Microorganisms living within the rhizospheres of wetland plants significantly contribute to greenhouse gas emissions. Understanding how microbes produce these gases under conditions that have been imposed by human activities (i.e., nitrogen pollution) is important to the development of future management strategies. Our results illustrate that within the rhizosphere of the wetland plant Juncus acutiflorus, physiological differences associated with nitrogen availability can influence microbial activity linked to greenhouse gas production. By pairing taxonomic information and environmental conditions like nitrogen availability with functional outputs of a system such as greenhouse gas fluxes, we present a framework to link certain taxa to both nitrogen load and greenhouse gas production. We view this type of combined information as essential in moving forward in our understanding of complex systems such as rhizosphere microbial communities.
KEYWORDS: Acidobacteria, Juncus acutiflorus, Opitutales, Sphingobacteriales, greenhouse gas, metagenomics, microbial community function, nitrogen, nitrogen metabolism, wetlands
ABSTRACT
Wetland ecosystems are important reservoirs of biodiversity and significantly contribute to emissions of the greenhouse gases CO2, N2O, and CH4. High anthropogenic nitrogen (N) inputs from agriculture and fossil fuel combustion have been recognized as a severe threat to biodiversity and ecosystem functioning, such as control of greenhouse gas emissions. Therefore, it is important to understand how increased N input into pristine wetlands affects the composition and activity of microorganisms, especially in interaction with dominant wetland plants. In a series of incubations analyzed over 90 days, we disentangled the effects of N fertilization on the microbial community in bulk soil and the rhizosphere of Juncus acutiflorus, a common and abundant graminoid wetland plant. We observed an increase in greenhouse gas emissions when N is increased in incubations with J. acutiflorus, changing the system from a greenhouse gas sink to a source. Using 16S rRNA gene amplicon sequencing, we determined that the bacterial orders Opitutales, subgroup 6 Acidobacteria, and Sphingobacteriales significantly responded to high N availability. Based on metagenomic data, we hypothesize that these groups are contributing to the increased greenhouse gas emissions. These results indicated that increased N input leads to shifts in microbial activity within the rhizosphere, altering N cycling dynamics. Our study provides a framework for connecting environmental conditions of wetland bulk and rhizosphere soil to the structure and metabolic output of microbial communities.
IMPORTANCE Microorganisms living within the rhizospheres of wetland plants significantly contribute to greenhouse gas emissions. Understanding how microbes produce these gases under conditions that have been imposed by human activities (i.e., nitrogen pollution) is important to the development of future management strategies. Our results illustrate that within the rhizosphere of the wetland plant Juncus acutiflorus, physiological differences associated with nitrogen availability can influence microbial activity linked to greenhouse gas production. By pairing taxonomic information and environmental conditions like nitrogen availability with functional outputs of a system such as greenhouse gas fluxes, we present a framework to link certain taxa to both nitrogen load and greenhouse gas production. We view this type of combined information as essential in moving forward in our understanding of complex systems such as rhizosphere microbial communities.
INTRODUCTION
Wetlands are globally impacted by agricultural industry through the leaching of nitrogen (N), mainly in the form of nitrate (NO3−), and by increased N deposition as a result of high emissions from fossil fuel burning and agriculture (1). Furthermore, due to reduced oxidation under stagnant, waterlogged conditions, these systems show increased availability of ammonium (NH4+) (2). The strongly increased anthropogenic N input influences ecosystem degradation by contributing to biodiversity loss and altering (mostly increasing) greenhouse gas fluxes such as nitrous oxide (N2O), methane (CH4), and carbon dioxide (CO2) (3–6).
The abundance, composition, and activity of microorganisms strongly influence the biogeochemical cycling of wetland nutrients, particularly those resulting in emissions of greenhouse gases (7, 8). Specifically, N2O emission may increase due to lowering of pH, which affects the activity of incomplete denitrifiers (4, 5, 9). CH4 emissions can increase due to competitive inhibition of the key enzyme of aerobic methanotrophs, methane monooxygenase (MMO), by elevated NH4+, osmotic stress of methanotrophs, or through the stimulation of methanogenic archaea (10–12). Finally, the rate of soil C loss can increase as a result of N addition through the stimulation of heterotrophic respiration (13). Although it is well established that microbial processes are important drivers of ecosystem functions, such as controls on greenhouse gas emissions and nutrient cycling, there is a lack of understanding of how these functions are linked, both to the environmental conditions and to the composition of the microbial community (8).
Wetland plant roots influence the soil region surrounding the root, known as the rhizosphere, by altering the availability of oxygen, organic matter, and organic plant exudates (14–16). The total area of soil influenced by roots can be considerable, meaning that this definition of the rhizosphere may extend to the vast majority of the upper soil layer (17). The rhizosphere is an active, complex ecosystem where viruses, bacteria, archaea, fungi, and protozoa interact with plant roots (18). These microorganisms significantly contribute to nutrient cycling and ecosystem structure by channeling energy into higher trophic levels (19, 20).
While the rhizosphere has been studied for decades, the effects of eutrophication on the plant-microbe interactions are of more recent interest. Specifically, it is important to understand how N availability influences plant physiology and ultimately C and N cycling in the rhizosphere. On the global scale, soil microbial communities differ, depending on the regional and local N regimes, although the diversity of these communities does not seem to vary much (21). Interestingly, variation in microbial community composition seems to be predictable based on local nutrient regimes (22, 23). Even though these studies demonstrate the link between nutrient loading and community structure, they do not demonstrate how changes in the microbial community are functionally relevant to the ecosystem.
To build dynamic models of plant-microbe interactions, it is necessary to gain a robust understanding of the connection between environmental conditions (i.e., N availability) and microbial community structure and function (i.e., the bulk biological processes resulting in greenhouse gas emissions). In this study, we aimed at assessing the impact of increased N input into wetland systems on the rhizosphere microbial community and its functions related to greenhouse gas production. To achieve this, we used Juncus acutiflorus (sharp-flowered rush), a very common graminoid plant in European wetlands that forms a dense vegetation and is known for radial oxygen loss (ROL) from roots (7). Furthermore, it has a high tolerance for increased N inputs (24). In the present report, a longitudinal study was used to determine that greenhouse gas emissions increase as a result of N addition in incubations with J. acutiflorus, but not in incubations with only bulk wetland soil, under controlled stable experimental conditions. Additionally, functional responses were linked to shifts in the dominant members of the microbial community. We hypothesize that certain key microbial groups contribute to greenhouse gas emissions, either directly or indirectly through the food web. Our study takes the first steps toward a predictive understanding of microbial dynamics within the rhizosphere, linking nutrient load, microbial community structure, and function.
RESULTS
Plant physiology.
J. acutiflorus and bulk soil were incubated over a course of 90 days under either a high-N treatment (800 kg N·ha−1·year−1) or a low-N treatment (40 kg N·ha−1·year−1). The soil collected from the Ravenvennen site and used in the incubations was a sandy soil with low organic matter content. Soil samples were taken at an initial time point (time zero [T0]), a midpoint (Tm; t = 45 days), and final time point (Tf; t = 90 days) (see Table S1 in the supplemental material). By Tm, J. acutiflorus incubations had significant root development throughout the incubated soil such that all soil was dominated by root biomass. Thus, all soil sampled corresponded to the rhizosphere. To determine the N utilization of the plants and to identify growth responses to N inputs, the total dry weight biomass of roots, rhizomes, and shoots and total N and C contents of J. acutiflorus tissue were measured from plants at Tf. Although there was no significant difference in total biomass and root/shoot ratio of J. acutiflorus between incubations, the average total N content of plant tissue (65 mg·g−1) was approximately twice as high in incubations with a high N input (P = 0.037) (see Table S2 in the supplemental material). Correspondingly, the total C/N ratio (averaged across the whole plant) was significantly higher in J. acutiflorus incubations with a low N input (P = 0.007) (Table S2). Interestingly, this change in C/N ratio was observed only for rhizome and shoot tissue, while the root C/N ratio did not significantly differ between incubations (Table S2).
Sample overview containing the time of sampling, N-load treatment, and whether the sample was bulk or rhizosphere soil. Additionally, the number of post-quality-filtered reads that were produced and the number of OTU found in each sample are shown. Greenhouse gas fluxes are reported in micromoles per square meter per day. Finally, Shannon diversity (H′) is reported. Download TABLE S1, XLSX file, 0.1 MB (14.5KB, xlsx) .
Copyright © 2018 Hester et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plant average dry weight (DW) and C/N ratio were determined in different sections of the plant, including the roots, shoots, and rhizomes. Biomass weight was determined as dry weight. The mean values from plants receiving high N and low N are reported (Mean High and Mean Low). The P value is reported as a result of a t test comparing mean values from high- and low-N treatments. Stdev, standard deviation. Download TABLE S2, XLSX file, 0.1 MB (11.5KB, xlsx) .
