ABSTRACT
Small-animal models have been used to obtain many insights regarding the pathogenesis and immune responses induced following infection with human respiratory syncytial virus (hRSV). Among those described to date, infections in cotton rats, mice, guinea pigs, chinchillas, and Syrian hamsters with hRSV strains Long and/or A2 have been well characterized, although clinical isolates have also been examined. Ferrets are also susceptible to hRSV infection, but the pathogenesis and immune responses elicited following infection have not been well characterized. Here, we describe the infection of adult ferrets with hRSV Long or A2 via the intranasal route and characterized virus replication, as well as cytokine induction, in the upper and lower airways. Virus replication and cytokine induction during the acute phase of infection (days 0 to 15 postinfection) were similar between the two strains, and both elicited high levels of F glycoprotein-specific binding and neutralizing antibodies following virus clearance (days 16 to 22 postinfection). Importantly, we demonstrate transmission from experimentally infected donor ferrets to cohoused naive recipients and have characterized virus replication and cytokine induction in the upper airways of infected contact animals. Together, these studies provide a direct comparison of the pathogenesis of hRSV Long and A2 in ferrets and highlight the potential of this animal model to study serological responses and examine interventions that limit transmission of hRSV.
IMPORTANCE Ferrets have been widely used to study pathogenesis, immunity, and transmission following human influenza virus infections; however, far less is known regarding the utility of the ferret model to study hRSV infections. Following intranasal infection of adult ferrets with the well-characterized Long or A2 strain of hRSV, we report virus replication and cytokine induction in the upper and lower airways, as well as the development of virus-specific humoral responses. Importantly, we demonstrate transmission of hRSV from experimentally infected donor ferrets to cohoused naive recipients. Together, these findings significantly enhance our understanding of the utility of the ferret as a small-animal model to investigate aspects of hRSV pathogenesis and immunity.
KEYWORDS: animal models, ferret, respiratory syncytial virus, respiratory viruses, transmission
INTRODUCTION
Human respiratory syncytial virus (hRSV) belongs to the family Pneumoviridae and is an enveloped virus with a negative-sense single-stranded RNA genome (1). hRSV is the most common cause of bronchiolitis and pneumonia in young children (2, 3) and a significant cause of morbidity and mortality in both elderly and immunocompromised individuals (4). In older children and healthy adults, hRSV can cause mild cold-like symptoms that resolve without major complications or specific treatment.
There are currently no licensed hRSV vaccines or specific antiviral therapies, although monoclonal antibody (MAb) prophylaxis is effective in reducing hRSV-associated hospitalizations in infants at risk of severe disease (5). While a number of hRSV vaccine candidates have progressed to clinical trials (6), the recent failure of the only vaccine to complete phase III highlights the challenge of providing protective efficacy in humans (7). Animal models of hRSV are critical for preclinical testing; however, these often fail to recapitulate particular aspects of human disease. Animal models used to study hRSV include nonhuman primates and chimpanzees as well as small-animal models, including cotton rats, ferrets, chinchillas, hamsters, guinea pigs, and mice (reviewed in references 6 and 8). Most small-animal models tend to be semipermissive for hRSV infection, requiring large inoculum doses for experimental infection and accompanied by little or no clinical sign of disease. Despite these limitations, mice and cotton rats in particular have been utilized to provide important insights regarding pathogenesis and immunity to hRSV and were instrumental in the development of some treatments (e.g., development of MAb prophylaxis for infants at high risk of severe hRSV infection [9, 10]).
Ferrets have been widely used to study human influenza virus infection. Ferrets are susceptible to human influenza viruses without requiring prior adaptation of the viruses, develop clinical signs similar to those in human infections (such as fever, sneezing, and lethargy), and possess a respiratory tract physiology similar to that of humans, making them an ideal model for human influenza (11). Infected ferrets can also transmit human influenza viruses to other naive ferrets through direct contact or by the airborne route. Currently, far less is known regarding the utility of the ferret model to study aspects of pathogenesis and immunity to hRSV infections. In this regard, intranasal (i.n.) inoculation of neonatal or adult ferrets with hRSV was shown to achieve high levels of virus replication in nasal tissues, although replication in the lungs was only observed in infant ferrets (12–14). Recent studies utilizing intratracheal (i.t.) inoculation with a low-passage-number clinical isolate of hRSV demonstrated virus replication in the upper (nasal turbinates and trachea) and lower (bronchus and lungs) airways, with hRSV antigens localized to epithelial cells in the trachea and bronchi (15). Higher titers and prolonged shedding of hRSV was demonstrated following i.t. inoculation of ferrets that were immunosuppressed with a combination of drugs used in transplantation (15).
The majority of studies in different animal models have used hRSV Long and/or A2, as well as a range of clinical isolates. Since their derivation from the lower respiratory tract of infants over 50 years ago (Long, 1956, Maryland, USA; A2, 1961, Melbourne, Australia), hRSV strains Long and A2 have been propagated in cell culture and used extensively in in vitro and in vivo studies. Therefore, the pathogenesis and immune responses elicited by experimental infection with Long and A2 can be compared between different animal models. Here, we infected adult ferrets by the i.n. route with an equivalent infectious dose of hRSV strain Long or A2 and assessed virus shedding in the upper respiratory tract (URT) using nasal wash (NW) samples and in the lower respiratory tract (LRT). In addition, we report induction of a range of cytokines, chemokines, and other inflammatory mediators in the URT and LRT of hRSV-infected ferrets, as well as the induction of serum antibodies, including neutralizing antibodies. Finally, we provide evidence of contact transmission from experimentally infected donor ferrets to cohoused naive recipients for both strains of hRSV.
RESULTS
Characterizing experimental i.n. infection of ferrets with Long and A2 strains of hRSV.
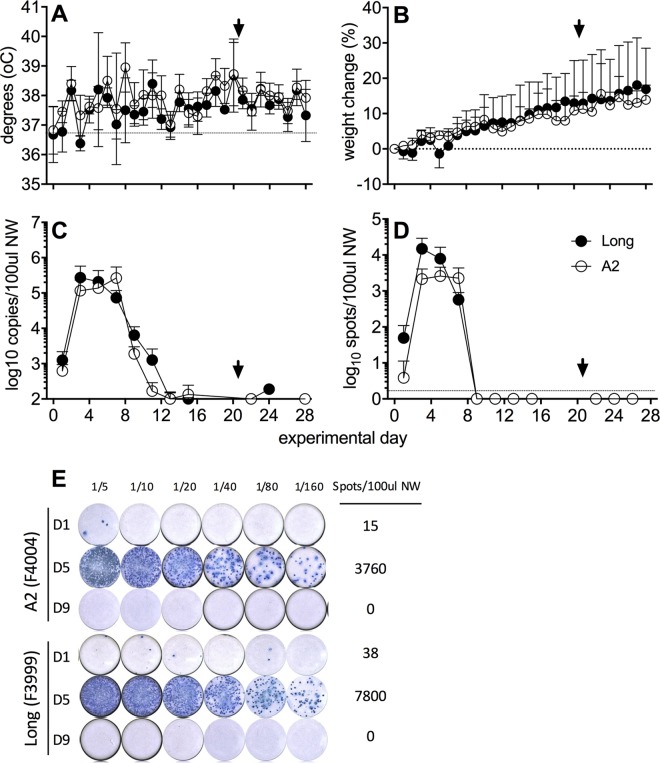
Ferrets (n = 4/group) were infected by the i.n. route with 105 PFU of hRSV strain Long or A2 and clinical disease and virus shedding were assessed at various times postinfection (p.i.). On day 21 p.i., ferrets were rechallenged via the i.n. route with an identical dose of homologous virus and monitored for an additional 7 days. A modest increase in temperature was observed 2 days after primary infection but no other trends were apparent (Fig. 1A). Ferrets maintained or increased weight over the 28-day monitoring period (Fig. 1B) and clinical signs were minimal, consistent with previous studies (14).
FIG 1.
Viral replication in the upper respiratory tract following experimental i.n. infection with hRSV. Ferrets (n = 4 per group) were infected i.n. with hRSV Long (closed circles) or A2 (open circles) virus, and NW samples were collected every second day. Temperature (A) and weight (B) were measured daily. (A and B) Means ± standard deviations from 4 ferrets per group are shown. (C) Viral shedding was quantified by real-time qPCR detection of the hRSV nucleoprotein (N) gene. (E) A ViroSpot (VS) assay was developed to measure titers of infectious virus in NW. Representative VS images of NW samples from two ferrets are shown at days 1, 5, and 9 p.i. (D) Titers of infectious virus in NW samples as determined by VS assay. Means ± standard deviations from 4 ferrets per group are shown in panels A and C. An arrowhead indicates reinfection with homologous virus strain. Statistical significance for virus shedding between viruses was analyzed by multiple t test with a Bonferroni-Dunn correction (alpha of 0.05), without assuming a consistent standard deviation. No significant differences were observed.
