Abstract
For the implementation of cellulosic ethanol technology, the maximum use of lignocellulosic materials is important to increase efficiency and to reduce costs. In this context, appropriate use of the pentose released by hemicellulose hydrolysis could improve de economic viability of this process. Since the Saccharomyces cerevisiae is unable to ferment the pentose, the search for pentose-fermenting microorganisms could be an alternative. In this work, the isolation of yeast strains from decaying vegetal materials, flowers, fruits and insects and their application for assimilation and alcoholic fermentation of xylose were carried out. From a total of 30 isolated strains, 12 were able to assimilate 30 g L−1 of xylose in 120 h. The strain Candida tropicalis S4 produced 6 g L−1 of ethanol from 56 g L−1 of xylose, while the strain C. tropicalis E2 produced 22 g L−1 of xylitol. The strains Candida oleophila G10.1 and Metschnikowia koreensis G18 consumed significant amount of xylose in aerobic cultivation releasing non-identified metabolites. The different materials in environment were source for pentose-assimilating yeast with variable metabolic profile.
Keywords: Xylose, Yeasts, Ethanol
Introduction
Bioethanol or first generation ethanol in Brazil is obtained by fermenting glucose from sugar cane juice and molasses (a residue from sugar making) using Saccharomyces cerevisiae. While this technology is a consolidated industrial process, production of second generation ethanol remains a challenge. One of the bottlenecks is the utilization of pentoses released by hydrolysis of hemicellulose that correspond to around 30% of the lignocellulosic feedstock. Yeasts able to convert xylose into ethanol have been described like as Scheffersomyces (Pichia) stipitis, S. (Candida) shehatae and Pachysolen tannophilus.1, 2 However the yield has not reached a satisfactory level for industrial applications On the other hand, pentose-assimilating yeasts can be used for obtainment of high aggregate value products. The pentose-assimilating capacity of yeasts Pseudozyma antarctica PYCC 5048T, P. aphidis PYCC 5535T and P. rugulosa PYCC 5537T and production of a glycolipid biosurfactant was demonstrate3 as well as the production of xylitol and arabitol by Debaryomyces hansenii from pentose.4
A number of recent studies have been focused on the genetic engineering of S. cerevisiae, aimed at making it able to produce ethanol from glucose and xylose.5 Nevertheless, the yield achieved resembles the xylose-fermenting species, the xylose utilization is slow and occurs only after glucose exhaustion.3, 4, 6, 7 The search for genetically modified strains is the focus of several studies. Thus, the study of microorganisms pentose-fermenting and isolation of genes involved in pentose fermentation can afford support for these studies.
Another research tendency is the use of pentose to produce alternative compounds like organic acids,8, 9, 10, 11, 12 xylitol,13 lipids for biodiesel production,14, 15, 16 isobutanol17 and hydrogen18 all using mainly yeast and bacteria. Besides compounds already cited, new alternative products with high aggregate value, are still appearing, increasing the options for the utilization of these sugars in biorefineries system.19
Considering the importance of the use of pentoses from lignocellulosic materials, the present work aimed at isolating yeast strains with the ability to assimilate and/or ferment xylose for future use for obtainment of products with biotechnological interest. Thus, this study contributes by increasing the information on xylose-fermenting strains and offers genomes for future studies of the metabolism of microorganisms.
Materials and methods
Isolation and identification of strains
The strains were isolated from fruits, flowers, insects and grape residues. About 0.5 g of material was suspended in 4 mL of YEPX medium (1% of yeast extract, 2% of peptone and of 2% xylose) and incubated at 30 °C. A loop of the homogenized culture was then streaked on the solid YEPX medium in Petri dishes to isolate the colonies. The strain identification was made using sequences of the D1/D2 domains of the rDNA.20
Xylose assimilation assay
To evaluate the potential of xylose assimilation, the strains were pre-cultivated in a YEP medium (1% yeast extract, 2% peptone and 2% glucose) and the biomass was centrifuged, watched twice with sterile distilled water and a suspension of them used as inoculum at 1 g of dry biomass per liter of culture medium composed of KH2PO4 (2.0 g L−1), (NH4)2SO4 (2.0 g L−1), MgSO4 ·7H2O (1.0 g L−1), urea (0.3 g L−1), CaCl2 (0.3 g L−1) and xylose (30.0 g L−1). The incubation was carried out at 30 °C and 150 rpm. The assays were done in duplicate and with three repetitions.
