Abstract
Significance: Since the discovery and structural characterization of bacillithiol (BSH), the biochemical functions of BSH-biosynthesis enzymes (BshA/B/C) and BSH-dependent detoxification enzymes (FosB, Bst, GlxA/B) have been explored in Bacillus and Staphylococcus species. It was shown that BSH plays an important role in detoxification of reactive oxygen and electrophilic species, alkylating agents, toxins, and antibiotics.
Recent Advances: More recently, new functions of BSH were discovered in metal homeostasis (Zn buffering, Fe-sulfur cluster, and copper homeostasis) and virulence control in Staphylococcus aureus. Unexpectedly, strains of the S. aureus NCTC8325 lineage were identified as natural BSH-deficient mutants. Modern mass spectrometry-based approaches have revealed the global reach of protein S-bacillithiolation in Firmicutes as an important regulatory redox modification under hypochlorite stress. S-bacillithiolation of OhrR, MetE, and glyceraldehyde-3-phosphate dehydrogenase (Gap) functions, analogous to S-glutathionylation, as both a redox-regulatory device and in thiol protection under oxidative stress.
Critical Issues: Although the functions of the bacilliredoxin (Brx) pathways in the reversal of S-bacillithiolations have been recently addressed, significantly more work is needed to establish the complete Brx reduction pathway, including the major enzyme(s), for reduction of oxidized BSH (BSSB) and the targets of Brx action in vivo.
Future Directions: Despite the large number of identified S-bacillithiolated proteins, the physiological relevance of this redox modification was shown for only selected targets and should be a subject of future studies. In addition, many more BSH-dependent detoxification enzymes are evident from previous studies, although their roles and biochemical mechanisms require further study. This review of BSH research also pin-points these missing gaps for future research. Antioxid. Redox Signal. 28, 445–462.
Keywords: : Bacillus subtilis, Staphylococcus aureus, bacillithiol, BSH biosynthesis, metal homeostasis, methylglyoxal, S-bacillithiolation, bacilliredoxin
Historical Context: Glutathione and the Discovery of Alternative Low-Molecular-Weight Thiols
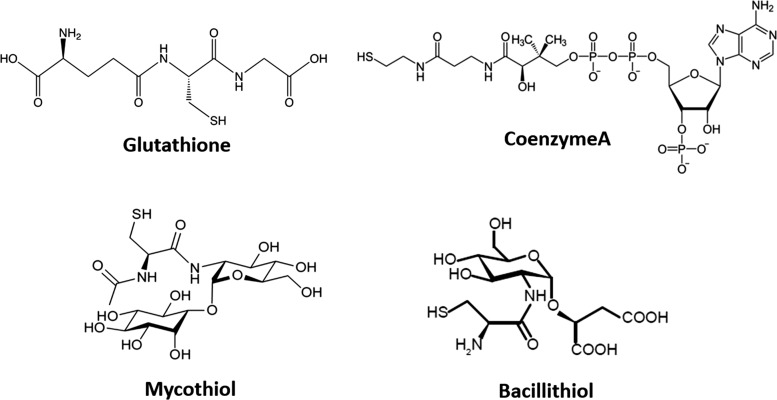
Low-molecular-weight (LMW) thiols serve a critical protective role in cells by helping maintain cytosolic proteins in their reduced state and as protection against reactive oxygen species (ROS) and reactive electrophilic species, antibiotics, as well as heavy metals. Glutathione (GSH), a tripeptide with composition γ-L-glutamyl-L-cysteinylglycine (Fig. 1), is, by far, the best studied member from this class of compounds.
FIG. 1.
Structures of major LMW thiols in bacteria. Glutathione is utilized as the major LMW thiol in eukaryotes and Gram-negative bacteria, mycothiol in Actinomycetes, and BSH in Firmicutes. Coenzyme A (CoASH) also serves as an LMW thiol in Staphylococcus aureus and Bacillus anthracis. BSH, bacillithiol; LMW, low molecular weight.
Although thiols were detected in tissues in the late 19th century, the discovery of GSH is appropriately attributed to the celebrated biochemist Sir Fredrick Hopkins in 1921, co-winner (with Christiaan Eijkman) of the 1929 Nobel Prize in Physiology or Medicine for his discovery of vitamins. In his seminal paper on GSH (53), Hopkins begins by giving credit to the studies of Joseph de Rey-Pailhade (20), several decades prior (1888), that led to the description of “philothion” as a cellular reductant. Despite uncertainty regarding the precise chemical composition, Hopkins proposed the name glutathione for what he surmised was a dipeptide containing Glu and Cys:
Provisionally, for easy reference, the name Glutathione will perhaps be admissible. It leaves a link with the historic Philothion, has the same termination as Peptone, which has long served as a name for the simpler peptides, and is a sufficient reminder that the dipeptide contains glutamic acid linked to a sulphur compound (53).
Doubts about the proposed chemical composition soon emerged. Hopkins acknowledged this, and in 1929 published evidence of the general tripeptide composition of GSH as well as methods for its large-scale purification (54). These studies revealed that GSH does not contain a thione (C = S) moiety, but instead has a thiol (C-SH) (Fig. 1), but the name glutathione has since been retained.
By the 1950s, the chemistry of thiols had been thoroughly investigated, and Barron (6) could conclude that GSH, as the major LMW thiol in cells, “...protects essential -SH groups from oxidation, and it protects the tissues from the toxic effects of heavy metals.” By this time, the impression had emerged that GSH was a universal constituent of cells (6). Despite the proclaimed ubiquity of GSH, by the mid-1970s, Robert Fahey's group had developed evidence pointing to the absence of GSH in several species of bacteria (28).
Following up on this insight, the alternative thiol designated mycothiol (MSH) was purified and structurally characterized from Streptomyces sp. AJ 9463 in the disulfide form (119), from Mycobacterium bovis (125) and from Streptomyces clavuligerus (94), and shown to be the major LMW thiol in most actinomycetes (Fig. 1) (92). As discussed later, the biosynthetic pathways leading to MSH and to bacillithiol (BSH) are closely related. The discovery and properties of MSH have been recently reviewed (27, 59).
In addition to MSH, some mycobacteria also produce another LMW thiol, ergothioneine (EGT; 2-mercaptohistidine trimethylbetaine), originally discovered in the ergot fungus as reviewed in (26). Chemically, EGT is distinct from other LMW thiols in that a major fraction exists as the thione tautomer. In addition to MSH, other alternative thiols have also become appreciated for their roles as alternative LMW thiols. One of the first to be described, trypanothione [T(SH)2; Bis-glutathionylspermidine], is found in trypanosomatid parasites and consists of two GSH molecules linked by spermidine (29). Trypanosomes also contain ovothiol A (N1-methyl-4-mercaptohistidine).
Bacillithiol: Discovery, Structure Determination, and Biosynthetic Pathway
Although the absence of GSH in many bacteria was appreciated by 1978 (28), nearly 20 years would elapse before bacillithiol (BSH) would be identified as the major LMW thiol in many Firmicutes (low GC, Gram-positive bacteria). This phylum is represented by the model organism Bacillus subtilis and includes several important human pathogens. BSH has, to date, been documented in Bacillus and Staphylococcus spp., Streptococcus agalactiae, and Deinococcus radiodurans (97). In the interim, it was speculated that the function of GSH in these organisms might have been subsumed by Cys (10) or coenzyme A (CoASH) (21). The presence of a specific CoA-disulfide reductase in Staphylococcus aureus and Bacillus anthracis supports the notion that CoASH is reversibly oxidized in vivo and may serve as an LMW thiol (21).
BSH was initially detected by HPLC analysis, together with CoASH, as an abundant 398 Da LMW thiol during studies of thiol-dependent enzymes in B. anthracis (99). Independently, a thiol of this same mass was detected by mass spectrometry as the major adduct for in vivo oxidized OhrR protein in B. subtilis (64). OhrR is a DNA-binding protein with a single redox-active Cys residue in each monomer and responds to thiol oxidants by formation of mixed disulfides, including, as a dominant product, the S-bacillithiolated protein.
Following up on this initial discovery, rapid progress was made in a coordinated multi-laboratory effort that led to the determination of the chemical structure for BSH, and insights into its distribution (97) and biosynthetic pathway (40). The identification of the biosynthetic pathway enabled the generation of mutant strains lacking BSH, and initial insights into its physiological role. Working on the general assumption that BSH has likely supplanted GSH as an enzyme cofactor in these organisms, it was also possible to predict the presence of various different types of BSH-dependent enzymes (e.g., bacillithiol-S-transferases [BST], analogous to glutathionyl-S-transferases) and redox partners (e.g., bacilliredoxins [Brx], analogous to glutaredoxins). Even at this early stage (only two prior research papers on BSH had appeared), this enabled the presentation of an early preview of how the BSH field might evolve, and readers are referred to this prior review for a more detailed discussion of these early steps, which provides a context for the present review (50).
The structure of BSH was determined, after purification of the S-bimane derivative from D. radiodurans, to be the α-anomeric glycoside of L-cysteinyl-D-glucosamine with L-malic acid (97). Based on the chemical similarities between MSH and BSH, it was possible to identify candidate genes for the first two steps in BSH biosynthesis by homology (40). This led to the identification of a cluster of co-transcribed genes in B. subtilis that includes bshA (encoding a L-malic acid glycosyltransferase) and bshB1 (one of two partially redundant deacetylases for conversion of GlcNAc-Mal to GlcN-Mal). This same operon also includes mgsA (a methylglyoxal synthase), which is of interest since (as discussed below) BSH plays a major role in methylglyoxal (MG) detoxification (11). This gene cluster is immediately upstream of and co-directional with the panBCD operon encoding enzymes in the CoASH biosynthetic pathway.
The bshC gene could not be identified by homology-based searches, but was revealed by phylogenomic comparisons (using the EMBL STRINGS web-based tool) as a gene with a high co-occurrence (and occasional co-localization) with bshA in bacterial genomes (40). The bshC gene is transcribed both from its own promoter and also as part of a two-gene operon with another predicted pantothenate biosynthesis gene, panE (ylbQ) (37).
The significance of this gene clustering and possible coordinate regulation is not yet understood, but an obvious suggestion is that the synthesis of CoASH and BSH may be positively correlated (50). Indeed, in B. subtilis, all of the genes required for BSH synthesis are upregulated in response to disulfide stress (e.g., diamide) through the action of the Spx transcription factor (37). A similar induction has also been documented in S. aureus (111). More broadly, genes for both BSH synthesis and BSH-dependent detoxification reactions are upregulated by several reactive oxidants and reactive electrophiles, as noted in several studies in B. subtilis, B. anthracis, and S. aureus [reviewed in Perera et al. (107)].
In addition to providing a candidate gene for the last and missing step in BSH biosynthesis (bshC), phylogenomic profiling also provided an intriguing list of genes encoding proteins that are likely to function in core BSH metabolism. These included several genes of previously unassigned function (so-called y-genes). The ypdA gene encodes a putative thioredoxin reductase (TrxR) homolog and is postulated to function as a bacillithiol disulfide (BSSB) reductase, although experimental evidence is still lacking. Two others (yqiW and yphP) encode proteins with DUF1094 domains (domain of unknown function containing a conserved Cys-x-Cys motif), and YphP was shown to be active as a disulfide isomerase (22). This led to the prediction that these proteins might function as Brx for the reduction of S-bacillithiolated proteins, as described later. Finally, ytxJ was found to encode another putative redox-active protein related to Trx that also presumably functions in BSH metabolism.
Identification of the biosynthetic genes for BSH enabled the generation of mutants lacking this thiol, confirming that BSH is non-essential. Mutants lacking the ability to synthesize BSH are sensitive to a variety of oxidative and electrophilic stress conditions in B. subtilis and S. aureus (14, 15, 40, 111, 114). Moreover, it quickly became apparent that a previous suggestion that the B. subtilis FosB fosfomycin resistance protein might utilize Cys as a cofactor (10) was incorrect. FosB (as described in more detail later) is a BSH-dependent enzyme, and cells lacking either fosB or BSH biosynthesis are equally sensitive to fosfomycin in B. subtilis and S. aureus (40, 111). In practice, this provides a very convenient (although indirect) assay for BSH levels in B. subtilis (40).
Chemical Properties of BSH
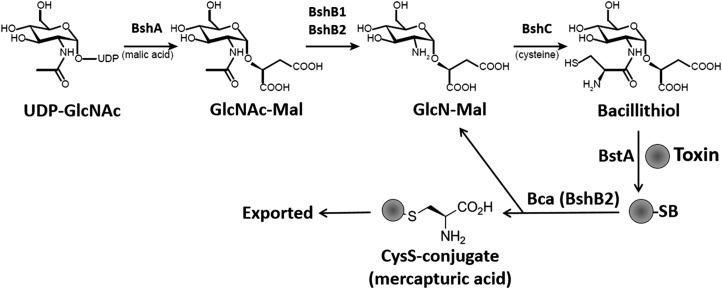
A key challenge for the further investigation of BSH was the need to obtain quantities that are sufficient for chemical and biochemical characterization. The development of methods for the chemical synthesis of BSH ultimately met this need. Chris Hamilton reported both a complete chemical synthesis and a hybrid chemoenzymatic route taking advantage of the ability of purified BshA to provide the D-GlcNAc-L-Mal precursor (121). These approaches provided access to not only BSH but also BSSB and to the biosynthetic intermediates GlcNAc-Mal and GlcN-Mal (Fig. 2). A complete chemical synthesis was also reported by Richard Armstrong's laboratory in which BSH could be obtained in 8–9% yield after 11 chemical steps (62). Purified BSH has facilitated the development of a detailed understanding of the redox chemistry and ionization behavior of BSH (120), the generation of specific BSH antibodies (an important tool for the study of S-bacillithiolation) (15), and provides a necessary cofactor for the study of BSH-dependent enzymes.
