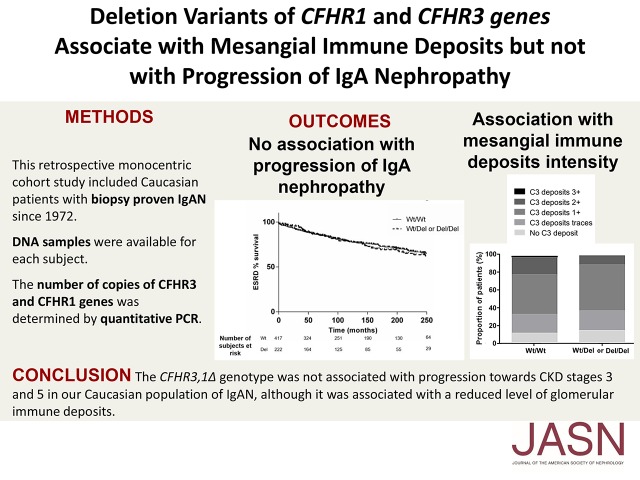
Abstract
Activation of complement through the alternative pathway has a key role in the pathogenesis of IgA nephropathy (IgAN). Large, international, genome-wide association studies have shown that deletion of complement factor H–related genes 1 and 3 (CFHR3,1Δ) is associated with a reduced risk of developing IgAN, although the prognostic value of these deletions in IgAN remains unknown. Here, we compared the renal outcomes of patients with IgAN according to their CFHR3,1Δ genotype. This retrospective, monocentric cohort study included 639 white patients with biopsy-proven IgAN since 1979 (mean age at diagnosis, 40.1 years; median follow-up, 132 months). We determined the number of CFHR3 and CFHR1 gene copies by quantitative PCR and collected clinical and biologic data by reviewing the patients’ medical records. In all, 30.5% of the patients were heterozygous and 4% were homozygous for CFHR3,1Δ. We did not detect an association between CFHR3,1Δ and age, eGFR, urinary protein excretion rate, or the presence of hypertension or hematuria at the time of diagnosis. The mean intensities of immune IgA, IgG, and C3 deposits were lower in the group with heterozygous or homozygous gene deletions than in those with no deletion. However, CFHR3,1Δ did not associate with progression to stage 3 CKD or renal death. In conclusion, the CFHR3,1Δ genotype did not associate with progression toward CKD stages 3 and 5 in our white population of patients with IgAN, although it did associate with a reduced level of glomerular immune deposits.
Keywords: glomerular disease, histopathology, human genetics, IgA nephropathy, complement
IgA nephropathy (IgAN) is the most common type of GN worldwide, leading to ESRD in up to 40% of patients within 20 years of diagnosis.1 The pathophysiology of the disease is considered to be a four-hit mechanism2: The first hit is characterized by increased levels of circulatory polymeric IgA1 with aberrant O-glycosylation of the hinge region (galactose-deficient IgA1 [Gd-IgA1]).3 The second hit corresponds to the presence or excess of a Gd-IgA1–binding element, which could be a unique anti-glycan antibody4 and/or a soluble form of Fc α-receptor (sCD89),5 along with complement elements such as activated complement component 3 (C3).6 The third hit is the formation of Gd-IgA1–containing circulating immune complexes, some of which may be deposited in the glomeruli, thereby triggering glomerular inflammation and injury (hit four), leading to clinical CKD.
The complement alternative pathway (AP) is thought to be an important player in the pathogenesis of IgAN.7 First, key AP components are codeposited with IgA in the mesangium. Indeed, C3 is detected in the immunodeposits in kidney tissue in up to 90% of patients,8 as are properdin (in 75%–100% of patients) and factor H (FH) (in 30%–90% of patients).9,10 IgA can activate AP in vitro, especially while immobilized on a surface in a polymeric form.11 Moreover, urinary membrane attack complex (MAC), FH, and properdin levels, as well as circulating levels of activated C3 products, have been reported to be higher in patients with IgAN than in healthy controls.12–14
Large, international, genome-wide association studies (GWAS) have recently identified several genomic regions associated with the risk of IgAN.15 Among the genome-wide significant single nucleotide polymorphisms (SNP) associated with IgAN, one signal (rs6677604) emerged from the genomic region 1q32, corresponding to an intronic variant of the FH gene. This variant was in strong linkage disequilibrium with the large deletion of FH-related genes 1 and 3 (CFHR1/3 genes). This association was protective in regard to the occurrence of IgAN and this has been confirmed in a larger international GWAS meta-analysis of 20,612 individuals, comprising 7658 patients with IgAN.16 A careful study of the inter-relationship between the SNP rs6677604 and the actual CFHR3,1Δ (assessed by multiplex ligation-dependent probe amplification) by conditional analysis revealed that the protective effect in a Chinese Han cohort was mainly due to just the CFHR3,1 deletion.17 The frequency of the deleted variant CFHR3,1Δ is very low in Asian populations (roughly 5%) compared with white populations (about 20%),18 making it more difficult to achieve sufficient power in these Asian studies to demonstrate differences.
This protective effect of CFHR3,1Δ in IgAN suggests a functional role of CFHR1 and/or CFHR3 proteins in the pathogenesis of the disease. These proteins are encoded by genes located in the FH gene cluster.19 CFHR genes comprise five family members, thought to be derived from internal duplications of the FH gene. CFHR1 and CFHR3 proteins display high sequence homology with FH in their C-terminal short consensus repeats, corresponding to a region that binds to surfaces and C3b. These proteins can inhibit the AP (e.g., CFHR1 can block C5 and terminal complement complex), but they lack some regulatory functions exhibited by FH, such as C3 convertase decay acceleration and factor I cofactor activity (in other words, CFHR3 has a low cofactor activity).20 Because CFHR 1 and 3 bind C3b and surfaces in competition with FH, their presence is associated with a less potent in vitro complement inhibition and their absence leads to a greater degree of regulation by FH. This mechanism could explain why the deletion of CFHR 1 and CFHR3 genes can lead to stronger inhibition of AP, resulting in a protective effect against a disease for which the pathogenesis is driven by AP activation, such as IgAN.