Ethanol production assay
The alcoholic fermentation was assayed in 125 mL Erlenmeyer flasks adapted for alcoholic fermentation, closed with a valve containing sodium metabisulfite solution at 1 g L−1 to ensure that no oxygen got, and containing 60 mL of medium described above but with yeast extract at 10 g L−1 and xylose at 100.0 g L−1. The incubation was at 30 °C. The assays were done in duplicate with three repetitions.
Analytical methods
To evaluate the dry cell mass, samples of fermented medium were centrifuged at 10,000 × g, by 15 min, the supernatant was discarded and the precipitated cells were dried at 60 °C until they reached constant weight.
The reducing sugar concentration was determined using the dinitrosalicylic (DNS) acid method, based on Miller (1959).21
The ethanol concentration was measured by a gas chromatograph (HP 5890) with an FFAP capillary column (polyethylene glycol – 30 mm × 0.22 mm × 0.3 μm) and a flame ionization detector, one split/splitless injector. Nitrogen was utilized as a carrier gas at 30 mL min−1. Temperatures at injector and detector were 250 °C.
The xylitol quantification was performed by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD). All samples were filtered (0.22 μm membrane) and injected (20 μL) in HPAEC-PAD System (ICS, Dionex Corporation, USA) equipped with automatic sampler AS40. The form of wave pattern used was the standard quadruple with the following potential pulse and durations: E1 = 0.10 V (t1 = 0.40 s); E2 = −2.00 V (t2 = 0.02 s); E3 = 0.60 V (t3 = 0.01 s); E4 = −0.10 V (t4 = 0.06 s). An Isocratic run was performed with 10 mM NaOH at a flow rate of 1 mL min−1 and 35 °C.
Results
Isolation, identification of yeasts and evaluation of xylose assimilation
A total of 30 strains were isolated and able to grow on the liquid YEPX medium, but, among those, four were not able to grow in liquid medium with xylose as the sole carbon source (Table 1). In aerobic cultivation, nine strains consumed all the xylose present in the medium (30 g L−1) in 120 h.
Table 1.
Yeasts isolated and xylose consumption during aerobic cultivation using a basal medium and xylose (30 g L−1) as the sole carbon source.
| Isolated strains | Samples | Xylose consumption |
Maximum biomass production |
Yield (biomass/xylose) | ||
|---|---|---|---|---|---|---|
| (g L−1) | Time (h) | (g L−1) | Time (h) | (g g−1) | ||
| Aureobasidium pullulans G19 | Flor (Nyctaginaceae) | 9.9 | 120 | 9.7 | 120 | 0.9 |
| Candida tropicalis E2 | Grape residue | 30.0 | 120 | 8.0 | 120 | 0.2 |
| C. tropicalis FP | Anthill | 30.0 | 72 | 7.2 | 120 | 0.2 |
| C. tropicalis S2 | Grape leaf | 10.0 | 120 | 3.3 | 96 | 0.3 |
| C. tropicalis S3 | Grape | 10.0 | 120 | 3.1 | 120 | 0.3 |
| C.tropicalis S4 | Grape leaf | 30.0 | 72 | 7.3 | 120 | 0.2 |
| C.tropicalis T | Wine must | 20.1 | 120 | 7.1 | 120 | 0.3 |
| C. oleophila G10.1 | Leaf (Asteraceae) | 30.0 | 96 | 8.2 | 120 | 0.3 |
| Hanseniaspora guilliermondii G9.1 | Leaf (Asteraceae) | 7.6 | 120 | 2.6 | 96 | 0.3 |
| Hanseniaspora sp. G1.3 | Fruit (Anarcadiaceae) | 19.4 | 120 | 3.7 | 72 | 0.2 |
| Hanseniaspora sp. G11.1 | Fruit (Rutaceae) | 7.6 | 120 | 3.5 | 120 | 0.5 |