Copyright © 2018 Hester et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Greenhouse gas fluxes.
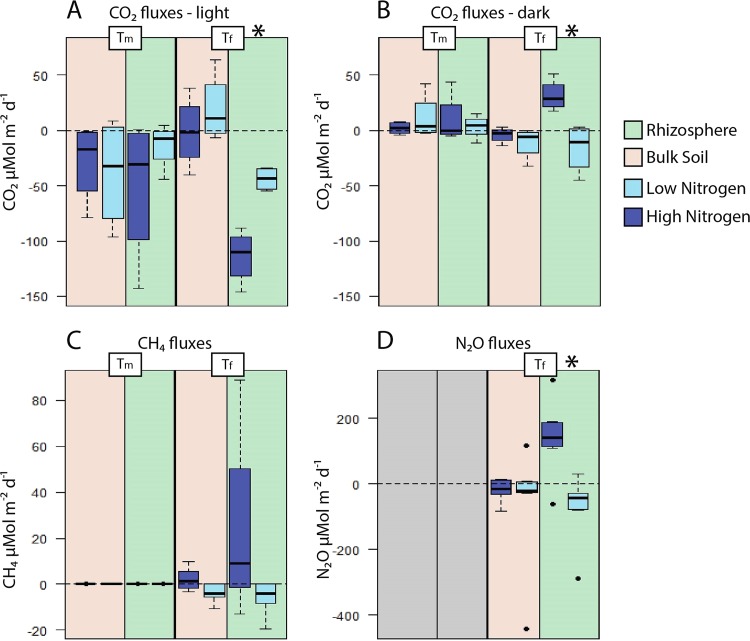
To link greenhouse gas fluxes with microbial community structure, gas flux measurements were performed at the same time points as soil sampling. Greenhouse gases were measured under both light and dark conditions, at Tm and Tf for CO2 and CH4 and at Tf for N2O (Fig. 1). Bulk soils generally did not have significant greenhouse gas fluxes (fluxes were not significantly different from 0) and will not further be discussed here. In the J. acutiflorus incubations, CO2 fluxes followed a day-night rhythm. Daytime CO2 fluxes were generally negative, indicating net CO2 fixation, with the largest rates significantly higher in high-N J. acutiflorus incubations at Tf (t = −5.28; P = 0.005) (Fig. 1A). Under dark conditions, CO2 fluxes were positive (net CO2 emission) only under the high-N treatment, while other treatments were not significantly different from 0 (t = 3.52; P = 0.01) (Fig. 1B). CH4 and N2O emissions did not vary between dark and light conditions, and therefore these conditions will not be compared. CH4 fluxes increased from Tm to Tf, and emissions tended to be highest in the J. acutiflorus incubations with a high N input; however, there was large variability in this group (t = 2.165; P = 0.064) (Fig. 1C). N2O emissions were highest in the high-N treatment (t = 2.56; P = 0.04) (Fig. 1D), while a negative N2O flux was observed in J. acutiflorus incubations receiving a low N input (Fig. 1D), indicating that this system can function as an N2O sink under N-limited conditions.
FIG 1 .
CO2, CH4, and N2O fluxes. Greenhouse gas fluxes were measured at a midpoint (Tm) and the final time point (Tf) during the 90-day incubation experiment. (A) CO2 light conditions, (B) CO2 dark conditions, (C) CH4, and (D) N2O. Asterisks denote significant differences (P < 0.05).
Denitrification potential.
To understand how increased N input influenced N cycling within bulk and J. acutiflorus rhizosphere soils, soil slurries were taken at Tf and their denitrification potential was measured. While we observed no significant difference in the N2 production between high- or low-N treatments (t = 0.32; P = 0.75), there was significantly higher N2O production from slurries originating from high-N-treatment soils (t = 2.41; P = 0.045) (see Fig. S2 in the supplemental material). This increased N2O production resulted in an approximately 10 times lower average N2/N2O ratio in high-N slurries (0.58 ± 0.61) compared to low-N-input slurries (5.36 ± 7.39), although not significantly different at P < 0.05 (t = −1.84; P = 0.11).
Microbial community structure.
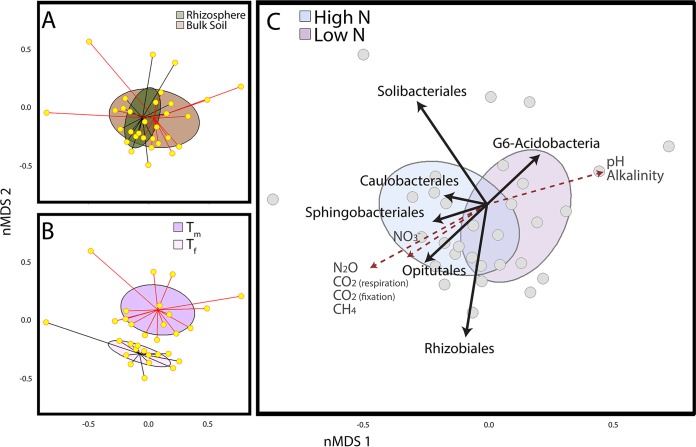
The v3-v4 fragment of the 16S rRNA gene was amplified and sequenced resulting in, on average, over 1,100 post-quality control (post-QC) sequences per sample. Each sample contained on average 264 ± 136 (mean ± standard deviation [SD]) operational taxonomic units (OTU). Rarefaction curves (see Fig. S3 in the supplemental material) suggest that sequencing depth was insufficient to capture the complete diversity of the communities. However, sampling depths of individual communities were not statistically linked to particular experimental groups, suggesting that there was a minimal effect of sampling effort on the group comparisons (N, P = 0.46; time, P = 0.19; rhizosphere, P = 0.69; t test) and that the observed community changes were caused by the different incubation conditions. In addition, over the course of the experiment the overall composition of communities changed (P < 0.001; permutational multivariate analysis of variance [PERMANOVA]) (Fig. 2B). There were also significant differences in the community composition of high- and low-N-treatment incubations (P = 0.003; PERMANOVA) (Fig. 2C) as well as rhizosphere and bulk soil incubations (P = 0.02) (Fig. 2A).
FIG 2 .
Microbial community structure and diversity. Nonmetric multidimensional scaling (nMDS) ordination plots of 16S rRNA samples show (A) rhizosphere or bulk soil, (B) the midpoint (Tm) and the final time point (Tf), and (C) high- and low-N treatment. The two-dimensional (2D) stress value was 0.19. Ellipses show the 95% confidence interval in the 2D space of samples in the respective treatment group. Red dashed lines indicate vectors for environmental parameters, while the black lines are for taxonomic groups.
On average, microbial diversity increased between Tm and Tf (t = 2.516; P = 0.0176; Shannon diversity index [H′]) (Table S1). Within each time point, diversity did not differ significantly between J. acutiflorus and bulk soil incubations, nor did N input have an impact (Table S1).
Linking microbial community members to function.
In order to understand how the microbial community members were linked to environmental conditions and greenhouse gas emissions, a random forest classifier was used to identify microbial taxa whose abundance was affected by N input, time of sampling, or presence of J. acutiflorus. Additionally, random forest was also used for regression to determine connections between abundance of these groups and environmental conditions or greenhouse gas fluxes, and these associations were further analyzed by fitting linear models.
The top three microbial groups that significantly responded to N input were the Opitutales (Verrucomicrobia) and Sphingobacteriales (Bacteroidetes), which were more abundant in the high-N-treatment group, and group 6 (G6) Acidobacteria, which were more abundant in the low-N-treatment group (Fig. 2C; Table 1). More specifically, the relative abundances of these three orders could be linked to N2O emissions (Table 1). Opitutales and Sphingobacteriales were positively associated with N2O fluxes, while a negative association was observed for the G6 Acidobacteria. In addition, Sphingobacteriales were correlated to CO2 fixation (Table 1).
TABLE 1 .
