Abstract
p62, as a scaffolding/adaptor protein, is involved in multiple physiological processes include inflammation, autophagy and mitosis. However, the influence of p62 in cancer patients has not been comprehensively investigated. Moreover, the prognostic value of p62 for the survival of patients with solid tumors remains controversial. In this present meta-analysis, twenty suitable articles were identified from PubMed, EMBASE and Web of Science, Nature databases, including 4271 patients. A random-effect or fixed-effect model was adopted to correlate p62 expression with different outcome measured in entire tumors. Combined with results of hazard ratios (HRs) and 95% confidence intervals (CIs), we concluded that higher expression of p62 is associated with poorer overall survival (OS) (HR: 2.22, 95% CI: 1.82–2.71, P < 0.05), disease-free survival (DFS) (HR = 2.48, 95% CI: 1.78–3.46, P < 0.05) and even certain clinicopathological parameters, such as lymph node metastasis (RR = 1.21, 95% CI: 1.06–1.37) and clinical stages (RR = 1.27, 95% CI: 1.12–1.45), in cancer patients. Consequently, our data showed that p62 might be an effective poor prognostic factor for patients with various solid tumors.
Keywords: solid tumors, p62, prognosis, meta-analysis
INTRODUCTION
p62 (sequestosome-1) was first identified as an interaction protein with human p56lck SH domain [1]. Distinct from human, murine p62 homolog ZIP was independently identified as a binding partner of atypical protein kinase C (PKC-ζ) [2]. A plenty of studies have found that p62 is a scaffold protein and is involved in several important signal pathways like NF-κB signaling, autophagy, mitosis to influence inflammation, deoxidization, cell growth, and cell cycle, which may affect tumorigenesis [3, 4].
p62 serves as a crucial factor during the process of tumorigenesis. Firstly, p62 down-regulates the level of reactive oxygen species (ROS) to promote tumorigenesis. p62 consists of multiple domains: the Phox1 and Bem1p (PB1) domain, that can interact with PKC-ζ, the ZZ-type zinc finger (ZZ) domain, that interact with receptor-interacting protein (RIP), the TB domain, that interact with tumor necrosis factor receptor-associated factor 6 (TRAF6), the light chain 3-interacting region (LIR) domain that interact with light chain 3 (LC3) and the ubiquitin-associated (UBA) domain that binding ubiquitin. The interactions between p62 with PKC-ζ, RIP and TRAF6 can activate NF-κB signaling [5, 6], which leads to down-regulation of ROS, thus avoiding the initiation of apoptosis pathway and promoting carcinogenesis [7]. Additionally, Keap1-Nrf2 complex is a key component to response to cellular oxidative stress [8], p62 directly interacts with Keap1 to prevent Nrf2 from being degraded by ubiquitin-proteasome system, subsequently allow Nrf2 to activate the downstream antioxidant genes to suppress ROS level [9–11]. Secondly, p62 exhibit non-liner, complicate interaction with autophagy to influence carcinogenesis process [3]. p62 can recruit Raptor and Rags proteins to activate mTORC1 and consequently inhibit autophagy [12]. Additionally, p62 is also a substrate of autophagy [13], thus the up-regulation of autophagy will decrease p62 level, which further activates autophagy. This feed forward loop may ensure the irreversible activation of autophagy under nutrient deprivation. Researchers have also found impaired autophagy can induce the accumulation of p62 to promote carcinogenesis [14]. In mitosis, constitutive phosphorylation of p62 T269 and S272 residues by Cdk1 can prevent cell from carcinogenesis. Mutant p62 that cannot be phosphorylated on those two residues will faster exit mitosis and increase cell proliferation, which promotes tumorigenesis in Ras-transformed cells [15].
Although much relationship between p62 and tumorigenesis has been unveiled in laboratory, it is unclear the actual prognosis value of this adaptor protein in cancer patients. Does p62, a hub of NF-κB, autophagy and mitosis pathway, can serve as a stable cancer biomarker? The aim of our meta-analysis is to give a quantitative assessment to the prognostic value of p62 in various type of cancer.
