Abstract
Many countries in the Americas have detected local transmission of multiple arboviruses that cause febrile illnesses. Therefore, laboratory testing has become an important tool for confirming the etiology of these diseases. The present study aimed to compare the sensitivity and specificity of three different Zika virus detection assays. One hundred serum samples from patients presenting with acute febrile symptoms were tested using a previously reported TaqMan® RT-qPCR assay. We used a SYBR® Green RT-qPCR and a conventional PCR methodologies to compare the results. Of the samples that were determined to be negative by the TaqMan® RT-qPCR assay, 100% (Kappa = 0.670) were also found to be negative by SYBR® Green RT-qPCR based on Tm comparison; however, 14% (Kappa = 0.035) were found to be positive by conventional PCR followed by agarose gel electrophoresis. The differences between the ZIKV strains circulating worldwide and the low viremia period can compromise diagnostic accuracy and thereby the accuracy of outbreak data. Therefore, improved assays are required to improve the diagnosis and surveillance of arbovirus.
Keywords: Zika virus, Detection, Sensitivity, Specificity
Introduction
Many countries in the Americas have detected local transmission of multiple arboviruses that cause febrile illnesses accompanied by rash, myalgia, or arthralgia. Therefore, laboratory testing has become an important tool for confirming the etiology of these diseases. Patients with the above symptoms should be considered suspected cases of Zika fever, chikungunya, and dengue virus infections, especially if they have traveled to within the previous two weeks or are living in an area with recent reports of arbovirus transmission.1
Laboratory evidence of recent infection with chikungunya (CHIKV), dengue (DENV), or Zika (ZIKV) virus is generally established by testing patient serum to detect viral nucleic acid or virus specific immunoglobulins and neutralizing antibodies.2 However, serological cross-reactivity may occur between ZIKV and other flaviviruses (DENV, yellow fever virus, St. Louis encephalitis virus, Japanese encephalitis virus, and West Nile virus). Hence, emphasis should be placed on molecular diagnostic assays (e.g., RT-PCR) for acute specimens collected from individuals with clinically compatible illnesses.3, 4, 5, 6
The current diagnostic tests for ZIKV have several limitations.7 Therefore, this work aimed to compare the sensitivity and specificity of the previously reported TaqMan® RT-qPCR for ZIKV detection5 with those of SYBR® Green RT-qPCR, and conventional PCR.
Material and methods
Sample collection
A total of 100 serum samples from patients presenting with acute febrile disease for ≤5 days from January to May 2016 were tested for ZIKV infection by TaqMan® RT-qPCR,5 SYBR® Green real-time RT-PCR, and conventional PCR (unpub. data). The patients were examined by the São José do Rio Preto Health Service, and the illness was diagnosed as Zika fever based only on clinical-epidemiological data (signs compatible with ZIKV infection), provided by the Brazilian Ministry of Health. Information on the clinical presentation of the disease was collected and registered in the National System of Injury of Notifications (SINAN). All patients were examined by the Public Health Office and were considered autochthonous cases. This work was part of an ongoing surveillance project approved by the FAMERP's Ethical Review Board (CEP n° 02078812.8.0000.5415).
Sample preparation
Blood samples were collected, and the sera were separated and stored at −80 °C. Viral RNA was extracted from 140 μL of each serum using the QIAamp Viral RNA Mini kit (QIAGEN, Germany) according to the manufacturer's instructions, and the RNAs were tested by the various PCR-based assays.
ZIKV detection by TaqMan®
The RNA was initially tested for ZIKV by a TaqMan® RT-qPCR, as previously described,5 using the GoTaq® Probe 1-Step RT-qPCR System (Promega). The same samples were also screened for DENV and CHIKV by Multiplex-nested-PCR8 and RT-qPCR9 respectively. The qPCRs were performed using the Thermocycler StepOne Real-Time PCR System (Applied Biosystems) and the multiplex-nested-PCR was performed using the Veriti 96 well Thermal Cycler (Applied Biosystem).
