Abstract
Hospitalized infants have the highest rates of invasive S. aureus disease of any population, and infection control strategies such as decolonization have been insufficient. For decades, researchers began studying the microbiome in search of new prevention strategies. The resident microbiota was found to be closely associated with susceptibility and at times, resistance to S. aureus colonization. The evolution of nucleic acid based techniques has enhanced our understanding of the complex relationship between the nasal microbiota and S. aureus colonization. We review what is known about bacterial communities in the nasal cavity of infants and discuss how future microbiome studies may help identify novel interventions to protect high-risk infants from S. aureus disease.
I. Introduction
Staphylococcus aureus is a leading cause of infectious morbidity and mortality in hospitalized infants and children1. Despite decades of research, S. aureus remains the second most common cause of nosocomial infections in neonates and children, including central-line associated bloodstream infection, surgical site infection, and ventilator-associated pneumonia 1, 2, 3, 4. S. aureus can colonize the nasal cavity, and colonization is associated with an increased risk of subsequent infection5, 6, 7. Intranasal antibiotics and antiseptic agents have been used to eradicate colonization in an attempt to prevent S. aureus disease8, 9. The agents used for decolonization do not solely target S. aureus, but eradicate many bacterial communities with unknown long-term consequences10. Unfortunately, comprehensive infection control strategies including decolonization have been insufficient at preventing S. aureus disease in hospitalized patients, especially in neonates, who have the highest rates of invasive S. aureus disease of any age group 11, 12, 13, 14, 15.
Over the last 10 years, the field of microbiome science has exploded using nucleic acid based tools to characterize organisms, including S. aureus, in their ecologic niche. A more comprehensive understanding of the nasal microbiota may lead to the identification of bacterial communities, interactions, and functional characteristics that are associated with S. aureus colonization. As clinicians and researchers, we are hopeful that these findings will uncover new strategies for the prevention of S. aureus disease. Below we review 1) early studies manipulating the nasal microbiome to eradicate existing S. aureus colonization or prevent colonization following exposure, 2) how nucleic acid based techniques have enhanced our understanding of the nasal microbiota and S. aureus colonization, 3) major differences between the nasal microbiome of adults and children, and 4) how future microbiome studies may identify new strategies for the prevention of S. aureus disease.
II. Manipulating the microbiome: evolution of a strategy to prevent S. aureus disease
Some bacterial communities may produce an environment in the nasal cavity that is inhospitable to S. aureus colonization. For decades, researchers have been investigating the role that these commensal bacterial species play on a human host’s susceptibility to S. aureus colonization and disease. As a result, bacterial interference emerged as a strategy to use commensal bacterial species to eliminate or prevent the colonization of an anatomic niche by a pathogenic species 16.
IIa. Early trials of bacterial interference
In the 1960s, the clinical application of bacterial interference was spurred by several nursery outbreaks of highly virulent S. aureus strain 80/81 9, 17, 18, 19, 20. Light et al. found that topical decolonization with intranasal neomycin and hexachlorophene bathing failed to effectively eradicate S. aureus 80/81 carriage in these nurseries. They subsequently began inoculating all neonates admitted to the nursery with an attenuated S. aureus strain, hypothesizing that the attenuated strain 502A would occupy the nasal cavity and inhibit invasion by competing S. aureus strains 20, 21. After two inoculation cycles with the attenuated S. aureus strain, only 1 out of 202 cultured infants tested positive for strain 80/81 18. This method, though successful in reducing the rates of colonization by a highly virulent strain of S. aureus, was discontinued after several neonates developed invasive infections with what was perceived as the non-virulent strain 502A 9, 17, 18, 19, 20. Since the 1960s, there has been continued interest in the concept of nasal microbiome manipulation to eliminate existing S. aureus colonization or prevent S. aureus colonization following exposure.
