Abstract
Solid progress has occurred over the last decade in our understanding of the molecular genetic basis of neurodevelopmental disorders, and of schizophrenia and autism in particular. Although the genetic architecture of both disorders is far more complex than previously imagined, many key loci have at last been identified. This has allowed in vivo and in vitro technologies to be refined to model specific high-penetrant genetic loci involved in both disorders. Using the DISC1/NDE1 and CYFIP1/EIF4E loci as exemplars, we explore the opportunities and challenges of using animal models and human-induced pluripotent stem cell technologies to further understand/treat and potentially reverse the worst consequences of these debilitating disorders.
This article is part of a discussion meeting issue ‘Of mice and mental health: facilitating dialogue between basic and clinical neuroscientists’.
Keywords: Schizophrenia, autism, mouse and iPSC models
1. Introduction
Schizophrenia (SCZ) and autism (ASD) are two of the most important neurodevelopmental disorders encountered in routine clinical psychiatric practice. Both are diagnosed on the basis of clinical history, symptoms and behaviour. These include SCZ-positive symptoms, such as hallucinations, delusions and thought disorder, and SCZ-negative symptoms such as social withdrawal anhedonia and poverty of thought: there are also a range of cognitive abnormalities especially of attention, memory and executive function. ASD is characterized by abnormalities of social communication and interaction and repetitive patterns of interests and behaviour. Unfortunately, in spite of intensive efforts spanning several decades, there are still no objective tests (biomarkers) in routine clinical psychiatric practice to assist with diagnosis of any psychiatric disorders including SCZ and ASD [1]. Although ages of clinical presentation of SCZ and ASD are normally early adult life and early infancy, respectively, both have at least in part neurodevelopmental origins, namely antecedents affecting brain development, and, in turn, predisposition to one or both disorders can occur at any point in the life cycle probably from conception onwards. There are also pre-conceptual intergenerational effects, the most studied being parental and grandparental age. Antecedents may be environmental or genetic/epigenetic or the effects of gene–environment (G × E) interactions. Environmental risk factors are often discussed in the context of a ‘stress-vulnerability’ aetiological model where early biological and psychological insults, occurring in both the pre- and postnatal periods, result in changes of gene and protein expression, and/or changes in the intracellular and extracellular milieu of the developing brain. For recent relevant reviews of environmental risk factors, see [2,3]. However, perhaps, the most intriguing finding to emerge from epidemiological studies is that SCZ and ASD appear to share a remarkable number of environmental risk factors [4,5]. A similar pattern of overlapping genetic risk profiles for SCZ and ASD will be discussed below.
2. Familiarity
SCZ and ASD are both strongly familial neuropsychiatric disorders. The evidence in SCZ comes from multiple twin family and adoption studies and points to a heritability of up to 80% with monozygotic concordance of 40–50% [6,7]. There have been many fewer twin and family studies of ASD and surprisingly no adoption studies. Earlier twin studies suggested heritability as high as 80–90% for ASD with little contribution from the environment [8]. Newer studies of MZ twins have yielded concordance rates of less than 50%, with lower concordance for dizygotic twins, suggesting that both genes and environment play roles in the development of ASD [9]. The current consensus is that up to 40–50% of variance is determined by environmental factors [2].
Early linkage and candidate gene mapping studies of SCZ and ASD have yielded little in the way of findings that have stood the test of time. The most studied are disrupted in schizophrenia one (DISC1 gene) identified by cloning the breakpoints of a balanced 1 : 11 chromosomal rearrangement associated with multiple cases of mental illness including SCZ in a large Scottish pedigree [10], chromosome 22 deletion syndrome associated with a range of severe neurodevelopmental disorders [11], fragile × syndrome [12], Rett syndrome [13] and rare cases of ASD with mutations of neuroligin genes [14].
Our understanding of the genetic architecture of neurodevelopmental disorders expanded enormously with the advent of methods for systematic interrogation of DNA across the whole genome, first through genome-wide association studies (GWAS) and more recently whole exome and whole genome sequencing. It allows us to detect association with rare (less than 1%) high-penetrant genetic lesions including copy number variants (CNVs) and association with common low-penetrant genetic risk factors identified using single-nucleotide polymorphism (SNP) microarrays. These latter common low-penetrant risk factors have odds ratios approximately 1.0–1.2. Rare low-penetrant genetic lesions also exist, but sample sizes required for their detection with reasonable statistical supporting evidence are far beyond those currently available worldwide; a similar problem confounds potential genome-wide studies of gene/gene interactions/epistasis [15]. It is also possible to examine non-statistically significant common variants for association as a whole so-called polygenic liability risk [16]. This latter approach does not, however, help with selection of individual gene targets for in vivo or in vitro modelling.
(a). Rare variants
The genetic findings in both SCZ and ASD are broadly similar. In both disorders, there is enormous genetic heterogeneity, but only a small proportion (5–10%) of the overall genetic risk results from rare high-penetrant genetic mutations including CNVs. Many of these latter loci (causing deletions or duplications of stretches of DNA) show pleiotropy, i.e. they display a range of clinical phenotypic abnormalities that include ASD, intellectual impairment, SCZ and epilepsy [17]. This means that there is considerable overlap of high-penetrant loci between SCZ and ASD. Many mutations, especially in ASD, have arisen de novo and are not found in the parents of the affected proband [18–21]. This reflects the fact that they are heavily selected against due to the reduced fecundity associated with neurodevelopmental disorders. In ASD, this is so pronounced that it is almost impossible to find families with ASD in more than two generations. In SCZ, familial cases are more common but with high-penetrant loci, the effects of reduced fecundity are also clinically observable. This was elegantly demonstrated in the Icelandic population where it was possible to examine formally family inheritance patterns of recurrent non-de novo SCZ-associated CNVs. Although recurrent CNVs have high mutation rates due to non-allelic homologous recombination, they are eliminated fast by negative selection and seldom survive more than two or three generations [22]. Around 800 rare loci are reported in ASD (far fewer in SCZ), but the evidence to support their causal involvement varies enormously and in only a few dozen including recurrent CNVs is there statistical evidence of genetic association [19]. Among these genes are NGLN4X [14], SHANK3 [23,24], NRXN1 [25,26], SHANK2 [27], CNTN4 [28–30] and CNTNAP2 [31,32]. The findings in SCZ are broadly similar [33–35].
(b). Common variants
Initial genome-wide SCZ SNP association studies, involving several thousand cases and controls, yielded only two or three loci that meet statistical significance (p < 1 × 10−7.5–8), the precise significance level depending on the number of tests performed [16,22]. However with increased sample sizes to around 150,000 individuals, over 100 loci were reported to meet genome wide significance [36] with additional loci being subsequently reported [37]. Although multiple common low-risk variants are reported associated with ASD, to date no loci for ASD have consistently met criteria for genome-wide significant association; this is probably the result of inadequate sample sizes. There are a number of excellent articles discussing gene/gene interactions/epistasis [15], SCZ epigenetics [38] and modelling of polygenic risk [16,39]. In particular, two earlier studies highlight the potential of being able to elucidate a better understanding of the effects of regulatory polymorphism on the expression of genes essential to mental health [40,41]. Furthermore, the identification of these regulatory determinants will, in turn, permit critical insights into the role of epigenetic factors such as DNA methylation that are known to influence gene expression. To date, however, there are very few instances where specific low penetrance loci for ASD or SCZ have been deemed worthy of modelling in animals or human-induced pluripotent stem cell (hiPSC).
(c). Missing heritability
The majority of genetic risk for both SCZ and ASD is still to be elucidated and is likely to involve many more rare high- and low-risk factors, common low-risk factors, epistasis and epigenetic interactions, the so-called missing heritability. Their tiny effect sizes represent a formidable challenge: what sort of clinical or behavioural phenotype if any should one expect to find? A recent ‘omnigenic model’ has proposed that gene regulatory networks are sufficiently interconnected, such that all genes expressed in disease-relevant cells are liable to affect the functions of core disease-related genes and that most heritability can be explained by effects on genes outside core pathways [42]. The genes we have chosen to discuss here are likely to affect the function of core pathways and so are likely to provide insights into wider populations of patients with these disorders, even although the majority of patients are not enriched for high-impact variants.
3. Modelling: from mice to men
Two of the most important methods for attempting to model neurodevelopmental disorders are genetically modified animals, especially rodent models, and in vitro modelling using hiPSCs differentiated into neuronal precursors and, in turn, to three-dimensional organoid systems. The advantages and disadvantages of the two methods are elegantly described elsewhere [43], but are summarized below with modifications. It cannot be stressed enough however that the full benefits of modelling studies are predicated on knowing what phenotype to expect and this depends on careful and deep phenotyping of patients and individuals with mutations at the specific loci under investigation. The enormous genetic heterogeneity encountered in SCZ and ASD as well as locus pleiotropy makes predictions of expected phenotype from population findings alone much less satisfactory.
(a). Animal models: the pros and cons
Animal models of disruption exist for almost all human genes. Coding regions of the genome are especially well preserved and easier to model, whereas non-coding DNA, including regulatory elements, show poor conservation across species. The mouse genome is almost as well characterized as the human and murine models have become relatively cost effective, straightforward to produce, and amenable to study at molecular, cellular, circuit and behavioural levels. The advantages of rat models are usually outweighed by the costs of their generation and maintenance. The highest-throughput and least-expensive models include zebrafish (Danio rerio) and fruit fly (Drosophila melanogaster) [44], but obviously these are unsuitable for modelling more complex human behaviours. Care must be given also to which mouse strain is used as genetic background effects are potential confounders. However, there are a number of very obvious limitations and drawbacks when using such models to study neuropsychiatric disorders. Although there are established batteries to phenotype core features of ASD in mice [45], mice exhibit profound differences in social behaviour from humans and furthermore, even within mouse studies, variations in laboratory environments impose further variance. How these truly reflect the human condition is debatable. Interpreting SCZ like phenotypes in mice, including complex symptoms such as paranoia and delusional beliefs, is even more challenging: they can only be inferred indirectly from disordered mice behaviour, a major limitation of modelling schizophrenia in animals.