ZIKV detection by SYBR®
To identify ZIKV by SYBR Green® RT-qPCR, primers described by Lanciotti et al.,5 that target the Flavivirus envelope protein region (ZIKV 1086 and ZIKV 1162c) were used. The reactions were performed using the SuperScript® III Platinum® SYBR® Green One-Step qRT-PCR Kit (Invitrogen). The RT-PCR mixture contained 10 μL of 2× Sybr Green buffer, 0.3 μL of 10 μM of each primer,5 0.4 μL of ROX Reference Dye (25 μM), 0.4 μL of SuperScript® III/Platinum® Taq Mix, 5 μL of the RNA sample and water to make the volume up to 20 μL. The RT-PCR mixture was incubated for 3 min at 42 °C, 5 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C, and a final single cycle for 1 min at 42 °C. The amplification step was, followed by a melting curve analysis. The thermal cycling was performed in the thermocycler StepOne Real-Time PCR System (Applied Biosystems). Positivity was determined by Tm comparison between the positive controls and the samples.
ZIKV detection by conventional PCR
To perform the conventional PCR, four primers targeting the ZIKV NS5 protein were designed. The primers were based on infectious clones from the African ZIKV strains (GenBank accessions LC002520.1, KF268948.1 and KF268950.1) and were kindly provided by Prof. Ricardo Parreira, the Tropical Medicine and Hygiene Institute, Universidade NOVA de Lisboa, Portugal. The methodology was based on nested RT-PCR. The RT mixture contained 8 μL of the RNA template, 4 μL of 5× first strand buffer (250 mM Tris–HCl [pH 8.3], 375 mM KCl, 15 mM MgCl2), 1.4 μL of dithiothreitol (0.1 M), 1 μL of reverse primer (5′10097CATGTCCTCAGTRGTCATCC101163′; 15 μM), 1.6 μL of dNTP mixture (250 μM each dNTP), 200 U of RNase inhibitor (RNaseOUT; Invitrogen), 200 U of reverse transcriptase (Superscript; Invitrogen), and water to make the volume up to 20 μL. The mixture was incubated at 50 °C for 50 min and at 70 °C for 5 min to inactivate the reverse transcriptase. The PCR mixture contained 8 μL of cDNA, 5 μL of 10× PCR buffer (200 mM Tris–HCl [pH 8.4], 500 mM KCl), 2 μL of MgCl2 (50 mM), 1 μL of forward primer (5′9104CCATCTGGTACATGTGG91203′) at 15 μM, 4 μL of dNTP mixture (250 μM each dNTP), 1 U of Taq DNA polymerase (Platinum Taq DNA polymerase; Invitrogen), and water to make the volume up to 50 μL. The mixture was subjected to 30 cycles at 94 °C for 1 min, 53 °C for 1 min, and 72 °C for 2 min, followed by a final extension step at 72 °C for 5 min.
The nested-PCR mixture contained 1 μL of the mixture from the first amplification, 5 μL of PCR buffer (200 mM Tris–HCl [pH 8.4], 500 mM KCl), 2 μL of MgCl2 (50 mM), 1 μL of each primer (5′9671GTGGAGATGACTGCGTTGTGAAGCC96953′ and 5′9990CCATCAGTCGAAGGTCTCTTCTGTGG100153′; 15 μM), 4 μL of dNTP mixture (250 μM each dNTP), 1 U of Taq DNA polymerase (Platinum Taq DNA polymerase; Invitrogen) and water to make the volume up to 50 μL. The mixture was subjected to 25 cycles at 94 °C for 1 min, 53 °C for 1 min, and 72 °C for 2 min. A final extension step was carried out at 72 °C for 5 min. The nested RT-PCR was performed in the Veriti 96 well Thermal Cycler (Applied Biosystems).
PCR products were loaded onto a 1.5% agarose gel and visualized under ultraviolet light. Amplicon sizes were determined by comparison with a 100 bp DNA ladder (Invitrogen). Precautions to avoid contamination were taken; positive (Zika virus strain MR 766) and negative (water + all PCR components) controls were used in all reactions/amplification protocols, and the procedure was repeated 3 times.10
Statistical analyses
The accuracy of any diagnostic method can be determined by calculating its positive predictive value (PPV) and negative predictive value (NPV). The effect of prevalence on predictive values was calculated according to Bayes's theorem.11 Differences in the results of the tests were assessed using Fisher's exact test12, 13 and Kappa test.14 The confidence interval of the estimates was computed using the ‘exact method’ developed by Clopper and Pearson (GraphPad Prism package 2.01; Graph-Pad).