Following primary infection, low levels of viral RNA (vRNA) were detected in NW samples at day 1 p.i., and shedding peaked at day 3 and day 7 for animals infected with Long or A2, respectively (Fig. 1C). vRNA was detected for up to 11 days in some animals. We developed a ViroSpot (VS) assay to measure titers of infectious hRSV (see Materials and Methods) and confirmed that its sensitivity was similar to that of plaque assay (data not shown). An example of a VS assay measuring titers of infectious virus in NW from hRSV-infected ferrets is shown (Fig. 1E). Similar to vRNA, low levels of infectious virus were detected at day 1 p.i., and shedding peaked on day 3 in animals infected with the Long strain and at days 3 to 7 in animals infected with the A2 strain. Infectious virus was cleared by day 9 (Fig. 1D). Statistical analyses showed no significant differences between shedding of vRNA or infectious virus between the two virus strains on any of the days tested (data not shown). Note that the persistence of vRNA shedding in the absence of infectious virus is also seen in the cotton rat model (16). Throat swabs were measured alongside NW samples for a subset of ferrets, with equivalent results (data not shown); thus, NW sampling was used for the remaining experiments.
Induction of inflammatory mediators in the upper respiratory tract following experimental i.n. infection of ferrets with hRSV.
We next used real-time quantitative PCR (qPCR) to determine the expression of a range of inflammatory mediators in NW samples collected at various times after direct i.n. infection of ferrets with the Long or A2 strain of hRSV. Expression of interleukin-1α (IL-1α) and IL-1β mRNAs peaked on days 1 to 5, beta interferon (IFN-β) peaked on days 3 to 5, and IFN-α mRNA levels peaked on day 7 (Fig. 2B to E). Increased expression of these proinflammatory cytokines coincided with maximum levels of hRSV N mRNA (Fig. 2A) on days 5 to 7 for hRSV Long and days 5 to 9 for hRSV A2. Expression of IFN-α and IFN-β mRNAs correlated with expression of hRSV N A2 mRNA (IFN-α, r = 0.6395, P = 0.0057; IFN-β, r = 0.6447, P = 0.0052; Pearson correlation), while expression of IFN-β mRNA strongly correlated with hRSV Long N mRNA (r = 0.6466, P < 0.0001; Pearson correlation). Expression of granzymes A and B peaked from days 9 to 11 as viral RNA was clearing (Fig. 2F to H). Compared to uninfected controls, infection of ferrets with either Long or A2 induced a range of additional chemokines and cytokines, including mRNAs for IL-6, IL-8, IL-10, IL-12p40, IL-17, monocyte chemoattractant protein 1 (MCP-1), and tumor necrosis factor alpha (TNF-α), although no clear peaks in expression were observed (Fig. 2I to O). Compared to levels in A2-infected ferrets, slightly higher levels of mRNA for some genes were observed in animals infected with hRSV Long (e.g., IL-1α, IFN-α, IL-10, and IL-12p40), but these differences generally were not significant, although levels of IFN-α and IFN-β mRNA were significantly higher on day 9 in A2-infected ferrets.
FIG 2.
Expression of inflammatory mediator genes in nasal wash samples from ferrets after i.n. infection with hRSV. Ferrets were infected via the i.n. route with hRSV Long or A2 virus. NW samples collected at various times p.i. were assayed for mRNA for the indicated genes using real-time qPCR assays. Reference samples from uninfected ferrets are indicated at day 0, with a fold change of 1. Data are from a single experiment (n = 4 animals/group). Statistical significance for cytokine expression between viruses was analyzed by a multiple t test with a Bonferroni-Dunn correction (alpha of 0.05) without assuming a consistent standard deviation (*, P < 0.05; **, P < 0.01, ***, P < 0.001).
hRSV can replicate throughout the respiratory tract following experimental i.n. infection of ferrets.
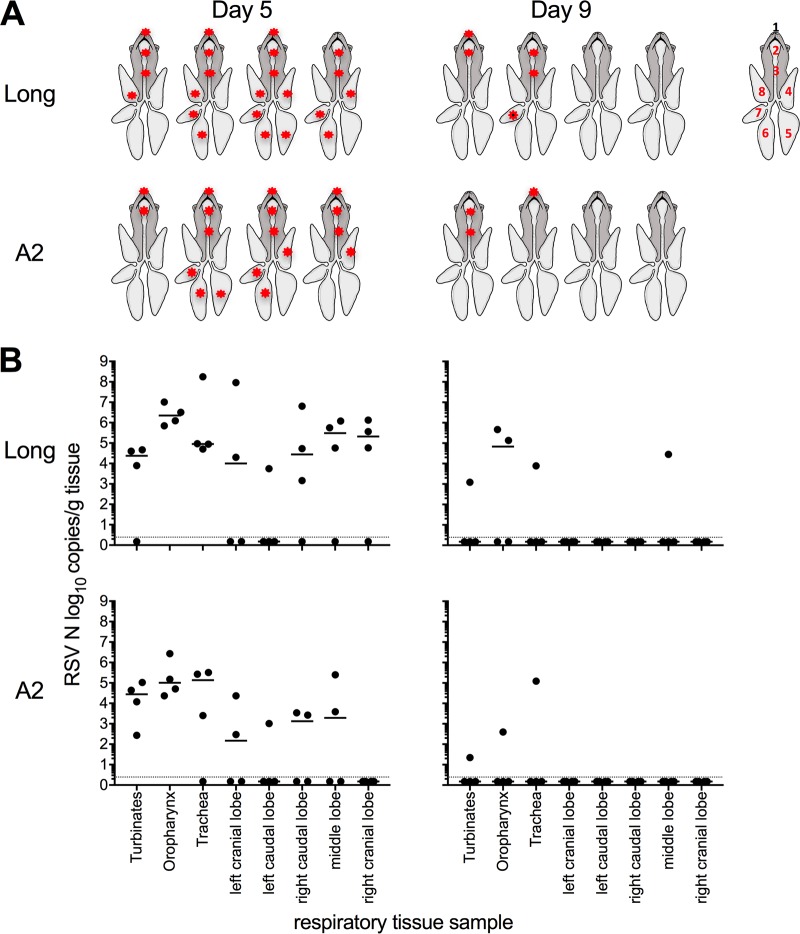
We assessed whether hRSV Long and A2 replicated throughout the respiratory tract of ferrets following i.n. infection. As hRSV was detected at high levels on days 3 to 7 in the NW of i.n.-infected animals (Fig. 1), we hypothesized that day 5 p.i. represented an appropriate time to sample for virus replication throughout the airways. We included a later time point (day 9) to assess if replication kinetics were delayed in the lower airways and/or virus had been cleared at this time point. At day 5 p.i., hRSV was detected throughout the upper and lower respiratory tracts of infected ferrets. Both Long and A2 strains were detected in the turbinates, oropharynx, and trachea from the majority of animals (Fig. 3A). hRSV Long was detected in the lungs of all infected animals, including in all 5 lobes of one ferret, in 3 lobes of two ferrets, and in 1 lobe of the remaining ferret (Fig. 3A). Infection was more restricted in animals infected with A2 virus, with virus detected in lung tissues of 3/4 infected ferrets. Virus was detected in 3 lobes in two ferrets and in 1 lobe of the third ferret (Fig. 3A). Note that viral titers were generally similar between the two strains and when comparing individual tissues (Fig. 3B), and no statistical differences were observed.
FIG 3.
hRSV strains Long and A2 can replicate throughout the respiratory tract of ferrets after i.n. inoculation. Ferrets were infected via the i.n. route with hRSV Long or A2 virus and tissues collected on day 5 or 9 p.i. Levels of vRNA for the viral N gene were determined in tissue homogenates by real-time quantitative RT-PCR assay. (A) Diagrammatic representation of virus spread (indicated by an asterisk) throughout the respiratory tract: 1, turbinates; 2, oropharynx; 3, trachea; 4, left cranial lobe; 5, left caudal lobe; 6, right caudal lobe; 7, middle lobe; 8, right cranial lobe. Figure adapted from reference 37. (B) Virus load in different tissues from the respiratory tract of ferrets. Results for individual animals are shown (n = 4/group), and the horizontal line represents the median value. The dotted line indicates the limit of detection. Titers for each tissue were compared between virus strains using the Mann-Whitney U test.
