Abstract
Cancer metastasis risk increases in older individuals, but the mechanisms for this risk increase are unclear. Many peritoneal cancers, including ovarian cancer, preferentially metastasize to peritoneal fat depots. However, there is a dearth of studies exploring aged peritoneal adipose tissue in the context of cancer. Because adipose tissue produces signals which influence several diseases including cancer, proteomics of adipose tissue in aged and young mice may provide insight into metastatic mechanisms. We analyzed mesenteric, omental, and uterine adipose tissue groups from the peritoneal cavities of young and aged C57BL/6J mouse cohorts with a low-fraction SDS-PAGE gelLC-MS/MS method. We identified 2308 protein groups and quantified 2167 groups, among which several protein groups showed 2-fold or greater abundance differences between the aged and young cohorts. Cancer-related gene products previously identified as significant in another age-related study were found altered in this study. Several gene products known to suppress proliferation and cellular invasion were found downregulated in the aged cohort, including R-Ras, Arid1a, and heat shock protein β1. In addition, multiple protein groups were identified within single cohorts, including the proteins Cd11a, Stat3, and Ptk2b. These data suggest that adipose tissue is a strong candidate for analysis to identify possible contributors to cancer metastasis in older subjects. The results of this study, the first of its kind using uterine adipose tissue, contribute to the understanding of the role of adipose tissue in age-related alteration of oncogenic pathways, which may help elucidate the mechanisms of increased metastatic tumor burden in the aged.
Keywords: Adipose, Mass spectrometry, Proteomics, GelLC-MS/MS, Peritoneal metastasis
Graphical abstract
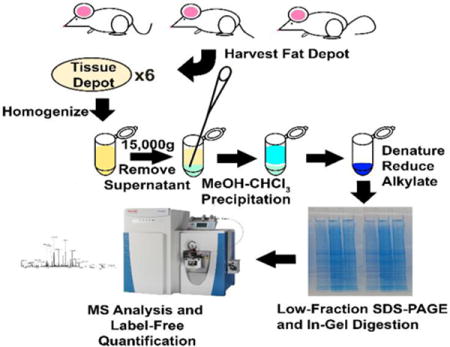
Introduction
Adipose tissue is a valuable, yet understudied, functional tissue in the body. Adipose tissue provides not only energy storage, but also important endocrine responses [1, 2]. Historically, adipose tissue was thought to act solely as an inert fat depot, providing energy in lean times to help an organism survive [3, 4]. More recently, adipose tissue has been connected to several metabolic processes, and has been shown to be highly active [3, 5]. In both animal models and humans, individuals with excess adipose tissue tend to develop cardiovascular disease, fatty livers, insulin resistance, pre-diabetes, and type 2 diabetes mellitus, as well as other pathologies [6–12]. Several fat depots are common to both humans and model organisms, and many of these depots excrete proteins and hormones that influence the immune system and vascular processes [3, 5, 8, 13–16].
The study of proteins altered in adipose tissue has become more prominent due to several global health concerns, such as the increased rates of obesity and type 2 diabetes mellitus [1, 2, 17, 18]. Proteomic analysis of adipose tissue has focused almost exclusively on human subjects and animal models of diabetes and other metabolic diseases [19]. The main tissue groups used for analysis in proteomic studies are subcutaneous and omental (visceral) fat depots, which are heavily affected in metabolic diseases [1, 20]. However, adipose tissue function affects several other medical conditions, including inflammatory diseases, parasitic infections, immune responses, and cancer [8, 21–23]. The various fat depots respond to select hormones and release specific adipokines. Adipokines can affect paracrine, autocrine, and endocrine responses [9, 12, 24–26]. Visceral adipose tissue releases adipokines known to regulate lipid metabolism, which in turn can affect bodily inflammation [25, 26]. Quantitative proteomics of adipose tissue is advantageous for examining protein expression affected by adipose tissue hormone secretions. By identifying and quantifying proteins in adipose tissue that affect inflammatory responses and metabolic processes, we can probe the mechanisms by which inflammation aids tumor infiltration into adipose tissue depots and develop methods to target specific pathways for inflammation and cancer prevention.
This study uses tissue from common ovarian cancer peritoneal metastatic sites in a murine ovarian cancer model system, but is relevant to other malignancies as ovarian cancer shares these sites with multiple other cancers. Peritoneal adipose tissue is a known metastatic site for ovarian cancer [23, 27], as well as mesothelioma and gastric, pancreatic, colorectal, and liver cancers. Ovarian cancer (OvCa) preferentially metastasizes to fat depots within the peritoneal cavity, especially the omental fat band and mesenteric fat in humans, and including the uterine fat depot and periovarian fat depot in mouse models [27–31]. The fat depots within the peritoneal cavity can provide a rich source of energy to cancer cells, allowing them to divide and thrive [28]. OvCa occurrence and metastasis is also correlated with increasing age and body fat (Loughran et al, In Preparation). The majority of OvCa cases occur over the age of 63 [32], and a BMI over 30 increases the chances of OvCa [33]. OvCa metastasis is a major factor in mortality. Of all new OvCa cases per year, 69% are diagnosed after age 55. In total, 60% of OvCa cases are diagnosed with distant metastasis, and the 5-year survival rate for a woman with distant metastasis is only 28.9% [32]. The microenvironment is known to affect cancer signaling and cancer metastasis [28]. Conversely, cancer cells also have influence on their microenvironments [34]. Some factors that affect OvCa in adipose tissue are known, but no proteomic analyses of adipose tissue in relation to OvCa or other peritoneal cancers have been reported. The effects of aging on the proteome in the absence of tumors are of interest, as factors in healthy aging may play a part in enhancing the disease state. This study relates both age and adipose tissue location to possible metastasis-enhancing factors in three tissues known for peritoneal cancer metastasis.
Proteomic analysis of adipose tissue is very challenging. Triglycerides compose more than 50% of the tissue by volume and a high lipid content in tissue is disadvantageous for extracting and solubilizing protein [35]. There are several sub-categories of adipose tissue, including white and brown adipose tissue [36–39]. Both white and brown adipose tissue are difficult to analyze and have distinct proteomic profiles [35–39]. Our study uses isolated white adipose tissue from several fat depots within the murine peritoneal cavity. Vascularization within these fat depots is variable, and multiple cell types exist within the tissue, including adipocytes, pre-adipocytes, macrophages, and leukocytes, as well as connective tissue matrices and blood vessels [13, 22, 40, 41]. Previously published proteomic studies of adipose tissue focus on secreted proteins, generally from adipocyte cell cultures, although several studies have been published investigating whole tissue lysates [1, 12, 42, 43]. Due to the challenges of analyzing proteins in the presence of lipids, most of the techniques used to analyze adipose tissue proteomes focus on gel-based approaches [1, 2, 15, 35].
In this study, adipose tissue from separate, age-matched cohorts of C57BL/6J mice were subjected to mass spectrometry-based proteomic analysis. The mouse model used in this study provides data for age-related conditions that would normally be difficult to study in humans, as mice reach full maturity and reproductive age in a short period of time. Age-related diseases are easier to study in mice, and the ages of mice can be roughly correlated to human age ranges. The age ranges of the mice used in our study correspond to human ages of roughly 20–30 years for the younger cohort and 56–69 years for the older cohort [44]. The age ranges we studied are appropriate for correlation to low and high OvCa metastasis occurrences in women. A previous study of more than 4000 protein groups found that the mouse tissue proteome does not dramatically change with age, either quantitatively or in terms of protein identifications [45]. However, another study determined age-related hallmarks in certain tissues [46]. With a cohort study of young and aged groups and direct comparisons of tissues, we aim to discover possible age-related OvCa or peritoneal metastasis factors in adipose tissue that are due to healthy aging. Using a low-fraction count in-gel digestion protocol, we identify over 2300 protein groups in total, and describe differences in the adipose tissue proteome when comparing different fat depot locations and cohort groups. The low-fraction gelLC-MS used in this study is similar in concept to the short-range gel, which has been helpful in microproteomic analyses of complex samples [47]. Although the adipose tissues used in this study do not fall into the microproteomics category, the low-fraction gel has some advantages compared to traditional gelLC-MS. Fewer fractions increase sample throughput on the mass spectrometer, increasing the speed of our analysis compared with a traditional number of gel fractions. With fewer fractions, a poorer separation is achieved by the gel, but overall less time is spent on preparation, allowing for a larger volume of analyzed samples. The low-fraction gelLC-MS still removes the majority of contaminating substances from the proteins and provides sufficient separation before LC-MS/MS analysis.