Correlations of microbial community members to environmental conditions and greenhouse gas fluxesa
| Microbial community and parameter |
t | P value | Mean relative abundance | Correlate | Adjusted R2 value |
Coefficient | P value | |
|---|---|---|---|---|---|---|---|---|
| High versus low N | High N | Low N | ||||||
| Opitutales | 4.17 | <0.001 | 0.040 | 0.010 | N2O | 0.11 | 3.50E−4 | 0.012 |
| G6 Acidobacteria | −4.22 | <0.001 | 0.007 | 0.020 | N2O | 0.19 | −3.18E−5 | 0.058 |
| Sphingobacteriales | 2.88 | 0.008 | 0.010 | 0.005 | N2O | 0.32 | 3.10E−5 | 0.016 |
| CO2 (fixation) | 0.29 | 7.07E−5 | 0.011 | |||||
| Rhizosphere vs bulk soil |
Rhizosphere | Bulk | ||||||
| Caulobacterales | −3.46 | 0.002 | 0.052 | 0.032 | NO3− | 0.21 | −8.50E−5 | 0.003 |
| Tm vs Tf | Tm | Tf | ||||||
| Rhizobiales | 6.66 | <0.001 | 0.099 | 0.184 | CO2 (respiration) | 0.27 | −6.40E−4 | 0.001 |
| Solibacterales | −4.76 | <0.001 | 0.179 | 0.116 | Alkalinity | 0.26 | −2.00E−2 | 0.002 |
The mean relative abundances of the top bacterial families distinguishing high versus low N, rhizosphere versus bulk soil or Tm versus Tf sampling time points are indicated, as are the t test results and statistics. Additionally, the top environmental or functional traits correlated with these groups are reported along with linear model statistics.
The top bacterial order distinguishing microbial communities from rhizosphere and bulk soil were the alphaproteobacterial order Caulobacterales, which were more abundant in the rhizosphere than in bulk soil and had a negative association with elevated NO3− concentrations (Fig. 2A and C; Table 1). The Rhizobiales and Solibacterales orders of the Alphaproteobacteria class and Acidobacteria phylum, respectively, were most distinctive for the microbial communities sampled at Tm versus Tf (Fig. 2B; Table 1). Rhizobiales abundance was negatively associated with CO2 fluxes under dark conditions, while the Solibacterales were correlated to pore water alkalinity, which is a proxy for anaerobic decomposition (25) (Fig. 2; Table 1).
Soil metagenomics.
In addition to sequencing the 16S rRNA genes, which do not allow inference of an organism’s functional traits on their own, total DNA was sequenced from 5 soils derived from T0 and rhizosphere and bulk soil samples at Tm and Tf from the high-N treatment. The goal of the metagenomic sampling was to survey the genetic potential of organisms that were most strongly influenced by N loading (i.e., those identified in the 16S rRNA analysis). In particular, we wanted to find support for the roles the above taxa have in the rhizosphere of J. acutiflorus. These libraries resulted in on average 1 million post-QC reads per library (see Table S3 in the supplemental material). Coassembly of the metagenomic reads from all sequencing libraries yielded over 130,000 contigs with a mean contig length of 1,053 bp and a maximum length of over 46 kbp. The contigs were binned to obtain metagenome-assembled genomes (MAGs), with subsequent taxonomic assignment and genome completeness estimation. Bins with taxonomic affiliations matching with the taxa identified above as being associated with different N treatments were used for further analysis. Of the three selected bins, the Acidobacteria bin consisted of 261 contigs (assembled with 5,198 mapped reads), the Opitutales bin 374 contigs (3,979 reads), and the Sphingobacteriales bin 164 contigs (10,111 reads), with genome completeness estimates of 2.04, 12.25, and 2.35%, respectively. The Acidobacteria, Opitutales, and Sphingobacteriales contigs had N50 scores of 2,221, 1,897, and 2,856 bp, respectively.
Metagenome library overview, including the time of sampling (Time), number of post-QC reads (Reads), average length (Avg_len), and the standard deviation in read length (Sd_len). Download TABLE S3, XLSX file, 0.1 MB (11.3KB, xlsx) .
Copyright © 2018 Hester et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Although estimated to be highly incomplete, all bins were annotated to identify the functional potential of these species. The Acidobacteria bin contained carbon metabolism-associated genes involved in polysaccharide degradation and in the anaerobic degradation of aromatic compounds. Other than a nitrate-nitrite transporter, no nitrogen cycling genes were detected. The Opitutales bin contained a diverse set of genes related to oligosaccharide degradation and fermentation (acetoin and butyryl-coenzyme A [CoA] dehydrogenase), an amylomaltase for polysaccharide degradation, and genes for organic acid utilization. In addition, multiple fatty acid-, lipid-, and isoprenoid biosynthesis-related genes were detected. Among the nitrogen cycling genes detected were genes associated with nitrogen fixation (nitrogenase), denitrification (nitrous oxide reductase), and hydroxylamine reduction. The Sphingobacteriales bin contained carbon metabolism genes associated with di- and oligosaccharide degradation and fermentation (sugar/maltose fermentation stimulation protein homolog), a xylanase, and genes involved in the utilization of xylose as well as other plant-associated one-carbon metabolism-related genes.
As is reflected by the low completeness estimations and highly fragmented nature of our MAGs, retrieval of high-quality genomes from soil metagenomic data sets is highly challenging. To circumvent these challenges, we additionally applied a gene-centric approach to survey genetic potential for N and C cycling in N amended samples. Custom databases of genes involved in N and C cycling processes (26) were used to identify metagenomic reads of major N cycling genes (amoA and hao, involved in NH4+ oxidation; narG, nirK, nirS, norB, and nosZ, involved in denitrification; nrfA, involved in dissimilatory nitrite reduction to ammonia; and nifH, involved in N fixation) and CH4 cycling genes (pmoA and mmoX, involved in CH4 oxidation; and phnGHI and mcrA, involved in methanogenesis) and their abundance in the high-N incubations (abbreviations found in Table S4 in the supplemental material). There were no nirS genes detected in the data set, and only two reads annotated as mcrA were detected in the metagenomes. All other N and CH4 cycling genes were present (see Table S5 in the supplemental material).
N and C cycling gene abbreviations. Download TABLE S4, XLSX file, 0.1 MB (9.8KB, xlsx) .
Copyright © 2018 Hester et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The number of reads for each N and C cycling marker gene and their respective relative abundances (expressed in parentheses) in soil/rhizosphere metagenomes. Download TABLE S5, XLSX file, 0.1 MB (9.8KB, xlsx) .
Copyright © 2018 Hester et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Greenhouse gas emissions remain a global challenge. A thorough understanding of the factors that alter microbial community structure and function, such as increased N input, is important in developing management strategies for greenhouse gas emissions. This is particularly important in ecosystems as extensive as wetlands. With an estimated area of up to 12.8 million km2 worldwide, wetlands considerably contribute to the total terrestrial carbon storage (27, 28). Here we studied the impact of increased N input on the microbial community and greenhouse gas fluxes from the rhizosphere of J. acutiflorus, a very common plant in European wetland ecosystems and a model for other Juncus species globally. We found characteristic shifts in the microbial community structure and a stimulation of greenhouse gas fluxes in J. acutiflorus incubations in response to N input.
Plant physiological shifts as a response to high-N inputs.
The plant plays a prominent role in the maintenance of the rhizosphere microbial community (29). Roots release oxygen through radial oxygen loss, providing an oxic niche in otherwise anoxic wetland soils (30). Plants also release labile organic matter in the form of organic acids, neutral sugars, and amino acids (31, 32). The composition of this organic matter structures the microbial community within the rhizosphere by providing different substrates for heterotrophic microorganisms (33). The exuded organic acids acidify the surrounding soil, preventing many microbial species from thriving within the rhizosphere, but also modifying nutrient availability (34, 35). The quantity of organic matter released is closely associated with photosynthetic activity (36). As plants are often N limited in natural systems, relieving this limitation promotes plant growth (37). In this study, we observed that when incubated under high-N input, J. acutiflorus showed increased C fixation rates (Fig. 1A) and plant tissue becomes saturated with N (Table S2). This also suggests that J. acutiflorus without N limitation excretes larger amounts of labile carbon into the surrounding soil, which is also evident from the observed decreases in pore water pH in the high-N incubations (see Fig. S4 in the supplemental material). Additionally, due to root-derived oxygen, increased nitrification rates could contribute to this observed drop in pH (7). Together, higher N input could result in higher photosynthetic rates in J. acutiflorus specimens, likely depositing larger amounts of organic matter into surrounding soil, stimulating the heterotrophic microbial community in return (Fig. 2 and 3).
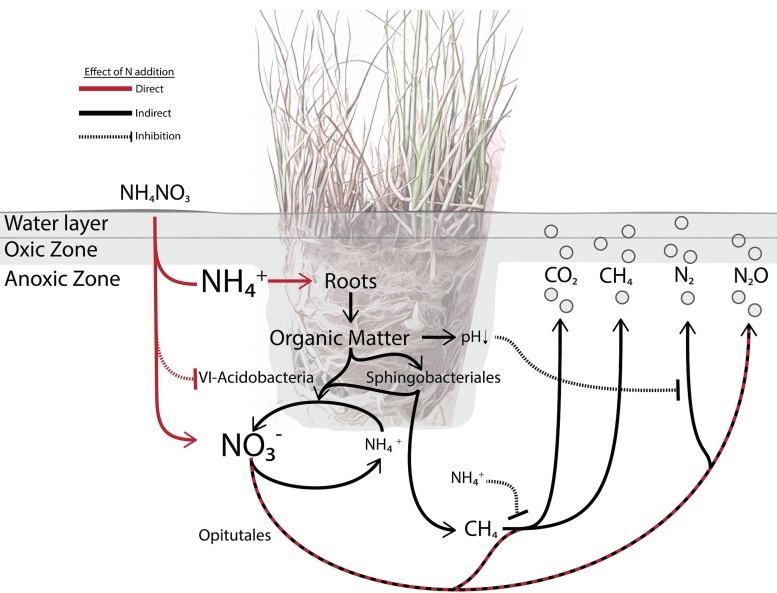
FIG 3 .