(b). Human in vitro stem cell models: advantages and limitations
Human iPSC technologies are allowing researchers to interrogate human cortical development in health and disease and provide unlimited platforms of mature neuronal and glial cellular subtypes and co-cultures for downstream studies such as cellular physiology, phenotypic screening, and for drug development and screening. Such human iPSC models confer a number of advantages including the fact that it is possible to model for both coding and non-coding variants and, in fact, it is also possible to model for disease without actually knowing the causal genetic factor [46]. Clearly, though knowing the causal/contributory variants confers an advantage to translational studies and a greater understanding of putative mechanisms of disease [47]. It is possible to study the effects of genomic mutations on brain development and in neuropsychiatric disorders using clustered regularly interspaced short palindromic repeats (CRISPR) gene-editing technologies. Proteomics, transcriptomics, signalling and cell biology analysis of isogenic-mutant paired lines at the neuronal stem cell and differentiated neuron cell state offer unique opportunities. However, limitations include heterogeneity and reproducibility issues arising from multiple sources, including culture methodology and differences in lines and clones used. Furthermore, these hiPSC cultures produce immature fetal-like neurons, limiting their potential to properly model later developmental stages. This, however, has become less of an issue as it is now possible to mature cells by co-culture and also using advanced organoid cultures [48,49], discussed further below.
(c). Towards three-dimensional cellular systems: growing brain organoids
An organoid is a multicellular collection of cells that self-organizes and develops from stem cell progenitors to resemble the structure and function of an organ in vivo [50]. In vitro models of the developing brain such as three-dimensional brain organoids offer an unprecedented opportunity to study aspects of human brain development and disease, in particular the ability to follow development over time. Neuronal migration, cortical lamination, projection patterns and circuit-level organization are difficult to model in two-dimensional cultures. Tissue engineering and three-dimensional organoid cultures will enable the study of some of these phenotypes. As mentioned earlier, rodent models have been heavily used to study the cellular function of many of the genes implicated in these disorders, especially those genes which are proposed to have an important role in fundamental neurodevelopmental processes such as cerebral cortex organization. However, cortex development and organization is very different in rodents compared to humans, so unsurprisingly neurodevelopmental diseases cannot be consistently recapitulated in animal models. This is all about to change as over the past few years there have been further cutting-edge advances in developmental neurobiology: we can now grow three-dimensional cerebral organoid cultures from patient-derived stem cells to study the early events of human brain development. Proof-of-principle studies using human pluripotent stem cell-derived three-dimensional organoid cultures have allowed researchers to model human brain development and microcephaly in a dish [51]. These ‘cerebral organoids’ develop various discrete brain regions including a cerebral cortex that produces functional cortical neuron subtypes capable of displaying spontaneous synaptic transmission and producing action potentials. Subsequent studies have also shown that it is possible to develop region-specific identities, including neocortex [52], telencephalon [53], cerebellum [54], neural tube [55], pituitary [56], hippocampus [57], optic-cup [58] and retina [59]. Through altering specific culture conditions, it is possible to differentiate iPSC and embryonic stem cells (ESC) into a range of neuronal [60] and glial subtypes, including GABAergic interneurons and glutamatergic neurons [61,62], dopaminergic neurons [63], motoneuron [64] and glial progenitors [65,66].
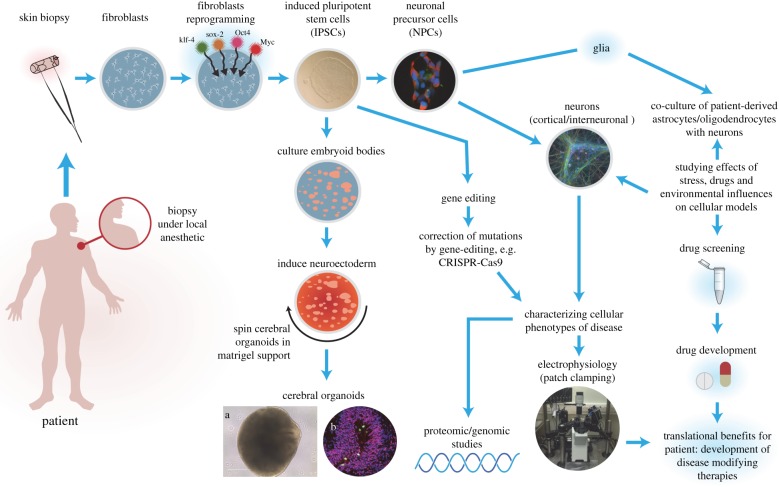
Most protocols adopted to generate cerebral organoids depend on step-wise establishment of spatio-temporal strategies using human ESC or iPSC (figure 1). The first stage depends on the re-aggregation of iPSCs or ESCs in low-adhesion conditions such as those provided by serum-free embryoid body (EB) protocols, and allowing the cells enough time to proliferate and expand [67,68]. During this initial stage, the stem cells maintain pluripotency and the EBs that form exhibit all three germ layers (ectoderm, mesoderm and endoderm). The next stage involves neural induction where the goal is to drive differentiation to neuroectoderm formation. During these early stages, the initial organoids formed display apical–basal and dorsal–ventral polarity and further induction can promote regional identity such that it is possible to produce region-specific organoids [49]. The human cerebral cortex is a well-defined structure with six layers of neurons: superficial and deeper layers are connected to one another, yet have distinct structural and functional projections and fates [69,70]. One of the greatest challenges in the development of the human cerebral cortex is the assembly of circuits composed of glutamatergic neurons, generated in the dorsal forebrain (pallium), and GABAergic interneurons arising in the ventral forebrain (subpallium). However, it has recently been shown for the first time using a three-dimensional differentiation approach using hiPSC to specify neural spheroids and assemble these in vitro to model salutatory migration of human interneurons towards the cerebral cortex and functionally integrate into microcircuits [49].
Figure 1.
Overview of human iPSC model systems to study SCZ and ASD. Human iPSCs are generated by reprogramming fibroblasts from skin biopsies from volunteers using a variety of techniques, most commonly using standard Yamanaka factors, delivered in non-integrative episomal vectors. Other starting cellular materials can be used such as keratinocytes from hair or from peripheral blood mononuclear cells. Once generated and extensively tested, the hiPSCs can be used to either make neuronal precursor cells or glial precursor cells (e.g. oligodendrocyte precursors) or grown and lifted to make three-dimensional organoids as shown in frames (a) phase bright image of a cerebral organoid at two months of age and frame (b) shows an organoid that has been sectioned and stained with antibodies to Pax6 and phospho-histone H3, clearly demonstrating a ventricular zone. The cellular platforms generated can then be used for further downstream studies including electrophysiology, transcriptomic and proteomic studies, drug screening as well as morphological studies, and co-culturing with other cell types. In addition, the hiPSCs can be gene-edited using CRISR–Cas9 to attempt to rescue phenotypes observed.
Organoid cultures are, however, not without limitations: spontaneous self-organization of cerebral organoids in culture generates significant heterogeneity in cell type and structure, with prolonged neotany in development and differentiation limits their utility to early studies of brain development. There are also challenges with scalability. However, with modifications to culture systems such as the use of mini-reactors [71] and microfluidics [72] combined with improved seeding technologies (e.g. laminin-coated nanoparticles) [73], it is possible to scale-up, improve consistency and robustness, and reduce associated costs plus provide higher throughput for drug screening. In this regard, iPSC-derived two-dimensional and three-dimensional model systems hold potential in future to screen drug targets for pharmaceutical development (figure 1). Fundamentally, however, it is a human in vitro system, and as such in vivo connectivity and external milieu are not preserved, thus findings may not precisely translate to in vivo biology experienced during human fetal brain development. It is also important to remember that the human brain develops both in utero and during the postnatal period in an environment with inputs via sensory systems as well as from neighbouring brain areas which collectively helps to shape the cellular environment and circuits that develop. Obviously, in vitro culture systems cannot recapitulate this degree of complexity other than ambient fluctuations in temperature, pH and chemical gradients. Furthermore, the lack of neuromodulatory inputs to synaptic function may preclude our ability to precisely study the effects of systems such as the monoaminergic system in neuropsychiatric disorders and limit their utility in drug development. Until recently, another major drawback of organoids to model neurodevelopmental disorders was the unanswered questions as to what extent they truly modelled regional complexity, cellular diversity and circuit functionality of the brain. Gene expression analysis in over 80 000 individual cells isolated from 31 human brain organoids has shown that organoids generate a broad diversity of cells, which are related to endogenous classes, including cells from the cerebral cortex and retina [74]. Some caution should be held, however, as to the relative quantities of the different cell types generated in these organoid systems and to what extent this reflects the quantities in the human developing embryonic and fetal brain. In the Quadrato et al.'s study [74], only two of 10 cell clusters analysed were found to contain neurons from the cerebral cortex, accounting for approximately 20% of cells examined, somewhat less than what might be expected in vivo. Furthermore, these two cell clusters were found in only 32% and 52% of all organoids examined and within these populations approximately half of the cells expressed the radial glial marker PAX6 after six months, reflecting that they could not truly be classified as wholly mature neurons. However, this study has allayed fears that organoid cultures are limited by immaturity as a proportion of the cells do appear more mature than has been seen previously in culture. This team also elegantly demonstrated that neuronal activity, within the organoid, could be controlled using light stimulation of photosensitive cells which provides further opportunity for the coupled use of optogenetics to probe the functionality of human neuronal circuits and specifically model higher-order functions of the human brain, such as cellular interactions and neural circuit dysfunctions related to neurodevelopmental and neuropsychiatric pathologies.
