Abstract
Efficient delivery of siRNA remains one of the primary challenges of RNA interference therapy. PEGylated polycationic carriers have been widely used for the condensation of DNA and RNA molecules into complex-core micelles. The PEG corona of such nanoparticles can significantly improve their colloidal stability in serum, but PEGylation of the carriers also reduces their condensation capacity, hindering the generation of micellar particles with sufficient complex stability. This presents a particularly significant challenge for packaging siRNA into complex micelles, as it has a much smaller size and more rigid chain structure than DNA plasmids. Here, we report a new method to enhance the condensation of siRNA with PEGylated linear polyethylenimine (lPEI) using organic solvent and to prepare smaller siRNA nanoparticles with a more extended PEG corona and consequently higher stability. As a proof of principle, we have demonstrated the improved gene knockdown efficiency resulting from the reduced siRNA micelle size in mouse liver following intravenous administration.
Keywords: gene therapy, siRNA delivery, nanoparticle, solvent polarity, size control
Introduction
RNA interference (RNAi) has been demonstrated to be a potent and highly specific post-translational gene regulation process [1]. As it specifically targets the gene of interest through the use of short interfering RNA (siRNA), RNAi holds great potential as a therapeutic agent for the treatment of numerous disorders [2, 3]. However, delivery of naked siRNA molecules via intravenous injection has failed to yield significant gene knockdown due to their poor pharmacokinetic profile, resulting from a high susceptibility to nuclease degradation and rapid renal clearance [3]. Thus, a critical challenge in realizing the full therapeutic potential of siRNA-mediated gene knockdown centers on the development of a safe and effective delivery system. Among the currently explored delivery strategies [4], polycationic nanoparticles have gained significant attention owing to their versatility and ease of formulation [5]. Polyethylenimine (PEI) is the most commonly used polycationic carrier, due to its high buffering capacity that facilitates endosomal escape of nanoparticles and siRNA into the cytoplasm [6]. However, PEI/siRNA nanoparticles typically carry positive residual charges on their surface, which lead to significant aggregation in the presence of serum proteins, thereby greatly reducing their efficacy in vivo [7]. Furthermore, they often elicit a high degree of inflammatory responses and toxicity, and are prone to clearance by macrophages following systemic administration, particularly for branched PEI [8, 9].
Decorating the particle surface with hydrophilic polyethylene glycol (PEG) has often been employed as an effective strategy to alleviate aggregation, reduce opsonization and inflammation response, and prolong nanoparticle circulation time [10]. PEGylation can be achieved by grafting PEG chains onto the polycation backbone. The resulting PEGylated polycations retain their ability to complex with siRNA, forming micellar nanoparticles with a polycation/siRNA core and a PEG corona, where the steric shielding effect of the corona has been demonstrated to lessen aggregation of PEI-g-PEG/siRNA nanoparticles [11, 12]. However, whereas a higher degree of PEG grafting density favors the colloidal stability and biocompatibility of the micelles, it reduces the RNA-condensation capacity of the PEI-g-PEG carrier [13]. Thus, it is a considerable challenge to balance PEI-g-PEG/siRNA complex stability with colloidal stability and compatibility.
Materials and Methods
Experimental Methods
Preparation of lPEI-g-PEG/siRNA micelles
The siRNA (1.33 µg, Qiagen, Valencia, CA) was first dissolved in 50 µL of DI water or a 7:3 (v/v) DMF–water mixture and then added to an equal volume of lPEI-g-PEG polymer solution at an N/P ratio of 20 prepared in the same mixing solvent. The mixture was vortexed and then incubated for 30 min at room temperature before further characterization. Crosslinked micelles were prepared and purified according to a protocol that we established previously [14]. Micelle purification and characterization methods are provided in the Supplementary Information.
