ABSTRACT
Exosomes are small membrane-enclosed vesicles produced by various cells and actively released into the extracellular space. They participate in intercellular communication and transfer of biologically active proteins, lipids, and nucleic acids. Accumulating evidence suggests that exosomes derived from cells infected by some viruses selectively encapsulate viral proteins, genetic materials, or even virions to mediate cell-to-cell communication and/or virus transmission. Porcine reproductive and respiratory syndrome virus (PRRSV) is an Arterivirus that has been devastating the global swine industry since the late 1980s. Recent studies have shown that major proteins secreted from PRRSV-infected cells are exosomal proteins and that the serum-derived exosomes from PRRSV-infected pigs contain viral proteins. However, the role of exosomes in PRRSV infection remains unclear. In this study, purified exosomes isolated from PRRSV-infected cells were shown with reverse transcription-PCR and mass spectrometry to contain viral genomic RNA and partial viral proteins. Furthermore, exosomes from PRRSV-infected cells established productive infection in both PRRSV-susceptible and -nonsusceptible cells. More importantly, exosome-mediated infection was not completely blocked by PRRSV-specific neutralizing antibodies. In summary, this study demonstrated that exosomes can mediate PRRSV transmission and are even resistant to antibody neutralization, identifying a potential immune evasion mechanism utilized by PRRSV.
IMPORTANCE Exosomes have recently been characterized as bioactive vesicles that function to promote intercellular communication. The exosomes from virally infected cells containing altered compositions confer numerous novel functionalities. A study of the secretome of cells infected with PRRSV indicated that the exosomal pathway is strongly activated by PRRSV infection. Here, we demonstrate that PRRSV can utilize host exosomes to infect naive healthy cells. Furthermore, exosome-mediated viral transmission is largely resistant to PRRSV-specific neutralizing antibodies. Our study provides novel insights into an alternative mechanism of PRRSV transmission that can compromise the host's anti-PRRSV immune response.
KEYWORDS: exosome, infection, intercellular transmission, porcine reproductive and respiratory syndrome virus
INTRODUCTION
Porcine reproductive and respiratory syndrome (PRRS) was first recognized in the United States in 1987 (1). The prevailing clinical symptoms are failures in pregnant sows, including late-term abortions, stillborn piglets, mummified piglets, and respiratory distress in piglets (2, 3). Since then, PRRS has become one of the most economically significant swine diseases in the world. The causative agent, porcine reproductive and respiratory syndrome virus (PRRSV), is a positive-sense single-stranded enveloped RNA virus of the family Arteriviridae. Its 15.4-kb genome encodes at least 10 open reading frames (ORFs). ORF1a and ORF1b encode two long polypeptides that produce 14 mature nonstructural proteins (nsp's) after enzymatic cleavage. ORF2 to ORF7, located at the 3′ terminus, encode the structural proteins glycoprotein 2 (GP2), envelope (E) protein, GP3, GP4, GP5, ORF5a protein, matrix (M) protein, and nucleocapsid (N) protein (4–6). Evidence indicates that PRRSV can compromise the innate immune response and establish a persistent infection (7–9). Although several kinds of vaccines have been developed to combat PRRSV, most of them have had only limited success in protecting pigs against PRRSV infection and disease (10–12). Neutralizing antibodies (NAbs) are believed to be crucial for anti-PRRSV immunity (13, 14). However, apparent paradoxes have been reported. For example, Lopez et al. reported that pigs passively immunized with PRRSV-specific NAbs achieved a nonviremic status, but PRRSV still displayed disseminated infection and replicated in the peripheral pig tissues (15). Furthermore, PRRSV was isolated from the sera of PRRSV-infected pigs despite the presence of high levels of PRRSV-specific NAbs (16). These paradoxes suggest the existence of other mechanisms of PRRSV dissemination and/or its evasion of NAbs.
We previously analyzed the secretome of cells infected with PRRSV and found that approximately 72.4% of all the secretory proteins identified were listed in the ExoCarta database (17), suggesting that the release of exosomal proteins was strongly activated by PRRSV infection. A recent study also identified viral proteins of PRRSV in serum-derived exosomes from pigs actively or previously infected with PRRSV (18).
Exosomes are small lipid bilayer vesicles secreted by most cell types that originate from late endosomal compartments called “multivesicular bodies” (MVBs) and are 30 to 150 nm in size (19). Various biological materials, including proteins, RNAs, and lipids, can be carried by exosomes (20, 21). Once released into the extracellular space, exosomes can mediate cell-to-cell communication through the transmission of signaling-competent proteins or functional RNAs, playing important roles in intercellular communication and signal transduction (22–24). Because of the importance of exosomes in intercellular communication, it is not surprising that some viruses have evolved mechanisms to hijack them to promote the viruses' survival and replication (25). For example, exosomes derived from human immunodeficiency virus (HIV)-infected cells contain viral transactivating response (TAR) RNA transcribed from the integrated provirus, which acts as a decoy in the recipient cells to make them more susceptible to HIV infection (26). Exosomes isolated from hepatitis C virus (HCV)-infected cells contain viral RNA and can mediate the viral-receptor-independent transmission of HCV (27). These studies highlight the potential role of exosomes in viral infection and transmission. Although preliminary studies have shown that PRRSV infection may promote exosome release and partial viral proteins have been found in exosomes (17, 18), the functions of exosomes in PRRSV infection remain unclear.
In this study, we examined exosomes derived from PRRSV-infected cells and found that the purified exosomes contained viral genomic RNA and partial viral proteins. PRRSV appears to exploit the cellular exosomal delivery system to transmit viral components that can establish a productive infection. Exosome-mediated PRRSV transmission is also largely resistant to NAbs.
RESULTS
Isolation and characterization of exosomes from PRRSV-infected cells.
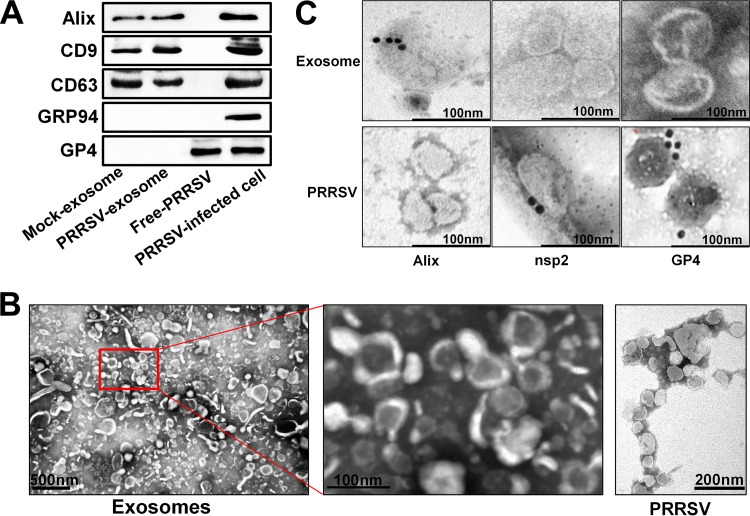
Because PRRSV virions and exosomes share similar sizes and buoyant densities, the conventional ultracentrifugation method cannot efficiently separate exosomes from viral contamination. To obtain exosomes with greater purity, we used a stringent isolation method to extract and purify exosomes from PRRSV-infected cells, with polyethylene glycol (PEG) enrichment precipitation (28) and ultracentrifugation combined with CD63 immunomagnetic bead affinity purification. To this end, PK-15CD163 cells (a porcine kidney cell line stably expressing the PRRSV receptor CD163, gifted by En-min Zhou at Northwest A&F University, China) were infected with PRRSV strain WUH3 at a multiplicity of infection (MOI) of 1.0 and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 2% exosome-depleted fetal bovine serum (FBS). At 36 h postinfection (hpi), the supernatants were collected to isolate and purify exosomes. The purified exosomes were characterized by an analysis of exosomal markers with Western blotting. Three representative exosome markers, Alix, CD9, and CD63, were detected, and the absence of GRP94 (endoplasmin marker) and GP4 (PRRSV structural protein) excluded contamination with endoplasmic vesicles and PRRSV particles (Fig. 1A). Transmission electron microscopy (TEM) showed that the purified exosomes display a cup-shaped appearance ranging from about 30 to 150 nm in size, while the purified PRRSV particles are enveloped, have a spherical appearance, and range from 50 to 70 nm in diameter (Fig. 1B). The purified exosomes were further characterized by immunogold labeling with antibodies against the exosome marker Alix, viral structural protein GP4, and nsp2, a viral nonstructural protein that has been demonstrated to be incorporated into PRRSV virions (29). The results showed that the Alix marker protein but not viral nsp2 and GP4 could be probed in the exosomes isolated from PRRSV-infected cells (Fig. 1C). Taken together, these results indicate that there is no free-PRRSV-virion contamination in the purified exosomes.