RESULTS
Study selection and characteristics description
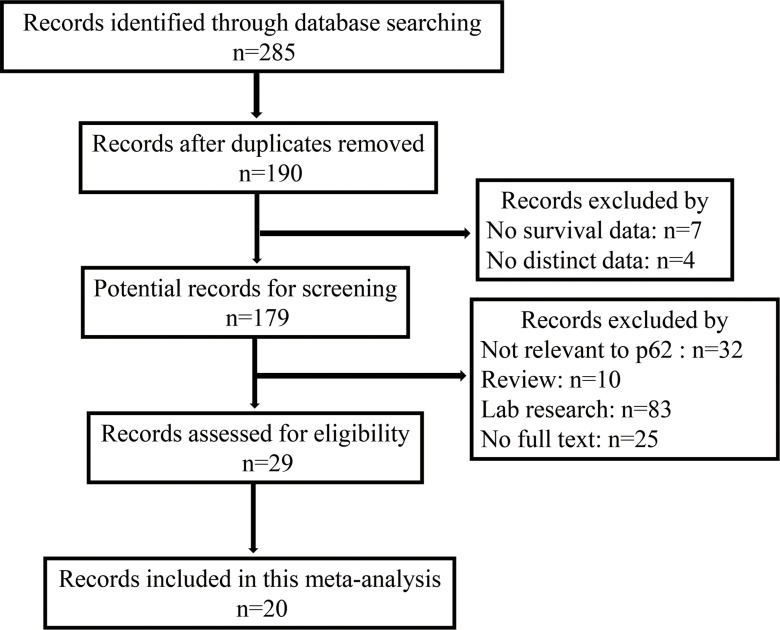
The databases including PubMed, EMBASE and ISI Web of Science, were originally searched for a total of 285 articles containing keywords p62 and prognosis. 265 of those were excluded, due to repetitive researches (n = 95), without full texts (n = 25), laboratory studies (n = 103), reviews (n = 10), studies not relevant to the current analysis (n = 32). 20 publications [16–35] were selected for this meta-analysis containing 4271 patients. The flow chart of the study search and selection process is reported in Figure 1. The articles to be selected were collected as of September 2017. Immunohistochemistry (IHC) was the only method to evaluate p62 expression in human specimens. The main tumor types of these patients are breast cancer (n = 1474), non-small lung cancer (n = 659) and melanoma (n = 196). In addition, > 10% positive tumor cells and scores greater than one were the most suitable cut-off values for overall survival, > 10% positive tumor cells were a more suitable cut-off value for disease-free survival at the same time. The main characteristics of the 20 included studies are summarized in Table 1. This meta-analysis was performed with the guideline of Cochrane.
Figure 1. Flow diagram of the selection of eligible studies.
Table 1. Main characteristics of studies exploring the relationship between p62 expression and tumor prognosis.
| Author | Year | Region | Cancer Type | No. of Patients | Follow-up Time Median (range) | Detection Method | Cut-off | Outcomes | NOS Score |
|---|---|---|---|---|---|---|---|---|---|
| Shun Nakayama [16] | 2017 | Japan | Colorectal Carcinoma | 118 | 69.8 m (2–131) | IHC (MBL) | ≥ 10% | OS | 7 |
| Akihito Arai [17] | 2017 | Japan | Hypopharyngeal Carcinoma | 54 | NR | IHC (MBL) | NR | DFS | 5 |
| Diana Y. L. Tang [18] | 2016 | UK | Melanoma | 75 | 5 y | IHC (NR) | ≥ 20% | OS, DFS | 6 |
| Xifeng Wang [19] | 2015 | China | Non-Small Cell Lung Cancer | 104 | 48.5 m (3–96.5) | IHC (Abcam) | IRS ≥ 4 | OS | 7 |
| Reiko Iwadate [20] | 2015 | Japan | Endometrial Cancer | 194 | 22.0 m (2.0–58.0) | IHC (Santa Cruz) | ≥ 10% | OS | 8 |
| Mingfei Zhao [21] | 2015 | China | Gliomas | 75 | 24 m (6–60) | IHC (Santa Cruz) | IRS ≥ 3 | OS, DFS | 6 |
| Xianhan Jiang [22] | 2015 | China | Prostate Cancer | 149 | 10 y | IHC (Santa Cruz) | IRS ≥ 4 | OS | 7 |
| J-L Liu [23] | 2014 | China | Oral Squamous Cell Carcinoma | 195 | 47.08 ± 32.37 m | IHC (Abcam) | IRS ≥ 4 | OS, DFS | 8 |
| Reiko Iwadate [24] | 2014 | Japan | Epithelial Ovarian Cancer | 266 | 59 m (1–120) | IHC (Santa Cruz) | ≥ 10% | OS | 8 |