In vitro cell uptake and gene silencing of lPEI-g-PEG/siRNA micelles
HepG2 cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 U/mL Penicillin/100 µg/mL Streptomycin at 37°C and 5% CO2. At 24 h prior to the experiment, cells were seeded in 24-well plates at a density of 5 × 104 cells/well. For cell-uptake studies, micelles were prepared with Alexa Fluor 488-modified siRNA (Qiagen, Valencia, CA). An aliquot of 50 µL micelles equivalent to 100 nM siRNA was added to each well followed by 4 h incubation at 37°C, after which the cells were washed with PBS, trypsinized, and fixed with 2% paraformaldehyde. Fluorescence associated with individual cells was analyzed with a BD FACSCalibur flow cytometer (BD Biosciences, San Jose, CA) fitted with a 488-nm excitation source and detected using a 515–545 nm filter. A minimum of 10,000 events per sample was collected for analysis. Gene-silencing studies were performed using a previously reported protocol [15].
In vivo gene knockdown efficiency of siRNA micelles via intravenous administration
Animal studies were conducted under a protocol that was approved by the Johns Hopkins School of Medicine Institutional Animal Care and Use Committee (IACUC # RA09A447). A liver-specific gene-knockdown model was developed by modification of previously published protocols [16, 17]. Wistar rats (female, 6–8 weeks, 200–300 g) were transfected with a firefly luciferase plasmid DNA (20 µg DNA in PBS with a volume corresponding to 9 vol/wt% of the rat’s body weight) via hydrodynamic infusion administered through the tail vein over 15 s according to a published procedure [18]. After 5 days, micelles containing 80 µg siRNA in 1 mL PBS were injected via the tail vein. At 24 and 48 h, rats were anesthetized and given 1 mL of D-luciferin solution (i.p. 30 mg/mL). The rat was then imaged on an IVIS Spectrum Imaging System. The bioluminescence signal was collected for 1 min, and the level of luciferase expression was expressed as the total photon count per section in the region of interest and was normalized to PBS control to determine knockdown efficiency.
Statistical Analysis
All data were expressed as mean ± SD unless otherwise noted. Statistical comparisons were carried out using a one-way analysis of variance (ANOVA) followed by Turkey’s post-hoc test for groups with equal variance or Games–Howell test for groups with unequal variance (SPSS software, version 21, IBM Inc., Armonk, NY). All data were considered to be significant at p < 0.05.
Computational Methods
Modeling of lPEI-g-PEG/siRNA micelles
Molecular dynamics simulations were performed using coarse-grained models. lPEI-g-PEG was represented as a bead–spring polymer model with bead size 7.35 Å and charge density 35%. The siRNA molecule was coarse-grained as a 24-bead rigid body using the VMD Shape-Based Coarse-Graining (SBCG) tool [19, 20], based on a 22-bp RNA molecule isolated from the Protein Data Bank file 2F8S [21]. To increase the computational efficiency, we scaled down each coarse-grained model to one fourth of its original length. The solvent was simulated implicitly using a Langevin thermostat, and different solvent compositions were represented through variation of the attractive strength of a Lennard-Jones potential. Monovalent counterions were included to maintain global charge neutrality, and electrostatic interactions were computed using the Ewald method. To accelerate the dissociation and reformation of aggregates (to improve sampling of aggregate conformations), we employed the parallel tempering method [22], in which 24 copies of the same system were simulated in parallel, at closely spaced temperatures. This approach exploits the larger degree of fluctuations at higher temperatures to provide pathways that permit the simulation at the original temperature to transition between different states of low free energy that are separated by free-energy barriers.