Results
We selected 100 serum samples collected between January and May 2016 from patients with suspected cases of ZIKV disease, based on the presence of macular or papular rash with two or more of the following signs and symptoms: fever or conjunctival hyperemia without secretion and pruritus, polyarthralgia or joint edema. Of these, 59 samples (59%) were determined positive (Ct < 38.5) and 41 samples (41%) were determined negative (Ct > 38.5) using TaqMan® RT-qPCR.5
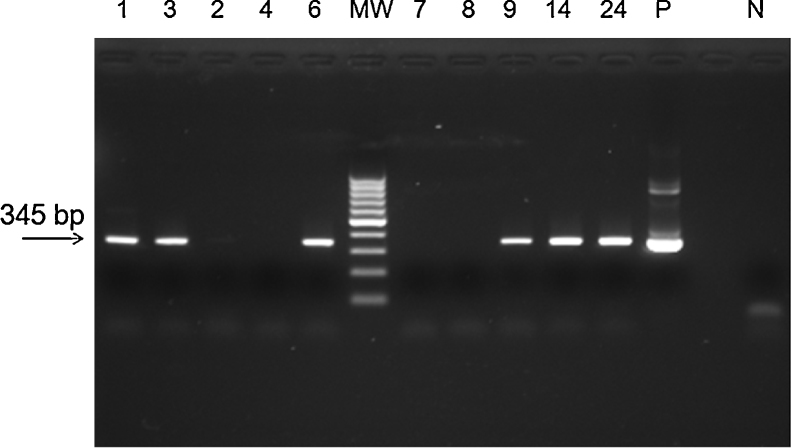
The sensitivity and specificity of the methodology were compared with those of other laboratory methods (SYBR® Green RT-qPCR and conventional PCR) (Table 1). From the samples that were found to be negative by TaqMan® RT-qPCR, 41 (100%) (Kappa = 0.670) were also found to be negative by SYBR® Green RT-qPCR based on Tm comparison (Supplementary Table S1), but 6 (14%) (Kappa = 0.035, p < 0.0001) were found to be positive by conventional PCR followed by agarose gel electrophoresis (Fig. 1). To confirm the results, the 6 positive samples were inoculated on Vero cells. RNA was extracted and ZIKV identification was performed by a TaqMan® RT-qPCR, as previously described.5
Table 1.
Performance of TaqMan®, SYBR-green® and conventional PCR assay for ZIKV detection in one hundred sera samples.
| Laboratory methods | Number of samples | ||
|---|---|---|---|
| TaqMan® RT-qPCR |
|||
| Positive | Negative | Total | |
| SYBR®Green RT-qPCR | |||
| Positive | 42 | 0 | 42 |
| Negative | 17 | 41 | 58 |
| Total | 59 | 41 | 100 |
| Conventional PCR | |||
| Positive | 11 | 6 | 17 |
| Negative | 48 | 35 | 83 |
| Total | 59 | 41 | 100 |
Fig. 1.
Agarose gel eletrophoresis of amplicons from PCR assay for ZIKV. MW, molecular weight marker (100 bp); positive samples, 1, 3, 6, 9, 14, 24; negative samples: 2, 4, 7, 8; P, positive control for ZIKV (Zika virus strain MR 766); N, negative control. PCR assay showing the amplicon of 345 bp of ZIKV.
