Abstract
The quenching interaction of atomoxetine (ATX) with bovine serum albumin (BSA) was studied in vitro under optimal physiological condition (pH=7.4) by multi-spectroscopic techniques. The mechanism of ATX-BSA system was a dynamic quenching process and was confirmed by the fluorescence spectra and lifetime measurements. The number of binding sites, binding constants and other binding characteristics were computed. Thermodynamic parameters ∆H° and ∆S° indicated that intermolecular hydrophobic forces predominantly stabilized the drug-protein system. The average binding distance between BSA and ATX was studied by Försters theory. UV-absorption, Fourier transform infrared spectroscopy (FT-IR), circular dichroism (CD), synchronous spectra and three-dimensional (3D) fluorescence spectral results revealed the changes in micro-environment of secondary structure of protein upon the interaction with ATX. Displacement of site probes and the effects of some common metal ions on the binding of ATX with BSA interaction were also studied.
Keywords: Atomoxetine, Bovine serum albumin, 3D fluorescence spectra, FT-IR, Energy transfer, Lifetime measurement
1. Introduction
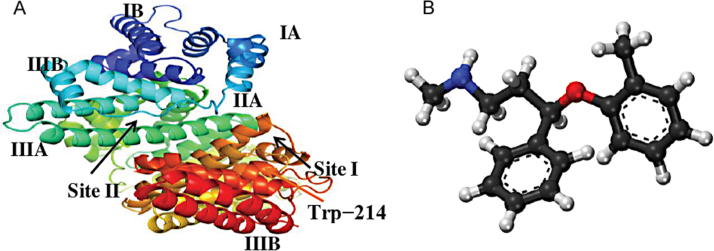
Protein plays a significant role in the living organisms by performing various biological activities. Serum albumin is employed as a model for studying drug–protein interaction in vitro. Many drugs and other bioactive small molecules bind reversibly to albumin and other serum components so that they function as carriers [1]. This type of interaction can also influence the drug stability and toxicity during the chemotherapeutic process [2]. They play a dominant role in the transport and deposition of endogenous and exogenous functional groups in blood, since serum albumins often increase the apparent solubility of hydrophobic drugs in plasma and modulate their delivery to cells in vivo and in vitro. They also play an important role in storage and transport of energy [3]. The binding ability of drug-protein in blood stream may have a significant impact on distribution, free concentration and metabolism of drug. Bovine serum albumin (BSA) (Fig. 1A) is well suited to these initial studies, since it has been extensively characterized [4]. BSA consists of 582 amino acids with 2 tryptophans located at position 134 (located on the surface of domain I) and 214 (located within the hydrophobic pocket of domain II). Tryptophan residues are the main intrinsic fluorophores that are extremely sensitive to their microenvironment. The knowledge on the mechanism of interaction between the drug and plasma protein is of crucial importance to understanding the pharmacodynamics and pharmacokinetics of a drug [5]. Therefore, the studies on the interactions of a bioactive compound with BSA assume significance in chemistry, life sciences and clinical medicine.
Fig. 1.
(A) Pictorial structure of bovine serum albumin and (B) chemical structure of atomoxetine.
Atomoxetine (ATX) hydrochloride is known chemically as N-methyl-γ-(2-methylphenoxy)-benzene propanamine hydrochloride (Fig. 1B) (Trade name: Strattera). ATX is a potent norepinephrine reuptake inhibitor of the presynaptic norepinephrine transporter with minimal affinity for other monoamine transporters or receptors [6], [7]. ATX has been used as a first-line therapeutic agent for the treatment of attention deficit/hyperactivity disorder (ADHD) in children, adolescents and adults [8].
The literature survey revealed that attempts have not been made so far to investigate the binding mechanism of ATX with BSA by spectroscopic techniques. The purpose of this study was to understand the interaction mechanism of ATX with BSA by investigating the binding parameters such as binding constants, the number of binding sites, thermodynamic parameters, the effect of some common metal ions, site probes and the conformational change of BSA with ATX using multiple spectroscopic techniques.
2. Experimental
2.1. Reagents and chemicals
BSA and ATX were purchased from Sigma Aldrich Bangalore, India and used as such. Site probes warfarin, ibuprofen and digitoxin received from Sigma Chemical Company, Bangalore, India, were initially dissolved in 5% methanol-water and then diluted with distilled water. The solutions of BSA, site probes and metal ions were prepared in 0.1 M phosphate buffer (0.1 M NaH2PO4 and 0.1 M Na2HPO4) of physiological pH 7.4. BSA solution was prepared based on the molecular weight 65,000 and ATX solution was prepared in methanol. All other materials were of analytical reagent grade and Millipore water was used throughout the experiment.
2.2. Instrumentation
Fluorescence spectra were recorded using an RF-5301 PC Hitachi Spectrofluorometer Model F-2000 (Tokyo, Japan) with a 150 W Xenon lamp, a 1 cm quartz cell and thermostatic cuvette holder. The excitation and emission bandwidths were both 5 nm and the scan rate was 1200 nm/s. The temperature of the sample was maintained by recycling water throughout the experiment. Circular dichroism (CD) measurements were recorded on a Jasco J-715 spectropolarimeter (Tokyo, Japan) with a 0.2 cm quartz cell. The absorption spectra were recorded on a double beam CARY 50-BIO UV–visible spectrophotometer (Victoria, Australia) with a scan rate of 600 nm/min. Fourier transform infrared (FT-IR) spectroscopic measurements were made at room temperature on a Nicolet 5700 FT-IR spectrometer equipped with a germanium attenuated total reflection (ATR) accessory, a DTGS KBr detector and a KBr beam splitter. The pH measurements were performed with an Elico LI120 pH meter (Elico Ltd., India). Fluorescence lifetime measurements were carried out in an ISS', Chronos BH fluorescence lifetime spectrometer (ISS Inc, USA).
2.3. Procedure
2.3.1. Fluorescence quenching of ATX–protein interaction study
A stock solution of 250 µM BSA and ATX were prepared in phosphate buffer solution (pH=7.4). An appropriate concentration of the BSA solution (5 µM from 250 µM stock) and ATX solution (5 µM from 250 µM stock) were transferred into a 5 mL conical flask containing 2 mL of phosphate buffer solution (pH 7.4) and then were shaken. On the basis of preliminary experiments, BSA concentration was fixed at 5 µM and drug concentration was varied from 5 µM to 45 µM. Fluorescence spectra were recorded at three different temperatures (288, 298 and 308 K) in the range of 290–550 nm upon excitation at wavelength of 296 nm in each case.
2.3.2. Fluorescence lifetime measurement
The fluorescence lifetime measurements of BSA in the presence and absence of ATX were recorded by fixing 296 nm as the excitation wavelength and 343 nm as the emission wavelength. The BSA concentration was fixed at 5 µM while the drug concentration was varied from 5 µM to 15 µM in the presence of phosphate buffer at room temperature.
