In the present report, we show that the human fecal microbiota contains promising and universal biomarkers for the noninvasive evaluation of inflammatory bowel disease severity and IFX treatment efficacy, emphasizing the potential ability to mine the gut microbiota as a modality to stratify IBD patients and apply personalized therapy for optimal outcomes.
KEYWORDS: gut microbiota, infliximab treatment, disease activity, inflammatory bowel disease
ABSTRACT
Gut microbiota dysbiosis contributes to the onset and perpetuation of inflammatory bowel disease (IBD). Given that gut microbiotas vary across geography and ethnicity, it remains obscure whether any universal microbial signatures for IBD diagnosis and prognosis evaluation exist irrespective of populations. Here we profiled the fecal microbiota of a series of Chinese IBD patients and combined them with two Western IBD cohorts, PRISM and RISK, for meta-analyses. We found that the gut microbial alteration patterns in IBD are similar among Chinese and Westerners. Our prediction model based on gut microbiome for IBD diagnosis is robust across the cohorts, which showed 87.5% and 79.1% prediction accuracy in Crohn’s disease (CD) and ulcerative colitis (UC) patients, respectively. A relative increase in the levels of Actinobacteria and Proteobacteria (Enterobacteriaceae) and a relative decrease in the levels of Firmicutes (Clostridiales) were strongly correlated with IBD severity (P < 0.05). Additionally, restoration of gut microbiota diversity and a significant increase in Clostridiales relative abundance were found in patients responding to infliximab (IFX [Remicade]) treatment compared to those in relapse. Moreover, certain microbes, mainly Clostridiales, predicted the treatment effectiveness with 86.5% accuracy alone and 93.8% accuracy in combination with calprotectin levels and Crohn’s disease activity index (CDAI). Taking the results together, we conclude that gut microbiota can offer a set of universal biomarkers for diagnosis, disease activity evaluation, and infliximab treatment response prediction in IBD.
IMPORTANCE In the present report, we show that the human fecal microbiota contains promising and universal biomarkers for the noninvasive evaluation of inflammatory bowel disease severity and IFX treatment efficacy, emphasizing the potential ability to mine the gut microbiota as a modality to stratify IBD patients and apply personalized therapy for optimal outcomes.
INTRODUCTION
Inflammatory bowel diseases (IBD) are chronic relapsing inflammatory disorders that have been categorized into two main clinical phenotypes: ulcerative colitis (UC) and Crohn’s disease (CD). The global prevalence of IBD has been rising significantly, largely paralleling industrialization, with a concurrent increase in health care costs (1). Although their etiology and pathogenesis are not fully understood, genetic polymorphisms associated with IBD suggest an underlying role of aberrant immune responses to imbalanced gut microbiota as a key mechanism involved in disease pathogenesis (2, 3). Genome-wide association studies (GWAS) have identified more than 160 single nucleotide polymorphisms (SNPs) associated with IBD, many of which are involved in pathways that modulate the host response to microbial stimuli. The NOD2 gene, the first gene identified to have an association with IBD susceptibility, recognizes components of the bacterial cell membrane (4, 5). In patients with IBD, the diversity of the fecal microbiome is reduced compared to that of healthy controls (HC) (6). The major changes in the diversity of gut microbiota in the context of new-onset CD (before treatment is initiated) are correlated strongly with disease status (7–9). A broad pattern that differentiates IBD patients from healthy individuals has begun to emerge: reduced biodiversity, decreased abundance of several taxa in the Firmicutes phylum, and increased abundance of Gammaproteobacteria (10–14). Such alterations are consistent with the response of a complex microbial community to environmental stressors introduced by the host inflammatory response such as the production of alternative electron acceptors that promote nitrate respiration (15), as well as of oxygen radicals leading to oxidative stress (11). However, most of the aforementioned studies have been conducted in North American, European, and Japanese populations, whose genetics, ethnic backgrounds, environments, dietary habits, and lifestyles differ from those in China (16). It is well known that the gut microbiotas of human populations residing across different geographical locations are associated with significant differences in microbiota composition (17). However, it is currently unknown whether a disease state such as IBD would modify the composition of the gut microbiota in a consistent fashion independently of these geographical influences.
The natural history of CD is characterized by poorly predictable phases of quiescence and activity (18). In the absence of reliable predictors at the individual level, optimal medical strategies (e.g., top-up versus top-down management) remain debatable. The use of Infliximab (IFX [Remicade]), a human/mouse chimeric monoclonal antibody targeting tumor necrosis factor alpha (TNF-α) (19), is an effective treatment for patients with refractory moderate to severe CD. Nevertheless, some patients do not respond to IFX or else relapse after initial response. Our previous studies had suggested that the relapse rates were 10.53% in CD (20) and 25.0% in UC (21) at week 22 and were much higher after 30 weeks posttreatment. The inability to effectively predict the long-term efficacy of IFX treatment prevents adoption of a more targeted approach to the use of this class of biological agents with the intent of reducing both costs and the potential incidence of adverse events.
The choice of therapy is currently driven primarily by clinical predictive factors (22–24). Fecal calprotectin, a cytosolic protein of mucosal neutrophils that are extruded into the gastrointestinal (GI) tract when they undergo apoptosis (25), is useful to predict relapse in IBD patients (23, 26). Patients with high C-reactive protein (CRP) levels are more likely to achieve and maintain a response to biological therapy than those with low or normal CRP levels (27). Recently, the gut microbiota has gained attention as a reservoir of numerous microbial markers. It was found that reduced Firmicutes abundance is correlated with a shorter time to relapse after IFX withdrawal (28). The abundance of six clades of bacteria, including Eubacterium rectale and Bifidobacterium spp., predicted the response to anti-TNF-α medication in pediatric IBD patients (29). Thus, the gut microbiota may provide potential biomarkers for monitoring and predicting IBD treatment outcomes.
A global rise in IBD has been reported, especially in countries with previously low incidence rates, including China. To our knowledge, only a few studies have reported the characteristics of gut microbiota diversity in Chinese IBD patients (30, 31) and have mainly described correlations between shifts in microbial composition and disease phenotypes. Quantitative real-time PCR or denaturing gradient gel electrophoresis (DGGE), each targeting the 16S rRNA gene of selected bacteria, was used in those two Chinese studies. Due to the low throughput and low resolution of these methods, some key players of the microbial dysbiosis in IBD may not be discovered.
Using bar-coded 16S rRNA amplicon sequencing, we examined the gut microbiota of Chinese healthy individuals and patients with new-onset UC and CD before treatment initiation. We also included subjects representing a variety of phenotypes with respect to disease locations and activities. These data sets were compared with results from the RISK and PRISM IBD cohorts in the United States (9). This multicenter association study of gut microbiota and IBD, in which over 1,000 treatment-naive patients were included, represents the most comprehensive cross-cohort and cross-ethnic analysis involving this disease state performed to date. Additionally, we characterized the composition of fecal microbiotas from prospectively recruited patients with CD prior to and after receiving IFX treatment. The aims of this study were to identify gut microbiome patterns in Chinese IBD patients with different disease activities and statuses, to discover homogeneity and heterogeneity of IBD gut microbiota patterns in different populations, and to find out if there are any universal and specific biomarkers in gut microbes which can indicate and predict disease progression and IFX treatment responses.
RESULTS
Dysbiosis of gut microbiota patterns in Chinese IBD patients.
We recruited 72 CD patients, 51 UC patients, and 73 healthy volunteers who were members of the Han ethnic group living in China. The clinical characteristics of the participants are shown in Table S1 in the supplemental material. Sixteen patients with active CD received IFX treatment and were followed for up to 30 weeks posttreatment.
Baseline clinical characteristics of the subjects. Download TABLE S1, DOCX file, 0.1 MB (57.4KB, docx) .