| Hanseniaspora sp. G13 | Fruit (Rutaceae) | 10.4 | 120 | 2.7 | 72 | 0.2 |
| Hanseniaspora sp. G14 | Flower (Myrtaceae) | 9.9 | 120 | 2.0 | 120 | 0.2 |
| Hanseniaspora sp. G15 | Fruit (Malpighiaceae) | 3.5 | 120 | 2.2 | 96 | 0.6 |
| Hanseniaspora sp. G17 | Flower (Rubiaceae) | 5.4 | 120 | 2.8 | 48 | 0.5 |
| Hanseniaspora sp. G2 | Flower (Amaryllidaceae) | 10.2 | 120 | 5.6 | 48 | 0.5 |
| Hanseniaspora sp. G4.1 | Flower (Nyctaginaceae) | 23.0 | 120 | 8.2 | 120 | 0.3 |
| Hanseniaspora sp. G7.1 | Fruit (Myrtaceae) | 23.9 | 120 | 8.1 | 120 | 0.3 |
| Issatchenkia terricola G20 | Fruit (Myrtaceae) | 5.1 | 120 | 5.0 | 96 | 0.9 |
| Metschnikowia koreensis G18 | Flower (Amaryllidaceae) | 30.0 | 120 | 6.7 | 96 | 0.2 |
| Pichia guilliermondii G1.2 | Fruit (Anarcadiaceae) | 30.0 | 48 | 10.8 | 120 | 0.3 |
| P.guilliermondii G4.2 | Fruit (Rosaceae) | 30.0 | 72 | 10.6 | 120 | 0.3 |
| P. guilliermondii B1 | Wine must | nda | – | nd | – | – |
| P. guilliermondii B2 | Grape | nd | – | nd | – | – |
| P. kluyveri G1.1 | Fruit (Anarcadiaceae) | 20.5 | 120 | 7.2 | 120 | 0.3 |
| Rhodotorula mucilaginosa G7.2 | Flower (Euphorbiaceae) | 29.4 | 120 | 11.5 | 120 | 0.4 |
| R. mucilaginosa G11.2 | Flower (Plantaginaceae) | 17.3 | 120 | 6.7 | 120 | 0.4 |
| Rhodotorula sp. G10.2 | Flower (Plantaginaceae) | 27.8 | 120 | 8.1 | 120 | 0.3 |
| Yarrowia lipolitica CO1 | Grape | nd | – | nd | – | – |
| Yarrowia lipolítica CO2 | Grape | nd | – | nd | – | – |
Identification based on sequences of the D1/D2 domains of the rDNA (at least 98% similarity, with reference to homologous sequences from GenBank).
Not quantified, the strains were not able to grow with xylose as the sole carbon source.
The strain Pichia guilliermondii G1.2 was the most efficient in the xylose consumption (maximum at 48 h) while strains Candida tropicalis FP, C. tropicalis S4 and P. guilliermondii G4.2 exhausted the sugar in 72 h.
Species or genera isolated in this work, such as Aureobasidium pullulans, Issatchenkia terricola, I. orientalis, Metschnikowia bicuspidata, M. chrysoperlae, M. zobellii, P. guilliermondii, P. stipitis, Rhodotorula minuta and R. mucilaginosa are mentioned in the literature as being able to produce ethanol from xylose.22, 23, 24, 25, 26, 27, 28, 29 The genera Hanseniaspora and the specie I. terricola are glucose fermenting, but there is contradicting information about the xylose assimilation by these yeasts.22, 25 The most efficient xylose-assimilating yeasts were chosen to continue the experiments and used for fermentation for ethanol production in subsequent assays.
Ethanol production by yeasts
The ethanol production was evaluated for 30 strains and four of them were able to produce ethanol from xylose (from 3 to 6 g L−1) (data not shown). The three strains of C. tropicalis showed different profiles of consumption of xylose and ethanol production. The highest ethanol production was observed with C. tropicalis S4 with 6 g L−1 of ethanol and whose xylose assimilation was 56 g L−1 resulting in a yield of 0.1 g g−1, following by C. tropicalis FP that produced 4.6 g L−1 of ethanol from 84 g L−1 of xylose (yield of 0.05 g g−1).