IIb. Studies of bacterial interference to eliminate existing S. aureus colonization
Until recently, studies of bacterial interference to eliminate existing S. aureus colonization relied on culture-dependent techniques to characterize organism interactions. Researchers identified cultivable bacterial species that were associated with S. aureus in nasal cultures22, 23, 24. Species cultured from people without S. aureus colonization were then studied for their potential to eliminate existing S. aureus colonization or prevent S. aureus colonization following exposure. For example, Corynebacteria are some of the most frequently identified organisms in the nares of individuals who are not colonized by S. aureus 25. As common commensals of the skin and mucosal surfaces, Corynebacteria have various characteristics that confer ecological fitness, which may contribute to the prevention or elimination of colonization by competing species. Uehara et al. found that Corynebacterium strain Co304 had a higher affinity to nasal mucin than S. aureus for example, which they suggest may contribute to its ability to outcompete S. aureus and eliminate carriage 25. Some species of Corynebacteria have also been shown to decrease the gene expression of factors that are important for S. aureus colonization and virulence like surface protein A, which allows S. aureus to evade detection and elimination by a host’s immune system26. In one study, Corynebacterium pseudodiphtheriticum was inoculated in six MRSA colonized men and led to a decrease in abundance of S. aureus. However, as the abundance of C. pseudodiphtheriticum decreased, MRSA abundance returned to baseline level 27. Similarly, when Corynebacterium strain Co304 was inoculated in the nasal cavities of over a dozen adults, S. aureus was eradicated in 71% of subjects and maintained in follow up periods of up to 3 years 25.
Staphylococcus epidermidis is another common commensal species found in the human nasal cavity23. A recent study in HIV-positive adults found that those colonized with S. epidermidis in the nose and throat were 80% less likely to be colonized with S. aureus at the same site 28. Iwase et al. also observed that individuals who were not colonized with S. aureus were twice as likely to be colonized by S. epidermidis. This group inoculated adult S. aureus carriers with a serine protease-producing S. epidermidis strain and reported significantly reduced rates of S. aureus colonization23. These studies postulate that S. epidermidis may have a functional impact on the environment that makes the nasal cavity inhospitable to S. aureus. The exact mechanisms by which these common commensal organisms eliminate S. aureus colonization remain uncertain. Bacterial interference trials in humans, though few, have suggested that the bacterial species commonly found in the nasal cavity may help eliminate S. aureus colonization.
IIc. Studies of bacterial interference to prevent S. aureus colonization following exposure
The potential to prevent S. aureus colonization after a known exposure using certain bacterial strains, however, has only been studied in mouse models. Researchers have deliberately exposed naïve mice to S. aureus to assess the protective effects of other organisms. Park et al. assessed the potential of S. epidermidis to prevent acquisition of S. aureus colonization. They first treated mice with antibiotics to clear the resident microbiota, hoping that the absence of competing species would improve their success of inoculating the mice with S. epidermidis. After antibiotic pretreatment, mice were inoculated with S. epidermidis. Mice whose nasal cavities were inoculated with S. epidermidis prior to being exposed to MRSA showed reduced rates of S. aureus colonization compared to controls that were not initially inoculated with S. epidermidis, suggesting that S. epidermidis may alter its environment in the nasal cavity and make it unfavorable for S. aureus acquisition 29. Similarly, Barbagelata et al. inoculated mice with an attenuated mutant strain of S. aureus (NK41) before exposing them to several virulent S. aureus strains isolated from humans22. Prior exposure to NK41 significantly reduced the abundance of virulent strains, suggesting that the mutant strain could exclude competing strains from the nasal cavity.
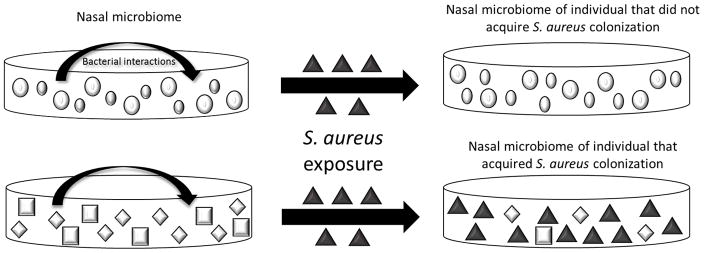
While the results of bacterial interference studies conducted to date are promising, most of the microorganisms that colonize humans cannot be cultivated, so many organisms in the nasal microbiota may be overlooked30. Although these prior studies to eradicate existing colonization or prevent colonization following exposure are limited by their reliance on conventional cultures, they suggest that the resident microbiota may play a significant role in regulating S. aureus colonization (fig 1). Nucleic acid based approaches will help to determine 1) the role of non-cultivatable organisms in resistance to S. aureus colonization and 2) whether a community of organisms, rather than a single species, is necessary to resist S. aureus colonization.
