Abstract
The PD-L1/PD-1 pathway is a critical component of the immunosuppressive tumor microenvironment in acute myeloid leukemia (AML), but little is known about its regulation. We investigated the role of the MUC1 oncoprotein in modulating PD-L1 expression in AML. Silencing of MUC1 in AML cell lines suppressed PD-L1 expression without a decrease in PD-L1 mRNA levels, suggesting a post-transcriptional mechanism of regulation. We identified the microRNAs miR-200c and miR-34a as key regulators of PD-L1 expression in AML. Silencing of MUC1 in AML cells led to a marked increase in miR-200c and miR-34a levels, without changes in precursor microRNA, suggesting that MUC1 might regulate microRNA-processing. MUC1 signaling decreased the expression of the microRNA-processing protein DICER, via the suppression of c-Jun activity. NanoString (Seattle, WA, USA) array of MUC1-silenced AML cells demonstrated an increase in the majority of probed microRNAs. In an immunocompetent murine AML model, targeting of MUC1 led to a significant increase in leukemia-specific T cells. In concert, targeting MUC1 signaling in human AML cells resulted in enhanced sensitivity to T-cell-mediated lysis. These findings suggest MUC1 is a critical regulator of PD-L1 expression via its effects on microRNA levels and represents a potential therapeutic target to enhance anti-tumor immunity.
INTRODUCTION
Acute myeloid leukemia (AML) is a lethal hematological malignancy in which the tumor microenvironment is characterized by an immunosuppressive milieu that fosters disease progression.1,2 The PD-L1/PD-1 pathway confers a critical negative co-stimulatory signal that induces T-cell exhaustion and supports immune evasion by malignant cells.3–6 In contrast, antibody blockade of PD-L1/PD-1 signaling results in the reversal of tumor-mediated immune suppression and durable responses in subsets of patients with solid tumors7–9 and hematological malignancies.10 Although PD-L1 expression in AML is dynamic, little is known about the mechanism(s) responsible for regulating PD-L1 expression in AML.
MUC1 is a heterodimeric oncoprotein aberrantly expressed in solid tumors and hematological malignancies including AML, that supports critical aspects of the malignant phenotype including cell proliferation, self-renewal and resistance to apoptosis.11–16 MUC1 interacts with the WNT/-catenin pathway and promotes the activation of WNT target genes,17,18 NFΚ -B19–21 and STAT1/3,22,23 pathways critical for the proliferation and survival of tumor cells. In addition, MUC1 regulates pathways responsible for autonomous self-renewal24 and is uniquely expressed on leukemia stem cells as compared to normal hematopoietic stem cells.25 Inhibition of MUC1 using a cell-penetrating peptide (GO-203) that blocks MUC1-C homodimerization necessary for downstream signaling,26,27 abrogates leukemia engraftment and eradicates established disease in a xenogeneic leukemia model.25 Given the critical function of MUC1, in supporting the malignant phenotype of AML blasts and stem cells, we sought to explore the role of MUC1 in mediating the immunosuppressive milieu of the tumor microenvironment. Here, we demonstrate that silencing of MUC1 markedly suppresses PD-L1 expression in AML cells. However, MUC1 suppression is associated with a paradoxical increase in PD-L1 mRNA, suggesting that MUC1 regulation of PD-L1 expression in AML occurs at the post-transcriptional level.
Noncoding RNAs epigenetically regulate critical aspects of the oncogenic phenotype through the disruption of protein translation via selective binding and degradation of target mRNAs.28 The microRNAs miR-200c and miR-34a demonstrate homology with the 3′-UTR section of PD-L1 mRNA.4,29 MiR-200c was recently shown to downregulate the expression of PD-L1 protein in a lung cancer model,29 and miR-34a was shown to target PD-L1 in AML cell lines.4 In the present study, we demonstrate that MUC1 negatively regulates expression of miR-200c and miR-34a, which in turn controls PD-L1 expression in AML cells. Consistent with these findings, upregulation of miR-200c or miR-34a via lentiviral transduction results in a corresponding decrease in PD-L1 expression. Of note, silencing of MUC1 results in increased levels of mature miR-34a and miR-200c while precursor-microRNAs are unaffected. Consistent with this observation, MUC1 inhibition resulted in increased expression of DICER protein, which mediates the final splicing of precursor miRNAs to their active form. Indeed, microRNA array of MUC1-silenced AML cells demonstrated a profound global upregulation of microRNAs, consistent with an increase in DICER expression. These findings strongly suggest MUC1 as a key regulator of microRNA expression and demonstrate a critical mechanism by which MUC1 signaling exploits noncoding RNAs to elicit an immunosuppressive milieu in the bone marrow microenvironment (BM).