FIG. 2.
Biosynthesis pathway of BSH and BSH-dependent detoxification. In the BSH synthesis pathway of Bacillus subtilis, the glycosyltransferase BshA first adds GlcNAc to malate-producing GlcNAc-Mal. Then, the paralog N-deacetylases BshB1 and BshB2 catalyze deacetylation of GlcNAc-Mal, and BshC adds cysteine (presumably in an unidentified activated form) to GlcN-Mal, producing BSH. Detoxification of toxins, xenobiotics, or electrophiles involves their conjugation with BSH by the BSH-S-transferase BstA, generating BS-conjugates that are cleaved by the deacetylase BshB2 (Bca) to CysS-conjugates and GlcN-Mal used for BSH recycling. The CysS-conjugates are exported from the cells as mercapturic acid derivatives. In S. aureus, only one BshB-like enzyme is present that functions both as deacetylase and amidase and is essential for BSH biosynthesis.
A detailed chemical study of BSH redox and ionization properties indicates that the BSH thiol is more acidic than Cys or GSH, with a higher fraction in the reactive thiolate state at physiological pH (120). For example, at pH 7.7, an estimated 22% of BSH is in the bacillithiolate (BS−) form compared with 14.5% for Cys. The standard thiol redox potential for the BSH, BSSB couple (−221 mV) is much closer to that of free Cys (−223 mV) compared with GSH (−240 mV). Considering the relative abundance of Cys, CoASH, and BSH in the cell, it was concluded that BSH (which is present at levels near 1 mM during growth in B. subtilis and S. aureus) is the dominant LMW thiol, with levels of the BSH thiolate exceeding those of CoASH and Cys thiolates by ∼10–100-fold (111, 120).
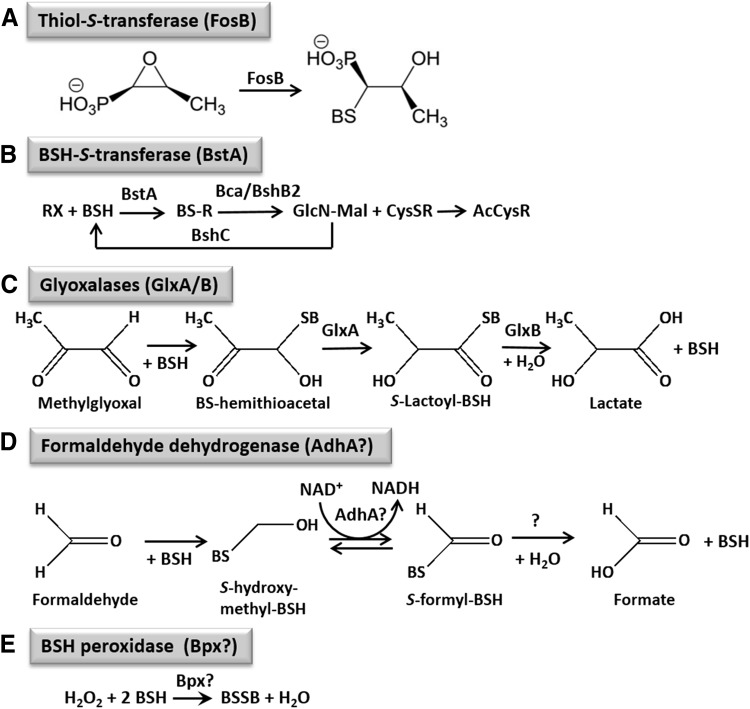
The chemistry of LMW thiol redox buffers is dominated by the interconversion of the reduced (thiol) and oxidized (disulfide) forms, and this chemistry accounts for their role in maintaining proteins in their reduced states in the cytosol. However, the thiolate anion can also serve as a nucleophile in conjugate addition reactions as commonly employed in detoxification pathways. For BSH, these reactions are mediated by BST enzymes, analogous to the well-characterized GST enzymes (Fig. 3A, B). The best characterized representative for this type of enzyme to date is FosB, as noted earlier.
FIG. 3.
The functions of BSH-dependent detoxification enzymes in B. subtilis and S. aureus. BSH functions in detoxification of reactive oxygen and electrophilic species, HOCl, and antibiotics, such as fosfomycin in B. subtilis and S. aureus. (A) BSH is a cofactor for the thiol S-transferase FosB that adds BSH to fosfomycin for its detoxification. (B) Electrophiles, xenobiotics, and toxins (RX) are conjugated to BSH by the BSH S-transferase BstA to form BS-conjugates (BSR), which are cleaved by the BSH S-conjugate amidase BshB2 (Bca) to CysSR and a mercapturic acid (AcCySR) that is exported from the cell. (C) BSH functions in methylglyoxal detoxification as a cofactor for the glyoxalases I/II (GlxA/B) in B. subtilis. GlxA converts BS-hemithioacetal to S-lactoyl-BSH that is hydrolyzed by GlxB to D-lactate. (D) AdhA is a thiol-dependent formaldehyde dehydrogenase that is induced under FA stress (98), likely converting S-hydroxymethyl-BSH to S-formyl-BSH. In the final step, BSH and formate are released by an unidentified S-formyl-BSH hydrolase. (E) Unknown thiol-dependent peroxidases or peroxiredoxins (Bpx) might function in peroxide detoxification. Question marks indicate uncharacterized reactions.
Oxidized disulfides can also react with hydrogen sulfide (H2S), a metabolite produced by sulfate-reducing bacteria and also serving as a secondary messenger in mammalian systems (7). H2S exists predominantly as the hydrosulfide anion (HS−), which reacts with BSSB to generate BSH and BSSH (bacillithiol persulfide). S. aureus was recently shown to contain an inducible system that functions to detoxify reactive persulfides. The CstB protein, a persulfide dioxygenase sulfurtransferase, oxidizes BSSH (as well as other LMW thiol persulfides) in the presence of sulfite to generate reduced thiols (BSH) and thiosulfate (122). These authors also suggest that previously noted enzymes with a homology to CoASH disulfide reductases (CoA disulfide reductase-rhodanese homology domain protein; CoADR-RHD) may actually function as reductases for the CoASSH persulfide. The general prevalence of persulfides in bacterial physiology, and the various means for their detoxification, is not yet fully understood.
Biochemical Mechanism of BSH Biosynthesis
Since BSH plays a critical role in antibiotic detoxification and oxidative stress responses, the enzymes involved in BSH biosynthesis are attractive targets for novel antibiotic development. In fact, S. aureus strains unable to produce BSH are compromised for survival in the presence of macrophages and neutrophils (111). BshA, BshB, and BshC catalyze the enzymatic synthesis of BSH (Fig. 2). BshA is homologous to Mycobacterium tuberculosis MshA and is a GT-4 class glycosyltransferase that is required for the first committed step in BSH biosynthesis. BshA catalyzes the addition of UDP-N-acetylglucosamine (UDP-GlcNAc) to L-malate through a metal-independent SN1-like mechanism, forming N-acetylglucosaminyl-malate (GlcNAc-Mal) and free UDP (130, 133). Structural and functional studies suggest that the release of the UDP-leaving group and nucleophilic attack by L-malate occur on the same face of the hexose sugar and are asynchronous, resulting in the formation of a short-lived oxocarbenium intermediate (133).
The second step of BSH biosynthesis is catalyzed by BshB, an N-acetylhydrolase, that generates glucosamine malate (GlcN-Mal) from GlcNAc-Mal. Genetic studies indicate the presence of one or more functionally redundant proteins in B. subtilis, B. anthracis, and B. cereus (31, 40). In B. subtilis, strains lacking BshB1 still produced detectible levels of BSH (40). However, BSH is completely absent on inactivation of both BshB1 and a second deacetylase BshB2, encoded by bshB2 (40). The same overlapping roles in BSH synthesis were observed for the two BshB-paralogs BA1557 and BA3888 in B. anthracis as well as for BC1534 and BC3461 in B. cereus (31). Biochemical evidence was provided that both BshB-paralogs (BA1557 and BA3888) and the orthologs in B. cereus have BshB activity and catalyze the N-deacetylation of GlcNAc-Mal (31). In contrast, S. aureus only encodes a single BshB-like protein that is essential for BSH synthesis and has a dual function as BSH conjugate amidase (Bca) (114).
Redundancy has been observed also for MSH biosynthesis, where the deacetylation can be carried out by either the BshB-like enzyme MshB or the “moonlighting” enzyme Mca (mycothiol-S-conjugate amidase). The primary role of Mca is in detoxification of xenobiotic compounds through the cleavage of MSH conjugates, resulting in GlcN-myo-inositol and the Cys-S-conjugate (mercapturic acid) that is exported from the cell (93).
Biochemical studies of the BshB-paralogs of B. anthracis and B. cereus identified only BA3888 as a bacillithiol-S-conjugate amidase (Bca) able to hydrolyze the amide linkage of bacillithiol-S-bimane (BSmB) to liberate GlcNAc-Mal (31). The reaction proceeds via a Zn2+ dependent acid-base catalysis. The active site is highly conserved, the Zn2+ is coordinated by two histidine residues and one aspartate (His12, His113, and Asp15), and the residues involved in catalysis await identification. Bca activity is highly specific to the methyl aglycone, as suggested by its low enzymatic activity when tested with BSH analogs where the methyl aglycone was replaced with an uncharged methyl or benzyl group. The in vivo contribution of Bca to BSH biosynthesis and the physiological substrates of Bca remain to be identified.
The addition of Cys to GlcN-Mal, the final step in BSH biosynthesis, is catalyzed by the putative cysteine ligase, BshC. BshC was identified by a phylogenomic analysis for genes whose presence was correlated with the presence of BshA and BshB utilizing the STRING protein interaction network tool (40). Strains lacking BshC are unable to produce BSH and accumulate elevated levels of the BSH precursor GlcN-Mal (40). Interestingly, the reaction catalyzed by BshC is unknown and has yet to be reconstituted in vitro, possibly due to the absence of a required cofactor, substrate, or protein. Structural and functional studies suggest that BshC forms a dimer in solution through interactions between an extended coiled-coil domain from each subunit and that the BshC active sites are solvent exposed, which may allow for access by an additional protein involved in catalysis (132).
BSH and Detoxification
Fosfomycin and other antibiotics
LMW thiols play an important role in the detoxification of xenobiotic compounds and antibiotics. The most well understood LMW-based detoxification mechanism involves thiol-S-transferases, which catalyse the reaction of LMW thiols to a variety of substrates. The first characterized BST was FosB, involved in the detoxification of fosfomycin (Fig. 3A) (62, 115, 128). Fosfomycin is a broad-spectrum antibiotic that is also used in clinical practice to treat methicillin-resistant S. aureus (MRSA) infections (84, 127). Fosfomycin inhibits the first step in cell wall synthesis through covalent modification of the active site cysteine of the MurA enzyme.
To counter the action of this antibiotic, bacteria have evolved fosfomycin detoxification enzymes. B. subtilis and S. aureus encode FosB, an Mn2+-dependent BST. FosB inactivates fosfomycin by catalyzing the nucleophilic addition of BSH to the C2 position of the fosfomycin epoxide ring, resulting in an inactive BS-fosfomycin complex (115). Strains lacking FosB or BSH are hypersensitive to fosfomycin and the increase in sensitivity is comparable to strains lacking both FosB and BSH, indicating that FosB utilizes BSH as a thiol substrate (40). In support of this hypothesis, biochemical studies demonstrated that S. aureus FosB is significantly more active in the presence of BSH than other LMW thiols (115).
A second class of BSTs comprised members of the DinB/YfiT superfamily of thiol transferases, which utilize a thiol cofactor to detoxify reactive electrophiles and xenobiotics (Fig. 3B). B. subtilis YfiT was identified as a putative BST by structural homology to the mycothiol-S-transferase (RV0443) found in M. tuberculosis (96). In vitro studies monitoring the reaction of the electrophilic compound monochlorobimane with BSH demonstrated that YfiT (96) and BstA (106), a structural homolog found in S. aureus, indeed, function as BSTs and are highly specific for BSH as a thiol substrate. The relevant compounds detoxified by BSTs in vivo are currently unknown. The only DinB/YfiT family protein with an identified function in vivo is the MSH-dependent maleylpyruvate isomerase (ngcl2918) from Corynebacterium glutamicum, which is essential for growth on aromatic compounds as a carbon source (32).