Figure 1.
III. Nucleic acid based approaches provide insight into the nasal microbiota and S. aureus colonization
Until recently, researchers relied on culture-dependent techniques to identify organisms in people that did not have S. aureus nasal colonization, then tested those organisms for their potential to eliminate existing S. aureus colonization. Nucleic acid based approaches, such as 16S rRNA gene amplification and sequencing techniques, can provide a more complete and detailed characterization of the nasal microbiota and its potential for colonization resistance against S. aureus.
High-throughput sequencing studies in adults suggest that the nasal microbiota plays an important but complex role in S. aureus colonization. The abundance of certain bacterial species in the nasal microbiota is strongly associated with S. aureus carriage. A study conducted by Liu et al. demonstrated that the absolute abundance of Corynebacterium species is a predictor of S. aureus carriage in a threshold-dependent manner 31. As the abundance of Corynebacterium species increases, the likelihood of S. aureus colonization decreases. In fact, multiple studies have found a higher relative abundance of Corynebacterium species in people not colonized with S. aureus32, 33, 34, 35. Other organisms such as the gram-positive facultative anaerobic cocci of genus Dolosigranulum, have been shown to negatively correlate with S. aureus carriage. The same group also demonstrated that there was a 16% rate of S. aureus colonization among individuals with a certain threshold abundance of Dolosigranulum species. Corynebacterium and Dolosigranulum species often co-colonize non-carriers, suggesting that the presence or absence of a single organism alone may not determine susceptibility or resistance to S. aureus colonization.
In addition to correlating S. aureus with other organisms in the nasal microbiota, culture-independent sequencing methods have evaluated organisms’ distribution and diversity metrics to suggest that resistance to S. aureus colonization may be multifactorial. Frank et al. showed that the nasal microbiota of healthy adults was more than twice as diverse as those of hospitalized patients and more evenly distributed33. They also found that the microbiota of hospitalized adults was enriched with S. epidermidis or S. aureus but lacking in Actinobacteria, such as Corynebacterium and Propionibacterium33. In addition, the microbiota of S. aureus carriers, has a lower biodiversity and more unevenly distributed bacterial communities than that of non-carriers; however, it remains unclear whether certain commensal species foster the dynamics often found in healthy adults or whether these dynamics favor colonization by commensal species over pathogenic bacteria 32, 36. Microbiome studies can further inform the relative importance of biodiversity and organism composition in contributing to S. aureus colonization.
IV. Nucleic acid based approaches reveal important differences in the nasal microbiota of children and adults
Studies conducted in adults have revealed that a diverse nasal microbiota composed of certain beneficial bacterial communities appears to provide colonization resistance against S. aureus in exposed individuals36. However, there is a notable lack of similar data on the microbiota of neonates and children. Findings in adults cannot be extrapolated to a neonatal population for several reasons. First, the mature microbiome is more diverse than the naïve microbiome, and this diversity contributes significantly to its stability and resistance to pathogen invasion 37.
Although the mature microbiota is highly variable between individuals, it is more resistant to environmental exposures than that of infants and young children 38. Second, the nasal microbiota of adults is composed of different organisms compared to newborns and children 39. Both community-dwelling and hospitalized adults host a nasal microbiota dominated by organisms of the phyla Actinobacteria and Firmicutes and rarely by gram negative organisms of the phylum Proteobacteria found commonly in infants 33, 34, 35, 36, 40. Third, studies in adults have revealed the importance of certain commensal species in the protection against colonization by pathogens like S. aureus 28, 31, 32, 33, 34, 36, 41, 42. Infants, however, lack these commensals initially, and during the first years of life the microbiome is still dynamic and susceptible to extrinsic exposures 43. Further studies are needed to characterize potentially beneficial communities and whether they can foster the same protective effects in neonates.
IVa. Emergence of a personalized microbiota in neonates
From birth, neonates are exposed to and subsequently colonized with a wide array of microbes. These organisms play a pivotal role in determining the succession of bacterial communities into mature stable configurations44. Neonates initially harbor a microbiota that is homogenous across different anatomic sites and significantly impacted by whether they are born by vaginal delivery or Caesarian-section45. Infants delivered by Caesarian-section are initially colonized by their mother’s skin microbiota and therefore are more likely to possess a higher abundance of Corynebacterium, Propionibacterium, and Staphylococcus species45. These initial colonizers interact with a neonate’s tolerant immune system, leading to the activation and suppression of different inflammatory responses46. The long-term impact of the resident microbiota on the immature immune system may also play a part in dictating when an infant will acquire important commensals and if they will acquire pathogens such as S. aureus 46, 47.