Copyright © 2018 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A total number of 1,376,142 high-quality 16S rRNA gene sequences were obtained for 196 samples from the cross-sectional study, with an average of 14,042 ± 7,304 (mean ± standard deviation [SD]) sequences per sample.
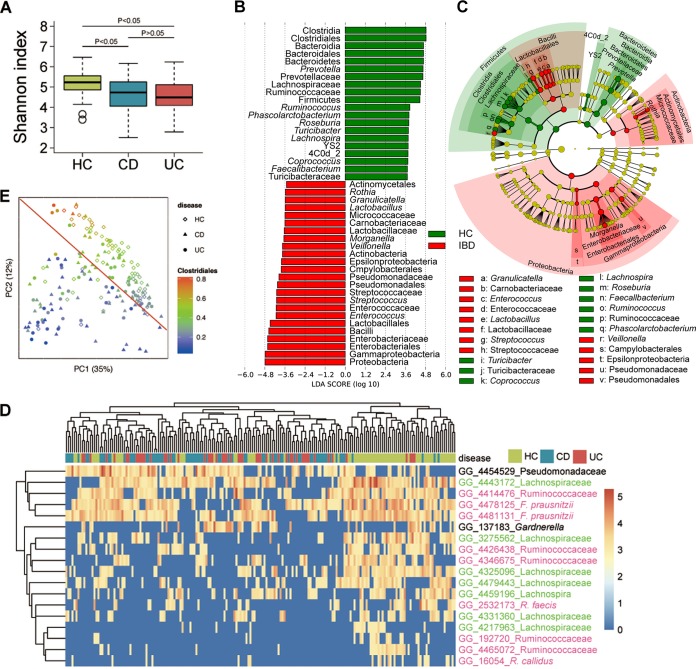
Consistent with previous reports (6, 32), the levels of alpha diversity of both CD and UC in our cohort were markedly reduced compared to the levels seen with healthy controls as indicated by the Shannon index (Fig. 1A). Firmicutes, Bacteroidetes, and Proteobacteria were the most abundant phyla, together accounting for up to 95% of the sequences on average, while Actinobacteria, Fusobacteria, Verrucomicrobia, Tenericutes, Synergistetes, and Cyanobacteria each accounted for 0.1% to 5% of sequences (see Fig. S1A in the supplemental material). Genus-level characterization is more complex, as the 20 most abundant genera observed in our study constituted only up to 60% of the total microbiome, with Bacteroides dominating the composition (Fig. S1B). At both the phylum and genus levels, we observed that the microbial composition seen with both CD and UC patients was different from that seen with the HC group (Fig. 1B and C; see also Fig. S2A and B). We used Kruskal-Wallis analysis combined with Bonferroni adjustment for multiple comparisons to screen the gut microbiome differences at the operational taxonomic unit (OTU) level between HC, CD, and UC. A total of 18 OTUs were significantly enriched in healthy controls and reduced in both CD and UC patients. These OTUs distinguished healthy controls from IBD patients, although they did not enable a distinction between patients with CD and UC (Fig. 1D and Fig. S2C), and the same groups were similarly indistinguishable in the Gevers RISK cohort (Fig. S2D). Most of these OTUs belonged to the order Clostridiales. This was also confirmed by the use of a linear discriminant analysis effect size (LEfSe) algorithm, showing that it was the most significantly enriched taxon in the healthy individuals (Fig. 1C and D). Specifically, levels of members of the family Lachnospiraceae, including the genera Roseburia and Coprococcus, were depleted under IBD conditions. This result accords with previous studies showing reduction of Clostridiales levels in IBD microbiota (33). Unsupervised clustering using principal-coordinate analysis (PCoA) based on weighted UniFrac distance data (34) also showed that the gut microbiotas of IBD differed significantly from those of healthy controls (HC) (analysis of similarity [ANOSIM] test, P = 0.001) and that the HC samples showed greater Clostridiales enrichment (Fig. 1E).
FIG 1 .
Dysbiosis of gut microbiota patterns in Chinese patients with IBD. (A) Comparison of Shannon index values between HC, CD, and UC groups. (B) Taxa listed according to their linear discriminant analysis (LDA) values determined from comparisons between the HC and IBD groups as computed by the use of the LEfSe algorithm. (C) Taxa illustrated according to their taxonomic relationship using a cladogram, showing the discriminative patterns in taxonomic lineages. The LDA cutoff values for panels A and B were set at 3.5. (D) Heat map showing abundance distributions for the 18 operational taxonomic units (OTUs) identified as key variables using the Kruskal-Wallis test (after Bonferroni correction) among the HC, CD, and UC groups. Lachnospiraceae OTUs are colored in green, and those of Ruminococcaceae are colored in red. (E) Weighted UniFrac PCoA data showing grouping patterns of HC, CD, and UC. Each dot represents a sample, with shapes indicating groups and colors the abundance of Clostridiales.
Microbial composition at the phylum level (A) and genus level (B) of HC, CD, and UC groups. Download FIG S1, TIF file, 0.7 MB (754.6KB, tif) .
Copyright © 2018 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Discriminative taxa determined by LEfSe (the LDA cutoff value was set at 3.5) between HC versus CD groups (A) and HC versus UC groups (B). The supervised classification analysis was performed using random forest analysis to distinguish between the UC and CD groups in the present Chinese cohort (C) and in the RISK cohort of Gevers et al. (D). Download FIG S2, TIF file, 1.7 MB (1.7MB, tif) .
Copyright © 2018 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The alteration of gut microbiota in Chinese IBD patients is consistent with that of Westerners.
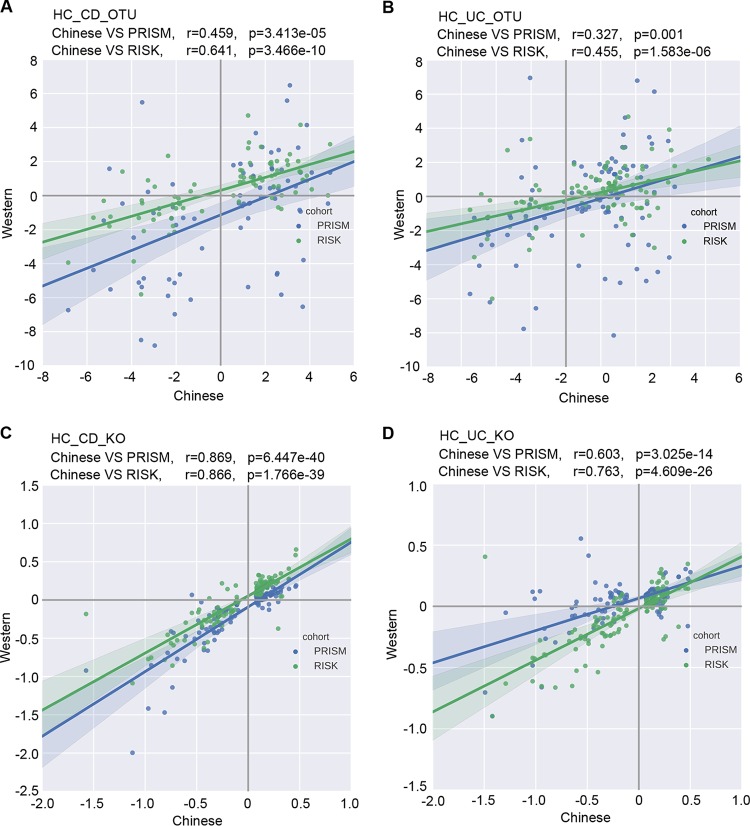
It is well known that host lifestyle affects gut microbiota. The gut microbiotas harbored by the Chinese population are different from those harbored by the Western population (35). Additionally, samples from different studies of gut microbiota are generally clustered by study due to the technical variations in sample collecting and processing. Thus, it is not surprising that the Chinese samples were separated from the Western samples in the PCoA when we combined data from this study with data from the cohorts studied by Gevers et al. (9) (Fig. S3). Despite the overall microbial difference across the studies as shown in the PCoA plot, the results of the differential abundance analyses described above suggest that the microbial shift in Chinese IBD patients, compared to HC patients, may resemble that in Westerners. To examine this further, we selected OTUs that differed in relative abundances between HC and CD in our Chinese cohort, on the basis of a permutation test performed with a false-discovery rate (FDR) of less than 0.1 (the criterion was mildly relaxed to include more OTUs for this analysis). The log2-fold changes in these OTU abundances between the CD and HC groups were computed and plotted against those from biopsy samples of RISK and PRISM cohorts. As shown in Fig. 2A, these OTU abundance changes were highly correlated across these cohorts (Spearman correlation coefficient r = 0.459, P value = 3.41e−5 for PRISM; r = 0.641, P value = 3.47e−10 for RISK). A similar universal pattern was also seen in the UC cohorts (r = 0.327, P value = 0.001 for PRISM; r = 0.455, P value = 1.58e−6 for RISK) (Fig. 2B). However, stool samples from the two Western cohorts showed much less resemblance (Fig. S4A and B). The reasons for these differences are unclear; they could even have arisen from different practices in sample collection and processing. Samples were collected from the midstream stool in the present study, which is less convenient than swabbing but may retain the signal of microbial changes in IBD patients better, due to the biogeographic heterogeneity in the stool.