2.3.3. UV measurements
The UV measurements of BSA in the presence and absence of ATX were made in the range of 200–400 nm. BSA concentration was fixed at 5 µM while the drug concentration was varied from 5 µM to 45 µM in the presence of phosphate buffer as a solvent.
2.3.4. Synchronous fluorescence measurements
The synchronous fluorescence spectral measurements of ATX–BSA were recorded at different scanning intervals of ∆λ (∆λ=∆λex −∆λem). The spectral behavior of tyrosine and tryptophan residues of BSA was observed at ∆λ=15 nm and ∆λ=60 nm, respectively. The spectra were recorded in the range of 200–500 nm. The concentration of BSA was fixed at 5 µM whereas the concentration of ATX was varied from 5 µM to 45 µM.
2.3.5. CD measurements
The CD spectra of BSA in the presence and absence of ATX at 298 K were recorded in the range of 200–250 nm for three concentrations of ATX (5 µM, 10 µM and 15 µM) and the BSA concentration was fixed at 5 µM in the presence of phosphate buffer.
2.3.6. 3D fluorescence spectra
The 3D fluorescence spectra of BSA were recorded with and without ATX. Protein solution at 5 μM was transferred to a quartz cell, diluted with 2.0 mL phosphate buffer and mixed well. To this, 5 µM of ATX was added and the 3D fluorescence spectra were recorded by scanning excitation wavelength in the range of 200–500 nm and emission wavelength from 200 to 600 nm at an interval of 10 nm. The scanning parameters were the same as in the fluorescence quenching experiments.
2.3.7. FT-IR measurements
The FT-IR spectra of BSA in the presence and absence of ATX at 298 K were recorded in the range of 1200–1800 cm−1. The concentrations of BSA and ATX were 5 µM in the presence of phosphate buffer.
2.3.8. Competitive binding studies
The competitive binding studies were performed using different site probes, viz., warfarin, ibuprofen and digitoxin for site I, II and III, respectively [9] by keeping the concentration of BSA and the probe constant (5 µM each). The fluorescence quenching titration was used as before to determine the binding constant of BSA–ATX in the presence of above site probes.
2.3.9. Effects of some common metal ions
The effects of some common metal ions, viz., Co2+(CoCl2), Cu2+(CuCl2), Ni2+(NiCl2), Ca2+(CaCl2) and Zn2+(ZnCl2) were investigated on ATX-BSA interactions. The fluorescence spectra of ATX-BSA system were recorded in the range of 290–550nm upon the excitation at 296 nm. The overall concentration of BSA and that of the common ions was fixed at 5 µM.
3. Results and discussion
3.1. Fluorescence quenching of BSA by ATX
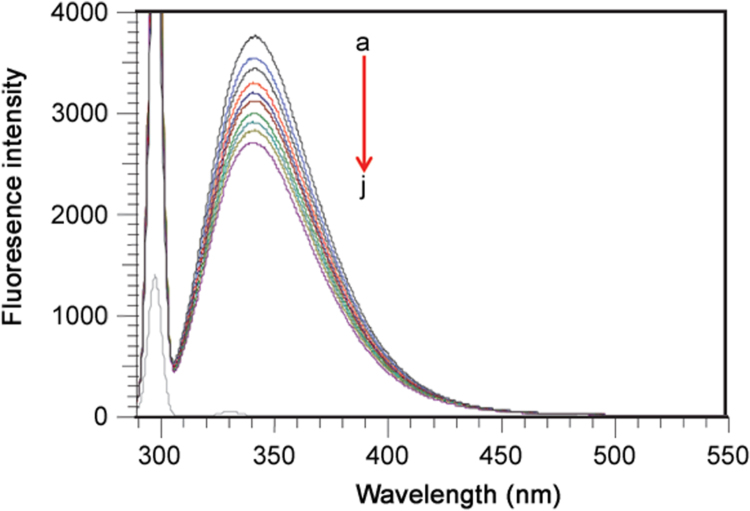
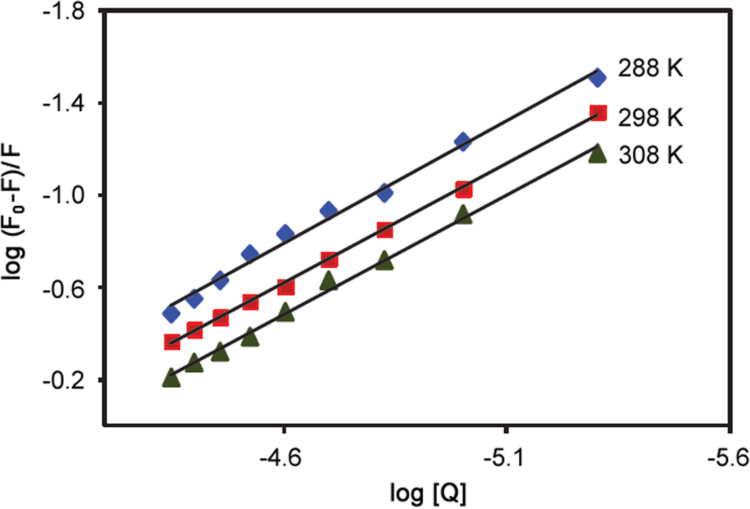
Fluorescence methods have been widely used to investigate the interaction between ligands and proteins and can give information about the quenching mechanism, binding constants and binding sites. We utilized the technique to study the interaction between ATX and BSA and the fluorescence spectra of BSA at increasing concentrations of ATX are shown in Fig. 2. The fluorescence intensity of BSA decreased regularly with an increased concentration of ATX, with an emission wavelength shift from 342 nm to 338 nm (blue shift), which indicated that ATX could bind to BSA. The different mechanisms of fluorescence quenching are usually classified as dynamic quenching or static quenching [10]. Dynamic quenching results from interaction through collision between the fluorophore and the quencher and static quenching is caused by formation of a non-fluorescent ground state fluorophore–quencher complex. Dynamic quenching and static quenching can be distinguished by their different dependence upon the temperature of quenching constants or viscosity, or by lifetime measurements [11], [12]. The quenching rate constants decreased with increased temperature for static quenching, but the reverse effect is observed in the case of dynamic quenching [13]. In both cases, molecular contact is required between the flurophore and the quencher for fluorescence quenching [14]. To confirm the mechanism and a possible quenching mechanism, the fluorescent quenching data were subjected to well-known Stern-Volmer [15] Eq. (1)
| (1) |
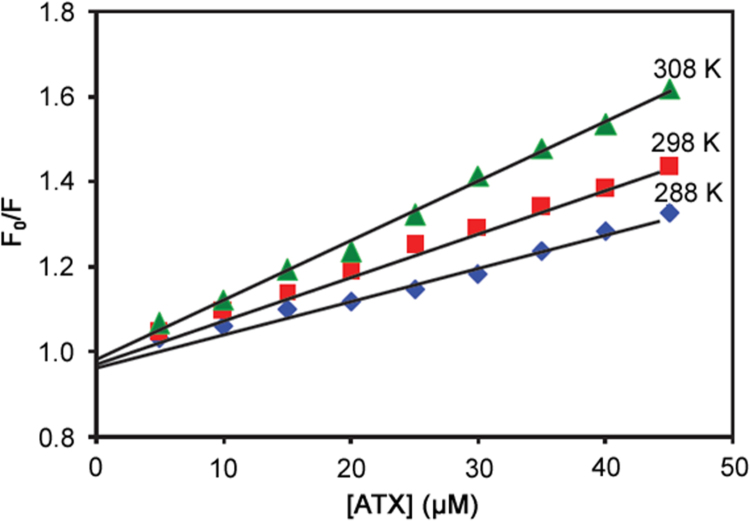
where F0 and F are the fluorescence intensities of BSA in the absence and presence of the quencher, KSV is the Stern-Volmer quenching constant, [Q] is the concentration of the quencher, kq is the quenching rate constant of the BSA, and τ0 is the average lifetime of the biomolecule BSA without quencher [16]. The Stern-Volmer plots of interactions carried out at different temperatures (288, 298 and 308 K) were observed to be linear (Fig. 3). The Stern-Volmer quenching constant KSV and the correlation coefficient of each curve were calculated from the slope of the regression curves. The values of KSV for ATX-BSA system at 288, 298, and 308 K were found to be 0.73×104 L/mol (r ≥0.983, SD ≤0.005), 0.97×104 L/mol (r ≥0.999, SD ≤0.025), and 1.43×104 L/mol (r ≥0.995, SD ≤0.005), respectively; this indicates that the quenching mechanism between ATX and BSA was a dynamic type [17]. The quenching rate constant can be calculated using the following Eq. (2)
| (2) |
Fig. 2.