FIG 1.
Isolation and characterization of exosomes from PRRSV-infected cells. (A) Purified exosomes derived from mock- or PRRSV-infected cells were analyzed on Western blots probed with antibody directed against Alix, CD9, CD63, GRP94, or GP4. The PRRSV-infected cells and purified virions (free PRRSV) were used as controls. (B) Transmission electron microscopy observations of negatively stained purified exosomes from PRRSV-infected cells and free PRRSV. Purified exosomes with both higher and lower magnification are shown. (C) Immunoelectron microscopy images of purified exosomes or virions from PRRSV-infected cells. Immunogold labeling (10-nm gold particles) was performed with antibodies against exosome marker protein Alix and viral proteins nsp2 and GP4, respectively.
Exosomes derived from PRRSV-infected cells contain viral components.
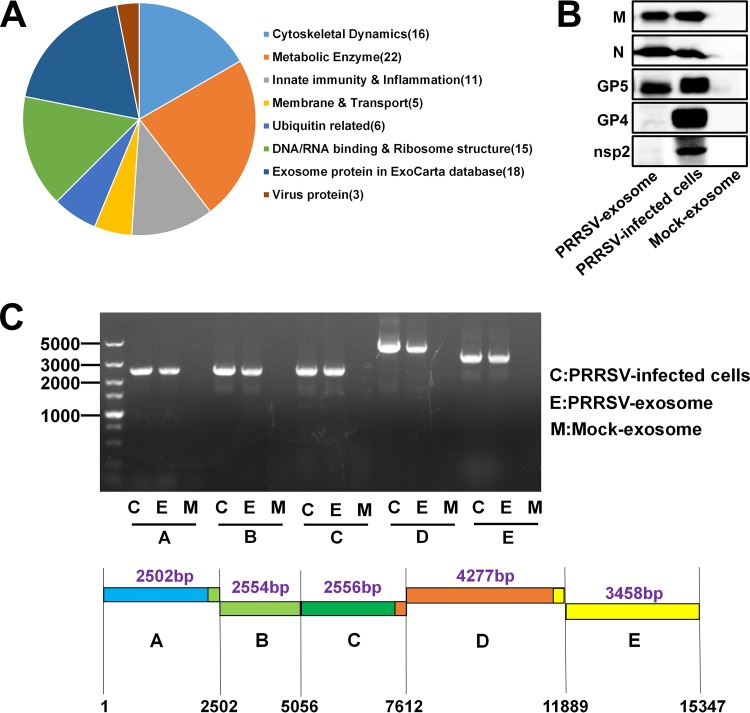
To characterize the contents of exosomes purified from PRRSV-infected cells, a liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis was performed. In total, 216 host proteins were identified within the purified exosomes. Gene ontology (GO) analysis showed important enrichment of membrane or transport proteins, cytoskeletal dynamics proteins related to exosome composition and innate immunity, and metabolic enzyme proteins related to exosome functions (Fig. 2A). Among the proteins identified, 92% were listed in the Vesiclepedia database (http://www.microvesicles.org/), and only 16 proteins were not documented in that database. LC-MS/MS analysis also revealed the presence of three viral proteins: GP5, M, and N. Western blots probed with monoclonal antibodies (MAbs) directed against these viral proteins confirmed that all three proteins, but not nsp2 and GP4, were present in the purified exosomes (Fig. 2B). Previous studies have suggested that viral nucleic acids are present in exosomes from cells infected by several viruses, including HCV and hepatitis B virus (30, 31). We also used reverse transcription (RT)-PCR to investigate whether the exosomes derived from PRRSV-infected cells contain viral RNAs. Because it is difficult to amplify the complete genomic RNA by a single PCR, the whole genome of PRRSV was divided into five overlapping fragments (A, B, C, D, and E) to be amplified. As shown by the results in Fig. 2C, all five overlapping fragments could be amplified from the exosomes isolated from PRRSV-infected cells, indicating that the exosomes contained the complete PRRSV genomic RNA. Taken together, these results indicate that exosomes isolated from the supernatants of PRRSV-infected cells contain viral genomic RNAs and partial viral proteins.
FIG 2.
Components of exosomes derived from PRRSV-infected cells. (A) Purified exosomes derived from PRRSV-infected cells were analyzed with LC-MS/MS to determine the host proteins and viral proteins present. Classification of host proteins according to function is shown. (B) PRRSV proteins in exosomes were confirmed on Western blots probed with antibody directed against M, N, GP5, GP4, or nsp2. (C) PRRSV genomic RNAs in exosomes isolated from PRRSV-infected cells were detected with RT-PCR. Five overlapping fragments (A, B, C, D, and E) were designed based on the genome sequence of PRRSV strain WUH3.
Exosomes transmit PRRSV and establish productive infections in susceptible and nonsusceptible cells.
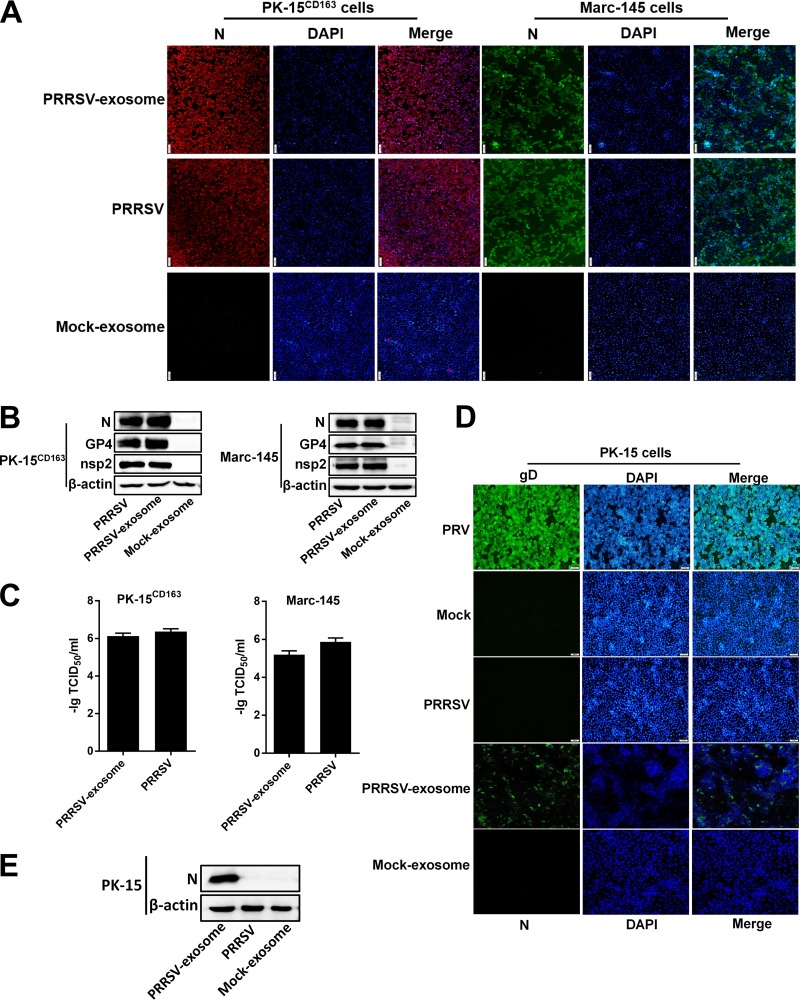
The presence of PRRSV components in the purified exosomes prompted us to investigate whether exosomes from PRRSV-infected cells can transmit infection to naive cells. To this end, the exosomes purified from PRRSV-infected cells were incubated with naive PK-15CD163 cells and indirect immunofluorescence assay (IFA) for viral N protein was performed at 36 hpi to confirm infection. Cells infected with cell-culture-derived PRRSV (free PRRSV) were used as the positive control, and cells inoculated with exosomes isolated from mock-infected cell supernatants with the same method were used as the negative control. As shown by the results in Fig. 3A, obvious cytopathic effects were observed in the cells treated with PRRSV-positive exosomes or infected with free PRRSV. The IFA performed with an MAb directed against PRRSV N protein also confirmed that a productive infection was established in the naive PK-15CD163 cells inoculated with PRRSV-positive exosomes. We also tested Marc-145 cells, another cell line permissive of PRRSV that is used extensively for PRRSV studies and vaccine production. A similar productive infection was established after treatment with PRRSV-positive exosomes, whereas no cytopathic effect was observed in cells treated with the control exosomes (Fig. 3A). The expression of viral proteins N, GP4, and nsp2 could also be detected with Western blotting in PK-15CD163 and Marc-145 cells treated with PRRSV-positive exosomes (Fig. 3B), further confirming the establishment of productive infections. Fifty-percent tissue culture infective dose (TCID50) assays were performed to determine the viral titers in the exosome-treated PK-15CD163 and Marc-145 cells, and cell-culture-derived PRRSV was used as the control. As shown by the results in Fig. 3C, the purified PRRSV-positive exosomes generated high viral titers in both PK-15CD163 and Marc-145 cells that were similar to the levels seen in cells infected with the cell-culture-derived PRRSV.