At day 9 p.i., virus was detected in respiratory tissues from two animals infected with Long and two animals infected with A2. Long virus was detected in URT tissues (turbinates and oropharynx) from two animals and in the lung of one animal (middle lobe), while A2 virus was detected only in the URT of two animals (Fig. 3A and B). In future experiments, it would be useful to also determine titers of infectious virus to correlate levels of vRNA with titers of infectious virus in the lungs of RSV-infected ferrets.
Induction of inflammatory mediators in the lungs of ferrets following experimental i.n. infection with hRSV.
We next assessed expression of inflammatory mediators in the lower airways. For these analyses, we assessed cytokine levels in individual lung lobes obtained from (i) uninfected animals or (ii) animals at day 5 p.i. with hRSV. Note that data from A2/Long were combined and that virus-positive and virus-negative lobes (as seen in Fig. 3A) were assessed independently. First, we confirmed that mRNA for IFN-α, IL-1α, IL-1β, granzyme A, granzyme B, IFN-γ, IL-10, and IL-17 were significantly higher in virus-positive lobes than in lobes from uninfected animals (Fig. 4, *). In contrast, no significant differences were noted in levels of IFN-β, IL-6, IL-12p40, MCP-1, TNF-α, or IL-8 (data not shown). Of note, IFN-α, granzyme A, granzyme B, and IL-17 were also significantly upregulated in virus-negative lobes from infected animals compared to uninfected animals, whereas IL-1α, IL-1β, IFN-γ, and IL-10 were not (Fig. 4, ÷). Levels of IL-1α and IL-1β were significantly higher in virus-positive lobes than virus-negative lobes from infected animals, suggesting that localized virus replication is important for expression of these cytokines (Fig. 4, ±). Note that independent analysis of virus-positive lobes from A2- or Long-infected animals showed no significant differences between virus strains in levels of IFN-α, IL-1α, IL-1β, granzyme A, granzyme B, IFN-γ, IL-10, and IL-17 (data not shown).
FIG 4.
Detection of inflammatory mediators in the lung after hRSV infection. Ferrets were infected via the i.n. route with hRSV Long or A2 virus. Lung lobes were collected on day 5 after infection and assayed for mRNA for the indicated genes by real-time qPCR. Note that data from A2/Long were combined and samples were classified as virus positive or virus negative based on vRNA results described in Fig. 3. Circles represent individual lung lobes from up to 4 ferrets, and the horizontal line represents the mean value for each mediator. For each graph, qPCR data first were expressed relative to values for lobes from uninfected animals and then normalized to ATF4, HPRT, and GAPDH housekeeping genes. Values for virus-positive and virus-negative lobes from hRSV-infected animals then were expressed as fold change relative to the corresponding lobes from uninfected animals. For statistical analyses, inflammatory mediators were compared between virus-positive lobes and lobes from uninfected animals (*, P < 0.05; **, P < 0.01, ***, P < 0.001; ****, P < 0.0001), between virus-negative lobes from infected animals and lobes from uninfected animals (÷, P < 0.05; ÷÷, P < 0.01; ÷÷÷, P < 0.001; ÷÷÷÷, P < 0.0001), and between virus-positive and virus-negative lobes from infected animals (±, P < 0.05; ±±, P < 0.01; ±±±, P < 0.001; ±±±, P < 0.0001). Note that the data did not show a normal distribution; therefore, ROUT could not be used to remove outliers. Instead, Kruskal-Wallis one-way analysis of variance (ANOVA) with Dunn's multiple-comparison test was used.
Characterization of humoral responses elicited in ferrets following experimental i.n. infection and rechallenge with hRSV.
We developed two assays to characterize humoral responses elicited in ferrets following i.n. infection with hRSV Long or A2 (as seen in Fig. 1 and 2). First, an enzyme-linked immunosorbent assay (ELISA) was used to determine titers of serum antibodies capable of binding the recombinant F glycoprotein of hRSV. Antibodies were measured by ELISA at baseline (i.e., day 0 relative to infection) and at day 16 or 21 p.i. in 2 independent experiments. Titers of F glycoprotein-specific antibodies were low at day 0 but increased markedly by days 16 and 21 (Fig. 5A). When expressed as a fold change relative to the level on day 0 (Fig. 5B), titers had increased 1,000- to 10,000-fold by day 21 p.i. While there was a tendency for the fold changes to be higher in sera obtained at day 21 than at day 16, this was not significant for either virus strain. Together, these data demonstrate that experimental i.n. infection with either A2 or Long strain induces high levels of F glycoprotein-specific antibodies by day 21 p.i.
FIG 5.
Humoral responses elicited following hRSV infection of ferrets by i.n. inoculation. Sera were collected from animals immediately prior to i.n. infection (day 0) or on day 16 (experiment 1) or 21 (experiment 2) after infection and assayed by ELISA (to detect antibodies capable of binding hRSV F glycoprotein) (A and B) or by ViroSpot microneutralization (VS MN) assay for neutralizing antibodies (C, D, and E). (C) A VS MN assay was developed to measure titers of neutralizing antibodies in ferret serum. Representative images of a VS MN assay using day 0 and day 21 serum samples from a single ferret are shown, including a cell control (CC; 1/4 are shown) and virus control (VC; 1/4 are shown). For panels A, B, D, and E, results from individual animals infected with hRSV Long (closed circles) or A2 (open circles) are shown. For panels A and D, individual titers are shown and the horizontal line represents the GMT. For panels B and E, fold change compared to day 0 are shown and the horizontal line indicates median value.
We next developed a microneutralization (MN) assay based on the VS assay used to measure infectious virus (Fig. 1B). First, we used the MN assay to assess the neutralizing activity of control samples from BEI Resources against A2 and Long strains of RSV. We obtained MN titers for positive-control (NR-21973 immune globulin; A2 titer, 2,560; Long titer, 5,120) and negative-control (NR-49447 human serum IgG depleted; A2 titer, <20; Long titer, <20) samples. We next examined MN titers in ferrets at different times after hRSV infection. A representative example of the MN assay with results using serum from a single ferret on days 0 and 21 is shown (Fig. 5C). This MN assay was used to determine titers of neutralizing antibodies against the homologous challenge strain of hRSV.
While MN titers (geometric mean titers [GMT] with 95% confidence intervals [95% CI]) were generally low at day 0, they were significantly higher for the Long strain than for the A2 strain (GMT [95% CI] for Long, 25.9 [19.2, 35.0]; A2, 19.3 [18.9, 19.6]; P = 0.0033 by Mann-Whitney test) (Fig. 5D). Low-level neutralizing activity observed in naive sera from some animals may reflect the presence of serum inhibitors (rather than antibodies), as reported for influenza and other viruses (17). Despite this, neutralizing antibodies were induced following experimental infection of ferrets, with titers to the Long strain reaching similar high levels at 16 or 21 days p.i. (GMT [95% CI] for day 16, 452.5 [45.6, 4,496]; day 21, 640 [134.5, 3,045]) (Fig. 5D), while titers to the A2 strain were lower on day 16 than day 21 p.i. (GMT [95% CI] for day 16, 223.4 [13.4, 3,724]; day 21, 1,522 [876.9, 2,642]) (Fig. 5D). This difference in kinetics was also reflected in the fold change over time for each ferret, with a median fold change of 24 at days 16 and 21 for ferrets infected with the Long strain and a median fold change of 32 at day 16 and 128 at day 21 for ferrets infected with the A2 strain (Fig. 5E). Of note, one ferret infected with A2 did not seroconvert (Fig. 5D and E).
Given there were no major differences between antibody responses elicited following experimental i.n. infection with either hRSV Long or A2, data were pooled for further analyses. First, we noted a high level of correlation between the titers for the ELISA and VS MN assay (r = 0.9464, P < 0.001; data not shown). We next assessed the relationship(s) between the antibody titer achieved after infection and the kinetics of virus shedding for ferrets infected by the i.n. route with hRSV. No significant correlations were observed between (i) the peak virus load or (ii) the number of days of shedding >104 copies/100 μl NW and titers of F glycoprotein-specific or neutralizing antibodies (data not shown). Note that the longer the time from the start of virus shedding until the collection of serum, the higher the titer of F glycoprotein-specific (ELISA) or neutralizing (VS MN) antibodies achieved (r = 0.5639 and P = 0.0229 for ELISA and r = 0.6049 and P = 0.0131 for VS MN; data not shown).