Fluorescence quenching spectra of BSA–ATX system with increased concentration of ATX (a) 0 µM, (b) 5 µM, (c)10 µM, (d) 15 µM, (e) 20 µM, (f) 25 µM, (g) 30 µM, (h) 35 µM, (i) 40 µM, and (j) 45 µM while BSA concentration was fixed at 5 µM in physiological pH 7.4 PBS buffer at 298 K.
Fig. 3.
The Stern-Volmer curves for quenching of ATX with BSA at different temperatures. λex =296 nm; λem =355 nm and [BSA] =5 µM.
The fluorescence lifetime of the biopolymer is 10−8 s [18]. The quenching rate constant at 298 K was calculated to be 0.97×1012 L/mol s and the values at different temperatures are listed in Table 1. The quenching rate constant values increased with increased temperature, which supports the dynamic quenching interaction between ATX and BSA. However, the maximum scatter collision quenching constant, k՛q of various quenchers with the biomolecule is 2×1010 L/mol s [16], [19] Hence the quenching rate constant of ATX-BSA system is greater than quenching constant of the biomolecule.
Table 1.
Stern-volmer quenching constant, binding constant and thermodynamic parameters of ATX-BSA system.
| Temp (K) | Ksv (×10−4) (L/mol) | kq (×10–12) (L/mol s) | K (×10–4) (L/mol) | n | ∆H0 (kJ/mol) | ∆S0 (J/K mol) | ∆G0298 (kJ/mol) |
|---|---|---|---|---|---|---|---|
| 288 | 0.73 | 0.73 | 1.23 | 1.06 | 16.1 | 134.1 | −23.86 |
| 298 | 0.97 | 0.97 | 1.49 | 1.04 | |||
| 308 | 1.43 | 1.43 | 1.90 | 1.03 |
KSV : Stern-Volmer quenching constant.
kq : Quenching rate constant.
K : Binding constant.
n : Number of binding sites.
3.2. Fluorescence lifetime measurement studies
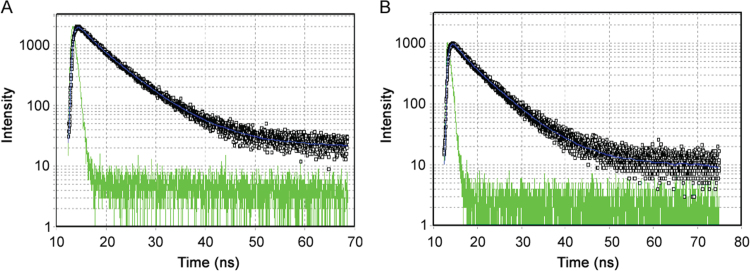
Time-resolved fluorescence lifetime measurement is an ideal nanoscale probe detection method. Here the emission of a fluorophore can be highly influenced by its environment or the presence of other interacting molecules [20]. The steady state emission spectrum lifetime decay of the excited state to the ground state is in homogeneous environment and quenching process. For these measurements the sample is exposed to a pulse of light, where the pulse width is typically shorter than the decay time of the molecules. Lifetime based measurements are rich in information and provide unique insights into the systems under investigation [19]. Time-resolved fluorescence lifetime measurements were carried out for BSA in the absence and presence of ATX (Fig. 4). Time-resolved fluorescence spectroscopy was used to determine the decay times, thus enabling the differentiation between the presence of static and dynamic quenching. The dynamic quenching constant was determined by lifetime measurements using the Eq. (3).
| (3) |
where τ0 and τ are the fluorescence lifetimes of BSA in the absence and presence of ATX, respectively; and KD is the dynamic quenching constant. The fluorescence lifetime (τ), average lifetime (<τ>), intensity fraction (f), Chi-square (χ2) and their relative amplitudes (α) are listed in Table 2. From the plot of τ0/τ versus [Q], the KD value was found to be 1.67×103 M. The value of static quenching constant, KS, was calculated [19] using the Eq. (4).
| (4) |
Fig. 4.
Time-resolved fluorescence decay for lifetime measurement spectra of (A) BSA and (B) ATX-BSA system, (pH=7.4, 298 K).
Table 2.
Lifetimes of fluorescence decay of BSA (5 µM) in phosphate buffer solution of pH 7.4 at 298 K at different concentrations of ATX.
| [ATX] (µM) | τ 1 (ns) | τ 2 (ns) | f 1 | f 2 | <τ> | α 1 | α 2 | χ2 |
|---|---|---|---|---|---|---|---|---|
| 0 | 6.54 | 2.20 | 0.866 | 0.114 | 6.17 | 13.3 | 5.62 | 1.02 |
| 5 | 6.39 | 2.04 | 0.872 | 0.128 | 6.04 | 13.4 | 6.32 | 1.04 |
| 10 | 6.32 | 1.84 | 0.876 | 0.124 | 5.86 | 13.7 | 7.74 | 1.05 |
| 15 | 6.29 | 1.75 | 0.844 | 0.156 | 5.64 | 13.9 | 8.96 | 1.06 |
τ : Fluorescence lifetime.
<τ> : Average lifetime.
f : Intensity fractions.
α : Relative amplitudes.
χ2 : Chi-square.