| Robert A Ellis [25] | 2014 | UK | Melanoma | 121 | 7 y | IHC (NR) | ≥ 20% | DFS | 7 |
| Sang Kyum Kim [26] | 2013 | Korea | Phyllodes Tumor | 190 | NR | IHC (Abcam) | IRS ≥ 2 | OS, DFS | 7 |
| Rong-Zhen Luo [27] | 2013 | China | Breast Cancer | 163 | 112 m (15–145) | IHC (Santa Cruz) | IRS ≥ 2 | OS, DFS | 7 |
| Jae Myung Park [28] | 2012 | USA | Colon Carcinoma | 178 | 4 y | IHC (MBL) | ≥ 50% | OS | 7 |
| Junjeong Choi [29] | 2012 | Korea | Breast Cancer | 489 | 82.0 ± 36.5 m | IHC (Abcam) | IRS ≥ 2 | OS, DFS | 8 |
| Sewha Kim [30] | 2012 | Korea | Breast Cancer | 119 | 59.2 ± 27.9 m | IHC (Abcam) | IRS ≥ 2 | OS, DFS | 6 |
| Daisuke Inoue [31] | 2012 | Japan | Lung Adenocarcinoma | 109 | 1626 d (17–3366) | IHC (Santa Cruz) | ≥ 10% | OS | 6 |
| Phil Rolland [32] | 2007 | UK | Breast Cancer | 523 | 76 m | IHC (INC) | ≥ 5% | OS | 8 |
| L-Z Xu [33] | 2016 | China | Breast Cancer | 369 | NR | IHC (NR) | NR | OS, DFS | 8 |
| Ji-Ye Kim [34] | 2014 | Italy | Breast Cancer | 334 | NR | IHC (Abcam) | ≥ 30% | DFS | 7 |
| Anna M. Schläfli [35] | 2016 | Switzerland | Non-Small Cell Lung Cancer | 446 | NR | IHC (MBL) | ≥ 25% | OS, DFS | 7 |
NR: Not Reported; y: year; m: month; d: day; OS: Overall Survival; DFS: Disease-Free Survival; IRS: Immunoreactive Score.
Correlations between p62 expression and overall survival (OS)
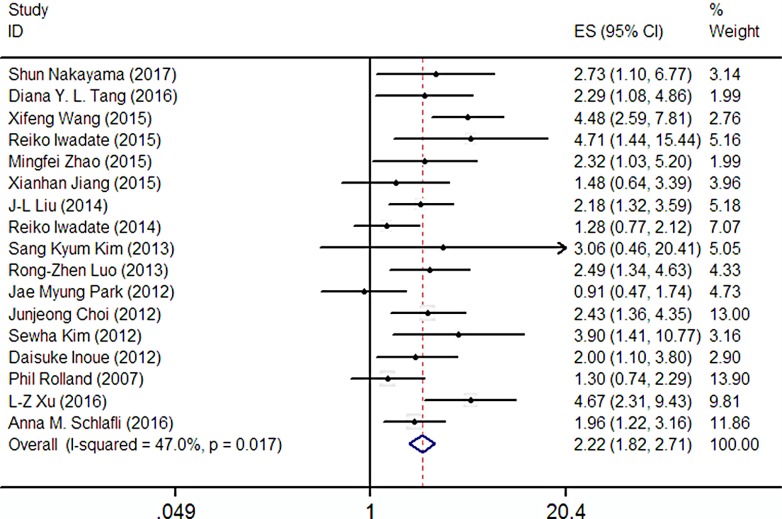
17 articles included 3762 patients were selected to evaluate the relationship between p62 expression and OS. In addition, this test was analyzed using a random-effect model due to high heterogeneity (I2 = 47.0%). The pooled HR revealed that there was a clear correlation between the high expression of p62 and the worse OS (HR: 2.22, 95% CI: 1.82–2.71, P < 0.05; Figure 2) in multivariate analysis. These discoveries indicate that p62 is a prognostic factor for various types of cancer.
Figure 2. Forest plot describing the association between p62 expression and OS.
To minimize heterogeneity, the subgroup analyses were performed according to the ethnics (Asian or not), case number (≥ 150 or not), NOS score (≥ 7 or not), antibodies (various company), cut-off value (various scoring criteria). The pooled HRs and heterogeneities according to all these factors were presented in Table 2. Unfortunately, all these subgroup analyses demonstrated that there were no significant lower I2 value when the P < 0.05. Therefore, subgroup analysis were failed to find the origin of high heterogeneity.
Table 2. Associations between p62 expression and OS stratified according to the ethnics, case number, NOS score, antibodies and cut-off value.