FIG 2 .
The similar shifts of gut microbiota in IBD across cohorts. Comparisons of OTUs (A and B) and predicted KOs (C and D) differentiated between healthy controls and subjects with IBD in the current study and the RISK and PRISM cohorts with biopsy samples. Each dot represents an OTU or KO that differed significantly between healthy and disease samples in Chinese cohorts. Axes indicates the log2-fold changes of the levels of these OTU/KO abundances between subjects with disease and healthy individuals in the Chinese cohort (x axis) and the Western cohorts (y axis), with the RISK cohort indicated in green and the PRISM cohort in blue. The correlation coefficients and the P values determined from comparisons between the cohorts are labeled on the plot.
PCoA based on unweighted UniFrac distance indicates that the Chinese samples (including HC, CD, and UC) are separated from the cohorts of Gevers et al. Download FIG S3, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2018 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The similar shifts of gut microbiota in IBD across ethnicities. Comparison of OTUs (A and B) and predicted KOs (C and D) differentiated between the healthy and IBD subjects in the current study and the study by Gevers et al. performed with stool samples. Each dot represents an OTU or KO statistically significantly different between healthy and disease samples in Chinese cohorts. Axes indicates the log2-fold changes of these OTU/KO abundances between disease subjects and healthy individuals in the Chinese cohort (x axis) and Western cohorts (y axis), with RISK data indicated in green and PRISM data in blue. The correlation coefficients and the P values determined from comparisons between the cohorts are labeled on the plot. Download FIG S4, TIF file, 0.7 MB (686.9KB, tif) .
Copyright © 2018 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Furthermore, we predicted KEGG orthology (KO) data from 16S rRNA amplicon taxonomic profiles using PICRUSt and found that the KO abundances changed similarly across cohorts, reflecting the patterns that we saw at the OTU level (Fig. 2C and D; see also Fig. S4C and D). These patterns were even more consistent in the KEGG orthologues than the OTUs, suggesting that there are some variations among cohorts and ethnic groups in OTU composition, while those OTUs seem to provide similar functions. Specifically, the pathways that increased in members of both the CD and UC groups included xenobiotic degradation (caprolactam degradation, limonene and pinene degradation, and toluene degradation), amino acid metabolism (tryptophan metabolism and lysine degradation), and electron transfer carriers; in contrast, the decreased pathways included microbial motility (bacterial chemotaxis, bacterial motility proteins, and flagellar assembly), germination, and sporulation.
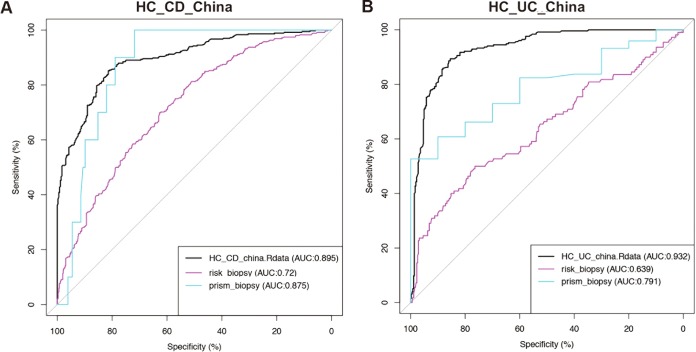
To explore the possibility of using the observed OTU changes to identify IBD, we built supervised classification models based on Chinese samples and evaluated the accuracies of the models with 5 repeats of 10-fold cross-validation. The gut microbiota is informative enough to distinguish HC samples from CD and UC samples with model accuracy of 89.5% and 93.2%, respectively. Similarly, the model built from RISK and PRISM biopsy samples achieved high prediction accuracies as well, although Western fecal samples are less informative for classification of IBD from HC (Fig. S5), in concordance with the findings in the correlation analysis described above. Additionally, to investigate whether the model can be applied across cohorts, we tested a model trained by the use of Chinese samples on the RISK and PRISM samples. The prediction accuracy across cohorts was reduced only marginally. For example, the predictive model constructed using Chinese CD stool has an 87.5% accuracy level in predicting PRISM CD biopsy samples and the model trained using Chinese UC stool has a 79.1% accuracy level in predicting PRISM UC biopsy samples (Fig. 3). The RISK cohort is less well predicted than the PRISM cohort, likely because the RISK samples were mainly from children and adolescents instead of adults. Consistent with Fig. 2 and Fig. S4, the biopsy samples are better predicted with the Chinese model than the fecal samples (Fig. S6A and B). Taken together, these findings suggest that there are consistent changes in gut microbiota of IBD patients across populations and that they can serve as universal biomarkers for the classification of IBD states (32).
FIG 3 .
Gut microbiotas distinguish diseases from health similarly across cohorts. The black ROC curve indicates the accuracy of the classification model built on the Chinese cohort for classifying HC versus CD (A) and HC versus UC (B). Colored curves are the classification accuracies when these models are applied to the other cohorts.
Supervised classification of the HC group and the CD and UC groups using biopsy or stool samples from the RISK cohorts. The PRISM cohort does not have sufficient samples to build a robust classification model through cross-validation. Download FIG S5, TIF file, 0.5 MB (515.5KB, tif) .
Copyright © 2018 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gut microbiotas distinguish IBD subjects from healthy subjects similarly across cohorts. The black ROC curve indicates the accuracy of the classification model built on data from the Chinese cohort for classifying the HC data versus the CD data (A) and the HC data versus the UC data (B). Colored curves represent the prediction accuracies seen in applying this model to the other cohorts. Download FIG S6, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2018 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gut microbiota signatures associated with disease activities.
