Abstract
Phyllanthus species plants are a rich source of phenolics and widely used due to their medicinal properties. A liquid chromatography–tandem mass spectrometry (LC–MS/MS) method was developed using high-pressure liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry (HPLC-ESI-QTOF-MS/MS) for the identification and characterization of quercetin, kaempferol, ellagic acid and their derivatives in ethanolic extracts of Phyllanthus species. The chromatographic separation was carried out on Thermo Betasil C8 column (250 mm×4.5 mm, 5 µm) using 0.1% formic acid in water and 0.1% formic acid in methanol as the mobile phase. The identification of diagnostic fragment ions and optimization of collision energies were carried out using 21 reference standards. Totally 51 compounds were identified which include 21 compounds identified and characterized unambiguously by comparison with their authentic standards and the remaining 30 were tentatively identified and characterized in ethanolic extracts of P. emblica, P. fraternus, P. amarus and P. niruri.
Keywords: Phyllanthus species, HPLC-ESI-QTOF-MS/MS, Phenolics
1. Introduction
Phyllanthus species (Euphorbiaceae) is widely distributed throughout the tropical and subtropical countries of Africa, Asia, South America and West Indies. The plants of genus Phyllanthus such as P. emblica, P. fraternus, P. amarus and P. niruri are extensively used in Indian System of Medicine (Ayurveda and Siddha) and Traditional Chinese Medicine due to their medicinal properties for the treatment of jaundice, asthma, malaria, eczema, wart, diarrhea and headache [1], [2], [3], [4], [5]. The extracts of Phyllanthus species have been reported to show several biological activities such as antioxidant, hepatoprotective, hypotensive, analgesic, antihepatotoxic, antiviral, antimicrobial, anticancer, antiamnesic, antiulcer, analgesic, antiinflammatory, antiallodynic, and anti-HIV/AIDS ones [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17]. Genus Phyllanthus is a rich source of phenolics and also contains alkaloids and terpenoids [14]. Phenolics can act as protective agents, inhibitors, natural animal toxicants and pesticides against invading organisms such as herbivores, nematodes, phytophagous insects, and fungal and bacterial pathogens. Phenolics are also important elements in the flavor of wine and dietary supplements due to their potent antioxidant activity [18].
Most of the qualitative and quantitative analyses of phenolics are commonly reported by traditional methods such as high performance thin layer chromatography (HPTLC) and high performance liquid chromatography (HPLC) in Phyllanthus species [17], [19], [20], [21], [22], [23], [24], [25], [26], [27]. There are few reports on the comparative identification and characterization of compounds in crude extracts of Phyllanthus species by liquid chromatography–mass spectrometry (LC–MS) [16], [17], [28], [29], [30], [31], [32], [33], [34], [35], gas chromatography–mass spectrometry (GC–MS) [36] and high performance liquid chromatography-solid phase extraction-nuclear magnetic resonance (HPLC-SPE-NMR) [37], [38], [39]. Published LC–MS methods either had very long run time [30], [35] and identified few compounds with unit mass [29], or targeted or studied only one species [31], [32], [33].
The aim of this study was to develop an LC–MS/MS method for identification, characterization and distribution of phenolics and terpenoids in ethanolic extracts of P. emblica, P. fraternus, P. amarus and P. niruri using high-pressure liquid chromatography coupled with quadrupole time-of-flight mass spectrometry (HPLC-ESI-QTOF-MS/MS).
2. Experimental
2.1. Chemicals and reagents
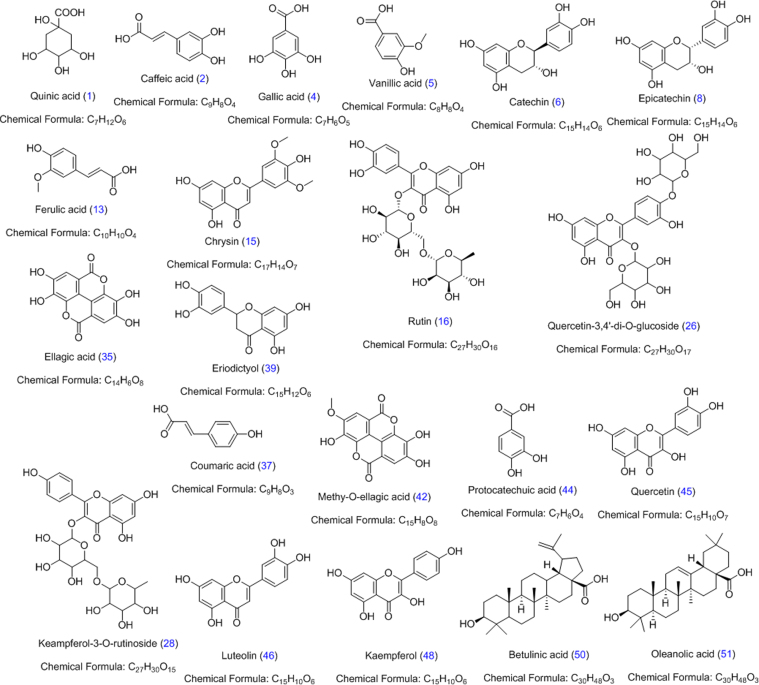
Standards quinic acid (1), caffeic acid (2), gallic acid (4), vanillic acid (5), catechin (7), epicatechin (9), ferulic acid (13), chrysin (15), rutin (16), quercetin-3,4′-di-O-glucoside (26), kaempferol-3-O-rutinoside (28), ellagic acid (35), coumaric acid (37), eriodictyol (39), methy-O-ellagic acid (42), protocatechuic acid (44), quercetin (45), luteolin (46), kaempferol (48) betulinic acid (50) and oleanolic acid (51) were purchased from Sigma-Aldrich (St. Louis, MO, USA) (Fig. 1). LC–MS grade solvents (acetonitrile methanol and formic acid) were also purchased from Sigma–Aldrich (St. Louis, MO, USA) and used throughout the study. Ultra-pure water was produced by Milli-Q Advantage system (Millipore, Milford, MA, USA). AR grade ethanol (Merck, Darmstadt, Germany) was used in the preparation of the ethanolic extracts.
Fig. 1.
Chemical structures of standard compounds.