The prognosis with IgAN is highly variable,21 and little is known about the determinants of this heterogeneity. No GWAS have been carried out to date to identify the genetic susceptibility for disease progression. Complement activation has been reported to be associated with the severity of IgAN evolution.22–25 We hypothesized that, by altering AP activation, the CFHR3,1Δ could prevent the progression of IgAN.
In this study, we aimed to assess the protective prognostic effect of the deletion of CFHR1 and CFHR3 genes in a large, well characterized, monocentric cohort of white patients with IgAN.
Results
Study Population
A total of 901 patients with biopsy-proven primary IgAN were recruited between 1972 and 2013 in our center. Of these, 262 patients without an available DNA sample or without an available clinical follow-up were discarded. Six hundred thirty-nine patients were ultimately retained for the final analysis.
The population characteristics are presented in Table 1. Briefly, the study population was 75% male and the mean age at the time of diagnosis was 40.1 years of age. The median total follow-up was 132 months. Of note, the remaining patients without an available DNA sample exhibited a less severe manifestation of the disease compared with the experimental group (e.g., lower mean proteinuria [0.8 g/d versus 1.3 g/d, respectively]; higher mean eGFR [84.1 ml/min per 1.73 m2 versus 72.4 ml/min per 1.73 m2, respectively]; lower systolic arterial pressure [128.7 mmHg versus 136.6 mmHg, respectively]; and a shorter follow-up [7.5 years versus 11.1 years, respectively]).
Table 1.
Population characteristics at the time of diagnosis and at the last follow-up
| Characteristic | At Renal Biopsy | Last Follow-Up |
|---|---|---|
| n | 639 | |
| Males, n (%) | 479 (75) | |
| Age, yr | 40.1 (±15.7) | 52.1 (±15.7) |
| Body mass index, kg/m2 | 24.6 (±4.9) | 25.6 (±4.8) |
| Proteinuria, g/d | 0.7 (0.2–1.8) | 0.2 (0–0.9) |
| Proteinuria categories (g/d), n (%) | ||
| <0.3 | 173 (28.6) | 274 (51.9) |
| 0.3–1 | 182 (30.1) | 125 (23.7) |
| 1–3 | 165 (27.3) | 81 (15.3) |
| >3 | 84 (13.9) | 48 (9.1) |
| Hypertension, % | 41 | 63 |
| eGFR, ml/min per 1.73 m2 | 74 (50–94) | 61 (8–84) |
| CKD stages, n (%) | ||
| Stage 1 | 188 (30.5) | 108 (16.9) |
| Stage 2 | 225 (36.5) | 215 (33.7) |
| Stage 3 | 123 (19.9) | 109 (17.1) |
| Stage 4 | 46 (7.5) | 33 (5.2) |
| Stage 5 | 35 (5.7) | 173 (27.1) |
| Serum IgA, mg/dl | 310 (229–419) | |
| Corticosteroids, % | 0 | 33 |
| Follow-up, mo | 132 (56–216) | |
Data are means (±SD) or medians (interquartile range), or percentages when indicated.
At the time of diagnosis, the median eGFR was 74 ml/min per 1.73 m2, the median proteinuria was 0.7 g/d, and 41% of the population were hypertensive or required antihypertensive drugs. Overall, renin-angiotensin system blockade and corticosteroids were used in 41% and in 33%, respectively, during follow-up.
Out of the entire cohort of patients, information about C3 mesangial deposit intensities was available for 566 patients, and Oxford classification scoring was achievable for 475 patients. M, E, S, T, and C items were significantly associated with progression to ESRD, and only a trend linking C3 deposits to renal outcomes was detected (Supplemental Figure 1, Table 2). C3 deposits correlated with more severe Oxford classification scores (Supplemental Table 1).
Table 2.
Prediction of renal outcomes according to the presence of C3 deposits (0 versus ≥traces) or the Oxford classification (univariate Cox model)
| Outcome | Stage 3 CKD | ESRD | |||||
|---|---|---|---|---|---|---|---|
| n | HR | 95% CI | P Value | HR | 95% CI | P Value | |
| C3 deposits | 566 | 1.43 | 0.97 to 2.1 | 0.07 | 1.60 | 0.89 to 2.9 | 0.12 |
| M | 475 | 1.25 | 0.88 to 1.8 | 0.21 | 1.65 | 1.03 to 2.64 | 0.04 |
| E | 475 | 2.08 | 1.6 to 2.8 | <0.001 | 3.00 | 2.0 to 4.5 | <0.001 |
| S | 475 | 3.36 | 2.5 to 4.6 | <0.001 | 4.16 | 2.5 to 6.9 | <0.001 |
| T>0 | 475 | 8.60 | 6.4 to 11.6 | <0.001 | 11.5 | 7.6 to 17.5 | <0.001 |
| C>0 | 475 | 1.64 | 1.13 to 2.37 | <0.01 | 2.16 | 1.3 to 3.5 | 0.002 |
95% CI, 95% confidence interval; M, mesangial hypercellularity; E, endocapillary hypercellularity; S, segmental glomerulosclerosis; T, tubular atrophy/interstitial fibrosis; C, crescents.
Three hundred and eight patients (48%) developed stage 3 CKD, whereas 172 patients (27%) developed ESRD during the follow-up.
CFHR3,1Δ Distribution
The CFHR3,1Δ genotype of 639 patients was determined by Taqman quantitative real-time PCR.