Discussion
ZIKV, which emerged after DENV and CHIKV in the city of São José do Rio Preto, represents a new public health concern in this region, and continues to be misdiagnosed.15
Laboratory diagnosis of acute ZIKV infection currently relies upon the detection of ZIKV RNA in biological fluids include serum, plasma, urine, saliva, and amniotic fluid.16 The ability of RT-PCR to detected of ZIKV RNA in blood is limited, because ZIKV viremia is usually low and limited to the third and fourth day after disease onset.5 Future studies with larger numbers of positive specimens will be required to further characterize the dynamics of ZIKV levels in blood and to determine whether virus load is correlated with disease severity and immune responses.16
Accurate diagnosis of acute ZIKV infection may require testing of multiple specimen types. According to the literature, the mean days of illness were not significantly different between patients with ZIKV RNA detectable only in the saliva and those with ZIKV RNA detectable only in the serum.16, 17 Higher ZIKV RNA loads are detectable in the urine and semen after longer periods (10–20 and 27–62 days after onset of symptoms, respectively), suggesting that the detection of the ZIKV RNA in other specimen types could be a potential alternative diagnostic technique.4, 18
The current diagnostic tests for ZIKV includes the conventional one-step RT-PCR assay targeting the E gene19, 20 and the nonstructural proteins 1 (NS1),20 3 (NS3)21 and 5 (NS5),5 respectively. A previous genetic study using nucleotide sequences derived from the NS5 gene indicated three ZIKV lineages: East African, West African, and Asian.5 Genetic analyses have revealed close relationships between the virus strains in the South American and Pacific Island outbreaks, suggesting that the Pacific islands may have been the springboard for the South American epidemic.22, 23 The sequences of these strains are different from those used for the detection of ZIKV, which were designed by the CDC using primer/probe sets based on ZIKV MR766 GenBank accession no. AY632535 (country: Uganda).5 Interestingly, in the present study, the Brazilian samples (Asian lineage) were negative for ZIKV in the TaqMan® RT-qPCR and SYBR® Green RT-qPCR assay, whereas 12% of the samples were positive for ZIKV in the conventional PCR assay; for both methodologies, primers based on African ZIKV isolates were used.
In addition to the benefits common to any qPCR reaction, the statistical parameters (Kappa = 0.670) prove optimized SYBR Green method has similar performances to TaqMan method and data analysis was show with the help of high performance primer designing software and by utilization proper protocols and material we can achieve high quality and precise data by SYBR Green as the TaqMan method.24
Future work will involve reducing the limitations of current diagnostic tests for ZIKV. As this pandemic evolves, further development, evaluation, and widespread implementation of ZIKV diagnostics will be critical to monitoring, preventing, and treating Zika fever.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
We thank the FAPESP (grant #2013/21719-3 to MLN) for financial support.
Associate Editor: Roxane Piazza
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bjm.2017.04.011.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.CDC . 2016. Division of Vector-Borne Diseases. Revised Diagnostic Testing for Zika, Chikungunya, and Dengue Viruses in US Public Health Laboratories. Available at: http://www.cdc.gov/zika/pdfs/denvchikvzikv-testing-algorithm.pdf Accessed 26.06.16. [Google Scholar]
- 2.World Health Organization (WHO) World Health Organization; 2016. Laboratory Testing for Zika Virus Infection. Available at: http://www.who.int/csr/resources/publications/zika/laboratory-testing/en Accessed 26.06.16. [Google Scholar]
- 3.Dejnirattisai W., Supasa P., Wongwiwat W. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat Immunol. 2016 doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gourinat A., O’Connor O., Calvez E., Goarant C., Dupont-Rouzeyrol M. Detection of Zika virus in Urine. Emerg Infect Dis. 2015;21:84–86. doi: 10.3201/eid2101.140894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lanciotti R.S., Kosoy O.L., Laven J.J. Genetic and serologic properties of zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis. 2008;14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gyurech D., Schilling J., Schmidt-Chanasit J., Cassinotti P., Kaeppeli F., Dobec M. False positive dengue NS1 antigen test in a traveller with an acute Zika virus infection imported into Switzerland. Swiss Med Wkly. 2016;146:w14296. doi: 10.4414/smw.2016.14296. [DOI] [PubMed] [Google Scholar]