2.2. Plant materials
The plant parts of P. emblica (leaf, bark and fruit) were obtained from the campus of CSIR-Indian Institute of Integrated Medicine (CSIR-IIIM), Jammu, India, and its voucher specimen (P. emblica-IIIM 52949) was deposited in Biodiversity and Applied Botany Division, CSIR-IIIM, Jammu. P. fraternus (leaf, bark and twigs) was collected from Aizawl, Mizoram, India, and voucher specimen (P. fraternus-MZU/BT/18) was deposited in Department of Forestry, Mizoram University. Certified whole plants of P. niruri (Batch No. 10PN-1442) and P. amarus (Reference no. PCA/PA/778) were purchased from Tulsi Amrit Pvt. Ltd (Indore, India) and Natural Remedies Private Limited (Bangalore, India), respectively. Plant parts of P. emblica and P. fraternus were washed thoroughly with normal tap water followed by Milli-Q water and dried at room temperature (26–28 °C). All dried plants were crushed into powder using grinding machine (Decibel, Lab Willey Griender, and Model No. DB 5581-4, New Delhi, India) and stored in airtight container at room temperature until analysis.
2.3. Extraction
Each sample (5 g) was dipped with ethanol (15 mL) followed by 30 min sonication at 30 °C and kept for 48 h at the room temperature. The ethanol extracts were filtered by Whitman No. 1 filter paper and filtrate was concentrated under reduced pressure at 20–50 kPa at 40 °C using a Buchi rotary evaporator [22]. This procedure was applied three times with fresh solvent. All extracts were stored in the refrigerator at –20 °C until analysis. Each extract (approximately 1 mg) was weighed accurately and dissolved in methanol accordingly to prepare 1 mg/mL stock solution.
2.4. HPLC-ESI-QTOF-MS/MS conditions
Analyses were carried out using an Agilent 1200 HPLC system interfaced with Agilent 6520 hybrid quadrupole time of flight mass spectrometer (Agilent technologies, USA). 1200 HPLC system was equipped with quaternary pump (G1311A), online vacuum degasser (G1322A), autosampler (G1329A), column compartment (G1316C) and diode-array detector (G1315D).
2.4.1. Chromatographic conditions
Chromatographic separations were performed using a Thermo Betasil C8 column (250 mm×4.5 mm, 5 µm) operated at 25 °C employing a gradient elution using 0.1% formic acid in water (A) and 0.1% formic acid in methanol (B) as mobile phase at a flow rate of 0.4 mL/min. The elution consisted of a gradient of 35%–90%, 0–7 min, 90%–90%, 7–25 min, 90%–35%, 25–35 min and initial condition was maintained for 5 min. The sample injection volume was 1 μL.
2.4.2. Mass spectrometric condition
Mass spectrometer was operated in negative electrospray ionization mode and spectra were recorded by scanning the mass range from m/z 50 to 1500 in both MS and MS/MS modes. Nitrogen was used as drying, nebulising and collision gas. Drying gas flow rate was 12 L/min. The heated capillary temperature was set at 350 °C and nebulizer pressure at 45 psi. The source parameters such as capillary voltage (VCap), fragmentor, skimmer and octapole voltages were set at 3500 V, 175 V, 65 V and 750 V, respectively. For the MS/MS analysis, collision energies were set at 15, 20, 25, 30, 35 and 40 eV. The accurate mass data of the molecular ions were processed through the Mass Hunter Workstation (version B 04.00) software.
3. Results
3.1. LC–MS/MS analysis of flavonoids
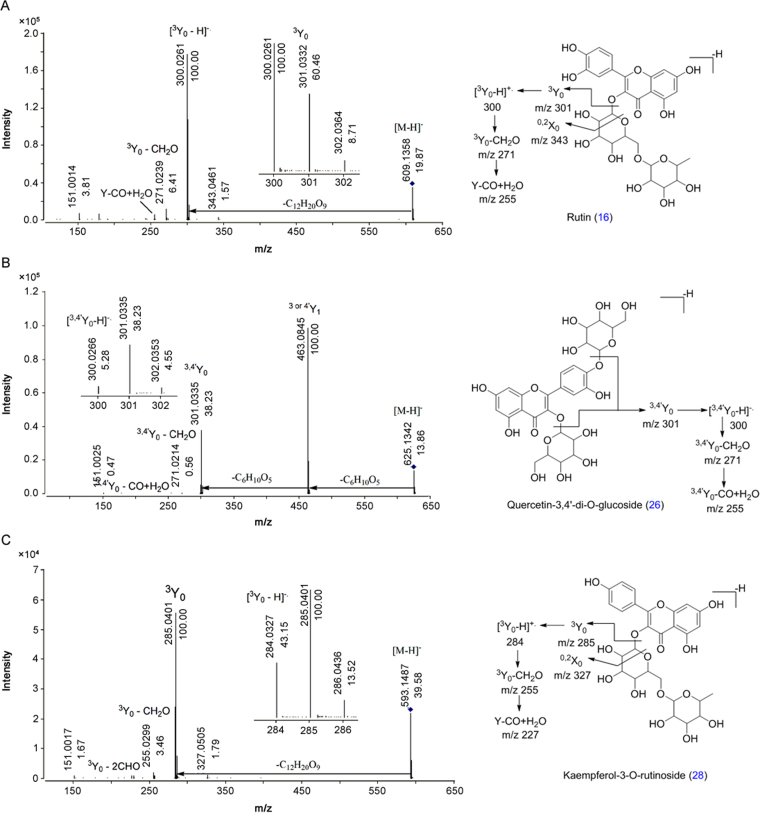
MS/MS spectra of selected flavonol-O-glucosides were analyzed at different collision energies (5–50 eV) shown in Fig. 2 and Fig. S1. Rutin (16), quercetin-3,4-di-O-glucoside (26) and kaempferol-3-O-rutinoside (28) were selected as templates. Compounds 16, 26 and 28 showed abundant [Y]− ions at collision energies of 35, 20 and 30 eV, respectively, in MS/MS analysis. The abundance of [Y1]− ion was decreased with increased abundance of [Y-H]− ions at high collision energies (Fig. S1). Thus, flavonol-O-glucosides also showed abundant [Y-H]− product ion at high collision energy [40], [41], [42], [43].
Fig. 2.
MS/MS spectra of standards (A) rutin (16), (B) quercetin-3,4-di-O-glucoside (26) and (C) kaempferol-3-O-rutinoside (28) at collision energy 35, 20 and 30 eV, respectively.