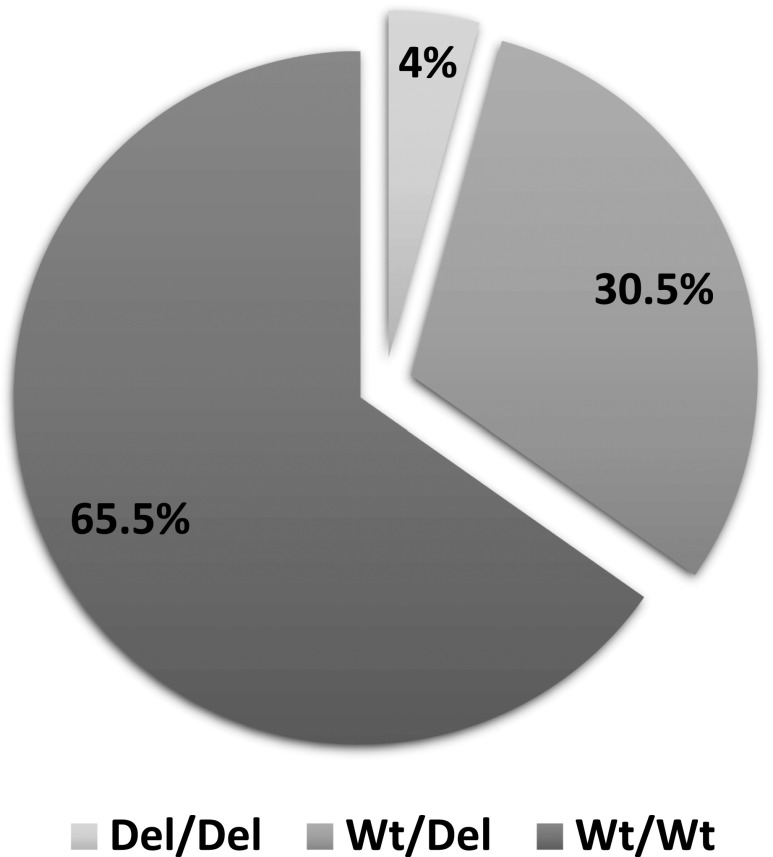
The homozygous deletion (no copy) of both the CFHR1 gene and the CFHR3 gene was detected in 27 patients (4%), whereas 184 patients (29%) had only a single copy of each gene (CFHR1 and CFHR3). Four hundred seventeen individuals (65%) had at least two copies of CFHR1 and CFHR3.
Some patients (11), representing 1.7% of the entire cohort, exhibited discrepancies in the number of copies of the CFHR1 and CFHR3 genes. All of these divergent genotypes were assessed twice. Nine of them had a CFHR1 gene deletion only, one patient had no CFHR1 and a single copy of CFHR3, and one patient had a single copy of CFHR1 and three copies of CFHR3. To simplify the analysis, these patients were considered to be heterozygous for CFHR3,1Δ, because they had one copy of either CFHR3 or CFHR1.
The absence of CFHR1 protein in the serum of randomly selected homozygous CFHR3,1Δ patients was confirmed by SDS-PAGE and crossreactive anti-H western blot (Supplemental Figure 2).
The final distribution is shown in Figure 1. The patients with one deletion of at least one of the genes (i.e., CFHR1 or CFHR3) were considered Del/Wt (n=195). Patients with a deletion of CFHR1 and a deletion of CFHR3 were considered to be Del/Del (n=27). Patients with at least two copies of both genes were considered to be Wt/Wt in terms of the CFHR3,1Δ genotype (n=417). The genotype frequencies followed the Hardy–Weinberg equilibrium (Χ2=0.48, P=0.49). The total proportion of patients with at least one deletion was 34.7% and the minor allele frequency was 19.5%.
Figure 1.
Distribution of the CFHR3,1Δ genotypes. Del/Del, deletion/deletion; Del/Wt, deletion/wild type; Wt/Wt, wild type/wild type.
Genotype-Phenotype Correlation
We next evaluated genotype-phenotype correlations so as to explore the clinical implications of CFHR3,1Δ. We first compared clinical and biologic data at the time of diagnosis among patients with or without CFHR3,1Δ. The age at onset was similar for the two groups, and no significant differences between clinical, biologic, and histologic characteristics were noted (Table 3). The absolute renal risk of dialysis/death21 and the proportion of deaths did not differ according to the genotype (data not shown).
Table 3.
Baseline characteristics according to the CFHR3,1∆ genotype
| Characteristic | CFHR3,1 Genotype | P Value | |
|---|---|---|---|
| Wt/Wt (n=417) | Wt/Del or Del/Del (n=222) | ||
| Age at onset, yr | 34.9 (±16.0) | 35.9 (±16.3) | 0.49 |
| eGFR, ml/min per 1.73 m2 | 72.9 (47.0–93.5) | 75.8 (54.5–95.7) | 0.16 |
| CKD stages, n (%) | 0.48 | ||
| Stage 1 | 124 (30.1) | 64 (31.2) | |
| Stage 2 | 145 (35.2) | 80 (39.0) | |
| Stage 3 | 84 (20.4) | 39 (19.0) | |
| Stage 4 | 31 (7.5) | 15 (7.3) | |
| Stage 5 | 28 (6.8) | 7 (3.4) | |
| Proteinuria, g/d | 0.6 (0.2–1.6) | 0.7 (0.1–2) | 0.89 |
| Proteinuria categories (g/d), n (%) | 0.51 | ||
| <0.3 | 109 (27.0) | 64 (31.8) | |
| 0.3–1 | 131 (32.4) | 51 (25.4) | |
| 1–3 | 114 (28.2) | 51 (25.4) | |
| >3 | 50 (12.3) | 35 (17.4) | |
| Hypertension, % | 42.6 | 39.6 | 0.49 |
| Microscopic hematuria, % | 80.2 | 86.0 | 0.27 |
| Macroscopic hematuria, % | 36.9 | 27.1 | 0.1 |
Data are means (±SD) or medians (interquartile range), or percentages when indicated. Wt/Wt, wild type/wild type; Wt/Del, wild type/deletion; Del/Del, deletion/deletion.