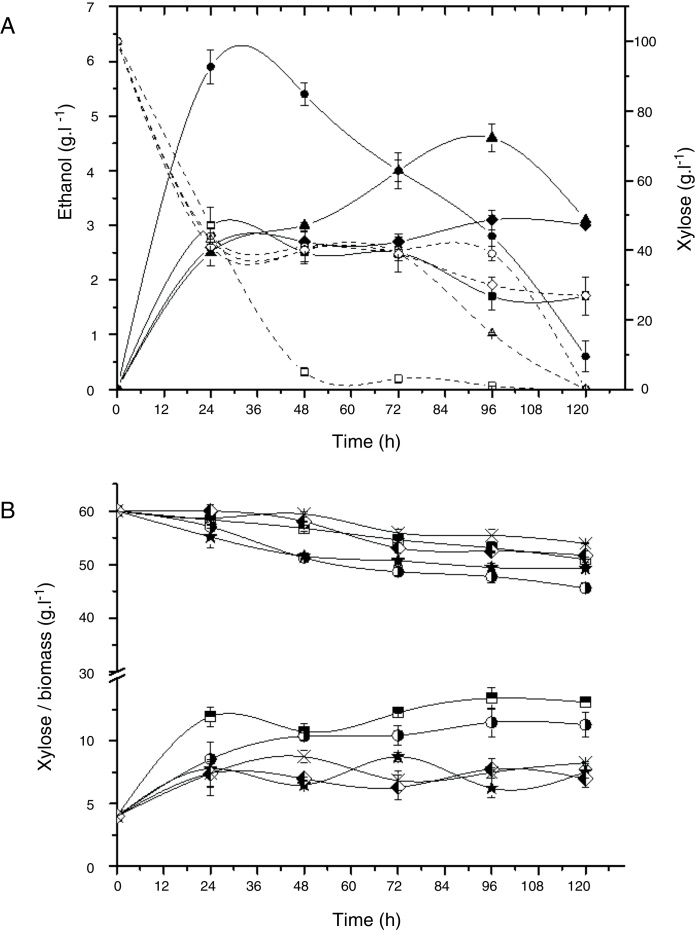
The strains Rhodotorula sp. G10.2 and C. tropicalis E2 showed similar production of ethanol (around 3 g L−1) to each other but Rhodotorula sp. G10.2 consumed 73 g L−1 of xylose with yield of 0.04 g g−1 and C. tropicalis E2 consumed 53 g L−1 with yield of 0.05 g g−1 (Fig. 1A).
Fig. 1.
Alcoholic fermentation assay in a basal medium with xylose (100 g L−1) and yeast extract (10 g L−1). (A) Ethanol production and xylose consumption by C. tropicalis E2 (squares), C. tropicalis S4 (circles), C. tropicalis FP (triangle), and Rhodotorula sp. G10.2 (diamond); Open symbols: xylose; Full symbols: ethanol. (B) Consumption of xylose and biomass by P. guilliermondii G1.2 (squares half up) and G4.2 (circle half right), C. oleophila G10.1 (star), M. koreensis G18 (diamond half left), R. mucilaginosa G7.2 (cross).
Fig. 1B shows the xylose assimilation and biomass production by strains that not were able to produce of ethanol. P. guilliermondii G4.2, C. oleophila G10.1 and M. koreensis G18 showed yields of biomass of 0.7 g g−1 meanwhile P. guilliermondii G1.2 and R. mucilaginosa G7.2 had yields of 1.1 up to1.3 g g−1, suggesting that P. guilliermondii G4.2, C. oleophila G10.1 and M. koreensis G18 produced not identified metabolite.
Since C. tropicalis has been reported as a xylitol-producing species30 and taking into account its consumption of xylose, the xylitol quantification in the fermentation media was carried out (Table 2).
Table 2.
Xylitol productivity by C. tropicalis FP strain. Fermentation realized in a basal medium with xylose (100 g L−1) and yeast extract (10 g L−1) at 30 °C in 120 h.
| Parameters evaluated | Xylitol |
Ethanol |
||||
|---|---|---|---|---|---|---|
| E2 | S4 | FP | E2 | S4 | FP | |
| Sugar consumed (g L−1) | 100 | 100 | 100 | 100 | 100 | 100 |
| Production (g L−1) | 22 | 15.5 | 4.5 | 3.0 | 5.9 | 4.6 |
| Yield – Yp/s (g g−1) | 0.22 | 0.15 | 0.04 | 0.03 | 0.06 | 0.04 |
| Volumetric productivity – Qp (g L−1 h−1) | 0.18 | 0.12 | 0.03 | 0.12 | 0.24 | 0.05 |
| Conversion efficiency – η (%)a | 24 | 16 | 4 | 6 | 12 | 9 |
The strain C. tropicalis E2 reached a production of 22 g L−1 of xylitol from 100 g L−1 of xylose (yield of 0.2 g g−1) while S4 produced 15.5 g L−1 of xylitol (0.15 g g−1) and C. tropicalis FP produced 4.5 g L−1 of xylitol (0.04 g g−1). The comparison between xylitol and ethanol fermentative parameters shows that the strain C. tropicalis E2 exhibits more potential for xylitol production while C. tropicalis FP produced similar concentrations of both alcohols but with low yield. In the culture medium of strain C. tropicalis S4 a higher concentration of xylitol than ethanol was measured.