BSTs may also be involved in the detoxification of endogenously produced toxins that are byproducts or intermediates during secondary metabolite biosynthesis. In fact, toxic metabolites that are produced during actinorhodin biosynthesis by Streptomyces coelicolor appear to be detoxified by formation of an MSH-S-conjugate (126). The formation of this conjugate may be dependent on a DinB/YfiT-like protein.
Toxic electrophiles
The discovery of BSH as the major LMW thiol in B. subtilis led us to postulate that BSH may serve a protective function against the toxic dicarbonyl compound MG in a manner that is functionally analogous to GSH (Fig. 3C). The first hint came from the observation that the gene encoding MGS, mgsA, is co-transcribed with the genes encoding the first two enzymes in the BSH biosynthetic pathway, bshA and bshB1. In fact, strains lacking BSH are more sensitive to added MG (40).
MG, an α,β unsaturated aldehyde, is a toxic, endogenous byproduct of glycolysis, synthesized by MGS under conditions of excess carbon or phosphate limitation due to an imbalance between the rate of carbon acquisition and the lower segment of glycolysis (129). The main role of MGS is to restore inorganic phosphate levels. As an electrophile, MG can modify guanine bases in DNA, leading to DNA damage and an increased rate of mutation in surviving cells (33). Furthermore, MG can react with arginine, lysine, and cysteine residues in proteins, resulting in protein inactivation (72).
The major mechanism of protection from MG in Escherichia coli is the GSH-dependent acidification of the cytoplasm mediated by the KefGB and KefFC K+ efflux systems (34). Exposure to MG leads to the spontaneous formation of the GSH adduct hemithioacetal (HTA). Glyoxalase I (GlxI) catalyzes the formation of S-lactoylglutathione (SLG) from HTA (77). Subsequently, glyoxalase II converts SLG to D-lactate and regenerates GSH (102). KefGB and KefFC K+ efflux systems are directly inhibited by GSH and activated by GSH adducts. Thus, the SLG intermediate is critical for protection from MG stress as it is required for the full activation of the KefGB and KefFC K+ efflux pumps (77, 87). The H+ influx that accompanies the KefGB and KefFC-mediated K+ efflux leads to the cytoplasmic acidification that is sufficient to confer resistance to MG (34). Interestingly, protection from MG by cytoplasmic acidification does not increase the rate of MG detoxification (33, 34). Rather, cytoplasmic acidification likely protects cells from MG and other electrophiles by protonating nucleophilic residues on DNA, thereby preventing alkylation of DNA by electrophiles (33).
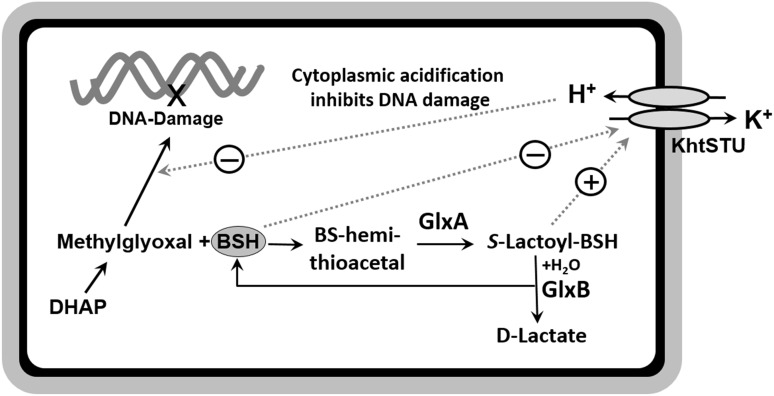
B. subtilis encodes a BSH-dependent MG-detoxification pathway (Figs. 3C and 4) (11). Null mutants of the glyoxalase I and II homologs, GlxA (formerly YwbC) and GlxB (formerly YurT), respectively, are more sensitive to MG and function in the same genetic pathway as BSH and a putative BSH-gated K+/H+ antiporter, KhtSTU (formerly YhaSTU). Upon MG stress, intracellular pH decreases ∼0.4 U, as measured by an intracellular GFP-based pH reporter (Fig. 4). This cytoplasmic acidification is sufficient for protection against MG, since cells treated with the weak membrane-permeant acid sodium acetate before MG exposure are less sensitive to MG.
FIG. 4.
BSH-dependent detoxification of methylglyoxal, leading to cytoplasmic acidification. Methylglyoxal can be produced as a byproduct of the glycolysis from DHAP. Methylglyoxal reacts spontaneously with BSH, forming BS-hemithioacetal, which is converted to S-lactoyl-BSH by the glyoxalase-I (GlxA) and to lacatate by the glyoxalase-II (GlxB). S-lactoyl-BSH activates the potassium proton antiporter KhtSTU for K-efflux and proton import, leading to cytoplasmic acidification that likely inhibits interaction of methylglyoxal with the DNA to prevent DNA damage. BSH inhibits the antiporter KhtSTU. DHAP, dihydroxyacetone phosphate.
A GSH-independent MG detoxification pathway is also present in E. coli, in which MG is converted directly to D-lactate by the glyoxalase III enzyme, HchA. Using the Phyre2 structural homology search tool, a glyoxalase III homolog, GlxC (formerly YdeA) in B. subtilis was identified (11). GlxC null mutants are more sensitive to MG than wild-type cells, although to a lesser extent than a BSH null mutant. The MG sensitivity of the glcX bshC double mutant was additive when compared with the glxC and bshC single mutants, which suggests that GlxC is a BSH-independent glyoxalase III enzyme and that the major detoxification pathway is BSH dependent (11).
Methanotrophic and methylotrophic bacteria generate formaldehyde (FA) as an intermediate during the oxidation of methane to carbon dioxide. FA is a toxic carbonyl compound that, similar to MG, is a reactive electrophile and can react with nucleophilic groups in proteins and DNA, leading to protein-protein and protein-DNA crosslinking. In these bacteria, FA is either assimilated by the serine or ribose monophosphate pathway (RuMP) to be used as a major source of cellular carbon or oxidized to generate NADH and formate by LMW thiol-dependent formaldehyde dehydrogenases (Fdh).
Recently, a metabolomics approach identified a role for BSH in FA detoxification in Bacillus methanolicus (90) (Fig. 3D). B. methanolicus is a thermotolerant, facultative methylotroph, making it a useful strain for the large-scale production of amino acids from methanol. When grown in the presence of methanol, an S-formyl-BSH intermediate was detected in cell lysates by LC-MS. The hydrolysis of related S-formyl-GSH is usually catalyzed by S-formyl-GSH-hydrolases that are homologs of human esterases and participate in FA oxidation in GSH-producing bacteria (46). The putative esterase that is specific for hydrolysis of S-formyl-BSH is unknown in B. subtilis. The BSH-dependent FA oxidation pathway appears to be the most important under conditions where the RuMP and THF pathways are overwhelmed, such as on a rapid shift in carbon source from mannitol to methanol. Non-methanotrophs, such as B. subtilis, also use the RuMP pathway to detoxify FA. In addition, transcriptomic and proteomic characterization of the B. subtilis response to FA stress led to the identification of AdhA as a putative LMW thiol-dependent Fdh that is induced under FA and MG stress, although its precise role has not been confirmed (Fig. 3D) (98).
BSH and Metal Homeostasis
Metal ions are essential for life, yet many fundamental questions regarding the size of the cellular metal quota, how it varies with growth conditions, and how it is modulated in response to stress still remain. Metal deficiency leads to a genome-wide response that serves to increase metal import, decrease metal demand, mobilize stored metals, redistribute metals from lower-priority enzymes to support higher-priority needs, and replace metal-dependent enzymes with pathways that are dependent on other cofactors (82). Conversely, metal excess induces the expression of metal efflux systems to prevent the mismetallation of essential enzymes and the production of ROS (100). Both metal deficiency and excess are utilized by the host immune system in response to pathogens (23, 103).
The chemical speciation of metals within cells is largely unexplored. Metallation of metalloenzymes, and detection of cellular metal status by metalloregulatory proteins, is governed by the labile metal pool, which is defined as those metal ions that are hydrated or otherwise in a rapidly exchanging form. LWM thiols in eukaryotic and GSH-producing bacteria are known to participate in buffering the labile pool and in detoxifying metal ions. GSH was shown to function together with glutaredoxins in iron-sulfur (FeS)-cluster trafficking since an FeS cluster could be assembled and bridged between the active site Cys of monothiol Grx3/4 and GSH, as demonstrated both in vitro and in vivo in yeast cells (67, 89, 108).
In plants and fungi, GSH is necessary for heavy-metal sequestration as it is the major substrate for the synthesis of the heavy metal-binding peptide phytochelatin (44). In yeast, resistance to cadmium requires GSH (69). In contrast, the contribution of LWM thiols to metal ion homeostasis is poorly understood in non-GSH-producing bacteria (45, 48, 49). In the next section, we discuss the emerging role of BSH as a key player in metal homeostasis.
Zinc buffering
Between 5% and 10% of proteins require Zn2+ as either a structural or catalytic cofactor (3). Unlike Fe2+, which generates cell-damaging hydroxyl radicals in the presence of hydrogen peroxide (H2O2), Zn2+ is not redox reactive, allowing it to be adopted as a structural cofactor in a number of proteins. Zn2+ must be kept at a high enough concentration to ensure that the Zn2+ quota is sufficient for Zn2+ to perform its essential roles. However, since Zn2+, in general, binds with higher avidity than most other metals (except for Cu2+), Zn2+ must not be present in excess so that proteins utilizing other metals are not mismetallated (58). Thus, the free steady-state Zn2+ levels are highly regulated at multiple levels such that they are buffered in with a total intracellular concentration of ∼1 mM and a free Zn2+ concentration in the ∼pM range (101).
The narrow range of intracellular free Zn2+ is set by Zn2+ sensing transcription factors (101). In B. subtilis, Zur acts as a sensor of Zn2+ limitation and CzrA is a sensor of Zn2+ excess (39, 88). These metalloregulators sense the labile Zn(II) pool, consisting of Zn(II) bound reversibly to small molecules, nucleotides, and proteins in a rapidly exchanging form. Under conditions of Zn2+ sufficiency, the Zn2+-sensing transcription factor Zur represses transcription of the Zn2+ uptake systems. When Zn2+ is in excess, CzrA is inactivated, leading to derepression of transcription of the cadA and czcD efflux pumps and Zn2+ efflux. Organisms have also evolved Zn2+-independent paralogs that can functionally replace Zn2+ requiring proteins under Zn2+ starvation conditions (1, 91). In addition, Zn2+ may be mobilized from a labile Zn2+ pool in response to stress under non-steady state conditions. Work from the Maret lab demonstrated that metallothionein (MT), a cysteine-rich, Zn2+-binding LMW protein, serves as a zinc “sink” in eukaryotes and is able to control the availability of kinetically available Zn2+ in response to oxidative stress (80).
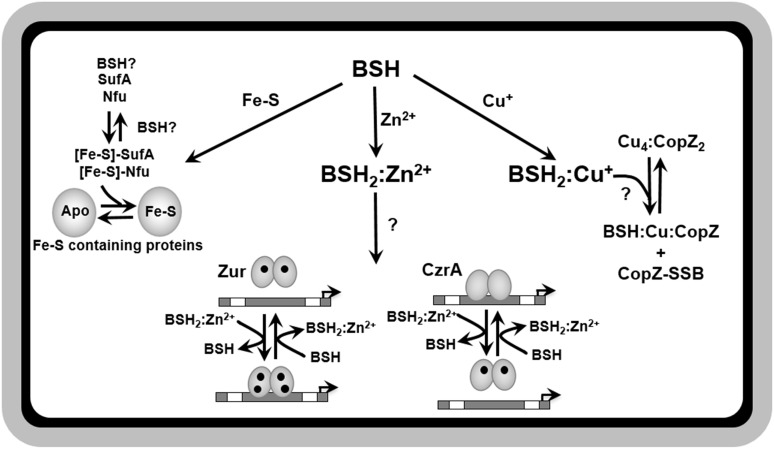
Since B. subtilis utilizes BSH as the major LMW thiol and does not produce MT or GSH, BSH could serve a similar function in metal buffering. BSH has several potential metal coordinating ligands, including a sulfur-containing thiolate, a primary amine, and two carboxylates (97). Genetic, physiological and biochemical evidence suggests that, in B. subtilis and related low G + C Firmicutes, BSH serves as the major buffer of the labile Zn2+ pool (Fig. 5) (75). BSH binds Zn2+ as a (BSH)2:Zn2+ complex with an affinity (Ka) of 1.9 × 1012 M−2 (75). Given that the intracellular concentration of BSH can range from 1 to 5 mM (120), depending on the growth phase, it is reasonable to suggest that BSH can account for ∼1/3 of the total Zn2+ pool under Zn2+ excess conditions (75).
FIG. 5.