The M, E, S, and T items of the Oxford classification were also similar between the groups, as were the crescent proportions. The interobserver accuracy was considered to be adequate because the κ coefficient (95% confidence interval) was 0.83 (0.78 to 0.89). The mean intensities of immune mesangial deposits were lower for the gene deletion group (Table 4).
Table 4.
Oxford classification, crescents proportion, and immune deposits intensity according to the CFHR3,1∆ genotype
| Variable | CFHR3,1 Genotype | P Value | |
|---|---|---|---|
| Wt/Wt | Wt/Del or Del/Del | ||
| Morphologic evaluation, % | n=324 | n=151 | |
| M1 | 15.7 | 20.5 | 0.24 |
| E1 | 20.6 | 25.3 | 0.28 |
| S1 | 52.2 | 54.3 | 0.69 |
| T≥1 | 20.4 | 20.5 | >0.99 |
| Crescents | 2.2 | 2.5 | 0.70 |
| Immune deposits intensity | n=369 | n=191 | |
| IgA deposits | 1.77 (±0.73) | 1.64 (±0.70) | 0.04 |
| IgG deposits | 0.05 (±0.20) | 0.02 (±0.11) | 0.01 |
| C3 deposits | 1.00 (±0.68) | 0.85 (±0.56) | <0.01 |
| C4 deposits | 0.03 (±0.17) | 0.01 (±0.07) | 0.08 |
After Bonferroni correction, only C3 deposit intensities were significantly associated with the genotype. Immune deposits were scored 0 for absent, 0.5 for traces, 1 for 1+, 2 for 2+, and 3 for 3+. Values are percentages or means (±SD). Wt/Wt, wild type/wild type; Wt/Del, wild type/deletion; Del/Del, deletion/deletion; M1, mesangial hypercellularity; E1, Endocapillary hypercellularity; S1, Segmental glomerulosclerosis; T≤1, tubular atrophy/interstitial fibrosis.
As shown in Figure 2, patients with CFHR3,1∆ had less intense mesangial C3 deposits compared with patients without this deletion (0, traces, 1+, 2+, and 3+: 12.7%, 21.7%, 43.9%, 19.8%, and 1.9%, respectively, in the Wt/Wt gene group versus 15.2%, 22.9%, 51.0%, 10.4%, and 0.5%, respectively, in the Wt/Del or Del/Del CFHR3 gene group). Strong C3 staining (>1+) was 50% less common in the CFHR3,1∆ group (10.9% versus 21.7%, respectively, P=0.03). The deposition of C4 was similar for the two groups.
Figure 2.
Intensity of mesangial C3 deposits is lower in the deleted group. Intensity of mesangial C3 deposits depending on the presence/absence of CFHR3,1∆ allele (A) or genotypes (B). Del/Del, deletion/deletion; Del/Wt, deletion/wild type; Wt/Wt, wild type/wild type.
Association between Genotype and Renal Outcomes
To determine the prognostic value of CFHR3,1∆, progression toward stage 3 CKD and ESRD were recorded retrospectively for the entire cohort.
The median follow-up of patients was comparable for the two groups (131 and 133 months, respectively, for the Wt/Wt group and the Wt/Del or Del/Del group). The CFHR3,1∆ genotype was not associated with the risk of progressing toward stage 3 CKD (univariate Cox model, hazard ratio [HR], 0.95; 0.78 to 1.14; P value=0.9) or ESRD (HR, 0.96; 0.74 to 1.23; P value=0.83). On the basis of our data, the detectable effect sizes at 80% power (HR for protective effect of the deleted allele) were 0.68 and 0.59 for stage 3 CKD and ESRD, respectively. Kaplan–Maier survival analyses are shown in Figure 3. CFHR3,1∆ does not affect the long-term prognosis for IgAN.
Figure 3.
CFHR3,1∆ genotype is not associated with renal prognosis. Association between the CFHR3,1∆ genotype and the development of stage 3 CKD and ESRD. The left part of the figure is the Kaplan–Maier survival curves according to the presence/absence of the deletion and the right part is the genotype-based analysis. Del, deletion; Del/Del, deletion/deletion; Del/Wt, deletion/wild type; Wt, XXX; Wt/Wt, wild type/wild type.
The effect of the gene deletion on the risk of death was NS (Cox model, adjusted on age, HR, 0.68; 0.4 to 1.17; P=0.17).
Discussion
Our study concluded that there was no prognostic effect of CFHR3,1∆ on IgAN progression in our white patient cohort.
The frequency of the gene deletion was particularly high in our population (19.5%), compared with other published French cohorts (8.6%),26 despite the fact that this frequency could be expected to be even lower in an IgAN population.15 Nevertheless, these frequencies have been poorly assessed in French populations, and they have been reported to be highly variable within a given country.18 In this regard, our study represents the largest population sample in France to have undergone evaluation of the actual frequency of CFHR3,1 deletion. The high frequency conferred sufficient statistical power to our study to safely assume that CFHR3,1∆ appears to have no effect on IgAN progression. Conversely, the frequency of the deletion is very low in Asian populations (5%), making it more difficult to eliminate a prognostic effect of CFHR3,1∆ in studies of this population.
The assessment of the genotyping was ensured by several measures. The copy number of these genes was obtained by direct measurement, which is preferable to the genotyping of a related SNP, even in strong linkage disequilibrium, as recently demonstrated by Xie et al.17 The quantification of genomic DNA by quantitative PCR compared with a reference gene is a valid method, as is multiplex ligation-dependent probe amplification. The high concordance between the number of copies of CFHR1 and CFHR3 and the corresponding protein levels further underscores the robustness of our results.