| Categories | Subgroups | Ref | HR (95% CI) | Heterogeneity test (I2, P-value) |
|---|---|---|---|---|
| Ethnics | Asian | [16, 19–24, 26, 27, 29–31, 33] | 2.69 (2.08–3.48) | 37.3%, 0.085 |
| Not Asian | [18, 28, 32, 35] | 1.48 (1.08–2.04) | 39.4%, 0.176 | |
| Case Number | ≥ 150 | [20, 23, 24, 26–29, 32, 33, 35] | 2.15 (1.69–2.72) | 56.5%, 0.014 |
| <150 | [16, 18, 19, 21, 22, 30, 31] | 2.53 (1.84–3.47) | 15.4%, 0.312 | |
| NOS Score | ≥ 7 | [16, 19, 20, 22–24, 26–29, 32, 33, 35] | 2.18 (1.75–2.71) | 58.1%, 0.004 |
| <7 | [18, 21, 30, 31] | 2.61 (1.70–4.00) | 0.0%, 0.681 | |
| Antibody | Santa Cruz | [20–22, 24, 27, 31] | 2.11 (1.50–2.96) | 23.5%, 0.258 |
| Abcam | [19, 23, 26, 29, 30] | 2.77 (1.77–4.32) | 9.9%, 0.350 | |
| MBL | [16, 28, 35] | 1.72 (1.20–2.46) | 59.3%, 0.086 | |
| NR | [18, 32, 33] | 2.21 (1.47–3.34) | 74.2%, 0.021 | |
| Cut-off Value | IRS | [19,21–23,26, 27, 29, 30] | 2.55 (1.80–3.61) | 0.0%, 0.448 |
| Percentage | [16, 18, 20, 24, 28, 31, 32, 35] | 1.73 (1.34–2.23) | 34.9%, 0.150 |
IRS: Immunoreactive Score; NR: Not Reported.
Correlations between p62 expression and disease-free survival (DFS)
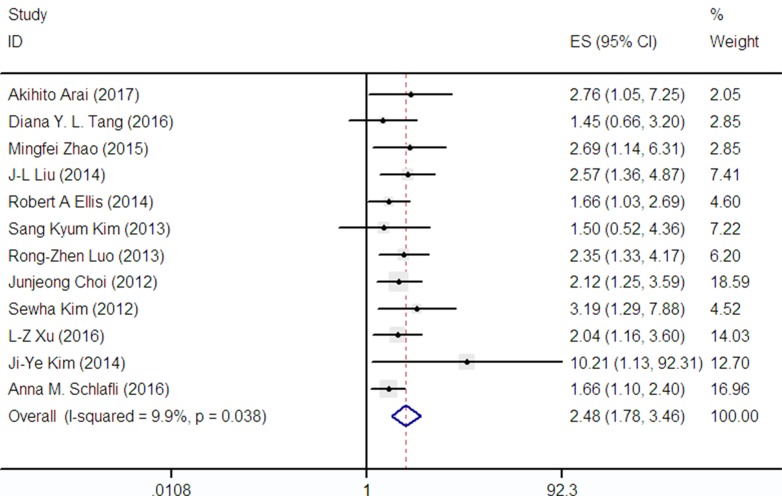
DFS was reported in 12 publications covering 2630 patients. A low heterogeneity (I2 = 9.9%) was observed among these studies, so we adopt a fixed-effect model to analysis. Nonetheless, the combined HR for these articles assessing p62 amplification on DFS was 2.48 (95% CI: 1.78–3.46) as shown in Figure 3, demonstrating that p62 overexpression was an indicator of poor prognosis in cancer patients.
Figure 3. Forest plot describing the association between p62 expression and DFS.
Correlations between p62 expression and clinicopathological parameters
As shown in Supplementary Figure 1, 14 eligible articles were used to collect the clinical and pathological parameters. Meanwhile, pooled results of the correlations were identified between the over-expressed p62 and clinicopathological features of patients with solid tumors. Our results showed that p62 overexpression was related to lymph node metastasis (RR = 1.21, 95% CI: 1.06–1.37) and clinical stages (RR = 1.27, 95% CI: 1.12–1.45), which was independent of gender (RR = 1.00, 95% CI: 0.78–1.29), tumor differentiation (RR = 0.86, 95% CI: 0.67–1.11) and tumor status (RR = 1.00, 95% CI: 0.78–1.29) (see Table 3 and Supplementary Figure 1).
Table 3. Meta-analysis results of the associations of p62 expression with clinicopathological parameters.