Materials and Methods
Tissue Collection
Healthy C57BL/6J mice were anesthetized in a chamber using isoflurane vapors and sacrificed by cervical dislocation immediately prior to harvesting adipose tissue. Adipose tissue was obtained from six female mice from two age cohorts: three young mice (five months in age) and three aged mice (21 months of age). Mice were sacrificed, tissues were examined and found devoid of gross pathological defects, and adipose tissue was harvested from the omental fat band, uterine fat depot, and the mesenteric fat (Figure 1A). Samples were immediately flash-frozen in liquid nitrogen post-harvest. Before lysis, the tissue was thawed on ice. To remove blood and serum proteins, tissue was washed twice; ice-cold phosphate buffered saline was added to the tissue, gently agitated by vortexing, and centrifuged at 1000 g for five minutes, after which the solution was removed by pipette.
Fig. 1.
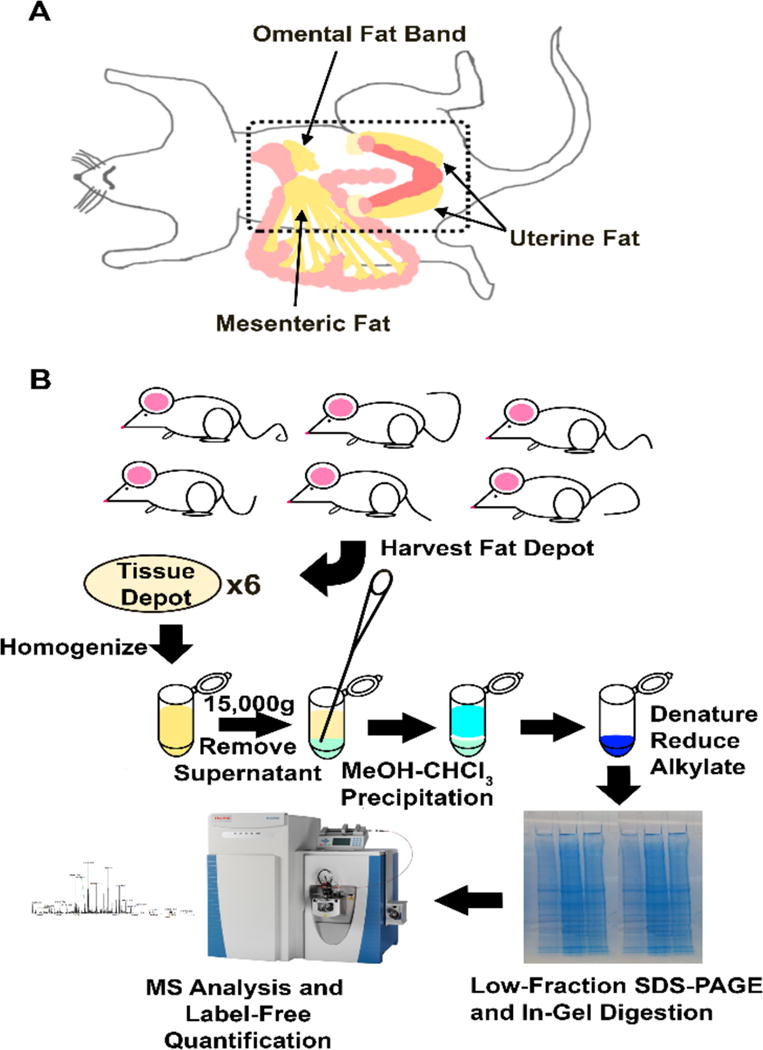
Tissue harvest locations and proteomic workflow. (A) All adipose tissues harvested for this experiment originate in the peritoneal cavity (dotted outline). The omental fat band, mesenteric fat depot, and the left uterine fat depot were harvested from six mice, three old and three young. (B) The 18 tissues were subjected to the proteomic workflow shown. Tissue depots were lysed in SDS and sonicated. Tissues were centrifuged, and the aqueous supernatant was removed and precipitated by methanol-chloroform. Samples were denatured, reduced, and alkylated in gel running buffer. In-gel digestion was performed, resulting in three fractions of peptides per tissue per animal for mass spectrometry analysis
Protein Extraction and Preparation
A 2% SDS/100 mM ammonium bicarbonate lysis buffer was added to the tissue to a volume of 1 mL, and tissue was homogenized using a spinning blade tissue homogenizer (OMNI International, TH Tissue Homogenizer). The homogenate was kept on ice for 30 minutes and then sonicated twice with a Branson 450 Sonifier for complete lysis and protein extraction. After sonication, samples were centrifuged at 15,000 g for 30 minutes at room temperature, in order to separate lipid and aqueous phases. The bottom aqueous phase was carefully removed with a gel loading tip and aliquoted in 100 μL volumes.
A methanol-chloroform extraction was performed to concentrate and purify proteins before analysis. Briefly, 400 μL MS-grade methanol was added to the 100 μL aliquot and vortexed for 1 minute. Then, 100 μL chloroform was added. Vortexing was repeated, then 300 μL MS-grade water was added. The solution clouded, and then vortexing was repeated for 3 minutes, after which vortexing for 1 minute, and the solution was centrifuged for 3 minutes at 15,000 g to pellet the protein. The methanol-chloroform mixture was removed and samples were dried in a hood for ~3 minutes, until all liquid was removed.
Sample pellets were dissolved in 10 μL of either 8M urea or 8M urea/2M thiourea buffer. Protein quantity was determined from 8M urea samples only using the BCA assay, as thiourea is a major interfering substance for BCA. Samples in 8M urea/2M thiourea (~30 μg protein, 10 μL volume) were reduced with 2.5 μL 500 mM dithiothreitol at 60°C, then alkylated with 2.5 μL 100 mM iodoacetamide at room temperature in the dark. A 5 μL volume of 2% SDS was mixed into the samples, and then 5 μL gel loading buffer was added and the solutions vortexed.
SDS-PAGE
A commercial bis-Tris mini-SDS-PAGE gel (4–12%, NuPAGE, Invitrogen) was loaded with sample, grouped by tissue type (one tissue type per gel). The gel was run for 1 hr on standard NuPAGE settings (200V constant). After electrophoresis, the gel was fixed and stained overnight with Coomassie Colloidal Brilliant Blue staining kit. The gel was destained with distilled water for 3 hours until the background was clear. The gel was divided into six bands of roughly equal protein amount for each sample lane, and in-gel digestion proceeded according to Shevchenko et al. [48]. After digestion, the six fractions per lane were combined into three fractions (1 and 2, 3 and 4, and 5 and 6). Samples were desalted using C18 Ziptips (Millipore) and resuspended in 20 μL 0.1% formic acid in MS grade water.
UPLC/nESI-MS/MS analysis
A Waters nanoACQUITY Ultra-performance LC system (Milford, MA, USA) was used for separation of gel fractions. Buffer A (0.1% formic acid in water) and buffer B (0.1% formic acid in acetonitrile) were used as the mobile phase for the separation. For all runs, 1.5 μL peptides were loaded onto a C18 reverse-phase column (Waters, 100 μm × 100 mm, 1.7 μm particle size, BEH130C18) held at 40°C. Samples were run with 2% buffer B for 12 minutes at 1 μL/min, followed by 65 minutes from 8% buffer B to 30% buffer B at 0.7 μL/min. The column was washed at 80% buffer B at 1 μL/min, followed by equilibration for 12 minutes at 2% buffer B at 1 μL/min. The total UPLC method time was 95 minutes. The eluted peptides were pumped through a nano-ESI spray emitter and analyzed with a Q-Exactive instrument (Thermo Fisher Scientific, Waltham, MA, USA). The electrospray voltage was 2.2 kV. Each fraction was run in technical duplicate. A top 20 data dependent acquisition (DDA) method was used. Full MS scans were acquired in the Orbitrap mass analyzer over m/z 350–1800 range with resolution 70,000 (m/z 400). The target value was 1.00E+06. The twenty most intense peaks with charge state ≥ 2 were selected for sequencing and fragmented in the ion trap with normalized collision energy of 30, isolation window of 2 m/z units and one microscan. Dynamic exclusion was enabled, and peaks selected for fragmentation more than once within 30 seconds were excluded from selection for 60 seconds.