FIG 3.
Exosomes transmit PRRSV and establish productive infections in both susceptible and nonsusceptible cells. (A) IFAs demonstrated productive infection of Marc-145 and PK-15CD163 cells after treatment with PRRSV-positive exosomes. Marc-145 and PK-15CD163 cells were cocultured with PRRSV-positive exosomes or infected with PRRSV at an MOI of 1.0. At 36 h after treatment or infection, the cells were fixed and an IFA was performed with MAb directed against PRRSV N protein. Fluorescence was observed with an Olympus IX73 inverted microscope. (B) Western blot analysis of PRRSV N, GP4, and nsp2 expression in Marc-145 and PK-15CD163 cells infected with PRRSV or treated with PRRSV-positive exosomes. (C) PK-15CD163 and Marc-145 cells were infected with PRRSV or treated with purified PRRSV-positive exosomes, and the viral titers were determined with TCID50 assay. (D) PK-15 cells were incubated with PRRSV-positive exosomes or infected with PRRSV at an MOI of 1.0. Pseudorabies virus (PRV) was used as the positive control for infection of PK-15 cells. At 36 h after treatment or infection, cells were fixed and IFAs were performed with MAb directed against PRRSV N protein or PRV gD. (E) Western blot analysis of PRRSV N protein expression in PK-15 cells infected with PRRSV or purified PRRSV-positive exosomes. DAPI, 4',6-diamidino-2-phenylindole.
Lack of the receptor CD163 makes PK-15 cells nonsusceptible to PRRSV infection (32). We investigated whether a productive infection could be established in PK-15 cells by PRRSV-positive exosomes. Pseudorabies virus (PRV), which can efficiently infect PK-15 cells, was used as the positive infection control. After coculture with PK-15 cells, IFA with an N protein-specific MAb showed that PRRSV-positive exosomes established a productive infection in PK-15 cells, whereas no specific immunofluorescence was observed in PK-15 cells infected with free PRRSV (Fig. 3D). In the control cells infected with PRV, bright immunofluorescence was detected with an antibody directed against PRV glycoprotein D (gD) (Fig. 3D). Western blot analyses used to detect the expression of PRRSV N protein were consistent with the results of IFAs (Fig. 3E). Taken together, these results suggest that PRRSV-positive exosomes can establish productive infections in both susceptible and nonsusceptible cells.
Inhibition of exosome release impairs PRRSV transmission mediated by exosomes.
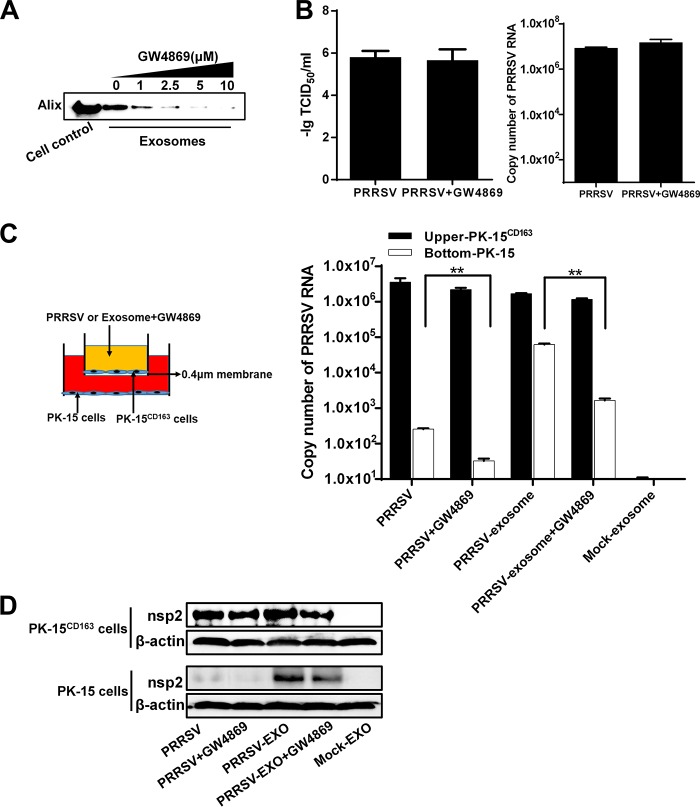
To further investigate the involvement of the exosomal pathway in PRRSV transmission, we examined the effects of an inhibitor of exosome release, GW4869 {N,N′-bis[4-(4,5-dihydro-1H-imidazol-2-yl)phenyl]-3,3′-p-phenylene-bis-acrylamide dihydrochloride}. GW4869, a neutral sphingomyelinase inhibitor (33) that is known to inhibit ceramide biosynthesis, contributes to exosome secretion by triggering the budding of exosomes into MVBs. MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay demonstrated no obvious cytotoxicity at any of the concentrations of GW4869 tested in PK-15CD163 cells (data not shown). The effect of GW4869 on exosome biogenesis was assessed with Western blotting to quantitate the expression of exosome biomarker Alix. As shown by the results in Fig. 4A, as the concentration of GW4869 increased, the amount of exosomes gradually and continuously declined. To test whether GW4869 affects PRRSV replication, PK-15CD163 cells were infected with PRRSV and then treated with medium containing GW4869 at the concentrations indicated in Fig. 4A. TCID50 and real-time RT-PCR assays were performed to determine the viral titers and propagation. Compared with the mock-treated group, treatment with 10 μM GW4869 did not cause a significant reduction in PRRSV infection or replication (Fig. 4B). At this concentration of GW4869, which effectively inhibits exosome release but has no effect on PRRSV propagation, we mimicked the exosome transfer from PK-15CD163 cells to PK-15 cells using a coculture model (Transwell system) in the presence of GW4869, as shown by the schematic diagram in Fig. 4C (left). PRRSV RNAs from the cells in both the top and bottom chambers were quantified with real-time RT-PCR. As shown by the results in Fig. 4C (right), PRRSV RNAs were detected in the PK-15 cells in the bottom chamber when they were cocultured with PK-15CD163 cells infected with free PRRSV or PRRSV-positive exosomes. These results confirm the transmission of PRRSV-positive exosomes through the membrane when free PRRSV- or PRRSV-exosome-infected PK-15CD163 cells from the top chamber released exosomes containing PRRSV components. However, the presence of GW4869 in the top chamber inhibited the release of exosomes, and the PRRSV RNA copy numbers were markedly lower in the PK-15 cells (Fig. 4C, right). At the same time, cell samples from both top and bottom chambers were collected and subjected to Western blotting with antibody against PRRSV nsp2. The protein expression level (Fig. 4D) of each group remained in accordance with the PRRSV RNA level (Fig. 4C). These results suggest that GW4869 has a direct effect on the transmission of PRRSV mediated by exosomes, indirectly supporting our conclusion that exosomes derived from PRRSV-infected cells can establish a productive infection in nonsusceptible cells.
FIG 4.