By 6 weeks of age, infants have developed a site-differentiated and personalized microbiota44, 48. At this stage, the resident microbiota varies more between different individuals than in the same individual over time44. Infants share many of the same organisms in their nasal microbiomes, but in variable proportions. For this reason, the nasal microbiome is often classified by the organism with the highest relative abundance, whether Streptococcus, Moraxella, Staphylococcus, Corynebacterium and/or Dolosigranulum 49. The most abundant bacterial species indicate the overall stability and diversity of other bacterial communities in the nasal cavity50. A high relative abundance of certain commensal organisms, such as Corynebacterium, is associated with more diverse communities and lower overall bacterial density 47. These characteristics promote the stability of the microbiota over time and help maintain homeostasis in the face of external disruptions37, 47.
As the nasal microbiome develops, commensal species may play a direct role in host protection by eliminating or excluding pathogens from their niche 16. Children whose microbiota contains commensals Corynebacterium or Dolosigranulum, for example, are less likely to harbor potential pathogens of genera Streptococcus, Staphylococcus, and Moraxella44. Although the specific mechanisms by which commensals exclude other organisms from the nasal cavity remain unclear, studies conducted on the immature nasal microbiota suggest that early acquisition of certain commensal species may impart long-term resistance to colonization by pathogens in newborns16, 51.
IVb. Plasticity of the neonatal microbiome
Lower biodiversity and higher bacterial burden characterize the immature microbiota and predispose it to disruption, including dysbiosis 42, 52. Dysbiosis, or disequilibrium, has been shown to increase the risk of colonization and potential domination of a niche by pathogenic species 53. These changes in the microbiota composition may increase an infant’s risk of infection by a colonizing organism. A study by Hilty et al. explored the association between the nasal microbiota and acute otitis media (AOM) in infants. They show that the majority of infants with AOM caused by S. pneumoniae also had a nasal microbiota dominated by S. pneumoniae. These infants had a significantly higher bacterial burden overall and lower bacterial diversity in their nasal cavities than infants colonized by only commensal streptococcal species47, 54. In fact, the dominance of any potential respiratory pathogen, whether Moraxella Catarrhalis, Haemophilus influenzae, or S. aureus, in the nasal microbiome, significantly increased the likelihood of AOM with the same organism 47.
The same qualities that contribute to a vulnerability to pathogen invasion also represent an opportunity for successful colonization by protective commensal species. The nasal microbiome of infants is dynamic and receptive to new organisms 55. A high relative abundance of certain commensal species is associated with greater biodiversity and lower bacterial burden 47. Both are characteristics shown to promote stability and subsequently, colonization resistance.
A newborn’s naïve nasal microbiome is highly sensitive to environmental exposures 55. Breastfeeding, for example, is associated with a nasal microbiome dominated by commensal Corynebacterium and Dolosigranulum species. Formula-fed infants are significantly more likely to harbor profiles dominated by potential pathogens such as S. aureus 48. Adopting habits that increase rates of colonization by potentially protective bacterial species is not always feasible, however. Antibiotics, for instance, are among the many clinical exposures related to a significant reduction in colonizing commensal species in hospitalized infants and children 47. The identification of species and the mechanisms by which they exclude S. aureus from the immature nasal microbiome could potentially make other interventions available for high-risk infants.