Data analysis
Data analysis was performed using MaxQuant (version 1.5.6.5) [49], and the Andromeda search engine was used for database searching against the UniProt mouse database containing forward and reversed sequences (number of sequences: 82,838). The database also contains common contaminants. MaxQuant default mass tolerances were used. Analysis included an initial search with a precursor mass tolerance of 20 ppm, main search precursor mass tolerance of 4.5 ppm and fragment mass tolerance of FTMS 20 ppm and ITMS 0.5 Da. The search included trypsin as the enzyme, variable modifications of methionine oxidation, protein N-terminal and lysine acetylation and deamidation (NQ), and fixed modification of carbamidomethyl cysteine. The minimal peptide length was set to seven amino acids and the maximum number of missed cleavages was set to two. The match-between-runs function was used. The label-free quantitation (LFQ) function [50] was also enabled for the 6 mice in order to quantitatively evaluate the protein expression abundance between single mice and tissue groups. The FDR was set to 0.01 for both peptide and protein identifications. The Perseus data analysis suite was used to evaluate and filter LFQ data.
Discussion
Methodology
SDS-PAGE can generally remove lipid contaminants, and for this reason, gel-based approaches are more common in studies of adipose tissue. In this study, gel-based analysis was combined with a methanol-chloroform precipitation for maximum lipid removal and protein concentration prior to SDS-PAGE. The methanol-chloroform precipitation was described by Wessel and Flugge [51]; it removes lipids from samples and is one of the preferred methods for isolating membrane proteins [52, 53]. It has excellent recovery, even in dilute solutions, and allows rapid clean-up and concentration of protein for most proteomic preparations.
Gel-based approaches have been reasonably successful for adipose proteomics, although the number of proteins identified from gel-based approaches is significantly lower in adipose tissue and culture samples compared to other sample types. The largest gel-based proteomics study using adipose to date was performed by Adachi et al., in which 3T3-L1 adipocytes were subcellularly fractionated, and then the resulting fractions were further separated by SDS-PAGE and subjected to LC-MS/MS. In total, Adachi identified 3287 protein groups from 45 total fractions [54]. In another study, Xie et al. used 20 molecular weight fractions and LC-MS/MS analysis of human subcutaneous adipose tissue on an FT-ICR instrument to identify a total of 1493 protein groups [55]. Comparable SDS-PAGE in-gel digestions for non-adipose cultures have produced over 5000 protein group identifications from far fewer fractions [56, 57], and for tissues, over 3000 identifications have been reported [57, 58].
Recently, non-gel-based procedures have been introduced, although, to our knowledge, currently only six proteomic studies have been performed with gel-free approaches. Kim et al. performed a filter-based approach on human adipose tissue in a clinical study of type 2 diabetes. After delipidation with high-speed centrifugation, the authors identified over 4700 protein groups from 59 total samples [17]. Ma et al. also used a filter-based approach. Delipidating the samples with high-speed centrifugation, they analyzed omental adipose tissue samples of twelve human subjects and identified 3528 protein groups [59]. Pasing et al. performed in-solution digestion of adipose tissue. They compared several extraction buffers for low-cost adipose tissue proteomic analysis, resulting in a total of 2564 protein groups from a single subject [35]. Fang et al. performed immune-depletion of albumin proteins after delipidation. They identified 637 protein groups in subcutaneous fat and 604 protein groups in omental fat via in-solution digests and single LC-MS runs on an LTQ-XL mass spectrometer [20]. Gomez-Serrano et al. obtained adipose tissue from 16 subjects and lysed in modified RIPA buffer. Using FASP, iTRAQ, and Oasis cation exchange cartridge fractionation, they identified 2525 protein groups. In a comparison of gender, age, and type 2 diabetes, about 250 differentially abundant proteins (DAPs) showed significant abundance differences [2]. Ke et al. used high-pH reverse-phase fractionation after delipidation and 18O labeling of C57BL/6J and KKAy mouse epididymis adipose tissue. They identified 329 proteins, among which 121 were differentially quantified [60]. The recently developed gel-free methods described in this report are distinct in the sense that they generally require less overall sample preparation time, focusing instead on LC-MS/MS instrument time.
For gelLC-MS/MS studies with the goal of maximum proteomic coverage, the usual approach requires from five to twenty gel fractions [48]. Large numbers of fractions require more mass spectrometry analysis time. This study focused instead on sample throughput, without sacrificing proteomic coverage within the gel. By running just three fractions per animal per tissue group, the entire set of tissues was analyzed within ten days of mass spectrometry time. The study included three tissues analyzed from six healthy animals (three aged animals and three young) (Figure 1A). The tissues were separated into three fractions. A 95 minute LC-MS/MS gradient was used with technical duplicate analysis. From a total of 108 LC-MS/MS runs, we identified 2308 protein groups and quantified 2167 of those groups using label-free quantification; complete MaxQuant results can be found in the Electronic Supplementary Material (ESM) Table S1. The experimental setup used in this study (Figure 1B) provides some of the advantages of gel-based procedures, while allowing generally higher throughput than a typical gelLC-MS/MS experiment.
The low-fraction gel does not inhibit coverage of the proteome or quantification of identified proteins. In this study, most gel lanes identified over 1000 protein groups individually from three fractions, although the mesentery tissue group had fewer identifications. The quantified proteins were normally distributed in each fraction (ESM Fig. S1). Comparatively, Adachi et al. performed a 105-minute gradient on eleven or twelve fractions from four cellular components (total of 45 fractions), and identified 3287 proteins from four cellular fractions [54]. Xie et al. ran three sets of 20 fractions in triplicate with 80 minute gradients, identifying 1210, 1312, and 1003 protein groups from their individual subjects [55]. Tomé-Carneiro et al. performed a gel-based Super-SILAC experiment with mouse adipose tissue, identifying 88 protein groups and quantifying 39 protein groups [61]. This study analyzed more subjects than either Xie et al. or Adachi et al. with similar time frames and comparable results per gel lane, and outperformed a similar study by Tomé-Carneiro in terms of identification and quantification.
Our gelLC-MS/MS procedure provides a simple, low-cost method for mass spectrometry-based analysis of adipose tissue. Pasing et al. introduced several low-cost alternatives for proteomic analysis of adipose tissue. Their in-solution digestion strategies provide excellent results, and they cite time-consuming gel fractionation as a problem with previous gel-based analyses [35]. However, without any fractionation, proteomics may rely on long LC gradients (greater than three hours) and instrument time, which may be at a premium for some laboratories. Several other non-gel-based procedures rely on filter-based methods as well as increased instrument time [2, 17, 59], neither of which are especially low-cost. Following the advice of Pasing et al., our study provides a more rapid gel-based fractionation method than described previously, with lowered instrument time requirements. Table 1 provides a comparison of previous gel-based and gel-free approaches in terms of identifications, instrument time, and study methods. Our method compares favorably to these methods, in that we maintain balance between identification and instrument time. The main advantage we see using low-fraction gelLC-MS for adipose tissue is its efficient use of instrument time while maintaining clean samples. Future studies on low-fraction gelLC-MS using different tissues or cell cultures may give more insights on how useful the low-fraction gelLC-MS technique may be in terms of proteomic coverage, and whether its usefulness may be limited to adipose tissue.
Table 1. Comparison of MS-Based Adipose Tissue Proteomics.
