: 100–106, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ta P, Buchmeier N, Newton GL, Rawat M, and Fahey RC. Organic hydroperoxide resistance protein and ergothioneine compensate for loss of mycothiol in Mycobacterium smegmatis mutants. J Bacteriol 193: 1981–1990, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tanret C. Sur une base nouvelle retiree du seigle ergote, l'ergothioneine. Compt Rende 49: 22–224, 1909 [Google Scholar]
- 64.Thon M, Al Abdallah Q, Hortschansky P, Scharf DH, Eisendle M, Haas H, and Brakhage AA. The CCAAT-binding complex coordinates the oxidative stress response in eukaryotes. Nucleic Acids Res 38: 1098–1113, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vilcheze C, Av-Gay Y, Attarian R, Liu Z, Hazbon MH, Colangeli R, Chen B, Liu W, Alland D, Sacchettini JC, and Jacobs WR., Jr. Mycothiol biosynthesis is essential for ethionamide susceptibility in Mycobacterium tuberculosis. Mol Microbiol 69: 1316–1329, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vit A, Mashabela GT, Blankenfeldt W, and Seebeck FP. Structure of the ergothioneine-biosynthesis amidohydrolase EgtC. Chembiochem 16: 1490–1496, 2015 [DOI] [PubMed] [Google Scholar]
- 67.Vit A, Misson L, Blankenfeldt W, and Seebeck FP. Crystallization and preliminary X-ray analysis of the ergothioneine-biosynthetic methyltransferase EgtD. Acta Crystallogr F Struct Biol Commun 70: 676–680, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoshida S, Shime H, Funami K, Takaki H, Matsumoto M, Kasahara M, and Seya T. The anti-oxidant ergothioneine augments the immunomodulatory function of TLR agonists by direct action on macrophages. PLoS One 12: e0169360, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao Q, Wang M, Xu D, Zhang Q, and Liu W. Metabolic coupling of two small-molecule thiols programs the biosynthesis of lincomycin A. Nature 518: 115–119, 2015 [DOI] [PubMed] [Google Scholar]