A decreasing in the ethanol concentration during the fermentative route was observed (Fig. 1A), probably because the yeast can assimilate ethanol and, in this case, the ethanol determined in the medium is, perhaps, not real.
Discussion
In general, the yeasts strains presented xylose assimilation but low ethanol production. C. tropicalis S4 produced the highest amount of ethanol (6 g L−1), with a yield of 0.1 g g−1. This strain showed good sugar consumption and the species is not among the most promising yeasts for ethanol production. In fact, the literature shows that the most promising species for ethanol production are Scheffersomyces stipitis, S. shehatae and Pachysolen tannophilus, while C. tropicalis is related to xylitol production.
The strains C. oleophila G10.1, M. koreensis G18 and P. guilliermondii G4.2 showed smaller yields of biomass production than P. guilliermondii G1.2 and R mucilaginosa G7.2. This sugar consumption with minor growth suggests the production of metabolites which were not ethanol or xylitol. Thus, these strains need further investigation about alternative compounds like organic acids.
P. guilliermondii is frequently cited for production of xylitol and/or ethanol, but C. oleophila and M. koreensis have not been reported in the literature for the fermentation of hydrolysate lignocellulosic material. C. oleophila has been reported as an agent of biologic control due to its ability to suppress the growth of some fungi that cause fruit spoilage like Botrytis cinerea and, Penicillium digitatum.32, 33 M. koreensis was described more recently27 and was reported to be a carbonyl reductase34 and an L-talitol producer.35
The specie A. pulluluans, a yeast-like, is able to assimilate glucose and xylose but is reported as unable to ferment glucose. This feature makes this yeast interesting in a possible co-cultivation with S. cerevisiae. Furthermore, it is reported as being producer of high xylanase activity22, 28 allowing the hydrolysis of derived from hemicellulose in the production of ethanol from lignocellulosic material.
The strain C. tropicalis S4 showed slower xylose assimilation but consumed all the sugar of the medium in 120 h and produced the most ethanol. Furthermore, the decrease in the ethanol in the fermentation media over the course of time indicates that, perhaps, it is able to assimilate the ethanol, which would justify the slow xylose utilization.36
Under the conditions tested, C. tropicalis FP consumed all the xylose. Nevertheless, it presented a low yield for both ethanol and xylitol. Also, during the fermentation process, the strain did not present a large increase in the biomass (maximum biomass was 7 g L−1), that suggests the presence of other coproducts. Meanwhile, C. tropicalis E2 presented high xylose assimilation with a higher yield for xylitol than for ethanol, showing its promise for research into xylitol production.
The amount of xylitol produced by C. tropicalis strains shows the importance of the individual study of the characteristics of each strain, since differences may occur in the production of compounds, although belonging to same species. While the strain E2 produced a small quantity of ethanol and a larger quantity of xylitol, the strain FP produced the same quantity of both compounds.
The ethanol and xylitol production is strongly influenced by the fermentation conditions such as pH, temperature, oxygen and nutrients which were not studied in this work. Maybe productivity could be increased in different fermentation conditions, mainly controlling the oxygen supply.
The missing of ethanol production from xylose by the most of yeasts is marked by a redox imbalance that occurs between cofactors NAD and NADP during the formation of xylitol and xylulose. The xylose reductase enzyme reduces D-xylose to xylitol using NADPH, whereas xylitol dehydrogenase requires NAD+ mainly, which hampers the recycling of cofactors. This imbalance causes xylitol accumulation in the medium and the pathway does not proceed to produce ethanol. The recycling of NADP+ can be relieved through respiratory chain. However, depending on the oxygen level, cell growth may occur instead of ethanol production.37, 38
Xylitol is industrially interesting for its applicability as a sweetener for diabetics and is utilized in the food, pharmaceutical, oral health and cosmetics industries. In addition, its production from several lignocellulosic materials, such as corncob and cotton stalk, has been tested.30, 39, 40 Furthermore, to increase the xylitol yield from lignocellulosic materials, some studies looked at strategies such as hydrolysate detoxification or yeast adaptation, cell immobilization and varying the process conditions.41, 42, 43, 44, 45, 46
In conclusion, the assimilating yeasts are common in the environment but the understanding of the absorption and assimilation of the sugar is needed so that biotechnological utilization of this pathway is possible. Our results showed the importance of the individual study of each strain, since they have different characteristics concerning response to external factors, even if the strains belong to the same species. The study of the production of alternative compounds to ethanol is important because the application of these strains in industrial processes has not been well studied.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
The authors would like to thanks Fundação de Amparo à Pesquisa do Estado de São Paulo (Proc n° 2010/12624-0), Conselho Nacional de Desenvolvimento Científico e Tecnológico and Coordenação de Aperfeiçoamento de Pessoal em Nível Superior for their financial support and Daniela Correia de Oliveira Lisboa for the technician support.