The functions of BSH in metal homeostasis. A role for BSH in metal homeostasis has been identified for iron, zinc, and copper. BSH is required for the full activity of FeS requiring proteins in S. aureus, possibly as a key component in the assembly and delivery of FeS clusters. BSH is believed to function in an independent, yet overlapping role with the FeS carrier proteins, SufA and Nfu. BSH can also bind zinc with a high affinity and serves as a major cytosolic zinc buffer as demonstrated in B. subtilis, allowing the cell to avoid zinc intoxication under conditions of excess. Biochemical evidence also suggests a role for BSH in facilitating the delivery and removal of zinc from the zinc-sensing metalloregulators, Zur and CzrA. Lastly, BSH may also work in concert with CopZ in interacellular copper buffering and delivery to metalloproteins and may protect CopZ from overoxidation through S-bacilliothiolation. FeS, iron-sulfur.
BSH may also have direct effects on Zn2+ sensing by Zur and CzrA. In vitro experiments monitoring the binding of CzrA to its operator site as a proxy for CzrA metallation status demonstrated that Zn2+ dissociation from holo-CzrA was much faster in the presence of BSH. This suggests that BSH can facilitate Zn2+ loading and removal into CzrA, presumably through a ligand-exchange mechanism that was analogous to that described for Cu+ chaperones (116). This may provide a mechanism for rapid re-repression of Zn2+ efflux if cells experience a sudden shift from Zn2+ excess to starvation. Thus, BSH influences Zn2+ homeostasis at multiple levels as a significant intracellular Zn2+ buffer under conditions of Zn2+ excess to prevent intoxication, and as a facilitator of transcription regulation in response to fluctuations in Zn2+ levels.
Iron-sulfur cluster assembly
The major cytosolic requirements for Fe2+ are for utilization in heme-containing enzymes and the assembly of FeS clusters. FeS cluster-containing proteins are involved in a wide range of cellular functions (83). Many key enzymes in amino acid biosynthesis, carbon metabolism, cofactor biosynthesis, and antibiotic resistance are dependent on FeS cluster-containing enzymes. Thus, inactivation of these enzymes under conditions of Fe2+ starvation or oxidative stress is detrimental for proliferation (57).
A broad phenotypic survey of a S. aureus strain lacking BSH revealed many phenotypes that are consistent with defects in FeS biogenesis (Fig. 5) (30, 118). S. aureus strains lacking BSH are severely impaired for growth in media lacking leucine (Leu) or isoleucine (Ile). This growth defect could be suppressed by supplementation with either amino acid or Fe2+. The activity of key FeS requiring dehydratase enzymes in Leu or Ile biosynthesis is decreased in a BSH null mutant strain. In addition, aconitase activity, an unrelated FeS-dependent enzyme that catalyzes the conversion of citrate to isocitrate, also decreased, suggestive of an overall defect in cellular FeS cluster biogenesis.
The FeS biosynthetic machinery in S. aureus is encoded by the suf operon, which utilizes cysteine as a sulfur source and an unidentified Fe2+ source. On assembly, the FeS clusters are transferred to FeS carrier proteins (Nfu and/or SufA) for delivery to FeS-containing proteins (81). Genetic analysis suggests that the role of BSH in FeS biogenesis is independent of Nfu and SufA (118). Thus BSH, Nfu and SufA perform independent, yet overlapping roles in FeS biogenesis.
In yeast, monothiol glutaredoxins together with GSH play a key role in FeS cluster biogenesis and trafficking (17, 67, 68, 89, 108). Grx3/4 were shown to bind a bridging [2Fe-2S] cluster in vitro that is coordinated by the active-site Cys residue of the Grx domain and GSH as ligands (67, 108). The deficiency of Grx affected the synthesis of Fe/S clusters, heme, and di-iron centers and thus, this Grx FeS center has a crucial role in iron trafficking and sensing in yeast cells. It remains an interesting question for future studies to unravel whether BSH chelates Fe2+ directly or is involved in FeS cluster assembly via a BSH-containing Brx FeS center that perhaps could function in iron trafficking and sensing.
BSH may also have a role in maintaining the labile Fe2+ pool. In bacteria, Fe2+ sufficiency is sensed by the Fe2+-specific metalloregulator, Fur (63). On Fe2+ sufficiency, B. subtilis Fur binds Fe2+ at two different sensing sites, leading to DNA binding and repression of its regulon (76). Fur controls a large and complex regulon, including many iron uptake systems and an iron efflux pump (PfeT) (4). In addition, Fur indirectly regulates many more genes by repression of a small noncoding RNA (FsrA) and putative RNA chaperones (FbpABC) that are analogous in function to Hfq from enterobacteria, which together mediate an Fe2+-sparing response (36, 123, 124). A S. aureus BSH null strain contains lower levels of Fur-accessible Fe2+, as judged by slower transcriptional repression on Fe2+ sufficiency of the Fur-regulated isdB promoter, which is involved in Fe2+ acquisition from heme (118).
Copper trafficking
Copper plays a critical role in many cellular processes, yet Cu+ is highly toxic in excess due to enzyme mismetallation (78, 79). Thus, bacteria have evolved mechanisms to limit intracellular Cu+ toxicity by keeping it tightly bound to proteins, thereby limiting deleterious side reactions. Delivery of Cu+ into Cu+-containing proteins is often mediated by Cu+ chaperones, such as Atx1 in yeast (70) or possibly CopZ in B. subtilis (5). CopZ is a Atx1-like protein that may sequester excess intracellular Cu+ and deliver Cu+ to the CopA CPx-type ATP-ase for efflux. Recent in vitro studies identified S-bacillithiolated forms of apo-CopZ as well as CopZ loaded with BSH:Cu+ adducts (Fig. 5) (60). Further biochemical analysis revealed that BSH binds Cu+ with a relatively high affinity of β2(BSH) = 4 × 1017 M−2 and a stoichiometry of 2 BSH to 1 Cu+, indicating a possible role for BSH in intracellular Cu+ buffering. Cells lacking BSH are no more sensitive to Cu+ than wild-type cells. Interestingly, expression of the CsoR-regulated, Cu+-responsive copZA operon is induced in the absence of BSH, suggesting elevated levels of labile Cu+ and a role for BSH in Cu+ buffering (75). However, the precise in vivo role of BSH in Cu+ homeostasis remains to be determined.
Functions of BSH in S. aureus Virulence
BSH also plays an important role under infection-like conditions in the S. aureus clinical isolates, as shown in two phenotype studies using macrophage infection assays (111, 112). BSH-deficient mutants in clinical MRSA strains COL and USA300 showed a decreased survival in human whole-blood survival assays (111). In microarray analyses, the biosynthetic operon for staphyloxanthin was upregulated and the staphyloxanthin level was strongly decreased in the bshA mutant, suggesting lower radical scavenging ability in the absence of BSH. Staphyloxanthin is a carotenoid pigment that is produced by some S. aureus strains and is responsible for the yellow color from which the species name is derived. Staphyloxanthin provides protection against oxidative stress in neutrophil-infection assays and enhances the fitness of S. aureus (16).
Among the various S. aureus clonal complexes, it was found that natural BSH-deficient S. aureus strains have evolved that belong to the S. aureus NCTC8325 lineage (e.g., strain SH1000). They carry a bshC null mutation due to an 8 bp duplication in the bshC gene as a possible remnant of a transposon insertion (95, 111, 112). The natural S. aureus SH1000 bshC mutant was compromised in survival during infection assays using murine macrophages and human epithelial cell lines in comparison to the repaired and bshC-complemented S. aureus SH1000 strain (111, 112). Thus, BSH is an important virulence factor contributing to pathogen fitness and provides protection against the host-immune system in S. aureus clinical MRSA isolates under infection-related conditions.
This makes the BSH biosynthesis enzymes attractive drug targets for the treatment of emerging MRSA infections. In particular, BshB inhibitors would be good candidates for anti-infectives that block biosynthesis and salvage of BSH. Future studies should be directed toward unraveling the mechanisms of virulence control by BSH in S. aureus, which could involve S-bacillithiolation of key enzymes or regulators.
Protein S-Bacillithiolation and Its Reversal by Brx
Targets of protein S-bacillithiolation in B. subtilis and S. aureus
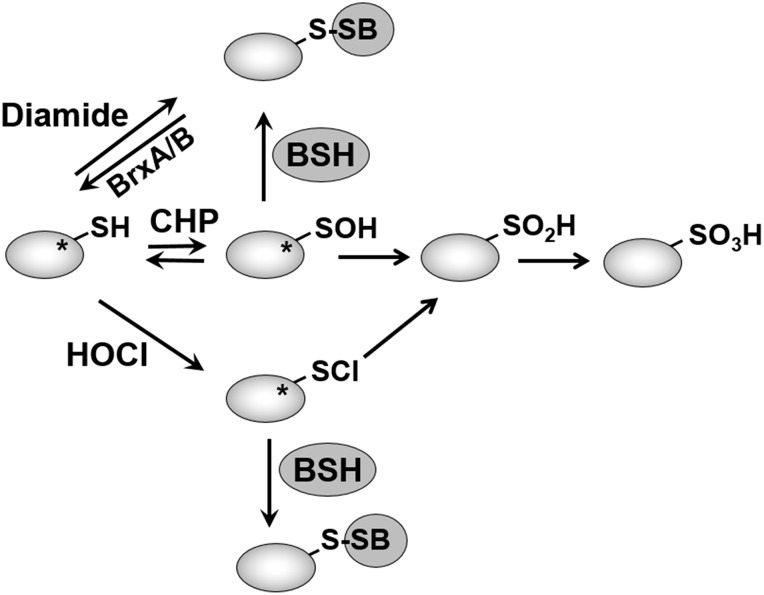
In eukaryotes, protein S-glutathionylation functions as an important thiol-protection and redox-regulatory mechanism and is implicated in many physiological and pathophysiological processes, such as neurodegenerative and cardiovascular diseases, cancer, and diabetes (18, 42, 43). In Firmicutes, BSH plays a related role in a redox modification termed protein S-bacillithiolation (Fig. 6) (74). Protein S-bacillithiolation was first discovered as redox modification of the organic hydroperoxide repressor OhrR under cumene hydroperoxide (CHP) stress (Fig. 7). OhrR is inactivated due to S-bacillithiolation at its lone Cys15 residue under CHP stress, leading to upregulation of the OhrA peroxiredoxin that is involved in CHP detoxification (35, 64). However, OhrR is also involved in redox sensing of hypochloric acid and is inactivated by S-bacillithiolation under NaOCl stress, leading to ohrA derepression that confers resistance against NaOCl stress in B. subtilis (14). Thus, S-bacillithiolation functions as a redox-switch mechanism to control the activity of the redox-sensing transcription factor OhrR in B. subtilis in response to both CHP and NaOCl treatment.
FIG. 6.
Mechanisms of S-bacillithiolation and its reversal. Proteins are S-bacillithiolated under CHP, HOCl, and diamide stress in Bacillus and Staphylococcus species. Diamide is a reactive electrophile species leading directly to the formation of mixed BSH disulfides. CHP and HOCl activate thiols to a sulfenic acid (-SOH) and sulfenylchloride (-SCl) intermediates, respectively, that react further with BSH to form S-bacillithiolated proteins. In the absence of proximal thiols, -SOH and -SCl are overoxidized to sulfinic or sulfonic acids. Thus, S-bacillithiolation serves to protect vulnerable thiols against irreversible overoxidation. The asterisk indicates that often active site Cys residues are targets for S-bacillithiolation. The reversal of S-bacillithiolation is catalyzed by the BrxA/B. Brx, bacilliredoxin; CHP, cumene hydroperoxide.
FIG. 7.
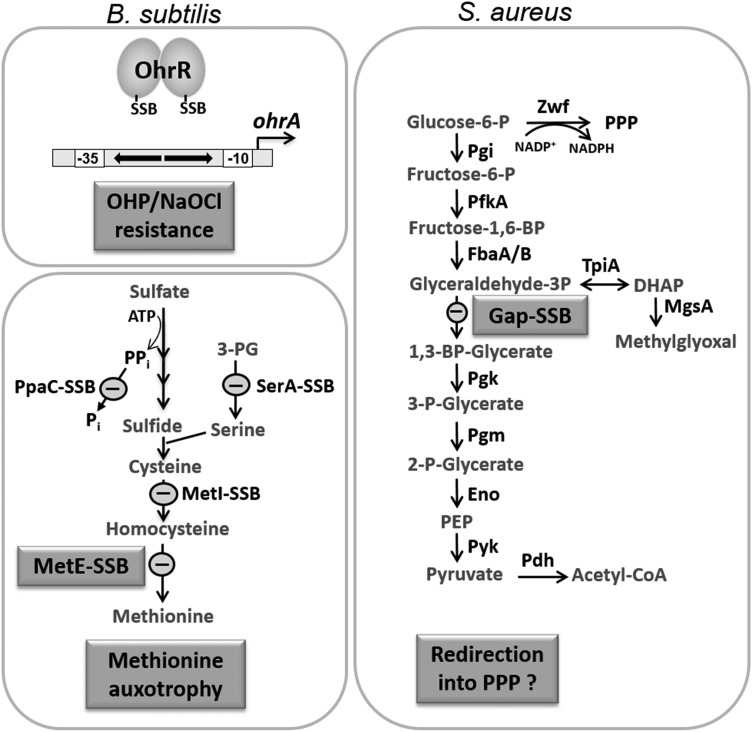
Physiological roles of S-bacillithiolations for OhrR and MetE in B. subtilis and for Gap of S. aureus. NaOCl leads to S-bacillithiolation of OhrR and MetE as main targets in B. subtilis that have regulatory roles under NaOCl stress. S-bacillithiolation inactivates the OhrR repressor, leading to induction of the OhrA peroxiredoxin that confers resistance to NaOCl and OHP. S-bacillithiolation of the methionine synthase MetE at its active site Cys730 and of other enzymes of the Cys and Met biosynthesis pathway (YxjG, PpaC, SerA, MetI) leads to methionine auxotrophy. In S. aureus, the glycolytic Gap is the main target for S-bacillithiolation under NaOCl stress. S-bacillithiolation of the Gap active site Cys151 leads to reversible Gap inactivation and prevents its overoxidation to Cys sulfonic acid. Gap inactivation under oxidative stress might redirect the glycolytic flux into the PPP for NADPH regeneration, as shown in yeast cells. Gap, glyceraldehyde-3-phosphate dehydrogenase; OHP, organic hydroperoxide; PPP, pentose phosphate pathway.