4. Selection of loci for modelling of neurodevelopmental disorders
(a). Single-nucleotide polymorphism-associated loci
In spite of their large numbers and widespread involvement in SCZ and ASD, there have been few attempts to model individual common low-penetrant SNP-associated loci using either animals or human iPSC technologies. This is unsurprising. The vast majority of SNPs significantly associated in GWAS are located outside gene-coding regions and, in many instances, often a considerable distance from the nearest coding region. Most effort has, therefore, concentrated on attempting to fine map putative functional variants presumed to be in linkage disequilibrium with the GWAS-associated SNPs. This is paralleled by in silico bioinformatic investigations using pathway analyses/gene ontology studies to try to obtain further corroboration of their functional significance. To date, success has been very limited [33]. Fortunately, successful studies designed to ascribe regulatory functionality to directly associated SNPs, or those in linkage disequilibrium, using comparative genomics and CRISPR-modified preclinical mouse models are well underway. These studies promise to develop a better understanding of the effects of regulatory polymorphism on the expression of genes essential to mental health. Furthermore, the identification of these regulatory determinants will, in turn, permit critical insights into the role of epigenetic factors such as DNA methylation that are known to influence gene expression.
Two modelling attempts are worthy of note.
(1) The very strong allelic association of SCZ to the major histocompatibility complex region of chromosome 6 prompted detailed exploration of the putative involvement of complex variation at the complement component 4 candidate gene. In mice, the authors showed that some patterns led to excess synaptic pruning [75], a dynamic process proposed to rid the brain during the development of wasteful neural connections and strengthen others, and proposed to be a reason why brains from patients with SCZ have fewer synaptic connections in multiple brain regions [76].
(2) The strong association of SNPs within CACNA1C with autism, bipolar disorder and schizophrenia [36,77] has been investigated functionally.
The risk-associated genotypes appear to affect RNA abundance but results are inconclusive. CACNA1C has several dozen exons, with multiple transcripts and promoters. The locus is also independently associated with de novo Mendelian dominant exonic mutations responsible for Timothy syndrome (TS), a neurodevelopmental disorder which has features of ASD. The most interesting findings have emerged from studying hiPSCs from individuals with TS [49,78]. Building on previous work that showed in rodents that L-type calcium channel (LTCC) genes play a critical role in interneuron migration [79]. Birey et al. [49] found that cortical interneurons derived from patients with TS display a cell-autonomous migration defect whereby they move more frequently but less efficiently [49]. What is more the TS interneuron defect is rescued by pharmacologically manipulating LTCCs.
(b). Rare highly penetrant genetic mutations
The selection of which high-penetrant genetic mutations to model in mice or using hiPSC technologies poses separate challenges from common low-risk SNP-associated loci.
Causative or non-causative? Often the mutations are so rare that statistical evidence of association with the disorder is lacking. This is less of a problem in SCZ where linkage with the mutation in multiplex families is often available for additional corroboration. Also through PGC and other consortia, DNA from many thousands of cases is available for interrogation to try to identify additional mutations at the locus of interest. In ASD, where de novo mutation is more common, corroborating data from multiplex families is usually not available. It can be argued that de novo mutation itself may support a causative role in a disorder where the absence of familial cases is due to negative selection. A word of caution is merited. It must be borne in mind that each individual harbours approximately 60–100 de novo events [80], and deciding which/if any are causative is not a trivial problem, especially if it has implications for genetic counselling. Often, therefore, one of the main purposes of modelling is to try to demonstrate a causative mechanism that may result in the disorder under investigation. This especially applies where the gene is not an obvious candidate for the disorder under investigation, e.g. complement component 4 discussed above. In the case of rare variants, biology does have a role in both establishing a genetic association and later in understanding its role [81].
In some cases, the statistical and/or circumstantial evidence for involvement of the locus with the disorder is sufficiently compelling that modelling in mice and/or hiPSCs justifies the time and cost. Many such loci are currently being examined using animal modelling and hiPSC technologies. In these circumstances, the main questions concern what sort of phenotype to expect at the different levels of analysis and obviously also how to decide the precise nature of the modelling itself. The authors have been fortunate to have been involved with the identification and/or analysis of several loci that meet such criteria. These we discuss in more detail below. They are (i) DISC1 and a key interactor NDE1 and (ii) CYFIP1 and EIF4E genes which, with FXMR, encode for a single molecular complex responsible for translation including at the synapses in the brain.
5. Disrupted in schizophrenia one
DISC1 is a major vulnerability factor for a wide range of chronic mental illnesses, including SCZ [82]. DISC1 was first isolated by cloning the breakpoints of a 1 : 11 balanced translocation co-segregating with major psychiatric disorders in a large Scottish pedigree [10,83]. Within this one family, the logarithm of the odds (LOD) score for SCZ alone met stringent genome-wide significance, while for SCZ plus bipolar disorder and major depressive disorder, it substantially exceeded genome-wide significance (multipoint logarithm of odds = 7.1). A second wave of follow-up confirmed these original findings [84]. Further evidence supporting the involvement of DISC1 in mental disorders has been more recently debated [81,85].
DISC1 expression in the brain is particularly high in the hippocampus during neurogenesis and remains high in the adult dentate gyrus, olfactory bulb and limbic regions [86,87], and it appears that DISC1 regulates important developmental processes such as neuronal migration, integration [88], synapse formation and neuronal stem cell maturation [87,89–91]. DISC1 is thus critical for neurodevelopment and normal adult neuronal function. In addition, transgenic or mutant mice with impaired DISC1 function show brain morphological changes, deficits in neural circuits, working memory impairment and behavioural traits related to SCZ and also bipolar disorder [92]. One of the more interesting of the mice transgenic models, denoted Disc1tr, expresses two copies of truncated Disc1 encoding the first eight exons generated using a bacterial artificial chromosome (BAC) [93]. With this partial simulation of the human situation, they discovered a range of phenotypes including a series of novel features not previously reported. Disc1tr transgenic mice display enlarged lateral ventricles, reduced cerebral cortex, partial agenesis of the corpus callosum and thinning of layers II/III with reduced neural proliferation at mid-neurogenesis [93]. Parvalbumin (PV+) GABAergic neurons are reduced in the hippocampus and medial prefrontal cortex, and displaced in the dorsolateral frontal cortex. In culture, transgenic neurons grow fewer and shorter neurites. Behaviourally, these transgenic mice exhibit increased immobility and reduced vocalization in depression-related tests, and impairment in conditioning of latent inhibition. The BAC mouse model uses the full mouse genomic sequence and natural promoters. This may be responsible for the considerable SCZ reminiscent brain pathology observed in this study compared to other studies using more artificial constructs.
It is still not clear the mode of action of the t1 : 11 mutation. Haploinsufficiency seems most likely. No truncated DISC1 protein has ever been identified, suggesting elimination of mutated RNA by non-sense-mediated decay. It has also been shown that transient knockdown of DISC1 by in utero electroporation in mouse, in the pre- and perinatal stages, specifically in a lineage of pyramidal neurons mainly in the prefrontal cortex, leads to selective abnormalities in postnatal mesocortical dopaminergic maturation and behavioural abnormalities associated with disturbed cortical neurocircuitry after puberty [94]. Nevertheless, a dominant negative mode of action from mutated DISC1 protein dimerizing with the wild-type cannot be ruled out. What is clear is that the mutations reported in Disc1 do seem to alter the structural organization of the DISC1 protein [95].
The molecular and genetic mechanisms that are involved in biological alterations can often be modelled in Drosophila or zebrafish [44]. DISC1 causes associative memory and developmental defects and disruption of sleep rhythms in Drosophila [96,97]. In zebrafish studies, DISC1 variants were first identified from patient pools and tested in Disc1 loss-of-function (LOF) mouse embryos to determine which could and which could not rescue neuronal progenitor proliferation. When they were injected in disc1 LOF zebrafish embryos, the variants that showed maintenance or loss of activity in mice exhibited similar patterns in rescuing or not, respectively, brain ventricle and axon tract defects in zebrafish embryos [98]. These results emphasize the conservation of variant function between species, and indicate that a much higher number of variants can be analysed in zebrafish than is feasible in the mouse.
It has also been possible to generate isogenic hiPSCs with an engineered disease-relevant disruption of DISC1, which affects neural progenitor cells (NPCs) proliferation, baseline wingless-type mouse mammary tumor virus integration site signalling and expression of NPC fate markers such as FOXG1 and Tbr2 [99]. Ming & Song's group have since generated hiPSCs from four members of an American family in which a frameshift mutation of DISC1 co-segregated with major psychiatric disorders [100] and furthermore produced different isogenic iPS cell lines via gene editing [101]. In an elegant series of experiments, they showed that mutant DISC1 causes synaptic vesicle release deficits in hiPSC-derived forebrain neurons [102–104]. Mutant DISC1 depleted wild-type DISC1 protein and, furthermore, dysregulated expression of many genes related to synapses and psychiatric disorders in human forebrain neurons, providing new insights into the molecular and synaptic etiopathology of psychiatric disorders [101]. Although similar studies have not yet been published from the Scottish DISC1 family, it will be interesting to see whether synaptic dysregulation is also evident in neurons derived from these hiPSCs.