We rechallenged some of the ferrets to determine if the level of antibodies achieved after primary infection could be boosted. Two ferrets were rechallenged with Long virus and two others were rechallenged with A2 virus 21 days after a primary infection. Levels of vRNA were very low, none of the ferrets shed detectable infectious virus after rechallenge, and no clinical signs were detected (Fig. 1A to D, arrowheads). There was no change in antibody titers from the time of rechallenge to 7 days after rechallenge, when animals were killed (GMT [95% CI] at day 21 for Long of 1,280 [0] and A2 of 2,560 [0]; GMT [95% CI] at day 28 for Long of 1,280 [0] and A2 of 2,560 [0]) (data not shown). These data suggest that neutralizing antibody titers of ≥1,280 are sterilizing following i.n. infection with hRSV in the ferret.
Transmission between cohoused ferrets following experimental i.n. infection of donor ferrets with A2 or Long strain of hRSV.
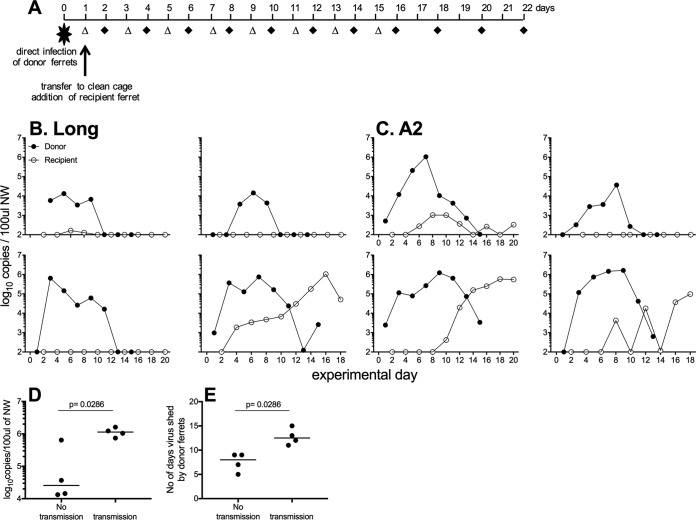
We next investigated the ability of hRSV to be transmitted between cohoused ferrets. Naive ferrets (recipients) were cohoused for up to 3 weeks, with ferrets inoculated 1 day previously via the i.n. route with hRSV (donor) (Fig. 6A), and virus shedding was assessed in all animals. hRSV Long was shed by all infected donor ferrets for at least 5 days yet transmitted to only one of the four recipient ferrets. Shedding of the virus in the recipient ferret was detected 3 days after the animals were cohoused (Fig. 6B). hRSV A2 was shed for at least 9 days by all of the donor ferrets and was transmitted to 3 of 4 recipient ferrets. Shedding of the virus in the recipient ferrets was detected from 5 to 9 days after the animals were cohoused (Fig. 6C). Overall, transmission occurred when donor animals shed higher peak amounts of virus (median peak shedding of virus for donors that transmitted virus was 106.1 copies/100 μl NW compared to 104.4 copies/100 μl NW for donors that did not transmit virus; P = 0.0286) (Fig. 6D) and shed virus for a longer period of time (median shedding of 12.5 days compared to 8 days; P = 0.0286) (Fig. 6E). No recipient animals that were infected and shed hRSV displayed weight loss or clinical signs (data not shown).
FIG 6.
hRSV can transmit between cohoused ferrets. (A) Experimental plan. Donor ferrets were infected via the i.n. route with hRSV Long (B) or A2 (C) virus. One day later, donor animals were placed in clean cages and a naive recipient ferret was added. NW were collected on alternate days (donor, open triangles; recipients, black diamonds). (B and C) Virus shedding was measured in NW samples by real-time qPCR assay detecting vRNA. (D and E) The peak of virus shedding (D) and the number of days that detectable virus was shed in NW (E) was compared for donor ferrets that did or did not transmit virus to naive contacts. Horizontal bars represent median values, and significance was determined by Mann-Whitney test (P values are shown). Data are pooled from two independent experiments, with 4 donors and 4 recipients for each virus.
Inflammatory mediators were also detected in NW from recipient ferrets that were cohoused with infected donor ferrets and shed detectable levels of hRSV vRNA (Fig. 7). For these analyses, data were pooled from A2 (3/4) and Long (1/4) recipient ferrets. Note that samples were normalized to the first day that the recipient ferret shed detectable levels of virus in NW (i.e., irrespective of one animal first shedding detectable vRNA on experimental day 3 or 5, these are both adjusted to day 1 p.i. for analysis). Inflammatory mediators induced following transmission of hRSV to cohoused recipients were generally lower than those detected in donor ferrets (Fig. 2). Trends in cytokine induction were not as distinct as those for donor ferrets; however, induction of a number of inflammatory mediators (e.g., IL-1α, IL-1β, ΙL6, IL-8, MCP-1, and TNF-α [Fig. 7]) showed an overall pattern similar to levels of vRNA (Fig. 7A). In contrast to data from ferrets experimentally infected via the i.n. route with hRSV (Fig. 2F and G), distinct peaks in granzymes A and B were not observed at days 11 to 13 p.i. in recipient ferrets (Fig. 7F and G).
FIG 7.
Expression of inflammatory mediator genes in nasal wash samples from ferrets after hRSV infection by contact transmission. Naive recipients were cohoused with donor ferrets that had been infected via the i.n. route with hRSV A2 or Long virus. Only recipient animals that shed detectable hRSV were examined, and data have been adjusted such that the first day that vRNA was detected in NW has been designated day 1 p.i. Real-time qPCR assays were used to assess levels of different inflammatory mediators using mRNA derived from NW samples. Data are from two independent experiments (n = 4 animals/group).
Note that while intranasal infection of donor ferrets with either hRSV strain induced high levels of F glycoprotein-specific (Fig. 5A and B) and neutralizing antibodies (Fig. 5D and E), animals were not kept alive long enough to confirm seroconversion in recipients.
DISCUSSION
A recent study using a low-passage-number hRSV A clinical isolate to inoculate ferrets via i.n./i.t. routes (105 50% tissue culture infectious doses [TCID50]/animal) resulted in virus replication in the upper airways as well as in the trachea and lungs. Virus titers were elevated and shedding was prolonged in hRSV-infected ferrets that were treated with a combination of immunosuppressive compounds used in transplant recipients (15). Our study focusing on the well-characterized A2 and Long laboratory-adapted strains provides a number of important advances in our understanding of the ferret model of hRSV infection. First, we confirm that after i.n. inoculation, both strains replicate efficiently in the URT of ferrets, and sporadic evidence of virus replication can also be detected in the lungs. Second, we demonstrate that i.n. infection of ferrets induces a range of cytokines and chemokines in both the upper and lower airways. Third, we describe the induction of similar levels of F glycoprotein-specific and neutralizing antibodies following i.n. infection of ferrets with either hRSV strain. Finally, we demonstrate for the first time that donor ferrets experimentally infected with hRSV can transmit virus to cohoused naive recipient animals. Of interest, the A2 strain transmitted more efficiently between cohoused ferrets (3/4) than the Long strain (1/4). Together, these findings significantly enhance our understanding of the utility of the ferret as a small-animal model to investigate aspects of hRSV pathogenesis and immunity.
Currently, there are not enough data to determine if specific hRSV strains induce differential disease, immunity, or transmissibility in the ferret model. Of note, variable disease and differential immune responses have been noted following i.n. infection of mice with laboratory strains (e.g., A2 and Long), as well as with different clinical isolates (18, 19). Our data examining A2/Long virus load in the URT show titers and kinetics similar to those recently reported following i.n. inoculation of immunocompetent ferrets with a low-passage-number clinical isolate (subgroup A). However, no direct comparison can be made for LRT viral replication, as the previous study only examined replication within the lung after i.t., not i.n., inoculation (15). Of note, previous studies using i.n. inoculation to inoculate ferrets with the Long strain reported virus replication in the URT of both adult and infant ferrets but only in the lungs of infant animals (14). The LRT replication of i.n.-delivered hRSV that we observed in adult ferrets may be a consequence of differences in inoculum volume between the studies, as we used 0.5 ml compared to a maximum of 0.2 ml in the previous work (14). Consistent with this, larger inoculum volumes were associated with more uniform replication of influenza viruses in the LRT of ferrets (20). Overall, our data using the well-characterized laboratory strains A2 and Long provide a baseline for comparing pathogenesis, immunity, and transmission of different hRSV strains, including recent clinical isolates, in the ferret model.