3.2. Screening of bioactive compounds
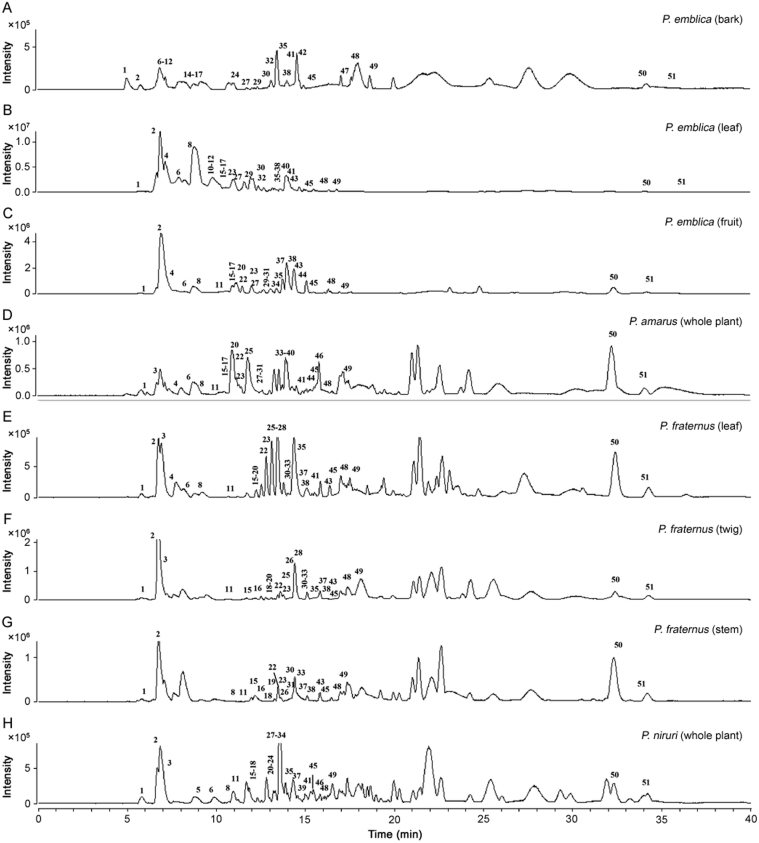
To achieve satisfactory separation, the ethanolic extracts were analyzed using gradient mobile phase consisting of 0.1% formic acid in methanol and aqueous formic acid (0.1% formic acid) after optimization. Different column types, column temperature, mobile phase, elution conditions, flow rates and MS conditions were also optimized. Base peak chromatograms (BPCs) of P. emblica (A, B and C), P. amarus (D), P. fraternus (E, F and G), and P. niruri (H) in negative ionization mode are shown in Fig. 3. Retention time (RT), observed [M-H]−, molecular formula, error (Δppm), major fragment ions and their relative abundance and distribution along with assignment are presented in Table 1, Table 2, Table 3.
Fig. 3.
Base peak chromatograms of (A) P. emblica (bark), (B) P. emblica (leaf), (C) P. emblica (fruit), (D) P. amarus, (E) P. fraternus (leaf), (F) P. fraternus (leaf), (G) P. fraternus (leaf), and (H) P. niruri.
Table 1.
Chromatographic and spectrometric data of quercetin and its derivatives identified compounds in ethanolic extracts of Phyllanthus species.
| S. No. | Retention time (min) | Observed. [M-H]− | Error (ppm) | Molecular formula | Fragment ions (Relative intensity %) |
Compounds | Distribution |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [Y]− | [Y-H]−. | [Y-CH2O]− | [Y-CO+H2O]− | PEB | PEF | PEL | PA | PFL | PFT | PFS | PN | ||||||
| 3 | 7.87 | 447.0947 | −3.21 | C21H20O11 | 301.0323 (93) | 300.0283 (100) | 271.0248 (13) | 255.0283 (28) | Quercetin 3-O-hexosde | – | – | – | + | + | + | + | + |
| 16 | 12.26 | 609.1463 | 0.03 | C27H30O16 | 301.0342 (42) | 300.0277 (100) | 271.0251 (8) | 255.0312 (5) | Rutin† | + | + | + | + | + | + | + | + |
| 18 | 12.52 | 755.2039 | 1.02 | C33H40O20 | 301.0294 (94) | 300.0287 (100) | 271.0235 (11) | 255.0367 (3) | Quercetin 3-isorhamninoside (FT) | – | – | – | – | + | + | + | + |
| 19 | 12.75 | 623.1252 | 0.25 | C27H28O17 | 301.0328 (68) | 300.0277 (100) | 271.0222 (19) | 255.0330 (8) | Quercetin derivative | – | – | – | – | + | + | + | – |
| 20 | 13.00 | 593.1514 | 0.27 | C27H30O15 | 301.0324 (84) | 300.0356 (100) | 271.0345 (6) | 255.0377 (5) | Quercetin derivative | – | – | + | + | + | + | – | + |
| 21 | 13.04 | 625.1412 | 0.02 | C27H30O17 | 301.0344 (62) | 300.0282 (100) | 271.0228 (7) | 255.0310 (3) | Quercetin-di-O-hexoside (isomer) | – | – | – | – | – | – | – | + |
| 24 | 13.12 | 595.1282 | 3.87 | C26H28O16 | 301.0318 (61) | 300.0251 (100) | 271.0228 (7) | 255.0429 (2) | Quercetin 3-sambubioside | – | – | – | – | – | – | – | + |
| 26 | 13.42 | 625.1413 | 0.03 | C27H30O17 | 301.0328 (100) | 300.0274 (54) | 271.0245 (5) | 255.0367 (2) | Quercetin-3,4-di-O-glucoside† | – | – | – | – | + | + | + | – |
| 30 | 13.79 | 463.0944 | 1.02 | C21H20O12 | 301.0327 (63) | 300.0257 (100) | 271.0236 (10) | 255.0271 (8) | Quercetin-O-hexoside | + | + | + | + | + | + | + | + |
| 32 | 13.91 | 433.0772 | 0.65 | C20H18O11 | 301.0290 (49) | 300.0286 (100) | 271.0300 (15) | 255.0231 (7) | Quercetin 3-arabinoside | + | + | – | – | + | + | – | + |
| 33 | 13.96 | 477.0662 | 2.63 | C21H18O13 | 301.0336 (100) | 300.0283 (100) | 271.0248 (13) | 255.0271 (1) | Quercetin 3-O-glucuronide | – | – | – | + | + | + | + | + |
| 45 | 15.35 | 301.0358 | 0.03 | C15H10O7 | – | – | 271.0413 (1) | 255.0271 (2) | Quercetin† | + | + | + | + | + | + | + | + |
PEB: P. emblica bark; PEF: P. emblica fruit; PEL: P. emblica leaf; PA: P. amarus; PFL: P. fraternus leaf; PFT: P. fraternus twig; PFS: P. fraternus stem; PN: P. niruri
Table 2.