Unlike in ASD, SCZ psychosis can be thought of as a neurodevelopmental disorder with psychosis as a late stage of illness, even though several population-based studies indicate that the problems are evident much earlier [105]. In this model of SCZ, Insel proposes that reduced myelination could alter connectivity in SCZ. There are multiple studies showing white matter changes in SCZ (reviewed in [106,107]) and specifically in the DISC1 family [108]. It will be possible using hiPSC from the DISC1 family to generate oligodendrocyte and astrocytes to study the impact of glia on the pathophysiology. Insel also argues that the trajectory of cognitive development in children developing SCZ could include reduced elaboration of inhibitory pathways and excessive pruning of excitatory pathways leading to an altered excitatory–inhibitory balance in the prefrontal cortex. In this regard, it will also be interesting to now use hiPSC-derived GABAergic interneurons from the DISC1 families to specifically look for deficits in inhibitory interneuron activity and N-methyl-d-aspartate receptor expression. Protocols are now available to generate GABAergic inhibitory interneurons from hiPSC [109], which can be matured in culture to generated PV+ interneurons for the electrophysiological study of these cell types in vitro. Furthermore, as discussed earlier, Birey et al. [49] have recently generated three-dimensional spheroids from hiPSC that resemble either the dorsal or ventral forebrain and contain cortical glutamatergic or GABAergic neurons [49]. This is a seminal study as it demonstrates for the first time that it is now possible to generate organoids/spheroids with network activity: these subdomain-specific forebrain spheroids can be assembled in vitro to recapitulate the salutatory migration of interneurons observed in the fetal forebrain. These protocols will open the gates for the generation and studies of human forebrain spheroids from hiPSC from patients with other disease-associated mutations of SCZ and ASD.
6. Nuclear distribution E homologue 1
NDE1 (nudE Nuclear Distribution E homologue 1) is a gene in which different mutations result in a wide range of human brain diseases including microcephaly [110], intellectual disability [111], ASD [112], attention-deficit hyperactivity disorder (ADHD) [113] and SCZ [114–116]. NDE1 encodes a cytoskeletal protein localizing to the centrosome that participates in essential neurodevelopmental processes, including neuronal precursor proliferation and differentiation, neuronal migration and neurite outgrowth [117]. NDE1 is part of the lissencephaly-1/cytoplasmic dynein complex and as such participates in regulation of cell proliferation, migration and intercellular transport [118–132]. Cytoplasmic dynein is the main molecular motor moving towards the minus ends of microtubules, and is therefore responsible for carrying vesicles and other entities from axon tips towards the cell bodies of neurons (retrograde transport) [133–135].
Through protein–protein interaction, DISC1 regulates NDE1 function: evidence supports a shared binding domain for NDE1 and NDEL1 to DISC1, with opposite effects of the DISC1 Ser704Cys mutation on binding patterns [129]. NDE1 and NDEL1 localize to the centrosome, and mutations in both genes result in defective neurogenesis and neuronal migration. This is proposed to arise from decoupling of the centrosome from the nucleus as a result of defective microtubule bundles connecting both organelles, and also from the proposed role, all three genes have in regulating the cell cycle and mitosis [122,136–138]. Furthermore, it has been shown that familial mutations in NDE1 caused both severe failure of neurogenesis and a deficiency of cortical lamination (microlissencephaly) [110,139]. Elegant mouse studies have shown that while cortical lamination is mostly preserved, the mutant cortex has fewer neurons and very thin superficial cortical layers (II– IV) [138]. BrdU birthdating revealed retarded and modestly disorganized neuronal migration; however, more dramatic defects on mitotic progression, mitotic orientation and mitotic chromosome localization in cortical progenitors were observed in Nde1 mutant embryos. Another Nde1 mutant mouse study has demonstrated catastrophic DNA double-strand breaks concurrent with DNA replication, leading to p53-dependent apoptosis and reduced neurons in cortical layer II/III, and that this stalling of DNA replication in the Nde1 mutants specifically occurred in mid-late S-phase [140]. More recently, knockdown in rat using in utero electroporation confirmed these findings and shows that Nde1 effects are pronounced on premitotic nuclear migration with specific effects on radial glial progenitor cells and on primary cilia [141]. These studies elegantly demonstrate some of the mechanisms whereby haploid reduction of Nde1 expression may cause more subtle neurodevelopmental phenotypes.
It should be highlighted that although NDE1 does not appear as a top GWAS ‘hit’, deletions and duplications spanning NDE1 (on Chromosome 16p13.11) are among the most common CNVs in SCZ. CNVs in NDE1 have also been found by others to associate with a range of phenotypically different neurodevelopmental disorders including intellectual disability [111], ASD [112], ADHD [113] and SCZ [111,114], which suggest that the locus contains dosage-sensitive gene(s) that may play a critical role in neurodevelopment. The deCODE genetics study of 4345 SCZ patients and 35 079 controls from eight European populations found a threefold excess of duplications and deletions at the 16p13.1 locus in SCZ cases, compared with controls with duplications being far more commonly found [115]. In a Scottish population sample, we found a fourfold excess of duplications at the 16p13.1 locus in SCZ patients compared with controls [116]. Significant sex differences in prevalence, course and severity have been described for a number of these conditions, but the biological and environmental factors underlying such sex-specific features remain unclear [142]. Rare SNPs in NDE1 have also been shown to associate with SCZ susceptibility [143]. NDE1 has also been identified as associating with psychosis proneness in a large Finnish birth cohort upon re-analysis of GWAS linkage data conditioned on a DISC1-associating risk haplotype [144]. Thus, consistent with current neurodevelopmental concepts in SCZ, the genetic and biological evidence for DISC1 and NDE1 provides evidence for a shared ‘risk’ pathway.
The underlying molecular mechanisms of the 16p13.11 microduplication, which despite being conserved across mice and human species, have remained elusive. Ingason et al. [115] subdivided the 16p13.1 region between 14.66 and 18.70 Mb (Human Genome Build 36) into three single-copy sequence intervals, denoted intervals I, II and III, each of which is flanked by sequences rich in low-copy repeats (LCRs). All duplications and deletions so far reported are contained within this region, with the most common breakpoints in the LCR clusters distal to interval I and proximal to interval II (so-called Dup I + II carriers) [115]. Dup I + II carriers showed the highest common odds ratio of all 16p13.11 microduplication carriers. The functional implication of these variants in mental illness and the mechanism of disease causation remain unknown, although the potential of investigating this in neuronal cell types derived from hiPSCs from specific patients hold much promise as has been shown in a proof-of-principle studies modelling SCZ and ASD using hiPSCs [43,46,47].
Despite the importance of studying neurodevelopmental disorders and because data from human embryonic tissue are scarce, there is a real challenge of finding an adequate model system. Rodent models have been heavily used to study the cellular function of Nde1, which revealed an important role of NDE1 protein in regulation of proliferation of neuronal progenitors and neuronal migration retardation. However, cortex development and organization is very different in animal models and humans. In particular, the outer subventricular zone, which is only present to a limited degree in rodents, is populated by a unique stem cell subset termed outer radial glia [145,146] that allow for the striking expansion in neuronal output and brain size seen in humans. Therefore, it is not surprising that neurodevelopment diseases cannot be consistently recapitulated in animal models. However, as discussed earlier, the beauty and utility of hiPSC-derived cerebral organoid will present a wealth of new possibilities to thoroughly study the role of NDE1 in cellular proliferation, migration and differentiation, in real time, in the human cerebral cortex and allow the interrogation of genetic risk factors hypothesized to play important roles in human corticogenesis.
7. Cytoplasmic fragile X mental retardation 1–interacting protein
Chromosome 15q11.2 CNVs have emerged as prominent risk factors for various neuropsychiatric disorders, including SCZ, autistic spectrum disorder and intellectual disability [147]. 15q11.2 microdeletion (15q11.2 del) was identified as one of the most frequent CNVs associated with increased risk for SCZ [22], a finding subsequently confirmed in additional cohorts [114,148,149]. 15q CNVs are not as penetrant as other recurrent CNVs associated with neurodevelopmental disorders. They are, however, under negative selection [22] and even in normal subjects, and 15q11.2 del is associated with cognitive variation and changes in structural measures on MRI scanning [150]. 15q11.2 CNVs encompass four genes: non-imprinted in Prader–Willi and Angelman 1 and 2 (NIPA1 and NIPA2), cytoplasmic fragile X mental retardation 1–interacting protein (CYFIP1) and TUBGCP5. While little is known about functions of these genes in mammalian neural development, CYFIP1 has been shown to interact with Rac1 [151], FMRP [152] and eIF4E [153]. Biochemical studies have also identified CYFIP1 as a regulator of the WAVE complex, consisting of WAVE1, WAVE2, Nap1 and Abi1, a complex known to regulate Arp2/3- mediated actin polymerization and membrane protrusion formation in non-neuronal cell lines [151,154,155]. The function of WAVE signalling in mammalian neurogenesis is not well understood. However, an elegant study has been published using stem cells from patients with 15q11.2 CNVs [47]. Yoon et al. [47] took a multifaceted approach to investigate why 15q11.2 CNVs are prominent risk factors for SCZ and ASD. Even in normal control subjects, carriers of the 15q11.2 deletion have cognitive deficits and structural changes on MRI scanning raising questions about how this genetic variant brings about these changes in the carriers. They showed that hiPSC-derived neural progenitor cells carrying 15q11.2 microdeletions exhibited deficits in adherens junctions and apical polarity resulting from haploinsufficiency of CYFIP1 [47]. Furthermore, they showed that deficiency in CYFIP1 and WAVE in the developing mouse cortex affects radial glial cell migration causing ectopic localization outside of the ventricular zone [47]. Targeted human genetic association analyses revealed an epistatic interaction between CYFIP1 and WAVE signalling mediator actin-related protein 2 and risk for SCZ. Therefore, by integrating human neural stem cells, in vivo animal modelling and targeted human genetic association studies, a mechanistic understanding of how 15q11.2 microdeletions affect neural development has been uncovered.