Associate Editor: Solange I. Mussatto
References
- 1.Bajwa P.K., Phaenark C., Grant N. Ethanol production from selected lignocellulosic hydrolysates by genome shuffled strains of Scheffersomyces stipitis. Bioresour Technol. 2011;102(21):9965–9969. doi: 10.1016/j.biortech.2011.08.027. [DOI] [PubMed] [Google Scholar]
- 2.Webb S.R., Lee H. Regulation of d-xylose utilization by hexoses in pentose-fermenting yeasts. Biotechnol Adv. 1990;8(4):685–697. doi: 10.1016/0734-9750(90)91991-o. [DOI] [PubMed] [Google Scholar]
- 3.Faria N.T., Santos M.V., Fernandes P., Fonseca L.L., Fonseca C., Ferreira F.C. Production of glycolipid biosurfactants, mannosylerythritol lipids, from pentoses and d-glucose/d-xylose mixtures by Pseudozyma yeast strains. Process Biochem. 2014;49(11):1790–1799. [Google Scholar]
- 4.Gírio F.M., Amaro C., Azinheira H., Pelica F., Amaral-Collaço M.T. Polyols production during single and mixed substrate fermentations in Debaryomyces hansenii. Bioresour Technol. 2000;71(3):245–251. [Google Scholar]
- 5.Moon J., Lewis Liu Z., Ma M., Slininger P.J. New genotypes of industrial yeast Saccharomyces cerevisiae engineered with YXI and heterologous xylose transporters improve xylose utilization and ethanol production. Biocatal Agric Biotechnol. 2013;2(3):247–254. [Google Scholar]
- 6.Hector R.E., Dien B.S., Cotta M.A., Mertens J.A. Growth and fermentation of D-xylose by Saccharomyces cerevisiae expressing a novel D-xylose isomerase originating from the bacterium Prevotella ruminicola TC2-24. Biotechnol Biofuels. 2013;6(1):84. doi: 10.1186/1754-6834-6-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo C., Jiang N. Physiological and enzymatic comparison between Pichia stipitis and recombinant Saccharomyces cerevisiae on xylose fermentation. World J Microbiol Biotechnol. 2013;29(3):541–547. doi: 10.1007/s11274-012-1208-x. [DOI] [PubMed] [Google Scholar]
- 8.Kato H., Matsuda F., Yamada R. Cocktail δ-integration of xylose assimilation genes for efficient ethanol production from xylose in Saccharomyces cerevisiae. J Biosci Bioeng. 2013;116(3):333–336. doi: 10.1016/j.jbiosc.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 9.Wen S., Liu L., Nie K.L., Deng L., Tan T.W., Fang W. Enhanced fumaric acid production by fermentation of xylose using a modified strain of Rhizopus arrhizus. BioResources. 2013;8(2):2186–2194. https://ojs.cnr.ncsu.edu/index.php/BioRes/article/view/BioRes_08_2_2186_Wen_Fumaric_Acid_Fermentation Accessed 01.05.16. [Google Scholar]
- 10.Zhao J., Xu L., Wang Y. Homofermentative production of optically pure L-lactic acid from xylose by genetically engineered Escherichia coli B. Microb Cell Fact. 2013;12(1):57. doi: 10.1186/1475-2859-12-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye L., Zhou X., Hudari M.S., Li Z., Wu J.C. Highly efficient production of L-lactic acid from xylose by newly isolated Bacillus coagulans C106. Bioresour Technol. 2013;132:38–44. doi: 10.1016/j.biortech.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Chen T., Zhu N., Xia H. Aerobic production of succinate from arabinose by metabolically engineered Corynebacterium glutamicum. Bioresour Technol. 2014;151:411–414. doi: 10.1016/j.biortech.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 13.Jaros A.M., Rova U., Berglund K.A. Acetate adaptation of clostridia tyrobutyricum for improved fermentation production of butyrate. SpringerPlus. 2013;2(1):47. doi: 10.1186/2193-1801-2-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iverson A., Garza E., Zhao J. Increasing reducing power output (NADH) of glucose catabolism for reduction of xylose to xylitol by genetically engineered Escherichia coli AI05. World J Microbiol Biotechnol. 2013;29(7):1225–1232. doi: 10.1007/s11274-013-1285-5. [DOI] [PubMed] [Google Scholar]