There are few studies using adipose and mass spectrometry-based proteomics. Our study (last row) is comparable to other studies, while using more tissues, less instrument time, or both.
| Authors | Protein group IDs | Subject | MS time | Method |
|---|---|---|---|---|
| Kim et al. (2014) | 4707 | Human omental tissue | ~8.5 days | FASP |
| Adachi et al. (2007) | 3287 | 3T3-L1 Adipocytes | 3.6 days/rep | GelLCMS |
| Pasing et al. (2016) | 2564 | Human adipose | ~12 days | In-solution/FASP |
| Xie et al. (2010) | 1493 | Human subcutaneous | ~10 days | GelLCMS |
| Ma et al. (2016) | 3528 | Human omental tissue | 4h+/run | FASP |
| Fang et al. (2015) | 637/604 | Human subQ/omental | ~2h/run | In-solution |
| Gomez-serrano et al. (2016) | 2525 | Human omental tissue | 28 hrs | iTRAQ/FASP |
| Meierhofer et al. (2014) | ~3500 | Mouse visceral adipose (also contained liver) | 2 days/rep | GelLCMS |
| Tomé-Carneiro et al. (2017) | 88 | Mouse visceral adipose | ~2hr/run | GelLCMS |
| Ke et al. (2017) | 329 | Mouse Epididymis fat pad | 31.5 hrs | In-solution |
| Our study | 2308 | Mouse Omental/Uterine/Mesenteric tissue | 10 days | GelLCMS |
Proteomic Results
Within the body, adipose tissue depots differ in location and function, but there is a large overlap in protein groups identified among tissue groups. We analyzed two distinct visceral adipose depots in our study, which were largely similar in the protein groups identified. The third adipose depot, uterine fat, differs from the other two depots despite being localized in the peritoneal microenvironment and similarly colonized by metastasizing cells in mice [29]. Uterine fat is sometime classified as gonadal fat, and at other times as a visceral fat depot, leading to inconsistency in the literature [14]. Ours is the only study to use uterine fat for proteomic analysis so far, and so no definitive conclusions can be drawn; however, our data suggest that uterine fat may occupy a distinct class of adipose tissue compared to the two visceral fat depots, considering the number of exclusive identifications and the similar totals to the omental fat band.
The lack of variability between tissue groups provides insight into the similarities in visceral adipose tissue groups. The proteins identified in the omental fat band overlapped about 93%, and the mesentery fat had about 98% overlap with at least one other tissue group (Figure 2). The omental fat band and the mesentery occupy similar spaces in the abdominal cavity, although their structural roles are distinct within the body [14]. The omental fat band covers and supports the lower abdominal organs. The mesenteric fat connects to the intestines and other peritoneal organs. Uterine fat was the most distinct among the three tissue groups in terms of exclusive protein identifications (ESM Fig. S2). Previous studies have substituted the term “visceral adipose tissue” for any adipose tissue within the abdominal cavity [17, 62]; usually, this means the omental fat, but mesenteric fat has also been analyzed previously. We determined that the mesenteric fat produced the fewest number of identifications by roughly 600 protein groups. This fact may be due to the lower protein complexity in an organ that serves mainly as structural support. Further studies on the mesenteric fat may elucidate more reasons for the low identification numbers found here.
Fig. 2.
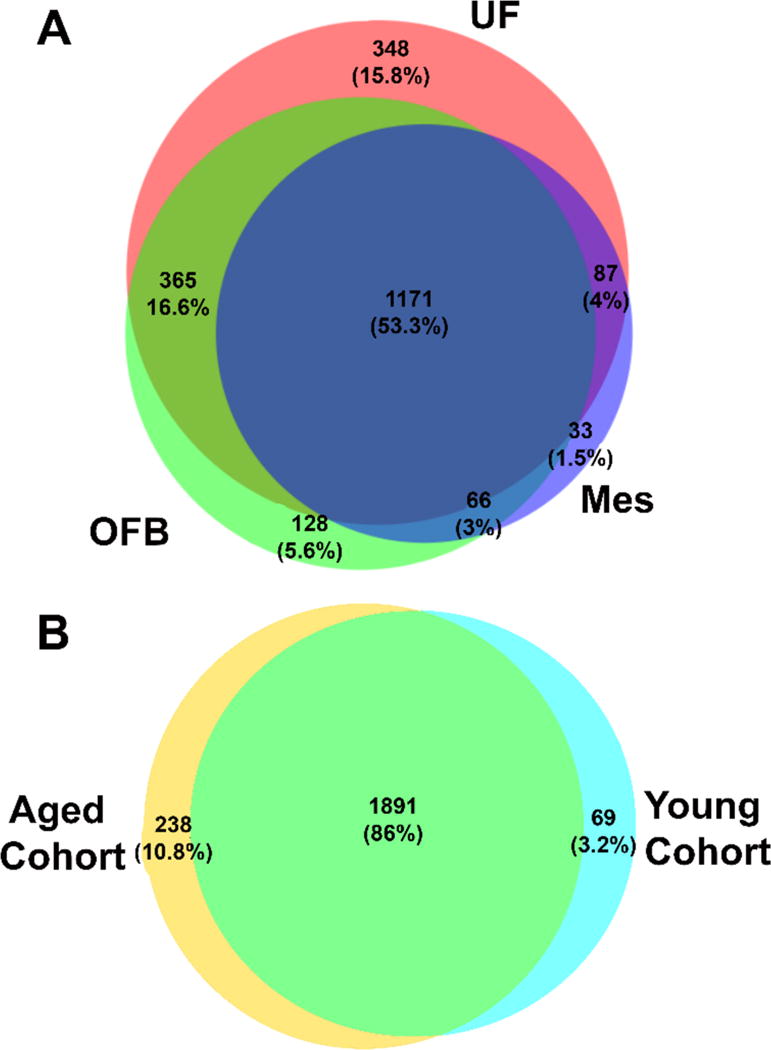
Protein identification breakdown by tissue and animal. Protein groups were exclusively identified by tissue group (A) and cohort (B). The omental fat band (OFB) and the mesenteric fat (Mes) were highly similar in identifications. About 15% of the identifications were exclusively identified in uterine fat (UF) Old and young tissues were similar in the protein groups identified, with 86% held in common. Among the protein groups identified solely in either aged or young, multiple cancer relevant protein groups were found with interesting biological functions. Several cytokines and apoptotic regulators were found in the aged cohort, while cell adhesion molecules were found in the young cohort.
Our work demonstrates the first successful MS-based proteomic analysis of uterine adipose tissue. Uterine adipose tissue was included in our study because of its propensity for OvCa metastasis in murine models. The uterine fat depot (in mice only) and uterine tissue are often metastatic sites for ovarian tumors, possibly due to their proximity to the ovaries within the peritoneal cavity [14]. However, in our data we did not find large numbers of proteins that correlated between age and OvCa, but instead found small differences that may have an effect on the tissue microenvironment. The uterine adipose tissues analyzed in our study showed remarkable proteomic similarity among all six subjects, with most animals having greater than 95% correlation in protein abundances observed in a multi-scatter plot with Pearson correlations (Figure 3). Uterine adipose tissue is extremely consistent in its proteomic constituents (Figure 3), suggesting that it changes very little from animal to animal.
Fig. 3.
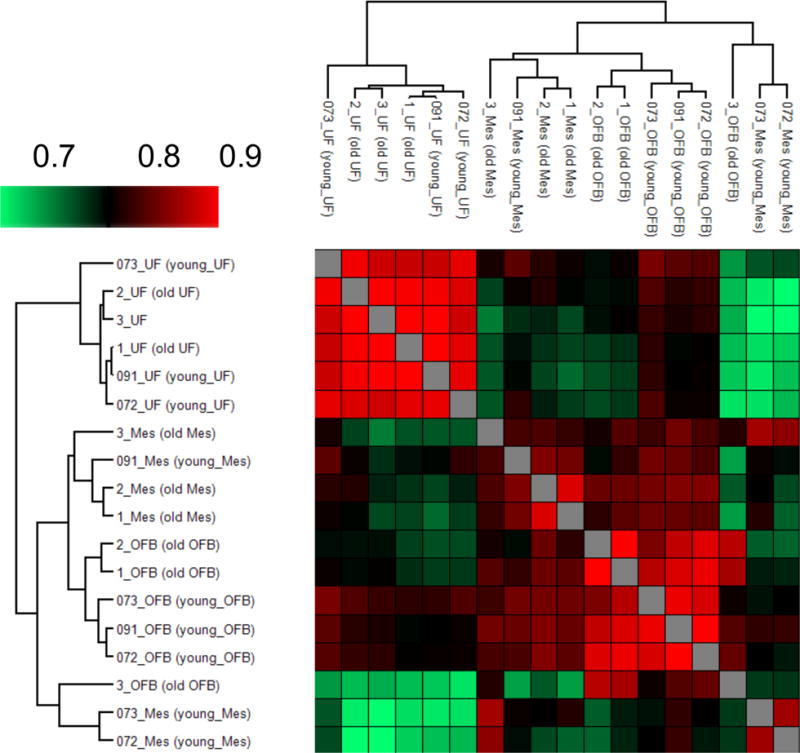
Pearson correlations for comparison of similarity between animals. Pearson correlations of the log2-transformed LFQ intensity show similarities among and between the cohorts of animals. The correlations are consistent within tissue groups, showing consistency in the quantification data. The samples are clustered using a simple k-means preprocessing algorithm in the Perseus data analysis suite. Among the three tissue groups, uterine fat showed the highest similarity in both the clustering and Pearson correlations (> 0.9), suggesting little change in function between aged and young animals with regard to uterine fat.