A Juncus acutiflorus rhizosphere microbial food web model. In the model, microbial processes are directly (red lines) or indirectly (black lines) influenced by N deposition. J. acutiflorus preferentially takes up NH4+, which stimulates plant productivity and rhizodeposition of organic matter and oxygen (24). Released oxygen and labile organic matter contribute to soil acidification, in addition to stimulating complex polymer degradation (Sphingobacteriales) and heterotrophic denitrifiers (Opitutales). The production of N2 can be affected by a drop in pH, which influences the activity of complete denitrifiers. The group 6 Acidobacteria are outcompeted at higher N availability. Recalcitrant organic matter degraded by Sphingobacteriales can enter the microbial food web and be fermented by fermenters, which in turn provide substrates for methanogens (mcr). The activity of phosphonate lyases (phn) might also stimulate the production of methane, while anaerobic methane oxidation also contributes to methane consumption. Additionally, methane consumption by aerobic methanotrophs through methane monooxgenases (pmo) could be inhibited by excess NH4+ (12).
Greenhouse gas fluxes as a result of N input.
N availability has previously been shown to alter greenhouse gas emission dynamics (8). Here we observed that greenhouse gas fluxes, both positive and negative, in J. acutiflorus incubations were stimulated by increased N input (Fig. 1). CO2 fixation rates were highest in J. acutiflorous incubations with high-N input under the light conditions, likely due to increased photosynthetic activity of the plant and photosynthetic microorganisms. In the dark, the same J. acutiflorus incubations showed elevated CO2 emissions, likely due to increased plant and microbial respiration (Fig. 1A and B). The highest CH4 emissions were observed in J. acutiflorus incubations with high-N input, although with large variability (Fig. 1C). Still, the elevated emission rates suggest that the J. acutiflorus rhizosphere could become a net source of CH4 under high-N input. The total amount of CH4 released reflects the sum of CH4 production (methanogenesis) and consumption (methanotrophy). In the present study, both mcrA and phnGHI genes, which are involved in the production of methane, as well as pmoA and mmoX, involved in methane oxidation, were detected (Table S5). Methanogenesis has been linked to plant productivity, thought to be due to increased availability of labile organic carbon from photosynthate exudates (38, 39). Furthermore, methanogens can be stimulated through an indirect priming mechanism. Labile organic matter from plant exudation can stimulate microbial activity responsible for degrading recalcitrant organic matter, which in turn makes this carbon source available to methanogens (40–43). Alternatively, net CH4 emissions can be increased by inhibiting CH4 consumption—for instance, through the competitive inhibition of the key enzyme methane monooxygenase by NH4+ (44, 45).
The reduction of NOx to N2 is often incomplete, resulting in the production of the greenhouse gas N2O. Incomplete denitrification occurs when microbial species do not utilize N2O as an electron acceptor either due to physiological constraints or induced by certain environmental conditions (46, 47). It has been observed that N fertilization has the largest impact on N2O emission, with NO3− availability being the main driver of emission rates (4). As denitrification is largely a microbial process, the composition of the microbial community plays an important role in the total amount of gaseous N forms emitted from soils. Representatives from a diverse set of phyla are known to denitrify (8, 46), and denitrification rates are therefore considered to be robust to changes in the microbial community composition (48). Here we observed elevated N2O emissions in J. acutiflorus incubations under high-N input, whereas they acted as an N2O sink in the low-N incubations (Fig. 1D). Interestingly, N2O emissions by bulk soil were not significantly influenced by the tested N regimes, despite the microbial community containing the full suite of denitrification-associated genes, indicating that J. acutiflorus plays a substantial role in stimulating N-reducing microbial species, probably by supplying labile carbon (Fig. 1D; Table S5). Elevated N loading additionally caused an almost 10-fold increase in the production of N2O relative to N2, suggesting that high-N input can shift the microbial rhizosphere community toward partial denitrifiers, which has important implications given the strong greenhouse potential of N2O.
Shifts in microbial community structure as a response to N input.
Association of microbial metabolisms (i.e., those resulting in greenhouse gas emission) with the structure of microbial communities and abiotic factors defined by the environment is essential to predict how the structure and function of these microbial ecosystems may adapt to future conditions. Bulk and rhizosphere soils contain diverse microbial communities with equally diverse metabolisms (49, 50). It remains a challenge to understand the role that key groups play in these systems and how they affect their environment.
Using 16S rRNA gene amplicon sequencing, we were able to identify three bacterial orders that were associated most strongly with N input and greenhouse gas emissions (Fig. 2C; Table 1). We further investigate the potential functional roles of these species in the J. acutiflorus rhizosphere by utilizing metagenomic data. Due to the immense diversity of the soil ecosystem, it is challenging to recover high-quality genomes from these systems. As a result, we were only able to obtain partial genomes of the organisms identified in the amplicon data set. We therefore built a conceptual model of the J. acutiflorus rhizosphere by first utilizing our metagenomic data, followed by conservatively drawing support from available literature on these organisms.
The verrucomicrobial Opitutales were associated with high-N input and elevated N2O emissions. Members of this order are diversely associated with different rhizospheres, ranging from sugarcane to wetland plants (51, 52). They have been physiologically described as anaerobic polysaccharide-utilizing bacteria that are capable of reducing NO3− to NO2− (53). In the present study, the Opitutales bin contained a diverse array of carbohydrate-degrading and fermentation-associated genes, including genes encoding an amylomaltase, which catalyzes the transglycosylation of maltodextrins, and butyryl-CoA and acetoin dehydrogenases. Apart from the O2 derived from the plant roots, which is quickly consumed by aerobic heterotrophs, wetland soils are waterlogged systems resulting in an anoxic environment. Opitutales may take advantage of the abundant organic carbon in the rhizosphere and anoxic niches, possibly fermenting some fraction of this carbon pool. While many of the carbon-related genes detected in the Opitutales bin and the nitrous oxide reductase are consistent with the literature, we did not detect nitrate or nitrite reductase as expected. A lack of genome completeness prevents conclusive statements about the role this particular Opitutales group plays in the J. acutiflorus rhizosphere; however, it is probable that expected denitrification genes were not retrieved. Based on a combination of literature describing the physiology of Opitutales from other rhizosphere environments with genomic evidence from the present study, it is probable that members of this group are taking advantage of plant-derived organic matter and have a denitrifying potential (Fig. 3).
The Sphingobacteriales from the phylum Bacteroidetes were also overrepresented in the high-N-input incubations (Fig. 2; Table 1). Described Sphingobacteriales are understood as copiotrophic bacteria, referring to their ability to metabolize a wide array of carbon sources and being present at great abundances in soils with high carbon availability (54, 55). Consistent with this, the Sphingobacteriales bin from the present study contained a wide array of genes encoding enzymes involved in carbohydrate utilization, particularly sugars that would originate from the plant, such as xylose. In the present study, the majority of organic matter would originate from the plant as the sandy soil used had low organic matter content. Rhizodeposition in this case would be very important for microbial groups such as Sphingobacteriales, not only as a carbon source but also as an O2 source, since Sphingobacteriales have been reported to be particularly sensitive to O2 availability. When tested for cellulolytic activity in oxic or anoxic environments, they were exclusively active in oxic treatments, suggesting that this group may require oxygenated environments for at least some forms of carbon degradation (56). Interestingly, the Sphingobacteriales bin contains some fermentation-associated genes (i.e., sugar/maltose fermentation stimulation protein homolog), indicating possible flexibility in oxygen requirements. Considering findings from this study and the literature, we hypothesize that Sphingobacteriales within the J. acutiflorus rhizosphere could be benefiting from the elevated carbon input from the roots under higher N input, cycling this carbon and possibly making it available to other community members (Fig. 3).