- 15.Babau M., Cescut J., Allouche Y. Towards a microbial production of fatty acids as precursors of biokerosene from glucose and xylose. Oil Gas Sci Technol – Rev d’IFP Energies Nouv. 2013;68(5):899–911. [Google Scholar]
- 16.Gao D., Zeng J., Zheng Y., Yu X., Chen S. Microbial lipid production from xylose by Mortierella isabellina. Bioresour Technol. 2013;133:315–321. doi: 10.1016/j.biortech.2013.01.132. [DOI] [PubMed] [Google Scholar]
- 17.Mondala A., Hernandez R., Holmes W. Enhanced microbial oil production by activated sludge microorganisms via co-fermentation of glucose and xylose. AIChE J. 2013;59(11):4036–4044. [Google Scholar]
- 18.Brat D., Boles E. Isobutanol production from D-xylose by recombinant Saccharomyces cerevisiae. FEMS Yeast Res. 2013;13(2):241–244. doi: 10.1111/1567-1364.12028. [DOI] [PubMed] [Google Scholar]
- 19.Zhao C., Lu W., Wang H. Simultaneous hydrogen and ethanol production from a mixture of glucose and xylose using extreme thermophiles. II. Effect of hydraulic retention time. Int J Hydrogen Energy. 2013;38(22):9131–9136. [Google Scholar]
- 20.Isern N.G., Xue J., Rao J.V., Cort J.R., Ahring B.K. Novel monosaccharide fermentation products in Caldicellulosiruptor saccharolyticus identified using NMR spectroscopy. Biotechnol Biofuels. 2013;6(1):47. doi: 10.1186/1754-6834-6-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melo W.G.P., Arcuri S.L., Rodrigues A., Morais P.B., Meirelles L.A., Pagnocca F.C. Starmerella aceti f.a., sp. nov., an ascomycetous yeast species isolated from fungus garden of the leafcutter ant Acromyrmex balzani. Int J Syst Evol Microbiol. 2014;64(PART 4):1428–1433. doi: 10.1099/ijs.0.058818-0. [DOI] [PubMed] [Google Scholar]
- 22.Miller G.L. Use of dinitrosalicylic acid reagent for fetermination of reducing sugar. Anal Chem. 1959;31(3):426–428. [Google Scholar]
- 23.Kurtzman C.P., Fell J.W. In: The Yeasts. 5th ed. Kurtzman C.P., Fell J.W., editors. Elsevier; London: 2011. [Google Scholar]
- 24.Gong C., Chen L., Flickinger M.C., Chiang L., Tsao G.T. Production of ethanol from D-xylo se by using D-xylose isomerase and yeasts. Appl Environ Microbiol. 1981;41(2):430–436. doi: 10.1128/aem.41.2.430-436.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toivola A., Yarrow D., van den Bosch E., van Dijken J.P., Scheffers W.A. Alcoholic fermentation of d-xylose by yeasts. Appl Environ Microbiol. 1984;47(6):1221–1223. doi: 10.1128/aem.47.6.1221-1223.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nigam J.N., Ireland R.S., Margaritis A., Lachance M.A. Isolation and screening of yeasts that ferment d-xylose directly to ethanol. Appl Environ Microbiol. 1985;50(6):1486–1489. doi: 10.1128/aem.50.6.1486-1489.1985. http://aem.asm.org/content/50/6/1486.abstract Accessed 01.05.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sreenath H., Jeffries T. Production of ethanol from wood hydrolyzate by yeasts. Bioresour Technol. 2000;72(3):253–260. [Google Scholar]
- 28.Hong S.G., Chun J., Oh H.W., Bae K.S. Metschnikowia koreensis sp nov., a novel yeast species isolated from flowers in Korea. Int J Syst Evol Microbiol. 2001;51:1927–1931. doi: 10.1099/00207713-51-5-1927. WOS:000171162800039. [DOI] [PubMed] [Google Scholar]
- 29.Bhadra B., Rao R.S., Singh P.K., Sarkar P.K., Shivaji S. Yeasts and yeast-like fungi associated with tree bark: diversity and identification of yeasts producing extracellular endoxylanases. Curr Microbiol. 2008;56(5):489–494. doi: 10.1007/s00284-008-9108-x. [DOI] [PubMed] [Google Scholar]