We report for the first time the induction of inflammatory mediators in the upper and lower airways of healthy adult ferrets following i.n. infection with hRSV. Note that the overall patterns and magnitude of cytokine/chemokine expressed in the URT was very similar for ferrets infected with either the A2 or Long strain of virus. Our previous studies showed that particular immune mediators (e.g., IL-6, IFN-α, IFN-β, MCP-1, IFN-γ, granzyme A, and granzyme B) showed clear peaks of induction in the URT of ferrets following i.n. infection of ferrets with type A (A/H1pdm, A/H3) or B (B/Yam) human influenza viruses, while others (e.g., IL-12p40, IL-10, IL-17, and IL-8) were induced but showed no clear peak (11). Moreover, only influenza A/H1pdm replicated in the LRT of infected ferrets, inducing IL-6, IFN-α, IFN-β, MCP-1, IFN-γ, granzyme A, and granzyme B. The inability of influenza A/H3 and type B viruses to replicate in the lung correlated with a lack of detectable cytokines and/or chemokines at this site (11). Compared to influenza infections, the patterns of chemokine and cytokine induction in the URT following hRSV infection were not as clear, although the induction of IL-1α, IL-1β, IFN-α, and IFN-β mRNAs generally correlated with the kinetics of viral replication in the URT. As for influenza virus, some mediators (e.g., granzyme A and B and IFN-γ) were expressed in the URT at times that correlated with clearance of hRSV from this site. As the hRSV inoculum was prepared in Hep2 cells, it may contain inflammatory mediators that also could contribute to the induction of chemokines and cytokines in experimentally infected animals. Moreover, this could be an additional factor contributing to the different patterns of inflammatory mediators induced in animals infected experimentally or following cohousing with infected donor ferrets. In future studies, we will examine the ability of UV-inactivated hRSV to induce inflammatory mediators following experimental infection of ferrets.
In the LRT, hRSV replication tended to be more sporadic, with vRNA often detected in some, but not all, lung lobes of infected ferrets at day 5 p.i. Consistent with this, a range of cytokines and chemokines could be detected in the lung, generally in lung lobes from infected ferrets where vRNA was also detected. Like ferrets, the utility of the cotton rat model is restricted by the limited pool of immunological reagents available compared with those available for mice. However, qPCR demonstrated that following i.n. inoculation of cotton rats, the Long strain rapidly induced inflammatory mediators in the LRT (e.g., IFN-α, IFN-γ, IL-1β, IL-6, IP-10, MIP-1β, RANTES, TNF-α, and GRO [the equivalent of human IL-8]), and their peak and decline tended to correlate with virus peak (days 3 to 5 p.i.) and clearance (day 10 p.i.), respectively (13, 14).
Cotton rats have become a standard model for the evaluation of potential vaccines and antivirals for hRSV, as well as for preclinical testing for anti-hRSV therapeutics. Experimental infection of cotton rats with hRSV results in virus replication in the upper and lower respiratory tract and induction of high titers of serum-neutralizing antibodies, although infected animals do not tend to develop clinical signs of disease (8, 21). Of interest, i.n. inoculation of ferrets with either Long or A2 induced high titers of antibodies (F glycoprotein specific by ELISA or neutralizing by VS MN assay) by day 21 p.i. These findings were surprising, given that a previous study reported neutralizing antibody titers to be weak or absent in 0-, 3-, 7-, 14-, or 28-day-old ferrets (with serum taken at various time points, including 7, 14, 21, and 28 days p.i.) following i.n. infection with the Long strain at an inoculum dose sufficient to induce high titers (>105 PFU/g) of virus replication in the URT of all age groups and the LRT of infant ferrets (14). In fact, in our study the titers of neutralizing antibodies induced in adult ferrets were similar to those induced following i.n. infection of cotton rats with the Long strain (22). While the potential of the ferret model to assess the effectiveness of potential vaccines and antiviral agents has not been well explored, intranasal delivery of adenovirus-vectored vaccines expressing hRSV F and/or G glycoproteins was reported to induce neutralizing antibody responses and provided protection following challenge with hRSV A2 in a dose-dependent manner (13).
In small-animal models of hRSV infection, inoculum doses used for experimental infections generally (i) are in the range of 104 to 108 PFU/animal, (ii) are delivered by the i.n. or i.t. route, and (iii) result in moderate to high levels of virus replication in the upper airways and moderate to low levels in the LRT (reviewed in reference 5). Our results using 105 PFU/ferret of hRSV Long or A2 for experimental i.n. infection are consistent with earlier studies. Furthermore, we report that hRSV, in particular the A2 strain, transmitted to naive cohoused ferrets (transmission to 3/4 and 1/4 recipients for A2 and Long, respectively). In previous studies, 1/7 mothers of i.n.-infected infant ferrets developed antibodies to hRSV (14), suggesting that transmission occurs by close contact. Our data extend these observations and provide clear evidence of transmission between cohoused adult ferrets, allowing us to assess viral kinetics in contact animals following infection acquired from a donor ferret with an inoculum dose and route of infection that are likely to be more representative of natural hRSV infections in humans (23). Note that viral titers in the URT of some recipient animals reached peak levels similar to those in experimentally infected donor animals, implying that a high dose of virus inoculum is not necessarily required to achieve efficient replication in the upper airways of ferrets. In future studies, it will be of interest to determine if transmission of hRSV to naive cohoused recipients results in any virus replication in the LRT, as seen following experimental i.n. infection with Long or A2. While animal-to-animal transmission of hRSV (originally termed chimpanzee coryza agent) has been reported in chimpanzees (24), our findings highlight the potential of ferrets as a small-animal model to explore the ability of vaccines and antivirals to block hRSV transmission.
Further refinement and investigation of hRSV transmission in ferrets is required, including investigation of noncontact (or aerosol) transmission, as described for influenza (reviewed in references 11 and 25), as well as investigating contact and noncontact transmission by hRSV isolates. While experimental i.n. infection was associated with high levels of anti-RSV antibodies by day 21 p.i., it will be important to examine a number of later time points to understand the timing of the peak and the decay of anti-RSV antibodies in experimentally infected animals. Moreover, contact ferrets showed little evidence of seroconversion by day 18 to 22 p.i. despite shedding peak titers of vRNA similar to those of experimentally infected animals, possibly because the interval between contact infection and serum sampling was too short. Further studies are required to more thoroughly investigate the timing and kinetics of anti-RSV antibody induction in recipient animals infected with hRSV. Overall, the ferret model of hRSV infection and transmission has the potential to offer significant advantages over other small-animal models, in particular in assessing novel therapeutic (e.g., treatment of infected donors) or prophylactic (e.g., treatment of naive recipients prior to introduction of infected donor animals) interventions against hRSV, as well as the potential for novel vaccines to limit transmission to vaccinated recipient animals.
MATERIALS AND METHODS
Ferrets.
Ferrets (mean weight, 986 g; range, 780 to 1,220 g) were housed in the Bioresources Facility at the Peter Doherty Institute for Infection. Experiments were conducted with approval from the University of Melbourne Biochemistry & Molecular Biology, Dental Science, Medicine, Microbiology & Immunology, and Surgery Animal Ethics Committee, in accordance with the NHMRC Australian code of practice for the care and use of animals for scientific purposes.
Cells and viruses.
Human epithelial type 2 (HEp-2) cells, a derivative of HeLa cells (26), were passaged in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% (vol/vol) fetal calf serum (FCS) (Bovogen Biologicals Pty Ltd.), 2 mM l-glutamine (GlutaMAX; Gibco), nonessential amino acids (Gibco), 0.55% (vol/vol) sodium bicarbonate (Gibco), 20 mM HEPES (Gibco), 200 U/ml penicillin (Gibco), and 200 μg/ml streptomycin (Gibco). The Long strain of hRSV (VR-26) was purchased from the American Type Culture Collection (ATCC), and the A2 strain was obtained from the Department of Microbiology and Immunology (DMI) at the University of Melbourne.
Growth of hRSV in HEp-2 cells.