G6 Acidobacteria were overrepresented in the low-N-input incubations, and there was no significant difference in their abundance between bulk and rhizosphere soils. Unlike Opitutales and Sphingobacteriales, they were negatively correlated with N2O emissions (Fig. 2; Table 1). While the G6 Acidobacteria group is not well studied, one genome (GenBank accession no. CP015136.1) was recently published (57) and was shown to contain nitric and nitrous oxide reductases. In the Acidobacteria bin, a nitrate-nitrite transport gene was detected; however, no denitrification-associated genes were present. Genomic and physiological studies of a closely related group (group 1 Acidobacteria) showed that they were anaerobic organoheterotrophs capable of utilizing NO3− for respiration (58), and other Acidobacteria have also been described as important soil carbon and N cyclers. However, many N-cycling reactions are restricted to particular clades, indicating that these functions are heterogeneously represented across the Acidobacteria phylum (59, 60). In addition to N cycling, Acidobacteria are known for their utilization of C derived from autotrophic microorganisms in anoxic environments (61). They have been reported to utilize various plant- and microbe-derived polysaccharides, like xylan, cellobiose, and gellan (60, 62), and thrive in various soils and rhizospheres, including anoxic soils with low pH (54, 63). We detected multiple genes associated with carbohydrate metabolism in the Acidobacteria bin, namely, those for maltose and maltodextrin utilization. The cultured representatives of Acidobacteria have low growth rates and appear to be adapted to oligotrophic environments (54, 64). Thus, G6 Acidobacteria may not be competitive under high N availability with fast-growing (partial) denitrifiers. Together, in the low-N-input rhizosphere, the G6 Acidobacteria may be involved in a slower turnover of organic carbon, either from other bacteria in the community or from plant biomass. Increased N availability might reduce this group’s abundance by altering competitive advantages (Fig. 3).
A model microbial food web within bulk soil and the J. acutiflorus rhizosphere.
Increased N input poses a distinct threat to wetland ecosystems, contributing to the degradation of biodiversity and altering greenhouse gas emissions (3, 8). Plants such as J. acutiflorus influence the abundance and composition of microorganisms living in the rhizosphere by exuding organic matter and releasing oxygen from their roots (29). In the present study, N addition resulted in increased productivity of J. acutiflorus, stimulating the effect of the plant on the microbial community but also directly affecting microbial metabolism. Based on our observations and published knowledge, we built a conceptual model of the J. acutiflorus microbial food web, indicating how N input impacts the soil microbial community (Fig. 3).
N fertilization can influence the soil microbial community by providing excess NH4+ and NO3−. Previous studies have shown that J. acutiflorus prefers NH4+ over NO3− as an N source, leading to a surplus of NO3− in the rhizosphere (24) (Fig. S4). This alters N cycling dynamics in the rhizosphere, favoring microbial species and metabolisms reducing NO3− to N2O rather than to N2 (65, 80) (Fig. 1; Fig. S2). The combined effect of enhanced plant-derived carbon input and higher N availability stimulates heterotrophic activity, resulting in increased N2O and CO2 emissions (Fig. 1 and 3). While excess NO3− spurs anaerobic respiration, increased NH4+ concentrations can lead to an inhibition of methane oxidation, possibly contributing to the heterogeneity observed in CH4 emissions (Fig. 1C). High N availability can also have an indirect effect by influencing plant physiology. The observed increased rates of carbon fixation by J. acutiflorus under high-N input may result in augmented release of organic matter (including organic acids) and oxygen from the roots. This acidifies the rhizosphere soil, which can alter the activity of nosZ-containing microbes (65). Additionally, elevated oxygen availability stimulates heterotrophic activity in an otherwise anoxic environment, leading to higher CO2 emissions. Thus, altered N input in the J. acutiflorus rhizosphere leads to increased greenhouse gas fluxes directly by altering the abundance of N-cycling species and indirectly through the stimulation of plant primary productivity (Fig. 3).
Conclusions.
With continued anthropogenic inputs of nitrogen into wetlands, it is critical to understand how this activity may affect globally relevant carbon and nitrogen cycling within wetlands. The results here support that under high N input, greenhouse gas emissions from the J. acutiflorus rhizosphere increase, shifting the system from a greenhouse gas sink to a source. Three bacterial orders, the Opitutales, G6 Acidobacteria, and Sphingobacteriales, respond to increased N availability, and genomic evidence supports their involvement in processes leading to changes in greenhouse gas fluxes. Our view is that understanding interactions within the rhizosphere that result in increased greenhouse gas emissions is essential for creating management solutions aimed to address emission goals, efficient agricultural practices, and conservation efforts. To move forward in our understanding of the complex dynamics within ecosystems such as the rhizosphere, future effort needs to be made in building extensive data sets that can be used to build predictive models of how these microbial ecosystems might respond under altered environmental conditions. We propose that conceptual models, such as our J. acutiflorus rhizosphere plant-microbial food web model, should be used to set the framework for building such data sets.
MATERIALS AND METHODS
Sample collection and experimental setup.
Plants and sandy soil were sampled from the Ravenvennen (51.4399°N, 6.1961°E) in Limburg, The Netherlands (August 2015), and returned to the Radboud University greenhouse facilities for conditioning. The Ravenvennen is a protected marshy area consisting of sandy soil rich in vegetation with a high prevalence of Juncus spp. Plants were removed from soil, and rhizomes were cut into eight 2-cm fragments and reconditioned on hydroculture in a nutrient-rich medium as described by Hoagland and Arnon (66). After sufficient root development (to approximately 25 cm after 2 weeks), eight plants and eight bulk soil incubations were randomly assigned to high- or low-nitrogen experimental groups, totaling 16 incubations (see Table S1 and Fig. S1 in the supplemental material). Soil collected from the field was homogenized and sieved to remove any contaminating roots and potted. The reconditioned plants were transferred to pots with diameters of 19 cm at the base and 26 cm at the top and a height of 19 cm containing the prepared soil, moved to an indoor water bath set to 15°C with a cryostat (Neslab Thermoflex 1400; Thermo Electron Corp., Breda, The Netherlands), and cultivated with a day/night cycle of 16 h of light and 8 h of dark (Master Son-T PiaPlus; Philips, Eindhoven, The Netherlands). Pots were kept waterlogged with a 2-cm water layer on top. A drip-percolation-based system ensured a constant supply of nutrients. The low-N-input nutrient solution contained 12.5 µM NH4NO3, corresponding to an N loading rate of 40 kg N·ha−1·year−1. The high-N-input solution contained 250 µM NH4NO3, corresponding to 800 kg N·ha−1·year−1. These rates fall within N loading of wetlands in agricultural catchments and thus represent contrasting extremes (67).
Experimental design schema depicting sample replicates per treatment in either rhizosphere or bulk soil. Additionally, the sampling points and types are denoted by colored boxes. GHG, greenhouse gas. Download FIG S1, EPS file, 1.3 MB (1.3MB, eps) .
Copyright © 2018 Hester et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Denitrification potential from soil slurries. N2 and N2O production rates were estimated to determine potential denitrification of the soil and rhizosphere microbial communities. Download FIG S2, EPS file, 2.7 MB (2.8MB, eps) .
Copyright © 2018 Hester et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Rarefaction curves with number of species observed as a function of sequencing effort (sample depth). Download FIG S3, EPS file, 1.8 MB (1.8MB, eps) .
Copyright © 2018 Hester et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pore water inorganic nutrients, pH, and alkalinity. Shown are the concentrations of inorganic nutrients, pH, and alkalinity in pore water sampled throughout the incubation. Download FIG S4, EPS file, 2.8 MB (2.8MB, eps) .
Copyright © 2018 Hester et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Incubation measurements.
Five representative J. acutiflorus specimens were harvested for initial measurements of plant dry weight and C/N ratios. At the final time point (Tf, 90 days), all plants were harvested to measure dry weight and C/N ratios of roots, shoots, and rhizomes. Pore water was extracted using 0.15-µm porous soil moisture samplers (SMS rhizons, Rhizosphere Research Products, Wageningen, The Netherlands) and measured over the course of the experiment to determine inorganic nutrients as well as metals using an AutoAnalyzer (AutoAnalyzer 3; Bran+Luebbe, Germany) and ICP-OES (iCAP6000; Thermo Scientific, Waltham, MA). To reduce the impact of soil heterogeneity, samples were extracted in duplicate and mean values were calculated.
Greenhouse gas measurements.
To determine greenhouse gas fluxes, a cylindrical transparent collection chamber (7.5 by 30 cm) was used to measure accumulation or depletion of CO2, CH4, and N2O in the headspace. CO2 and CH4 fluxes were measured at Tm (45 days), and Tf and N2O fluxes were measured at Tf. Fluxes were measured using a Picarro G2308 NIRS-CRD greenhouse gas analyzer (Picarro, Inc., Santa Clara, CA). Fluxes were determined by fitting a smoothed spline to the time series using the R function sm.spline from the pspline package, and the average rate of change was calculated (68).
Denitrification potential.