Despite the negative outcome in terms of the correlation with disease progression, our study nonetheless answered an important question raised by (1) the recent GWAS, (2) the putative role of complement on the progression of IgAN, and (3) a number of histologic hallmarks common with C3 nephropathy27 and thrombotic microangiopathy.28 The absence of CFHR1 and CFHR3 was demonstrated to allow for a more potent in vitro regulation by FH.20 In age-related macular degeneration, CFHR3,1∆ is also protective in regard to the occurrence of this disease.29,30 This genotype also affected the severity of this pathology, because the frequency of the deletion was 18% in unilateral geographic atrophy compared with 7% in the corresponding bilateral form.31
Our study has some limitations that warrant mention. Although DNA samples were available for most of the patients of the entire cohort, thus allowing genotype analysis, 262 patients could nonetheless not be included. This aspect represents a possible selection bias, possibly rendering our analyzed cohort less than fully representative of the complete population with IgAN. In light of the inherent limitations of such a retrospective cohort, a well controlled prospective cohort would theoretically be needed to clearly establish the role of this deletion on outcomes. However, potential issues in regard to practical aspects of such a prospective study should not be underestimated, with an absolute requirement for a long follow-up in addition to the need for a large study population so as to be in a position to establish the consequences of a deletion that is present in only a small proportion of patients.
Our results are in line with previous Asian studies. Zhu et al.32 have provided a detailed report regarding the effect of rs6677604, which is closely related to deletion of CFHR3,1 genes, in a IgAN cohort. This study found reduced levels of C3 deposits in the A/G genotype compared with the G/G population, which is in good agreement with the results of our study. They reported that this reduced level of mesangial deposits was associated with higher levels of circulating C3, higher levels of FH, and lower levels of C3a, thus reflecting the more potent systemic inhibition of the AP in the genotype corresponding to the deletion of CFHR3,1. Although this study lacked long-term clinical follow-up, it also did not find a matching correlation with clinical or histologic severity. Xie et al.17 have published their detailed findings regarding the concurrent protective effect of rs6677604 and CFHR3,1∆ on the occurrence of IgAN. Their results, on the basis of conditional analysis, clearly indicate that the large deletion drives this effect by itself.17 This study revealed an effect of the deletion on the Oxford classification of histologic severity, with significantly lower S and T scores, which is inconsistent with the findings of our and the study by Zhu et al. Nevertheless, no effect on eGFR, proteinuria, or hypertension was noted, and once again no longitudinal clinical evaluation was available.
CFHR3,1∆ seems to influence the initiation of the IgAN pathology, rather than progression to more severe stages of kidney damage. The absence of a correlation with the Oxford classification of elementary lesions may corroborate the fact that the deletion does not affect the induction of glomerular inflammatory lesions triggered by deposited immune complexes. Nevertheless, this finding could be specific to whites (an association with the S and T score has been reported in one Chinese study), or it was not detected in our population in light of its limited power (detectable effect size of 0.276 at 80% power). The weaker intensities of immune deposits could suggest that the deletion alters the formation and/or the solubility of IgA-immune complexes. This result raises the issue of the effect of C3 deposits on renal outcomes. The study by Kim et al.24 suggests an effect of C3 deposit intensities on the prognosis of IgAN. However, in this study, these deposits were correlated with glomerular lesions, which were not controlled for in multivariate analysis. Thus, C3 could be associated with histologic severity, without it having a bona fide deleterious effect. The findings of Zhu et al.32 corroborate our findings that (1) the genotype correlates with C3 deposits; (2) C3 deposits alone correlate with histologic lesions, but in fine; and (3) the genotype did not correlate with disease activity.
In an in vitro model of engineered immune complexes associating Gd-IgA1 and recombinant anti-glycan IgG in the presence of Ig-depleted normal human serum, C3 cleavage products were found to be bound to these immune complexes.6 In this model, these immune complexes induced mesangial proliferation in vitro, unless the plasma was heated to 56°C before the formation of the immune complexes.33 On the other hand, several authors have ascribed a role to AP in determining the solubility of immune complexes, as a result of the coating of these complexes with C3b,34–36 which can then interact with Complement Receptor 1 (CR1).37 Altogether, these data suggest that in the presence of CFHR1 and/or 3, in immune complexes, the level of AP activation could be higher, thus increasing the formation and solubility of nephritogenic immune complexes.
Our data do not rule out complement regulation as being a significant player in the progression of the disease. Zhai et al.38 recently reported enrichment of rare CFHR5 variants in a Chinese population of patients with IgAN. In light of all of these observations, we hypothesize that complement AP regulation could be modified in IgAN by the combined effect of several polymorphisms and rare variants that lead to a higher level of AP activation. In order to thoroughly test this hypothesis, large-scale sequencing of AP regulation genes in a large sample of patients is required, with the ultimate goal of evaluating the cumulative effect of variants on the course of the disease. In this regard, we plan to develop large-scale sequencing of CFHR5, FB, FI, FH, MCP, and C3 genes in our cohort to address this hypothesis. Our data do not indicate that there is a significant effect of CFHR3,1 deletion on the risk of progression to ESRD. The deletion was associated with a lower level of deposited IgA, IgG, and C3 in the mesangium, thus supporting a role for deletion of these genes in the formation and deposition of immune complexes rather than in inducing local inflammatory responses and fibrosis processes.
Our results do not rule out a critical role for the regulation of AP on patient outcomes, and a more extensive evaluation of the variants of genes implicated in AP is warranted in order to explore a promising regulatory pathway that could potentially be targeted by novel biotherapies.