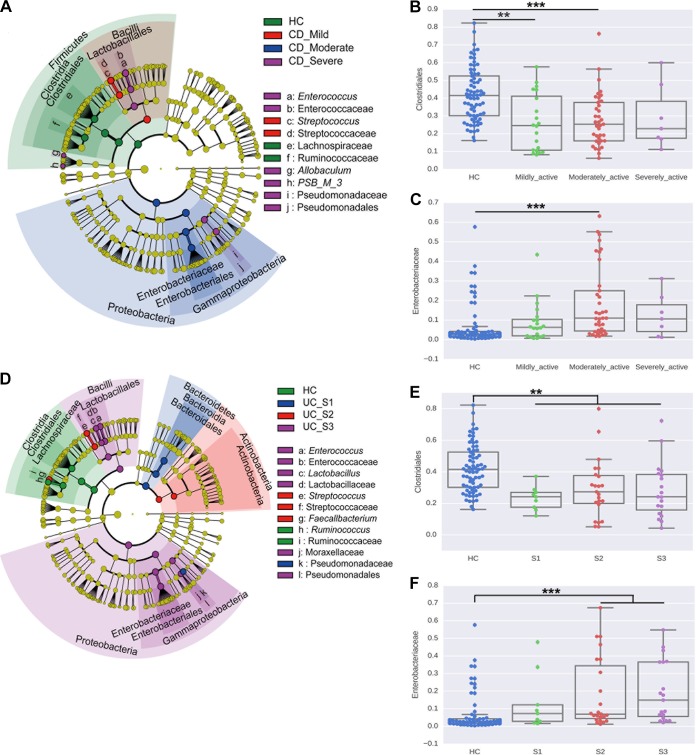
We further analyzed the characteristics of gut microbiota in different disease activity subgroups of IBD patients. LEfSe results showed larger proportions of Bacilli, represented by Streptococcus, in patients with mild CD compared to other groups. Significant enrichment in Proteobacteria and Enterococcaceae (Fig. 4A and C) and depletion in Ruminococcaceae and Clostridiales (Fig. 4A and B) were seen in patients with moderate to severe CD. Levels of Bacteroidetes, represented by Bacteroidia, and Pseudomonadaceae were enriched in patients with mild UC (Montreal classification of severity of ulcerative colitis score, S1). Streptococcus levels were increased in patients with moderate UC (score, S2), resembling mild CD. Species of the Proteobacteria phylum and Bacilli class were enriched in patients with severe UC (score, S3) (Fig. 4D and F). Clostridiales levels were decreased in all active UC patients (Fig. 4E). Notably, a majority of these differences in microbiota with regard to disease activity were related to the Firmicutes, Bacteroidetes, and Proteobacteria phyla (Fig. 4A and D).
FIG 4 .
Bacterial biomarkers associated with disease severity. (A) Cladogram representing taxa with different abundances for CD activity. The size of each circle is proportionate to the abundance of the taxon. (B and C) Relative abundances of Clostridiales (B) and Enterobacteriaceae (C) in CD activity groups are shown. (D) Cladogram representing taxa with different abundances for UC activity. (E and F) Relative abundances of Clostridiales (E) and Enterobacteriaceae (F) in UC activity groups are shown. The statistical significance was determined with the Wilcoxon test and was adjusted using the false-discovery rate (FDR). **, P value < 0.01; ***, P value < 0.001.
Crohn’s disease may lead to a stricture phenotype and penetrating complications, which indicate disease progression and impact the efficacies of treatments (27). In an advanced stage, CD can induce fistulas, i.e., abnormal passageways created between the bowel and other body parts. They often cause severe impairment in the patient’s quality of life (36). To determine whether any of the microbes were associated with these disease behaviors, we used the LEfSe algorithm for analysis and found that Enterobacteriaceae and Pseudomonadaceae were enriched in stricturing CD (CD_B2 [Montreal classification of stricturing behavior of Crohn’s disease]), while levels of Aeromonadaceae in the Proteobacteria phylum were enriched in penetrating CD (CD_B3 [Montreal classification of penetrating behavior of Crohn’s disease]) (Fig. S7A). Enterococcaceae and Pseudomonadaceae were the key taxa enriched in fistulizing CD patients (Fig. S7B). These results also show that the increase in Proteobacteria (Enterobacteriaceae) was strongly correlated with CD severities.
Biomarkers determined for CD behavior (A) and for CD behavior complicated by the presence of fistulas (B) using LEfSe and illustrated by cladogram. The LDA cutoff value was set at 2. Download FIG S7, TIF file, 0.9 MB (942.3KB, tif) .
Copyright © 2018 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The gut microbiota is restored during disease remission, and certain microbes, especially Clostridiales, enabled predictions of the response of IFX treatment in CD.
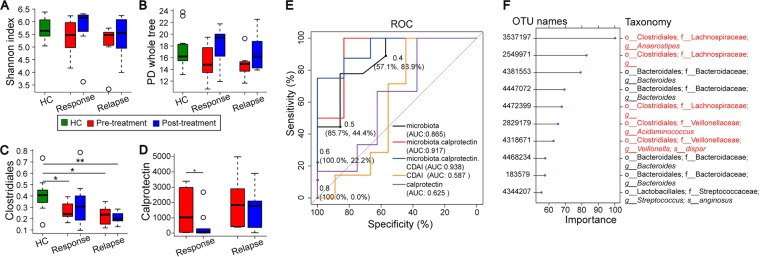
We followed 16 CD patients treated with IFX to week 30 to explore if the gut microbiotas were restored after IFX treatment and whether there were any microbial differences between IFX response and IFX relapse patients. A total number of 1,646,642 sequences were obtained from 27 fecal samples (including the 11 HC samples described above) for this longitudinal analysis, with an average of 15,106 ± 6,902 (mean ± SD) sequences. After initial IFX-induced remission, relapse occurred in 43.75% (7/16) of patients when reexamined at the end of week 30. The IFX treatment alleviated disease activity and increased microbial alpha diversity, measured by both Shannon index and PD whole tree (Fig. 5A and B), in the response group and, to a lesser extent, in the relapse group. The level of Clostridiales, the reduction of which was found as a signature of IBD (Fig. 1D and E; see also Fig. 4B and E), was not detected to be statistically significantly different from that of HC after IFX treatment in the response group, indicating its restoration after the IFX-induced response (Fig. 5C). These results imply that the Clostridiales reenrichment was correlated with disease remission after treatment and could potentially be used as a biomarker to guide treatment. The level of calprotectin, which has been recommended as a biomarker for IBD activity and prognosis, was also decreased more in the response group than in the relapse group after treatment (Fig. 5D).
FIG 5 .
The microbiotas differ in the response and the relapse groups of CD patients treated with infliximab (IFX). The alpha diversity, measured by Shannon index (A) and PD whole tree (B), Clostridiales abundance (C), and calprotectin abundance (D), was restored in the response group after treatment compared to the relapse group. Stars (*) indicate P values of <0.05. (E and F) Supervised prognostic prediction of Crohn’s disease progression (IFX response or relapse). (E) The accuracy of prediction was best using the combination of microbiota, calprotectin, and CDAI (93.8%). The use of the microbiota alone (86.5%) still outperformed the results seen with the traditional clinic markers of CDAI (58.7%) or calprotectin (62.5%). (F) The top informative OTUs that contribute to the classification model. Clostridiales OTUs, colored in orange, are the most informative OTUs.
To further test whether the gut microbiota provides biomarkers for prognosis of IFX treatment for CD patients, we derived and evaluated a model trained on the gut microbiota at baseline (at week 0) to predict the IFX-induced outcome (response or relapse) at week 30. The use of the microbiota alone improved the prediction to 86.5% accuracy, compared with that determined with the Crohn’s disease activity index (CDAI) (58.7%) and the level of calprotectin (62.5%), both of which are conventionally used to assess treatment effectiveness in clinic. The use of microbiota data in combination with calprotectin and CDAI data can further improve the accuracy of prediction of the prognosis (to 93.8%) (Fig. 5E). The most informative features that contribute to the prognosis model include multiple Clostridiales OTUs (Fig. 5F). These results highlight the advantage of using gut microbiota to stratify IBD patients and to apply personalized treatment for optimal outcomes, although the data warrant further verification in a larger cohort(s).