| Clinicopathological parameter | Ref | Overall OR (95% CI) | Heterogeneity test (I2, P-value) |
|---|---|---|---|
| Gender (male vs female) | [16, 17, 19, 21, 23, 31, 35] | 1.00 (0.78–1.29) | 0.0%, 0.663 |
| Tumor Differentiation (poor VS well) | [16, 17, 19, 20, 29–32] | 0.86 (0.67–1.11) | 71.8%, 0.001 |
| Tumor Size (T3-4 vs T1-2) | [17, 20, 23, 27, 29, 30, 32, 33, 35] | 1.13 (0.96–1.33) | 60.6%, 0.009 |
| Lymph Node Metastasis (yes vs no) | [16, 19, 20, 27, 29–33] | 1.21 (1.06–1.37) | 78.6%, < 0.001 |
| Clinical Stage (III-IV vs I-II) | [16, 17, 19–21, 23, 24, 27, 29–31, 33, 35] | 127 (1.12–1.45) | 84.3%, < 0.001 |
Assessment of heterogeneity and sensitivity analysis
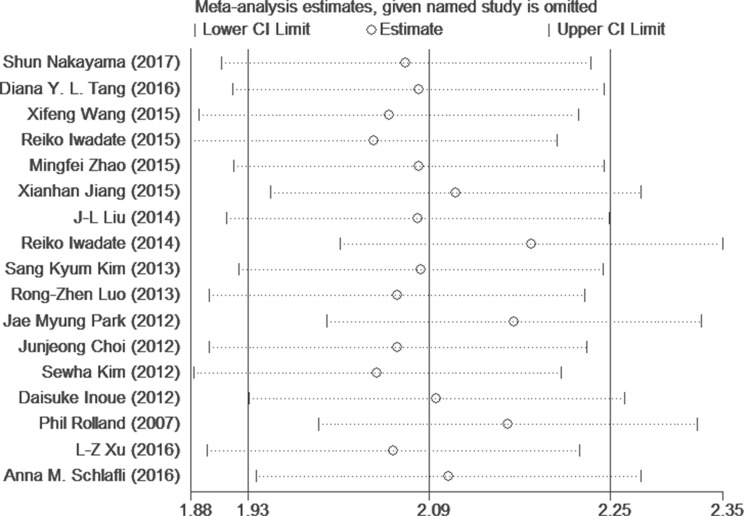
There was significant heterogeneity (I2 > 30%) among studies in OS and clinical pathological parameters analyses. Therefore, a random-effect model was adopted in these studies. A meta-regression analysis with published country, case number (≥150 or not), NOS Score (≥7 or not), antibodies (used for different companies) and cut-off value (IRS scores or Percentage) as covariates was conducted. All covariates were fit into the meta-regression model one at a time to identify potential sources of heterogeneity. However, none of these covariates were verified as a significant source of heterogeneity (Table 4). Moreover, to determine whether modifications of the included criteria affected the results, we tested this meta-analysis by a sensitivity analysis (Figure 4). The results indicated that the pooled estimates of the effect of over-expressed p62 on OS in solid tumors did not vary significantly with the exclusion of any individual studies. Also it meant that the results of this meta-analysis were stable after using the leave-one-out method.
Table 4. Results of meta-regression analysis exploring the source of heterogeneity with OS.
| Covariates | OS | ||
|---|---|---|---|
| Coef. | S.E. | P value | |
| Country | -0.126 | 0.067 | 0.079 |
| Case Number | -0.272 | 0.237 | 0.269 |
| NOS | -0.118 | 0.297 | 0.698 |
| Antibody | -0.004 | 0.110 | 0.970 |
| Cut-off value | -0.510 | 0.192 | 0.081 |
Coef.: Coefficient; S.E.: Standard Error.
Figure 4. Sensitivity analysis of the OS in the meta-analysis.
Publication bias
We constructed Begg’s funnel plot with pseudo 95% confidence limits and Egger’s test to assess the publication bias of these applicable studies. The shapes of the funnel plots for OS, DFS and clinicopathological parameters showed no evidences of obvious asymmetry, and Egger’s test indicated the absence of publication bias (p > 0.05). The above results indicated that this meta-analysis was statistically reliable. Furthermore, these findings were other strong evidences to verify that p62 was a prognostic factor for cancer patients (see Figure 5 and Supplementary Figure 2).
Figure 5.
Funnel plot for the assessment of potential publication bias regarding OS (A) and DFS (B) in the meta-analysis.
DISCUSSION
The meta-analysis presented herein is the first to describe all the reported studies investigating the impact of p62 expression in human tumors on prognosis. Furthermore, this analysis aimed to examine the association between p62 expression with OS and DFS of cancer patients. We combined the outcomes of 4271 cancer patients from 20 individual studies, suggesting that p62 high-expression significantly predicted poor OS (HR: 2.22, 95% CI: 1.82–2.71, P < 0.05) and DFS (HR: 2.48, 95% CI: 1.78–3.46, P < 0.05). Moreover, obvious correlations were observed between p62 overexpression and clinicopathological characteristics including lymph node metastasis (RR = 1.21, 95% CI: 1.06–1.37) and clinical stages (RR = 1.27, 95% CI: 1.12–1.45), which indicated general roles of p62 in cancer prognosis.
Our study has several strengths. The included original articles were all prospective, which greatly reduces the likelihood of selection bias and reverse causation. In addition, a large number of cases had been collected from different studies, and 4271 participants represented a large number, significantly increasing the statistical capacity of the analysis. Lastly, funnel plot and Begg’s analysis didn’t discover any publication bias, suggesting that the results has a high degree of credibility. Nevertheless, this meta-analysis also had an inherent potential limitation that should be considered. This potential limitation is the high heterogeneity between OS and different clinicopathological parameter analyses. High heterogeneity may come from the following sources: first, there were no uniform criteria for IHC evaluation and cutoff point now. The assessment of p62 expression and determination of cutoff values were based on personal judgment. Second, the results might vary with age, gender, territory, tumor grade and staging. Third, diverse methods of survival data analysis in various studies were considered as potential sources of heterogeneity.