To measure denitrification potential, two soil slurries were made from each experimental pot by mixing 50 g soil with 100 ml Milli-Q water, divided into control and experimental bottles, and made anoxic by flushing with argon gas. Bottles were preincubated overnight at 15°C to allow for residual unlabeled NO3− to be consumed. A 15N-labeled NaNO3 solution was added to the experimental bottles to a final concentration of 500 µM, and a KCl solution was added to the control bottles to a final concentration of 500 µM. Production of N2O and N2 was measured by taking samples 2, 7, and 22 h after adding substrate on a gas chromatography-mass spectrometry (GC-MS) device (5975C; Agilent Technologies, Santa Clara, CA).
DNA extraction and 16S rRNA amplicon and metagenomic sequencing.
Soil was collected from each of the 16 incubations at three time points: one initial soil sample from the site and Tm and Tf samples. A single core per pot was taken using a 1- by 7-cm corer. DNA was extracted using the PowerSoil DNA isolation kit (MoBio, Carlsbad, CA). From all 16 incubations, 16S rRNA genes (variable regions 3 to 4) were amplified in triplicate reactions using IonTorrent sequencing adapter barcoded primers 341F (CCATCTCATCCCTGCGTGTCTCCGACTCAGxxxxxxxxxxGATCCTACGGGNGGCWGCAG) and 785R (CCACTACGCCTCCGCTTTCCTCTCTATGGGCAGTCGGTGATGACTACHVGGGTATCTAATCC) and pooled. The pooled amplicons were cleaned with Ampure beads (Beckman Coulter, Inc., Fullerton, CA) and subsequently prepared for sequencing on the IonTorrent PGM using the manufacturer’s instructions (Life Technologies, Inc., Carlsbad, CA).
From the same DNA samples, total DNA from 5 representative incubations (4 Tm, 4 Tf, 16 Tm, and 16 Tf samples and the initial soil sample) was sheared into approximately 400-bp fragments via sonication. The resulting fragments were prepared for sequencing following the manufacturer’s instructions with the Ion Plus Fragment Library kit (Life Technologies, Inc., Carlsbad, CA).
Data analysis.
16S rRNA gene amplicons were filtered for quality (Q > 25) and size (>200 bp) using QIIME v1.9 (69). Quality-controlled reads were then clustered into OTU at a 97% identify level and phylogenetically classified by utilizing the NINJA-OPS v1.3 pipeline (70). The reference database used for taxonomic assignment was the SILVA database version 123 (71). The resulting OTU table was used for downstream analysis in R (72). Count data were normalized to relative abundances to account for differing sequence depth between samples, and a square root transformation was applied. The vegan R package was used to calculate Shannon diversity with the diversity function, Bray-Curtis dissimilarity matrices with the vegdist function, permutational multivariate analysis with the adonis2 function, and nonmetric multidimensional scaling (nMDS) with the metaMDS function (73). The Bray-Curtis dissimilarity was used in calculating the nMDS. The RandomForest R package was used for classification and regression (74). Linear models were fit with the glm function in the stats package.
Metagenomic reads were quality filtered (Q > 25), and small fragments (<100 bp) were removed using PrinSeq (75). Quality-filtered reads were assembled using metaSPAdes (version 3.7 [76]). The Resulting contigs (>1 kbp) were binned using BinSanity, and taxonomic assignments and bin quality were checked with CheckM (77, 78). Annotations of bins were performed using the SEED database (79).
Accession number(s).
Raw reads were submitted to NCBI and archived under SRA accession no. SRP099838.
ACKNOWLEDGMENTS
We thank Theo van Alen for sequencing support, Jeroen Frank for assistance with binning, and Sebastian Krosse, Paul van der Ven, and General Instrumentation at Radboud University for support with elemental analysis.
Funding was provided by the European Research Council (ERC AG 339880 ECOMOM) to M.S.M.J. and the Netherlands Organization for Scientific Research (NWO) through Gravitation Grants SIAM (024.002.002) and NESSC (024.002.001) to M.S.M.J. and VENI grant 863.14.019 to S.L.
E.R.H., S.F.H., J.M.H.V.D., L.L.L., M.S.M.J., C.L., S.L., and C.U.W. designed research. E.R.H., S.F.H., and J.M.H.V.D. performed research. E.R.H., S.F.H., and J.M.H.V.D. analyzed data. E.R.H., S.L., and C.U.W. wrote the paper. All authors reviewed and agreed with the final version of the manuscript.
REFERENCES
- 1.Galloway JN, Townsend AR, Erisman JW, Bekunda M, Cai Z, Freney JR, Martinelli LA, Seitzinger SP, Sutton MA. 2008. Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320:889–892. doi: 10.1126/science.1136674. [DOI] [PubMed] [Google Scholar]
- 2.Britto DT, Kronzucker HJ. 2002. NH4+ toxicity in higher plants: a critical review. J Plant Physiol 159:567–584. doi: 10.1078/0176-1617-0774. [DOI] [Google Scholar]
- 3.Bobbink R, Hornung M, Roelofs JGM. 1998. The effects of air-borne nitrogen pollutants on species diversity in natural and semi-natural European vegetation. J Ecol 86:717–738. doi: 10.1046/j.1365-2745.1998.8650717.x. [DOI] [Google Scholar]
- 4.Liu L, Greaver TL. 2009. A review of nitrogen enrichment effects on three biogenic GHGs: the CO 2 sink may be largely offset by stimulated N2O and CH4 emission. Ecol Lett 12:1103–1117. doi: 10.1111/j.1461-0248.2009.01351.x. [DOI] [PubMed] [Google Scholar]
- 5.Van den Heuvel RN, Bakker SE, Jetten MSM, Hefting MM. 2011. Decreased N2O reduction by low soil pH causes high N2O emissions in a riparian ecosystem. Geobiology 9:294–300. doi: 10.1111/j.1472-4669.2011.00276.x. [DOI] [PubMed] [Google Scholar]
- 6.Soons MB, Hefting MM, Dorland E, Lamers LPM, Versteeg C, Bobbink R. 2017. Nitrogen effects on plant species richness in herbaceous communities are more widespread and stronger than those of phosphorus. Biol Conserv 212:390–397. doi: 10.1016/j.biocon.2016.12.006. [DOI] [Google Scholar]
- 7.Lamers LPM, van Diggelen JMH, Op den Camp HJM, Visser EJW, Lucassen EC, Vile MA, Jetten MSM, Smolders AJP, Roelofs JGM. 2012. Microbial transformations of nitrogen, sulfur, and iron dictate vegetation composition in wetlands: a review. Front Microbiol 3:156. doi: 10.3389/fmicb.2012.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Philippot L, Hallin S, Börjesson G, Baggs EM. 2009. Biochemical cycling in the rhizosphere having an impact on global change. Plant Soil 321:61–81. doi: 10.1007/s11104-008-9796-9. [DOI] [Google Scholar]
- 9.Brenzinger K, Dörsch P, Braker G. 2015. pH-driven shifts in overall and transcriptionally active denitrifiers control gaseous product stoichiometry in growth experiments with extracted bacteria from soil. Front Microbiol 6:961. doi: 10.3389/fmicb.2015.00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King GM, Schnell S. 1998. Effects of ammonium and non-ammonium salt additions on methane oxidation by Methylosinus trichosporium OB3b and Maine forest soils. Appl Environ Microbiol 64:253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bodelier PLE, Laanbroek HJ. 2004. Nitrogen as a regulatory factor of methane oxidation in soils and sediments. FEMS Microbiol Ecol 47:265–277. doi: 10.1016/S0168-6496(03)00304-0. [DOI] [PubMed] [Google Scholar]
- 12.Dunfield P, Knowles R. 1995. Kinetics of inhibition of methane oxidation by nitrate, nitrite, and ammonium in a humisol. Appl Environ Microbiol 61:3129–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bragazza L, Freeman C, Jones T, Rydin H, Limpens J, Fenner N, Ellis T, Gerdol R, Hájek M, Hájek T, Iacumin P, Kutnar L, Tahvanainen T, Toberman H. 2006. Atmospheric nitrogen deposition promotes carbon loss from peat bogs. Proc Natl Acad Sci U S A 103:19386–19389. doi: 10.1073/pnas.0606629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith CJ, Delaune RD. 1984. Influence of the rhizosphere of Spartina alterniflora Loisel. On nitrogen loss from a Louisiana Gulf Coast salt marsh. Environ Exp Bot 24:91–93. doi: 10.1016/0098-8472(84)90064-9. [DOI] [Google Scholar]
- 15.Abou Seada MNI, Ottow JCG. 1985. Effect of increasing oxygen concentration on total denitrification and nitrous oxide release from soil by different bacteria. Biol Fertil Soils 1:31–38. doi: 10.1007/BF00710968. [DOI] [Google Scholar]
- 16.Bardgett RD, van der Putten WH. 2014. Belowground biodiversity and ecosystem functioning. Nature 515:505–511. doi: 10.1038/nature13855. [DOI] [PubMed] [Google Scholar]