DISCUSSION
Conventionally, IBD is regarded as a Western disease. However, following the path of Western countries, the IBD incidence in Asian populations has been increasing, and IBD has increasingly become a global health care problem over the past decade (37). Although the exact etiology of IBD remains elusive, it is widely accepted that various factors, including host genetic background, gut microbiome, and environmental triggers, contribute to the onset of IBD symptoms (2, 3). People who have certain variant alleles of genes (such as NOD2 and interleukin-23 receptor [IL23R]) are more prone than others to developing IBD (4, 5, 38). Epidemiology evidence has also shown that smoking, diet, appendectomies, and stress have a complicated impact on IBD (1). The distinct genetic backgrounds of emerging IBD populations without risk gene alleles emphasize the role that environmental factors play in IBD pathogenesis. There is no doubt that the human gut microbiome is a key player in this process as a consequence of interacting with the immune system (1). For example, Bacteroides fragilis can secrete capsular polysaccharide A to induce expression of interleukin-10 from regulatory T cells and protect mucous from colitis in a NOD2- and ATG16L1-dependent way (39).
In this study, we characterized dysbiosis in a Chinese IBD population. We found that gut microbial diversity was reduced in IBD patients compared with healthy controls, with a nonsignificant trend toward a greater reduction of diversity in UC patients than in CD patients. These findings are coherent with those of previous research on colonic mucosa-associated bacterial microbiota (32). Our results comparing the gut microbiota from different populations demonstrated that the microbial alteration patterns of both Chinese and Western IBD patients are consistent with each other, as shown by the cross-cohort and cross-ethnicity meta-analyses. To the best of our knowledge, this was the first attempt to compare microbiota communities in Chinese and Western IBD populations. This report conceptually proves the potential of the use of the gut microbiome in one cohort to help diagnose and evaluate IBD status in other cohorts. It will therefore be of great value in clinical trials across multiple populations in IBD management, a major unsolved challenge.
Infliximab has been proven to be more effective in the treatment of CD and UC than some treatments using traditional medicines such as corticosteroids and thiopurines in previous studies (40, 41), but some issues still need to be addressed, including which population benefits most, when the therapy should be stopped, and whether the therapy is still effective if clinical relapse occurs (27). There are many factors associated with disease relapse or response such as demographic variables (including smoking, old age, and long duration of steroids), clinical variables (including CDAI scores and longer duration of disease), laboratory variables (including CRP and calprotectin), and IFX-related variables (IFX doses, serum IFX concentration, and IFX antibodies). However, these factors are post hoc or retrospective. In this study, we followed up the CD patients who received scheduled infliximab and analyzed their fecal microbiota before and after treatment to explore the potential predictors for CD clinical relapse based on gut microbial composition. We found that imbalanced microbial diversity and reduced Clostridiales abundance in CD patients were restored in patients who responded to infliximab treatment. Moreover, the use of the gut microbiota, alone or together with calprotectin and CDAI data, enabled more-effective prediction of infliximab treatment outcomes, although more samples are needed to confirm and improve this model before it can serve in clinical practice. These findings may help establish a set of microbiota-based biomarkers for predicting treatment efficacy for IBD, which may pave the way to the usage of gut microbiota to stratify IBD patients and apply personalized therapy for optimal outcomes.
Interestingly, although species of Clostridiales are depleted in IBD patients, CD patients with a relatively higher abundance of Clostridiales respond better to IFX treatment than those with lower abundance. During remission, Clostridiales is restored to close to the abundance level of healthy individuals. This indirectly suggests the protective role of the taxa in IBD pathogenesis. Many commensal Clostridiales species are well-known defensive symbionts. They can suppress proinflammatory bacteria (42), produce short-chain fatty acids (SCFAs) (43), and induce an immune response (44). The suppression of these fermentation-related bacteria causes a decline in SCFA production, resulting in increased colonic pH and ammonia production and absorption in the intestine (45). For example, Faecalibacterium prausnitzii is a well-described anti-inflammatory organism that is considered to be a health-promoting bacterium (46). Reduced abundance of Faecalibacterium prausnitzii has been associated with a higher rate of IBD recurrence (46). However, it is still unknown what other strains protect against IBD in what capacity, which needs further mechanistic investigation.
In conclusion, our report reveals congruence in the gut microbiome dysbiosis in IBD patients in cross-cohort and cross-ethnicity groups. These findings may aid the establishment of principles guiding IBD treatment. Our results reinforce the idea that the gut microbiota contains promising biomarkers for the noninvasive evaluation of IBD activity and assessment of therapeutic responses. The identification of disease activity-associated microbiome is a step toward establishing a set of microbiota-based biomarkers for the assessment of treatment and progression of inflammatory bowel disease.
MATERIALS AND METHODS
Ethics statement.
The Ethics Committee of Nanfang Hospital, Southern Medical University, approved this study (NHMEC2013-081). Patients were included in the study after providing written consent.
Patients and samples.
Patients with CD or UC who had not received any treatments for those conditions were recruited for this study between June 2012 and July 2013 in the Department of Gastroenterology of Nanfang Hospital, Southern Medical University, China. Healthy volunteers at age 20 to 40 (to match the age and gender of patients with CD) were recruited from the adjacent community. Exclusion criteria were receipt of IBD treatment, age <18 years, receipt of antibiotics or probiotics within the previous 4 weeks, other known chronic disease, and pregnancy or breastfeeding status. Sixteen patients with active CD who received treatment with IFX (Remicade; Cilag AG, Schaffhausen, Switzerland) (5 mg/kg of body weight) at weeks 0, 2, 6, 14, 22, and 30 were followed up for 30 weeks.
All enrolled patients underwent colonoscopies for diagnostic purposes. Fecal samples (from midstream stool; both the first-stream stool and the last-stream stool were discarded to toilet) were collected from all enrolled subjects at hospital and stored at −80°C before further processing.
Diagnostics of IBD diseases.
Diagnoses of UC and CD were based on the internationally accepted Lennard-Jones criteria (47). According to the Montreal classification (48), UC was classified as ulcerative proctitis (E1), left-sided (distal) UC (E2), or extensive UC (pancolitis; E3), based on the extent of the disease. CD was classified as located in the ileum (L1), colon (L2), or ileocolon (L3). Disease behavior (B) for CD was also classified as B1 (nonstricturing, nonpenetrating), B2 (stricturing), and B3 (penetrating).
For the evaluation of disease activity, the Mayo score (41) for UC and the Crohn’s disease activity index (CDAI) score (49) for CD were determined to estimate UC and CD activity (mild [S1], moderate [S2], or severe [S3]).
Evaluation of clinical outcome following infliximab treatment.
Patients receiving IFX treatment underwent endoscopy at baseline and after 30 weeks of treatment. For the evaluation of disease activity and response to IFX therapy, the CDAI was determined prior to each IFX infusion through the last follow-up visit (at week 30). CRP level, erythrocyte sedimentation rate, white blood cell count, and neutrophil ratio were also determined.
Clinical response was defined as a reduction of ≥70 points in the CDAI after infusion. Clinical remission was defined as a CDAI value of <150. Clinical relapse during follow-up was defined as worsening of symptoms and a CDAI value of >150, with an increase of ≥70 points compared with the CDAI value at remission; the need for an additional steroid or IFX course; or the need for surgical resection. All other outcomes were defined as nonresponse (50).
Fecal calprotectin assay.
Fecal calprotectin concentrations were measured with a quantitative PhiCal enzyme-linked immunosorbent assay (ELISA) kit (Immundiagnostik AG, catalog no. K6927) according to the manufacturer’s instructions. Fecal specimens were diluted 1:2,500. ELISA plates were read by the use of a Thermo Scientific microplate reader (Multiskan FC; optical density at 450 nm against 620 nm). Samples containing ≥100 μg of calprotectin per 1 g of feces were considered calprotectin positive (51).
Total bacterial genomic DNA extraction.