Chromatographic and spectrometric data of kaempferol and its derivatives identified compounds in ethanolic extracts of Phyllanthus species.
| S. No. | Retention time (min) | Observed. [M-H]− | Error (ppm) | Molecular formula | Fragment ions (Relative intensity %) |
Compounds | Distribution |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [Y]− | [Y-H]−. | [Y-CH2O]− | [Y-2CHO]− | PEB | PEF | PEL | PA | PFL | PFT | PFS | PN | ||||||
| 25 | 13.13 | 739.2092 | 0.20 | C33H40O19 | 285.0385 (65) | 284.0319 (100) | 255.0315 (35) | 227.0332 (10) | Robinin | – | – | – | + | + | + | – | – |
| 28 | 13.53 | 593.1551 | −0.65 | C27H30O15 | 285.0303 (100) | 284.0331 (78) | 255.0299 (12) | 227.0327 (3) | Kaempferol-3-O-rutinoside† | – | – | – | + | + | + | – | + |
| 29 | 13.64 | 609.1462 | 0.05 | C27H30O16 | 285.0348 (100) | 284.0304 (15) | 255.0427 (25) | 227.0542 (5) | Kaempferol-diglucoside | + | + | + | + | – | – | – | + |
| 38 | 14.36 | 447.0930 | 0.65 | C21H20O11 | 285.0368 (43) | 284.0315 (100) | 255.0276 (23) | 227.0332 (18) | Kaempferol 3-O-hexoside | + | + | + | + | + | + | ||
| 40 | 14.70 | 417.0828 | 0.06 | C20H18O10 | 285.0348 (30) | 284.0304 (100) | 255.0275 (36) | 227.0329 (17) | Kaempferol derivatives | – | + | – | – | – | – | – | – |
| 41 | 14.84 | 461.0722 | −0.25 | C21H18O12 | 285.0390 (100) | 284.0323 (14) | 255.0453 (3) | 227.0590 (5) | Kaempferol 3-glucuronide | + | + | – | + | + | – | – | + |
| 43 | 15.14 | 431.0983 | 0.18 | C21H20O10 | 285.0437 (61) | 284.0370 (100) | 255.0299 (42) | 227.0327 (19) | Kaempferol 3-hexoside | – | + | + | – | + | + | + | – |
| 48 | 16.31 | 285.0477 | 0.54 | C15H10O6 | – | – | 255.0219 (32) | 227.0356 (45) | Kaempferol† | + | + | + | + | + | + | + | + |
PEB: P. emblica bark; PEF: P. emblica fruit; PEL: P. emblica leaf; PA: P. amarus; PFL: P. fraternus leaf; PFT: P. fraternus twig; PFS: P. fraternus stem; PN: P. niruri
Table 3.
Chromatographic and spectrometric data of other identified classes of compounds in ethanolic extract of Phyllanthus species.
| S. No. | Retention time (min) | Observed. [M-H]− | Error (ppm) | Molecular formula | Fragment ions (Relative intensity %) | Compounds | Distribution |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PEB | PEF | PEL | PA | PFL | PFT | PFS | PN | |||||||
| 1 | 6.01 | 191.0565 | 0.26 | C7H12O6 | 127.0428, 109.0315, 93.0369, 85.0318 (100), 81.0366, 59.0165, 43.0214 | Quinic acid† | + | + | + | + | + | + | + | + |
| 2 | 7.27 | 179.0380 | 0.12 | C9H8O4 | 135.0444 (100) | Caffeic acid† | + | + | + | + | – | – | + | |
| 4 | 9.16 | 169.0130 | 1.02 | C7H6O5 | 125.0232 (100) | Gallic acid† | – | + | + | + | + | + | – | – |
| 5 | 10.28 | 167.0370 | 0.41 | C8H8O4 | 151.0003 (9), 123.0441 (67), 107.0105 (9), 95.0472 (11), 83.012 (67), 81.0337 (100), 63.0250 (6), 57.0366 (13) | Vanillic acid† | – | – | – | – | – | – | – | + |
| 6 | 10.53 | 315.0713 | 3.19 | C13H16O9 | 152.0093 (100), 108.0215 (55) | Gentisic acid-O-hexoside | + | + | + | + | + | – | – | + |
| 7 | 10.55 | 289.0718 | C15H14O6 | 247.0240 (100), 205.0494 (9), 151.0391 (6), 125.0231 (10) | (+)-catechin† | + | + | – | – | – | – | – | – | |
| 8 | 11.68 | 291.0148 | −0.72 | C13H8O8 | 247.0228 (100), 219.0282 (11), 191.0336 (23), 175.0336 (6) | Brevifolincarboxylic acid | + | + | + | + | + | – | – | + |
| 9 | 11.74 | 289.0720 | 0.54 | C15H14O6 | 247.0229 (72), 245.0812 (32), 221.0795 (22), 203.0695 (62), 161.0584 (28), 151.0375 (49), 137.0221 (35), 125.0226 (67), 123.0434 (69), 109.0281 (100) | Epicatechin† | + | – | – | – | – | – | – | – |
| 10 | 11.85 | 625.1046 | 0.23 | C26H26O18 | 300.9990 (100) | Ellagic acid-O-dihexoside | + | + | – | – | – | – | – | – |
| 11 | 11.99 | 463.0518 | 0.12 | C20H16O13 | 300.9979 (100), 299.9909 (51), 243.9948 (3) | Ellagic acid-O-hexoside | + | + | + | + | + | + | + | + |
| 12 | 12.15 | 477.0310 | 1.02 | C10H10O4 | 300.9967 (86) | Ellagic acid-O-glucuronide | + | + | – | – | – | – | – | – |
| 13 | 12.18 | 194.0578 | −0.12 | C10H10O4 | 178.0266 (45), 134.0362 (100) | Ferulic acid† | – | – | – | – | – | – | – | + |
| 14 | 12.22 | 441.0829 | 0.12 | C22H18O10 | 289.0686 (34), 245.0787 (16), 169.021 (100), 125.0214 (23) | Catechin 3-gallate | + | – | – | – | – | – | – | – |
| 15 | 12.23 | 253.0506 | 0.23 | C15H10O4 | 152.0124 (100), | Chrysin† | + | + | + | + | + | + | + | + |
| 17 | 12.29 | 183.0299 | −0.02 | C8H8O5 | 169.016 (14), 124.0131 (100) | Methyl gallate | + | + | + | + | + | – | – | + |
| 22 | 13.05 | 305.0305 | 0.81 | C14H10O8 | 273.0022 (3), 245.0091 (33), 217.0137 (100), 201.0193 (15), 189.0189 (58), 161.0243 (82), 145.0286 (44), 133.0292 (63) | Methyl brevifolincarboxylate | – | – | + | + | + | + | + | + |
| 23 | 13.11 | 433.0411 | 0.42 | C19H14O12 | 300.9964 (100), 299.9934 (81), 271.9887 (2), 243.9948 (3) | Ellagic acid-O-arabinoside | + | + | + | + | + | + | + | + |