To analyze the variability amongst tissue groups, individual animals, and cohorts, the LFQ data from MaxQuant were analyzed with the Perseus data analysis suite. The data were first filtered to remove contaminants and reverse database sequences. LFQ intensities from each of the six mice were then grouped according to tissue type and age cohort. Data were log2 transformed, and filtering was applied. Any protein group not appearing in at least two of the three animals within a tissue group was filtered out. Pearson correlations between each sample were calculated and plotted in a heat map for comparison of separate tissue groups and cohorts (Figure 3), similar in function to scatter plots described by Cox, et al. [50]. The student‟s T-test was also performed in Perseus between the aged and young cohort for each tissue group (p < 0.05). After Benjamini - Hochberg correction for false discovery rate, few protein groups showed a statistically significant (q < 0.05) difference between aged and young cohorts. Despite the lack of statistical significance, several cancer-relevant protein groups had at least 2-fold abundance differences between the aged and young cohorts (Table 2), which suggests a link between known biology and proteomic profile in aging.
Table 2. Selected cancer pathways, proteins identified, and their fold-change in abundance.
Several protein groups are increased or decreased in the aged cohort, with multiple protein groups at least 2-fold altered. These cancer-relevant protein groups provide information on multiple simultaneous changes that occur with age that may affect the microenvironment and increase metastasis susceptibility.
| Pathway | Involved Protein Groups | Log2 Fold-Change |
|---|---|---|
| Chemo/cytokine inflammation (P00031) | Myosin 9; Biglycan | 1.09; 1.05 |
| p53 pathway (P00059) | Glucosidase II subunit β | 1.41 |
| Angiogenesis (P00005) | High mobility group proteinB1B2/HMG-I | 2.06 |
| RhoGTPase cytoskeletal regulation (P00016) | Rho-related GTP-binding protein RhoC | 1.07 |
| Ras pathway (P04393) | Galectin 1/12 | 1.00 |
| Cell cycle (P00013) | Proteasome activator complex subunit 1/2 | 1.57, 1.24 |
| FAS signaling pathway (P00020) | Prelamin A/C | − 0.58 |
| Wnt signaling pathway (P00057) | Arid1a | − 0.44 |
| CCKR signalling map (P06959) | Heat Shock Protein 27 | − 1.06 |
| Integrin signalling pathway (P00034) | Filamin A/B/C, R-Ras | − 0.63, − 0.83 |
In addition to LFQ data analysis, Perseus was also used to determine which protein groups, if any, were solely identified in the aged or young cohort. Identifications were first filtered to remove common contaminants and reverse database identifications. Intensities from each of the six mice were then grouped according to tissue type and age cohort. Data were log2 transformed, and filtering was applied. Any protein group not appearing in at least two of the three animals within a tissue group was filtered out. Matrices were exported by tissue group as text files, which were then loaded into Excel. Intensity values were summed within cohorts. Any identification with non-zero summed intensity in both cohorts was removed, leaving only protein groups differentially identified between cohorts.
The PANTHER (Protein ANalysis THrough Evolutionary Relationships) Classification System (http://www.pantherdb.org) was designed to classify proteins and their genes in order to facilitate large-scale analysis [63, 64]. Using PANTHER, we classified and compared the protein groups from each adipose tissue group. We observed that the molecular functions in the three tissue classifications are very similar, although the numbers of proteins used for the analysis are different (Figure 4). We also compared the pathway lists of the proteins whose quantity was altered between the two age groups with lists from previous studies on aging. The older cohort showed an up-regulation of several proteins in pathways associated with cancer metastasis, including inflammatory cytokines and protein kinases (Table 2; ESM Fig. S3).
Fig. 4.
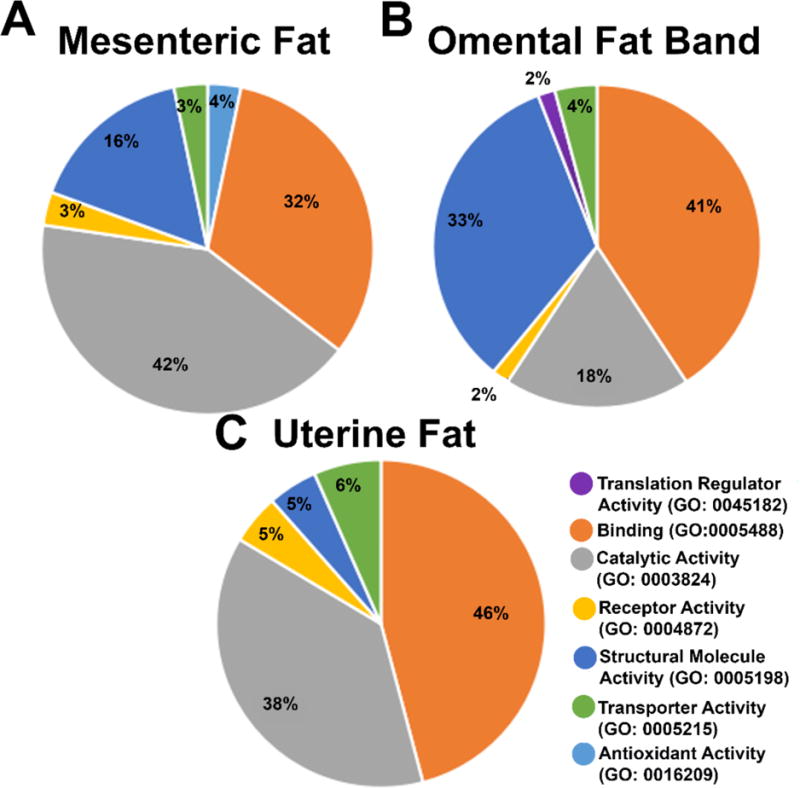
Molecular function analysis of significantly altered protein groups from three adipose tissue depots. The three tissue depots show similarities between the molecular functions of the protein groups with significant abundance differences. However, the percentages of the genes identified with these functions suggests that the number of protein groups identified corresponding to these functions, as well as the function of the tissue depot, plays a role in the regulation of these tissues in aging.
Age-related alteration of the proteome has been investigated previously, both within adipose tissue [2] and in other fatty tissues such as brain [45]. The interpretation of the results in these manuscripts differed, likely due to the varying approaches used for the data analysis. Of the two, Gomez-Serrano et al. focused on a systems biology approach. Using the weighted spectrum, peptide and protein (WSPP) model, they identified 155 differentially abundant proteins (out of 2,371 total proteins quantified) between non-diabetic human female age groups (mean ages 50 years and 32 years) [2]. The sample types analyzed are similar to ours in respect to age groups and disease state, and in fact we found homologous proteins with non-significant abundance changes that follow the trends described by Gomez-Serrano et al, such as collagen VI, which was lowered in aged mice. Overall, however, less than 7% of the proteome quantified by Gomez-Serrano et al. showed a statistically significant alteration with age. The statistical approaches presented here more closely align with Walther and Mann, in that traditional t-tests and false discovery rate correction were used in conjunction with filtering of quantifications [45]. Using SILAC mouse tissues, Walther and Mann quantified over 4000 proteins in various organs, and found very little alteration in the proteome between aged and young mice. The interpretation of their data aligns with our results in that the aged group did not vary significantly from the young group. Our analysis of uterine fat agrees with this conclusion, as the uterine fat was extremely consistent among all six animals in identifications and quantifications. The differing results in the manuscripts described here illustrate the effect of distinct approaches to the same problem, and each study points to a relatively stable proteome with small age-related changes. Determining the small, key alterations in the proteome that affect disease susceptibility is not just a matter of the mass spectrometer used or the sample preparation and separations, but also of the statistical approaches.