8. Eukaryotic translation initiation factor 4E
Eukaryotic translation initiation factor 4E (EIF4E) is the rate-limiting component of eukaryotic translation initiation and plays a key role in learning and memory through its control of translation within the synapse. EIF4E-mediated translation is the final common process modulated by the mammalian target of rapamycin (mTOR), phosphatase and tensin homologue (PTEN) and fragile X mental retardation protein (FMRP) pathways, all of which are implicated in ASD [156,157]. Germline mutations in PTEN human homologue are present in 1–5% of patients with ASD, and PTEN knockout (KO) mice exhibit cognitive impairment and deficits in social interaction which are rescued by rapamycin [158]. Similarly, mutations in two tuberous sclerosis genes (TSC1 and TSC2) cause ASD in a subset of patients with tuberous sclerosis. Mice with deletions of one copy of TSC1 or TSC2 genes also display deficits in synaptic plasticity and memory that are rescued by rapamycin. The mTOR/Eif4E pathway is hyperactivated in fragile × syndrome (F×S) patients, one of the leading genetic causes for ASD spectrum disorder. In F×S, a full mutation (greater than 200 repeats) leads to hypermethylation of FMR1, an epigenetic mechanism that silences FMR1 gene expression and reduces levels of the FMR1 gene product, FMRP. The absence of FMRP upregulates synaptic translation through failure of recruitment of CYFIP1, the EIF4E-binding protein [159]. The most well-characterized rodent model is the Fmr1 KO mouse, which lacks FMRP protein due to a disruption in its Fmr1 gene. These mice display a range of molecular, cellular, tissue and behavioural abnormalities consistent with the human phenotype, but the pattern and severity is variable depending among other things upon the strain of mouse [160].
Linkage of ASD to the EIF4E region on chromosome 4q was reported in genome-wide linkage studies [161] and was subsequently directly implicated in ASD [162]. In a boy with classic ASD, the authors observed a de novo balanced chromosome translocation between 4q and 5q and mapped the breakpoint site to within a proposed alternative transcript of EIF4E [162]. They then screened 120 ASD families for mutations in EIF4E and found two unrelated families where in each case both autistic siblings and one of the parents harboured the same single-nucleotide insertion at position 225 in the basal element of the EIF4E promoter. Electrophoretic mobility shift assays and reporter gene studies show that this mutation enhances binding of a nuclear factor and EIF4E promoter activity. These genetic observations implicate EIF4E, and more specifically control of EIF4E activity, directly in ASD. They raised the exciting possibility that pharmacological manipulation of EIF4E may provide therapeutic benefit for those with ASD caused by disturbance of the converging pathways controlling EIF4E activity.
These studies have been paralleled by molecular/cellular and animal studies aimed at elucidating the key downstream regulatory mechanisms responsible for so many upstream forms of ASD as well as mutations in EIF4E itself. In the brain, EIF4E activity is fundamental to the regulation of lasting alterations in synaptic strength or plasticity, and of long-term potentiation: these are important in learning and memory. Increased activity in these systems can lead to repetitive, perseverative behaviour patterns. In mice KO of EIF4E-binding protein (4E-BP2), an inhibitor of EIF4E leads to increased translation of neuroligins, also genetically implicated in ASD [14], as well as pathophysiological and behavioural abnormalities similar to those found in ASD. The phenotype was rescued by pharmacological inhibition [163]. In a separate study, direct overexpression of eIF4e in mice results in exaggerated cap-dependent translation and a range of repetitive and perseverative behaviours and social interaction deficits reminiscent of autism. They are accompanied by synaptic pathophysiology in medial prefrontal cortex, striatum and hippocampus. The autistic behaviours are corrected by intracerebral infusion of cap-dependent translation inhibitor 4EG1-1 [164]. In both studies, pharmacological normalization of EIF4E activity rectified many of the abnormalities observed in the mice [163,164]. These findings indicate that behavioural defects caused by exaggerated cap-dependent translation are not irrevocable and may be corrected well into adulthood.
9. Conclusion
The remarkable complexity of the genetic architecture of SCZ and ASD poses formidable challenges for clinicians and scientists aiming to find methods to diagnose, sub-classify, prevent and treat what were until recently considered incurable neurodevelopmental disorders. Over the last 10 years, however, a quiet revolution has been in progress: our understanding of key molecular pathways associated with SCZ and ASD has increased in leaps and bounds as have methods for modelling neurodevelopmental disorders in animals; this has been paralleled by the new opportunities presented by hiPSC technologies, especially when combined with CRISPR editing, three-dimensional organoid development and engraftment of in vitro technologies on to in vivo models; several instances now exist where the worst symptoms of human neurodevelopmental phenotypes can be arrested and/or reversed at least in non-human animal models and with in vitro hiPSC studies. This must surely be one of the most promising areas of current psychiatric research.
Acknowledgements
We are grateful to the Royal Society for their support of the costs of attending the meeting ‘Of mice and mental health: facilitating dialogue between basic and clinical neuroscientists' convened by Amy Milton and Emily A. Holmes.
Data accessibility
This article has no additional data.
Competing interests
We declare we do not have competing interest.
Funding
M.J. is funded by a Wellcome Trust Clinical Postdoctoral Research Fellowship, the Sackler Foundation and the RS Macdonald Trust.
References
- 1.Kapur S, Phillips AG, Insel TR. 2012. Why has it taken so long for biological psychiatry to develop clinical tests and what to do about it? Mol. Psychiatry 17, 1174–1179. ( 10.1038/mp.2012.105) [DOI] [PubMed] [Google Scholar]
- 2.Modabbernia A, Velthorst E, Reichenberg A. 2017. Environmental risk factors for autism: an evidence-based review of systematic reviews and meta-analyses. Mol. Autism 8, 13 ( 10.1186/s13229-017-0121-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Opler M, Charap J, Greig A, Stein V, Polito S, Malaspina D. 2013. Environmental risk factors and schizophrenia. Int. J. Mental Health 42, 23–32. ( 10.2753/IMH0020-7411420102) [DOI] [Google Scholar]
- 4.Chisholm K, Lin A, Abu-Akel A, Wood SJ. 2015. The association between autism and schizophrenia spectrum disorders: a review of eight alternate models of co-occurrence. Neurosci. Biobehav. Rev. 55, 173–183. ( 10.1016/j.neubiorev.2015.04.012) [DOI] [PubMed] [Google Scholar]
- 5.Hamlyn J, Duhig M, McGrath J, Scott J. 2013. Modifiable risk factors for schizophrenia and autism–shared risk factors impacting on brain development. Neurobiol. Dis. 53, 3–9. ( 10.1016/j.nbd.2012.10.023) [DOI] [PubMed] [Google Scholar]
- 6.Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF, Hultman CM. 2009. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet 373, 234–239. ( 10.1016/S0140-6736(09)60072-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan PF, Kendler KS, Neale MC. 2003. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry 60, 1187–1192. ( 10.1001/archpsyc.60.12.1187) [DOI] [PubMed] [Google Scholar]
- 8.Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. 1995. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol. Med. 25, 63–77. ( 10.1017/S0033291700028099) [DOI] [PubMed] [Google Scholar]
- 9.Ronald A, Hoekstra RA. 2011. Autism spectrum disorders and autistic traits: a decade of new twin studies. Am. J. Med. Genet. B 156B, 255–274. ( 10.1002/ajmg.b.31159) [DOI] [PubMed] [Google Scholar]
- 10.Millar JK, et al. 2000. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 9, 1415–1423. ( 10.1093/hmg/9.9.1415) [DOI] [PubMed] [Google Scholar]
- 11.Schneider M, et al. 2014. Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome: results from the international consortium on brain and behavior in 22q11.2 deletion syndrome. Am. J. Psychiatry 171, 627–639. ( 10.1176/appi.ajp.2013.13070864) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards RI, Sutherland GR. 1992. Dynamic mutations: a new class of mutations causing human disease. Cell 70, 709–712. ( 10.1016/0092-8674(92)90302-S) [DOI] [PubMed] [Google Scholar]
- 13.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. 1999. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23, 185–188. ( 10.1038/13810) [DOI] [PubMed] [Google Scholar]
- 14.Jamain S, et al. 2003. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat. Genet. 34, 27–29. ( 10.1038/ng1136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei WH, Hemani G, Haley CS. 2014. Detecting epistasis in human complex traits. Nat. Rev. Genet. 15, 722–733. ( 10.1038/nrg3747) [DOI] [PubMed] [Google Scholar]
- 16.Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. 2009. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752. ( 10.1038/nature08185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conrad DF, et al. 2010. Origins and functional impact of copy number variation in the human genome. Nature 464, 704–712. ( 10.1038/nature08516) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy D, et al. 2011. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron 70, 886–897. ( 10.1016/j.neuron.2011.05.015) [DOI] [PubMed] [Google Scholar]