| 27 | 13.44 | 247.0248 | 2.23 | C12H8O6 | 219.0309 (38), 191.0352 (100), 173.0249 (52), 145.0297 (79) | Brevifolin | + | + | + | + | + | + | ||
| 31 | 13.83 | 197.0444 | 5.93 | C9H10O5 | 169.0131 (16), 124.0156 (100) | Ethyl gallate | – | – | + | + | + | + | + | + |
| 34 | 13.98 | 319.0459 | 2.29 | C15H12O8 | 273.0040 (62), 245.0099 (100), 229.0154 (10), 217.0142 (100), 201.0201 (11), 189.0194 (14) | Ethyl brevifolincarboxylate | – | – | + | + | – | – | – | – |
| 35 | 14.07 | 300.999 | 0.33 | C14H6O8 | 283.9975 (66), 245.0085 (36), 229.0135 (45), 200.0103 (58), 185.0242 (39), 173.0232 (60), 145.0299 (100) | Ellagic acid† | + | + | + | + | + | + | – | + |
| 36 | 14.12 | 289.0718 | 1.23 | C15H14O6 | 247.0242 (100), 245.0817 (8), 221.0851 (52), 203.0660 (67), 151.0374 (68) | Propyl-O-methyl Brevifolin | – | + | + | – | – | – | – | |
| 37 | 14.34 | 163.0411 | 1.02 | C9H8O3 | 119.0414 (100) | Coumaric acid† | + | + | + | + | + | + | + | + |
| 39 | 14.56 | 287.0586 | 1.23 | C15H12O6 | 151.0029 (100), 135.0446 (86), 125.0240 (4), 107.0138 (19) | Eriodictyol† | – | – | – | + | – | – | – | + |
| 42 | 15.06 | 315.0149 | 0.93 | C15H8O8 | 299.9888 (100) | Methy-O-ellagic acid† | + | – | – | – | – | – | – | – |
| 44 | 15.19 | 153.0196 | 1.39 | C7H6O4 | 109.0284 (100) | Protocatechuic acid† | – | – | + | + | – | – | – | – |
| 47 | 16.08 | 329.0304 | −0.40 | C16H10O8 | 314.0060 (100), 298.9824 (24), 270.9910 (8) | Dimethyl-O-ellagic acid | + | – | – | – | – | – | – | – |
| 49 | 18.14 | 343.0416 | 0.23 | C17H12O8 | 328.0259 (17),312.9999 (100), 297.9739 (64), 285.0023 (18), 269.9803 (19) | Trimethyl-O-ellagic acid | + | + | + | + | + | + | + | + |
| 46 | 15.91 | 285.0405 | 0.12 | C15H10O6 | 151.0035 (31), 133.0289 (100), 121.0290 (4), 107.0149 (15) | Luteolin† | – | – | + | – | – | – | + | |
| 50 | 33.18 | 455.3535 | 0.21 | C30H48O3 | Betulinic acid† | + | + | + | + | + | + | + | + | |
| 51 | 34.03 | 455.3537 | 0.34 | C30H48O3 | 307.3314 (7) | Oleanolic acid† | + | + | + | + | + | + | + | + |
PEB: P. emblica bark; PEF: P. emblica fruit; PEL: P. emblica leaf; PA: P. amarus; PFL: P. fraternus leaf; PFT: P. fraternus twig; PFS: P. fraternus stem; PN: P. niruri
matched with reference compounds
Eleven compounds, namely, 3 (quercetin 3-O-hexoside), 16 (rutin), 18 (quercetin 3-isorhamninoside), 19 (quercetin derivative), 20 (quercetin derivative), 21 (quercetin-di-O-hexoside), 24 (quercetin 3-sambubioside), 26 (qauercetin-3,4-di-O-glucoside), 30 (quercetin-O-hexoside), 32 (quercetin 3-arabinoside) and 33 (quercetin 3-O-glucuronide), were identified as quercetin derivatives. All these compounds (3, 16, 18, 19, 20, 21, 24, 26, 30, 32 and 33) showed characteristic fragment ion at m/z 301 [Y]− due to elimination of C6H10O4, C12H20O9, C18H30O13, C12H18O10, C12H20O8, C12H20O10, C11H18O9, C12H20O10, C6H10O5 C5H8O4 and C6H8O6, respectively. Further loss of H radical from [Y]− ion generated radical ion [Y-H]−• at m/z 300 [44]. Similarly, all these compounds and 45 (quercetin) produced fragment ions at m/z 271 and 255 due to loss of [Y-CHO]− and [CO+H2O]−, respectively. Identification of compounds 16, 26 and 45 was also confirmed by comparison of RT and MS/MS spectra with the authentic standards. Compounds 21 and 26 were isomers which showed the same MS/MS fragment ions with different relative abundance at 13.04 and 13.42 min, respectively. All compounds also showed Retro Diels Alder (RDA) fragment ion at m/z 151 due to B-ring cleavage (Table 1).
Seven compounds, namely, 25 (robinin), 28 (kaempferol-3-O-rutinoside), 29 (kaempferol- hexoside), 38 (kaempferol-3-O- hexoside), 40 (kaempferol derivatives), 41 (kaempferol 3-O-glucuronide) and 43 (kaempferol-O-hexoside), were identified as kaempferol derivatives. The characteristic fragment ion at m/z 285 [Y]− was observed in all the compounds (25, 28, 29, 38, 40, 41 and 43) due to loss of C18H31O13, C6H10O5, C12H20O9, C6H10O5, C5H8O4, C6H8O6 and C6H10O4, respectively. Fragment ion [Y-H]− was observed as a radical anion at m/z 284 due to loss of H radical. All these compounds and 48 (kaempferol) produced fragment ions at m/z 255 and 227 due to loss of CHO and 2CHO. Compounds 28 and 48 were also confirmed with authentic standards (Table 2).
Compounds 1, 2, 4, 37 and 44 were identified as quinic acid, caffeic acid, gallic acid, coumaric acid and protocatechuic acid by comparison of RT and MS/MS with their standards. MS/MS spectra of compounds 2, 4, 37 and 44 showed fragment ions at m/z 135, 125, 119 and 109, respectively due to loss of CO2. Compound 5 was identified as vanillic acid which showed fragment ions at m/z 151 and 123 due to loss of CH3 and CO2, respectively. Fragment ions at m/z 151 and 123 produced common fragment ion at m/z 107 due to losses of HCO2 and CH4, respectively. Compound 6 was identified as gentisic acid-O-hexoside which showed fragment ion at m/z 152 due to loss of hexoside.