By plotting the graph of [(F0–F)/F]/[Q] versus [Q], the value of KS was found to be 1.08×103 M−1. It reveals that the value KD was greater than that of KS, which suggests that quenching mechanism of ATX-BSA is predominantly dynamic quenching compared with that of static quenching.
3.3. Binding parameters
When small molecules bind independently to a set of equivalent sites on a macromolecule [21], the binding constant (K) and the number of binding sites (n) can be obtained from the formula (5)
| (5) |
where F0 and F are the fluorescence intensities of protein in the absence and presence of drug, respectively; and [Q] is the concentration of drug. The values of K and n are obtained from the intercept and slope of the plot of log (F0-F)/F versus log [Q] (Fig. 5) and are shown in Table 1. The value of K increases with the increase in temperature, which also supports the dynamic quenching mechanism. The number of binding sites n is approximately equal to 1, indicating that there was one binding site in BSA for ATX during their interaction. The value of the binding constant at 288, 298, and 308 K were found to be 1.23 (r ≥0.991, SD ≤0.007), 149 (r ≥0.999, SD ≤0.005), 1.90 (r ≥0.983, SD ≤0.004) L/mol in the order of 104, indicating that a strong interaction exists between ATX and BSA. Even if a low concentration of ATX is present, ATX can interact with BSA easily.
Fig. 5.
The plots of log (F0–F)/F versus log [Q] for binding ATX to BSA at different temperatures.
3.4. Determination of the binding forces
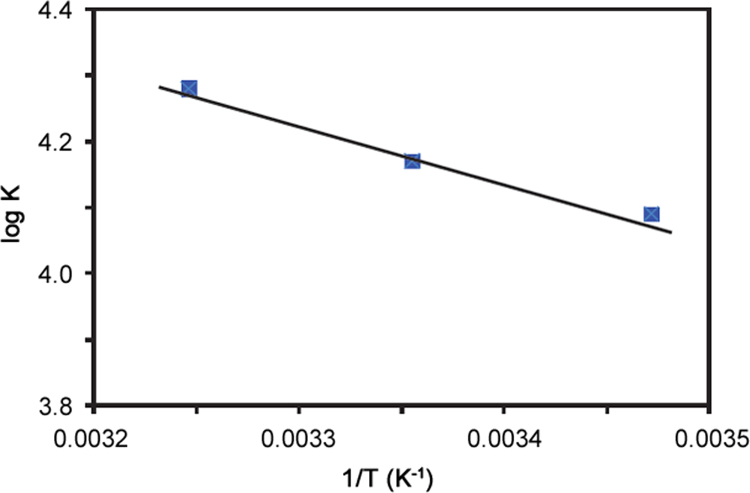
Thermodynamic measurements can help to determine the major binding forces between drug and BSA. There are four types of interactions between small molecule, ligands and biological macromolecules: hydrophobic forces, hydrogen bonds, Vander Waals’ interactions and electrostatic forces [22]. Thermodynamic parameters are important for confirming the non-covalent acting forces. Ross and Subramanian have summed up the thermodynamic laws to determine the types of binding forces [23]. The enthalpy change (∆H0), free-energy change (∆G0) and the entropy change (∆S0) for the interaction between ATX and BSA were calculated based on the van't Hoff Eq. (6)
| (6) |
where K is the binding constant at the corresponding temperature, R is the gas constant and T is the temperature. From the plot of logK vs 1/T (Fig. 6), the values of ΔH0 and ΔS0 for the binding process were obtained. The value of ΔG0 was calculated using the Eq. (7)
| (7) |
Fig. 6.
van't Hoff plot logK vs 1/T for binding of ATX with BSA.
The values of ΔH0, ΔS0 and ΔG0 are listed in Table 1. The negative value of ∆G0 indicates that the binding process is spontaneous. The positive values of ΔH0 and ΔS0 indicate that the binding process is mainly entropy driven and the enthalpy is unfavorable. The hydrophobic forces played an effective role in the reaction [24].
3.5. UV-absorption spectroscopic studies
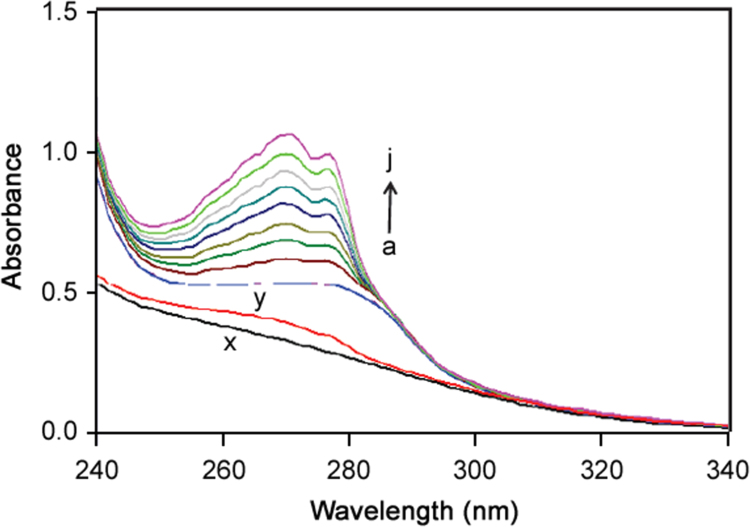
UV–visible absorption spectroscopy allows non-intrusive measurement of substances; it is a simple, useful technique to investigate conformational changes of proteins, even at the low concentrations. This method is applicable to knowing the change in hydrophobicity [25] and the interaction between the drug and protein [26]. Typically tryptophan has a wavelength of maximum absorption of 280 nm and an emission peak at the range from 300–350 nm depending on the polarity of the local environment [27], [28]. The λmax of BSA observed at around 280 nm was mainly due to the presence of amino acid residues of tryptophan and at 274 nm due to the amino acid residues of tyrosine in BSA. It was evident from the UV-spectrum that the absorption intensity of BSA at 280 nm increased regularly with increasing concentration of ATX (Fig. 7). The maximum peak position of ATX–BSA was shifted from 280 to 284 nm towards higher wavelength region. The change in λmax in the present study indicates the change in polarity around the tryptophan residue and the change in peptide strand of BSA molecules and hence the change in hydrophobicity.
Fig. 7.
Absorbance spectra of BSA–ATX system with increasing concentration of ATX (a) 0 µM, (b) 5 µM, (c)10 µM, (d) 15 µM, (e) 20 µM, (f) 25 µM, (g) 30 µM, (h) 35 µM, (i) 40 µM, and (j) 45 µM while BSA concentration was fixed at 5 µM in physiological pH 7.4 PBS buffer at 298 K. ((x) pH 7.4 PBS and (y) PBS+ATX (5 µM)).