We hypothesized in this study that proteins affecting metastatic susceptibility may be differentially expressed in aged individuals. Several cancer-related protein groups were found to be altered in abundance in the older cohort compared to the young. Notably, R-Ras, Arid1a, and heat shock protein β1 (HSP β1) were found to be lowered in abundance. These factors have all been noted previously to influence cancer progression within the tumor microenvironment. Lowered activity of R-Ras was found in over 60% of breast tumor tissues when compared to the normal adjacent tissues [65]. Decreased expression of Arid1a in Huh7 hepatocarcinoma cells was associated with metastasis, as well as poor prognosis in mice and humans [66]. Lowered expression of Arid1a may also affect E-cadherin signaling pathways. HSP β1, also known as heat shock protein 27 or hsp27, is inversely correlated with cellular proliferation in tumor cells [67]. Several other pathways were found altered in this study, and may influence the microenvironment to be more amenable to metastasis in older cohorts.
Several inflammation and cancer-related proteins were identified solely in aged adipose tissue, while multiple anti-cancer proteins were identified unique to the young cohort. In particular, integrin alpha-L (Itgal or Cd11a), signal transducer and activator of transcription 3 (Stat3), and protein-tyrosine kinase 2-beta (Pyk2 or Ptk2b) were identified in the old cohort. Cd11a increases the instance of adipose tissue T-cell macrophages during inflammation processes [68]. Stat3 acts to regulate a host of pro- and anti-inflammatory proteins through multiple cytokine inflammatory receptors, and may be necessary for malignant tumor progression [69]. Pyk2 induces leukocytic response in the immune system during inflammation signaling and can be regulated by C-terminal SRC kinase [70], another protein solely identified in the older cohort. Other identifications were unique to the younger cohort. Among them, four separate unconventional myosins were identified in the younger cohort. Unconventional myosins are often associated with adhesion pathways, and may act as tumor suppressors [71].
Among the adipose tissues examined in this study, omental adipose tissue is the best understood. Omental adipose tissue has been studied in OvCa metastasis [28–31], but the proteomics of the omental fat band have focused primarily on obesity. Many of the same factors for obesity could be related to OvCa and other peritoneal cancers due to the increase of cancer risk with increasing adipose tissue burden [72]. Of several differences between the young and aged cohort, several protein groups were found in the cholecystokinin receptor (CCKR) pathway, which is linked to cancer via invasion and inflammation pathways [73]. Previous research has linked omental fat tissue to influencing OvCa cells and increasing their migratory ability, either through adipose tissue mechanisms or hormones secreted by so-called „milky spots‟ [29]. Omental adipose tissue is also known to have increased response to secreted cytokines [14]. Combined with the overall increase of inflammation pathways found in our aged animals, this fact suggests a link between age and inflammation in cancer metastasis risk. An up-regulation of inflammation pathways suggests that these adipose tissue depots may become more inflamed with age, creating a more hospitable environment for metastasizing cells to invade and thrive. Further studies including targeted approaches for specific adipokines, receptors, and other inflammation-related proteins may clarify the role of adipose tissue in the mechanisms of peritoneal metastasis.
Conclusions
Adipose tissue is an organ that contributes to the pathology of cancer, with specific fat depots affecting immune responses and inflammation pathways. Using a low-fraction count gel, we identified 2308 protein groups and quantified 2167 of those proteins from three adipose tissue depots within the peritoneal cavity in an age cohort mouse model. Several proteins in aged mice represent pathways known to contribute to cancer, with roles in proliferation and invasion. Previously published data implicate several of these proteins in cancer pathophysiology. The CCKR pathway highlighted in this study has known associations with both cancer and obesity, and ovarian cancer metastasis and prognosis has been associated with increased body fat mass. The low-fraction gelLC-MS allowed for increased throughput of samples compared to larger numbers of fractions while maintaining proteomic depth. Further investigation of the low-fraction gel will hopefully lead to an optimized protocol for faster high-quality sample analysis. Investigation of the proteins and associated networks reported here may lead to better understanding of the role of adipose tissue in peritoneal metastasis and later, novel interventions in the prevention of metastatic ovarian cancer.
Supplementary Material
Acknowledgments
PF was supported by an Arthur J. Schmitt Presidential Fellowship. ABH was supported by the National Institutes of Health (R01GM110406), and the National Science Foundation (CAREER Award, CHE-1351595). EAL was supported by a National Science Foundation Graduate Research Fellowship Program grant DGE-1313583. MSS is supported by the National Institutes of Health (RO1CA109545) and the Leo and Anne Albert Charitable Trust. We gratefully acknowledge the assistance of the Notre Dame Mass Spectrometry and Proteomics Facility (MSPF).
This research includes animal research. All mouse procedures were carried out according to the regulations of the University of Notre Dame Animal Care and Use Committee [IACUC 14-02-1577] and with approval of the Notre Dame Institutional Review Board.
Footnotes
Compliance with Ethical Standards
Conflict of Interest:
The authors declare no conflicts of interest.
References
- 1.Kim EY, Kim WK, Oh K-J, Han BS, Lee SC, Bae K-H. Recent Advances in Proteomic Studies of Adipose Tissues and Adipocytes. Int J Mol Sci. 2015;16(3):4581–99. doi: 10.3390/ijms16034581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gómez-Serrano M, Camafeita E, García-Santos E, López JA, Rubio MA, Sánchez-Pernaute A, et al. Proteome-wide alterations on adipose tissue from obese patients as age-, diabetes- and gender-specific hallmarks. Scientific Reports. 2016;6 doi: 10.1038/srep25756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galic S, Oakhill JS, Steinberg GR. Adipose tissue as an endocrine organ. Mol Cell Endocrinol. 2010;316(2):129–39. doi: 10.1016/j.mce.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 4.Kershaw EE, Flier JS. Adipose tissue as an endocrine organ. J Clin Endocrinol Metab. 2004;89(6):2548–56. doi: 10.1210/jc.2004-0395. [DOI] [PubMed] [Google Scholar]
- 5.Coelho M, Oliveira T, Fernandes R. Biochemistry of adipose tissue: an endocrine organ. Arch Med Sci. 2013;9(2):191–200. doi: 10.5114/aoms.2013.33181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boden G. Effects of free fatty acids (FFA) on glucose metabolism: significance for insulin resistance and type 2 diabetes. Exp Clin Endocrinol Diabetes. 2003;111(3):121–4. doi: 10.1055/s-2003-39781. [DOI] [PubMed] [Google Scholar]
- 7.Duvnjak L, Duvnjak M. The metabolic syndrome - an ongoing story. J Physiol Pharmacol. 2009;60(Suppl 7):19–24. [PubMed] [Google Scholar]
- 8.Matsuzawa Y. Therapy Insight: adipocytokines in metabolic syndrome and related cardiovascular disease. Nat Clin Pract Cardiovasc Med. 2006;3(1):35–42. doi: 10.1038/ncpcardio0380. [DOI] [PubMed] [Google Scholar]
- 9.Fontana L, Eagon JC, Trujillo ME, Scherer PE, Klein S. Visceral fat adipokine secretion is associated with systemic inflammation in obese humans. Diabetes. 2007;56(4):1010–3. doi: 10.2337/db06-1656. [DOI] [PubMed] [Google Scholar]
- 10.Frayn KN. Visceral fat and insulin resistance–causative or correlative? Br J Nutr. 2000;83(Suppl 1):S71–7. doi: 10.1017/s0007114500000982. [DOI] [PubMed] [Google Scholar]