Results and Discussion
siRNA condensation and size control
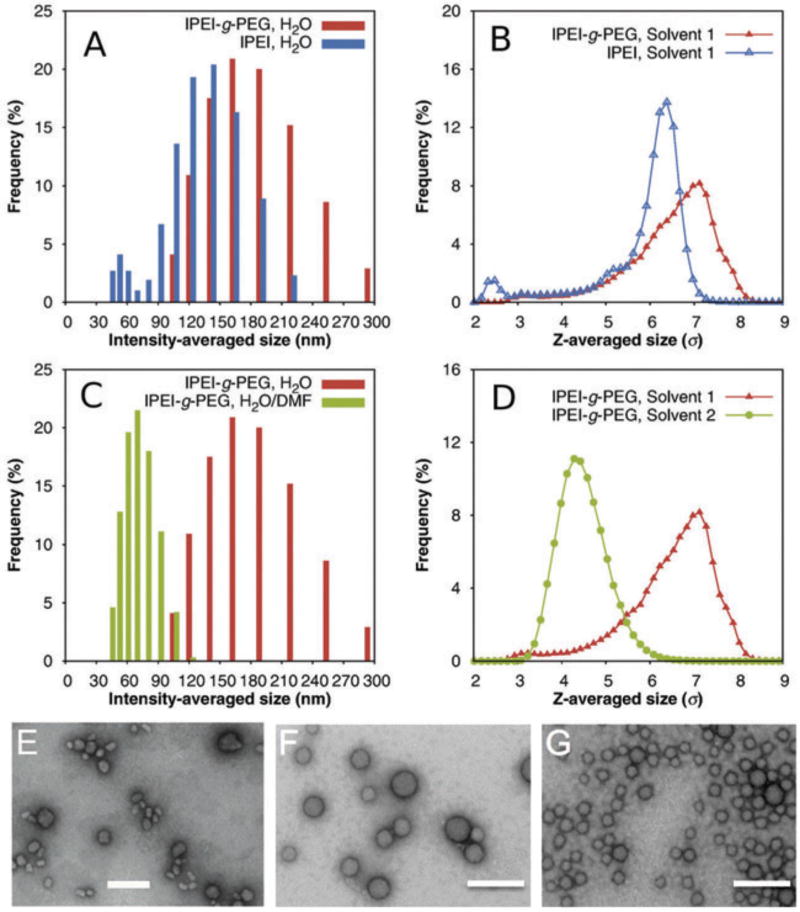
Our new approach for condensing siRNA employs a PEGylated linear PEI (lPEI) carrier, lPEI-g-PEG, which was synthesized by grafting PEG (Mn = 10 kDa) to the backbone of lPEI (Mn = 17 kDa; see Supplementary Material for copolymer synthesis and characterization). We prepared an lPEI-g-PEG copolymer with an average of 4.6 PEG grafts per lPEI, and identified a minimal N/P ratio of 20 for complete siRNA condensation by agarose gel retardation assay (Fig. S1). For all following experiments, we prepared siRNA micelles by mixing equal volumes of 200 µg/mL of lPEI-g-PEG solution and 20 µg/mL of siRNA solution (corresponding to an N/P ratio of 20) at room temperature. The intensity-averaged diameter of these lPEI-g-PEG/siRNA micelles prepared in water was 117 ± 2.3 nm, as measured by dynamic light scattering (DLS; Fig. 1, A and F). On the other hand, lPEI/siRNA nanoparticles exhibited a slightly smaller diameter of 96.8 ± 10.3 nm (Fig. 1, A and E). Whereas this size difference may be attributed to the PEG corona, it is nevertheless remarkable given our observations for plasmid DNA, where condensation with block copolymers significantly reduced the size of the micelles compared to condensation with polyelectrolytes alone [14].
FIG. 1.
Size distribution of lPEI-g-PEG/siRNA micelles in different solvents. (A) Size distribution of lPEI-g-PEG/siRNA micelles and lPEI/siRNA nanoparticles prepared in water, as determined by dynamic light scattering; (B) Simulation results for size distributions of lPEI-g-PEG/siRNA micelles and lPEI/siRNA nanoparticles in water; (C) Size distribution of lPEI-g-PEG/siRNA micelles prepared in pure water and 7:3 (v/v) DMF–water mixture; (D) Simulation results for size distribution of lPEI-g-PEG/siRNA micelles in pure water (labeled “Solvent 1”) and 7:3 (v/v) DMF–water mixture (labeled “Solvent 2”); (E–G) TEM images of lPEI/siRNA nanoparticles (E), lPEI-g-PEG/siRNA micelles prepared in pure water (F), and lPEI-g-PEG/siRNA micelles prepared in DMF–water mixture (G), respectively. All scale bars represent 200 nm.