3.6. Energy transfer between ATX and BSA
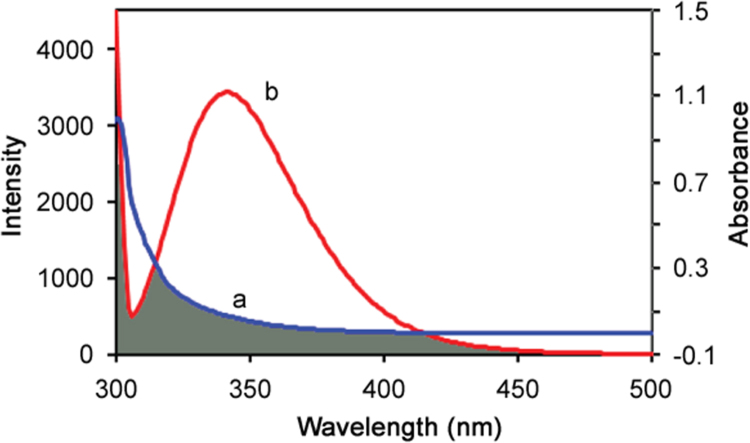
Fluorescence resonance energy transfer is an effective tool for the measurement of the distance between the drug and protein. The overlap of the UV-absorption spectra of ATX with the fluorescence emission spectra of BSA is shown in Fig. 8. The energy transfer process is very important in biochemistry; therefore, energy transfer has wide applications [29]. According to Förster's non-radiative energy transfer theory [30], the rate of energy transfer depends on (i) the relative orientation of the donor and acceptor dipoles, (ii) the extent of overlap of fluorescence emission spectrum of the donor with the absorption spectrum of the acceptor, and (iii) the distance between the donor and the acceptor. The energy transfer effect is related not only to the distance between the acceptor and donor, but also to the critical energy transfer distance R0 and the efficiency of energy transfer E. According to Förster's energy transfer theory, the energy transfer efficiency E can be calculated using the Eq. (8)
| (8) |
where F and F0 are the fluorescence intensities of BSA in the presence and absence of ATX, and the efficiency of energy transfer E was calculated using fluorescence intensities of BSA (5 µM) with and without ATX (5 µM) at 298 K. r is the distance between the acceptor and the donor, and R0 is the critical distance when the transfer efficiency is 50%. The value of R0 was evaluated using the Eq. (9)
| (9) |
where k2 is the spatial orientation factor of the dipole, N is the refractive index of the medium, Ф the fluorescence quantum yield of the donor and J is the overlap integral of the fluorescence emission spectrum of the donor and the absorption spectrum of the acceptor. J is given by the following Eq. (10)
| (10) |
where F(λ) is the fluorescence intensity of the fluorescent donor of wavelength, λ, and ε(λ) is the molar absorption coefficient of the acceptor at wavelength, λ. For ligand–BSA interaction, k2=2/3, N =1.336 and Ф=0.15 [31]. The values of J, R0, E and r were calculated to be J=2.49×10−15 cm3 L/mol, R0 =1.33 nm, E =0.042 and r=2.12 nm, respectively, at 298 K. The average distance of <7 nm between the donor and the acceptor indicated that the energy transfer from BSA to ATX occurred with high probability [32].
Fig. 8.
The overlap of (a) UV-absorption spectra of ATX (5 µM) with (b) fluorescence spectra of BSA (5 µM).
3.7. Synchronous fluorescence spectra
Synchronous fluorescence is a common spectral technique. This study is helpful for understanding the molecular environment in the vicinity of chromo sphere molecules, such as tyrosine or tryptophan residues, and has several merits, such as spectral simplification, reduction of the spectral bandwidth and avoidance of different perturbing effects [33].
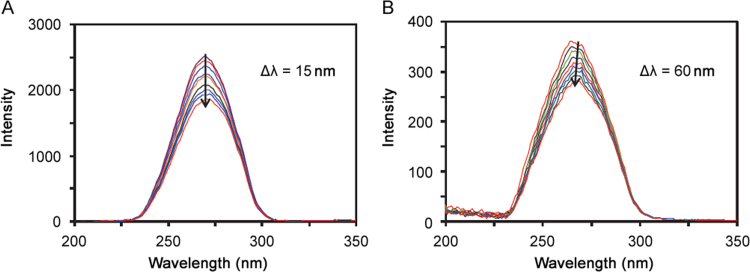
The synchronous fluorescence spectra can provide the characteristic interactions of tyrosine and tryptophan residues in BSA, when the difference between excitation and emission wavelengths (∆λ=λem–λex), was 15 nm and 60 nm respectively.
The emission peaks of tyrosine residues do not show any shift upon addition of ATX (Fig. 9A). However, tryptophan residues show a slight blue shift upon addition of ATX as shown in (Fig. 9B). This phenomenon expresses the conformational changes in tryptophan residue around which polarity is increased [34], [35].
Fig. 9.
Synchronous fluorescence spectra of BSA-ATX system with increased concentration of ATX (0–45 µM) and BSA concentration was fixed (5 µM) when (A) ∆λ=15 nm, and (B) ∆λ=60 nm, at pH 7.4 and 298 K.
3.8. CD measurements
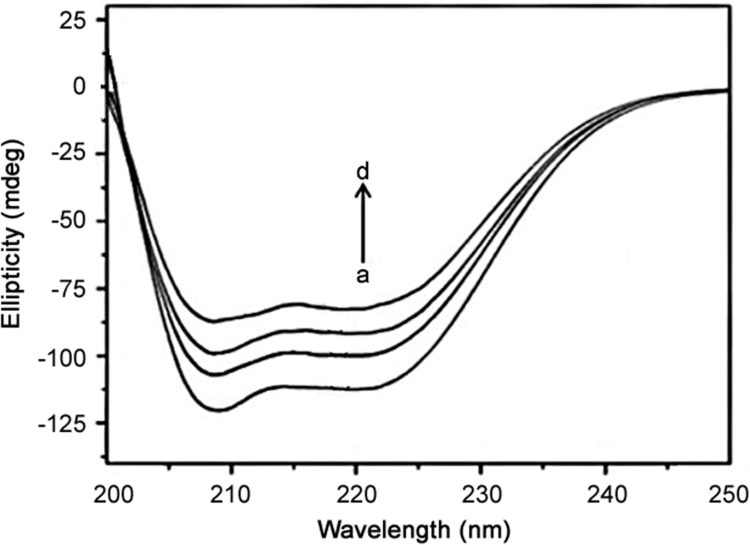
CD is a powerful tool in elucidating the modifications of the secondary structure of biopolymers as a result of interaction with small molecules [36]. The CD spectra of BSA in the absence and presence of ATX are shown in Fig. 10. The CD spectra of the protein exhibited two negative bands in UV region at 208 and 222 nm, which are characteristic of α-helical structure in a protein. The results of CD were expressed in terms of the mean residue ellipticity (MRE) according to the following Eq. (11)
| (11) |
where Cp is the molar concentration of the protein, n is the number of amino acid residues and l is the path length. The α-helical contents of free and combined protein were calculated [37] from mean MRE values at 208 nm using the Eq. (12).
| (12) |
where MRE208 is the observed MRE value at 208 nm, 4000 is the MRE of the form and random coil conformation cross at 208 nm and 33,000 is the MRE value of a pure helix at 208 nm. The CD spectra of BSA in the presence or absence of ATX showed similarity in shape.