- 7.Charrel R.N., Leparc-Goffart I., Pas S., Lamballerie X., Koopmans M., Reusken C. State of knowledge on Zika virus for an adequate laboratory response. Publ Bull World Heal Organ Type Res emergencies Artic ID BLT.16.171207. 2016:1–29. Available at: http://www.who.int/bulletin/online_first/16-171207.pdf. [Google Scholar]
- 8.Bronzoni R.V.M., Baleotti F.G., Nogueira R.M.R., Nunes M., Figueiredo L.T.M. Duplex reverse transcription-PCR followed by nested PCR assays for detection and identification of Brazilian alphaviruses and flaviviruses. J Clin Microbiol. 2005;43:696–702. doi: 10.1128/JCM.43.2.696-702.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanciotti R.S., Kosoy O.L., Laven J.J. Chikungunya virus in US travelers returning from India, 2006. Emerg Infect Dis. 2007;13:764–767. doi: 10.3201/eid1305.070015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borst A., Box A.T.A., Fluit A.C. False-positive results and contamination in nucleic acid amplification assays: suggestions for a prevent and destroy strategy. Eur J Clin Microbiol Infect Dis. 2004;23:289–299. doi: 10.1007/s10096-004-1100-1. [DOI] [PubMed] [Google Scholar]
- 11.Alberg A.J., Wan Park J., Hager B.H., Brock M.V., Diener-West M. The use of “overall accuracy” to evaluate the validity of screening or diagnostic tests. J Gener Intern Med. 2004;19:4690–4695. doi: 10.1111/j.1525-1497.2004.30091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colin C., Lanoir D., Touzet S., Meyaud-Kraemer L., Bailly F., Trepo C. Sensitivity and specificity of third-generation hepatitis C virus antibody detection assays: an analysis of the literature. J Viral Hepat. 2001;8:87–95. doi: 10.1046/j.1365-2893.2001.00280.x. [DOI] [PubMed] [Google Scholar]
- 13.Smedley R.C., Patterson J.S., Miller R. Sensitivity and specificity of monoclonal and polyclonal immunohistochemical staining for West Nile virus in various organs from American crows (Corvus brachyrhynchos) BMC Infect Dis. 2007;49:1–9. doi: 10.1186/1471-2334-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McHugh M.L. Interrater reliability: the kappa statistic. Biochem Med (Zagreb) 2012;22:276–282. [PMC free article] [PubMed] [Google Scholar]
- 15.Estofolete C.F., Terzian A.C.B., Parreira R. Clinical and laboratory profile of Zika virus infection in dengue suspected patients: a case series. J Clin Virol. 2016;81:25–30. doi: 10.1016/j.jcv.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 16.Waggoner J.J., Pinsky B.A. Zika virus: diagnostics for an emerging pandemic threat. J Clin Microbiol. 2016;54:860–867. doi: 10.1128/JCM.00279-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musso D., Roche C., Nhan T., Robin E., Teissier A. Detection of Zika virus in saliva. J Clin Virol. 2015;69:53–55. doi: 10.1016/j.jcv.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 18.Atkinson B., Hearn P., Afrough B. Detection of Zika virus in semen. Emerg Infect Dis. 2016;22:940. doi: 10.3201/eid2205.160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faye O., Faye O., Dupressoir A., Weidmann M., Ndiaye M., Alpha Sall A. One-step RT-PCR for detection of Zika virus. J Clin Virol. 2008;43:96–101. doi: 10.1016/j.jcv.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Pyke A.T., Daly M.T., Cameron J.N. Imported Zika virus infection from the Cook Islands into Australia, 2014. PLoS Curr. 2014;6 doi: 10.1371/currents.outbreaks.4635a54dbffba2156fb2fd76dc49f65e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tappe D., Rissland J., Gabriel M. First case of laboratory-confirmed Zika virus infection imported into Europe, November 2013. Euro Surveill. 2014;19 doi: 10.2807/1560-7917.es2014.19.4.20685. pii:20685. [DOI] [PubMed] [Google Scholar]
- 22.Gatherer D., Kohl A. Zika virus: a previously slow pandemic spreads rapidly through the Americas. J Gen Virol. 2016;97:269–273. doi: 10.1099/jgv.0.000381. [DOI] [PubMed] [Google Scholar]
- 23.Ai J.-W., Zhang Y., Zhang W. Zika virus outbreak: ‘a perfect storm’. Emerg Microb Infect. 2016;5:e21. doi: 10.1038/emi.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tajadini M., Panjehpour M., Javanmard S.H. Comparison of SYBR Green and TaqMan methods in quantitative real-time polymerase chain reaction analysis of four adenosine receptor subtypes. Adv Biomed Res. 2014;28(3):85. doi: 10.4103/2277-9175.127998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.