Bacterial DNA was extracted from the fecal samples using a Tiangen stool DNA kit (Tiangen Biotech, Beijing, China), according to the manufacturer’s instructions (52). DNA concentrations were determined using a NanoDrop 2000 BioAnalyser (Thermo Fisher Scientific, Inc., Waltham, MA), and the remaining samples were stored at −20°C before PCR was performed.
PCR amplification and Illumina sequencing.
We used bar-coded primers V4-515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and V4-806R (5′-GGACTACHVGGGTWTCTAAT-3′) to amplify the bacterial 16S rRNA V4 fragments. The PCR cycle conditions were as follows: initial denaturation at 94°C for 2 min; 30 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 40 s; and final extension at 72°C for 5 min. Each 25-μl reaction mixture consisted of 12.5 μl TaKaRa Premix Taq (D331A, version 2.0; TaKaRa Biotech, Dalian, China), 2 μl template DNA, 1 μl 10 μM bar-coded primer V4-515F, 1 μl 10 μM primer V4-806R, and 8.5 μl double-distilled water (ddH2O).
PCR products were gel purified using a QIAquick gel extraction kit (catalog no. 28704; Qiagen, Hilden, Germany) and sequenced using the 250-bp paired-ended strategy on an Illumina MiSeq system at Beijing Genomic Institute (BGI, Shenzhen, China).
Bioinformatics analysis.
The raw sequences were quality controlled using QIIME v1.9.1 (53) with default parameters. The closed-reference OTU clustering was done at 97% similarity level against the GreenGenes database (v13_8) (54). After the samples were rarefied to the same sequencing depth, alpha diversity, beta diversity, and differential OTU abundance analyses were performed with QIIME, PICRUSt (55), and LEfSe (56) tools.
To compare the IBD effects across ethnic groups, the sequences from RISK and PRISM cohorts of patients in the United States were downloaded from qiita.ucsd.edu (study identifier [ID]: 1939). All the sequences in these two studies were trimmed to the same length of 150 nucleotides (nt) on the same region of the 16S rRNA gene to minimize the technical variation. A single closed-reference OTU picking run was done on the combined sequences.
Random forest classification models were trained on features of the OTU data using the caret R package (57) with 5 repeats of 10-fold cross-validation, except for the IFX outcome classification, which was performed using leave-one-out cross-validation due to the small sample size. The model was evaluated with the area under the curve (AUC) derived from receiver operating characteristic (ROC) curve analysis. ROC analysis was used to compensate for the uneven distribution of the three sample types in this study (58). Importance scores of a model were determined for each feature based on the increase in prediction error when that feature was randomly permuted while all others were left unchanged (59).
The functional profile of KEGG orthology (KO) for each sample was predicted from 16S data with PICRUSt (55). The predicted KO abundances are collapsed to level 3 by grouping them into a higher level of functional categorization.
Data availability.
Data were deposited in ENA under accession number PRJEB22028.
ACKNOWLEDGMENTS
We thank Huimin Zheng for microbiota analysis assistance and figure preparation and Gail Ackermann for assisting with uploading our data to Qiita and EBI.
This work was supported by grants from the National Natural Science Foundation of China (NSFC31322003, 81570480, and 81700487) and the National High Technology Research and Development Program of China (“863” Program, 2015AA020701) and by the Crohn's and Colitis Foundation (New York, NY).
Y.Z. was responsible for the design of the study, recruitment of patients, statistical analysis and interpretation of the data, and drafting of the article. Z.Z.X. was responsible for the design of the study, analysis and interpretation of the data, and revision of the article. Y.H. was responsible for the design of the study, bioinformatics analysis and interpretation of the data, and revision of the article. Y.Y. was responsible for interpretation of the data and revision of the article. L.L. and Q.L. were responsible for recruitment of patients. Y.N. and M.L. were responsible for interpretation of the data and revision of the article. F.Z. and S.L. were responsible for interpretation of the data. A.A., A.G., and A.T. were responsible for analysis of the data. M.C. and G.D.W. were responsible for revision of the article. R.K. was responsible for interpretation of the data and revision of the article. H.Z. was responsible for bioinformatics analysis and interpretation of the data and revision of the article. Y.C. was responsible for the concept and design of the study, interpretation of the data, and revision of the article.
REFERENCES
- 1.Ng SC, Tang W, Leong RW, Chen M, Ko Y, Studd C, Niewiadomski O, Bell S, Kamm MA, de Silva HJ, Kasturiratne A, Senanayake YU, Ooi CJ, Ling KL, Ong D, Goh KL, Hilmi I, Ouyang Q, Wang YF, Hu P, Zhu Z, Zeng Z, Wu K, Wang X, Xia B, Li J, Pisespongsa P, Manatsathit S, Aniwan S, Simadibrata M, Abdullah M, Tsang SW, Wong TC, Hui AJ, Chow CM, Yu HH, Li MF, Ng KK, Ching J, Wu JC, Chan FK, Sung JJ; Asia-Pacific Crohn's and Colitis Epidemiology Study ACCESS Group . 2015. Environmental risk factors in inflammatory bowel disease: a population-based case-control study in Asia-Pacific. Gut 64:1063–1071. doi: 10.1136/gutjnl-2014-307410. [DOI] [PubMed] [Google Scholar]
- 2.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Buning C, Cohain A, Cichon S, D’Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, et al. 2012. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manichanh C, Borruel N, Casellas F, Guarner F. 2012. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol 9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 4.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. 2001. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 5.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nuñez G, Cho JH. 2001. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 6.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, Roca J, Dore J. 2006. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papa E, Docktor M, Smillie C, Weber S, Preheim SP, Gevers D, Giannoukos G, Ciulla D, Tabbaa D, Ingram J, Schauer DB, Ward DV, Korzenik JR, Xavier RJ, Bousvaros A, Alm EJ. 2012. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One 7:e39242. doi: 10.1371/journal.pone.0039242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ray K. 2014. IBD. Understanding gut microbiota in new-onset Crohn’s disease. Nat Rev Gastroenterol Hepatol 11:268. doi: 10.1038/nrgastro.2014.45. [DOI] [PubMed] [Google Scholar]
- 9.Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, González A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier RJ. 2014. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank DN, Robertson CE, Hamm CM, Kpadeh Z, Zhang T, Chen H, Zhu W, Sartor RB, Boedeker EC, Harpaz N, Pace NR, Li E. 2011. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis 17:179–184. doi: 10.1002/ibd.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. 2012. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andoh A, Imaeda H, Aomatsu T, Inatomi O, Bamba S, Sasaki M, Saito Y, Tsujikawa T, Fujiyama Y. 2011. Comparison of the fecal microbiota profiles between ulcerative colitis and Crohn’s disease using terminal restriction fragment length polymorphism analysis. J Gastroenterol 46:479–486. doi: 10.1007/s00535-010-0368-4. [DOI] [PubMed] [Google Scholar]