Inhibition of exosome release impairs PRRSV transmission mediated by exosomes. (A) PK-15CD163 cells were treated with the indicated concentrations of GW4869, and the exosomes isolated were subjected to Western blotting for Alix expression. (B) PK-15CD163 cells were infected with PRRSV at an MOI of 1 for 1 h and then maintained in medium containing 10 μM GW4869 for 36 h. TCID50 and real-time PCR assays were performed to determine the viral titers and RNA copy numbers, respectively. (C) Schematic diagram of the Transwell system used to coculture PK-15CD163 and PK-15 cells. The upper-chamber PK-15CD163 cells were infected with PRRSV or treated with purified PRRSV-positive exosomes for 3 h and then treated or not with GW4869 (10 μM). After 36 h, the total RNAs were extracted from cells in both chambers and analyzed for PRRSV RNA with real-time quantitative PCR. (D) Cell samples from both bottom and top chambers of the experiment diagrammed in panel C were collected and subjected to Western blotting with antibody against viral protein nsp2.
Exosome-mediated PRRSV infection is not blocked by PRRSV-specific NAbs.
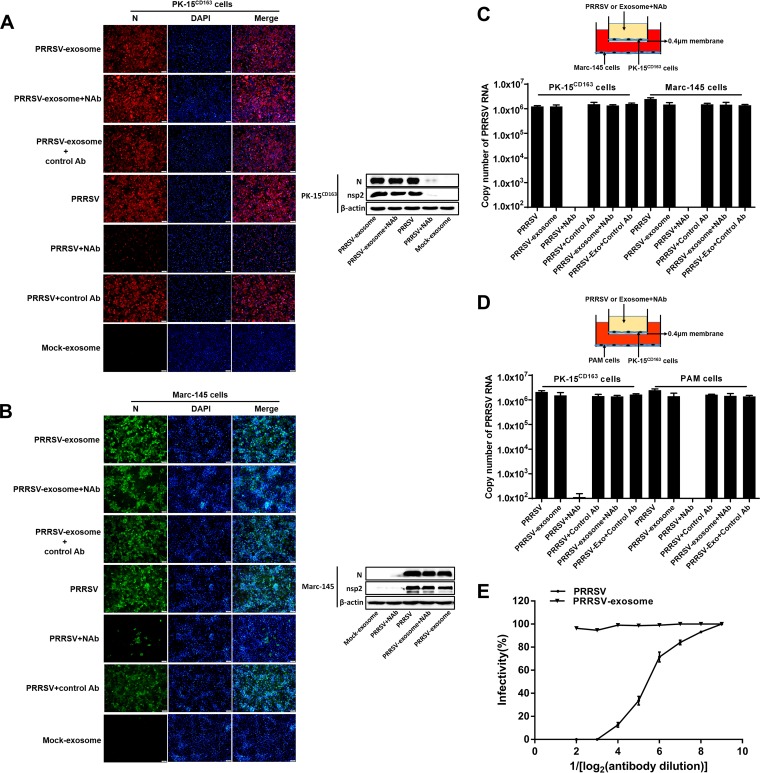
We also investigated whether exosome-mediated PRRSV transmission is blocked by PRRSV-specific NAbs. To avoid the interference of some unidentified factor(s) in hyperimmune sera, PRRSV-specific immunoglobulins (IgGs) were purified from the sera of pigs repeatedly immunized with PRRSV vaccine. PRRSV-positive exosomes or free PRRSV suspensions were incubated with the purified NAbs for 1 h and then transferred into a monolayer of PK-15CD163 cells. At 36 hpi, cells were fixed for IFAs with an MAb directed against PRRSV N protein. As expected, the number of infected cells decreased significantly more in the group treated with free PRRSV and NAbs (group PRRSV+NAbs) than in the group treated with PRRSV without NAbs (Fig. 5A, left), suggesting that the purified NAbs efficiently neutralized PRRSV infection. However, there was almost no difference between the PRRSV-exosome group (mock-treated PRRSV-positive exosomes) and the PRRSV-exosome+NAb group (NAb-treated PRRSV-positive exosomes) (Fig. 5A, left), indicating that NAbs had almost no blocking effect on PRRSV-positive exosomes. The results of Western blotting used to detect the expression of PRRSV N and nsp2 proteins confirmed the IFA results (Fig. 5A, right). Similar results were obtained in Marc-145 cells with both IFA (Fig. 5B, left) and Western blotting (Fig. 5B, right).
FIG 5.
Exosome-mediated PRRSV infection is not blocked by PRRSV-specific neutralizing antibodies (NAbs). (A, B) Purified PRRSV-positive exosomes or free PRRSV virions were incubated with PRRSV-specific NAbs for 1 h. Then, PK-15CD163 (A) or Marc-145 (B) cells were exposed to the antibody-treated exosomes or virus for 3 h. The exosomes or viruses were washed off, and the medium was replaced with fresh maintenance medium for a further 36 h. Immunofluorescent staining was performed with MAb against PRRSV N protein. Western blotting was performed with MAbs against PRRSV N and nsp2 proteins. (C) Schematic diagram of the Transwell system used to coculture PK-15CD163 and Marc-145 cells. PK-15CD163 cells in the upper chamber were incubated with purified PRRSV-positive exosomes or infected with PRRSV pretreated with PRRSV-specific NAbs (neutralization titer is 1:16) for 1 h. The viruses or exosomes were washed off, and cells were cultured in maintenance medium for a further 36 h. Total RNAs of the cells in both top and bottom chambers were extracted and analyzed for PRRSV genomic RNA with real-time quantitative PCR. (D) Schematic diagram of the Transwell system used to coculture PK-15CD163 cells and PAMs. PK-15CD163 cells from the upper chamber were treated with purified PRRSV-positive exosomes or infected with PRRSV pretreated with PRRSV-specific NAbs as described in the legend to panel C. Quantitative RT-PCR was performed to detect PRRSV genomic RNA in PK-15CD163 cells and PAMs. (E) The neutralizing effects of PRRSV-specific NAbs at different concentrations on exosomes and free PRRSV virions. The PRRSV-specific NAbs (neutralization titer is 1:16) were serially diluted, and the neutralizing effects on exosomes and free PRRSV were determined by using a rapid fluorescent focus neutralization assay.
To further confirm that exosome-mediated PRRSV infection cannot be blocked by PRRSV-specific NAbs, PK-15CD163 cells were cultured together with Marc-145 cells in a Transwell system to allow the transfer of exosomes but preclude any direct cell contact. Purified NAbs (neutralization titer is 1:16) were added to the medium in the top chamber containing PK-15CD163 cells, as shown in the schematic diagram in Fig. 5C (top). Real-time RT-PCR was used to analyze PRRSV RNAs in the top-chamber PK-15CD163 cells and the bottom-chamber Marc-145 cells. Consistent with the IFA and Western blotting results, the PRRSV RNA levels were significantly reduced in both PK-15CD163 cells and Marc-145 cells treated with PRRSV+NAbs but only barely reduced in cells treated with PRRSV-positive exosomes+NAbs (Fig. 5C, bottom). When we used porcine alveolar macrophages (PAMs) in the bottom chamber, similar results were observed (Fig. 5D).
We also compared the neutralizing effects of different concentrations of NAbs on free PRRSV and exosome. As shown by the results in Fig. 5E, the neutralizing effects on free PRRSV were gradually decreased with decreased concentrations of neutralizing antibodies; however, nearly no significant change could be detected for exosome-mediated infection after treatment with different concentrations of neutralizing antibodies. The results of all experiments (IFA, Western blotting, Transwell system, and neutralizing assay) support the conclusion that exosome-mediated PRRSV infection cannot be blocked by PRRSV-specific NAbs.
DISCUSSION
Exosomes were originally considered a vehicle to remove cellular waste from cytosol (34), but ever-increasing evidence supports the role of exosomes in intercellular communication by transferring functional cellular proteins, RNAs, and microRNAs (miRNAs), affecting the functional fates of recipient cells (35–37). Furthermore, many virally infected cells secrete exosomes that differ in content from those secreted from normal cells. However, they may also contain various viral proteins, RNAs, or even virions, so it is possible that viruses exploit exosomal pathways to spread infection and modify their target cells (38–40). In this study, we have demonstrated for the first time that exosomes derived from PRRSV-infected cells contain viral RNAs and transfer productive infections to naive cells, even in the presence of PRRSV-specific NAbs.