- 19.Sanders SJ, et al. 2012. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241. ( 10.1038/nature10945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neale BM, et al. 2012. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–245. ( 10.1038/nature11011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Rubeis S, et al. 2014. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215. ( 10.1038/nature13772) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stefansson H, et al. 2008. Large recurrent microdeletions associated with schizophrenia. Nature 455, 232–236. ( 10.1038/nature07229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durand CM, et al. 2007. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 39, 25–27. ( 10.1038/ng1933) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gauthier J, et al. 2009. Novel de novo SHANK3 mutation in autistic patients. Am. J. Med. Genet. B 150B, 421–424. ( 10.1002/ajmg.b.30822) [DOI] [PubMed] [Google Scholar]
- 25.Bucan M, et al. 2009. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 5, e1000536 ( 10.1371/journal.pgen.1000536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim HG, et al. 2008. Disruption of neurexin 1 associated with autism spectrum disorder. Am. J. Hum. Genet. 82, 199–207. ( 10.1016/j.ajhg.2007.09.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berkel S, et al. 2010. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat. Genet. 42, 489–491. ( 10.1038/ng.589) [DOI] [PubMed] [Google Scholar]
- 28.Fernandez T, Morgan T, Davis N, Klin A, Morris A, Farhi A, Lifton RP, State MW. 2004. Disruption of contactin 4 (CNTN4) results in developmental delay and other features of 3p deletion syndrome. Am. J. Hum. Genet. 74, 1286–1293. ( 10.1086/421474) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandez T, Morgan T, Davis N, Klin A, Morris A, Farhi A, Lifton RP, State MW. 2008. Disruption of contactin 4 (CNTN4) results in developmental delay and other features of 3p deletion syndrome. Am. J. Hum. Genet. 82, 1385 ( 10.1016/j.ajhg.2008.04.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glessner JT, et al. 2009. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 459, 569–573. ( 10.1038/nature07953) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakkaloglu B, et al. 2008. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am. J. Hum. Genet. 82, 165–173. ( 10.1016/j.ajhg.2007.09.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, Stephan DA, Morton DH. 2006. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N. Engl. J. Med. 354, 1370–1377. ( 10.1056/NEJMoa052773) [DOI] [PubMed] [Google Scholar]
- 33.Need AC, Goldstein DB. 2014. Schizophrenia genetics comes of age. Neuron 83, 760–763. ( 10.1016/j.neuron.2014.08.015) [DOI] [PubMed] [Google Scholar]
- 34.Harrison PJ. 2015. GABA circuitry, cells and molecular regulation in schizophrenia: life in the graveyard. Schizophr. Res. 167, 108–110. ( 10.1016/j.schres.2015.02.003) [DOI] [PubMed] [Google Scholar]
- 35.Jablensky A. 2015. Schizophrenia or schizophrenias? The challenge of genetic parsing of a complex disorder. Am. J. Psychiatry 172, 105–107. ( 10.1176/appi.ajp.2014.14111452) [DOI] [PubMed] [Google Scholar]
- 36.Schizophrenia Working Group of the Psychiatric Genomics Consortium. 2014. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. ( 10.1038/nature13595) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, et al. 2017. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nature Genetics 49, 1576–1583. ( 10.1038/ng.3973) [DOI] [PubMed] [Google Scholar]
- 38.Hannon E, et al. 2016. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat. Neurosci. 19, 48–54. ( 10.1038/nn.4182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Purcell SM, et al. 2014. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506, 185–190. ( 10.1038/nature12975) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davidson S, et al. 2011. Differential activity by polymorphic variants of a remote enhancer that supports galanin expression in the hypothalamus and amygdala: implications for obesity, depression and alcoholism. Neuropsychopharmacology 36, 2211–2221. ( 10.1038/npp.2011.93) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hing B, Davidson S, Lear M, Breen G, Quinn J, McGuffin P, MacKenzie A. 2012. A polymorphism associated with depressive disorders differentially regulates brain derived neurotrophic factor promoter IV activity. Biol. Psychiatry 71, 618–626. ( 10.1016/j.biopsych.2011.11.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyle EA, Li YI, Pritchard JK. 2017. An expanded view of complex traits: from polygenic to omnigenic. Cell 169, 1177–1186. ( 10.1016/j.cell.2017.05.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de la Torre-Ubieta L, Won H, Stein JL, Geschwind DH. 2016. Advancing the understanding of autism disease mechanisms through genetics. Nat. Med. 22, 345–361. ( 10.1038/nm.4071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCammon JM, Sive H. 2015. Addressing the genetics of human mental health disorders in model organisms. Annu. Rev. Genomics Hum. Genet. 16, 173–197. ( 10.1146/annurev-genom-090314-050048) [DOI] [PubMed] [Google Scholar]
- 45.Silverman JL, Yang M, Lord C, Crawley JN. 2010. Behavioural phenotyping assays for mouse models of autism. Nat. Rev. Neurosci. 11, 490–502. ( 10.1038/nrn2851) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brennand KJ, et al. 2011. Modelling schizophrenia using human induced pluripotent stem cells. Nature 473, 221–225. ( 10.1038/nature09915) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoon KJ, et al. 2014. Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell 15, 79–91. ( 10.1016/j.stem.2014.05.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pasca AM, et al. 2015. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671–678. ( 10.1038/nmeth.3415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Birey F, et al. 2017. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59. ( 10.1038/nature22330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lancaster MA, Knoblich JA. 2014. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 9, 2329–2340. ( 10.1038/nprot.2014.158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lancaster MA, et al. 2013. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379. ( 10.1038/nature12517) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, Sasai Y. 2013. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. USA 110, 20 284–20 289. ( 10.1073/pnas.1315710110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe K, Kamiya D, Nishiyama A, Katayama T, Nozaki S, Kawasaki H, Watanabe Y, Mizuseki K, Sasai Y. 2005. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat. Neurosci. 8, 288–296. ( 10.1038/nn1402) [DOI] [PubMed] [Google Scholar]
- 54.Muguruma K, Nishiyama A, Kawakami H, Hashimoto K, Sasai Y. 2015. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 10, 537–550. ( 10.1016/j.celrep.2014.12.051) [DOI] [PubMed] [Google Scholar]
- 55.Ranga A, Girgin M, Meinhardt A, Eberle D, Caiazzo M, Tanaka EM, Lutolf MP. 2016. Neural tube morphogenesis in synthetic 3D microenvironments. Proc. Natl Acad. Sci. USA 113, E6831–E6839. ( 10.1073/pnas.1603529113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davis SW, Mortensen AH, Keisler JL, Zacharias AL, Gage PJ, Yamamura K, Camper SA. 2016. Beta-catenin is required in the neural crest and mesencephalon for pituitary gland organogenesis. BMC Dev. Biol. 16, 16 ( 10.1186/s12861-016-0118-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakaguchi H, Kadoshima T, Soen M, Narii N, Ishida Y, Ohgushi M, Takahashi J, Eiraku M, Sasai Y. 2015. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun. 6, 8896 ( 10.1038/ncomms9896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eiraku M, Sasai Y. 2011. Mouse embryonic stem cell culture for generation of three-dimensional retinal and cortical tissues. Nat. Protoc. 7, 69–79. ( 10.1038/nprot.2011.429) [DOI] [PubMed] [Google Scholar]
- 59.Kuwahara A, Ozone C, Nakano T, Saito K, Eiraku M, Sasai Y. 2015. Generation of a ciliary margin-like stem cell niche from self-organizing human retinal tissue. Nat. Commun. 6, 6286 ( 10.1038/ncomms7286) [DOI] [PubMed] [Google Scholar]
- 60.Tao Y, Zhou X, Liu D, Li H, Liang C, Li F, Chen Q. 2016. Proportion of collagen type II in the extracellular matrix promotes the differentiation of human adipose-derived mesenchymal stem cells into nucleus pulposus cells. Biofactors 42, 212–223. ( 10.1002/biof.1266) [DOI] [PubMed] [Google Scholar]
- 61.Marin O, Muller U. 2014. Lineage origins of GABAergic versus glutamatergic neurons in the neocortex. Curr. Opin. Neurobiol. 26, 132–141. ( 10.1016/j.conb.2014.01.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bartolini G, Ciceri G, Marin O. 2013. Integration of GABAergic interneurons into cortical cell assemblies: lessons from embryos and adults. Neuron 79, 849–864. ( 10.1016/j.neuron.2013.08.014) [DOI] [PubMed] [Google Scholar]
- 63.Perrier AL, Tabar V, Barberi T, Rubio ME, Bruses J, Topf N, Harrison NL, Studer L. 2004. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc. Natl Acad. Sci. USA 101, 12 543–12 548. ( 10.1073/pnas.0404700101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li XJ, Du ZW, Zarnowska ED, Pankratz M, Hansen LO, Pearce RA, Zhang SC. 2005. Specification of motoneurons from human embryonic stem cells. Nat. Biotechnol. 23, 215–221. ( 10.1038/nbt1063) [DOI] [PubMed] [Google Scholar]
- 65.Tabata H. 2015. Diverse subtypes of astrocytes and their development during corticogenesis. Front. Neurosci. 9, 114 ( 10.3389/fnins.2015.00114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang S, et al. 2013. Human iPSC-derived oligodendrocyte progenitor cells can myelinate and rescue a mouse model of congenital hypomyelination. Cell Stem Cell 12, 252–264. ( 10.1016/j.stem.2012.12.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eiraku M, et al. 2008. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3, 519–532. ( 10.1016/j.stem.2008.09.002) [DOI] [PubMed] [Google Scholar]
- 68.Parr CJC, Yamanaka S, Saito H. 2017. An update on stem cell biology and engineering for brain development. Mol. Psychiatry 22, 808–819. ( 10.1038/mp.2017.66) [DOI] [PubMed] [Google Scholar]
- 69.van den Ameele J, Tiberi L, Vanderhaeghen P, Espuny-Camacho I. 2014. Thinking out of the dish: what to learn about cortical development using pluripotent stem cells. Trends Neurosci. 37, 334–342. ( 10.1016/j.tins.2014.03.005) [DOI] [PubMed] [Google Scholar]