- 13.Andoh A, Kuzuoka H, Tsujikawa T, Nakamura S, Hirai F, Suzuki Y, Matsui T, Fujiyama Y, Matsumoto T. 2012. Multicenter analysis of fecal microbiota profiles in Japanese patients with Crohn’s disease. J Gastroenterol 47:1298–1307. doi: 10.1007/s00535-012-0605-0. [DOI] [PubMed] [Google Scholar]
- 14.Kabeerdoss J, Jayakanthan P, Pugazhendhi S, Ramakrishna BS. 2015. Alterations of mucosal microbiota in the colon of patients with inflammatory bowel disease revealed by real time polymerase chain reaction amplification of 16S ribosomal ribonucleic acid. Indian J Med Res 142:23–32. doi: 10.4103/0971-5916.162091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winter SE, Lopez CA, Bäumler AJ. 2013. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep 14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng SC, Tang W, Ching JY, Wong M, Chow CM, Hui AJ, Wong TC, Leung VK, Tsang SW, Yu HH, Li MF, Ng KK, Kamm MA, Studd C, Bell S, Leong R, de Silva HJ, Kasturiratne A, Mufeena MN, Ling KL, Ooi CJ, Tan PS, Ong D, Goh KL, Hilmi I, Pisespongsa P, Manatsathit S, Rerknimitr R, Aniwan S, Wang YF, Ouyang Q, Zeng Z, Zhu Z, Chen MH, Hu PJ, Wu K, Wang X, Simadibrata M, Abdullah M, Wu JC, Sung JJ, Chan FK, Asia–Pacific Crohn's and Colitis Epidemiologic Study (ACCESS) Study Group . 2013. Incidence and phenotype of inflammatory bowel disease based on results from the Asia-Pacific Crohn’s and Colitis Epidemiology Study. Gastroenterology 145:158–165.e2. doi: 10.1053/j.gastro.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 17.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cosnes J, Gower-Rousseau C, Seksik P, Cortot A. 2011. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology 140:1785–1794. doi: 10.1053/j.gastro.2011.01.055. [DOI] [PubMed] [Google Scholar]
- 19.van Dullemen HM, van Deventer SJ, Hommes DW, Bijl HA, Jansen J, Tytgat GN, Woody J. 1995. Treatment of Crohn’s disease with anti-tumor necrosis factor chimeric monoclonal antibody (cA2). Gastroenterology 109:129–135. doi: 10.1016/0016-5085(95)90277-5. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Y, He H, Wang P, Zhang T, Lin M, Wang H, Nie Y, Chen Y. 2015. Infliximab for the treatment of Crohn’s disease. Eur J Gastroenterol Hepatol 27:1270–1275. doi: 10.1097/MEG.0000000000000447. [DOI] [PubMed] [Google Scholar]
- 21.Zhou YL, Xie S, Wang P, Zhang T, Lin MY, Tan JS, Zhi FC, Jiang B, Chen Y. 2014. Efficacy and safety of infliximab in treating patients with ulcerative colitis: experiences from a single medical center in southern China. J Dig Dis 15:483–490. doi: 10.1111/1751-2980.12161. [DOI] [PubMed] [Google Scholar]
- 22.Beaugerie L, Seksik P, Nion-Larmurier I, Gendre JP, Cosnes J. 2006. Predictors of Crohn’s disease. Gastroenterology 130:650–656. doi: 10.1053/j.gastro.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 23.Costa F, Mumolo MG, Ceccarelli L, Bellini M, Romano MR, Sterpi C, Ricchiuti A, Marchi S, Bottai M. 2005. Calprotectin is a stronger predictive marker of relapse in ulcerative colitis than in Crohn’s disease. Gut 54:364–368. doi: 10.1136/gut.2004.043406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis JD. 2011. The utility of biomarkers in the diagnosis and therapy of inflammatory bowel disease. Gastroenterology 140:1817–1826.e2. doi: 10.1053/j.gastro.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Røseth AG, Aadland E, Jahnsen J, Raknerud N. 1997. Assessment of disease activity in ulcerative colitis by faecal calprotectin, a novel granulocyte marker protein. Digestion 58:176–180. doi: 10.1159/000201441. [DOI] [PubMed] [Google Scholar]
- 26.Mao R, Xiao YL, Gao X, Chen BL, He Y, Yang L, Hu PJ, Chen MH. 2012. Fecal calprotectin in predicting relapse of inflammatory bowel diseases: a meta-analysis of prospective studies. Inflamm Bowel Dis 18:1894–1899. doi: 10.1002/ibd.22861. [DOI] [PubMed] [Google Scholar]
- 27.D’Haens GR, Panaccione R, Higgins PD, Vermeire S, Gassull M, Chowers Y, Hanauer SB, Herfarth H, Hommes DW, Kamm M, Lofberg R, Quary A, Sands B, Sood A, Watermeyer G, Lashner B, Lemann M, Plevy S, Reinisch W, Schreiber S, Siegel C, Targan S, Watanabe M, Feagan B, Sandborn WJ, Colombel JF, Travis S. 2011. The London Position Statement of the World Congress of Gastroenterology on Biological Therapy for IBD with the European Crohn’s and Colitis Organization: when to start, when to stop, which drug to choose, and how to predict response? Am J Gastroenterol 106:199–212; quiz 213. [DOI] [PubMed] [Google Scholar]
- 28.Rajca S, Grondin V, Louis E, Vernier-Massouille G, Grimaud JC, Bouhnik Y, Laharie D, Dupas JL, Pillant H, Picon L, Veyrac M, Flamant M, Savoye G, Jian R, Devos M, Paintaud G, Piver E, Allez M, Mary JY, Sokol H, Colombel JF, Seksik P. 2014. Alterations in the intestinal microbiome (dysbiosis) as a predictor of relapse after infliximab withdrawal in Crohn’s disease. Inflamm Bowel Dis 20:978–986. doi: 10.1097/MIB.0000000000000036. [DOI] [PubMed] [Google Scholar]
- 29.Kolho KL, Korpela K, Jaakkola T, Pichai MV, Zoetendal EG, Salonen A, de Vos WM. 2015. Fecal microbiota in pediatric inflammatory bowel disease and its relation to inflammation. Am J Gastroenterol 110:921–930. doi: 10.1038/ajg.2015.149. [DOI] [PubMed] [Google Scholar]
- 30.Chen L, Wang W, Zhou R, Ng SC, Li J, Huang M, Zhou F, Wang X, Shen B, A Kamm M, Wu K, Xia B. 2014. Characteristics of fecal and mucosa-associated microbiota in Chinese patients with inflammatory bowel disease. Medicine 93:e51. doi: 10.1097/MD.0000000000000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sha S, Xu B, Wang X, Zhang Y, Wang H, Kong X, Zhu H, Wu K. 2013. The biodiversity and composition of the dominant fecal microbiota in patients with inflammatory bowel disease. Diagn Microbiol Infect Dis 75:245–251. doi: 10.1016/j.diagmicrobio.2012.11.022. [DOI] [PubMed] [Google Scholar]
- 32.Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Fölsch UR, Timmis KN, Schreiber S. 2004. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walters WA, Xu Z, Knight R. 2014. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett 588:4223–4233. doi: 10.1016/j.febslet.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prideaux L, Kang S, Wagner J, Buckley M, Mahar JE, De Cruz P, Wen Z, Chen L, Xia B, van Langenberg DR, Lockett T, Ng SC, Sung JJ, Desmond P, McSweeney C, Morrison M, Kirkwood CD, Kamm MA. 2013. Impact of ethnicity, geography, and disease on the microbiota in health and inflammatory bowel disease. Inflamm Bowel Dis 19:2906–2918. doi: 10.1097/01.MIB.0000435759.05577.12. [DOI] [PubMed] [Google Scholar]
- 36.Fróes RDSB, Pugas Carvalho AT, Carneiro AJDV, de Barros Moreira AMH, Moreira JPL, Luiz RR, de Souza HS. 2017. The socio-economic impact of work disability due to inflammatory bowel disease in Brazil. Eur J Health Econ. doi: 10.1007/s10198-017-0896-4. [DOI] [PubMed] [Google Scholar]
- 37.Chi KR. 2016. Epidemiology: rising in the east. Nature 540:S100–SS102. doi: 10.1038/540S100a. [DOI] [PubMed] [Google Scholar]
- 38.Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, Abedian S, Cheon JH, Cho J, Dayani NE, Franke L, Fuyuno Y, Hart A, Juyal RC, Juyal G, Kim WH, Morris AP, Poustchi H, Newman WG, Midha V, Orchard TR, Vahedi H, Sood A, Sung JY, Malekzadeh R, Westra HJ, Yamazaki K, Yang SK; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium, Barrett JC, Alizadeh BZ, Parkes M, Bk T, Daly MJ, Kubo M, Anderson CA, Weersma RK. 2015. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 47:979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu H, Khosravi A, Kusumawardhani IP, Kwon AH, Vasconcelos AC, Cunha LD, Mayer AE, Shen Y, Wu WL, Kambal A, Targan SR, Xavier RJ, Ernst PB, Green DR, McGovern DP, Virgin HW, Mazmanian SK. 2016. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 352:1116–1120. doi: 10.1126/science.aad9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanauer SB, Feagan BG, Lichtenstein GR, Mayer LF, Schreiber S, Colombel JF, Rachmilewitz D, Wolf DC, Olson A, Bao W, Rutgeerts P; ACCENT I Study Group . 2002. Maintenance infliximab for Crohn’s disease: the ACCENT I randomised trial. Lancet 359:1541–1549. doi: 10.1016/S0140-6736(02)08512-4. [DOI] [PubMed] [Google Scholar]