Fig. 10.
CD spectra of (a) BSA (5 µM) (b) BSA 5 µM + ATX 5 µM, (c) BSA 5 µM + ATX 10 µM and (d) BSA 5 µM + ATX 15 µM.
The decreased content of the α-helical structure indicates that the ATX molecules bind with the amino acid residues of the main polypeptide chain and partly destroy the hydrogen bonding networks of the protein [38]. They differed from that of 60.41% (1:0) free BSA to 54.16% (1:1), 48.53% (1:2) and 42.29% (1:3) after ATX binding to BSA. This shows the binding between drug and protein with some conformational changes of BSA. The CD spectra of BSA in the presence and absence of ATX are similar in shape, which indicates that the structure of BSA after binding to ATX is α-helical in nature.
3.9. 3D fluorescence spectra
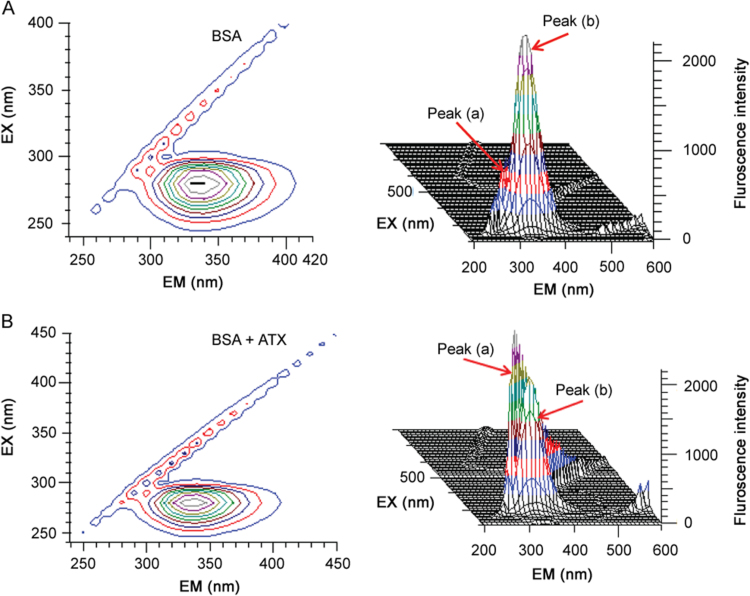
It is well known that 3D fluorescence spectra can provide more detailed information about the conformational changes of proteins. Fig. 11A presents the 3D fluorescence spectra and contour ones of BSA and BSA–ATX, respectively. The contour map displayed a bird's eye view of the fluorescence spectra. Further, from Fig. 11B, peak (a) is the Rayleigh scattering peak (λex=λem) [39]. With the addition of BSA–ATX, the fluorescence intensities of peak (a) increased. The reason for this is that when the BSA–ATX interaction occurred, the diameter of the macromolecule was increased, which in turn resulted in enhanced scattering effect. As for peak (b), it mainly reveals the spectral characteristic of tryptophan and tyrosine residues. The fluorescence intensity of the peak (b) decreased markedly and the maximum emission wavelengths of the peak (b) have obvious red shift following the addition of BSA–ATX, indicating the conformations of the tryptophan residues of BSA. Hence, the molecular micro-environment and conformational changes of protein occurred after interaction with ATX [40].
Fig. 11.
Three-dimensional fluorescence spectra of (A) BSA and (B) BSA–ATX system (pH 7.4, 298 K).
3.10. FT-IR spectroscopic measurements
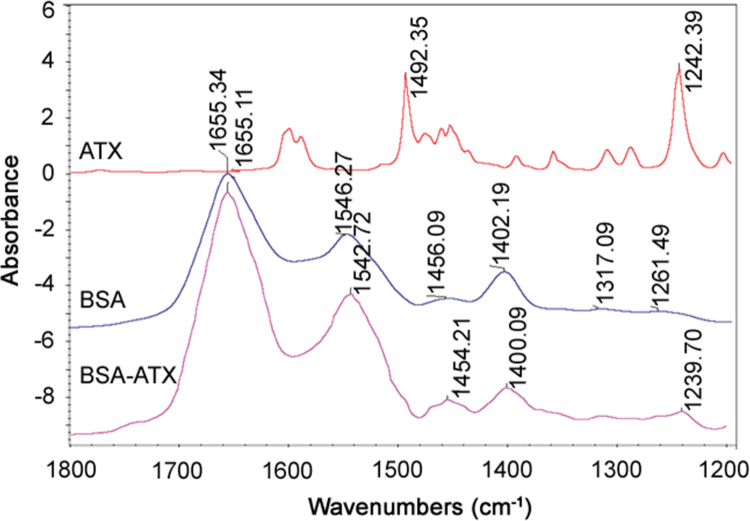
FT-IR spectroscopy has long been used as a powerful tool for investigating the secondary structure of proteins and their dynamics [41], [42]. Additional evidence for ATX–BSA interaction obtained from FT-IR spectra is shown in Fig. 12. Infrared spectrum of protein exhibited a number of amide bands due to different vibrations of the peptide moiety. Of all the amide modes of the peptide group, the most widely used in studies of protein secondary structure is the amide I and II. The amides I and II peaks occurred in the region of 1600–1700/cm and 1500–1600/cm, respectively. Hence, the amide bands are more useful for studies of secondary structure [43]. The FT-IR spectrum reveals that the peak position of amide II was shifted from 1546.27 cm−1 to 1542.72 cm−1 and the intensity also remarkably decreased. This indicated that the ATX interacted with BSA and the secondary structure of BSA was changed [44].
Fig. 12.
FT–IR spectra of ATX, BSA, and BSA–ATX at physiological pH 7.4 (298 K). Concentration of BSA and ATX was fixed at 5 µM.
3.11. Competitive binding studies
In order to know the binding probes in BSA for ATX, competitive displacement experiments were performed using site probes, warfarin, ibuprofen and digitoxin for sites I, II and III, respectively. The specific binding site of drug on BSA was found from the fluorescence quenching of BSA after adding a drug into the probe–BSA system [45], [46]. For this, emission spectra of BSA-site probe system at different concentrations of ATX were recorded. As evident from Table 3, calculated binding constant value of BSA–ATX system remains almost constant in the presence of ibuprofen and digitoxin. However, in the presence of warfarin the binding constant value decreased remarkably, suggesting that ATX is mainly located in site I of BSA.
Table 3.