V. Microbiome studies may identify new strategies to prevent S. aureus disease
To date, scientists’ understanding of the association between the nasal microbiome and S. aureus colonization is just the tip of the iceberg. Nucleic acid based studies are revealing that the relationship between the microbiota and S. aureus colonization is more complex than once thought. The microbiota seems to play an important role in colonization resistance, excluding S. aureus from colonizing the nasal cavity. Microbiome studies using nucleic acid based approaches extend the range of detectable organisms and facilitate genus and even species-level identification. Improving the ability to detect and distinguish bacterial species is essential for studying the association between S. aureus colonization and specific commensal species. The use of nucleic acid sequencing also allows for a more precise measure of organism composition and abundance. This has led to the development of new ways to measure biodiversity and population distribution that may help show the association between S. aureus colonization and overall dynamics of the nasal microbiota 56. In fact, some studies have suggested that microbiota can be categorized in community types based on the organisms with the highest relative abundance31. These community types may potentially identify people at a higher risk for S. aureus colonization and infection despite vast individual variations in the nasal microbiota. In addition to community structure, sequencing enables the detection of genes encoding certain functional attributes of bacterial communities such as the ability to synthesize anti-microbial substances, possibly elucidating the mechanisms behind colonization resistance. Future functional analyses may also reveal how the interdependence of gene expression in a microbial community may be employed to attenuate S. aureus virulence. Burnham et al. suggest that interventions aimed at producing colonization resistance to S. aureus in humans may need to involve more than just one bacterial interference mechanism57. Introducing a community of organisms found in healthy non-carriers to the nares of high-risk infants could potentially help establish a temporally stable microbiota less susceptible to colonization by pathobionts like S. aureus. While hospitalized neonates and infants are especially at risk for S. aureus disease, there is a lack of studies on the relationship between the immature nasal microbiota and S. aureus colonization. The increasing accessibility of nucleic acid sequencing technologies may help to inform such studies and to make safer and more targeted prevention strategies available for high-risk infants.
VI. Conclusion
S. aureus remains a leading cause of infectious morbidity in infants and children. Prior studies suggest that the nasal microbiota is closely associated with S. aureus colonization, and that certain resident commensal bacteria may be capable of excluding S. aureus from the nasal cavity. Sequencing technologies may help identify bacterial communities and the mechanisms by which they exclude S. aureus from the immature nasal microbiome and lead to novel targeted interventions to protect high-risk infants from S. aureus disease.
Acknowledgments
Financial support: This work was supported in part by the Agency for Healthcare Research and Quality R01HS022872.
Footnotes
Potential conflicts of interest: none
References
- 1.Laupland KB, Lyytikainen O, Sogaard M, Kennedy KJ, Knudsen JD, Ostergaard C, et al. The changing epidemiology of Staphylococcus aureus bloodstream infection: a multinational population-based surveillance study. Clinical microbiology and infection. 2013;19(5):465–471. doi: 10.1111/j.1469-0691.2012.03903.x. [DOI] [PubMed] [Google Scholar]
- 2.Hocevar SN, Edwards JR, Horan TC, Morrell GC, Iwamoto M, Lessa FC. Device-associated infections among neonatal intensive care unit patients: incidence and associated pathogens reported to the National Healthcare Safety Network, 2006–2008. Infect Control Hosp Epidemiol. 2012;33(12):1200–1206. doi: 10.1086/668425. [DOI] [PubMed] [Google Scholar]
- 3.Humphreys H, Becker K, Dohmen PM, Petrosillo N, Spencer M, van Rijen M, et al. Staphylococcus aureus and surgical site infections: benefits of screening and decolonization before surgery. J Hosp Infect. 2016;94(3):295–304. doi: 10.1016/j.jhin.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Larru B, Gong W, Vendetti N, Sullivan KV, Localio R, Zaoutis TE, et al. Bloodstream Infections in Hospitalized Children: Epidemiology and Antimicrobial Susceptibilities. Pediatr Infect Dis J. 2016;35(5):507–510. doi: 10.1097/INF.0000000000001057. [DOI] [PubMed] [Google Scholar]