Concise Methods
Study Population
All consecutive white patients with biopsy-proven primary IgAN at the department of nephrology of the Saint Etienne University Hospital (France) were included retrospectively from 1972 to 2013. Of these, patients without an available DNA sample or without an available clinical follow-up were discarded from the analysis. Other exclusion criteria included potential secondary causes of IgAN, such as liver or inflammatory bowel diseases, psoriasis, inflammatory chronic diseases, other autoimmune disorders, infections, active cancers, and Henoch–Schönlein purpura. All of the subjects provided informed consent to participate in the study. The study was approved by the local ethics committee.
Data Collection
Data regarding patient characteristics, disease activity, treatments, and outcomes were obtained by reviewing the medical records of the patients. At the time of the renal biopsy and at the last follow-up, body weight, BP, serum creatinine levels, and 24-hour urinary protein excretion were recorded. Hypertension was defined as a blood arterial pressure (BP) >140/90 mmHg and/or the need for antihypertensive drugs. The number of antihypertensive medications prescribed and the use of renin-angiotensin system blockade or corticosteroids were also recorded.
All of the biopsy specimens were reviewed by an experienced pathologist blinded to the clinical and biologic data, and they underwent standardized scoring using the recently developed Oxford MEST classification. One hundred randomly selected biopsy specimens were blindly rescored by a second physician to assess the interobserver reproducibility (κ score). The semiquantitative mesangial IgA, IgG, C3, C1q, and C4 immunofluorescence intensities were evaluated at the time of the biopsy and scored as being zero, one-half (traces), one (weak), two (moderate), or three (strong).
Renal Outcomes
The GFR was estimated using the Modification of Diet in Renal Disease formula. Stage 3 CKD was defined as a GFR (eGFR) that was <60 ml/min per 1.73 m2. ESRD was defined either as an eGFR<15 ml/min per 1.73 m2 (stage 5 CKD) or the need for dialysis or renal transplantation.
DNA Samples
Blood samples were harvested at the time of the diagnosis. Genomic DNA was extracted by the standard phenol-chloroform method to achieve a final concentration of 200 µg/ml before storage at −20°C.
CFHR3,1Δ Genotype Assessment
Quantitative PCR was used to determine the number of copies of the CFHR1 and CFHR3 genes for each patient, following manufacturer’s guidelines (Applied Biosystems). Quantitative real-time PCR was carried out on genomic DNA using the Taqman Copy Number Real-Time Detection System, involving four replicates for each patient. Three sets of predesigned primers and probes, including CFHR1 (FAM-labeled, intron 4, Assay ID Hs04197581_cn; Applied Biosystems), CFHR3 (FAM-labeled, intron 4, Assay ID Hs04206213_cn; Applied Biosystems), and RNaseP (VIC-labeled, standard reference; Applied Biosystems), were used. A duplex system was used, and quantitative real-time PCR was performed on the 7500 Real-Time PCR System (Applied Biosystems) with 40 cycles (95°C for 15 seconds followed by 60°C for 60 seconds). Copy number variation calls were determined for each patient using the 2−ΔΔCt method (Copy Caller Software; Applied Biosystems). The genotype result was confirmed on the basis of the absence of the CFHR1 protein by PAGE and western blot analysis of serum from a limited number of randomly selected patients. The serum was diluted 1:100, and the primary antibody was a goat anti-human FH (Complement Technology) diluted 1:5000.
Statistical Analyses
Continuous variables were presented as means±SD or medians±IQR, depending on the normality of the distribution. Categoric variables were expressed as percentages. Comparisons of means were performed with the t test and comparisons of medians were done using a Wilcoxon–Mann–Whitney test. The Fisher exact test was used to compare percentages. Survival curves were performed using the Kaplan–Maier method, and survival analyses were on the basis of a univariate Cox regression model. Statistical analyses were performed using R software (R Core Team [2015]. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria) and GraphPad Prism (GraphPad Software, La Jolla, CA). We calculated that a sample of 615 patients (including a minimum of 215 patients with at least one CFHR3,1Δ allele) would give the study 80% power to detect rates of progression toward stage 3 CKD (the first primary end point) of 40% in the group without the gene deletion and 30% in the deleted group (binomial test, package “pwr”). Survival-based power calculations were performed to assess the HR thresholds at 80% power using the “pwerSurvEpi” package.
Disclosures
None.
Supplementary Material
Acknowledgments
The authors acknowledge support from the Société Française de Néphrologie, Dialyse, Transplantation and Alexion for their research grant, and the university hospital of Saint Etienne for their grant (local tender 2015).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2017010019/-/DCSupplemental.