- 70.Anderson S, Vanderhaeghen P. 2014. Cortical neurogenesis from pluripotent stem cells: complexity emerging from simplicity. Curr. Opin. Neurobiol. 27, 151–157. ( 10.1016/j.conb.2014.03.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qian X, et al. 2016. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165, 1238–1254. ( 10.1016/j.cell.2016.04.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vadivelu RK, Kamble H, Shiddiky MJA, Nguyen N-T. 2017. Microfluidic technology for the generation of cell spheroids and their applications. Micromachines 8, 94 ( 10.3390/mi8040094) [DOI] [Google Scholar]
- 73.Lancaster MA, et al. 2017. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 35, 659–666. ( 10.1038/nbt.3906) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Quadrato G, et al. 2017. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545, 48–53. ( 10.1038/nature22047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sekar A, et al. 2016. Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–183. ( 10.1038/nature16549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boksa P. 2012. Abnormal synaptic pruning in schizophrenia: urban myth or reality? J. Psychiatry Neurosci. 37, 75–77. ( 10.1503/jpn.120007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ripke S, et al. 2013. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 45, 1150–1159. ( 10.1038/ng.2742) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pasca SP, et al. 2011. Using iPSC-derived neurons to uncover cellular phenotypes associated with timothy syndrome. Nat. Med. 17, 1657–1662. ( 10.1038/nm.2576) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bortone D, Polleux F. 2009. KCC2 expression promotes the termination of cortical interneuron migration in a voltage-sensitive calcium-dependent manner. Neuron 62, 53–71. ( 10.1016/j.neuron.2009.01.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gratten J, Visscher PM, Mowry BJ, Wray NR. 2013. Interpreting the role of de novo protein-coding mutations in neuropsychiatric disease. Nat. Genet. 45, 234–238. ( 10.1038/ng.2555) [DOI] [PubMed] [Google Scholar]
- 81.Porteous DJ, et al. 2014. DISC1 as a genetic risk factor for schizophrenia and related major mental illness: response to Sullivan. Mol. Psychiatry 19, 141–143. ( 10.1038/mp.2013.160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brandon NJ, Sawa A. 2011. Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat. Rev. Neurosci. 12, 707–722. ( 10.1038/nrn3120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Clair S, Blackwood D, Muir D, Carothers W, Walker A, Spowart M, Gosden G, Evans HJ. 1990. Association within a family of a balanced autosomal translocation with major mental illness. Lancet 336, 13–16. ( 10.1016/0140-6736(90)91520-K) [DOI] [PubMed] [Google Scholar]
- 84.Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. 2001. Schizophrenia and affective disorders—cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am. J. Hum. Genet. 69, 428–433. ( 10.1086/321969) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guha S, et al. 2013. Implication of a rare deletion at distal 16p11.2 in schizophrenia. JAMA Psychiatry 70, 253–260. ( 10.1001/2013.jamapsychiatry.71) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meyer KD, Morris JA. 2008. Immunohistochemical analysis of Disc1 expression in the developing and adult hippocampus. Gene Expr. Patterns 8, 494–501. ( 10.1016/j.gep.2008.06.005) [DOI] [PubMed] [Google Scholar]
- 87.Duan X, et al. 2007. Disrupted-in-schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell 130, 1146–1158. ( 10.1016/j.cell.2007.07.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lipska BK, et al. 2006. Expression of DISC1 binding partners is reduced in schizophrenia and associated with DISC1 SNPs. Hum. Mol. Genet. 15, 1245–1258. ( 10.1093/hmg/ddl040) [DOI] [PubMed] [Google Scholar]
- 89.Ozeki Y, et al. 2003. Disrupted-in-schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc. Natl Acad. Sci. USA 100, 289–294. ( 10.1073/pnas.0136913100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Miyoshi K, Honda A, Baba K, Taniguchi M, Oono K, Fujita T, Kuroda S, Katayama T, Tohyama M. 2003. Disrupted-in-schizophrenia 1, a candidate gene for schizophrenia, participates in neurite outgrowth. Mol. Psychiatry 8, 685–694. ( 10.1038/sj.mp.4001352) [DOI] [PubMed] [Google Scholar]
- 91.Kamiya A, et al. 2005. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat. Cell Biol. 7, 1167–1178. ( 10.1038/ncb1328) [DOI] [PubMed] [Google Scholar]
- 92.Tomoda T, Sumitomo A, Jaaro-Peled H, Sawa A. 2016. Utility and validity of DISC1 mouse models in biological psychiatry. Neuroscience 321, 99–107. ( 10.1016/j.neuroscience.2015.12.061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shen S, et al. 2008. Schizophrenia-related neural and behavioral phenotypes in transgenic mice expressing truncated Disc1. J. Neurosci. 28, 10 893–10 904. ( 10.1523/JNEUROSCI.3299-08.2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Niwa M, et al. 2010. Knockdown of DISC1 by in utero gene transfer disturbs postnatal dopaminergic maturation in the frontal cortex and leads to adult behavioral deficits. Neuron 65, 480–489. ( 10.1016/j.neuron.2010.01.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yerabham ASK, et al. 2017. A structural organization for the disrupted in schizophrenia 1 protein, identified by high-throughput screening, reveals distinctly folded regions, which are bisected by mental illness-related mutations. J. Biol. Chem. 292, 6468–6477. ( 10.1074/jbc.M116.773903) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sawamura N, et al. 2008. Nuclear DISC1 regulates CRE-mediated gene transcription and sleep homeostasis in the fruit fly. Mol. Psychiatry 13, 1138–1148, 1069 ( 10.1038/mp.2008.101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Furukubo-Tokunaga K, et al. 2016. Visualization of DISC1-Dysbindin interaction in glutamatergic synaptic termini in fruit flies. Mol. Psychiatry 21, 1157 ( 10.1038/mp.2016.142) [DOI] [PubMed] [Google Scholar]
- 98.Singh KK, et al. 2011. Common DISC1 polymorphisms disrupt Wnt/GSK3beta signaling and brain development. Neuron 72, 545–558. ( 10.1016/j.neuron.2011.09.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Srikanth P, et al. 2015. Genomic DISC1 disruption in hiPSCs alters Wnt signaling and neural cell fate. Cell Rep. 12, 1414–1429. ( 10.1016/j.celrep.2015.07.061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chiang CH, Su Y, Wen Z, Yoritomo N, Ross CA, Margolis RL, Song H, Ming GL. 2011. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Mol. Psychiatry 16, 358–360. ( 10.1038/mp.2011.13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wen Z, et al. 2014. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature 515, 414–418. ( 10.1038/nature13716) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Maher BJ, LoTurco JJ. 2012. Disrupted-in-schizophrenia (DISC1) functions presynaptically at glutamatergic synapses. PLoS ONE 7, e34053 ( 10.1371/journal.pone.0034053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tang W, Thevathasan JV, Lin Q, Lim KB, Kuroda K, Kaibuchi K, Bilger M, Soong TW, Fivaz M. 2016. Stimulation of synaptic vesicle exocytosis by the mental disease gene DISC1 is mediated by N-type voltage-gated calcium channels. Front. Synaptic Neurosci. 8, 15 ( 10.3389/fnsyn.2016.00015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Crabtree GW, et al. 2017. Alteration of neuronal excitability and short-term synaptic plasticity in the prefrontal cortex of a mouse model of mental illness. J. Neurosci. 37, 4158–4180. ( 10.1523/JNEUROSCI.4345-15.2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Insel TR. 2010. Rethinking schizophrenia. Nature 468, 187–193. ( 10.1038/nature09552) [DOI] [PubMed] [Google Scholar]
- 106.Davis KL, Stewart DG, Friedman JI, Buchsbaum M, Harvey PD, Hof PR, Buxbaum J, Haroutunian V. 2003. White matter changes in schizophrenia: evidence for myelin-related dysfunction. Arch. Gen. Psychiatry 60, 443–456. ( 10.1001/archpsyc.60.5.443) [DOI] [PubMed] [Google Scholar]
- 107.Kubicki M, McCarley RW, Shenton ME. 2005. Evidence for white matter abnormalities in schizophrenia. Curr. Opin. Psychiatry 18, 121–134. ( 10.1097/00001504-200503000-00004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Whalley HC, et al. 2015. Effects of a balanced translocation between chromosomes 1 and 11 disrupting the disc1 locus on white matter integrity. PLoS ONE 10, e0130900 ( 10.1371/journal.pone.0130900) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu Y, Liu H, Sauvey C, Yao L, Zarnowska ED, Zhang SC. 2013. Directed differentiation of forebrain GABA interneurons from human pluripotent stem cells. Nat. Protoc. 8, 1670–1679. ( 10.1038/nprot.2013.106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alkuraya FS, et al. 2011. Human mutations in NDE1 cause extreme microcephaly with lissencephaly [corrected]. Am. J. Hum. Genet. 88, 536–547. ( 10.1016/j.ajhg.2011.04.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mefford HC, et al. 2007. Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am. J. Hum. Genet. 81, 1057–1069. ( 10.1086/522591) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ullmann R, et al. 2007. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum. Mutat. 28, 674–682. ( 10.1002/humu.20546) [DOI] [PubMed] [Google Scholar]
- 113.Williams NM, et al. 2010. Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet 376, 1401–1408. ( 10.1016/S0140-6736(10)61109-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, Craddock N, Owen MJ, O'Donovan MC. 2009. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum. Mol. Genet. 18, 1497–1503. ( 10.1093/hmg/ddp043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ingason A, et al. 2011. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol. Psychiatry 16, 17–25. ( 10.1038/mp.2009.101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Johnstone M, et al. 2015. Copy number variations in DISC1 and DISC1-interacting partners in major mental illness. Mol. Neuropsychiatry 1, 175–190. ( 10.1159/000438788) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hennah W, et al. 2007. Families with the risk allele of DISC1 reveal a link between schizophrenia and another component of the same molecular pathway, NDE1. Hum. Mol. Genet. 16, 453–462. ( 10.1093/hmg/ddl462) [DOI] [PubMed] [Google Scholar]