HEp-2 cells grown to 90% confluence in tissue culture flasks (Corning) were inoculated with dilutions of hRSV (A2 or Long) and harvested when approximately 50% of cells showed syncytium formation (generally 3 to 5 days p.i.). Flasks then were frozen at −80°C and thawed rapidly at 37°C, and the cells and supernatant were collected and vortexed for 2 to 3 min. Cell debris was pelleted by centrifugation, and clarified supernatants were aliquoted, snap-frozen in liquid nitrogen, and then stored at −80°C. Virus copy number was determined by real-time reverse transcription-PCR (RT-PCR), and titers of infectious virus were quantified by plaque assay and/or ViroSpot assay as described below. Analysis of our A2 and Long virus stocks confirmed that there was no significant difference in the ratio of genome copy to infectious virus (mean ratios ± standard deviations [SD] were 36.5 ± 21.2 and 64.9 ± 56.0 for A2 and Long, respectively [P = 0.379 by Student's t test]).
Plaque assay and ViroSpot assay to determine titers of infectious hRSV.
Titers of infectious hRSV were determined by standard plaque assay on HEp-2 cells as described previously (27). Briefly, HEp-2 cells seeded into 6-well tissue culture plates were grown to 90 to 95% confluence, washed with serum-free maintenance medium (MM; serum-free DMEM supplemented with 2 mM GlutaMAX, nonessential amino acids [Gibco], 0.55% [vol/vol] sodium bicarbonate [Gibco], 20 mM HEPES [Gibco], 200 U/ml penicillin [Gibco], and 200 μg/ml streptomycin [Gibco]), and inoculated with 1 ml of virus dilutions prepared in MM. After incubation for 2 h at 37°C, virus inoculum was removed and 3 ml of overlay (MM supplemented with 2% FCS and 0.3% [wt/vol] agarose [Invitrogen]) and plates were incubated a further 7 days at 37°C. After incubation, cell monolayers were fixed in 1% (vol/vol) formalin (Sigma-Aldrich) and stained with 0.05% (wt/vol) neutral red (Sigma-Aldrich), and plaque numbers were counted to determine virus titer, expressed as PFU per milliliter.
A ViroSpot assay, modified from a similar assay developed to detect influenza virus (28, 29), was also developed to determine titers of infectious hRSV. Serial dilutions of samples containing hRSV were prepared in MM, and 100 μl was added to confluent Hep-2 cell monolayers in a 96-well plate. After incubation for 2 h at 37°C, samples were removed and replaced with 100 μl MM plus 100 μl infection medium (equal volumes of 6.4% [wt/vol] carboxymethyl cellulose [Sigma] and 2× minimal essential medium [Sigma]), and plates were incubated for 48 h at 37°C. Spots were detected using a chimeric monoclonal antibody produced in house based on the heavy and light chain variable regions of motavizumab (30) and the human backbone sequences using previously described methods (31). Chimeric human motavizumab was used at a concentration of 0.2 ng/well, followed by goat anti-human IgG (H+L) horseradish peroxidase (HRP) conjugate (Merck Millipore) diluted 1:500. After washing, 50 μl TrueBlue peroxidase substrate (SeraCare) was added to allow for color development, and the reaction was stopped with 200 μl H2O. Plates then were washed and dried, and spots were counted using a CTL-Immunospot S6 Macro analyzer with CTL Switchboard 2.6.0 (x86). Counts were used to determine infectious titers of hRSV, and results are expressed as infectious counts (IC) per milliliter or similar. When performed on 3 independent occasions, titers of infectious virus determined by plaque assay or by VS assay differed by less than 2-fold (data not shown).
Virus infection of ferrets and virus transmission.
Ferrets were anesthetized (12.5 mg/kg of body weight ketamine and 2.5 mg/kg Ilium Xylazil-20 in a 1:1 [vol/vol] mixture; Troy Laboratories) and inoculated by dropwise delivery of 105 PFU of hRSV in 0.5 ml into the nostrils. Ferrets were bled prior to primary virus infection (day 0) and at the termination of the experiment. Animals were weighed and visually inspected, and their temperature was measured daily using implanted temperature transponders fitted to identification chips (LifeChip Bio-Thermo; Digivet). For i.n. infection experiments, ferrets were housed in pairs in a HEPA-filtered isolation unit. For contact transmission experiments, the i.n.-infected (donor) ferrets were transferred into a new clean cage the day after infection and a naive (recipient) ferret added to the cage. The proportion of weight change was calculated as the percent difference from the weight at day of challenge for i.n.-infected ferrets or from the weight at day of addition to the cage for recipient ferrets.
Collection of samples from the respiratory tract of ferrets.
Nasal wash (NW), throat swab (TS), and respiratory tissue samples were collected from naive and hRSV-infected adult ferrets. NW and TS were collected under light sedation (5 mg/kg Ilium Xylazil-20; Troy Laboratories). NW were performed by delivery of 1 ml phosphate-buffered saline (PBS) supplemented with 50 U/ml penicillin, 50 μg/ml streptomycin (SAFC Biosciences), and 1% (wt/vol) bovine serum albumin (BSA; Sigma-Aldrich) (NW solution) into the nostril. Expelled liquid was collected and 4 separate aliquots prepared: 140 μl was immediately snap-frozen in liquid nitrogen and stored at −80°C for ViroSpot assay (see below), 140 μl was aliquoted into lysis buffer for vRNA extraction and quantification (see below), 200 μl was aliquoted into 1 ml RNAprotect saliva reagent (Qiagen, Germany) for cytokine and chemokine mRNA quantification (see below), and the remainder was snap-frozen and stored at −80°C. vRNA extraction was performed on the day of collection. For TS, the back of the throat and the upper palate were sampled using a sterile transport swab (Interpath Services, Copan). The tip only was placed in 1 ml NW solution and vortexed vigorously for 15 s, and the liquid was aliquoted as per NW samples. For collection of tissues, ferrets were anesthetized and then euthanized by cardiac injection of pentobarbitone. Respiratory tissues (individual lung lobes, nasal turbinates, oropharynx, and trachea) were incubated in RNAlater tissue collection RNA stabilization solution (Ambion, Life Technologies, Australia) overnight at 4°C. Following incubation, tissues were removed from solution and stored at −80°C until total RNA extraction could be performed.
Total RNA extraction.
Cells in ferret nasal wash samples treated with RNAprotect saliva reagent were pelleted and resuspended in 350 μl RLT buffer. Total RNA was extracted using the RNeasy Micro kit (Qiagen) according to the manufacturer's instructions and eluted with 14 μl RNase-free water. Total RNA was extracted from ferret respiratory tissue samples using the RNeasy Maxi kit (Qiagen) according to the manufacturer's instructions. Briefly, 3 ml RLT buffer containing 143 mM β-mercaptoethanol was added directly to the lung lobes, oropharynx, or trachea tissues in a gentleMACS M tube (Miltenyi Biotec). The sample was homogenized using the gentleMACS dissociator instrument (Miltenyi Biotec). For nasal turbinates, 5 ml RLT buffer containing 143 mM β-mercaptoethanol was added directly to the frozen tissue in a 50-ml tube and homogenized using a Polytron PT 2500E (ThermoFisher Scientific). The lysate was clarified twice by centrifugation at 3,000 × g for 10 min. RNA was extracted using the animal tissue protocol and eluted in 800 μl. RNA purity was assessed for all samples by spectrophotometry (A260/A280).
Real-time PCR assays to detect ferret cytokine and chemokine mRNA.
mRNA was isolated from ferret NW samples as previously described (32) and from lung tissues as describe above. mRNA for various ferret cytokines, chemokines, and housekeeping genes was quantified as previously described (32, 33).
Generation of DNA plasmids and RNA standards.
vRNA was isolated from cell-cultured A2 virus using the QIAamp viral RNA minikit (Qiagen) according to the manufacturer's instructions. The viral nucleoprotein (N) gene was amplified from 5 μl vRNA using the MyTaq one-step RT-PCR kit (BIOLi.n.E) with gene-specific primers F (5′CTCTTAGCAAAGTCAAGTTGAATGATACAC3′) and R (5′TCAAAGCTCTACATCATTATCTTTTGGAT3′) according to the manufacturer's instructions. The size of the product was confirmed by electrophoresis, purified using the QIAquick DNA gel extraction kit (Qiagen), and then cloned into the pCR4-Blunt-TOPO vector according to the manufacturer's instructions (Zero Blunt TOPO PCR cloning kit for sequencing, with One Shot TOP10 chemically competent Escherichia coli; Invitrogen). RNA standards were prepared from the pCR4-blunt-TOPO hRSV N plasmid using the Riboprobe in vitro transcription system (Promega). Transcribed RNA was purified using the RNeasy minikit (Qiagen) and treated with a Turbo DNA-free kit (Life Technologies) per the manufacturer's instructions. Tenfold RNA standards were prepared.