References
- 1.Berthoux FC, Mohey H, Afiani A: Natural history of primary IgA nephropathy. Semin Nephrol 28: 4–9, 2008 [DOI] [PubMed] [Google Scholar]
- 2.Suzuki H, Kiryluk K, Novak J, Moldoveanu Z, Herr AB, Renfrow MB, Wyatt RJ, Scolari F, Mestecky J, Gharavi AG, Julian BA: The pathophysiology of IgA nephropathy. J Am Soc Nephrol 22: 1795–1803, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moldoveanu Z, Wyatt RJ, Lee JY, Tomana M, Julian BA, Mestecky J, Huang W-Q, Anreddy SR, Hall S, Hastings MC, Lau KK, Cook WJ, Novak J: Patients with IgA nephropathy have increased serum galactose-deficient IgA1 levels. Kidney Int 71: 1148–1154, 2007 [DOI] [PubMed] [Google Scholar]
- 4.Suzuki H, Fan R, Zhang Z, Brown R, Hall S, Julian BA, Chatham WW, Suzuki Y, Wyatt RJ, Moldoveanu Z, Lee JY, Robinson J, Tomana M, Tomino Y, Mestecky J, Novak J: Aberrantly glycosylated IgA1 in IgA nephropathy patients is recognized by IgG antibodies with restricted heterogeneity. J Clin Invest 119: 1668–1677, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Launay P, Grossetête B, Arcos-Fajardo M, Gaudin E, Torres SP, Beaudoin L, Patey-Mariaud de Serre N, Lehuen A, Monteiro RC: Fcalpha receptor (CD89) mediates the development of immunoglobulin A (IgA) nephropathy (Berger’s disease). Evidence for pathogenic soluble receptor-Iga complexes in patients and CD89 transgenic mice. J Exp Med 191: 1999–2009, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maillard N, Boerma LA, Hall SD, Huang Z-Q, Mrug M, Moldoveanu Z, Julian BA, Renfrow MB, Novak J: Proteomic analysis of engineered IgA1-IgG immune complexes reveals association with activated complement C3. J Am Soc Nephrol 24: 490A, 2013 [Google Scholar]
- 7.Maillard N, Wyatt RJ, Julian BA, Kiryluk K, Gharavi A, Fremeaux-Bacchi V, Novak J: Current understanding of the role of complement in IgA nephropathy. J Am Soc Nephrol 26: 1503–1512, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas M: Histology and immunohistology of IgA nephropathy. J Nephrol 18: 676–680, 2005 [PubMed] [Google Scholar]
- 9.McCoy RC, Abramowsky CR, Tisher CC: IgA nephropathy. Am J Pathol 76: 123–144, 1974 [PMC free article] [PubMed] [Google Scholar]
- 10.Miyazaki R, Kuroda M, Akiyama T, Otani I, Tofuku Y, Takeda R: Glomerular deposition and serum levels of complement control proteins in patients with IgA nephropathy. Clin Nephrol 21: 335–340, 1984 [PubMed] [Google Scholar]
- 11.Hiemstra PS, Gorter A, Stuurman ME, Van Es LA, Daha MR: Activation of the alternative pathway of complement by human serum IgA. Eur J Immunol 17: 321–326, 1987 [DOI] [PubMed] [Google Scholar]
- 12.Onda K, Ohsawa I, Ohi H, Tamano M, Mano S, Wakabayashi M, Toki A, Horikoshi S, Fujita T, Tomino Y: Excretion of complement proteins and its activation marker C5b-9 in IgA nephropathy in relation to renal function. BMC Nephrol 12: 64, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wyatt RJ, Kanayama Y, Julian BA, Negoro N, Sugimoto S, Hudson EC, Curd JG: Complement activation in IgA nephropathy. Kidney Int 31: 1019–1023, 1987 [DOI] [PubMed] [Google Scholar]
- 14.Liu M, Chen Y, Zhou J, Liu Y, Wang F, Shi S, Zhao Y, Wang S, Liu L, Lv J, Zhang H, Zhao M: Implication of urinary complement factor H in the progression of immunoglobulin A nephropathy. PLoS One 10: e0126812, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gharavi AG, Kiryluk K, Choi M, Li Y, Hou P, Xie J, Sanna-Cherchi S, Men CJ, Julian BA, Wyatt RJ, Novak J, He JC, Wang H, Lv J, Zhu L, Wang W, Wang Z, Yasuno K, Gunel M, Mane S, Umlauf S, Tikhonova I, Beerman I, Savoldi S, Magistroni R, Ghiggeri GM, Bodria M, Lugani F, Ravani P, Ponticelli C, Allegri L, Boscutti G, Frasca G, Amore A, Peruzzi L, Coppo R, Izzi C, Viola BF, Prati E, Salvadori M, Mignani R, Gesualdo L, Bertinetto F, Mesiano P, Amoroso A, Scolari F, Chen N, Zhang H, Lifton RP: Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet 43: 321–327, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiryluk K, Li Y, Scolari F, Sanna-Cherchi S, Choi M, Verbitsky M, Fasel D, Lata S, Prakash S, Shapiro S, Fischman C, Snyder HJ, Appel G, Izzi C, Viola BF, Dallera N, Del Vecchio L, Barlassina C, Salvi E, Bertinetto FE, Amoroso A, Savoldi S, Rocchietti M, Amore A, Peruzzi L, Coppo R, Salvadori M, Ravani P, Magistroni R, Ghiggeri GM, Caridi G, Bodria M, Lugani F, Allegri L, Delsante M, Maiorana M, Magnano A, Frasca G, Boer E, Boscutti G, Ponticelli C, Mignani R, Marcantoni C, Di Landro D, Santoro D, Pani A, Polci R, Feriozzi S, Chicca S, Galliani M, Gigante M, Gesualdo L, Zamboli P, Battaglia GG, Garozzo M, Maixnerová D, Tesar V, Eitner F, Rauen T, Floege J, Kovacs T, Nagy J, Mucha K, Pączek L, Zaniew M, Mizerska-Wasiak M, Roszkowska-Blaim M, Pawlaczyk K, Gale D, Barratt J, Thibaudin L, Berthoux F, Canaud G, Boland A, Metzger M, Panzer U, Suzuki H, Goto S, Narita I, Caliskan Y, Xie J, Hou P, Chen N, Zhang H, Wyatt RJ, Novak J, Julian BA, Feehally J, Stengel B, Cusi D, Lifton RP, Gharavi AG: Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet 46: 1187–1196, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie J, Kiryluk K, Li Y, Mladkova N, Zhu L, Hou P, Ren H, Wang W, Zhang H, Chen N, Gharavi AG: Fine mapping implicates a deletion of CFHR1 and CFHR3 in protection from IgA nephropathy in Han Chinese. J Am Soc Nephrol 27: 3187–3194, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes LV, Strain L, Staniforth SJ, Moore I, Marchbank K, Kavanagh D, Goodship JA, Cordell HJ, Goodship THJ: Determining the population frequency of the CFHR3/CFHR1 deletion at 1q32. PLoS One 8: e60352, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skerka C, Chen Q, Fremeaux-Bacchi V, Roumenina LT: Complement factor H related proteins (CFHRs). Mol Immunol 56: 170–180, 2013 [DOI] [PubMed] [Google Scholar]