- 11.Gastaldelli A, Miyazaki Y, Pettiti M, Matsuda M, Mahankali S, Santini E, et al. Metabolic effects of visceral fat accumulation in type 2 diabetes. J Clin Endocrinol Metab. 2002;87(11):5098–103. doi: 10.1210/jc.2002-020696. [DOI] [PubMed] [Google Scholar]
- 12.Lehr S, Hartwig S, Lamers D, Famulla S, Müller S, Hanisch F-G, et al. Identification and validation of novel adipokines released from primary human adipocytes. Mol Cell Proteomics. 2012;11(1) doi: 10.1074/mcp.M111.010504. M111.010504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajer GR, van Haeften TW, Visseren FLJ. Adipose tissue dysfunction in obesity, diabetes, and vascular diseases. Eur Heart J. 2008;29(24):2959–71. doi: 10.1093/eurheartj/ehn387. [DOI] [PubMed] [Google Scholar]
- 14.Bjørndal B, Burri L, Staalesen V, Skorve J, Berge RK. Different adipose depots: their role in the development of metabolic syndrome and mitochondrial response to hypolipidemic agents. Journal of Obesity. 2011;2011:490650. doi: 10.1155/2011/490650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peral B, Camafeita E, Fernández-Real J-M, López JA. Tackling the human adipose tissue proteome to gain insight into obesity and related pathologies. Expert Rev Proteomics. 2009;6(4):353–61. doi: 10.1586/epr.09.53. [DOI] [PubMed] [Google Scholar]
- 16.Guerre-Millo M. Adipose tissue hormones. J Endocrinol Invest. 2002;25(10):855–61. doi: 10.1007/BF03344048. [DOI] [PubMed] [Google Scholar]
- 17.Kim S-J, Chae S, Kim H, Mun D-G, Back S, Choi HY, et al. A protein profile of visceral adipose tissues linked to early pathogenesis of type 2 diabetes mellitus. Mol Cell Proteomics. 2014;13(3):811–22. doi: 10.1074/mcp.M113.035501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma S, Jing F, Xu C, Zhou L, Song Y, Yu C, et al. Thyrotropin and Obesity: Increased Adipose Triglyceride Content Through Glycerol-3-Phosphate Acyltransferase 3. Scientific Reports. 2015;5 doi: 10.1038/srep07633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brockman D, Chen X. Proteomics in the characterization of adipose dysfunction in obesity. Adipocyte. 2012;1(1):25–37. doi: 10.4161/adip.19129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang L, Kojima K, Zhou L, Crossman DK, Mobley JA, Grams J. Analysis of the Human Proteome in Subcutaneous and Visceral Fat Depots in Diabetic and Non-diabetic Patients with Morbid Obesity. J Proteomics Bioinform. 2015;8(6):133–41. doi: 10.4172/jpb.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desruisseaux MS, Nagajyothi, Trujillo ME, Tanowitz HB, Scherer PE. Adipocyte, Adipose Tissue, and Infectious Disease. Infect Immun. 2007;75(3):1066–78. doi: 10.1128/IAI.01455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas LW. The Chemical Composition of Adipose Tissue of Man and Mice. Exp Physiol. 1962;47(2):179–88. doi: 10.1113/expphysiol.1962.sp001589. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Metzinger MN, Lewellen KA, Cripps SN, Carey KD, Harper EI, et al. Obesity Contributes to Ovarian Cancer Metastatic Success Through Increased Lipogenesis, Enhanced Vascularity, and Decreased Infiltration of M1 Macrophages. Cancer Res. 2015;75(23):5046–57. doi: 10.1158/0008-5472.CAN-15-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Booth A, Magnuson A, Fouts J, Foster M. Adipose tissue, obesity and adipokines: role in cancer promotion. Horm Mol Biol Clin Investig. 2015;21(1):57–74. doi: 10.1515/hmbci-2014-0037. [DOI] [PubMed] [Google Scholar]
- 25.Rytka JM, Wueest S, Schoenle EJ, Konrad D. The portal theory supported by venous drainage-selective fat transplantation. Diabetes. 2011;60(1):56–63. doi: 10.2337/db10-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozcelik F, Yuksel C, Arslan E, Genc S, Omer B, Serdar MA. Relationship between visceral adipose tissue and adiponectin, inflammatory markers and thyroid hormones in obese males with hepatosteatosis and insulin resistance. Arch Med Res. 2013;44(4):273–80. doi: 10.1016/j.arcmed.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Lengyel E. Ovarian Cancer Development and Metastasis. Am J Pathol. 2010;177(3):1053–64. doi: 10.2353/ajpath.2010.100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nature Medicine. 2011;17(11):1498–503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark R, Krishnan V, Schoof M, Rodriguez I, Theriault B, Chekmareva M, et al. Milky Spots Promote Ovarian Cancer Metastatic Colonization of Peritoneal Adipose in Experimental Models. Am J Pathol. 2013;183(2):576–91. doi: 10.1016/j.ajpath.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan SM, Funk HM, Thiolloy S, Lotan TL, Hickson J, Prins GS, et al. In vitro metastatic colonization of human ovarian cancer cells to the omentum. Clin Exp Metastasis. 2010;27(3):185–96. doi: 10.1007/s10585-010-9317-0. [DOI] [PubMed] [Google Scholar]
- 31.Pradeep S, Kim SW, Wu SY, Nishimura M, Chaluvally-Raghavan P, Miyake T, et al. Hematogenous metastasis of ovarian cancer: rethinking mode of spread. Cancer Cell. 2014;26(1):77–91. doi: 10.1016/j.ccr.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cancer of the Ovary - Cancer Stat Facts. NIH National Cancer Institute; https://seer.cancer.gov/statfacts/html/ovary.html. Accessed 10/20/2017 2017. [Google Scholar]
- 33.Leitzmann MF, Koebnick C, Danforth KN, Brinton LA, Moore SC, Hollenbeck AR, et al. Body mass index and risk of ovarian cancer. Cancer. 2009;115(4):812–22. doi: 10.1002/cncr.24086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol. 2017;17(9):559–72. doi: 10.1038/nri.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pasing Y, Colnoe S, Hansen T. Proteomics of hydrophobic samples: Fast, robust and low-cost workflows for clinical approaches. Proteomics. 2016 doi: 10.1002/pmic.201500462. [DOI] [PubMed] [Google Scholar]
- 36.Forner F, Kumar C, Luber CA, Fromme T, Klingenspor M, Mann M. Proteome differences between brown and white fat mitochondria reveal specialized metabolic functions. Cell Metab. 2009;10(4):324–35. doi: 10.1016/j.cmet.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 37.Okita N, Hayashida Y, Kojima Y, Fukushima M, Yuguchi K, Mikami K, et al. Differential responses of white adipose tissue and brown adipose tissue to caloric restriction in rats. Mechanisms of Ageing and Development. 2012;133(5):255–66. doi: 10.1016/j.mad.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 38.Li J, Zhao W-G, Shen Z-F, Yuan T, Liu S-N, Liu Q, et al. Comparative Proteome Analysis of Brown Adipose Tissue in Obese C57BL/6J Mice Using iTRAQ-Coupled 2D LC-MS/MS. PLOS ONE. 2015;10(3) doi: 10.1371/journal.pone.0119350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Müller S, Balaz M, Stefanicka P, Varga L, Amri E-Z, Ukropec J, et al. Proteomic Analysis of Human Brown Adipose Tissue Reveals Utilization of Coupled and Uncoupled Energy Expenditure Pathways. Scientific Reports. 2016;6:30030. doi: 10.1038/srep30030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riordan NH, Ichim TE, Min W-P, Wang H, Solano F, Lara F, et al. Non-expanded adipose stromal vascular fraction cell therapy for multiple sclerosis. J Transl Med. 2009;7:29. doi: 10.1186/1479-5876-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peinado JR, Jimenez-Gomez Y, Pulido MR, Ortega-Bellido M, Diaz-Lopez C, Padillo FJ, et al. The stromal-vascular fraction of adipose tissue contributes to major differences between subcutaneous and visceral fat depots. Proteomics. 2010;10(18):3356–66. doi: 10.1002/pmic.201000350. [DOI] [PubMed] [Google Scholar]
- 42.Lee MJ, Kim J, Kim MY, Bae Y-S, Ryu SH, Lee TG, et al. Proteomic Analysis of Tumor Necrosis Factor-α-Induced Secretome of Human Adipose Tissue-Derived Mesenchymal Stem Cells. Journal of Proteome Research. 2010;9(4):1754–62. doi: 10.1021/pr900898n. [DOI] [PubMed] [Google Scholar]
- 43.Alvarez-Llamas G, Szalowska E, Weening D, Landman K, Hoek A, et al. Characterization of the Human Visceral Adipose Tissue Secretome. Mol Cell Proteomics. 2007;6(4):589–600. doi: 10.1074/mcp.M600265-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Life span as a biomarker. https://www.jax.org/research-and-faculty/research-labs/the-harrison-lab/gerontology/life-span-as-a-biomarkerfiles/3/life-span-as-a-biomarker.html. Accessed 10/20/2017 2017.