Despite tremendous advances in the study of the interplay between exosome and virus, isolating pure exosomes free of contaminating viruses from virus-infected cells remains a huge challenge. Exosomes are conventionally isolated from virus-infected cells or bodily fluids with a sequential sucrose-gradient ultracentrifugation method (41). However, because some viral particles have sizes, buoyant densities, or sedimentation velocities similar to those of exosomes, it is difficult to separate the two populations completely. For example, exosomes derived from HIV- or HCV-infected cells cannot be readily distinguished or separated from infectious viral particles with sucrose-gradient ultracentrifugation (38, 42–45). Based on a recent review (46), CD63 or composite magnetic bead purification is by far the best method to completely separate exosomes and virions. In the present study, the exosomes isolated from PRRSV-infected cells were further purified by CD63 immunomagnetic bead affinity. Immunoelectron microscopy confirmed that there were no free PRRSV virions in the purified exosomes (Fig. 1C). LC-MS/MS analysis also showed that there were no minor structural proteins or nonstructural proteins of PRRSV in the purified exosomes, and the infectivity of exosomes isolated from PRRSV-infected cells was not blocked by PRRSV-specific NAbs. These results indicated that the purified exosomes were not contaminated with free PRRSV virions.
Although there were no PRRSV-encoded minor structural proteins or nonstructural proteins in the exosomes isolated from PRRSV-infected cells, three major structural proteins, GP5, M, and N, were identified in these exosomes with LC-MS/MS. Both GP5 and M proteins are envelope proteins encoded by PRRSV, and the two proteins form a disulfide-linked heterodimer in PRRSV-infected cells (47). It is surprising that envelope proteins are actively released through the exosomal pathway. We transfected HEK-293 cells with eukaryotic expression plasmids encoding GP5 or M protein and isolated exosomes from the transfected cells. Western blotting could detect GP5 and M proteins in the isolated exosomes (data not shown), indicating that GP5 and M are secreted through the exosomal pathway. Pleet et al. reported that the viral protein 40 (VP40) matrix protein of Ebola virus (EBOV) can be packaged into exosomes, and exosomes isolated from VP40-transfected cells are capable of inducing apoptosis in the recipient immune cells, which may allow the virus to replicate to high titers in the immunocompromised host (48). Previous studies have demonstrated that PRRSV induces apoptosis in pigs during infection and in cultured cells in vitro (49, 50), and GP5 has been reported to induce apoptosis in bystander cells. Whether the apoptosis induced by GP5 acts through the exosomal pathway requires further study. In addition, PRRSV N protein is a multifunctional nucleolar-cytoplasmic shuttling protein. Previous studies suggested that PRRSV N protein participates in the modulation of type I interferon and inflammatory responses (51, 52). Many exosomes derived from virus-infected cells contain cellular or viral proteins or RNAs that can affect the gene expression or immune response of recipient cells (53–55). Whether PRRSV N protein modulates innate immune responses via the exosomal pathway is under investigation in our laboratory.
As well as viral proteins, our LC-MS/MS analysis showed that 216 porcine proteins were present in the purified exosomes derived from PRRSV-infected cells, and most of these proteins were identified in the database of exosomes derived from sera of PRRSV-infected pigs in a previous study (18). GO analysis showed that these host proteins were associated with cytoskeletal structure, membrane transport, ubiquitin, and response to immune system processes. Viral infection may change the contents of exosomes, and several studies have demonstrated that the altered compositions of exosomes derived from virally infected cells confer novel functionalities, such as facilitating viral spread and viral evasion of host cell defenses (56, 57). Our previous quantitative secretome study also found that approximately 72.4% of secretory proteins that were differentially expressed in PRRSV-infected Marc-145 cells and mock-infected controls were exosomal proteins (17). These differentially expressed exosomal proteins may facilitate viral pathogenesis or host immune response. Unfortunately, we did not quantitatively analyze the contents of purified exosomes derived from PRRSV-infected cells in this study.
In recent years, the roles of exosomes released from infected cells have been investigated for several viruses, including HIV-1, HCV, hepatitis A virus (HAV), Dengue virus (DENV), human T-cell lymphotropic virus, the herpesviruses, and severe fever with thrombocytopenia syndrome virus (SFTSV). The findings of these studies suggested that the released exosomes contain viral particles, genomes, mRNAs, microRNAs (miRNAs), or proteins that play important roles in viral replication, pathogenesis, and transmission. For example, purified exosomes isolated from HCV- or HAV-infected cells were shown to contain complete viral particles and were capable of transmitting infection to naive cells (58). Cells infected with SFTSV also released exosomes containing virions, and when the exosomes were taken up by uninfected cells, viral replication was initiated (59). Epstein-Barr virus (EBV)-positive tumors secrete exosomes containing the viral protein latent membrane protein 1 (LMP1), which has inhibitory effects on immune responses, facilitating immune evasion (60). Some miRNAs encoded by Kaposi's sarcoma-associated herpesvirus are incorporated into exosomes to facilitate tumorigenesis (61). In this study, we found that exosomes from PRRSV-infected cells contained viral RNA and partial viral proteins. Similar to the previously reported exosome-mediated transmission of HCV, PRRSV also takes advantage of the exosomal pathway to transfer virus to both susceptible and nonsusceptible naive cells and establish productive infections. A recent study found that treatment with anti-HCV receptor antibody, which blocks free virus entry and infection, did not inhibit the exosomal transmission of HCV (27), indicating that exosomes can mediate virus transmission independently of a virus-specific receptor. This entry pathway may be why exosomes can transfer PRRSV infection to nonsusceptible cells.
An interesting finding of this study is that exosome-mediated PRRSV transmission is resistant to PRRSV-specific NAbs compared with their ability to block the transmission of the free virus. Previous studies have also shown that exosome-mediated transmission of HAV and enterovirus 71 (EV71) cannot be completely blocked by NAbs (62, 63), suggesting that viral exosomes can escape the humoral immune response. Using patient-derived IgGs, Cosset and Dreux demonstrated that exosomal HCV transmission escaped antibody neutralization and, surprisingly, that several IgGs enhanced the transmission of exosomes isolated from HCV subgenomic replicon cells (64). These studies and our present study suggest that exosomes containing virions or viral RNA may be less sensitive to NAbs, which represents a potential immune evasion mechanism. PRRSV viremia is clinically detected in the blood, even in those pigs with high NAb titers. Whether exosomes contribute to this phenomenon remains to be investigated.
As well as containing virions, viral RNA, or viral proteins, exosomes isolated from virus-infected cells often contain viral or host miRNAs, which may act as cofactors to increase the infectivity of viral exosomes or as immunoregulatory factors controlling immune responses in the recipient cells (65). It remains to be seen in future studies whether PRRSV-positive exosomes contain miRNAs and whether these miRNAs contribute to the exosome-mediated transmission of PRRSV.
MATERIALS AND METHODS
Cell culture and viruses.
Marc-145, PK-15CD163, and PK-15 cells were maintained in DMEM (Invitrogen, USA) supplemented with 10% exosome-depleted FBS (System Bioscience, USA) and 1% penicillin-streptomycin in a humidified incubator at 37°C with 5% CO2. PAMs were obtained from 5-week-old PRRSV-negative pigs as described previously (66) and maintained in RPMI 1640 medium (Invitrogen, USA) with 10% exosome-depleted FBS. The highly pathogenic PRRSV strain WUH3 (GenBank accession number HM853673), which was previously isolated from the brains of pigs suffering a high-fever syndrome in China (67), was used throughout this study. Pseudorabies virus (PRV) strain Ea, a wild virulent strain isolated in China (68), was propagated in PK-15 cells.
Exosome isolation and purification.
PRRSV- or mock-infected cell supernatants were collected and centrifuged for 5 min at 500 × g to remove the cells and larger debris. The samples were transferred to a new tube, centrifuged at 2,000 × g for 10 min to further remove cell debris, and filtered through a 0.2-μm filter. A 2-fold-concentrated (2×) stock solution of polyethylene glycol solution (16%) was prepared to precipitate extracellular vesicles, as reported previously (28). Once centrifuged and processed, the medium was added to an equal volume of a 2× PEG stock solution to achieve a final PEG concentration of 8% and incubated at 4°C for at least 12 h. The samples were then centrifuged at 3,214 × g for 1 h. The resulting pellets were suspended in 50 to 500 μl of particle-free phosphate-buffered saline (PBS) and centrifuged at 100,000 × g for 70 min to wash and re-pellet the vesicles. To purify the exosomes recovered, CD63-labeled Dynabeads (Invitrogen) were used according to the manufacturer's instructions. For electron microscopy observation, exosomes were eluted from the Dynabeads using ice-cold 100 nM glycine-HCl (pH 3.0) and immediately neutralized to pH 7.4 with neutralizing buffer (1 M Tris-HCl, pH 8.5).