- 5.Weidenmaier C, Goerke C, Wolz C. Staphylococcus aureus determinants for nasal colonization. Trends Microbiol. 2012;20(5):243–250. doi: 10.1016/j.tim.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 6.von Eiff C, Becker K, Machka K, Stammer H, Peters G. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N Engl J Med. 2001;344(1):11–16. doi: 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- 7.Kluytmans JA, Wertheim HF. Nasal carriage of Staphylococcus aureus and prevention of nosocomial infections. Infection. 2005;33(1):3–8. doi: 10.1007/s15010-005-4012-9. [DOI] [PubMed] [Google Scholar]
- 8.Popoola VO, Colantuoni E, Suwantarat N, Pierce R, Carroll KC, Aucott SW, et al. Active Surveillance Cultures and Decolonization to Reduce Staphylococcus aureus Infections in the Neonatal Intensive Care Unit. Infect Control Hosp Epidemiol. 2016;37(4):381–387. doi: 10.1017/ice.2015.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Light IJ, Walton R, Sutherland JM, Shinefield HR, Brackvogel V. Use of bacterial interference to control a staphylococcal nursery outbreak: Deliberate colonization of all infants with the 502a strain of staphylococcus aureus. American Journal of Diseases of Children. 1967;113(3):291–300. doi: 10.1001/archpedi.1967.02090180051001. [DOI] [PubMed] [Google Scholar]
- 10.Popoola VO, Milstone AM. Decolonization to prevent Staphylococcus aureus transmission and infections in the neonatal intensive care unit. Journal of perinatology. 2014;34(11):805–810. doi: 10.1038/jp.2014.128. [DOI] [PubMed] [Google Scholar]
- 11.Zingg W, Hopkins S, Gayet-Ageron A, Holmes A, Sharland M, Suetens C. Health-care-associated infections in neonates, children, and adolescents: an analysis of paediatric data from the European Centre for Disease Prevention and Control point-prevalence survey. The Lancet Infectious Diseases. 2017 doi: 10.1016/S1473-3099(16)30517-5. [DOI] [PubMed] [Google Scholar]
- 12.Dantes R, Mu Y, Belflower R, Aragon D, Dumyati G, Harrison LH, et al. National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA internal medicine. 2013;173(21):1970–1978. doi: 10.1001/jamainternmed.2013.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ericson JE, Popoola VO, Smith PB, Benjamin DK, Fowler VG, Benjamin DK, Jr, et al. Burden of Invasive Staphylococcus aureus Infections in Hospitalized Infants. JAMA Pediatr. 2015;169(12):1105–1111. doi: 10.1001/jamapediatrics.2015.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwamoto M, Mu Y, Lynfield R, Bulens SN, Nadle J, Aragon D, et al. Trends in invasive methicillin-resistant Staphylococcus aureus infections. Pediatrics. 2013;132(4):e817–824. doi: 10.1542/peds.2013-1112. [DOI] [PubMed] [Google Scholar]
- 15.Reich PJ, Boyle MG, Hogan PG, Johnson AJ, Wallace MA, Elward AM, et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus strains in the neonatal intensive care unit: an infection prevention and patient safety challenge. Clinical microbiology and infection. 2016;22(7):645.e641–648. doi: 10.1016/j.cmi.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen HW, Liu PF, Liu YT, Kuo S, Zhang XQ, Schooley RT, et al. Nasal commensal Staphylococcus epidermidis counteracts influenza virus. Sci Rep. 2016;6:27870. doi: 10.1038/srep27870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shinefield HR, Sutherland JM, Ribble JC, Eichenwald HF. Bacterial interference: its effect on nursery-acquired infection with Staphylococcus aureus. II. The Ohio epidemic. Am J Dis Child. 1963;105:655–662. [PubMed] [Google Scholar]
- 18.Shinefield HR, Ribble JC, Boris M, Eichenwald HF. Bacterial interference: its effect on nursery-acquired infection with Staphylococcus aureus. I. Preliminary observations on artificial colonzation of newborns. Am J Dis Child. 1963;105:646–654. [PubMed] [Google Scholar]
- 19.Houck PW, Nelson JD, Kay JL. Fatal septicemia due to staphylococcus aureus 502a: Report of a case and review of the infectious complications of bacterial interference programs. American Journal of Diseases of Children. 1972;123(1):45–48. [PubMed] [Google Scholar]
- 20.Light IJ, Sutherland JM, Schott JE. Control of a staphylococcal outbreak in a nursery: Use of bacterial interference. JAMA. 1965;193(9):699–704. doi: 10.1001/jama.1965.03090090005001. [DOI] [PubMed] [Google Scholar]