Although the role of the OhrA peroxiredoxin in the detoxification of organic hydroperoxides to their corresponding alcohols is well known in several bacteria (24), the function of OhrA in NaOCl detoxification has still to be demonstrated in vitro. Moreover, there are two OhrR homologs encoded in the genomes of S. aureus, SarZ and MgrA, controlling ohrA homologs and large regulons for antibiotics resistance determinants and virulence factors (12). Evidence for the redox control and structural changes of SarZ and MgrA at the conserved single Cys by S-thiolation was demonstrated by using a synthetic thiol, benzene thiol in vitro (110). Recently, the quantitative redox proteomics approach OxICAT revealed increased oxidation of both SarZ and MgrA under NaOCl stress in S. aureus USA300, suggesting their redox regulation by S-bacillithiolation in vivo (56).
Apart from OhrR, the methionine synthase MetE was identified as the most abundant S-bacillithiolated protein in Bacillus species under NaOCl stress (Fig. 7). S-bacillithiolation of MetE was observed when cells were grown in minimal medium under NaOCl stress and MetE-SSB could be visualized as a major band in BSH-specific Western blots under NaOCl and diamide stress. MetE was S-bacillithiolated at its Zn-binding active site Cys730 and at the non-essential surface-exposed Cys719, causing a methionine auxotrophy phenotype under NaOCl stress (12). MetE is also a main target for S-glutathionylation and the most susceptible protein for thiol-oxidation in E. coli under diamide treatment (52, 66). S-glutathionylation of MetE occurs at a non-conserved Cys645 that is located at the entrance to the active site, also leading to methionine auxotrophy in E. coli (52).
We have shown that the conserved Zn-binding active site forms the mixed disulfide with BSH in B. subtilis, which is also target for S-mycothiolation in C. glutamicum under NaOCl stress (13). Inactivation of MetE under NaOCl stress in B. subtilis is accompanied by an increased transcription of the S-box regulon controlling genes for methionine biosynthesis. It is hypothesized that MetE inactivation could lead to a translation arrest to enable detoxification of hypochlorite and to avoid further protein damage. Further support for this postulated translation stop is provided by the observation that other amino acid biosynthesis enzymes (AroA), protein translation factors (Tuf), and ribosomal proteins (RpsM) are also targets for S-thiolation across Gram-positive bacteria (14).
In S. aureus, S-bacillithiolation was recently shown to control the activity of the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (Gap) of S. aureus under NaOCl stress (Fig. 7) (56). Gap is S-bacillithiolated at the conserved active site Cys151, which is a well-known target for various post-translational thiol-modifications, also including S-glutathionylation in eukaryotes. The reactivity of the Gap active site was shown to depend on a specific H2O2-binding pocket, transition state stabilization, and a proton relay mechanism promoting leaving-group departure (51, 105).
In S. aureus, Gap contributes as the most abundant Cys protein in the proteome, with 4% to the total Cys proteome of S. aureus and the S-bacillithiolated Gap was observed as a major band in NaOCl-treated cells using BSH-specific Western blots (56). Using OxICAT, Gap showed the highest oxidation increase of 29% at the S-bacillithiolated active site Cys151 under NaOCl stress. Detailed Gap activity assays in the presence of H2O2 and NaOCl with or without BSH revealed that Gap inactivation is faster due to S-bacillithiolation compared with overoxidation. These results lead to the conclusion that S-bacillithiolation of the Gap active site can efficiently prevent its irreversible overoxidation under both H2O2 and NaOCl treatments. Molecular docking of BSH into the Gap active site was used to model the structure of the S-bacillithiolated active site. The model of the Gap-SSB structure suggests that BSH can undergo disulfide formation with Cys151 without major conformational changes. This may explain why the most abundant Cys-protein Gap is the preferred and dominant target for S-bacillithiolation inside S. aureus cells (56).
Protein S-bacillithiolation is a widespread thiol-protection and redox-regulatory mechanism in different Firmicutes under hypochlorite stress, including industrially important Bacillus and Staphylococcus species, such as B. subtilis, Bacillus amyloliquefaciens, Bacillus megaterium, Bacillus pumilus, and the meat-starter culture Staphylococcus carnosus (8, 14, 74). Eight common and 29 unique S-bacillithiolated proteins were identified in the different Firmicutes using shotgun proteomics based on the mass increase of 396 Da at Cys residues (14, 15). The complete set of S-bacillithiolated proteins (the S-bacillithiolome) includes antioxidant function proteins, such as peroxiredoxins (YkuU), thiol-disulfide oxidoreductases (YumC) and Brx (YphP YqiW, YtxJ), translation factors (Tuf), chaperones (DnaK, GrpE), and several metabolic enzymes involved in the biosynthesis of amino acids, cofactors, and nucleotides. The translation elongation factor TufA, the methionine synthase MetE and its homolog YxjG, the inosine monophosphate dehydrogenase GuaB, and the inorganic pyrophosphatase PpaC belong to the conserved targets for S-bacillithiolation across Firmicutes bacteria that are also modified by S-mycothiolation in Actinomycetes (13).
Surprisingly, the glycolytic Gap is the major target for S-bacillithiolation in S. aureus (56), but Gap is not S-bacillithiolated in Bacillus species. Instead, the Gap enzyme of B. subtilis is oxidized to an intramolecular disulfide in its highly conserved CTTNC motif under NaOCl stress, as confirmed by mass spectrometry (14). This intracellular disulfide was also shown for the E. coli Gap homolog under NaOCl stress, since both Cys residues showed increased oxidations in the OxICAT analysis (65). However, in S. aureus Gap, the second Cys in this CTTNC motif is replaced by a serine explaining the preference of Cys151 for S-bacillithiolation under NaOCl stress.
All global S-bacillithiolome studies to date identified S-bacillithiolated proteins in Firmicutes bacteria by mass spectrometry and non-reducing BSH-Western blot analyses under NaOCl and diamide stress, but not under H2O2 stress in vivo (15). Previously, strongly increased S-thiolations were also observed at the global level under diamide stress in B. subtilis and S. aureus (113). S-glutathionylation requires activation of thiols to sulfenic acid, sulfenylamides, thiyl radicals, thiosulfinate, or S-nitrosyl intermediates that, subsequently, react further with GSH to the mixed disulfide (2, 41, 85, 86). Hypochlorite activates the thiol group via chlorination, leading to an unstable sulfenylchloride intermediate that quickly reacts further to generate S-bacillithiolations (Fig. 6) (19, 47). Diamide is a reactive electrophile known to form mixed disulfides, including S-thiolations in several organisms (113). The second-order rate constant of the reaction of HOCl with thiols is seven orders of magnitude higher compared with H2O2 (19, 104), explaining why S-bacillithiolation was not observed under H2O2 stress in vivo. Moreover, the pathogen S. aureus exhibits a remarkable resistance to 200 mM H2O2 due to efficient detoxification mechanisms that allow the survival under infection conditions (55, 73).
Apart from H2O2, S. aureus encounters HOCl during infections by activated neutrophils that is produced by myeloperoxidase (134), and hence, S-bacillithiolation serves as a major thiol-protection mechanism of essential enzymes, such as Gap under infection-related HOCl stress conditions. It will be interesting to uncover the targets for S-bacillithiolation during infections, or in the anterior nares as the natural niche of S. aureus in future studies.
The BrxA and BrxB function in protein de-bacillithiolation
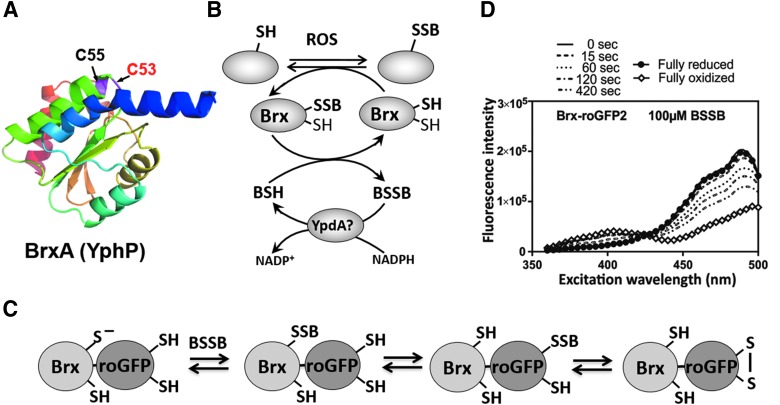
The Trx-fold proteins YtxJ, BrxA (YphP), and BrxB (YqiW) were identified as S-bacillithiolated in B. subtilis and S. carnosus that co-occur with the BSH biosynthesis enzymes (BshA, BshB, BshC) only in BSH-producing Firmicutes bacteria and were suggested to function as Brx in the reversal of S-bacillithiolations (Fig. 8A, B) (15). YtxJ is suggested as a monothiol Brx with the active site Cys located in a conserved TCPIS motif.
FIG. 8.
Structure of the BrxA (YphP) of B. subtilis (A), Brx redox pathway (B), principle of Brx-roGFP2 biosensor oxidation (C), and ratiometric change of excitation spectra of Brx-roGFP2 by BSSB in vitro (D). (A) The structure of the BrxA (YphP) with the CGC active site motif was generated by using the software Phyre2 and PyMol. (B) BrxA reduces S-bacillithiolated proteins, resulting in Brx-SSB formation. Recycling of BrxA may require BSH and an NADPH-dependent BSSB reductase that could be YpdA. (C) The Brx-roGFP2 biosensor reacts first with BSSB at the active site Cys of Brx, leading to Brx-SSB formation, subsequent transfer of the BSH moiety to the coupled roGFP2, and re-arrangement to the roGFP2 disulfide. The roGFP2 disulfide causes a change of the 405/488 nm excitation ratio. (D) Brx-roGFP2 reacts very fast with purified 100 μM BSSB, as shown by the ratiometric change in the excitation maxima at 405 and 488 nm. For fully oxidized and reduced probes, Brx-roGFP2 was treated with 5 mM diamide and 10 mM DTT, respectively. The Brx-roGFP2 fluorescence excitation spectra were monitored by using the Clariostar microplate reader. Adapted from a previous publication (73). BSSB, oxidized bacillithiol disulfide.
BrxA (YphP) and BrxB (YqiW) are DUF1094-family proteins with unusual CGC active site motifs, rather than the more common CxxC spacing seen in glutaredoxin (38, 40). BrxA was originally suggested to function as a thiol-disulfide isomerase based on its high standard redox potential of E0′ = −130 mV (22). However, both BrxA and BrxB function in the reduction of the S-bacillithiolated MetE and OhrR in vitro, thereby demonstrating Brx activity (38). The BrxBCGA-resolving Cys mutant could regenerate DNA-binding activity of S-bacillithiolated OhrR in vitro, but S-cysteinylated OhrR could not be reactivated. MetE de-bacillithiolation was catalyzed by the BrxBCGA mutant as revealed by non-reducing BSH-specific Western blot analysis and mass spectrometry, but MetE reactivation could not be demonstrated in vitro. In S. aureus, Gap is the major S-bacillithiolated protein under NaOCl stress and Gap activity could be fully restored by using the BrxA (YphP) homolog SAUSA300_1321 (Brx) and the BrxCGA-resolving Cys mutant in vitro (56).
In conclusion, the BrxA and BrxB have been characterized to catalyze the de-bacillithiolation of two essential metabolic enzymes (MetE and Gap) and of the redox-sensing regulator OhrR in B. subtilis or S. aureus. These results provide evidence for the function of glutaredoxin-like Brx in industrially and medically important BSH-producing bacteria. However, phenotypic analyses of brxA and brxB single and double mutants revealed no significant growth phenotypes under NaOCl stress in B. subtilis (38). Thus, the Brx pathway is not essential and the Trx pathway might be alternatively involved in the de-bacillithiolation in B. subtilis.