This study analyzed the expression of p62 in various kinds of tumor tissues and evaluated its prognostic value. The results showed that the high expression of p62 was related to poor prognosis. It is noteworthy that both autophagy and inflammatory pathway for cancer development have the duality in previous research, but our analysis shows the role of p62 in the prognosis of cancer development has been clear and simple. As a scaffold protein of multiple pathways, the simple relationship between p62 and cancer prognosis can be attributed to the alternate of following pathways under high level of p62: First, overexpression of p62 can inhibit autophagy to make the accumulation and aggregation of itself, which is toxic and can promote tumorigenesis. Second, high level of p62 can activate NF-κB signaling and Nrf2 to degrade ROS respectively, which may help cancer cell evade apoptosis to promote cancer progress. Furthermore, the role of p62 in autophagy pathways is worthy of widespread attention in cancer research. Autophagy is a crucial target for cancer therapy, and most autophagy-target anticancer drugs inhibit autophagy (like rapamycin and it analogs). In contrast, as shown in laboratory research and our meta-analysis, the high level of p62-a native autophagy inhibitor, can promote carcinogenesis. It is also worthy of note that most of the current autophagy-target drugs have poor efficacy and are susceptible to drug resistance [36]. Those facts remind us that p62-autophagy-cancer relationship is not only a valuable prognosis biomarker for cancer patients, but also help scientists to reevaluate the role of autophagy in cancer, and may contribute to the development of current autophagy-target cancer therapy.
MATERIALS AND METHODS
Search strategy and selection criteria
We searched the relevant studies from the PubMed, EMBASE and ISI Web of Science, Nature databases using the following keywords in all possible combinations: p62, prognosis, and tumor. The last systematical search was performed on September 20, 2017. Criteria for eligibility of each study included in this meta-analysis were: (1) the correlation between p62 expression and overall survival (OS) or disease-free survival (DFS) of cancer patients; (2) the expression of p62 was detected by immunohistochemistry; (3) to provide adequate information to assess hazard ratio (HR) and 95% confidence interval (CI); (4) pathological diagnosis of various tumor types or clinicopathological features were described; and (5) to be published as a full text in the English language. Reviews, letters, comments, repetitive researches, case reports, and personal communications were excluded. Laboratory studies were also excluded if they did not provide quantitative data regarding the primary outcome measure.
Data extraction
Two investigators (Haihua Ruan and Lingling Wang) independently reviewed each eligible publication and extracted data by a standardized data-extract form. The uniform information collected were as follows: first author’s name, publication date, the patient’s region, type of cancer, p62 detection method, number of cases, number of patients with p62-positive, follow-up times, cut-off values, and clinicopathological parameters. If the results were inconsistent, the third person would join the discussion and made the final decision [37].
Statistical method of meta-analyses and quality assessment
All the statistical analyses were performed using Stata 12.0 (Stata Corporation, College 216 Station, TX, USA) software. Pooled estimates of hazard ratios (HR) with their 95% confidence intervals (CIs) were used to assess the relevance between p62 expression and survival outcome and clinical parameters, including gender, tumor differentiation, lymph node metastasis, distant metastasis as well as clinical stage. Multivariate HR and 95% CI were employed when both univariate and multivariate results were provided. Moreover, each of the 20 eligible studies included in our meta-analysis was assessed for quality according to the Newcastle–Ottawa Scale (NOS). The NOS score ranged from 0 to 9, and studies with NOS score ≥7 were defined as high-quality studies. The heterogeneity within studies was tested with chi squared test (Cochrane’s Q test) and I-squared statistical test. The random-effects model was adopted when the result of a Q-test (I2 > 30% or P < 0.05, high heterogeneity). Furthermore, evidence of publication bias was used by Begg’s funnel plot and Egger’s test [38, 39]. P < 0.05 was considered to be statistically significant in this analysis.
SUPPLEMENTARY MATERIALS FIGURES
Footnotes
Author contributions
Haihua Ruan and Xichuan Li conceived and carried out the analysis. Haihua Ruan, Jingyue Xu and Lingling Wang performed literature search, study selection and data extraction. Zhenyu Zhao conducted statistical analysis and quality assessment. Lingqin Kong and Bei Lan prepared tables and figures. Haihua Ruan and Xichuan Li wrote the manuscript. All authors reviewed the manuscript.
CONFLICTS OF INTEREST
The authors declare no potential conflicts of interest.