To understand this apparent discrepancy, we performed molecular (MD) dynamics simulations of complexation between siRNA and lPEI-g-PEG copolymer. We emphasize that both the time scale and the number of particles involved in the complexation processes make coarse-graining (as described in the Supplementary Material) imperative. Whereas this approach omits atomistic details, prior work has shown it to be highly suitable for providing meaningful insight into the underlying mechanisms [14, 23]. In addition to the electrostatic interactions between siRNA and lPEI (taken into account via Ewald summation [24–26]), and the lPEI–solvent and PEG–solvent interactions [14], we also incorporated the interaction between lPEI and PEG arising from direct hydrogen bonding and from hydrogen bond bridges with water molecules, as lPEI is known to form strong inter- and intra-molecular hydrogen bonds in water [27]. Given that PEG not only has a molecular structure similar to lPEI, but also has a hydrogen bond acceptor in its repeat unit, we adopted the same effective interaction strength for PEG–lPEI as for lPEI–lPEI [27]. To compare to the intensity-averaged distribution of the hydrodynamic diameter measured by DLS, we report the z-averaged [28] radius of gyration of the lPEI-g-PEG/siRNA nanoparticles calculated in simulation (Fig. 1B). The modeling results indeed matched the experimental observation that the complexation of siRNA with lPEI-g-PEG copolymer results in a slight increase in particle size. This increase did not arise merely from the physical extent of the PEG corona: the average number of siRNA particles per micelle increased from 7.1 ± 1.3 to 9.4 ± 1.3. We hypothesized that this size increase can be attributed to the lPEI–PEG interaction. To test this conjecture, we performed a second set of simulations of the complexation of siRNA with lPEI-g-PEG copolymer, in which we artificially decreased the attractive interaction between lPEI and PEG (Fig. S4). We indeed observed that a weakening of the PEG–lPEI interaction significantly reduces the particle size.
Prompted by this observation, we aimed to condense lPEI-g-PEG/siRNA micelles into smaller nanoparticles by disrupting the interaction between lPEI and PEG mediated by hydrogen bonding with water molecules. We added dimethylformamide (DMF), a water-miscible solvent that has been shown to effectively reduce intermolecular hydrogen bonding in other polymeric micelle systems [29], to siRNA and lPEI-g-PEG solutions prior to micelle assembly. In a 7:3 (v/v) DMF–water mixture, we indeed observed a far smaller average particle size of 44.2 ± 6.6 nm (Fig. 1, C and G). This size reduction of micelles in response to a decrease in solvent polarity was confirmed by MD simulations (Fig. 1D). Arguably, the polarity reduction also leads to a decrease in siRNA solubility [30], which in turn could have resulted in stronger condensation and a growth in aggregate size [14]. We conclude that this effect is overshadowed by the reduction of PEG–lPEI and lPEI–lPEI hydrogen bonding which in turn permits the PEG corona to provide steric shielding.
Preservation of nanoparticle size following solvent removal
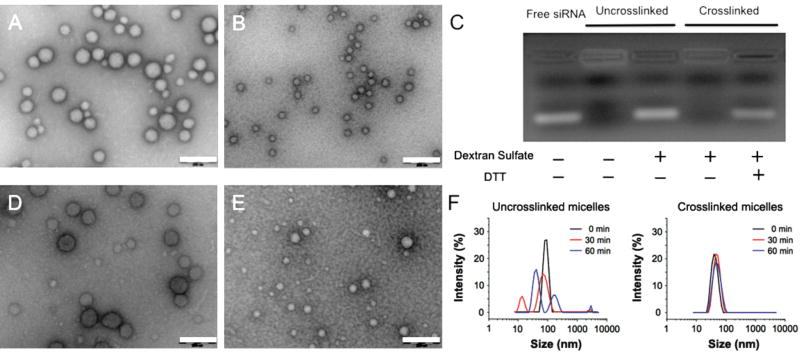
Whereas DMF as a co-solvent can significantly reduce particle size, it needs to be removed prior to nanoparticle transfection experiments in vitro or in vivo. However, DMF removal by dialysis resulted in nanoparticle swelling (data not shown). To preserve the nanoparticle size in aqueous media upon DMF removal, we employed a reversible disulfide crosslinking scheme previously tested in DNA/polymer nanoparticles [14]. We introduced thiol groups to the lPEI block and used the purified thiolated lPEI-g-PEG to condense siRNA using the protocol described above. Crosslinking was initiated by aerial oxidation over a 48-h incubation at room temperature followed by dialysis to remove the DMF. TEM micrographs (Fig. 2, A and B), confirm that no appreciable change in particle size or morphology occurred following solvent removal. The crosslinking was verified by gel electrophoresis (Fig. 2C): without disulfide crosslinks, nanoparticles released siRNA upon challenge with an excess amount of dextran sulfate, a polyanion that can effectively compete with siRNA to complex with lPEI. Conversely, crosslinked micelles released siRNA under the same conditions only after the disulfide bonds were reduced.