A Brx-roGFP2 biosensor monitors dynamic changes in the BSH redox potential in S. aureus
Recently, Brx (YphP) was fused to redox-sensitive GFP (roGFP2) to construct a genetically encoded Brx-roGFP2 biosensor to monitor dynamic changes in the BSH redox potential in S. aureus (Fig. 8C, D) (73). Brx-roGFP2 responds specifically only to physiological concentrations of BSSB in vitro, but it does not react with other thiol disulfides (cystine, GSSG, MSSM, CoASH disulfides). This further confirms the specificity of Brx for the reduction of BSH mixed disulfides in S. aureus.
The specificity of the Brx-roGFP2 biosensor to low doses of BSSB was shown to depend on the Brx active site in vitro, whereas direct responses of the roGFP2 to the oxidants in vivo could not be excluded. Brx-roGFP2 was shown to react very fast in S. aureus COL to low doses of 20–100 μM NaOCl in vivo, but high doses of a maximum of 100 mM H2O2 did not lead to complete biosensor oxidation. This confirms the high reactivity of NaOCl inside S. aureus that requires fast thiol protection by S-bacillithiolation to avoid the overoxidation of thiols. The weak biosensor response to H2O2 can be explained by the high level of peroxide resistance in S. aureus.
This Brx-roGFP2 biosensor was further applied to investigate the involvement of oxidative stress in the killing mode of antibiotics in S. aureus (73). The role of ROS in the mechanism of action of bactericidal antibiotics has been a subject of controversy (61, 71). Antibiotics that are commonly used to treat S. aureus infections were selected that target the RNA polymerase (rifampicin), cell-wall biosynthesis (fosfomycin, ampicillin, oxacillin, and vancomycin), protein biosynthesis (gentamycin, lincomycin, erythromycin, and linezolid), and DNA replication (ciprofloxacin). These different antibiotics classes did not lead to changes in the BSH redox potential at sub-lethal concentrations, indicating that these antibiotics do not impose strong oxidative stress in S. aureus.
These results are consistent with roGFP2 biosensor results obtained in Salmonella under antibiotic treatment (131). However, due to its high peroxide resistance, S. aureus might be able to quickly detoxify ROS generated during antibiotic treatment. In the case of fosfomycin, BSH is directly required as a cofactor for FosB for detoxification in S. aureus (111). However, previous studies found no correlation between the level of fosfomycin resistance and BSH levels in different S. aureus clinical isolates, and no increased BSH redox potential was measured under fosfomycin treatment in S. aureus COL.
The Brx-roGFP2 biosensor was further expressed in two MRSA isolates, S. aureus COL and USA300, and the latter is highly virulent community-acquired MRSA. An increased BSH redox potential was measured during the entry into stationary phase in both strains, ranging from −300 to −270 mV in COL and from −300 to −235 mV in USA300 along the growth curve. Our future studies are directed to apply this Brx-roGFP2 biosensor for screening of the BSH redox potential across S. aureus isolates of different clonal complexes to reveal the differences in pathogen fitness and in their ROS detoxification capacities as defense mechanisms against the host immune system.
Concluding Remarks
In a previous review in this journal (50), written soon after the discovery of BSH, readers were introduced to this novel thiol and given a preview of how the field might develop, guided by analogy with the prior extensive work on GSH and, to a lesser extent, MSH. Many of the predictions made at that time have held true, but there have also been many surprises as well as frustrations. Here, we briefly highlight some of the pressing questions in the field that have not yet been resolved, although ongoing efforts will surely work to address these challenges.
Although the biosynthetic pathway of BSH is now well established, and extensive structural and biochemical studies have led to a detailed understanding of this process, the actual reaction catalyzed by BshC is still undetermined. The analogous enzyme for MSH synthesis uses aminoacylated tRNACys as an activated form of Cys. We assume that BshC also requires an activated form of Cys, but the identity of this last substrate is unknown. Another key “missing” activity for our understanding of BSH biology is the BSSB reductase. Certainly, the B. subtilis YpdA protein (a TrxR homolog) is an attractive candidate, but attempts to demonstrate BSSB activity biochemically have been unsuccessful.
Although many of the proteins involved in the core metabolism of BSH are, by now, known, including the biosynthesis pathway and Brx, many BSH-utilizing enzymes and accessory functions are yet to be discovered. There are several peroxidases annotated as Gpx, thiol-peroxidase, or thiol-dependent peroxiredoxin in Bacillus and Staphylococcus species that could use BSH as a thiol cofactor for peroxide detoxification (Fig. 3E). It is also unclear as to whether BSH (or perhaps BSSB) is imported or exported from cells. In E. coli, GSH is exported to the periplasm (109), but an analogous process for BSH is not established. It is worth noting that B. subtilis has multiple importers assigned as having a role in cystine uptake (9), but it is not known as to whether one or more may, in addition, or instead, use BSSB as a substrate.
Although the role of BSH in thiol redox chemistry is well established, there are also other aspects that are still largely unexplored. As noted earlier, in some cases, BSH may play a key role in protecting cells against H2S stress (122), and by analogy with GSH a role in protection against reactive nitrogen species can be predicted. Although BSH has already been investigated for its role in buffering of Zn2+ (25, 75) and in the assembly of FeS clusters (30, 117, 118), many details remain to be discovered, such as the possible roles of BSH and other thiols together with Brx (glutaredoxin-like enzymes) in bridging FeS clusters, analogous to the role of GSH and Grx proteins in iron trafficking and sensing in GSH-producing eukaryotic organisms.
Similarly, proteomics studies have documented an abundance of protein S-bacillithiolation as a major post-translational modification under oxidative stress conditions (14, 15, 56, 64, 74, 107), but the pathways that reverse this modification (Brx) and the consequences for enzyme activity are still only now emerging. On a broader level, a clear picture has not yet emerged as to which bacteria synthesize BSH instead of GSH (or MSH), and whether one offers specific advantages over the other. We look forward to the continued rapid progress in this area and the resolution of these questions, and others not yet even imagined.
Abbreviations Used
- Bca
BSH S-conjugate amidase
- Brx
bacilliredoxin
- BSH
bacillithiol
- BshA
glycosyltransferase for GlcNAc-Mal biosynthesis
- BshB
deacetylase-producing GlcN-Mal
- BshC
cysteine ligase for BSH biosynthesis
- BSSB
oxidized bacillithiol disulfide
- Bst
BSH-S-transferases
- CA-MRSA
community-acquired MRSA
- CHP
cumene hydroperoxide
- CoASH
coenzymeA
- Cys
cysteine
- DHAP
dihydroxyacetone phosphate
- EGT
ergothioneine
- FA
formaldehyde
- Fdh
formaldehyde dehydrogenases
- FeS
iron-sulfur
- Gap
glycolytic glyceraldehyde −3-phosphate dehydrogenase
- GlcNAc
N-acetyl glucoseamine
- GlxA/B
glyoxalases A and B
- Grx
glutaredoxin
- GSH
glutathione
- GSSG
oxidized glutathione disulfide
- Gst
GSH-S-transferases
- H2O2
hydrogen peroxide
- H2S
hydrogen sulfide
- HTA
hemithioacetal
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- LMW
low molecular weight
- Mal
malate
- Met
methionine
- MG
methylglyoxal
- MgsA
methylglyoxal synthase
- MRSA
methicillin-resistant Staphylococcus aureus
- Mrx1
mycoredoxin1
- MSH
mycothiol
- MT
metallothionein
- NADH
nicotinamide adenine dinucleotide
- NADPH
nicotinamide adenine dinucleotide phosphate
- NaOCl
sodium hypochlorite
- OHP
organic hydroperoxide
- OhrR
organic hydroperoxide repressor
- PPP
pentose phosphate pathway
- protein-SSB
BSH protein mixed disulfide
- roGFP2
redox-sensitive green fluorescent protein
- ROH
organic alcohol
- ROS
reactive oxygen species
- RuMP
ribose monophosphate pathway
- SLG
S-lactoylglutathione
- T(SH)2
trypanothione
- Trx
thioredoxin
- TrxR
thioredoxin reductase
Acknowledgments
This work was supported by grants from the National Institutes of Health (NIGMS) under award numbers GM059323 and GM109993 (J.D.H.) and from the Deutsche Forschungsgemeinschaft (DFG) AN746/3-1 and AN746/4-1 within the DFG priority program SPP1710 (H.A.), by the DFG Research Training Group GRK1947, project (C1) (H.A.), and by an ERC Consolidator grant (GA 615585) MYCOTHIOLOME (H.A.).
References
- 1.Akanuma G, Nanamiya H, Natori Y, Nomura N, and Kawamura F. Liberation of zinc-containing L31 (RpmE) from ribosomes by its paralogous gene product, YtiA, in Bacillus subtilis. J Bacteriol 188: 2715–2720, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen EM. and Mieyal JJ. Protein-thiol oxidation and cell death: regulatory role of glutaredoxins. Antioxid Redox Signal 17: 1748–1763, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreini C, Banci L, Bertini I, and Rosato A. Zinc through the three domains of life. J Proteome Res 5: 3173–3178, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Baichoo N, Wang T, Ye R, and Helmann JD. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol Microbiol 45: 1613–1629, 2002 [DOI] [PubMed] [Google Scholar]
- 5.Banci L, Bertini I, Del Conte R, Markey J, and Ruiz-Duenas FJ. Copper trafficking: the solution structure of Bacillus subtilis CopZ. Biochemistry 40: 15660–15668, 2001 [DOI] [PubMed] [Google Scholar]
- 6.Barron ES. Thiol groups of biological importance. Adv Enzymol Relat Subj Biochem 11: 201–266, 1951 [DOI] [PubMed] [Google Scholar]
- 7.Barton LL, Fardeau ML, and Fauque GD. Hydrogen sulfide: a toxic gas produced by dissimilatory sulfate and sulfur reduction and consumed by microbial oxidation. Met Ions Life Sci 14: 237–277, 2014 [DOI] [PubMed] [Google Scholar]
- 8.Berney M, Weimar MR, Heikal A, and Cook GM. Regulation of proline metabolism in mycobacteria and its role in carbon metabolism under hypoxia. Mol Microbiol 84: 664–681, 2012 [DOI] [PubMed] [Google Scholar]
- 9.Burguiere P, Auger S, Hullo MF, Danchin A, and Martin-Verstraete I. Three different systems participate in L-cystine uptake in Bacillus subtilis. J Bacteriol 186: 4875–4884, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao M, Bernat BA, Wang Z, Armstrong RN, and Helmann JD. FosB, a cysteine-dependent fosfomycin resistance protein under the control of sigma(W), an extracytoplasmic-function sigma factor in Bacillus subtilis. J Bacteriol 183: 2380–2383, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandrangsu P, Dusi R, Hamilton CJ, and Helmann JD. Methylglyoxal resistance in Bacillus subtilis: contributions of bacillithiol-dependent and independent pathways. Mol Microbiol 91: 706–715, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen PR, Brugarolas P, and He C. Redox signaling in human pathogens. Antioxid Redox Signal 14: 1107–1118, 2011 [DOI] [PubMed] [Google Scholar]
- 13.Chi BK, Busche T, Van Laer K, Basell K, Becher D, Clermont L, Seibold GM, Persicke M, Kalinowski J, Messens J, and Antelmann H. Protein S-mycothiolation functions as redox-switch and thiol protection mechanism in Corynebacterium glutamicum under hypochlorite stress. Antioxid Redox Signal 20: 589–605, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chi BK, Gronau K, Mader U, Hessling B, Becher D, and Antelmann H. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol Cell Proteomics 10: M111 009506, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chi BK, Roberts AA, Huyen TT, Basell K, Becher D, Albrecht D, Hamilton CJ, and Antelmann H. S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in firmicutes bacteria. Antioxid Redox Signal 18: 1273–1295, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clauditz A, Resch A, Wieland KP, Peschel A, and Gotz F. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect Immun 74: 4950–4953, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Couturier J, Przybyla-Toscano J, Roret T, Didierjean C, and Rouhier N. The roles of glutaredoxins ligating Fe-S clusters: sensing, transfer or repair functions? Biochim Biophys Acta 1853: 1513–1527, 2015 [DOI] [PubMed] [Google Scholar]
- 18.Dalle-Donne I, Rossi R, Colombo G, Giustarini D, and Milzani A. Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem Sci 34: 85–96, 2009 [DOI] [PubMed] [Google Scholar]
- 19.Davies MJ. Myeloperoxidase-derived oxidation: mechanisms of biological damage and its prevention. J Clin Biochem Nutr 48: 8–19, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Rey Pailhade J. Sur un corps d’ origine organizue hydrogénant le soufre a’ froid. Compt Rend 106: 1683, 1888 [Google Scholar]