FUNDING
This work was supported by the National Natural Science Foundation of China (grants 81472681 to X.L. and 31540066 to H.R.).
REFERENCES
- 1.Park I, Chung J, Walsh CT, Yun Y, Strominger JL, Shin J. Phosphotyrosine-independent binding of a 62-kDa protein to the src homology 2 (SH2) domain of p56lck and its regulation by phosphorylation of Ser-59 in the lck unique N-terminal region. Proc Natl Acad Sci U S A. 1995;92:12338–12342. doi: 10.1073/pnas.92.26.12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puls A, Schmidt S, Grawe F, Stabel S. Interaction of protein kinase C ζ with ZIP, a novel protein kinase C-binding protein. Proc Natl Acad Sci U S A. 1997;94:6191–6196. doi: 10.1073/pnas.94.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moscat J, Diaz-Meco MT. p62: a versatile multitasker takes on cancer. Trends Biochem Sci. 2012;37:230–236. doi: 10.1016/j.tibs.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanz L, Sanchez P, Lallena MJ, Diaz-Meco MT, Moscat J. The interaction of p62 with RIP links the atypical PKCs to NF-κB activation. EMBO J. 1999;18:3044–3053. doi: 10.1093/emboj/18.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanz L, Diaz-Meco MT, Nakano H, Moscat J. The atypical PKC-interacting protein p62 channels NF-κB activation by the IL-1-TRAF6 pathway. EMBO J. 2000;19:1576–1586. doi: 10.1093/emboj/19.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, Moscat J. The signaling adaptor p62 is an important NF-κB mediator in tumorigenesis. Cancer Cell. 2008;13:343–354. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem. 2009;284:13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 10.Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Copple IM, Lister A, Obeng AD, Kitteringham NR, Jenkins RE, Layfield R, Foster BJ, Goldring CE, Park BK. Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1-Nrf2 cell defense pathway. J Biol Chem. 2010;285:16782–16788. doi: 10.1074/jbc.M109.096545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44:134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linares JF, Amanchy R, Diaz-Meco MT, Moscat J. Phosphorylation of p62 by cdk1 controls the timely transit of cells through mitosis and tumor cell proliferation. Mol Cell Biol. 2011;31:105–117. doi: 10.1128/MCB.00620-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakayama S, Karasawa H, Suzuki T, Yabuuchi S, Takagi K, Aizawa T, Onodera Y, Nakamura Y, Watanabe M, Fujishima F. p62/sequestosome 1 in human colorectal carcinoma as a potent prognostic predictor associated with cell proliferation. Cancer Med. 2017;6:1264. doi: 10.1002/cam4.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arai A, Chano T, Ikebuchi K, Hama Y, Ochi Y, Tameno H, Shimada T. p62/SQSTM1 levels predicts radiotherapy resistance in hypopharyngeal carcinomas. Am J Cancer Res. 2017;7:881. [PMC free article] [PubMed] [Google Scholar]
- 18.Tang DYL, Ellis RA, Lovat PE. Prognostic Impact of Autophagy Biomarkers for Cutaneous Melanoma. Front Oncol. 2016;6 doi: 10.3389/fonc.2016.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Du Z, Li L, Shi M, Yu Y. Beclin 1 and p62 expression in non-small cell lung cancer: relation with malignant behaviors and clinical outcome. Int J Clin Exp Pathol. 2015;8:10644–10652. [PMC free article] [PubMed] [Google Scholar]
- 20.Iwadate R, Inoue J, Tsuda H, Takano M, Furuya K, Hirasawa A, Aoki D, Inazawa J. High Expression of p62 Protein Is Associated with Poor Prognosis and Aggressive Phenotypes in Endometrial Cancer. Am J Pathol. 2015;185:2523. doi: 10.1016/j.ajpath.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 21.Zhao M, Xu H, Zhang B, Hong B, Yan W, Zhang J. Impact of nuclear factor erythroid-derived 2-like 2 and p62/sequestosome expression on prognosis of patients with gliomas. Hum Pathol. 2015;46:843. doi: 10.1016/j.humpath.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Jiang X, Zhong W, Huang H, He H, Jiang F, Chen Y, Yue F, Zou J, Li X, He Y. Autophagy defects suggested by low levels of autophagy activator MAP1S and high levels of autophagy inhibitor LRPPRC predict poor prognosis of prostate cancer patients. Mol Carcinog. 2014;54:1194–1204. doi: 10.1002/mc.22193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu JL, Chen FF, Lung J, Lo CH, Lee FH, Lu YC, Hung CH. Prognostic significance of p62/SQSTM1 subcellular localization and LC3B in oral squamous cell carcinoma. Br J Cancer. 