Compounds 9 (methyl gallate) and 23 (ethyl gallate) were identified as gallates of gallic acid which gave characteristic fragment ion at m/z 169 due to loss of CH3 and C2H5, respectively. MS/MS spectra of both compounds showed fragment ion at m/z 125 as base peak. Compound 19 was identified as brevifolin and other compounds 4 (brevifolincarboxylic acid), 14 (methyl brevifolincarboxylate), 26 (ethyl brevifolincarboxylate), and 28 (propyl-O-methyl brevifolin) were its derivatives. Compound 4 showed fragment ion at m/z 247 due to loss of CO2 whereas fragment ions at m/z 219, 191 and 175 were observed due to successive losses of CO. Fragment ion at m/z 273 was observed in compounds 14 and 26 due to loss of CH3OH and C2H5OH, respectively whereas other fragment ions were formed due to consecutive loss of CO. Compound 19 also showed major fragment ions at m/z 219 and 191 due to consecutive loss of CO. Similarly, compound 28 showed fragment ions at m/z 247 and 245 due to loss of C3H7 and CO2, respectively. Compounds 7, 9 and 14 were identified as catechin and epicatechin catechin 3-gallate, respectively. Compound 7 and 9 were isomers and showed the same fragment ions with different relative abundance. They were also confirmed by comparison with their standards.
Seven compounds, namely, 10 (ellagic acid-O-dihexoside), 11 (ellagic acid-O-hexoside), 12 (ellagic acid-O-glucuronide), 23 (ellagic acid-O-arabinoside), 42 (methy-O-ellagic acid), 47 (dimethyl-O-ellagic acid) and 49 (trimethyl-O-ellagic acid), were identified as ellagic acid derivatives. Compounds 10, 11, 12 and 23 showed characteristic fragment ion at m/z 300 due to loss of C12H20O10, C6H10O5, C6H8O6 and C5H8O4, respectively. Similarly, compounds 42, 47 and 49 showed fragment ions at m/z 299, 314 and 328, respectively, due to loss of CH3. Compound 35 showed fragment ions at m/z 283 and 245 due to loss of H2O and 2CO. Compounds 15, 35, 39, 42, 46, 50 and 51 were identified as chrysin, ellagic acid, eriodictyol, methy-O-ellagic acid, luteolin, betulinic acid and oleanolic acid, respectively and confirmed by comparison of RT and MS/MS spectra with their standards (Table 3).
4. Discussion
Most of the qualitative and quantitative analyses of phenolics in Phyllanthus species are reported by HPLC or HPTLC based on their RT and UV data [17], [18], [19]. Identification and distribution of 15 compounds are reported in P. amarus, P. stipulatus, P. niruri and P. tenellus in 60 min based on unit mass resolution only [29]. Yang et al. [30] have also identified hydrolysable tannins and other phenolic compounds in 65 min from P. emblica fruit using HPLC-DAD-ESI(−)-QTOF-MS/MS [30]. Recently, fingerprinting and identification in P. amarus and P. niruri using LC–MS/MS analysis have been reported by some authors independently [31], [32], [33]. In the our previous report, 11 compounds (gallic acid, protocatechuic acid, caffeic acid, quercetin, ellagicacid, rutin, keamferol-3-O-rutinoside, luteolin, kaempferol, quinic acid and ursolic acid) were unambiguously identified and characterized whereas the other 41 compounds were tentatively identified and characterized. Only five most abundant compounds were quantified in ethanolic extracts of P. amarus samples collected from three different locations [35].
HPLC-ESI-QTOF-MS/MS facilitates the identification and characterization of known and unknown compounds on the basis of their molecular formula, exact mass measurements and MS/MS fragmentations [44], [45]. It also differentiates isobaric compounds by exact masses with different elemental compositions. In addition, HPLC-ESI-QTOF-MS provides separation and targeted fragmentation of any particular ion of interest, which may contribute to structural elucidation and isomer distinction [44], [45], [46]. Analysis of phenolics is reported in positive and negative ionization modes [41], [42]. But negative ionization mode is found more sensitive for the analysis of these compounds [35], [40], [41], [42], [43]. In the present work, we have selected four Phyllanthus species plants or parts, namely, P. emblica, P. fraternus, P. amarus and P. niruri which are commonly used as medicine. Therefore, the comparative fingerprints of P. emblica, P. fraternus, P. amarus and P. niruri were generated using HPLC-ESI-QTOF-MS/MS in negative ionization mode. Twenty-one compounds were unambiguously identified and characterized by comparison (RT and MS/MS spectra) with authentic standards whereas 30 compounds were tentatively identified and characterized with the help of templates (reference compounds). Exact mass measurements and characteristic diagnostic fragment ions were used to identify the compounds which are more accurate and authentic than earlier reported methods. This method was initially developed on P. amarus extract and applied on other selected plants to test its suitability. Results proved the applicability of the developed method on various plants/parts of Phyllanthus species. Distribution of all the compounds is also reported according to the plant parts.
5. Conclusion
Optimization of suitable collision energies and identification of diagnostic fragment ions of rutin, quercetin-3,4-di-O-glucoside and kaempferol-3-O-rutinoside were successfully completed. HPLC-ESI-QTOF-MS/MS method was developed for the identification, characterization and distribution of phenolics on the basis of identified diagnostic fragment ions of flavonoides and reported diagnostic fragment ions of phenolic acids and other compounds in 35 min run time in the crude extracts of Phyllanthus species plants/parts. Total 51 compounds including 21 were unambiguously identified and characterized on comparison with their standards whereas remaining 30 were tentatively identified and characterized. Most of these compounds are reported for the first time in P. fraternus and P. niruri.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Acknowledgments
Grateful acknowledgments are made to SAIF CSIR-CDRI, Lucknow, India, where all mass spectral studies were done. Sunil Kumar is thankful to CSIR, New Delhi, India, for financial support and BK for NMPB grant GO/UP/03/09. The authors are also thankful to Dr. Bikarma Singh, Scientist (Biodiversity and Applied Botany Division, CSIR-Indian Institute of Integrative Medicine, Canal Road, Jammu-180001, India) for providing plant samples. CDRI communication number is 9420.