Exosome infection assays.
PK-15CD163, PK-15, and Marc-145 cells were seeded on 24-well plates at a density of 2 × 105 cells/well. About 5 × 104 TCID50 of free PRRSV and exosomes derived from PRRSV-infected cell culture were added to the cells, incubated for 2 h at 37°C in 5% CO2, and the medium was replaced with fresh maintenance medium. At 36 h postinfection, cell samples were fixed or collected for indirect immunofluorescence assay and Western blotting.
Electron microscopy and MS.
The purified exosomes or virions were spotted onto Formvar-coated copper grids (200 meshes). The adsorbed exosomes or purified virions were fixed in 2% (vol/vol) paraformaldehyde for 5 min at room temperature. After fixation, the grids were directly stained with uranyl acetate for contrast enhancement and then examined using a transmission electron microscope (Hitachi H-7000FA, Japan). For immunoelectron microscopy, the purified exosomes or virions resuspended in PBS were deposited on Formvar-carbon-coated copper grids, and the samples on the grids were blocked for 30 min using 5% bovine serum albumin (BSA)–PBS and incubated with primary antibody against Alix, nsp2, or GP4 and then with 10-nm-colloidal-gold-labeled secondary antibody (goat anti-mouse IgG). After fixation with glutaraldehyde, the samples were stained with uranyl acetate for contrast enhancement and processed for electron microscopy. For MS analysis, the purified exosomes were resuspended in 25 μl elution buffer (50 mM glycine, pH 2.8). Proteins were digested with the filter-aided sample preparation (FASP) procedure, as described by Wiśniewski et al. (69). Briefly, the protein pellet (about 30 μg) was solubilized in 30 μl SDT buffer (4% [mass/vol] SDS, 100 mM Tris-HCl, 1 mM dithiothreitol [DTT] [pH 7.6]) at 90°C for 5 min. The detergent, DTT, and other low-molecular-weight components were removed using 200 μl UA buffer (8 M urea, 150 mM Tris-HCl, pH 8.0) with repeated ultrafiltration (Microcon-30kDa centrifugal filter unit). Iodoacetamide (0.05 M, 100 μl) in UA buffer was then added to block the reduced cysteine residues, and the samples were incubated for 20 min in the dark. The filter was washed three times with 100 μl of UA buffer and then twice with 100 μl of 25 mM NH4HCO3. Finally, the protein suspension was digested with 2 μg of trypsin (Promega) in 40 μl of 25 mM NH4HCO3 overnight at 37°C. The resulting peptides were collected as the filtrate. Experiments were performed on a Q Exactive mass spectrometer coupled to an Easy-nLC liquid chromatograph (Proxeon Biosystems).
Indirect immunofluorescence assay (IFA).
Cells infected with PRRSV or treated with PRRSV-positive or mock exosomes were fixed with 4% paraformaldehyde for 15 min and immediately permeabilized with precooled methanol for 10 min. After blocking with 5% BSA solution, the cells were incubated with an anti-PRRSV N MAb (made in our laboratory) for 1 h at room temperature. The cells were then stained with Alexa Fluor 488- or 596-conjugated anti-mouse antibodies (Invitrogen) according to the manufacturer's protocol. Images were taken with an Olympus IX73 inverted microscope (Olympus).
Western blotting.
Western blotting was performed with the following established protocol. Briefly, predetermined concentrations of proteins were resolved on 12% SDS–PAGE gels. After electrophoresis, the separated proteins were transferred onto 0.2-μm polyvinylidene difluoride (PVDF) membranes (Millipore, USA). After protein transfer, the membranes were blocked for 1 h with Tris-buffered saline containing Tween 20 (TBST) with 10% nonfat dry milk. The blots were then incubated with a primary antibody at 4°C overnight. The primary antibodies used were directed against CD63 (Santa Cruz Biotechnology), CD9 (Proteintech), Alix (Cell Signaling Technology), GRP94 (Cell Signaling Technology), and PRRSV N, GP5, M, GP4, and nsp2 (made in our laboratory). The membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies that included a goat anti-mouse IgG and a goat anti-rabbit lgG (Beyotime Biotechnology). Finally, the proteins were visualized with Clarity ECL Western blotting substrate (Bio-Rad).
Analysis and quantification of PRRSV RNA.
For PCR detection of PRRSV RNA, total RNA from exosomes was extracted with a total exosome RNA and protein isolation kit (Life Technologies, USA) according to the manufacturer's instructions. To detect PRRSV genome RNA, the whole genome of PRRSV was divided into five overlapping fragments (A, B, C, D, and E) to be amplified. The primers for each fragment are listed in Table 1. To quantify the RNA copies of PRRSV in PRRSV-infected or exosome-treated cells, total RNAs from cell culture samples were isolated with the E.Z.N.A. total RNA kit I (Omega Bio-tek). Reverse transcription was performed with the Roche Transcriptor first-strand cDNA synthesis kit (Roche). The primers used to amplify the PRRSV nsp9 gene were 5′-GTTGATGGTGGTGTTGTGCT-3′ and 5′-AGACCAATTTTAGGCGCGTC-3′. Absolute real-time PCR was performed with the Applied Biosystems ViiA 7 real-time PCR system (Life Technologies). The quantity of PRRSV RNA was calculated based on the results for a standard plasmid diluted 10-fold (from 1010 to 101). Each sample was assayed at least three times.
TABLE 1.
Sequences of primers used for amplifying overlapping fragments of the PRRSV genome
| Fragment | Primer sequence (5′-3′) |
|
|---|---|---|
| Forward | Reverse | |
| A | ACCGTCATGACGTATAGGTGTTGGCT | CTCGTCGAGCCCGCTCGGCAG |
| B | CTTAAGGACCAGATGGAGGAGGATCTGCT | CCACAAGTGCTGTCAAGGGCAAGGTGA |
| C | TCACCTTGCCCTTGACAGCACTTGTGG | AGTTTAAACACTGCTCCTTAGTC |
| D | TAAACTGCTAGCCGCCAGCGGCTTG | CCCGAAACGCATCATTGTAATCCT |
| E | GTTTCGGGCGCGCCAGAAAGGGAA | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTAATTA |
Antibody-mediated neutralization of PRRSV.
PRRSV-specific NAb was isolated and purified from sera of pigs repeatedly immunized with commercial PRRSV vaccine against the highly pathogenic PRRSV. The titer of purified NAb was adjusted to 1:16, as determined by using a rapid fluorescent focus neutralization assay as described previously (70). Briefly, serial dilutions of antibodies were incubated with an equal volume of PRRSV strain WUH3 (200 focus-forming units) for 1 h at 37°C. The mixtures were added to 96-well plates containing confluent Marc-145 cells. After incubation for 24 h at 37°C in a humidified atmosphere containing 5% CO2, the cells were fixed with a solution of 50% methanol and 50% acetone for 5 min. After washing three times with PBS, the infected cells were detected with fluorescein isothiocyanate-conjugated MAb against PRRSV N protein. The neutralization titers were expressed as the reciprocal of the highest dilution that inhibited 90% of the fluorescent foci present in the control wells.
ACKNOWLEDGMENTS
We thank En-min Zhou for providing PK-15CD163 cells.
This work was supported by the Major Project of National Natural Science Foundation of China (grant number 31490602), the National Basic Research Program (973) of China (grant number 2014CB542700), the National Natural Sciences Foundation of China (grants number 31372467 and 31225027), and the Key Technology R&D Program of China (grant number 2015BAD12B02).