- 21.Shinefield HR, Ribble JC, Boris M. Bacterial Interference Between Strains of Staphylococcus aureus, 1960 to 1970. Amer J Dis Child. 1971;121(2):148–152. doi: 10.1001/archpedi.1971.02100130102013. [DOI] [PubMed] [Google Scholar]
- 22.Barbagelata MS, Alvarez L, Gordiola M, Tuchscherr L, von Eiff C, Becker K, et al. Auxotrophic mutant of Staphylococcus aureus interferes with nasal colonization by the wild type. Microbes Infect. 2011;13(12–13):1081–1090. doi: 10.1016/j.micinf.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465(7296):346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 24.Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature. 2016;535(7613):511–516. doi: 10.1038/nature18634. [DOI] [PubMed] [Google Scholar]
- 25.Uehara Y, Nakama H, Agematsu K, Uchida M, Kawakami Y, Abdul Fattah AS, et al. Bacterial interference among nasal inhabitants: eradication of Staphylococcus aureus from nasal cavities by artificial implantation of Corynebacterium sp. J Hosp Infect. 2000;44(2):127–133. doi: 10.1053/jhin.1999.0680. [DOI] [PubMed] [Google Scholar]
- 26.Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP. Staphylococcus aureus Shifts toward Commensalism in Response to Corynebacterium Species. Frontiers in microbiology. 2016;7:1230. doi: 10.3389/fmicb.2016.01230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiryukhina NV, Melnikov VG, Suvorov AV, Morozova YA, Ilyin VK. Use of Corynebacterium pseudodiphtheriticum for elimination of Staphylococcus aureus from the nasal cavity in volunteers exposed to abnormal microclimate and altered gaseous environment. Probiotics and Antimicrobial Proteins. 2013;5(4):233–238. doi: 10.1007/s12602-013-9147-x. [DOI] [PubMed] [Google Scholar]
- 28.Sullivan SB, Kamath S, McConville TH, Gray BT, Lowy FD, Gordon PG, et al. Staphylococcus epidermidis Protection Against Staphylococcus aureus Colonization in People Living With Human Immunodeficiency Virus in an Inner-City Outpatient Population: A Cross-Sectional Study. Open Forum Infectious Diseases. 2016;3(4):ofw234. doi: 10.1093/ofid/ofw234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park B, Iwase T, Liu GY. Intranasal Application of S. epidermidis Prevents Colonization by Methicillin-Resistant Staphylococcus aureus in Mice. PLoS ONE. 2011;6(10):e25880. doi: 10.1371/journal.pone.0025880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu CM, Price LB, Hungate BA, Abraham AG, Larsen LA, Christensen K, et al. Staphylococcus aureus and the Ecology of the Nasal Microbiome. Science Advances. 2015;1(5) doi: 10.1126/sciadv.1400216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alvarez AS, Remy L, Allix-Beguec C, Ligier C, Dupont C, Leminor O, et al. Patient nostril microbial flora: individual-dependency and diversity precluding prediction of Staphylococcus aureus acquisition. Clinical microbiology and infection. 2014;20(1):70–78. doi: 10.1111/1469-0691.12208. [DOI] [PubMed] [Google Scholar]
- 33.Frank DN, Feazel LM, Bessesen MT, Price CS, Janoff EN, Pace NR. The human nasal microbiota and Staphylococcus aureus carriage. PLoS One. 2010;5(5):e10598. doi: 10.1371/journal.pone.0010598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson RC, Ellis MW, Lanier JB, Schlett CD, Cui T, Merrell DS. Correlation between nasal microbiome composition and remote purulent skin and soft tissue infections. Infect Immun. 2015;83(2):802–811. doi: 10.1128/IAI.02664-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan M, Pamp SJ, Fukuyama J, Hwang PH, Cho DY, Holmes S, et al. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe. 2013;14(6):631–640. doi: 10.1016/j.chom.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bessesen MT, Kotter CV, Wagner BD, Adams JC, Kingery S, Benoit JB, et al. MRSA colonization and the nasal microbiome in adults at high risk of colonization and infection. J Infect. 2015;71(6):649–657. doi: 10.1016/j.jinf.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Petchey OL, Eklof A, Borrvall C, Ebenman B. Trophically unique species are vulnerable to cascading extinction. Am Nat. 2008;171(5):568–579. doi: 10.1086/587068. [DOI] [PubMed] [Google Scholar]