Real time RT-PCR quantification of viral load in ferret nasal wash and throat swab samples.
vRNA was extracted from NW and TS using the QIAamp viral RNA minikit (Qiagen) according to the manufacturer's instructions. Total RNA was extracted from lung tissues as described above. Four microliters of vRNA was used to amplify the hRSV N gene using the following combination of primers and probe with the SensiFAST probe Lo-ROX one-step kit (BIOLi.n.E): F(1) primer, 5′GAGTTGAAGGRA TYTTTGCAGGA3′; F(2) primer, 5′GAGTTGAAGGRATYTTTGCAGGAT3′; R(1) primer, 5′AAAACTCCCCATCTTAGCATTACTTG3′; R(2) primer, 5′CCCCCACCGTAACATC ACTT3′; probe, 6-carboxyfluorescein-5′TTATGAATGCCTATGGTKCAG3′-MGBNFQ. The RT-PCR consisted of 1 cycle of 45°C for 10 min and 95°C for 2 min and 45 cycles of 95°C for 5 s, 60°C for 30 s using the 7500 Fast real-time system (Applied Biosystems) and 7500 Fast system SDS software, version 1.4.0. The threshold was automatically set and threshold cycle (CT) determined. Samples, no-template controls, RNA standards, and positive (pCR4-blunt-TOPO hRSV N) and negative controls were included in all real-time RT-PCR assays. The detection limit was 10 copies.
ELISA to detect antibodies specific to the F glycoprotein of hRSV.
An ELISA was developed to measure titers of antibodies capable of binding to the F glycoprotein of hRSV. Prior to assay, ferret sera were pretreated with receptor-destroying enzyme II (RDE; 1:5, vol/vol; Denka Seiken Co. Ltd.) for 18 to 20 h at 37°C, and the reaction was stopped by addition of an equal volume of 1.6% (wt/vol) sodium citrate and incubation at 56°C for 30 min. Individual wells of 96-well flat-bottom plates (Nunc MaxiSorp; Invitrogen) next were coated with 50 μl of a 1-μg/ml preparation of recombinant hRSV F glycoprotein diluted in PBS. Note that a soluble recombinant F glycoprotein incorporating amino acids 1 to 106 and 144 to 511 from RSV F of laboratory strain A2 and a carboxyl-terminal trimerization domain was produced in Chinese hamster ovary cells and purified by immunoaffinity chromatography in a manner similar to that previously described for the Ebola GP ectodomain (34). Prefusion conformation was confirmed by reactivity with monoclonal antibodies D25 and MPE8 (35, 36). After overnight incubation at 4°C, plates were washed three times with 100 μl/well of PBS containing 0.05% (vol/vol) Tween (PBSTw) and then incubated with 200 μl/well blocking solution (5%, wt/vol, skim milk powder in PBSTw) at room temperature for >30 min. Serial 1/2-log dilutions of ferret sera were prepared in blocking solution, added to 96-well plates at 50 μl/well, and incubated for 60 min at 37°C. After washing, all wells received 50 μl/well of goat anti-ferret IgG (H+L) HRP conjugate (Abcam) diluted 1:10,000 and were incubated at 37°C for 60 min. Plates then were washed before addition of TMB 2-component microwell peroxidase substrate (SeraCare). The color reaction was stopped by addition of 50 μl/well of 1 M sulfuric acid before optical density was determined at a wavelength of 450 nm.
VS MN assay for hRSV.
A ViroSpot microneutralization (VS MN) assay was performed to detect neutralizing antibodies to hRSV. The assay was modified from a similar VS MN assay developed to detect neutralizing antibodies against influenza virus (28, 29). The following human serum samples were obtained from BEI Resources for testing in the MN assay: positive-control immune globulin (lot RSV-1; NR-21973) and negative-control human serum, IgG depleted (lot 63499869; NR-49447). Ferret sera were treated with RDE and then heat inactivated at 56°C for 30 min in human serum that was heat inactivated at 56°C for 30 min. Serial 2-fold dilutions of sera were prepared in MM and incubated with an equivalent volume of hRSV (diluted in MM to give 250 to 450 spots/well in virus-only control wells) for 60 min at 37°C before 100 μl/well of virus-serum mix was added to 96-well plates containing confluent HEp-2 cell monolayers. After incubation for 2 h at 37°C, the virus-serum mix was removed and replaced with 100 μl MM plus 100 μl infection medium (equal volumes of 6.4% [wt/vol] carboxymethyl cellulose [Sigma] and 2× minimal essential medium [Sigma]), and plates were incubated for 48 h at 37°C. After washing, 50 μl TrueBlue peroxidase substrate (SeraCare) was added to allow for color development, and the reaction was stopped with 200 μl H2O. The plates were washed once and resuspend in 200 μl MilliQ H2O for 30 min at room temperature, after which they were tapped dry on a paper towel and the plate left to dry upside down. Spots were detected and counted as described above for the infectious VS assay. VS MN titers were determined using the reciprocal of the highest serum dilution causing a 50% reduction in spots compared to virus-only control wells.
Statistical analysis.
Statistical analyses were conducted using Prism version 6.0g and are described in the figure legends.
ACKNOWLEDGMENTS
The Melbourne WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health. This study was supported in part by RMH Home Lottery grant GIA-035-2016.
We thank Philippa Saunders, University of Melbourne, for assistance with preparation of images in Fig. 3A.
REFERENCES
- 1.Afonso CL, Amarasinghe GK, Banyai K, Bao Y, Basler CF, Bavari S, Bejerman N, Blasdell KR, Briand FX, Briese T, Bukreyev A, Calisher CH, Chandran K, Cheng J, Clawson AN, Collins PL, Dietzgen RG, Dolnik O, Domier LL, Durrwald R, Dye JM, Easton AJ, Ebihara H, Farkas SL, Freitas-Astua J, Formenty P, Fouchier RA, Fu Y, Ghedin E, Goodin MM, Hewson R, Horie M, Hyndman TH, Jiang D, Kitajima EW, Kobinger GP, Kondo H, Kurath G, Lamb RA, Lenardon S, Leroy EM, Li CX, Lin XD, Liu L, Longdon B, Marton S, Maisner A, Muhlberger E, Netesov SV, Nowotny N, et al. 2016. Taxonomy of the order Mononegavirales: update 2016. Arch Virol 161:2351–2360. doi: 10.1007/s00705-016-2880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheltema NM, Gentile A, Lucion F, Nokes DJ, Munywoki PK, Madhi SA, Groome MJ, Cohen C, Moyes J, Thorburn K, Thamthitiwat S, Oshitani H, Lupisan SP, Gordon A, Sanchez JF, O'Brien KL, Group PS, Gessner BD, Sutanto A, Mejias A, Ramilo O, Khuri-Bulos N, Halasa N, de-Paris F, Pires MR, Spaeder MC, Paes BA, Simoes EAF, Leung TF, da Costa Oliveira MT, de Freitas Lazaro Emediato CC, Bassat Q, Butt W, Chi H, Aamir UB, Ali A, Lucero MG, Fasce RA, Lopez O, Rath BA, Polack FP, Papenburg J, Roglic S, Ito H, Goka EA, Grobbee DE, Nair H, Bont LJ. 2017. Global respiratory syncytial virus-associated mortality in young children (RSV GOLD): a retrospective case series. Lancet Glob Health 5:e984–e991. doi: 10.1016/S2214-109X(17)30344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. 2005. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med 352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 5.Cardenas S, Auais A, Piedimonte G. 2005. Palivizumab in the prophylaxis of respiratory syncytial virus infection. Expert Rev Anti Infect Ther 3:719–726. doi: 10.1586/14787210.3.5.719. [DOI] [PubMed] [Google Scholar]