- 41.Rutgeerts P, Sandborn WJ, Feagan BG, Reinisch W, Olson A, Johanns J, Travers S, Rachmilewitz D, Hanauer SB, Lichtenstein GR, de Villiers WJ, Present D, Sands BE, Colombel JF. 2005. Infliximab for induction and maintenance therapy for ulcerative colitis. N Engl J Med 353:2462–2476. doi: 10.1056/NEJMoa050516. [DOI] [PubMed] [Google Scholar]
- 42.Ramanan D, Bowcutt R, Lee SC, Tang MS, Kurtz ZD, Ding Y, Honda K, Gause WC, Blaser MJ, Bonneau RA, Lim YA, Loke P, Cadwell K. 2016. Helminth infection promotes colonization resistance via type 2 immunity. Science 352:608–612. doi: 10.1126/science.aaf3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van den Abbeele P, Belzer C, Goossens M, Kleerebezem M, De Vos WM, Thas O, De Weirdt R, Kerckhof FM, Van de Wiele T. 2013. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J 7:949–961. doi: 10.1038/ismej.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. 2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vince AJ, McNeil NI, Wager JD, Wrong OM. 1990. The effect of lactulose, pectin, arabinogalactan and cellulose on the production of organic acids and metabolism of ammonia by intestinal bacteria in a faecal incubation system. Br J Nutr 63:17–26. doi: 10.1079/BJN19900088. [DOI] [PubMed] [Google Scholar]
- 46.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottière HM, Doré J, Marteau P, Seksik P, Langella P. 2008. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lennard-Jones JE. 1989. Classification of inflammatory bowel disease. Scand J Gastroenterol Suppl 170:2–6; discussion 16–19. doi: 10.3109/00365528909091339. [DOI] [PubMed] [Google Scholar]
- 48.Satsangi J, Silverberg MS, Vermeire S, Colombel JF. 2006. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut 55:749–753. doi: 10.1136/gut.2005.082909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Assche G, Dignass A, Panes J, Beaugerie L, Karagiannis J, Allez M, Ochsenkühn T, Orchard T, Rogler G, Louis E, Kupcinskas L, Mantzaris G, Travis S, Stange E; European Crohn's and Colitis Organisation (ECCO) . 2010. The second European evidence-based Consensus on the diagnosis and management of Crohn's disease: definitions and diagnosis. J Crohns Colitis 4:7–27. doi: 10.1016/j.crohns.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 50.Lu C, Waugh A, Bailey RJ, Cherry R, Dieleman LA, Gramlich L, Matic K, Millan M, Kroeker KI, Sadowski D, Teshima CW, Todoruk D, Wong C, Wong K, Fedorak RN. 2012. Crohn’s disease genotypes of patients in remission vs relapses after infliximab discontinuation. World J Gastroenterol 18:5058–5064. doi: 10.3748/wjg.v18.i36.5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen CC, Huang JL, Chang CJ, Kong MS. 2012. Fecal calprotectin as a correlative marker in clinical severity of infectious diarrhea and usefulness in evaluating bacterial or viral pathogens in children. J Pediatr Gastroenterol Nutr 55:541–547. doi: 10.1097/MPG.0b013e318262a718. [DOI] [PubMed] [Google Scholar]
- 52.Peng X, Yu KQ, Deng GH, Jiang YX, Wang Y, Zhang GX, Zhou HW. 2013. Comparison of direct boiling method with commercial kits for extracting fecal microbiome DNA by Illumina sequencing of 16S rRNA tags. J Microbiol Methods 95:455–462. doi: 10.1016/j.mimet.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 53.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuhn M. 2008. Building predictive models in R using the Caret package. J Stat Softw 28:JSSv028i05. doi: 10.18637/jss.v028.i05. [DOI] [Google Scholar]
- 58.Fawcett T. 2006. An introduction to ROC analysis. Pattern Recognit Lett 27:861–874. doi: 10.1016/j.patrec.2005.10.010. [DOI] [Google Scholar]
- 59.Liaw A, Wiener M. 2002. Classification and regression by randomForest. R News 2:18–22. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Baseline clinical characteristics of the subjects. Download TABLE S1, DOCX file, 0.1 MB (57.4KB, docx) .
Copyright © 2018 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microbial composition at the phylum level (A) and genus level (B) of HC, CD, and UC groups. Download FIG S1, TIF file, 0.7 MB (754.6KB, tif) .
Copyright © 2018 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Discriminative taxa determined by LEfSe (the LDA cutoff value was set at 3.5) between HC versus CD groups (A) and HC versus UC groups (B). The supervised classification analysis was performed using random forest analysis to distinguish between the UC and CD groups in the present Chinese cohort (C) and in the RISK cohort of Gevers et al. (D). Download FIG S2, TIF file, 1.7 MB (1.7MB, tif) .
Copyright © 2018 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PCoA based on unweighted UniFrac distance indicates that the Chinese samples (including HC, CD, and UC) are separated from the cohorts of Gevers et al. Download FIG S3, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2018 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The similar shifts of gut microbiota in IBD across ethnicities. Comparison of OTUs (A and B) and predicted KOs (C and D) differentiated between the healthy and IBD subjects in the current study and the study by Gevers et al. performed with stool samples. Each dot represents an OTU or KO statistically significantly different between healthy and disease samples in Chinese cohorts. Axes indicates the log2-fold changes of these OTU/KO abundances between disease subjects and healthy individuals in the Chinese cohort (x axis) and Western cohorts (y axis), with RISK data indicated in green and PRISM data in blue. The correlation coefficients and the P values determined from comparisons between the cohorts are labeled on the plot. Download FIG S4, TIF file, 0.7 MB (686.9KB, tif) .
Copyright © 2018 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supervised classification of the HC group and the CD and UC groups using biopsy or stool samples from the RISK cohorts. The PRISM cohort does not have sufficient samples to build a robust classification model through cross-validation. Download FIG S5, TIF file, 0.5 MB (515.5KB, tif) .
Copyright © 2018 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gut microbiotas distinguish IBD subjects from healthy subjects similarly across cohorts. The black ROC curve indicates the accuracy of the classification model built on data from the Chinese cohort for classifying the HC data versus the CD data (A) and the HC data versus the UC data (B). Colored curves represent the prediction accuracies seen in applying this model to the other cohorts. Download FIG S6, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2018 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Biomarkers determined for CD behavior (A) and for CD behavior complicated by the presence of fistulas (B) using LEfSe and illustrated by cladogram. The LDA cutoff value was set at 2. Download FIG S7, TIF file, 0.9 MB (942.3KB, tif) .
Copyright © 2018 Zhou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Data were deposited in ENA under accession number PRJEB22028.