- 38.Arrieta M-C, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The Intestinal Microbiome in Early Life: Health and Disease. Frontiers in Immunology. 2014;5:427. doi: 10.3389/fimmu.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh J, Conlan S, Polley EC, Segre JA, Kong HH. Shifts in human skin and nares microbiota of healthy children and adults. Genome Medicine. 2012;4(10):77–77. doi: 10.1186/gm378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wos-Oxley ML, Plumeier I, von Eiff C, Taudien S, Platzer M, Vilchez-Vargas R, et al. A poke into the diversity and associations within human anterior nare microbial communities. ISME J. 2010;4(7):839–851. doi: 10.1038/ismej.2010.15. [DOI] [PubMed] [Google Scholar]
- 41.Cremers AJH, Zomer AL, Gritzfeld JF, Ferwerda G, van Hijum SAFT, Ferreira DM, et al. The adult nasopharyngeal microbiome as a determinant of pneumococcal acquisition. Microbiome. 2014;2:44. doi: 10.1186/2049-2618-2-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peterson SW, Knox NC, Golding GR, Tyler SD, Tyler AD, Mabon P, et al. A Study of the Infant Nasal Microbiome Development over the First Year of Life and in Relation to Their Primary Adult Caregivers Using cpn60 Universal Target (UT) as a Phylogenetic Marker. PLoS One. 2016;11(3):e0152493. doi: 10.1371/journal.pone.0152493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mueller NT, Bakacs E, Combellick J, Grigoryan Z, Dominguez-Bello MG. The infant microbiome development: mom matters. Trends in molecular medicine. 2015;21(2):109–117. doi: 10.1016/j.molmed.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014;190(11):1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 45.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belkaid Y, Hand T. Role of the Microbiota in Immunity and inflammation. Cell. 2014;157(1):121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hilty M, Qi W, Brugger SD, Frei L, Agyeman P, Frey PM, et al. Nasopharyngeal microbiota in infants with acute otitis media. J Infect Dis. 2012;205(7):1048–1055. doi: 10.1093/infdis/jis024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biesbroek G, Bosch AA, Wang X, Keijser BJ, Veenhoven RH, Sanders EA, et al. The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am J Respir Crit Care Med. 2014;190(3):298–308. doi: 10.1164/rccm.201401-0073OC. [DOI] [PubMed] [Google Scholar]
- 49.Biesbroek G, Wang X, Keijser BJF, Eijkemans RMJ, Trzciński K, Rots NY, et al. Seven-valent pneumococcal conjugate vaccine and nasopharyngeal microbiota in healthy children. Emerging Infectious Diseases. 2014;20(2):201–210. doi: 10.3201/eid2002.131220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bogaert D, Keijser B, Huse S, Rossen J, Veenhoven R, van Gils E, et al. Variability and diversity of nasopharyngeal microbiota in children: A metagenomic analysis. PLoS ONE. 2011;6(2) doi: 10.1371/journal.pone.0017035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sassone-Corsi M, Raffatellu M. No Vacancy: How beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. Journal of immunology. 2015;194(9):4081–4087. doi: 10.4049/jimmunol.1403169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stearns JC, Davidson CJ, McKeon S, Whelan FJ, Fontes ME, Schryvers AB, et al. Culture and molecular-based profiles show shifts in bacterial communities of the upper respiratory tract that occur with age. The ISME Journal. 2015;9(5):1246–1259. doi: 10.1038/ismej.2014.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu LC-H, Shih Y-A, Wu L-L, Lin Y-D, Kuo W-T, Peng W-H, et al. Enteric dysbiosis promotes antibiotic-resistant bacterial infection: systemic dissemination of resistant and commensal bacteria through epithelial transcytosis. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2014;307(8):G824–G835. doi: 10.1152/ajpgi.00070.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laufer AS, Metlay JP, Gent JF, Fennie KP, Kong Y, Pettigrew MM. Microbial Communities of the Upper Respiratory Tract and Otitis Media in Children. mBio. 2011;2(1):e00245–00210. doi: 10.1128/mBio.00245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bogaert D, De Groot R, Hermans PW. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis. 2004;4(3):144–154. doi: 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- 56.Kim Y, Koh I, Rho M. Deciphering the human microbiome using next-generation sequencing data and bioinformatics approaches. Methods (San Diego, Calif) 2015;79–80:52–59. doi: 10.1016/j.ymeth.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 57.Burnham CA, Hogan PG, Wallace MA, Deych E, Shannon W, Warren DK, et al. Topical Decolonization Does Not Eradicate the Skin Microbiota of Community-Dwelling or Hospitalized Adults. Antimicrobial agents and chemotherapy. 2016;60(12):7303–7312. doi: 10.1128/AAC.01289-16. [DOI] [PMC free article] [PubMed] [Google Scholar]