- 6.Neuzil KM. 2016. Progress toward a respiratory syncytial virus vaccine. Clin Vaccine Immunol 23:186–188. doi: 10.1128/CVI.00037-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rezaee F, Linfield DT, Harford TJ, Piedimonte G. 2017. Ongoing developments in RSV prophylaxis: a clinician's analysis. Curr Opin Virol 24:70–78. doi: 10.1016/j.coviro.2017.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor G. 2017. Animal models of respiratory syncytial virus infection. Vaccine 35:469–480. doi: 10.1016/j.vaccine.2016.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mejias A, Chavez-Bueno S, Rios AM, Saavedra-Lozano J, Fonseca Aten M, Hatfield J, Kapur P, Gomez AM, Jafri HS, Ramilo O. 2004. Anti-respiratory syncytial virus (RSV) neutralizing antibody decreases lung inflammation, airway obstruction, and airway hyperresponsiveness in a murine RSV model. Antimicrob Agents Chemother 48:1811–1822. doi: 10.1128/AAC.48.5.1811-1822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prince GA, Horswood RL, Chanock RM. 1985. Quantitative aspects of passive immunity to respiratory syncytial virus infection in infant cotton rats. J Virol 55:517–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enkirch T, von Messling V. 2015. Ferret models of viral pathogenesis. Virology 479-480:259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coates HV, Chanock RM. 1962. Experimental infection with respiratory syncytial virus in several species of animals. Am J Hyg (Lond.) 76:302–312. [DOI] [PubMed] [Google Scholar]
- 13.Hsu KH, Lubeck MD, Bhat BM, Bhat RA, Kostek B, Selling BH, Mizutani S, Davis AR, Hung PP. 1994. Efficacy of adenovirus-vectored respiratory syncytial virus vaccines in a new ferret model. Vaccine 12:607–612. doi: 10.1016/0264-410X(94)90264-X. [DOI] [PubMed] [Google Scholar]
- 14.Prince GA, Porter DD. 1976. The pathogenesis of respiratory syncytial virus infection in infant ferrets. Am J Pathol 82:339–352. [PMC free article] [PubMed] [Google Scholar]
- 15.Stittelaar KJ, de Waal L, van Amerongen G, Veldhuis Kroeze EJ, Fraaij PL, van Baalen CA, van Kampen JJ, van der Vries E, Osterhaus AD, de Swart RL. 2016. Ferrets as a novel animal model for studying human respiratory syncytial virus infections in immunocompetent and immunocompromised hosts. Viruses 8:E168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boukhvalova MS, Prince GA, Blanco JC. 2007. Respiratory syncytial virus infects and abortively replicates in the lungs in spite of preexisting immunity. J Virol 81:9443–9450. doi: 10.1128/JVI.00102-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krizanova O, Rathova V. 1969. Serum inhibitors of myxoviruses. Curr Top Microbiol Immunol 47:125–151. doi: 10.1007/978-3-642-46160-6_6. [DOI] [PubMed] [Google Scholar]
- 18.Lukacs NW, Moore ML, Rudd BD, Berlin AA, Collins RD, Olson SJ, Ho SB, Peebles RS Jr. 2006. Differential immune responses and pulmonary pathophysiology are induced by two different strains of respiratory syncytial virus. Am J Pathol 169:977–986. doi: 10.2353/ajpath.2006.051055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stokes KL, Chi MH, Sakamoto K, Newcomb DC, Currier MG, Huckabee MM, Lee S, Goleniewska K, Pretto C, Williams JV, Hotard A, Sherrill TP, Peebles RS Jr, Moore ML. 2011. Differential pathogenesis of respiratory syncytial virus clinical isolates in BALB/c mice. J Virol 85:5782–5793. doi: 10.1128/JVI.01693-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore IN, Lamirande EW, Paskel M, Donahue D, Kenney H, Qin J, Subbarao K. 2014. Severity of clinical disease and pathology in ferrets experimentally infected with influenza viruses is influenced by inoculum volume. J Virol 88:13879–13891. doi: 10.1128/JVI.02341-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byrd LG, Prince GA. 1997. Animal models of respiratory syncytial virus infection. Clin Infect Dis 25:1363–1368. doi: 10.1086/516152. [DOI] [PubMed] [Google Scholar]
- 22.Prince GA, Jenson AB, Horswood RL, Camargo E, Chanock RM. 1978. The pathogenesis of respiratory syncytial virus infection in cotton rats. Am J Pathol 93:771–791. [PMC free article] [PubMed] [Google Scholar]
- 23.Hall CB, Douglas RG Jr. 1981. Modes of transmission of respiratory syncytial virus. J Pediatr 99:100–103. doi: 10.1016/S0022-3476(81)80969-9. [DOI] [PubMed] [Google Scholar]
- 24.Blount RE Jr, Morris JA, Savage RE. 1956. Recovery of cytopathogenic agent from chimpanzees with coryza. Proc Soc Exp Biol Med 92:544–549. doi: 10.3181/00379727-92-22538. [DOI] [PubMed] [Google Scholar]
- 25.Belser JA, Eckert AM, Tumpey TM, Maines TR. 2016. Complexities in ferret influenza virus pathogenesis and transmission models. Microbiol Mol Biol Rev 80:733–744. doi: 10.1128/MMBR.00022-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen TR. 1988. Re-evaluation of HeLa, HeLa S3, and HEp-2 karyotypes. Cytogenet Cell Genet 48:19–24. doi: 10.1159/000132579. [DOI] [PubMed] [Google Scholar]
- 27.McKimm-Breschkin JL. 2004. A simplified plaque assay for respiratory syncytial virus–direct visualization of plaques without immunostaining. J Virol Methods 120:113–117. doi: 10.1016/j.jviromet.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 28.Leung VK, Carolan LA, Worth LJ, Harper SA, Peck H, Tilmanis D, Laurie KL, Slavin MA, Sullivan SG. 2017. Influenza vaccination responses: Evaluating impact of repeat vaccination among health care workers. Vaccine 35:2558–2568. doi: 10.1016/j.vaccine.2017.03.063. [DOI] [PubMed] [Google Scholar]
- 29.van Baalen CA, Jeeninga RE, Penders GH, van Gent B, van Beek R, Koopmans MP, Rimmelzwaan GF. 2017. ViroSpot microneutralization assay for antigenic characterization of human influenza viruses. Vaccine 35:46–52. doi: 10.1016/j.vaccine.2016.11.060. [DOI] [PubMed] [Google Scholar]
- 30.McLellan JS, Correia BE, Chen M, Yang Y, Graham BS, Schief WR, Kwong PD. 2011. Design and characterization of epitope-scaffold immunogens that present the motavizumab epitope from respiratory syncytial virus. J Mol Biol 409:853–866. doi: 10.1016/j.jmb.2011.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones ML, Seldon T, Smede M, Linville A, Chin DY, Barnard R, Mahler SM, Munster D, Hart D, Gray PP, Munro TP. 2010. A method for rapid, ligation-independent reformatting of recombinant monoclonal antibodies. J Immunol Methods 354:85–90. doi: 10.1016/j.jim.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Carolan LA, Rockman S, Borg K, Guarnaccia T, Reading P, Mosse J, Kelso A, Barr I, Laurie KL. 2015. Characterization of the localized immune response in the respiratory tract of ferrets following infection with influenza A and B viruses. J Virol 90:2838–2848. doi: 10.1128/JVI.02797-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carolan LA, Butler J, Rockman S, Guarnaccia T, Hurt AC, Reading P, Kelso A, Barr I, Laurie KL. 2014. TaqMan real time RT-PCR assays for detecting ferret innate and adaptive immune responses. J Virol Methods 205:38–52. doi: 10.1016/j.jviromet.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pyankov OV, Setoh YX, Bodnev SA, Edmonds JH, Pyankova OG, Pyankov SA, Pali G, Belford S, Lu L, La M, Lovrecz G, Volchkova VA, Chappell KJ, Watterson D, Marsh G, Young PR, Agafonov AA, Farmer JF, Volchkov VE, Suhrbier A, Khromykh AA. 2017. Successful post-exposure prophylaxis of Ebola infected non-human primates using Ebola glycoprotein-specific equine IgG. Sci Rep 7:41537. doi: 10.1038/srep41537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corti D, Bianchi S, Vanzetta F, Minola A, Perez L, Agatic G, Guarino B, Silacci C, Marcandalli J, Marsland BJ, Piralla A, Percivalle E, Sallusto F, Baldanti F, Lanzavecchia A. 2013. Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature 501:439–443. doi: 10.1038/nature12442. [DOI] [PubMed] [Google Scholar]
- 36.McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, Kumar A, Modjarrad K, Zheng Z, Zhao M, Xia N, Kwong PD, Graham BS. 2013. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lakdawala SS, Jayaraman A, Halpin RA, Lamirande EW, Shih AR, Stockwell TB, Lin X, Simenauer A, Hanson CT, Vogel L, Paskel M, Minai M, Moore I, Orandle M, Das SR, Wentworth DE, Sasisekharan R, Subbarao K. 2015. The soft palate is an important site of adaptation for transmissible influenza viruses. Nature 526:122–125. doi: 10.1038/nature15379. [DOI] [PMC free article] [PubMed] [Google Scholar]