The comparison of binding constants of BSA–ATX system upon addition of site probes (pH 7.4, 298 K).
| System | Binding constant (×10−4) (L/mol) | No. of binding sites |
|---|---|---|
| BSA–ATX | 1.49 | 1.04 |
| BSA–ATX+warfarin | 0.06 | 0.72 |
| BSA–ATX+ibuprofen | 1.25 | 0.96 |
| BSA–ATX+digitoxin | 1.04 | 0.97 |
3.12. Effect of some metal ions on the interactions of ATX with BSA
In plasma, there are some metal ions, which can affect the interactions of the drugs and serum albumins. Trace metal ions, especially the bivalent type are essential in the man body and play an important structural role in many proteins. It is reported that Ca2+, Co2+, Cu2+, Ni2+ and Zn2+ at 298 K and other metal ions can interact with serum albumins. Hence, the effects of some metal salt solutions, viz., CaCl2, CoCl2, CuCl2, NiCl2 and ZnCl2, on the binding of ATX with BSA were investigated in the present study. Under the experimental conditions, none of the cations gave the precipitate in phosphate buffer. The binding constants of ATX with BSA in the presence of the above ions were examined and the results are shown in Table 4. In the presence of all the above metal ions the binding constant of ATX-BSA system was decreased. This was likely to be caused by a conformational change in the vicinity of the binding site. The decrease in the binding constant in the presence of above metal ions would shorten the storage time of the drug in BSA and hence greater amounts of free drug would be available in BSA [22]. This shows the need for larger drug doses of drug to achieve the desired therapeutic effect in the presence of the above ions.
Table 4.
Effect of some common metal ions on binding constants of BSA–ATX system.
| System | Binding constant (×10−4) (L/mol) |
|---|---|
| BSA-ATX | 1.49 |
| BSA-ATX+Ca2+ | 0.294 |
| BSA-ATX+Co2+ | 0.387 |
| BSA-ATX+Cu2+ | 0.526 |
| BSA-ATX+Ni2+ | 0.379 |
| BSA-ATX+Zn2+ | 0.716 |
4. Conclusion
In this present study, the interaction of ATX with BSA has been investigated under the physiological conditions using different spectroscopic techniques. The interaction between BSA and ATX was dynamic quenching and further binding mechanism was confirmed by time resolved lifetime measurements. The binding interaction of BSA-ATX is mainly entropy driven and hydrophobic interaction forces are predominant. The results of UV-absorption spectra, FT-IR spectra, synchronous fluorescence spectra and 3D fluorescence spectra show the changes in secondary structure, molecular micro-environment and the conformational changes of protein. The binding of drugs to proteins is an important factor in determining their pharmacokinetics and pharmacological effects, where such work is useful for pharmaceutical industries and clinical laboratories.
Acknowledgment
Arunkumar T. Buddanavar thanks Karnatak University, Dharwad for providing UGC-UPE fellowship (146) and the authors thank UGC, New Delhi for the award of BSR Faculty Fellowship (F No.18-1/2011) to Prof. S.T, nandibewoor.
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
References
- 1.Yu J., Li B., Dai P., Ge S. Molecular simulation of the interaction between novel type rhodanine derivative probe and bovine serum albumin. Spectrochim. Acta A. 2009;74:277–281. doi: 10.1016/j.saa.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Hu Y.J., Liu Y., Shen X.S. Studies on the interaction between 1-hexylcarbamoyl-5-fluorouracil and bovine serum albumin. J. Mol. Struct. 2005;738:143–147. [Google Scholar]
- 3.Sandhya B., Hegde A.H., Kalanur S.S. Interaction of triprolidine hydrochloride with serum albumins: thermodynamic and binding characteristics, and influence of site probes. J. Pharm. Biomed. Anal. 2011;54:1180–1186. doi: 10.1016/j.jpba.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Zhao H.W., Ge M., Zhang Z.X. Spectroscopic studies on the interaction between riboflavin and albumins. Spectrochim. Acta A. 2006;65:811–817. doi: 10.1016/j.saa.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 5.Meti M.D., Nandibewoor S.T., Joshi S.D. Multi-spectroscopic investigation of the binding interaction of fosfomycin with bovine serum albumin. J. Pharm. Biomed. Anal. 2015;5:249–255. doi: 10.1016/j.jpha.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong D.T., Threlkeld P.G., Best K.L. A new inhibitor of norepinephrine uptake devoid of affinity for receptors in rat brain. J. Pharmacol. Exp. Ther. 1982;222:61–65. [PubMed] [Google Scholar]
- 7.Gehlert D.R., Gackenheimer S.L., Robertson D.W. Localization of rat brain binding sites for [3H] atomoxetine, an enantiomerically pure ligand for norepinephrine reuptake sites. Neurosci. Lett. 1993;157:203–206. doi: 10.1016/0304-3940(93)90737-6. [DOI] [PubMed] [Google Scholar]
- 8.Lee M.J., Yang K.C., Shyu Y.C. Attention deficit hyper activity disorder, its treatment with medication and the probability of developing a depressive disorder: a nationwide population-based study in Taiwan. J. Affect. Disord. 2016;189:110–117. doi: 10.1016/j.jad.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Chi Z., Liu R., Teng Y. Binding of oxytetracycline to bovine serum albumin: spectroscopic and molecular modeling investigations. J. Agric. Food Chem. 2010;58:10262–10269. doi: 10.1021/jf101417w. [DOI] [PubMed] [Google Scholar]
- 10.Gowda J.I., Nandibewoor S.T. Binding and conformational changes of human serum albumin upon interaction with 4-aminoantipyrine studied by spectroscopic methods and cyclic voltammetry. Spectrochim. Acta A. 2014;124:397–403. doi: 10.1016/j.saa.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 11.Enrico G., Jameson D.M., weber G. A model of dynamic quenching of fluorescence in globular proteins. J. Biophys. 1984;45:789–794. doi: 10.1016/S0006-3495(84)84223-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lakowicz J.R., Freshwater G., Weber G. Nanosecond segmental mobilities of tryptophan residues in proteins observed by lifetime resolved fluorescence anisotropies. J. Biophys. 1980;32:591–601. doi: 10.1016/S0006-3495(80)84992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naik K.M., Kolli D.B., Nandibewoor S.T. Elucidation of binding mechanism of hydroxyurea on serum albumins by different spectroscopic studies. SpringerPlus. 2014;3:360–373. doi: 10.1186/2193-1801-3-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y.Z., Zhou B., Zhang X.P. Interaction of malachite green with bovine serum albumin: determination of the binding mechanism and binding site by spectroscopic methods. J. Hazard. Mater. 2009;163:1345–1352. doi: 10.1016/j.jhazmat.2008.07.132. [DOI] [PubMed] [Google Scholar]