MATERIALS AND METHODS
Cell culture
The AML cells lines THP-1 and MOLM-14 and the murine cell line TIB-49 were purchased from ATCC, cultured at 37 °C in a humidified 5% CO2 incubator and maintained in RPMI 1640 media (Cellgro, Manassas, VA, USA) supplemented with heat-inactivated 10% human serum albumin (Sigma, St Louis, MO, USA) and 100 IU/ml penicillin, 100 µg/ml streptomycin (Cellgro). Cell lines were stably transduced with a lentiviral vector expressing a scrambled control shRNA (CshRNA, Sigma) or MUC1 shRNA (Sigma) in the presence of 4–8 g/ml polybrene (Sigma). Transduced cells were then selected using 2 µg/ml puromycin. For the CRISPR-edited cell line, sgRNAs targeting the first exon of the MUC1 gene were cloned into a lenti-plasmid (Genome Engineering Production Group, Harvard Medical School). MOLM-14 cells were transduced with viral vector containing the lenti-CRISPR plasmid and successfully transduced clones were selected for by limiting dilution and maintained in 2 µg/ml puromycin (Sigma). Alternatively, cells were stably transduced with lentiviral vectors expressing pHR-GFP, miR-200c or miR-34a with a GFP selection marker. Transduced cells were selected by flow cytometric sorting for GFP-positive cells. In a subset of experiments, tumor cells were obtained from BM aspirates or peripheral blood isolated from patients with AML at diagnosis. Patient characteristics are shown in Supplementary Table S1. AML cells were treated once daily with 2.5 µm MUC1-C inhibitor peptide (GO-203) or a control peptide (CP-3) (AnaSpec) for 3 days. The culture medium was not supplemented with additional cytokines. In specific experiments, AML cells were treated for 30 min with 100 or 400 µm of c-Jun peptide inhibitor (Tocris, Bristol, UK) or control (phosphate-buffered saline).
In vivo model
C57BL/6J mice were challenged with 100 × 103 GFP-transduced MUC1-C-silenced TIB-49 syngeneic AML cells via retro-orbital injection. TIB-49 cells underwent lentiviral transduction with shRNA hairpin against MUC1-C or a control shRNA. BM and spleen cells were isolated on day 14 and assessed for PD-L1 expression by multichannel flow cytometric analysis staining for GFP+ AML cells and PD-L1 expression using mouse antigen-presenting cell (APC) -conjugated anti-PD-L1 antibody. Alternatively, MUC1 signaling was inhibited via daily subcutaneous injection of the MUC1 inhibitor, GO-203, (14 mg/kg) initiated 24 h after tumor challenge.
AML-specific T cells were quantified in BM and spleen by flow cytometric analysis for intracellular IFN-γ expression following exposure to TIB-49 AML lysate. C57BL/6 mice were inoculated with 100 × 103 GFP-transduced syngeneic TIB-49 AML cells using retro-orbital injections. On day 14 following the inoculation BM cells were harvested and stimulated ex vivo with TIB-49 tumor lysate. Following 6 days of stimulation, CD4+ and CD8+ T cells underwent flow cytometric analysis for intracellular IFN-γ expression.
CTL assay
Lysis of AML cells by allogeneic T cells following MUC1 downregulation was assessed in a standard CTL flourochrome assay (OncoImmunin Inc., Gaithersburg, MD, USA). Target cells were incubated in APC labeled phosphate-buffered saline (1 µl of reconstituted TFL4 in phosphate-buffered saline at 1:3000 ratio) at 1 × 106 cells/ml for 30 min at 37 °C. Labeled cells were washed twice in phosphate-buffered saline. Using MUC1-silenced or control MOLM-14 cell lines, healthy donor peripheral blood mononuclear cells (PBMCs) were co-incubated with labeled target cells in the presence of a fluorogenic granzyme B substrate for 1–2 h at 37 °C. Cells were washed and analyzed by flow cytometry. Dead target cells are identified by cells that dually stain for granzyme B and APC label (right upper quadrant). As a negative control, Granzyme B positive tumor cells not co-cultured with T cells, were quantified.