2014;111:944–954. doi: 10.1038/bjc.2014.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwadate R, Inoue J, Tsuda H, Takano M, Furuya K, Hirasawa A, Aoki D, Inazawa J. High Expression of SQSTM1/p62 Protein Is Associated with Poor Prognosis in Epithelial Ovarian Cancer. Acta Histochem Cytochem. 2014;47:295–301. doi: 10.1267/ahc.14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellis RA, Horswell S, Ness T, Lumsdon J, Tooze SA, Kirkham N, Armstrong JL, Lovat PE. Prognostic impact of p62 expression in cutaneous malignant melanoma. J Invest Dermatol. 2014;134:1476–1478. doi: 10.1038/jid.2013.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sang KK, Jung WH, Koo JS. Expression of autophagy-related proteins in phyllodes tumor. Int J Clin Exp Pathol. 2013;6:2145–2156. [PMC free article] [PubMed] [Google Scholar]
- 27.Luo RZ, Yuan ZY, Li M, Xi SY, Fu J, He J. Accumulation of p62 is associated with poor prognosis in patients with triple-negative breast cancer. Onco Targets Ther. 2013;6:883. doi: 10.2147/OTT.S46222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park JM, Huang S, Wu TT, Foster NR, Sinicrope FA. Prognostic impact of Beclin 1, p62/sequestosome 1 and LC3 protein expression in colon carcinomas from patients receiving 5-fluorouracil as adjuvant chemotherapy. Cancer Biol Ther. 2013;14:100. doi: 10.4161/cbt.22954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi J, Jung W, Koo JS. Expression of autophagy-related markers beclin-1, light chain 3A, light chain 3B and p62 according to the molecular subtype of breast cancer. Histopathology. 2013;62:275–286. doi: 10.1111/his.12002. [DOI] [PubMed] [Google Scholar]
- 30.Kim S, Jung WH, Koo JS. Differences in autophagy-related activity by molecular subtype in triple-negative breast cancer. Tumor Biol. 2012;33:1681–1694. doi: 10.1007/s13277-012-0424-1. [DOI] [PubMed] [Google Scholar]
- 31.Inoue D, Suzuki T, Mitsuishi Y, Miki Y, Suzuki S, Sugawara S, Watanabe M, Sakurada A, Endo C, Uruno A. Accumulation of p62/SQSTM1 is associated with poor prognosis in patients with lung adenocarcinoma. Cancer Sci. 2012;103:760. doi: 10.1111/j.1349-7006.2012.02216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rolland P, Madjd Z, Durrant L, Ellis IO, Layfield R, Spendlove I. The ubiquitin-binding protein p62 is expressed in breast cancers showing features of aggressive disease. Endocr Relat Cancer. 2007;14:73–80. doi: 10.1677/erc.1.01312. [DOI] [PubMed] [Google Scholar]
- 33.Xu LZ, Li SS, Zhou W, Kang ZJ, Zhang QX, Kamran M, Xu J, Liang DP, Wang CL, Hou ZJ. p62/SQSTM1 enhances breast cancer stem-like properties by stabilizing MYC mRNA. Oncogene. 2016;36 doi: 10.1038/onc.2016.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JY, Jung WH, Koo JS. Expression of Autophagy-Related Proteins According to Androgen Receptor and HER-2 Status in Estrogen Receptor-Negative Breast Cancer. PLoS One. 2014;9:e105666. doi: 10.1371/journal.pone.0105666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schläfli AM, Adams O, Galván JA, Gugger M, Savic S, Bubendorf L, Schmid RA, Becker KF, Tschan MP, Langer R, Berezowska S. Prognostic value of the autophagy markers LC3 and p62/SQSTM1 in early-stage non-small cell lung cancer. Oncotarget. 2016;7:39544. doi: 10.18632/oncotarget.9647. https://doi.org/10.18632/oncotarget.9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Z, Wu Y, Zhou X, Qian J, Zhu W, Shu Y, Liu P. Clinical efficacy of mTOR inhibitors in solid tumors: a systematic review. Future Oncol. 2015;11:1687–1699. doi: 10.2217/fon.15.70. [DOI] [PubMed] [Google Scholar]
- 37.Yuan P, Cao JL, Abuduwufuer A, Wang LM, Yuan XS, Lv W, Hu J. Clinical Characteristics and Prognostic Significance of TERT Promoter Mutations in Cancer: A Cohort Study and a Meta-Analysis. PLoS One. 2016;11:e0146803. doi: 10.1371/journal.pone.0146803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Brit Med J. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duval S, Tweedie R. Trim and Fill: A Simple Funnel-Plot–Based Method of Testing and Adjusting for Publication Bias in Meta-Analysis. Biometrics. 2000;56:455. doi: 10.1111/j.0006-341x.2000.00455.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.