- 17.Robinson D, Hodge A, Fitter A. 2003. Constraints on the form and function of root systems, p 1–31. In Ecological Studies. Springer, Berlin, Germany. doi: 10.1007/978-3-662-09784-7_1. [DOI] [Google Scholar]
- 18.Buée M, De Boer W, Martin F, van Overbeek L, Jurkevitch E. 2009. The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 321:189–212. doi: 10.1007/s11104-009-9991-3. [DOI] [Google Scholar]
- 19.Curl EA, Harper JD. 1990. Fauna-microflora interactions, p 369–388. In Lynch JM (ed), The rhizosphere. John Wiley and Sons, Chichester, United Kingdom. [Google Scholar]
- 20.Hinsinger P, Bengough AG, Vetterlein D, Young IM. 2009. Rhizosphere: biophysics, biogeochemistry and ecological relevance. Plant Soil 321:117–152. doi: 10.1007/s11104-008-9885-9. [DOI] [Google Scholar]
- 21.Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R. 2012. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J 6:1007–1017. doi: 10.1038/ismej.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leff JW, Jones SE, Prober SM, Barberán A, Borer ET, Firn JL, Harpole WS, Hobbie SE, Hofmockel KS, Knops JMH, McCulley RL, La Pierre K, Risch AC, Seabloom EW, Schütz M, Steenbock C, Stevens CJ, Fierer N. 2015. Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc Natl Acad Sci U S A 112:10967–10972. doi: 10.1073/pnas.1508382112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramirez KS, Craine JM, Fierer N. 2012. Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob Chang Biol 18:1918–1927. doi: 10.1111/j.1365-2486.2012.02639.x. [DOI] [Google Scholar]
- 24.van Diggelen JMH, Smolders AJP, Visser EJW, Hicks S, Roelofs JGM, Lamers LPM. 2016. Differential responses of two wetland graminoids to high ammonium at different pH values. Plant Biol 18:307–315. doi: 10.1111/plb.12398. [DOI] [PubMed] [Google Scholar]
- 25.Berner RA, Scott MR, Thomlinson C. 1970. Carbonate alkalinity in the pore waters of anoxic marine sediments. Limnol Oceanogr 15:544–549. doi: 10.4319/lo.1970.15.4.0544. [DOI] [Google Scholar]
- 26.Lüke C, Speth DR, Kox MAR, Villanueva L, Jetten MSM. 2016. Metagenomic analysis of nitrogen and methane cycling in the Arabian Sea oxygen minimum zone. PeerJ 4:e1924. doi: 10.7717/peerj.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zedler JB, Kercher S. 2005. Wetland resources: status, trends, ecosystem services, and restorability. Annu Rev Environ Resour 30:39–74. doi: 10.1146/annurev.energy.30.050504.144248. [DOI] [Google Scholar]
- 28.Nahlik AM, Fennessy MS. 2016. Carbon storage in US wetlands. Nat Commun 7:13835. doi: 10.1038/ncomms13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinhold-Hurek B, Bünger W, Burbano CS, Sabale M, Hurek T. 2015. Roots shaping their microbiome: global hotspots for microbial activity. Annu Rev Phytopathol 53:403–424. doi: 10.1146/annurev-phyto-082712-102342. [DOI] [PubMed] [Google Scholar]
- 30.Armstrong W. 1971. Radial oxygen losses from intact rice roots as affected by distance from the apex, respiration and waterlogging. Physiol Plant 25:192–197. doi: 10.1111/j.1399-3054.1971.tb01427.x. [DOI] [Google Scholar]
- 31.Kamilova F, Kravchenko LV, Shaposhnikov AI, Azarova T, Makarova N, Lugtenberg B. 2006. Organic acids, sugars, and l-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol Plant Microbe Interact 19:250–256. doi: 10.1094/MPMI-19-0250. [DOI] [PubMed] [Google Scholar]
- 32.Jones DL. 1998. Organic acids in the rhizosphere—a critical review. Plant Soil 205:25–44. doi: 10.1023/A:1004356007312. [DOI] [Google Scholar]
- 33.Haichar FZ, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J, Heulin T, Achouak W. 2008. Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2:1221–1230. doi: 10.1038/ismej.2008.80. [DOI] [PubMed] [Google Scholar]
- 34.Marschner H, Romheld V, Cakmak I. 1987. Root-induced changes of nutrient availability in the rhizosphere. J Plant Nutr 10:1175–1184. doi: 10.1080/01904168709363645. [DOI] [Google Scholar]
- 35.Petersen W, Böttger M. 1991. Contribution of organic acids to the acidification of the rhizosphere of maize seedlings. Plant Soil 132:159–163. doi: 10.1007/BF00010396. [DOI] [Google Scholar]
- 36.Dilkes NB, Jones DL, Farrar J. 2004. Temporal dynamics of carbon partitioning and rhizodeposition in wheat. Plant Physiol 134:706–715. doi: 10.1104/pp.103.032045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reich PB, Hobbie SE, Lee T, Ellsworth DS, West JB, Tilman D, Knops JMH, Naeem S, Trost J. 2006. Nitrogen limitation constrains sustainability of ecosystem response to CO2. Nature 440:922–925. doi: 10.1038/nature04486. [DOI] [PubMed] [Google Scholar]
- 38.Whiting GJ, Chanton JP. 1993. Primary production control of methane emission from wetlands. Nature 364:794–795. doi: 10.1038/364794a0. [DOI] [Google Scholar]
- 39.Aulakh MS, Wassmann R, Bueno C, Rennenberg H. 2001. Impact of root exudates of different cultivars and plant development stages of rice (Oryza sativa L.) on methane production in a paddy soil. Plant Soil 230:77–86. doi: 10.1023/A:1004817212321. [DOI] [Google Scholar]
- 40.Jenkinson DS, Fox RH, Rayner JH. 1985. Interactions between fertilizer nitrogen and soil nitrogen—the so-called “priming” effect. J Soil Sci 36:425–444. doi: 10.1111/j.1365-2389.1985.tb00348.x. [DOI] [Google Scholar]
- 41.Kotsyurbenko OR. 2005. Trophic interactions in the methanogenic microbial community of low-temperature terrestrial ecosystems. FEMS Microbiol Ecol 53:3–13. doi: 10.1016/j.femsec.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 42.Kotsyurbenko OR, Nozhevnikova AN, Zavarzin GA. 1993. Methanogenic degradation of organic matter by anaerobic bacteria at low temperature. Chemosphere 27:1745–1761. doi: 10.1016/0045-6535(93)90155-X. [DOI] [Google Scholar]
- 43.Tveit AT, Urich T, Frenzel P, Svenning MM. 2015. Metabolic and trophic interactions modulate methane production by Arctic peat microbiota in response to warming. Proc Natl Acad Sci U S A 112:E2507–E2516. doi: 10.1073/pnas.1420797112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bosse U, Frenzel P, Conrad R. 1993. Inhibition of methane oxidation by ammonium in the surface layer of a littoral sediment. FEMS Microbiol Ecol 13:123–134. doi: 10.1111/j.1574-6941.1993.tb00058.x. [DOI] [Google Scholar]
- 45.Conrad R, Rothfuss F. 1991. Methane oxidation in the soil surface layer of a flooded rice field and the effect of ammonium. Biol Fertil Soils 12:28–32. doi: 10.1007/BF00369384. [DOI] [Google Scholar]
- 46.Philippot L. 2002. Denitrifying genes in bacterial and archaeal genomes. Biochim Biophys Acta 1577:355–376. doi: 10.1016/S0167-4781(02)00420-7. [DOI] [PubMed] [Google Scholar]
- 47.Wallenstein MD, Myrold DD, Firestone M, Voytek M. 2006. Environmental controls on denitrifying communities and denitrification rates: insights from molecular methods. Ecol Appl 16:2143–2152. [DOI] [PubMed] [Google Scholar]
- 48.Enwall K, Philippot L, Hallin S. 2005. Activity and composition of the denitrifying bacterial community respond differently to long-term fertilization. Appl Environ Microbiol 71:8335–8343. doi: 10.1128/AEM.71.12.8335-8343.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH. 2013. Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11:789–799. doi: 10.1038/nrmicro3109. [DOI] [PubMed] [Google Scholar]
- 50.Torsvik V, Øvreås L. 2002. Microbial diversity and function in soil: from genes to ecosystems. Curr Opin Microbiol 5:240–245. doi: 10.1016/S1369-5274(02)00324-7. [DOI] [PubMed] [Google Scholar]
- 51.Dedysh SN, Pankratov TA, Belova SE, Kulichevskaya IS, Liesack W. 2006. Phylogenetic analysis and in situ identification of bacteria community composition in an acidic sphagnum peat bog. Appl Environ Microbiol 72:2110–2117. doi: 10.1128/AEM.72.3.2110-2117.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Passel MWJ, Kant R, Palva A, Copeland A, Lucas S, Lapidus A, Glavina del Rio T, Pitluck S, Goltsman E, Clum A, Sun H, Schmutz J, Larimer FW, Land ML, Hauser L, Kyrpides N, Mikhailova N, Richardson PP, Janssen PH, de Vos WM, Smidt H. 2011. Genome sequence of the verrucomicrobium Opitutus terrae PB90-1, an abundant inhabitant of rice paddy soil ecosystems. J Bacteriol 193:2367–2368. doi: 10.1128/JB.00228-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chin KJ, Liesack W, Janssen PH. 2001. Opitutus terrae gen. nov., sp. nov., to accommodate novel strains of the division “Verrucomicrobia” isolated from rice paddy soil. Int J Syst Evol Microbiol 51:1965–1968. doi: 10.1099/00207713-51-6-1965. [DOI] [PubMed] [Google Scholar]