The impact of MUC1 inhibition on CTL mediated killing of patient-derived AML cells, by T cells stimulated with a dendritic cells (DCs)/AML fusion vaccine was examined. DC/AML fusions were generated as previously described.30 Briefly, autologous DCs were generated by culture of adherent peripheral mononuclear cells obtained from AML patients following remission, in the presence of GM-CSF, IL-4 and TNF-α. DCs were fused with autologous AML blasts, obtained at diagnosis, by coculture at a 1:1 ratio in the presence of polyethylene glycol. Autologous T cells were then stimulated with the DC/AML fusion vaccine for 6 days. Subsequently, vaccine stimulated T cells were co-cultured with control autologous AML blasts or AML blasts exposed to daily treatment with 2.5 µm GO-203 for 3 days. The lysis of AML blasts, with and without treatment with GO-203, by vaccine stimulated T cells was detected using a standard CTL assay as described above.
FACS analysis
Cells were analyzed for PD-L1 expression by multichannel flow cytometric analysis. Cells were first incubated with FcR blocking reagent (Miltenyi, Bergisch Gladbach, Germany) for 10 min at room temperature. Subsequently, cells incubated with monoclonal antibody (MAb) anti-PD-L1-APC (Biolegend, San Diego, CA, USA), anti-CD38-PE or anti-CD117-PE (BD Pharmigen, San Jose, CA, USA) or the appropriate isotype control. The cells were then analyzed by flow cytometry using FACS Aria (BD Biosciences, San Jose, CA, USA). Analysis of the obtained data were performed using Kaluza software (Beckman Coulter, Brea, CA, USA).
The expression of IFN-γ was analyzed by intracellular flow cytometry. T cells were pulsed with GolgiStop (1 µg/ml; BD Pharmingen) for 4–6 h at 37 °C before analysis. Cells were next harvested and labeled with CD4-PB and CD8-FITC. Cells were then permeabilized by incubation in Cyto-fix/Cytoperm plus (BD Pharmingen) containing formaldehyde and saponin for 30 min at 4 °C, washed twice in Perm/Wash solution (BD Pharmingen), and incubated with PE-conjugated IFN-γ (Invitrogen, Camarillo, CA, USA), or a matched isotype control antibody for 30 min. Cells were washed in 1 × Perm/Wash solution and fixed in 2% paraformaldehyde (Sigma, St Louis, MO, USA) becasue of the analysis. For Ki-67 analysis, AML cells were permeabilized by incubation in Cyto-fix/Cytoperm plus (BD Pharmingen) as described above and incubated with PE-conjugated Ki-67 (BD Pharmingen). Ki-67 expression was then assessed using flow cytometry.
Luciferase activity of 3′-UTR PD-L1
The full length of human PD-L1 3′-UTR was cloned into the Psicheck2 vector (Promega, Madison, WI, USA). MOLM-14 AML cells were transduced with GFP control vector, miR-34a and miR-200c. GFP expressing cells were subsequently isolated used flow cytometric sorting. 200 × 103 viable cells from each cell line were transiently transfected with the luciferase reporter Psicheck2 vector 3′-UTR PD-L1, to test the capacity of miR-34a and miR-200c to bind the 3′-UTR of PD-L1 and suppress the luciferase activity of the reporter. Each cell line was seeded in triplicates and transfected with DharmaFECT Duo transfection agent (Dharmacon, Lafayatte, CO, USA). Twenty-four hours post transfection, the samples were lysed according to the instructions of Dual luciferase Reporter Assay System (Promega). Firefly and renilla luciferase activity were measured with GloMax luminometer (Promega).