FIG. 2.
Preservation of size of lPEI-g-PEG/siRNA micelles via reversible disulfide crosslinking. (A–B) TEM images of crosslinked nanoparticles initially prepared in pure water (A) and in 7:3 (v/v) DMF–water mixture (imaged after removal of solvent) (B); (C) siRNA release from uncrosslinked and crosslinked lPEI-g-PEG/siRNA micelles in the presence of dextran sulfate and 50 mM dithiothreitol (DTT) in water; (D–E) TEM images of crosslinked micelles initially prepared in pure water (D) and 7:3 (v/v) DMF–water mixture (E), respectively, following 1 h incubation with 10% (v/v) FBS. (F) Size distributions of uncrosslinked and crosslinked lPEI-g-PEG/siRNA micelles following incubation with 0.15 M NaCl for 1 h. All scale bars represent 200 nm.
The PEG corona of lPEI-g-PEG/siRNA micelles significantly reduced the nanoparticle zeta potential from +30 mV to about +5 mV when condensed in water. Condensation in DMF–water (7:3, v/v) mixture followed by crosslinking and removal of solvent, reduced the zeta potential even further to −8 mV (Fig. S2), likely due to the increase of the PEG density at the surface and reduction of imines on the complex core that reacted with the crosslinkers. The near-neutral surface charge on these micelles enhances their colloidal stability in physiological media through suppression of serum protein-mediated agglomeration. Indeed, these crosslinked micelles showed no significant change in particle size in 10% serum (Fig. 2, D and E). Owing to the covalent crosslinks, these micelles also exhibited high complex stability; they did not show any size change when incubated in the presence of 150 mM sodium chloride (Fig. 2F). On the other hand, uncrosslinked lPEI-g-PEG/siRNA micelles were destabilized within 30 min of salt challenge.
siRNA delivery and knockdown efficiency
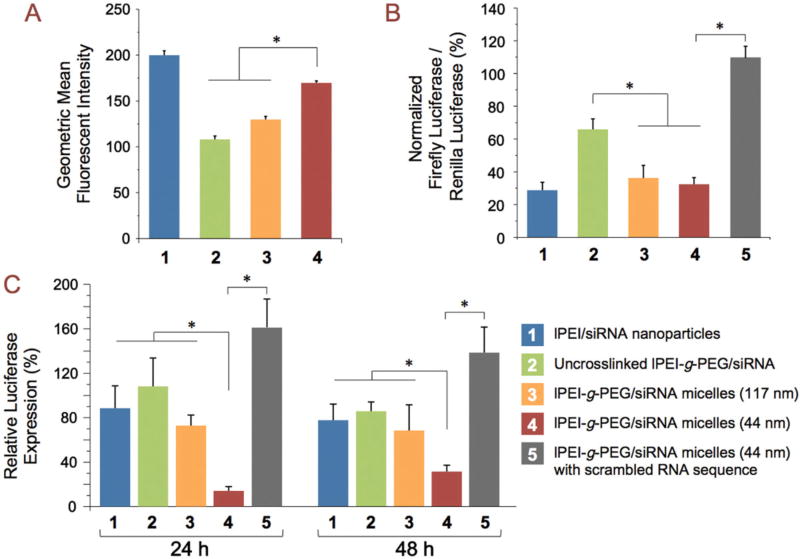
We anticipated a smaller average particle size to result in more efficient cellular uptake and transfection. To confirm this, we first measured the cellular uptake efficiency and gene knockdown efficiency in vitro. We also prepared 117-nm micelles that were crosslinked similarly to the 44-nm micelles to exclude differences resulting from the crosslinking. The crosslinked 44-nm micelles displayed 30% and 57% increase of uptake in a hepatocellular carcinoma cell line (HepG2) compared to the crosslinked and uncrosslinked 117-nm micelles, respectively (Fig. 3A). In vitro gene knockdown experiments showed that both the crosslinked 117-nm and 44-nm micelles achieved about 60% decrease in targeted protein expression, an efficiency similar to that of lPEI/siRNA nanoparticles. The uncrosslinked micelles only showed 30% knockdown efficiency, likely due to their limited stability in cell culture media (Fig. 3B). In addition, all tested particles maintained a high cell metabolic activity (>80% compared to untreated cells, Fig. S3). As an in vivo proof of concept, we assessed the effect of reduced nanoparticle size on gene-knockdown efficiency in the rat liver following intravenous administration (Fig. 3C). We first established high luciferase expression in the liver via hydrodynamic infusion of luciferase plasmid DNA [31]. After the transgene expression level stabilized at 5 d post-infusion, lPEI-g-PEG/siRNA micelles or lPEI/siRNA nanoparticles containing 80 µg luciferase siRNA were administered via tail vein injection. The 44-nm micelles displayed 85% and 