- 45.Walther DM, Mann M. Accurate Quantification of More Than 4000 Mouse Tissue Proteins Reveals Minimal Proteome Changes During Aging. Mol Cell Proteomics. 2011;10(2):M110.004523. doi: 10.1074/mcp.M110.004523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stauch KL, Purnell PR, Villeneuve LM, Fox HS. Data for mitochondrial proteomic alterations in the aging mouse brain. Data in Brief. 2015;4:127–9. doi: 10.1016/j.dib.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thakur D, Rejtar T, Wang D, Bones J, Cha S, Clodfelder-Miller B, et al. Microproteomic Analysis of 10,000 Laser Captured Microdissected Breast Tumor Cells Using Short-Range Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) and Porous Layer Open Tubular (PLOT) LC-MS/MS. J Chromatogr A. 2011;1218(45):8168–74. doi: 10.1016/j.chroma.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shevchenko A, Tomas H, Havli J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protocols. 2007;1(6):2856–60. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 49.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protocols. 2016;11(12):2301–19. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 50.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13(9):2513–26. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wessel D, Flügge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138(1):141–3. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 52.Di Girolamo F, Ponzi M, Crescenzi M, Alessandroni J, Guadagni F. A simple and effective method to analyze membrane proteins by SDS-PAGE and MALDI mass spectrometry. Anticancer research. 2010;30(4):1121–9. [PubMed] [Google Scholar]
- 53.Vertommen A, Panis B, Swennen R, Carpentier SC. Evaluation of chloroform/methanol extraction to facilitate the study of membrane proteins of non-model plants. Planta. 2010;231(5):1113–25. doi: 10.1007/s00425-010-1121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adachi J, Kumar C, Zhang Y, Mann M. In-depth Analysis of the Adipocyte Proteome by Mass Spectrometry and Bioinformatics. Mol Cell Proteomics. 2007;6(7):1257–73. doi: 10.1074/mcp.M600476-MCP200. [DOI] [PubMed] [Google Scholar]
- 55.Xie X, Yi Z, Bowen B, Wolf C, Flynn CR, Sinha S, et al. Characterization of the Human Adipocyte Proteome and Reproducibility of Protein Abundance by One-dimensional Gel Electrophoresis and HPLC-ESI-MS/MS. Journal of proteome research. 2010;9(9):4521–34. doi: 10.1021/pr100268f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weston LA, Bauer KM, Hummon AB. Comparison of bottom-up proteomic approaches for LC-MS analysis of complex proteomes. Anal Methods. 2013;5(18) doi: 10.1039/C3AY40853A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Piersma SR, Warmoes MO, de Wit M, de Reus I, Knol JC, Jiménez CR. Whole gel processing procedure for GeLC-MS/MS based proteomics. Proteome Sci. 2013;11:17. doi: 10.1186/1477-5956-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warmoes M, Jaspers JE, Pham TV, Piersma SR, Oudgenoeg G, Massink MPG, et al. Proteomics of Mouse BRCA1-deficient Mammary Tumors Identifies DNA Repair Proteins with Potential Diagnostic and Prognostic Value in Human Breast Cancer. Mol Cell Proteomics. 2012;11(7) doi: 10.1074/mcp.M111.013334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma Y, Gao J, Yin J, Gu L, Liu X, Chen S, et al. Identification of a Novel Function of Adipocyte Plasma Membrane-Associated Protein (APMAP) in Gestational Diabetes Mellitus by Proteomic Analysis of Omental Adipose Tissue. Journal of Proteome Research. 2016;15(2):628–37. doi: 10.1021/acs.jproteome.5b01030. [DOI] [PubMed] [Google Scholar]
- 60.Ke M, Wu H, Zhu Z, Zhang C, Zhang Y, Deng Y. Differential proteomic analysis of white adipose tissues from T2D KKAy mice by LC-ESI-QTOF. Proteomics. 2017;17(5) doi: 10.1002/pmic.201600219. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 61.Tomé-Carneiro J, Crespo MC, García-Calvo E, Luque-García JL, Dávalos A, Visioli F. Proteomic evaluation of mouse adipose tissue and liver following hydroxytyrosol supplementation. Food and Chemical Toxicology. 2017;107(Part A):329–38. doi: 10.1016/j.fct.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 62.Oliva K, Barker G, Rice GE, Bailey MJ, Lappas M. 2D-DIGE to identify proteins associated with gestational diabetes in omental adipose tissue. J Endocrinol. 2013;218(2):165–78. doi: 10.1530/JOE-13-0010. [DOI] [PubMed] [Google Scholar]
- 63.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. PANTHER: A Library of Protein Families and Subfamilies Indexed by Function. Genome Res. 2003;13(9):2129–41. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, et al. Applications for protein sequence–function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34(suppl_2):W645–W50. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song J, Zheng B, Bu X, Fei Y, Shi S. Negative association of R-Ras activation and breast cancer development. Oncol Rep. 2014;31(6):2776–84. doi: 10.3892/or.2014.3121. [DOI] [PubMed] [Google Scholar]
- 66.He F, Li J, Xu J, Zhang S, Xu Y, Zhao W, et al. Decreased expression of ARID1A associates with poor prognosis and promotes metastases of hepatocellular carcinoma. J Exp Clin Cancer Res. 2015;34(1) doi: 10.1186/s13046-015-0164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vargas-Roig LM, Fanelli MA, López LA, Gago FE, Tello O, Aznar JC, et al. Heat shock proteins and cell proliferation in human breast cancer biopsy samples. Cancer Detect Prev. 1997;21(5):441–51. [PubMed] [Google Scholar]
- 68.Jiang E, Perrard XD, Yang D, Khan IM, Perrard JL, Smith CW, et al. Essential Role of CD11a in CD8+ T-Cell Accumulation and Activation in Adipose Tissue. Arterioscler Thromb Vasc Biol. 2014;34(1):34–43. doi: 10.1161/ATVBAHA.113.302077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nguyen AV, Wu Y-Y, Liu Q, Wang D, Nguyen S, Loh R, et al. STAT3 in Epithelial Cells Regulates Inflammation and Tumor Progression to Malignant State in Colon. Neoplasia. 2013;15(9):998–1008. doi: 10.1593/neo.13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Di Cioccio V, Strippoli R, Bizzarri C, Troiani G, Cervellera MN, Gloaguen I, et al. Key role of proline-rich tyrosine kinase 2 in interleukin-8 (CXCL8/IL-8)-mediated human neutrophil chemotaxis. Immunology. 2004;111(4):407–15. doi: 10.1111/j.1365-2567.2004.01822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ouderkirk JL, Krendel M. Non-muscle myosins in tumor progression, cancer cell invasion and metastasis. Cytoskeleton (Hoboken) 2014;71(8):447–63. doi: 10.1002/cm.21187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat Rev Cancer. 2004;4(8):579–91. doi: 10.1038/nrc1408. [DOI] [PubMed] [Google Scholar]
- 73.Tripathi S, Flobak Å, Chawla K, Baudot A, Bruland T, Thommesen L, et al. The gastrin and cholecystokinin receptors mediated signaling network: a scaffold for data analysis and new hypotheses on regulatory mechanisms. BMC Systems Biology. 2015;9:40. doi: 10.1186/s12918-015-0181-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