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jpha.2017.01.005.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Sarin B., Verma N., Martín J.P. An overview of important ethnomedicinal herbs of Phyllanthus species: present status and future prospects. Sci. World J. 2014:1–12. doi: 10.1155/2014/839172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verma S., Sharma H., Garg M. Phyllanthus amarus: a review. J. Pharmacogn. Phytochem. 2014;3:18–22. [Google Scholar]
- 3.Mao X., Wu L.F., Guo H.L. The genus Phyllanthus: an ethnopharmacological, phytochemical, and pharmacological review. Evid. Based Complement. Altern. Med. 2016 doi: 10.1155/2016/7584952. 〈http://dx.doi.org/10.1155/2016/7584952〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagalkotkar G., Sagineedu S.R., Saad M.S. Phytochemicals from Phyllanthus niruri Linn. and their pharmacological properties: a review. J. Pharm. Pharmacol. 2006;58:1559–1570. doi: 10.1211/jpp.58.12.0001. [DOI] [PubMed] [Google Scholar]
- 5.Patel J.R., Tripathi P., Sharma V. Phyllanthus amarus: ethnomedicinal uses, phytochemistry and pharmacology: a review. J. Ethnopharmacol. 2011;138:286–313. doi: 10.1016/j.jep.2011.09.040. [DOI] [PubMed] [Google Scholar]
- 6.Tan W.C., Jaganath I.B., Manikam R. Evaluation of antiviral activities of four local Malaysian Phyllanthus species against herpes simplex viruses and possible antiviral target. Int. J. Med. Sci. 2013;10:1817–1829. doi: 10.7150/ijms.6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashim A., Khan M.S., Khan M.S. Antioxidant and α-amylase inhibitory property of Phyllanthus virgatus L.: an in vitro and molecular interaction study. Biomed. Res. Int. 2013:729393. doi: 10.1155/2013/729393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sulaiman S.F., Ooi K.L. Antioxidant and α-glucosidase inhibitory activities of 40 tropical juices from Malaysia and identification of phenolics from the bioactive fruit juices of Barringtonia racemosa and Phyllanthus acidus. J. Agric. Food Chem. 2014;62:9576–9585. doi: 10.1021/jf502912t. [DOI] [PubMed] [Google Scholar]
- 9.Ofuegbe S.O., Adedapo A.A., Adeyemi A.A. Anti-inflammatory and analgesic activities of the methanol leaf extract of Phyllanthus amarus in some laboratory animals. J. Basic Clin. Physiol. Pharmacol. 2014;25:175–180. doi: 10.1515/jbcpp-2013-0084. [DOI] [PubMed] [Google Scholar]
- 10.Amin Z.A., Alshawsh M.A., Kassim M. Gene expression profiling reveals underlying molecular mechanism of hepatoprotective effect of Phyllanthus niruri on thioacetamide-induced hepatotoxicity in Sprague Dawley rats. BMC Complement Altern. Med. 2013;13:1–10. doi: 10.1186/1472-6882-13-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh B., Sharma R.A. Antioxidant and antimicrobial activities of callus culture and fruits of Phyllanthus emblica L. J. Herbs Spices Med. Plants. 2015;21:230–242. [Google Scholar]
- 12.Mahata S., Pandey A., Shukla S. Anticancer activity of Phyllanthus emblica Linn. (Indian Gooseberry): inhibition of transcription factor AP-1 and HPV gene expression in cervical cancer cells. Nutr. Cancer. 2013;65:88–97. doi: 10.1080/01635581.2013.785008. [DOI] [PubMed] [Google Scholar]
- 13.Couto A.G., Kassuya C.A., Calixto J.B. Anti-inflammatory, antiallodynic effects and quantitative analysis of gallic acid in spray dried powders from Phyllanthus niruri leaves, stems, roots and whole plant. Rev. Bras. Farmacogn. 2013;23:124–131. [Google Scholar]
- 14.Qi W., Hua L., Gao K. Chemical constituents of the plants from the genus Phyllanthus. Chem. Biodiver. 2014;11:364–395. doi: 10.1002/cbdv.201200244. [DOI] [PubMed] [Google Scholar]
- 15.Saha H., Srikkanth A., Sikchi S. Comparative evaluation of antimicrobial and anti-inflammatory activities of Ocimum sanctum, Phyllanthus niruri and Cadaba fruticosa: an in vitro approach with emphasis on detection of their bioactive compounds using GC–MS. Int. J. Biol. Chem. 2015;9:235–248. [Google Scholar]
- 16.Maity S., Chatterjee S., Variyar P.S. Evaluation of antioxidant activity and characterization of phenolic constituents of Phyllanthus amarus root. J. Agric. Food Chem. 2013;61:3443–3450. doi: 10.1021/jf3046686. [DOI] [PubMed] [Google Scholar]
- 17.Tripathi A.K., Verma R.K., Gupta A.K. Quantitative determination of phyllanthin and hypophyllanthin in Phyllanthus species by high‐performance thin layer chromatography. Phytochem. Anal. 2006;17:394–397. doi: 10.1002/pca.936. [DOI] [PubMed] [Google Scholar]
- 18.Dey T.B., Chakraborty S., Jain K.K. Antioxidant phenolics and their microbial production by submerged and solid state fermentation process: a review. Trends Food Sci. Technol. 2016;53:60–74. [Google Scholar]
- 19.Dhalwal K., Biradar Y.S., Rajani M. High-performance thin-layer chromatography densitometric method for simultaneous quantitation of phyllanthin, hypophyllanthin, gallic acid, and ellagic acid in Phyllanthus amarus. J. AOAC Int. 2006;89:619–623. [PubMed] [Google Scholar]
- 20.Nayak P.S., Upadhyay A., Dwivedi S.K. Quantitative determination of phyllanthin in Phyllanthus amarus by high performance thin layer chromatography. Bol. Latinoam. Caribe Plant Med. Aromat. 2010;9:353–358. [Google Scholar]
- 21.Sparzak B., Krauze-Baranowska M., Pobocka-Olech L. High-performance thin-layer chromatography densitometric determination of beta-sitosterol in Phyllanthus species. J. AOAC Int. 2009;92:1343–1348. [PubMed] [Google Scholar]
- 22.Srivastava V., Singh M., Malasoni R. Original paper separation and quantification of lignans in Phyllanthus species by a simple chiral densitometric method. J. Sep. Sci. 2008;31:47–55. doi: 10.1002/jssc.200700282. [DOI] [PubMed] [Google Scholar]
- 23.Murugaiyah V., Chan K.L. Determination of four lignans in Phyllanthus niruri L. by a simple high-performance liquid chromatography method with fluorescence detection. J. Chromatogr. A. 2007;1154:198–204. doi: 10.1016/j.chroma.2007.03.079. [DOI] [PubMed] [Google Scholar]