70% efficiencies of transgene knockdown in the liver at 24 h and 48 h after injection, respectively. In contrast, the 117-nm micelles only showed ~30% knockdown at both 24 h and 48 h (p < 0.05). Control nanoparticles delivering a non-targeting sequence also did not show any reduction in luciferase expression. This significant increase in gene knockdown efficiency for the smaller nanoparticles may be attributed to improved nanoparticle deposition and increased cellular uptake of the smaller nanoparticles. On the other hand, the uncrosslinked micelles and lPEI/siRNA nanoparticles did not show significant knockdown at 24 h and yielded a low level (~15%) of transgene knockdown at 48 h after injection, suggesting that nanoparticle stability in medium is crucial to siRNA delivery in vivo. It is important to note that the gene knockdown activity mediated by the 44-nm siRNA nanoparticles is among the highest obtained via intravenous injection at such a relatively low siRNA dose (~0.4 mg/kg body weight), without employing any active targeting strategy [32–34].
FIG. 3.
Size-dependent transfection efficiency of lPEI-g-PEG/siRNA micelles. Groups 2 and 3 compare uncrosslinked lPEI-g-PEG/siRNA micelles (117 nm, Group 2) with crosslinked lPEI-g-PEG/siRNA micelles (117 nm, Group 3). Size comparison was conducted between Group 3 and Group 4 (crosslinked lPEI-g-PEG/siRNA micelles, 44 nm). Group 5 was the negative control using crosslinked lPEI-g-PEG/siRNA micelles with a scrambled RNA sequence (44 nm). Group 1 was the positive control using lPEI/siRNA nanoparticles. (A) In vitro cellular uptake of Alexa Fluor 488-labeled micelles in HepG2 cells. Bars represent mean ± SD (n = 3); (B) In vitro gene knockdown efficiency in HepG2 cells following 100 nM equivalent dose of siRNA. Bars represent mean ± SD (n = 3); (C) In vivo gene silencing in rat liver at 24 h and 48 h after administration of nanoparticles at a dose equivalent to 80 µg siRNA via tail vein injection. Bars indicate mean relative luciferease expression ± SD (n = 4–7). *p <0.05.
Conclusion
In conclusion, this study provides the first evidence of solvent-assisted condensation of lPEI-g-PEG/siRNA micelles to decrease nanoparticle size. By reducing solvent polarity, we decreased the average size of lPEI-g-PEG/siRNA micelles from 117 nm in water to 44 nm. Through molecular dynamics simulation we revealed the role of solvent quality and PEI–PEG hydrogen bonding in the assembly of lPEI-g-PEG/siRNA micelles. The micelle size was preserved after organic solvent removal by means of reversible disulfide crosslinking; we confirmed that the micelles maintained their size in water and physiological media. More importantly, our results have demonstrated size-dependent in vivo transfection efficiency following intravenous injection of the siRNA micelles in rats. The gene knockdown efficiency in rat liver achieved by the smaller siRNA micelles was significantly higher than for the larger micelles prepared from the same copolymer carrier. The condensation technique introduced here allows a simple and effective way to reduce siRNA particle size and provides a model platform for further study of the effect of particle size on in vivo cellular uptake, knockdown efficiency, biodistribution, and pharmacokinetics. As such, it can form the starting point for the development of an effective delivery system that harnesses the therapeutic potential of siRNA.