- 118.Feng Y, Olson EC, Stukenberg PT, Flanagan LA, Kirschner MW, Walsh CA. 2000. LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron 28, 665–679. ( 10.1016/S0896-6273(00)00145-8) [DOI] [PubMed] [Google Scholar]
- 119.Efimov VP, Morris NR. 2000. The LIS1-related NUDF protein of Aspergillus nidulans interacts with the coiled-coil domain of the NUDE/RO11 protein. J. Cell Biol. 150, 681–688. ( 10.1083/jcb.150.3.681) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kitagawa M, Umezu M, Aoki J, Koizumi H, Arai H, Inoue K. 2000. Direct association of LIS1, the lissencephaly gene product, with a mammalian homologue of a fungal nuclear distribution protein, rNUDE. FEBS Lett. 479, 57–62. ( 10.1016/S0014-5793(00)01856-1) [DOI] [PubMed] [Google Scholar]
- 121.Niethammer M, Smith DS, Ayala R, Peng J, Ko J, Lee MS, Morabito M, Tsai LH. 2000. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron 28, 697–711. ( 10.1016/S0896-6273(00)00147-1) [DOI] [PubMed] [Google Scholar]
- 122.Sasaki S, et al. 2005. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Mol. Cell. Biol. 25, 7812–7827. ( 10.1128/MCB.25.17.7812-7827.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yan X, Li F, Liang Y, Shen Y, Zhao X, Huang Q, Zhu X. 2003. Human Nudel and NudE as regulators of cytoplasmic dynein in poleward protein transport along the mitotic spindle. Mol. Cell. Biol. 23, 1239–1250. ( 10.1128/MCB.23.4.1239-1250.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shu T, Ayala R, Nguyen MD, Xie Z, Gleeson JG, Tsai LH. 2004. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron 44, 263–277. ( 10.1016/j.neuron.2004.09.030) [DOI] [PubMed] [Google Scholar]
- 125.Li J, Lee WL, Cooper JA. 2005. NudEL targets dynein to microtubule ends through LIS1. Nat. Cell Biol. 7, 686–690. ( 10.1038/ncb1273) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Stehman SA, Chen Y, McKenney RJ, Vallee RB. 2007. NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. J. Cell Biol. 178, 583–594. ( 10.1083/jcb.200610112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vergnolle MA, Taylor SS. 2007. Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Curr. Biol. 17, 1173–1179. ( 10.1016/j.cub.2007.05.077) [DOI] [PubMed] [Google Scholar]
- 128.Bradshaw NJ, Ogawa F, Antolin-Fontes B, Chubb JE, Carlyle BC, Christie S, Claessens A, Porteous DJ, Millar JK. 2008. DISC1, PDE4B, and NDE1 at the centrosome and synapse. Biochem. Biophys. Res. Commun. 377, 1091–1096. ( 10.1016/j.bbrc.2008.10.120) [DOI] [PubMed] [Google Scholar]
- 129.Burdick KE, et al. 2008. Elucidating the relationship between DISC1, NDEL1 and NDE1 and the risk for schizophrenia: evidence of epistasis and competitive binding. Hum. Mol. Genet. 17, 2462–2473. ( 10.1093/hmg/ddn146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lam C, Vergnolle MA, Thorpe L, Woodman PG, Allan VJ. 2010. Functional interplay between LIS1, NDE1 and NDEL1 in dynein-dependent organelle positioning. J. Cell Sci. 123, 202–212. ( 10.1242/jcs.059337) [DOI] [PubMed] [Google Scholar]
- 131.McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. 2010. LIS1 and NudE induce a persistent dynein force-producing state. Cell 141, 304–314. ( 10.1016/j.cell.2010.02.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shmueli A, et al. 2010. Ndel1 palmitoylation: a new mean to regulate cytoplasmic dynein activity. EMBO J. 29, 107–119. ( 10.1038/emboj.2009.325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vallee RB, McKenney RJ, Ori-McKenney KM. 2012. Multiple modes of cytoplasmic dynein regulation. Nat. Cell Biol. 14, 224–230. ( 10.1038/ncb2420) [DOI] [PubMed] [Google Scholar]
- 134.Allan VJ. 2011. Cytoplasmic dynein. Biochem. Soc. Trans. 39, 1169–1178. ( 10.1042/BST0391169) [DOI] [PubMed] [Google Scholar]
- 135.Kardon JR, Vale RD. 2009. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 10, 854–865. ( 10.1038/nrm2804) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hirotsune S, Fleck MW, Gambello MJ, Bix GJ, Chen A, Clark GD, Ledbetter DH, McBain CJ, Wynshaw-Boris A. 1998. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat. Genet. 19, 333–339. ( 10.1038/1221) [DOI] [PubMed] [Google Scholar]
- 137.Paylor R, Hirotsune S, Gambello MJ, Yuva-Paylor L, Crawley JN, Wynshaw-Boris A. 1999. Impaired learning and motor behavior in heterozygous Pafah1b1 (Lis1) mutant mice. Learn. Mem. 6, 521–537. ( 10.1101/lm.6.5.521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Feng Y, Walsh CA. 2004. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron 44, 279–293. ( 10.1016/j.neuron.2004.09.023) [DOI] [PubMed] [Google Scholar]
- 139.Bakircioglu M, et al. 2011. The essential role of centrosomal NDE1 in human cerebral cortex neurogenesis. Am. J. Hum. Genet. 88, 523–535. ( 10.1016/j.ajhg.2011.03.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Houlihan SL, Feng Y. 2014. The scaffold protein Nde1 safeguards the brain genome during S phase of early neural progenitor differentiation. eLife 3, e03297 ( 10.7554/eLife.03297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Doobin DJ, Kemal S, Dantas TJ, Vallee RB. 2016. Severe NDE1-mediated microcephaly results from neural progenitor cell cycle arrests at multiple specific stages. Nat. Commun. 7, 12551 ( 10.1038/ncomms12551) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tropeano M, et al. 2013. Male-biased autosomal effect of 16p13.11 copy number variation in neurodevelopmental disorders. PLoS ONE 8, e61365 ( 10.1371/journal.pone.0061365) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kimura H, et al. 2015. Identification of rare, single-nucleotide mutations in NDE1 and their contributions to schizophrenia susceptibility. Schizophr. Bull. 41, 744–753. ( 10.1093/schbul/sbu147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tomppo L, Ekelund J, Lichtermann D, Veijola J, Jarvelin MR, Hennah W. 2012. DISC1 conditioned GWAS for psychosis proneness in a large Finnish birth cohort. PLoS ONE 7, e30643 ( 10.1371/journal.pone.0030643) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fietz SA, et al. 2010. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat. Neurosci. 13, 690–699. ( 10.1038/nn.2553) [DOI] [PubMed] [Google Scholar]
- 146.Hansen DV, Lui JH, Parker PR, Kriegstein AR. 2010. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561. ( 10.1038/nature08845) [DOI] [PubMed] [Google Scholar]
- 147.Malhotra D, Sebat J. 2012. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell 148, 1223–1241. ( 10.1016/j.cell.2012.02.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tam GW, et al. 2010. Confirmed rare copy number variants implicate novel genes in schizophrenia. Biochem. Soc. Trans. 38, 445–451. ( 10.1042/BST0380445) [DOI] [PubMed] [Google Scholar]
- 149.Zhao Q, et al. 2013. Rare CNVs and tag SNPs at 15q11.2 are associated with schizophrenia in the Han Chinese population. Schizophr. Bull. 39, 712–719. ( 10.1093/schbul/sbr197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Stefansson H, et al. 2014. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature 505, 361–366. ( 10.1038/nature12818) [DOI] [PubMed] [Google Scholar]
- 151.Kobayashi K, et al. 1998. p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J. Biol. Chem. 273, 291–295. ( 10.1074/jbc.273.1.291) [DOI] [PubMed] [Google Scholar]
- 152.Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL. 2001. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc. Natl Acad. Sci. USA 98, 8844–8849. ( 10.1073/pnas.151231598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Napoli I, et al. 2008. The fragile × syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 134, 1042–1054. ( 10.1016/j.cell.2008.07.031) [DOI] [PubMed] [Google Scholar]
- 154.Kunda P, Craig G, Dominguez V, Baum B. 2003. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 13, 1867–1875. ( 10.1016/j.cub.2003.10.005) [DOI] [PubMed] [Google Scholar]
- 155.Steffen A, Rottner K, Ehinger J, Innocenti M, Scita G, Wehland J, Stradal TE. 2004. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 23, 749–759. ( 10.1038/sj.emboj.7600084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Auerbach BD, Osterweil EK, Bear MF. 2011. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature 480, 63–68. ( 10.1038/nature10658) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.O'Roak BJ, et al. 2012. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250. ( 10.1038/nature10989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhou J, Brugarolas J, Parada LF. 2009. Loss of Tsc1, but not Pten, in renal tubular cells causes polycystic kidney disease by activating mTORC1. Hum. Mol. Genet. 18, 4428–4441. ( 10.1093/hmg/ddp398) [DOI] [PubMed] [Google Scholar]
- 159.Hoeffer CA, et al. 2012. Altered mTOR signaling and enhanced CYFIP2 expression levels in subjects with fragile X syndrome. Genes Brain Behav. 11, 332–341. ( 10.1111/j.1601-183X.2012.00768.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kazdoba TM, Leach PT, Silverman JL, Crawley JN. 2014. Modeling fragile × syndrome in the Fmr1 knockout mouse. Intractable Rare Dis. Res. 3, 118–133. ( 10.5582/irdr.2014.01024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yonan AL, et al. 2003. A genomewide screen of 345 families for autism-susceptibility loci. Am. J. Hum. Genet. 73, 886–897. ( 10.1086/378778) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Neves-Pereira M, Muller B, Massie D, Williams JH, O'Brien PC, Hughes A, Shen SB, Clair DS, Miedzybrodzka Z. 2009. Deregulation of EIF4E: a novel mechanism for autism. J. Med. Genet. 46, 759–765. ( 10.1136/jmg.2009.066852) [DOI] [PubMed] [Google Scholar]
- 163.Gkogkas CG, et al. 2013. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature 493, 371–377. ( 10.1038/nature11628) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Santini E, Huynh TN, MacAskill AF, Carter AG, Pierre P, Ruggero D, Kaphzan H, Klann E. 2013. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature 493, 411–415. ( 10.1038/nature11782) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.