REFERENCES
- 1.Wensvoort G, Terpstra C, Pol JM, ter Laak EA, Bloemraad M, de Kluyver EP, Kragten C, van Buiten L, den Besten A, Wagenaar F, Broekhuijsen JM, Moonen PLJM, Zetstra T, de Boer EA, Tibben HJ, de Jong MF, van ‘t Veld P, Greenland GJR, van Gennep JA, Voets MT, Verheijden JHM, Braamskamp J. 1991. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet Q 13:121–130. doi: 10.1080/01652176.1991.9694296. [DOI] [PubMed] [Google Scholar]
- 2.Mengeling WL, Lager KM, Vorwald AC. 2000. The effect of porcine parvovirus and porcine reproductive and respiratory syndrome virus on porcine reproductive performance. Anim Reprod Sci 60-61:199–210. doi: 10.1016/S0378-4320(00)00135-4. [DOI] [PubMed] [Google Scholar]
- 3.Rossow KD. 1998. Porcine reproductive and respiratory syndrome. Vet Pathol 35:1–20. doi: 10.1177/030098589803500101. [DOI] [PubMed] [Google Scholar]
- 4.Fang Y, Snijder EJ. 2010. The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res 154:61–76. doi: 10.1016/j.virusres.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brar MS, Shi M, Hui RK, Leung FC. 2014. Genomic evolution of porcine reproductive and respiratory syndrome virus (PRRSV) isolates revealed by deep sequencing. PLoS One 9:e88807. doi: 10.1371/journal.pone.0088807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan S, Murtaugh MP, Schumann FA, Mickelson D, Faaberg KS. 2004. Characterization of heteroclite subgenomic RNAs associated with PRRSV infection. Virus Res 105:75–87. doi: 10.1016/j.virusres.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 7.Beura LK, Sarkar SN, Kwon B, Subramaniam S, Jones C, Pattnaik AK, Osorio FA. 2010. Porcine reproductive and respiratory syndrome virus nonstructural protein 1beta modulates host innate immune response by antagonizing IRF3 activation. J Virol 84:1574–1584. doi: 10.1128/JVI.01326-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Z, Chen Z, Lawson SR, Fang Y. 2010. The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism functions. J Virol 84:7832–7846. doi: 10.1128/JVI.00217-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wills RW, Doster AR, Galeota JA, Sur JH, Osorio FA. 2003. Duration of infection and proportion of pigs persistently infected with porcine reproductive and respiratory syndrome virus. J Clin Microbiol 41:58–62. doi: 10.1128/JCM.41.1.58-62.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lager KM, Mengeling WL, Brockmeier SL. 1999. Evaluation of protective immunity in gilts inoculated with the NADC-8 isolate of porcine reproductive and respiratory syndrome virus (PRRSV) and challenge-exposed with an antigenically distinct PRRSV isolate. Am J Vet Res 60:1022–1027. [PubMed] [Google Scholar]
- 11.Zuckermann FA, Garcia EA, Luque ID, Christopher-Hennings J, Doster A, Brito M, Osorio F. 2007. Assessment of the efficacy of commercial porcine reproductive and respiratory syndrome virus (PRRSV) vaccines based on measurement of serologic response, frequency of gamma-IFN-producing cells and virological parameters of protection upon challenge. Vet Microbiol 123:69–85. doi: 10.1016/j.vetmic.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Meng XJ. 2000. Heterogeneity of porcine reproductive and respiratory syndrome virus: implications for current vaccine efficacy and future vaccine development. Vet Microbiol 74:309–329. doi: 10.1016/S0378-1135(00)00196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osorio FA, Galeota JA, Nelson E, Brodersen B, Doster A, Wills R, Zuckermann F, Laegreid WW. 2002. Passive transfer of virus-specific antibodies confers protection against reproductive failure induced by a virulent strain of porcine reproductive and respiratory syndrome virus and establishes sterilizing immunity. Virology 302:9–20. doi: 10.1006/viro.2002.1612. [DOI] [PubMed] [Google Scholar]
- 14.Lopez OJ, Osorio FA. 2004. Role of neutralizing antibodies in PRRSV protective immunity. Vet Immunol Immunopathol 102:155–163. doi: 10.1016/j.vetimm.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Lopez OJ, Oliveira MF, Garcia EA, Kwon BJ, Doster A, Osorio FA. 2007. Protection against porcine reproductive and respiratory syndrome virus (PRRSV) infection through passive transfer of PRRSV-neutralizing antibodies is dose dependent. Clin Vaccine Immunol 14:269–275. doi: 10.1128/CVI.00304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vezina SA, Loemba H, Fournier M, Dea S, Archambault D. 1996. Antibody production and blastogenic response in pigs experimentally infected with porcine reproductive and respiratory syndrome virus. Can J Vet Res 60:94–99. [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao F, Fang L, Wang D, Song T, Wang T, Xin Y, Chen H, Xiao S. 2016. SILAC-based quantitative proteomic analysis of secretome of Marc-145 cells infected with porcine reproductive and respiratory syndrome virus. Proteomics 16:2678–2687. doi: 10.1002/pmic.201500486. [DOI] [PubMed] [Google Scholar]
- 18.Montaner-Tarbes S, Borras FE, Montoya M, Fraile L, Del Portillo HA. 2016. Serum-derived exosomes from non-viremic animals previously exposed to the porcine respiratory and reproductive virus contain antigenic viral proteins. Vet Res 47:59. doi: 10.1186/s13567-016-0345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thery C, Zitvogel L, Amigorena S. 2002. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 20.Thery C. 2011. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep 3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathivanan S, Simpson RJ. 2009. ExoCarta: a compendium of exosomal proteins and RNA. Proteomics 9:4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- 22.Thery C, Duban L, Segura E, Veron P, Lantz O, Amigorena S. 2002. Indirect activation of naive CD4+ T cells by dendritic cell-derived exosomes. Nat Immunol 3:1156–1162. doi: 10.1038/ni854. [DOI] [PubMed] [Google Scholar]
- 23.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 24.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky AM, Breakefield XO. 2008. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pelchen-Matthews A, Raposo G, Marsh M. 2004. Endosomes, exosomes and Trojan viruses. Trends Microbiol 12:310–316. doi: 10.1016/j.tim.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Narayanan A, Iordanskiy S, Das R, Van Duyne R, Santos S, Jaworski E, Guendel I, Sampey G, Dalby E, Iglesias-Ussel M, Popratiloff A, Hakami R, Kehn-Hall K, Young M, Subra C, Gilbert C, Bailey C, Romerio F, Kashanchi F. 2013. Exosomes derived from HIV-1-infected cells contain trans-activation response element RNA. J Biol Chem 288:20014–20033. doi: 10.1074/jbc.M112.438895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bukong TN, Momen-Heravi F, Kodys K, Bala S, Szabo G. 2014. Exosomes from hepatitis C infected patients transmit HCV infection and contain replication competent viral RNA in complex with Ago2-miR122-HSP90. PLoS Pathog 10:e1004424. doi: 10.1371/journal.ppat.1004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rider MA, Hurwitz SN, Meckes DG Jr. 2016. ExtraPEG: a polyethylene glycol-based method for enrichment of extracellular vesicles. Sci Rep 6:23978. doi: 10.1038/srep23978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kappes MA, Miller CL, Faaberg KS. 2013. Highly divergent strains of porcine reproductive and respiratory syndrome virus incorporate multiple isoforms of nonstructural protein 2 into virions. J Virol 87:13456–13465. doi: 10.1128/JVI.02435-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramakrishnaiah V, Thumann C, Fofana I, Habersetzer F, Pan Q, de Ruiter PE, Willemsen R, Demmers JA, Stalin Raj V, Jenster G, Kwekkeboom J, Tilanus HW, Haagmans BL, Baumert TF, van der Laan LJ. 2013. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci U S A 110:13109–13113. doi: 10.1073/pnas.1221899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y, Han Q, Hou Z, Zhang C, Tian Z, Zhang J. 2017. Exosomes mediate hepatitis B virus (HBV) transmission and NK-cell dysfunction. Cell Mol Immunol 14:465–475. doi: 10.1038/cmi.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calvert JG, Slade DE, Shields SL, Jolie R, Mannan RM, Ankenbauer RG, Welch SK. 2007. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J Virol 81:7371–7379. doi: 10.1128/JVI.00513-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu BX, Clarke CJ, Hannun YA. 2010. Mammalian neutral sphingomyelinases: regulation and roles in cell signaling responses. Neuromolecular Med 12:320–330. doi: 10.1007/s12017-010-8120-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. 1987. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem 262:9412–9420. [PubMed] [Google Scholar]