Supplementary Material
Acknowledgments
The authors acknowledge computing resources provided by the Quest High-Performance Computing Facility at Northwestern University. Funding for this study was provided by National Institute of Health grants R21EB015152 and U54CA151838 and National Science Foundation grants DMR-1310211 and DMR-1611076, as well as a pilot grant for the Institute of NanoBioTechnology at Johns Hopkins University.
Footnotes
Supplementary Material: Additional experimental methods, detailed computational model and methods, six figures, two tables, and references are available online.
Author Disclosure Statement: No competing financial interests exist.
References
- 1.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Bumcrot D, et al. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2(12):711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457(7228):426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khurana B, et al. siRNA Delivery Using Nanocarriers - An Efficient Tool for Gene Silencing. Curr Gene Ther. 2010;10(2):139–155. doi: 10.2174/156652310791111010. [DOI] [PubMed] [Google Scholar]
- 5.Howard KA. Delivery of RNA interference therapeutics using polycation-based nanoparticles. Adv Drug Deliv Rev. 2009;61(9):710–720. doi: 10.1016/j.addr.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Akinc A, et al. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7(5):657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 7.Nomoto T, et al. In situ quantitative monitoring of polyplexes and polyplex micelles in the blood circulation using intravital real-time confocal laser scanning microscopy. J Control Release. 2011;151(2):104–109. doi: 10.1016/j.jconrel.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 8.Owens DE, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int J Pharm. 2006;307(1):93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 9.Merkel OM, et al. In vitro and in vivo complement activation and related anaphylactic effects associated with polyethylenimine and polyethylenimine-graft-poly(ethylene glycol) block copolymers. Biomaterials. 2011;32(21):4936–42. doi: 10.1016/j.biomaterials.2011.03.035. [DOI] [PubMed] [Google Scholar]
- 10.Jokerst JV, et al. Nanoparticle PEGylation for imaging and therapy. Nanomedicine. 2011;6(4):715–728. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao S, et al. Influence of polyethylene glycol chain length on the physicochemical and biological properties of poly(ethylene imine)-graft-poly(ethylene glycol) block copolymer/SiRNA polyplexes. Bioconjug Chem. 2006;17(5):1209–1218. doi: 10.1021/bc060129j. [DOI] [PubMed] [Google Scholar]
- 12.Osada K, Christie RJ, Kataoka K. Polymeric micelles from poly(ethylene glycol)-poly(amino acid) block copolymer for drug and gene delivery. J R Soc Interface. 2009;6(Suppl 3):S325–39. doi: 10.1098/rsif.2008.0547.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merkel OM, et al. Nonviral siRNA delivery to the lung: investigation of PEG-PEI polyplexes and their in vivo performance. Mol Pharm. 2009;6(4):1246–60. doi: 10.1021/mp900107v. [DOI] [PubMed] [Google Scholar]
- 14.Jiang X, et al. Plasmid-Templated Shape Control of Condensed DNA-Block Copolymer Nanoparticles. Adv Mater. 2013;25(2):227–232. doi: 10.1002/adma.201202932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakanishi M, et al. Enhanced Stability and Knockdown Efficiency of Poly(ethylene glycol)-b-polyphosphoramidate/siRNA Micellar Nanoparticles by Co-condensation with Sodium Triphosphate. Pharm Res. 2011;28(7):1723–32. doi: 10.1007/s11095-011-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andaloussi SEL, et al. Design of a peptide-based vector, PepFect6, for efficient delivery of siRNA in cell culture and systemically in vivo. Nucleic Acids Res. 2011;39(9):3972–3987. doi: 10.1093/nar/gkq1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim SI, et al. Systemic and specific delivery of small interfering RNAs to the liver mediated by apolipoprotein A-I. Mol Ther. 2007;15(6):1145–1152. doi: 10.1038/sj.mt.6300168. [DOI] [PubMed] [Google Scholar]