- 54.Fierer N, Bradford MA, Jackson RB. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 55.Padmanabhan P, Padmanabhan S, DeRito C, Gray A, Gannon D, Snape JR, Tsai CS, Park W, Jeon C, Madsen EL. 2003. Respiration of 13C-labeled substrates added to soil in the field and subsequent 16S rRNA gene analysis of 13C-labeled soil DNA. Appl Environ Microbiol 69:1614–1622. doi: 10.1128/AEM.69.3.1614-1622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schellenberger S, Kolb S, Drake HL. 2010. Metabolic responses of novel cellulolytic and saccharolytic agricultural soil bacteria to oxygen. Environ Microbiol 12:845–861. doi: 10.1111/j.1462-2920.2009.02128.x. [DOI] [PubMed] [Google Scholar]
- 57.Huang S, Vieira S, Bunk B, Riedel T, Spröer C, Overmann J. 2016. First complete genome sequence of a subdivision 6 Acidobacterium strain. Genome Announc 4:e00469-16. doi: 10.1128/genomeA.00469-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dedysh SN, Kulichevskaya IS, Serkebaeva YM, Mityaeva MA, Sorokin VV, Suzina NE, Rijpstra WI, Damsté JS. 2012. Bryocella elongata gen. nov., sp. nov., a member of subdivision 1 of the Acidobacteria isolated from a methanotrophic enrichment culture, and emended description of Edaphobacter aggregans Koch et al. 2008. Int J Syst Evol Microbiol 62:654–664. doi: 10.1099/ijs.0.031898-0. [DOI] [PubMed] [Google Scholar]
- 59.Kielak AM, Scheublin TR, Mendes LW, van Veen JA, Kuramae EE. 2016. Bacterial community succession in pine-wood decomposition. Front Microbiol 7:231. doi: 10.3389/fmicb.2016.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koch IH, Gich F, Dunfield PF, Overmann J. 2008. Edaphobacter modestus gen. nov., sp. nov., and Edaphobacter aggregans sp. nov., Acidobacteria isolated from alpine and forest soils. Int J Syst Evol Microbiol 58:1114–1122. doi: 10.1099/ijs.0.65303-0. [DOI] [PubMed] [Google Scholar]
- 61.Meisinger DB, Zimmermann J, Ludwig W, Schleifer KH, Wanner G, Schmid M, Bennett PC, Engel AS, Lee NM. 2007. In situ detection of novel Acidobacteria in microbial mats from a chemolithoautotrophically based cave ecosystem (Lower Kane Cave, WY, USA). Environ Microbiol 9:1523–1534. doi: 10.1111/j.1462-2920.2007.01271.x. [DOI] [PubMed] [Google Scholar]
- 62.Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl Environ Microbiol 68:2391–2396. doi: 10.1128/AEM.68.5.2391-2396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pankratov TA, Dedysh SN. 2010. Granulicella paludicola gen. nov., sp. nov., Granulicella pectinivorans sp. nov., Granulicella aggregans sp. nov. and Granulicella rosea sp. nov., acidophilic, polymer-degrading Acidobacteria from sphagnum peat bogs. Int J Syst Evol Microbiol 60:2951–2959. doi: 10.1099/ijs.0.021824-0. [DOI] [PubMed] [Google Scholar]
- 64.Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N. 2009. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J 3:442–453. doi: 10.1038/ismej.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu B, Frostegård Å, Bakken LR. 2014. Impaired reduction of N2O to N2 in acid soils is due to a posttranscriptional interference with the expression of nosZ. mBio 5:e01383-14. doi: 10.1128/mBio.01383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoagland DR, Arnon DI. 1950. The water-culture method for growing plants without soil. University of California, College of Agriculture, Agricultural Experiment Station, Berkeley, CA. [Google Scholar]
- 67.Verhoeven JTA, Arheimer B, Yin C, Hefting MM. 2006. Regional and global concerns over wetlands and water quality. Trends Ecol Evol 21:96–103. doi: 10.1016/j.tree.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 68.Ramsay JO, Heckman N, Silverman BW. 1997. Spline smoothing with model-based penalties. Behav Res Methods Instrum Comput 29:99–106. doi: 10.3758/BF03200573. [DOI] [Google Scholar]
- 69.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Al-Ghalith GA, Montassier E, Ward HN, Knights D. 2016. NINJA-OPS: fast accurate marker gene alignment using concatenated ribosomes. PLoS Comput Biol 12:e1004658. doi: 10.1371/journal.pcbi.1004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.R Core Team 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 73.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2017. vegan: Community Ecology Package. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 74.Liaw A, Wiener M. 2002. Classification and regression by randomForest. R News 2:18–22. [Google Scholar]
- 75.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Graham ED, Heidelberg JF, Tully BJ. 2017. BinSanity: unsupervised clustering of environmental microbial assemblies using coverage and affinity propagation. PeerJ 5:e3035. doi: 10.7717/peerj.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saggar S, Jha N, Deslippe J, Bolan NS, Luo J, Giltrap DL, Kim DG, Zaman M, Tillman RW. 2013. Denitrification and N2O:N2 production in temperate grasslands: processes, measurements, modelling and mitigating negative impacts. Sci Total Environ 465:173–195. doi: 10.1016/j.scitotenv.2012.11.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sample overview containing the time of sampling, N-load treatment, and whether the sample was bulk or rhizosphere soil. Additionally, the number of post-quality-filtered reads that were produced and the number of OTU found in each sample are shown. Greenhouse gas fluxes are reported in micromoles per square meter per day. Finally, Shannon diversity (H′) is reported. Download TABLE S1, XLSX file, 0.1 MB (14.5KB, xlsx) .
Copyright © 2018 Hester et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plant average dry weight (DW) and C/N ratio were determined in different sections of the plant, including the roots, shoots, and rhizomes. Biomass weight was determined as dry weight. The mean values from plants receiving high N and low N are reported (Mean High and Mean Low). The P value is reported as a result of a t test comparing mean values from high- and low-N treatments. Stdev, standard deviation. Download TABLE S2, XLSX file, 0.1 MB (11.5KB, xlsx) .
Copyright © 2018 Hester et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metagenome library overview, including the time of sampling (Time), number of post-QC reads (Reads), average length (Avg_len), and the standard deviation in read length (Sd_len). Download TABLE S3, XLSX file, 0.1 MB (11.3KB, xlsx) .
Copyright © 2018 Hester et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
N and C cycling gene abbreviations. Download TABLE S4, XLSX file, 0.1 MB (9.8KB, xlsx) .
Copyright © 2018 Hester et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The number of reads for each N and C cycling marker gene and their respective relative abundances (expressed in parentheses) in soil/rhizosphere metagenomes. Download TABLE S5, XLSX file, 0.1 MB (9.8KB, xlsx) .
Copyright © 2018 Hester et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Experimental design schema depicting sample replicates per treatment in either rhizosphere or bulk soil. Additionally, the sampling points and types are denoted by colored boxes. GHG, greenhouse gas. Download FIG S1, EPS file, 1.3 MB (1.3MB, eps) .
Copyright © 2018 Hester et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Denitrification potential from soil slurries. N2 and N2O production rates were estimated to determine potential denitrification of the soil and rhizosphere microbial communities. Download FIG S2, EPS file, 2.7 MB (2.8MB, eps) .
Copyright © 2018 Hester et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Rarefaction curves with number of species observed as a function of sequencing effort (sample depth). Download FIG S3, EPS file, 1.8 MB (1.8MB, eps) .
Copyright © 2018 Hester et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pore water inorganic nutrients, pH, and alkalinity. Shown are the concentrations of inorganic nutrients, pH, and alkalinity in pore water sampled throughout the incubation. Download FIG S4, EPS file, 2.8 MB (2.8MB, eps) .
Copyright © 2018 Hester et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.