- 35.Wolfers J, Lozier A, Raposo G, Regnault A, Thery C, Masurier C, Flament C, Pouzieux S, Faure F, Tursz T, Angevin E, Amigorena S, Zitvogel L. 2001. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med 7:297–303. doi: 10.1038/85438. [DOI] [PubMed] [Google Scholar]
- 36.Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, Ratajczak MZ. 2006. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia 20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 37.Colombo M, Raposo G, Thery C. 2014. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 38.Meckes DG Jr, Raab-Traub N. 2011. Microvesicles and viral infection. J Virol 85:12844–12854. doi: 10.1128/JVI.05853-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alenquer M, Amorim MJ. 2015. Exosome biogenesis, regulation, and function in viral infection. Viruses 7:5066–5083. doi: 10.3390/v7092862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pegtel DM, van de Garde MD, Middeldorp JM. 2011. Viral miRNAs exploiting the endosomal-exosomal pathway for intercellular cross-talk and immune evasion. Biochim Biophys Acta 1809:715–721. doi: 10.1016/j.bbagrm.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Thery C, Amigorena S, Raposo G, Clayton A. 2006. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3:Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 42.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 43.Bess JW Jr, Gorelick RJ, Bosche WJ, Henderson LE, Arthur LO. 1997. Microvesicles are a source of contaminating cellular proteins found in purified HIV-1 preparations. Virology 230:134–144. doi: 10.1006/viro.1997.8499. [DOI] [PubMed] [Google Scholar]
- 44.Gluschankof P, Mondor I, Gelderblom HR, Sattentau QJ. 1997. Cell membrane vesicles are a major contaminant of gradient-enriched human immunodeficiency virus type-1 preparations. Virology 230:125–133. doi: 10.1006/viro.1997.8453. [DOI] [PubMed] [Google Scholar]
- 45.Chahar HS, Bao X, Casola A. 2015. Exosomes and their role in the life cycle and pathogenesis of RNA viruses. Viruses 7:3204–3225. doi: 10.3390/v7062770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raab-Traub N, Dittmer DP. 2017. Viral effects on the content and function of extracellular vesicles. Nat Rev Microbiol 15:559–572. doi: 10.1038/nrmicro.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Music N, Gagnon CA. 2010. The role of porcine reproductive and respiratory syndrome (PRRS) virus structural and non-structural proteins in virus pathogenesis. Anim Health Res Rev 11:135–163. doi: 10.1017/S1466252310000034. [DOI] [PubMed] [Google Scholar]
- 48.Pleet ML, Mathiesen A, DeMarino C, Akpamagbo YA, Barclay RA, Schwab A, Iordanskiy S, Sampey GC, Lepene B, Nekhai S, Aman MJ, Kashanchi F. 2016. Ebola VP40 in exosomes can cause immune cell dysfunction. Front Microbiol 7:1765. doi: 10.3389/fmicb.2016.01765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sirinarumitr T, Zhang Y, Kluge JP, Halbur PG, Paul PS. 1998. A pneumo-virulent United States isolate of porcine reproductive and respiratory syndrome virus induces apoptosis in bystander cells both in vitro and in vivo. J Gen Virol 79(Pt 12):2989–2995. doi: 10.1099/0022-1317-79-12-2989. [DOI] [PubMed] [Google Scholar]
- 50.Sur JH, Doster AR, Osorio FA. 1998. Apoptosis induced in vivo during acute infection by porcine reproductive and respiratory syndrome virus. Vet Pathol 35:506–514. doi: 10.1177/030098589803500605. [DOI] [PubMed] [Google Scholar]
- 51.Luo R, Fang L, Jiang Y, Jin H, Wang Y, Wang D, Chen H, Xiao S. 2011. Activation of NF-kappaB by nucleocapsid protein of the porcine reproductive and respiratory syndrome virus. Virus Genes 42:76–81. doi: 10.1007/s11262-010-0548-6. [DOI] [PubMed] [Google Scholar]
- 52.Sagong M, Lee C. 2011. Porcine reproductive and respiratory syndrome virus nucleocapsid protein modulates interferon-beta production by inhibiting IRF3 activation in immortalized porcine alveolar macrophages. Arch Virol 156:2187–2195. doi: 10.1007/s00705-011-1116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lundy SK, Klinker MW, Fox DA. 2015. Killer B lymphocytes and their fas ligand positive exosomes as inducers of immune tolerance. Front Immunol 6:122. doi: 10.3389/fimmu.2015.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dreux M, Garaigorta U, Boyd B, Decembre E, Chung J, Whitten-Bauer C, Wieland S, Chisari FV. 2012. Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe 12:558–570. doi: 10.1016/j.chom.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li J, Liu K, Liu Y, Xu Y, Zhang F, Yang H, Liu J, Pan T, Chen J, Wu M, Zhou X, Yuan Z. 2013. Exosomes mediate the cell-to-cell transmission of IFN-alpha-induced antiviral activity. Nat Immunol 14:793–803. doi: 10.1038/ni.2647. [DOI] [PubMed] [Google Scholar]
- 56.Nguyen DG, Booth A, Gould SJ, Hildreth JE. 2003. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J Biol Chem 278:52347–52354. doi: 10.1074/jbc.M309009200. [DOI] [PubMed] [Google Scholar]
- 57.Arenaccio C, Anticoli S, Manfredi F, Chiozzini C, Olivetta E, Federico M. 2015. Latent HIV-1 is activated by exosomes from cells infected with either replication-competent or defective HIV-1. Retrovirology 12:87. doi: 10.1186/s12977-015-0216-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Longatti A. 2015. The dual role of exosomes in hepatitis A and C virus transmission and viral immune activation. Viruses 7:6707–6715. doi: 10.3390/v7122967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Silvas JA, Popov VL, Paulucci-Holthauzen A, Aguilar PV. 2015. Extracellular vesicles mediate receptor-independent transmission of novel tick-borne bunyavirus. J Virol 90:873–886. doi: 10.1128/JVI.02490-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verweij FJ, van Eijndhoven MA, Hopmans ES, Vendrig T, Wurdinger T, Cahir-McFarland E, Kieff E, Geerts D, van der Kant R, Neefjes J, Middeldorp JM, Pegtel DM. 2011. LMP1 association with CD63 in endosomes and secretion via exosomes limits constitutive NF-kappaB activation. EMBO J 30:2115–2129. doi: 10.1038/emboj.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chugh PE, Sin SH, Ozgur S, Henry DH, Menezes P, Griffith J, Eron JJ, Damania B, Dittmer DP. 2013. Systemically circulating viral and tumor-derived microRNAs in KSHV-associated malignancies. PLoS Pathog 9:e1003484. doi: 10.1371/journal.ppat.1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mao L, Wu J, Shen L, Yang J, Chen J, Xu H. 2016. Enterovirus 71 transmission by exosomes establishes a productive infection in human neuroblastoma cells. Virus Genes 52:189–194. doi: 10.1007/s11262-016-1292-3. [DOI] [PubMed] [Google Scholar]
- 64.Cosset FL, Dreux M. 2014. HCV transmission by hepatic exosomes establishes a productive infection. J Hepatol 60:674–675. doi: 10.1016/j.jhep.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 65.Verweij FJ, van Eijndhoven MA, Middeldorp J, Pegtel DM. 2013. Analysis of viral microRNA exchange via exosomes in vitro and in vivo. Methods Mol Biol 1024:53–68. doi: 10.1007/978-1-62703-453-1_5. [DOI] [PubMed] [Google Scholar]
- 66.Bi J, Song S, Fang L, Wang D, Jing H, Gao L, Cai Y, Luo R, Chen H, Xiao S. 2014. Porcine reproductive and respiratory syndrome virus induces IL-1beta production depending on TLR4/MyD88 pathway and NLRP3 inflammasome in primary porcine alveolar macrophages. Mediators Inflamm 2014:403515. doi: 10.1155/2014/403515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li B, Fang L, Liu S, Zhao F, Jiang Y, He K, Chen H, Xiao S. 2010. The genomic diversity of Chinese porcine reproductive and respiratory syndrome virus isolates from 1996 to 2009. Vet Microbiol 146:226–237. doi: 10.1016/j.vetmic.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 68.Xiao S, Chen H, Fang L, Liu C, Zhang H, Jiang Y, Hong W. 2004. Comparison of immune responses and protective efficacy of suicidal DNA vaccine and conventional DNA vaccine encoding glycoprotein C of pseudorabies virus in mice. Vaccine 22:345–351. doi: 10.1016/j.vaccine.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 69.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. 2009. Universal sample preparation method for proteome analysis. Nat Methods 6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 70.Ostrowski M, Galeota JA, Jar AM, Platt KB, Osorio FA, Lopez OJ. 2002. Identification of neutralizing and nonneutralizing epitopes in the porcine reproductive and respiratory syndrome virus GP5 ectodomain. J Virol 76:4241–4250. doi: 10.1128/JVI.76.9.4241-4250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]