- 24.Dhooghe L., Meert H., Cimanga R.K. The quantification of ellagic acid in the crude extract of Phyllanthus amarus Schum. & Thonn. (Euphorbiaceae) Phytochem. Anal. 2011;22:361–366. doi: 10.1002/pca.1288. [DOI] [PubMed] [Google Scholar]
- 25.Viaene J., Goodarzi M., Dejaegher B. Discrimination and classification techniques applied on Mallotus and Phyllanthus high performance liquid chromatography fingerprints. Anal. Chim. Acta. 2015;877:41–50. doi: 10.1016/j.aca.2015.02.034. [DOI] [PubMed] [Google Scholar]
- 26.Bansal V., Sharma A., Ghanshyam C. Coupling of chromatographic analyses with pretreatment for the determination of bioactive compounds in emblica officinalis juice. Anal. Methods. 2014;6:410–418. [Google Scholar]
- 27.Shanker K., Singh M., Srivastava V. Simultaneous analysis of six bioactive lignans in Phyllanthus species by reversed phase hyphenated high performance liquid chromatographic technique. Acta Chromatogr. 2011;23:321–337. [Google Scholar]
- 28.Niu X., Qi L., Li W. Simultaneous analysis of eight phenolic compounds in Phyllanthus simplex Retz by HPLC–DAD-ESI/MS. J. Med. Plants Res. 2012;6:1512–1518. [Google Scholar]
- 29.da Fontoura-Sprenger R., Cass Q.B. Characterization of four Phyllanthus species using liquid chromatography coupled to tandem mass spectrometry. J. Chromatogr. A. 2013;1291:97–103. doi: 10.1016/j.chroma.2013.03.030. [DOI] [PubMed] [Google Scholar]
- 30.Yang B., Kortesniemi M., Liu P. Analysis of hydrolyzable tannins and other phenolic compounds in Emblic leaf flower (Phyllanthus emblica L.) fruits by high performance liquid chromatography–electrospray ionization mass spectrometry. J. Agric. Food Chem. 2012;60:8672–8683. doi: 10.1021/jf302925v. [DOI] [PubMed] [Google Scholar]
- 31.Sousa A.D., Maia A.I.V., Rodrigues T.H.S. Ultrasound-assisted and pressurized liquid extraction of phenolic compounds from Phyllanthus amarus and its composition evaluation by UPLC-QTOF. Ind. Crops Prod. 2016;79:91–103. [Google Scholar]
- 32.Guo J., Chen Q., Wang C. Comparison of two exploratory data analysis methods for classification of Phyllanthus chemical fingerprint: unsupervised vs. supervised pattern recognition technologies. Anal. Bioanal. Chem. 2015;407:1389–1401. doi: 10.1007/s00216-014-8371-x. [DOI] [PubMed] [Google Scholar]
- 33.Kaur B., Kaur N. Metabolic fingerprinting of different populations of Phyllanthus niruri L. from Punjab using electrospray ionization mass spectrometry (ESI–MS) Med. Chem. Res. 2016;25:2798–2821. [Google Scholar]
- 34.Avula B., Wang Y.H., Wang M. Simultaneous determination and characterization of tannins and triterpene saponins from the fruits of various species of Terminalia and Phyllantus emblica using a UHPLC-UV-MS method: application to Triphala. Planta Med. 2013;29:181–188. doi: 10.1055/s-0032-1328089. [DOI] [PubMed] [Google Scholar]
- 35.Kumar S., Chandra P., Bajpai V. Rapid qualitative and quantitative analysis of bioactive compounds from Phyllanthus amarus using LC/MS/MS techniques. Ind. Crops Prod. 2015;69:143–152. [Google Scholar]
- 36.Mamza U.T., Sodipo O.A., Khan I.Z. Gas chromatography-mass spectrometry (GC-MS) analysis of bioactive components of Phyllanthus amarus leaves. Int. J. Plant Sci. 2012;3:208–215. [Google Scholar]
- 37.Wang C.Y., Lee S.S. Analysis and identification of lignans in Phyllanthus urinaria by HPLC‐SPE‐NMR. Phytochem. Anal. 2005;16:120–126. doi: 10.1002/pca.830. [DOI] [PubMed] [Google Scholar]
- 38.Mediani A., Abas F., Khatib A. Phytochemical and biological features of Phyllanthus niruri and Phyllanthus urinaria harvested at different growth stages revealed by 1 H NMR-based metabolomics. Ind. Crops Prod. 2015;77:602–613. [Google Scholar]
- 39.Lam S.H., Wang C.Y., Chen C.K. Chemical investigation of Phyllanthus reticulatus by HPLC‐SPE‐NMR and conventional methods. Phytochem. Anal. 2007;18:251–255. doi: 10.1002/pca.979. [DOI] [PubMed] [Google Scholar]
- 40.Cuyckens F., Claeys M. Mass spectrometry in the structural analysis of flavonoids. J. Mass Spectrom. 2004;39:1–15. doi: 10.1002/jms.585. [DOI] [PubMed] [Google Scholar]
- 41.Cuyckens F., Rozenberg R., de Hoffmann E. Structure characterization of flavonoid O‐diglycosides by positive and negative nano‐electrospray ionization ion trap mass spectrometry. J. Mass Spectrom. 2001;36:1203–1210. doi: 10.1002/jms.224. [DOI] [PubMed] [Google Scholar]
- 42.Mercolini L., Protti M., Saracino M.A. Analytical profiling of bioactive phenolic compounds in argan (Argania spinosa) leaves by combined microextraction by packed sorbent (MEPS) and LC–DAD‐MS/MS. Phytochem. Anal. 2016;27:41–49. doi: 10.1002/pca.2585. [DOI] [PubMed] [Google Scholar]
- 43.Madala N.E., Piater L., Dubery I. Distribution patterns of flavonoids from three Momordica species by ultra-high performance liquid chromatography quadrupole time of flight mass spectrometry: a metabolomics profiling approach. Rev. Bras. Farm. 2016;26:507–513. [Google Scholar]
- 44.Kumar S., Singh A., Bajpai V. Structural characterization of monoterpene indole alkaloids in ethanolic extracts of Rauwolfia species by liquid chromatography with quadrupole time-of-flight mass spectrometry. J. Pharm. Anal. 2016;6:363–373. doi: 10.1016/j.jpha.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ling Y., Lin Z., Zha W. Rapid detection and characterisation of triterpene saponins from the root of Pulsatilla chinensis (bunge) regel by HPLC‐ESI‐QTOF‐MS/MS. Phytochem. Anal. 2016;27:174–183. doi: 10.1002/pca.2613. [DOI] [PubMed] [Google Scholar]
- 46.Ling Y., Fu Z., Zhang Q., Xu L. Identification and structural elucidation of steroidal saponins from the root of Paris polyphylla by HPLC-ESI–QTOF-MS/MS. Nat. Prod. Res. 2015;29:1798–1803. doi: 10.1080/14786419.2015.1007137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material