- 21.delCardayre SB, Stock KP, Newton GL, Fahey RC, and Davies JE. Coenzyme A disulfide reductase, the primary low molecular weight disulfide reductase from Staphylococcus aureus. Purification and characterization of the native enzyme. J Biol Chem 273: 5744–5751, 1998 [DOI] [PubMed] [Google Scholar]
- 22.Derewenda U, Boczek T, Gorres KL, Yu M, Hung LW, Cooper D, Joachimiak A, Raines RT, and Derewenda ZS. Structure and function of Bacillus subtilis YphP, a prokaryotic disulfide isomerase with a CXC catalytic motif. Biochemistry 48: 8664–8671, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Djoko KY, Ong CL, Walker MJ, and McEwan AG. The role of copper and zinc toxicity in innate immune defense against bacterial pathogens. J Biol Chem 290: 18954–18961, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubbs JM. and Mongkolsuk S. Peroxiredoxins in bacterial antioxidant defense. Subcell Biochem 44: 143–193, 2007 [DOI] [PubMed] [Google Scholar]
- 25.Eide DJ. Bacillithiol, a new role in buffering intracellular zinc. Mol Microbiol 94: 743–746, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fahey RC. Novel thiols of prokaryotes. Annu Rev Microbiol 55: 333–356, 2001 [DOI] [PubMed] [Google Scholar]
- 27.Fahey RC. Glutathione analogs in prokaryotes. Biochim Biophys Acta 1830: 3182–3198, 2013 [DOI] [PubMed] [Google Scholar]
- 28.Fahey RC, Brown WC, Adams WB, and Worsham MB. Occurrence of glutathione in bacteria. J Bacteriol 133: 1126–1129, 1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fairlamb AH, Blackburn P, Ulrich P, Chait BT, and Cerami A. Trypanothione: a novel bis(glutathionyl)spermidine cofactor for glutathione reductase in trypanosomatids. Science 227: 1485–1487, 1985 [DOI] [PubMed] [Google Scholar]
- 30.Fang Z. and Dos Santos PC. Protective role of bacillithiol in superoxide stress and Fe-S metabolism in Bacillus subtilis. Microbiologyopen 4: 616–631, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang Z, Roberts AA, Weidman K, Sharma SV, Claiborne A, Hamilton CJ, and Dos Santos PC. Cross-functionalities of Bacillus deacetylases involved in bacillithiol biosynthesis and bacillithiol-S-conjugate detoxification pathways. Biochem J 454: 239–247, 2013 [DOI] [PubMed] [Google Scholar]
- 32.Feng J, Che Y, Milse J, Yin YJ, Liu L, Ruckert C, Shen XH, Qi SW, Kalinowski J, and Liu SJ. The gene ncgl2918 encodes a novel maleylpyruvate isomerase that needs mycothiol as cofactor and links mycothiol biosynthesis and gentisate assimilation in Corynebacterium glutamicum. J Biol Chem 281: 10778–10785, 2006 [DOI] [PubMed] [Google Scholar]
- 33.Ferguson GP, Battista JR, Lee AT, and Booth IR. Protection of the DNA during the exposure of Escherichia coli cells to a toxic metabolite: the role of the KefB and KefC potassium channels. Mol Microbiol 35: 113–122, 2000 [DOI] [PubMed] [Google Scholar]
- 34.Ferguson GP, McLaggan D, and Booth IR. Potassium channel activation by glutathione-S-conjugates in Escherichia coli: protection against methylglyoxal is mediated by cytoplasmic acidification. Mol Microbiol 17: 1025–1033, 1995 [DOI] [PubMed] [Google Scholar]
- 35.Fuangthong M, Atichartpongkul S, Mongkolsuk S, and Helmann JD. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J Bacteriol 183: 4134–4141, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaballa A, Antelmann H, Aguilar C, Khakh SK, Song KB, Smaldone GT, and Helmann JD. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc Natl Acad Sci U S A 105: 11927–11932, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaballa A, Antelmann H, Hamilton CJ, and Helmann JD. Regulation of Bacillus subtilis bacillithiol biosynthesis operons by Spx. Microbiology 159: 2025–2035, 2013 [DOI] [PubMed] [Google Scholar]
- 38.Gaballa A, Chi BK, Roberts AA, Becher D, Hamilton CJ, Antelmann H, and Helmann JD. Redox regulation in Bacillus subtilis: the bacilliredoxins BrxA(YphP) and BrxB(YqiW) function in de-bacillithiolation of S-bacillithiolated OhrR and MetE. Antioxid Redox Signal 21: 357–367, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaballa A. and Helmann JD. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J Bacteriol 180: 5815–5821, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaballa A, Newton GL, Antelmann H, Parsonage D, Upton H, Rawat M, Claiborne A, Fahey RC, and Helmann JD. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc Natl Acad Sci U S A 107: 6482–6486, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallogly MM. and Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr Opin Pharmacol 7: 381–391, 2007 [DOI] [PubMed] [Google Scholar]
- 42.Ghezzi P. Regulation of protein function by glutathionylation. Free Radic Res 39: 573–580, 2005 [DOI] [PubMed] [Google Scholar]
- 43.Ghezzi P. Protein glutathionylation in health and disease. Biochim Biophys Acta 1830: 3165–3172, 2013 [DOI] [PubMed] [Google Scholar]
- 44.Grill E, Loffler S, Winnacker EL, and Zenk MH. Phytochelatins, the heavy-metal-binding peptides of plants, are synthesized from glutathione by a specific gamma-glutamylcysteine dipeptidyl transpeptidase (phytochelatin synthase). Proc Natl Acad Sci U S A 86: 6838–6842, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grosse C, Schleuder G, Schmole C, and Nies DH. Survival of Escherichia coli cells on solid copper surfaces is increased by glutathione. Appl Environ Microbiol 80: 7071–7078, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harms N, Ras J, Reijnders WN, van Spanning RJ, and Stouthamer AH. S-formylglutathione hydrolase of Paracoccus denitrificans is homologous to human esterase D: a universal pathway for formaldehyde detoxification? J Bacteriol 178: 6296–6299, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hawkins CL, Pattison DI, and Davies MJ. Hypochlorite-induced oxidation of amino acids, peptides and proteins. Amino Acids 25: 259–274, 2003 [DOI] [PubMed] [Google Scholar]
- 48.Helbig K, Bleuel C, Krauss GJ, and Nies DH. Glutathione and transition-metal homeostasis in Escherichia coli. J Bacteriol 190: 5431–5438, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helbig K, Grosse C, and Nies DH. Cadmium toxicity in glutathione mutants of Escherichia coli. J Bacteriol 190: 5439–5454, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Helmann JD. Bacillithiol, a new player in bacterial redox homeostasis. Antioxid Redox Signal 15: 123–133, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hildebrandt T, Knuesting J, Berndt C, Morgan B, and Scheibe R. Cytosolic thiol switches regulating basic cellular functions: GAPDH as an information hub? Biol Chem 396: 523–537, 2015 [DOI] [PubMed] [Google Scholar]
- 52.Hondorp ER. and Matthews RG. Oxidative stress inactivates cobalamin-independent methionine synthase (MetE) in Escherichia coli. PLoS Biol 2: e336, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hopkins FG. On an autoxidisable constituent of the cell. Biochem J 15: 286–305, 1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hopkins FG. On glutathione: a reinvestigation. J Biol Chem 84: 269–320, 1929 [Google Scholar]
- 55.Horsburgh MJ, Clements MO, Crossley H, Ingham E, and Foster SJ. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect Immun 69: 3744–3754, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Imber M, Huyen NT, Pietrzyk-Brzezinska AJ, Loi VV, Hillion M, Bernhardt J, Tharichen L, Kolsek K, Saleh M, Hamilton CJ, Adrian L, Grater F, Wahl M, and Antelmann H. Protein S-bacillithiolation functions in thiol-protection and redox regulation of the glyceraldehyde-3-phosphate dehydrogenase Gap in Staphylococcus aureus under hypochlorite stress. Antioxid Redox Signal 2016. [Epub ahead of print]; DOI: 10.1089/ars.2016.6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Imlay JA. Iron-sulphur clusters and the problem with oxygen. Mol Microbiol 59: 1073–1082, 2006 [DOI] [PubMed] [Google Scholar]
- 58.Irving H. and Williams RJP. 637. The stability of transition-metal complexes. J Chem Soc (Resumed) 1953: 3192–3210, 1953 [Google Scholar]
- 59.Jothivasan VK. and Hamilton CJ. Mycothiol: synthesis, biosynthesis and biological functions of the major low molecular weight thiol in actinomycetes. Nat Prod Rep 25: 1091–1117, 2008 [DOI] [PubMed] [Google Scholar]
- 60.Kay KL, Hamilton CJ, and Le Brun NE. Mass spectrometry of B. subtilis CopZ: Cu(i)-binding and interactions with bacillithiol. Metallomics 8: 709–719, 2016 [DOI] [PubMed] [Google Scholar]
- 61.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, and Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130: 797–810, 2007 [DOI] [PubMed] [Google Scholar]
- 62.Lamers AP, Keithly ME, Kim K, Cook PD, Stec DF, Hines KM, Sulikowski GA, and Armstrong RN. Synthesis of bacillithiol and the catalytic selectivity of FosB-type fosfomycin resistance proteins. Org Lett 14: 5207–5209, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee JW. and Helmann JD. Functional specialization within the Fur family of metalloregulators. Biometals 20: 485–499, 2007 [DOI] [PubMed] [Google Scholar]
- 64.Lee JW, Soonsanga S, and Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci U S A 104: 8743–8748, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leichert LI, Gehrke F, Gudiseva HV, Blackwell T, Ilbert M, Walker AK, Strahler JR, Andrews PC, and Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci U S A 105: 8197–8202, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leichert LI. and Jakob U. Protein thiol modifications visualized in vivo. PLoS Biol 2: e333, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H, Mapolelo DT, Dingra NN, Naik SG, Lees NS, Hoffman BM, Riggs-Gelasco PJ, Huynh BH, Johnson MK, and Outten CE. The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry 48: 9569–9581, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H. and Outten CE. Monothiol CGFS glutaredoxins and BolA-like proteins: [2Fe-2S] binding partners in iron homeostasis. Biochemistry 51: 4377–4389, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li ZS, Szczypka M, Lu YP, Thiele DJ, and Rea PA. The yeast cadmium factor protein (YCF1) is a vacuolar glutathione S-conjugate pump. J Biol Chem 271: 6509–6517, 1996 [DOI] [PubMed] [Google Scholar]
- 70.Lin SJ, Pufahl RA, Dancis A, O'Halloran TV, and Culotta VC. A role for the Saccharomyces cerevisiae ATX1 gene in copper trafficking and iron transport. J Biol Chem 272: 9215–9220, 1997 [PubMed] [Google Scholar]
- 71.Liu Y. and Imlay JA. Cell death from antibiotics without the involvement of reactive oxygen species. Science 339: 1210–1213, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lo TW, Westwood ME, McLellan AC, Selwood T, and Thornalley PJ. Binding and modification of proteins by methylglyoxal under physiological conditions. A kinetic and mechanistic study with N alpha-acetylarginine, N alpha-acetylcysteine, and N alpha-acetyllysine, and bovine serum albumin. J Biol Chem 269: 32299–32305, 1994 [PubMed] [Google Scholar]
- 73.Loi VV, Harms M, Muller M, Huyen NT, Hamilton CJ, Hochgrafe F, Pane-Farre J, and Antelmann H. Real-time imaging of the bacillithiol redox potential in the human pathogen Staphylococcus aureus using a genetically encoded bacilliredoxin-fused redox biosensor. Antioxid Redox Signal 2016. [Epub ahead of print]; DOI: 10.1089/ars.2016.6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Loi VV, Rossius M, and Antelmann H. Redox regulation by reversible protein S-thiolation in bacteria. Front Microbiol 6: 187, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma Z, Chandrangsu P, Helmann TC, Romsang A, Gaballa A, and Helmann JD. Bacillithiol is a major buffer of the labile zinc pool in Bacillus subtilis. Mol Microbiol 94: 756–770, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma Z, Faulkner MJ, and Helmann JD. Origins of specificity and cross-talk in metal ion sensing by Bacillus subtilis Fur. Mol Microbiol 86: 1144–1155, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.MacLean MJ, Ness LS, Ferguson GP, and Booth IR. The role of glyoxalase I in the detoxification of methylglyoxal and in the activation of the KefB K+ efflux system in Escherichia coli. Mol Microbiol 27: 563–571, 1998 [DOI] [PubMed] [Google Scholar]