- 15.Lakowicz J.R. 3rd ed. Plenum Press; New York: 2006. Principles of Fluorescence Spectroscopy; p. 280. [Google Scholar]
- 16.Ware W.R. Oxygen quenching of fluorescence in solution an experimental study of the diffusion process. J. Phys. Chem. 1962;66:455–458. [Google Scholar]
- 17.Punith R., Seetharamappa J. Spectral characterization of the binding and conformational changes of serum albumins upon interaction with an anticancer drug anastrozole. Spectrochim. Acta A. 2012;92:37–41. doi: 10.1016/j.saa.2012.02.038. [DOI] [PubMed] [Google Scholar]
- 18.Zhao X.C., Liu R.T., Chi Z.X. New insights into the behavior of bovine serum albumin adsorbed onto carbon nanotubes comprehensive spectroscopic studies. J. Phys. Chem. B. 2010;114:5625–5631. doi: 10.1021/jp100903x. [DOI] [PubMed] [Google Scholar]
- 19.Lakowicz J.R., Weber G. Quenching of fluorescence by oxygen. A probe for structural fluctuations in macromolecules. Biochemistry. 1973;12:4161–4170. doi: 10.1021/bi00745a020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maciejewski A., Demmer D.R., James D.R. Relaxation of the second excited singlet states of aromatic thiones: the role of specific solute-solvent interactions. J. Am. Chem. Soc. 1985;107:2831–2837. [Google Scholar]
- 21.Hu Y.J., Liu Y., Wang J.B. Study of the interaction between monoammonium glycyrrhizinate and bovine serum albumin. J. Pharm. Biomed. Anal. 2004;36:915–919. doi: 10.1016/j.jpba.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 22.Li Y., He W., Liu J. Binding of the bioactive component jatrorrhizine to human serum albumin. Biochim. Biophys. Acta. 2005;1722:15–21. doi: 10.1016/j.bbagen.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Ross P.D., Subramanian S. Thermodynamics of protein association reactions forces contributing to stability. Biochemistry. 1981;20:3096–3102. doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- 24.Lu J.Q., Jin F., Sun T.Q. Multi-spectroscopic study on interaction of bovine serum albumin with lomefloxacin–copper(II) complex. Int. J. Biol. Macromolec. 2007;40:299–306. doi: 10.1016/j.ijbiomac.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Guo X.J., Zhang L., Sun X.D. Spectroscopic studies on the interaction between sodium ozagrel and bovine serum albumin. J. Mol. Struct. 2009;928:114–120. [Google Scholar]
- 26.Zhang Y.Z., Zhang X.P., Hou H.N. Study on the interaction between Cu (phen)23þ and bovine serum albumin by spectroscopic methods. Biol. Trace Elem. Res. 2008;121:276–287. doi: 10.1007/s12011-007-8045-z. [DOI] [PubMed] [Google Scholar]
- 27.P. Held, Quantitation of Peptides and Amino Acids with a Synergy™HT using UV Fluorescence BioTek Instruments, Inc., Highland Park, Winooski, Vermont 05404-0998 USA, 〈www.biotek.com〉.
- 28.Schmid F.X. Macmillan Publishers Ltd, Nature Publishing Group, University of Bayreuth; Germany: 2001. Biological Macromolecules, UV-visible Spectrophotometry, Encyclopedia of Life Sciences. [Google Scholar]
- 29.Naik D.B., Moorty P.N., Priyadarsini K.I. Nonradiative energy transfer from 7-amino coumarin dyes to thiazine dyes in methanolic solutions. Chem. Phys. Lett. 1990;168:533–538. [Google Scholar]
- 30.Förster T. 10th Spiers memorial lecture. Transfer mechanisms of electronic excitation. Discuss. Faraday Soc. 1959;27:7–17. [Google Scholar]
- 31.Cyril L., Earl J.K., Sperry W.M. E&FN Epon Led. Press; London: 1961. Biochemist’s Hand Book. [Google Scholar]
- 32.Hu Y.J., Liu Y., Zhang L.X. Studies of interaction between colchicine and bovine serum albumin by fluorescence quenching method. J. Mol. Struct. 2005;750:174–178. [Google Scholar]
- 33.Lin H., Lan J., Guan M. Spectroscopic investigation of interaction between mangiferin and bovine serum albumin. Spectrochim. Acta A. 2009;73:936–941. doi: 10.1016/j.saa.2009.04.025. [DOI] [PubMed] [Google Scholar]
- 34.Jiang X.Y., Li W.X., Cao H. Study of the interaction between trans-resveratrol and BSA by the multi-spectroscopic method. J. Solut. Chem. 2008;37:1609–1623. [Google Scholar]
- 35.Brustein E.A., Vedenkina N.S., Irkova M.N. fluorescence and the location of tryptophan residues in protein molecules. Photochem. Photobiol. 1973;18:263–279. doi: 10.1111/j.1751-1097.1973.tb06422.x. [DOI] [PubMed] [Google Scholar]
- 36.Lu Z.X., Cui T., Shi Q.L. 1st ed. Science Press; Marrickville: 1987. Applications of Circular Dichroism and Optical Rotatory Dispersion in Molecular Biology; pp. 79–82. [Google Scholar]
- 37.Guo X.J., Hao A.J., Han X.W. The investigation of the interaction between ribavirin and bovine serum albumin by spectroscopic methods. Mol. Biol. Rep. 2011;38:4185–4192. doi: 10.1007/s11033-010-0539-7. [DOI] [PubMed] [Google Scholar]
- 38.Shaikh S.M.T., Seetharamappa J., Kandagal P.B. Spectroscopic investigations on the mechanism of interaction of bioactive dye with bovine serum albumin. Dyes Pigments. 2007;74:665–671. [Google Scholar]
- 39.Zhang H.X., Mei P. In vitro binding of furadan to bovine serum albumin. J. Sol. Chem. 2009;38:351–361. [Google Scholar]
- 40.Tian J.N., Liu J.Q., Hu Z.D. Interaction of wogonin with bovine serum albumin. Bioorg. Med. Chem. 2005;13:4124–4129. doi: 10.1016/j.bmc.2005.02.065. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y., Yang Z., Du J. Interaction of curcumin with intravenous immunoglobulin: a fluorescence quenching and Fourier transformation infrared spectroscopy study. Immunobiology. 2008;213:651–661. doi: 10.1016/j.imbio.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 42.Harding S.E., Chardhry B.Z. Oxford University Press; Oxford: 2001. Protein Ligand Interactions: Structure and Spectroscopy. [Google Scholar]
- 43.Brauner J.W., Flach C.R., Mendelsohn R. A quantitative reconstruction of the amide I contour in the IR spectra of globular proteins: from structure to spectrum. J. Am. Chem. Soc. 2005;127:100–109. doi: 10.1021/ja0400685. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y., Yang Y., Guo T. Genipin-crosslinked hydrophobical chitosan microspheres and their interactions with bovine serum albumin. Carbohydr. Polym. 2011;83:2016–2021. [Google Scholar]
- 45.Sudlow G., Birkett D.J., Wade D.N. Further characterization of specific drug binding sites on human serum albumin. Mol. Pharmacol. 1976;12:1052–1061. [PubMed] [Google Scholar]
- 46.Sjoholm I., Ekman B., Kober A. Binding of drugs to human serum albumin: XI. The specificity of three binding sites as studied with albumin immobilized in microparticles. Mol. Pharmacol. 1979;16:767–777. [PubMed] [Google Scholar]