- 30.Rao R.S., Bhadra B., Shivaji S. Isolation and characterization of ethanol-producing yeasts from fruits and tree barks. Lett Appl Microbiol. 2008;47(1):19–24. doi: 10.1111/j.1472-765X.2008.02380.x. [DOI] [PubMed] [Google Scholar]
- 31.Rafiqul I.S.M., Sakinah A.M.M. Processes for the production of xylitol—a review. Food Rev Int. 2013;29(2):127–156. [Google Scholar]
- 32.Gulati M., Kohlmann K., Ladisch M.R., Hespell R., Bothast R.J. Assessment of ethanol production options for corn products. Bioresour Technol. 1996;58(3):253–264. [Google Scholar]
- 33.Bar-Shimon M., Yehuda H., Cohen L. Characterization of extracellular lytic enzymes produced by the yeast biocontrol agent Candida oleophila. Curr Genet. 2004;45(3):140–148. doi: 10.1007/s00294-003-0471-7. [DOI] [PubMed] [Google Scholar]
- 34.Spadaro D., Gullino M.L. State of the art and future prospects of the biological control of postharvest fruit diseases. Int J Food Microbiol. 2004;91(2):185–194. doi: 10.1016/S0168-1605(03)00380-5. [DOI] [PubMed] [Google Scholar]
- 35.Singh A., Chisti Y., Banerjee U.C. Stereoselective biocatalytic hydride transfer to substituted acetophenones by the yeast Metschnikowia koreensis. Process Biochem. 2012;47(12):2398–2404. [Google Scholar]
- 36.Sasahara H., Izumori K. Production of L-talitol from L-psicose by Metschnikowia koreensis LA1 isolated from soy sauce mash. J Biosci Bioeng. 2005;100(3):335–338. doi: 10.1263/jbb.100.335. [DOI] [PubMed] [Google Scholar]
- 37.Carolan G., Catley B.J., Kelly P.J. Ethanol production during the growth cycle of Aureobasidium pullulans. Biochem Soc Trans. 1976;4(6):1021–1022. doi: 10.1042/bst0041021. [DOI] [PubMed] [Google Scholar]
- 38.Liang M., He Q.P., Jeffries T.W., Wang J. 2013 Am Control Conf. 2013. Elucidating xylose metabolism of Scheffersomyces stipitis by integrating principal component analysis with flux balance analysis; pp. 3777–3782. [Google Scholar]
- 39.Barbosa M.F.S., Medeiros M.B., Mancilha I.M., Schneider H., Lee H. Screening of yeasts for production of xylitol fromd-xylose and some factors which affect xylitol yield in Candida guilliermondii. J Ind Microbiol. 1988;3(4):241–251. [Google Scholar]
- 40.Van Vleet J.H., Jeffries T.W. Yeast metabolic engineering for hemicellulosic ethanol production. Curr Opin Biotechnol. 2009;20(3):300–306. doi: 10.1016/j.copbio.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 41.Ping Y., Ling H.-Z., Song G., Ge J.-P. Xylitol production from non-detoxified corncob hemicellulose acid hydrolysate by Candida tropicalis. Biochem Eng J. 2013;75:86–91. [Google Scholar]
- 42.Deng L.H., Tang Y., Liu Y. Detoxification of corncob acid hydrolysate with SAA pretreatment and xylitol production by immobilized Candida tropicalis. Sci World J. 2014;2014 doi: 10.1155/2014/214632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohamad N.L., Kamal S.M.M., Abdullah N., Ismail I. Evaluation of fermentation conditions by Candida tropicalis for xylitol production from sago trunk cortex. BioResources. 2013;8(2):2499–2509. [Google Scholar]
- 44.Soleimani M., Tabil L. Evaluation of biocomposite-based supports for immobilized-cell xylitol production compared with a free-cell system. Biochem Eng J. 2014;82:166–173. [Google Scholar]
- 45.Zhang Q., Li Y., Xia L., Liu Z., Pu Y. Enhanced xylitol production from statistically optimized fermentation of cotton stalk hydrolysate by immobilized Candida tropicalis. Chem Biochem Eng. 2014;28(1):87–93. [Google Scholar]
- 46.Li Z., Guo X., Feng X., Li C. An environment friendly and efficient process for xylitol bioconversion from enzymatic corncob hydrolysate by adapted Candida tropicalis. Chem Eng J. 2015;263:249–256. [Google Scholar]