- 78.Macomber L. and Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A 106: 8344–8349, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Macomber L, Rensing C, and Imlay JA. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J Bacteriol 189: 1616–1626, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maret W. Oxidative metal release from metallothionein via zinc-thiol/disulfide interchange. Proc Natl Acad Sci U S A 91: 237–241, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mashruwala AA, Pang YY, Rosario-Cruz Z, Chahal HK, Benson MA, Mike LA, Skaar EP, Torres VJ, Nauseef WM, and Boyd JM. Nfu facilitates the maturation of iron-sulfur proteins and participates in virulence in Staphylococcus aureus. Mol Microbiol 95: 383–409, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Merchant SS. and Helmann JD. Elemental economy: microbial strategies for optimizing growth in the face of nutrient limitation. Adv Microb Physiol 60: 91–210, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mettert EL. and Kiley PJ. How is Fe-S cluster formation regulated? Annu Rev Microbiol 69: 505–526, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Michalopoulos AS, Livaditis IG, and Gougoutas V. The revival of fosfomycin. Int J Infect Dis 15: e732–e739, 2011 [DOI] [PubMed] [Google Scholar]
- 85.Mieyal JJ. and Chock PB. Posttranslational modification of cysteine in redox signaling and oxidative stress: focus on S-glutathionylation. Antioxid Redox Signal 16: 471–475, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mieyal JJ, Gallogly MM, Qanungo S, Sabens EA, and Shelton MD. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid Redox Signal 10: 1941–1988, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Miller S, Douglas RM, Carter P, and Booth IR. Mutations in the glutathione-gated KefC K+ efflux system of Escherichia coli that cause constitutive activation. J Biol Chem 272: 24942–24947, 1997 [DOI] [PubMed] [Google Scholar]
- 88.Moore CM, Gaballa A, Hui M, Ye RW, and Helmann JD. Genetic and physiological responses of Bacillus subtilis to metal ion stress. Mol Microbiol 57: 27–40, 2005 [DOI] [PubMed] [Google Scholar]
- 89.Muhlenhoff U, Molik S, Godoy JR, Uzarska MA, Richter N, Seubert A, Zhang Y, Stubbe J, Pierrel F, Herrero E, Lillig CH, and Lill R. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab 12: 373–385, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Muller JE, Meyer F, Litsanov B, Kiefer P, and Vorholt JA. Core pathways operating during methylotrophy of Bacillus methanolicus MGA3 and induction of a bacillithiol-dependent detoxification pathway upon formaldehyde stress. Mol Microbiol 98: 1089–1100, 2015 [DOI] [PubMed] [Google Scholar]
- 91.Natori Y, Nanamiya H, Akanuma G, Kosono S, Kudo T, Ochi K, and Kawamura F. A fail-safe system for the ribosome under zinc-limiting conditions in Bacillus subtilis. Mol Microbiol 63: 294–307, 2007 [DOI] [PubMed] [Google Scholar]
- 92.Newton GL, Arnold K, Price MS, Sherrill C, Delcardayre SB, Aharonowitz Y, Cohen G, Davies J, Fahey RC, and Davis C. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J Bacteriol 178: 1990–1995, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Newton GL, Av-Gay Y, and Fahey RC. A novel mycothiol-dependent detoxification pathway in mycobacteria involving mycothiol S-conjugate amidase. Biochemistry 39: 10739–10746, 2000 [DOI] [PubMed] [Google Scholar]
- 94.Newton GL, Bewley CA, Dwyer TJ, Horn R, Aharonowitz Y, Cohen G, Davies J, Faulkner DJ, and Fahey RC. The structure of U17 isolated from Streptomyces clavuligerus and its properties as an antioxidant thiol. Eur J Biochem 230: 821–825, 1995 [DOI] [PubMed] [Google Scholar]
- 95.Newton GL, Fahey RC, and Rawat M. Detoxification of toxins by bacillithiol in Staphylococcus aureus. Microbiology 158: 1117–1126, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Newton GL, Leung SS, Wakabayashi JI, Rawat M, and Fahey RC. The DinB superfamily includes novel mycothiol, bacillithiol, and glutathione S-transferases. Biochemistry 50: 10751–10760, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Newton GL, Rawat M, La Clair JJ, Jothivasan VK, Budiarto T, Hamilton CJ, Claiborne A, Helmann JD, and Fahey RC. Bacillithiol is an antioxidant thiol produced in Bacilli. Nat Chem Biol 5: 625–627, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nguyen TT, Eiamphungporn W, Mader U, Liebeke M, Lalk M, Hecker M, Helmann JD, and Antelmann H. Genome-wide responses to carbonyl electrophiles in Bacillus subtilis: control of the thiol-dependent formaldehyde dehydrogenase AdhA and cysteine proteinase YraA by the MerR-family regulator YraB (AdhR). Mol Microbiol 71: 876–894, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nicely NI, Parsonage D, Paige C, Newton GL, Fahey RC, Leonardi R, Jackowski S, Mallett TC, and Claiborne A. Structure of the type III pantothenate kinase from Bacillus anthracis at 2.0 A resolution: implications for coenzyme A-dependent redox biology. Biochemistry 46: 3234–3245, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev 27: 313–339, 2003 [DOI] [PubMed] [Google Scholar]
- 101.Outten CE. and O'Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292: 2488–2492, 2001 [DOI] [PubMed] [Google Scholar]
- 102.Ozyamak E, Black SS, Walker CA, Maclean MJ, Bartlett W, Miller S, and Booth IR. The critical role of S-lactoylglutathione formation during methylglyoxal detoxification in Escherichia coli. Mol Microbiol 78: 1577–1590, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Palmer LD. and Skaar EP. Transition metals and virulence in bacteria. Annu Rev Genet 50: 67–91, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pattison DI. and Davies MJ. Absolute rate constants for the reaction of hypochlorous acid with protein side chains and peptide bonds. Chem Res Toxicol 14: 1453–1464, 2001 [DOI] [PubMed] [Google Scholar]
- 105.Peralta D, Bronowska AK, Morgan B, Doka E, Van Laer K, Nagy P, Grater F, and Dick TP. A proton relay enhances H2O2 sensitivity of GAPDH to facilitate metabolic adaptation. Nat Chem Biol 11: 156–163, 2015 [DOI] [PubMed] [Google Scholar]
- 106.Perera VR, Newton GL, Parnell JM, Komives EA, and Pogliano K. Purification and characterization of the Staphylococcus aureus bacillithiol transferase BstA. Biochim Biophys Acta 1840: 2851–2861, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Perera VR, Newton GL, and Pogliano K. Bacillithiol: a key protective thiol in Staphylococcus aureus. Expert Rev Anti Infect Ther 13: 1089–1107, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Picciocchi A, Saguez C, Boussac A, Cassier-Chauvat C, and Chauvat F. CGFS-type monothiol glutaredoxins from the cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe-2S] cluster. Biochemistry 46: 15018–15026, 2007 [DOI] [PubMed] [Google Scholar]
- 109.Pittman MS, Robinson HC, and Poole RK. A bacterial glutathione transporter (Escherichia coli CydDC) exports reductant to the periplasm. J Biol Chem 280: 32254–32261, 2005 [DOI] [PubMed] [Google Scholar]
- 110.Poor CB, Chen PR, Duguid E, Rice PA, and He C. Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus. J Biol Chem 284: 23517–23524, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Posada AC, Kolar SL, Dusi RG, Francois P, Roberts AA, Hamilton CJ, Liu GY, and Cheung A. Importance of bacillithiol in the oxidative stress response of Staphylococcus aureus. Infect Immun 82: 316–332, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pother DC, Gierok P, Harms M, Mostertz J, Hochgrafe F, Antelmann H, Hamilton CJ, Borovok I, Lalk M, Aharonowitz Y, and Hecker M. Distribution and infection-related functions of bacillithiol in Staphylococcus aureus. Int J Med Microbiol 303: 114–123, 2013 [DOI] [PubMed] [Google Scholar]
- 113.Pother DC, Liebeke M, Hochgrafe F, Antelmann H, Becher D, Lalk M, Lindequist U, Borovok I, Cohen G, Aharonowitz Y, and Hecker M. Diamide triggers mainly S Thiolations in the cytoplasmic proteomes of Bacillus subtilis and Staphylococcus aureus. J Bacteriol 191: 7520–7530, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rajkarnikar A, Strankman A, Duran S, Vargas D, Roberts AA, Barretto K, Upton H, Hamilton CJ, and Rawat M. Analysis of mutants disrupted in bacillithiol metabolism in Staphylococcus aureus. Biochem Biophys Res Commun 436: 128–133, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Roberts AA, Sharma SV, Strankman AW, Duran SR, Rawat M, and Hamilton CJ. Mechanistic studies of FosB: a divalent-metal-dependent bacillithiol-S-transferase that mediates fosfomycin resistance in Staphylococcus aureus. Biochem J 451: 69–79, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Robinson NJ. and Winge DR. Copper metallochaperones. Annu Rev Biochem 79: 537–562, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rosario-Cruz Z. and Boyd JM. Physiological roles of bacillithiol in intracellular metal processing. Curr Genet 62: 59–65, 2016 [DOI] [PubMed] [Google Scholar]
- 118.Rosario-Cruz Z, Chahal HK, Mike LA, Skaar EP, and Boyd JM. Bacillithiol has a role in Fe-S cluster biogenesis in Staphylococcus aureus. Mol Microbiol 98: 218–242, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sakuda S, Zhou ZY, and Yamada Y. Structure of a novel disulfide of 2-(N-acetylcysteinyl)amido-2-deoxy-alpha-D-glucopyranosyl-myo-inositol produced by Streptomyces sp. Biosci Biotechnol Biochem 58: 1347–1348, 1994 [DOI] [PubMed] [Google Scholar]
- 120.Sharma SV, Arbach M, Roberts AA, Macdonald CJ, Groom M, and Hamilton CJ. Biophysical features of bacillithiol, the glutathione surrogate of Bacillus subtilis and other firmicutes. Chembiochem 14: 2160–2168, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sharma SV, Jothivasan VK, Newton GL, Upton H, Wakabayashi JI, Kane MG, Roberts AA, Rawat M, La Clair JJ, and Hamilton CJ. Chemical and chemoenzymatic syntheses of bacillithiol: a unique low-molecular-weight thiol amongst low G + C gram-positive bacteria. Angew Chem Int Ed Engl 50: 7101–7104, 2011 [DOI] [PubMed] [Google Scholar]
- 122.Shen J, Keithly ME, Armstrong RN, Higgins KA, Edmonds KA, and Giedroc DP. Staphylococcus aureus CstB is a novel multidomain persulfide dioxygenase-sulfurtransferase involved in hydrogen sulfide detoxification. Biochemistry 54: 4542–4554, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Smaldone GT, Antelmann H, Gaballa A, and Helmann JD. The FsrA sRNA and FbpB protein mediate the iron-dependent induction of the Bacillus subtilis lutABC iron-sulfur-containing oxidases. J Bacteriol 194: 2586–2593, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Smaldone GT, Revelles O, Gaballa A, Sauer U, Antelmann H, and Helmann JD. A global investigation of the Bacillus subtilis iron-sparing response identifies major changes in metabolism. J Bacteriol 194: 2594–2605, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Spies HS. and Steenkamp DJ. Thiols of intracellular pathogens. Identification of ovothiol A in Leishmania donovani and structural analysis of a novel thiol from Mycobacterium bovis. Eur J Biochem 224: 203–213, 1994 [DOI] [PubMed] [Google Scholar]
- 126.Taguchi T, Yabe M, Odaki H, Shinozaki M, Metsa-Ketela M, Arai T, Okamoto S, and Ichinose K. Biosynthetic conclusions from the functional dissection of oxygenases for biosynthesis of actinorhodin and related Streptomyces antibiotics. Chem Biol 20: 510–520, 2013 [DOI] [PubMed] [Google Scholar]
- 127.Tang HJ, Chen CC, Cheng KC, Toh HS, Su BA, Chiang SR, Ko WC, and Chuang YC. In vitro efficacy of fosfomycin-containing regimens against methicillin-resistant Staphylococcus aureus in biofilms. J Antimicrob Chemother 67: 944–950, 2012 [DOI] [PubMed] [Google Scholar]
- 128.Thompson MK, Keithly ME, Harp J, Cook PD, Jagessar KL, Sulikowski GA, and Armstrong RN. Structural and chemical aspects of resistance to the antibiotic fosfomycin conferred by FosB from Bacillus cereus. Biochemistry 52: 7350–7362, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Totemeyer S, Booth NA, Nichols WW, Dunbar B, and Booth IR. From famine to feast: the role of methylglyoxal production in Escherichia coli. Mol Microbiol 27: 553–562, 1998 [DOI] [PubMed] [Google Scholar]
- 130.Upton H, Newton GL, Gushiken M, Lo K, Holden D, Fahey RC, and Rawat M. Characterization of BshA, bacillithiol glycosyltransferase from Staphylococcus aureus and Bacillus subtilis. FEBS Lett 586: 1004–1008, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.van der Heijden J, Vogt SL, Reynolds LA, Pena-Diaz J, Tupin A, Aussel L, and Finlay BB. Exploring the redox balance inside gram-negative bacteria with redox-sensitive GFP. Free Radic Biol Med 91: 34–44, 2015 [DOI] [PubMed] [Google Scholar]
- 132.VanDuinen AJ, Winchell KR, Keithly ME, and Cook PD. X-ray crystallographic structure of BshC, a unique enzyme involved in bacillithiol biosynthesis. Biochemistry 54: 100–103, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Winchell KR, Egeler PW, VanDuinen AJ, Jackson LB, Karpen ME, and Cook PD. A structural, functional, and computational analysis of BshA, the first enzyme in the bacillithiol biosynthesis pathway. Biochemistry 55: 4654–4665, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Winterbourn CC, Kettle AJ, and Hampton MB. Reactive oxygen species and neutrophil function. Annu Rev Biochem 85: 765–792, 2016 [DOI] [PubMed] [Google Scholar]