- 18.Maruyama H, et al. High-level expression of naked DNA delivered to rat liver via tail vein injection. J Gene Med. 2002;4(3):333–341. doi: 10.1002/jgm.281. [DOI] [PubMed] [Google Scholar]
- 19.Arkhipov A, Freddolino PL, Schulten K. Stability and Dynamics of Virus Capsids Described by Coarse-Grained Modeling. Structure. 2006;14(12):1767–1777. doi: 10.1016/j.str.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Arkhipov A, Yin Y, Schulten K. Four-Scale Description of Membrane Sculpting by BAR Domains. Biophys J. 2008;95(6):2806–2821. doi: 10.1529/biophysj.108.132563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan Y-R, et al. A Potential Protein-RNA Recognition Event along the RISC-Loading Pathway from the Structure of A. aeolicus Argonaute with Externally Bound siRNA. Structure. 2006;14(10):1557–1565. doi: 10.1016/j.str.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frenkel D, Smit B. Understanding Molecular Simulations. 2. San Diego, USA: Academic Press; 2002. [Google Scholar]
- 23.Wei Z, et al. Simulation and Experimental Assembly of DNA–Graft Copolymer Micelles with Controlled Morphology. ACS Biomater. Sci. Eng. 2015;1(6):448–455. doi: 10.1021/acsbiomaterials.5b00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guáqueta C, Luijten E. Polyelectrolyte Condensation Induced by Linear Cations. Phys Rev Lett. 2007;99(13):138302. doi: 10.1103/PhysRevLett.99.138302. [DOI] [PubMed] [Google Scholar]
- 25.Hsiao P-Y, Luijten E. Salt-Induced Collapse and Reexpansion of Highly Charged Flexible Polyelectrolytes. Phys Rev Lett. 2006;97(14):148301. doi: 10.1103/PhysRevLett.97.148301. [DOI] [PubMed] [Google Scholar]
- 26.Sanders LK, et al. Control of electrostatic interactions between F-actin and genetically modified lysozyme in aqueous media. Proc Natl Acad Sci USA. 2007;104(41):15994–15999. doi: 10.1073/pnas.0705898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gembitskii PA, et al. Properties of linear polyethylene imine and its oligomers. Polymer Science USSR. 1978;20(11):2932–2940. [Google Scholar]
- 28.Hallett FR, Watton J, Krygsman P. Vesicle sizing: Number distributions by dynamic light scattering. Biophys J. 1991;59(2):357–362. doi: 10.1016/S0006-3495(91)82229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen D, Jiang M. Strategies for Constructing Polymeric Micelles and Hollow Spheres in Solution via Specific Intermolecular Interactions. Acc Chem Res. 2005;38(6):494–502. doi: 10.1021/ar040113d. [DOI] [PubMed] [Google Scholar]
- 30.Jacobson GB, et al. Nanoparticle formation of organic compounds with retained biological activity. J Pharm Sci. 2010;99(6):2750–2755. doi: 10.1002/jps.22035. [DOI] [PubMed] [Google Scholar]
- 31.Herweijer H, Wolff JA. Gene therapy progress and prospects: Hydrodynamic gene delivery. Gene Ther. 2007;14(2):99–107. doi: 10.1038/sj.gt.3302891. [DOI] [PubMed] [Google Scholar]
- 32.Akinc A, et al. Development of Lipidoid-siRNA Formulations for Systemic Delivery to the Liver. Mol Ther. 2009;17(5):872–879. doi: 10.1038/mt.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siegwart DJ, et al. Combinatorial synthesis of chemically diverse core-shell nanoparticles for intracellular delivery. Proc Natl Acad Sci USA. 2011;108(32):12996–13001. doi: 10.1073/pnas.1106379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H-X, et al. N-acetylgalactosamine functionalized mixed micellar nanoparticles for targeted delivery of siRNA to liver. J Control Release. 2013;166(2):106–114. doi: 10.1016/j.jconrel.2012.12.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.