- 20.Fritsche LG, Lauer N, Hartmann A, Stippa S, Keilhauer CN, Oppermann M, Pandey MK, Köhl J, Zipfel PF, Weber BHF, Skerka C: An imbalance of human complement regulatory proteins CFHR1, CFHR3 and factor H influences risk for age-related macular degeneration (AMD). Hum Mol Genet 19: 4694–4704, 2010 [DOI] [PubMed] [Google Scholar]
- 21.Berthoux F, Mohey H, Laurent B, Mariat C, Afiani A, Thibaudin L: Predicting the risk for dialysis or death in IgA nephropathy. J Am Soc Nephrol 22: 752–761, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komatsu H, Fujimoto S, Hara S, Sato Y, Yamada K, Eto T: Relationship between serum IgA/C3 ratio and progression of IgA nephropathy. Intern Med 43: 1023–1028, 2004 [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Wang C, Tang Y, Peng H, Ye Z-C, Li C-C, Lou T-Q: Serum immunoglobulin A/C3 ratio predicts progression of immunoglobulin A nephropathy. Nephrology (Carlton) 18: 125–131, 2013 [DOI] [PubMed] [Google Scholar]
- 24.Kim SJ, Koo HM, Lim BJ, Oh HJ, Yoo DE, Shin DH, Lee MJ, Doh FM, Park JT, Yoo T-H, Kang S-W, Choi KH, Jeong HJ, Han SH: Decreased circulating C3 levels and mesangial C3 deposition predict renal outcome in patients with IgA nephropathy. PLoS One 7: e40495, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Espinosa M, Ortega R, Sánchez M, Segarra A, Salcedo MT, González F, Camacho R, Valdivia MA, Cabrera R, López K, Pinedo F, Gutierrez E, Valera A, Leon M, Cobo MA, Rodriguez R, Ballarín J, Arce Y, García B, Muñoz MD, Praga M; Spanish Group for Study of Glomerular Diseases (GLOSEN) : Association of C4d deposition with clinical outcomes in IgA nephropathy. Clin J Am Soc Nephrol 9: 897–904, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dragon-Durey M-A, Blanc C, Marliot F, Loirat C, Blouin J, Sautes-Fridman C, Fridman WH, Frémeaux-Bacchi V: The high frequency of complement factor H related CFHR1 gene deletion is restricted to specific subgroups of patients with atypical haemolytic uraemic syndrome. J Med Genet 46: 447–450, 2009 [DOI] [PubMed] [Google Scholar]
- 27.Cook HT, Pickering MC: Histopathology of MPGN and C3 glomerulopathies. Nat Rev Nephrol 11: 14–22, 2015 [DOI] [PubMed] [Google Scholar]
- 28.El Karoui K, Hill GS, Karras A, Jacquot C, Moulonguet L, Kourilsky O, Frémeaux-Bacchi V, Delahousse M, Duong Van Huyen J-P, Loupy A, Bruneval P, Nochy D: A clinicopathologic study of thrombotic microangiopathy in IgA nephropathy. J Am Soc Nephrol 23: 137–148, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubista KE, Tosakulwong N, Wu Y, Ryu E, Roeder JL, Hecker LA, Baratz KH, Brown WL, Edwards AO: Copy number variation in the complement factor H-related genes and age-related macular degeneration. Mol Vis 17: 2080–2092, 2011 [PMC free article] [PubMed] [Google Scholar]
- 30.Sawitzke J, Im KM, Kostiha B, Dean M, Gold B: Association assessment of copy number polymorphism and risk of age-related macular degeneration. Ophthalmology 118: 2442–2446, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cantsilieris S, White SJ, Richardson AJ, Guymer RH, Baird PN: Comprehensive analysis of Copy Number Variation of genes at chromosome 1 and 10 loci associated with late age related macular degeneration. PLoS One 7: e35255, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu L, Zhai Y-L, Wang F-M, Hou P, Lv J-C, Xu D-M, Shi S-F, Liu L-J, Yu F, Zhao M-H, Novak J, Gharavi AG, Zhang H: Variants in complement factor H and complement factor H-related protein genes, CFHR3 and CFHR1, affect complement activation in IgA nephropathy. J Am Soc Nephrol 26: 1195–1204, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanagihara T, Brown R, Hall S, Moldoveanu Z, Goepfert A, Tomana M, Julian BA, Mestecky J, Novak J: In vitro-generated immune complexes containing galactose-deficient IgA1 stimulate proliferation of mesangial cells. Results Immunol 2: 166–172, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takata Y, Tamura N, Fujita T: Interaction of C3 with antigen-antibody complexes in the process of solubilization of immune precipitates. J Immunol 132: 2531–2537, 1984 [PubMed] [Google Scholar]
- 35.Takahashi M, Tack BF, Nussenzweig V: Requirements for the solubilization of immune aggregates by complement: Assembly of a factor B-dependent C3-convertase on the immune complexes. J Exp Med 145: 86–100, 1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi M, Takahashi S, Brade V, Nussenzweig V: Requirements for the solubilization of immune aggregates by complement. The role of the classical pathway. J Clin Invest 62: 349–358, 1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cornacoff JB, Hebert LA, Smead WL, VanAman ME, Birmingham DJ, Waxman FJ: Primate erythrocyte-immune complex-clearing mechanism. J Clin Invest 71: 236–247, 1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhai Y-L, Meng S-J, Zhu L, Shi S-F, Wang S-X, Liu L-J, Lv J-C, Yu F, Zhao M-H, Zhang H: Rare variants in the complement factor H-related protein 5 gene contribute to genetic susceptibility to IgA nephropathy. J Am Soc Nephrol 27: 2894–2905, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.