Immunoblotting
Whole-cell lysates were prepared in RIPA lysis buffer and analyzed by immunoblotting with anti-MUC1-C (Thermo Scientific, Waltham, MA, USA,) anti-DICER, anti-Argo-2, anti-p-c-Jun, anti-c-Jun (Cell Signaling, Danvers, MA, USA) anti-PD-L1 (Abcam, Cambridge, MA, USA), anti-JNK, antiphospho- JNK, anti-ERK, anti-phospho-ERK, anti-GAPDH and anti-β-actin (Cell Signaling, Danvers, MA, USA) as described.31 Immune complexes were detected using horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence (GE Healthcare, Little Chalfont, UK).
Quantitative RT-PCR
Total RNA was isolated from cells using the RNeasy total RNA isolation kit (Qiagen, Hilden, Germany). cDNA synthesis was performed with 1 µg of total RNA using the MiScript RT-PCR system (Qiagen). The SYBR green quantitative PCR (qPCR) assay kit (Roche, Basel, Switzerland) was used with 2 µl of diluted cDNA for each sample. The forward and reverse primers for qPCR of murine MUC1, human PD-L1, DICER, c-Jun, miR-200c, miR-34a, premir-200c, pre-mir-34a and GAPDH are listed in Supplementary Table S2. Statistical significant was determined by the student’s t-test.
Chromatin immunoprecipitations (ChIP)
THP-1 cells were crosslinked for 10 min in 1% formaldehyde at room temperature, followed by 0.125 m final concentration of glycine to stop fixation. Aliquots containing 1 × 106 nuclear equivalents were subjected to chromatin immunoprecipitation-PCR for c-Jun and IgG as described previously.32 Crosslinked pellets were lysed for 10 min on ice and chromatin fragmented using a Branson 250 digital sonifier machine with 0.7 s 'On' and 1.3 s 'Off' pulses at 40% power amplitude for 2 × 2 min cycles. Solubilized chromatin was diluted and incubated with 1 µg antibody overnight at 4 °C. Immune complexes were captured with a 50/50 slurry of protein A and protein G Dynabeads and enriched chromatin was isolated by reverse-crosslinking and proteinase K digestion at 65 °C. AMPure XP beads (Beckman Coulter A63882) were used to clean up and isolate chromatin immunoprecipitation DNA for subsequent qPCR per manufactures protocol. The following antibodies were used for chromatin immunoprecipitation: c-Jun (Cell Signaling Technologies #9165) and IgG (Cell Signaling Technologies #2729). Binding sites for c-Jun in the DICER1 promoter were predicted using UCSC Genome browser33 and three sets of primers (DICER1, 2 and 3) were manufactured using ThermoFischer custom Oligo design. C-Jun is a described auto-regulator of c-Jun34 and was therefore used as a positive control. Primers are listed in Supplementary Table S2. The average Ct-value was determined from triplicate reactions and normalized against nonspecific IgG controls with standard curves for each primer pair.
mRNA Array
RNA was isolated from paired GO-203 treated or control peptide-treated AML samples from three subjects (2.5 µm GO-203 daily for 3 days). Affymetrix (Santa Clara, CA, USA) mRNA expression array, consisting of 47 321 probe features corresponding to 34 692 transcripts, was performed as described35 and all arrays were included in the differential expression analysis. Standard normalization methods were applied, followed by paired t-test analysis for Bonferroni-corrected significance, using the Limma Bioconductor package. Correction for multiple comparisons was performed using the false discovery rate of Benjamini and Hochberg.36
miRNA Array
RNA was isolated from AML cells as described and run in triplicate on a NanoString nCounter instrument using the human miRNA Expression Assay Kit v3, according to the manufacturer’s instructions. Data obtained were then normalized to positive miRNA-ligation reaction controls and background noise was subtracted. Correction for multiple comparisons was performed using the false discovery rate of Benjamini and Hochberg.36
Viability assays
AML cells were seeded in white flat-bottom 96-well plates at 200 000 cells/well. At 72 h of daily treatment with GO-203, cell viability was assessed using the CellTiter-Glo (CTG) Luminescent Cell Viability Assay. Raw luminescence values were obtained from each well using Infinite M200 Pro luminometer (Tecan, Mannedorf, Switzerland).
RESULTS
MUC1 inhibition leads to decrease in PD-L1 expression
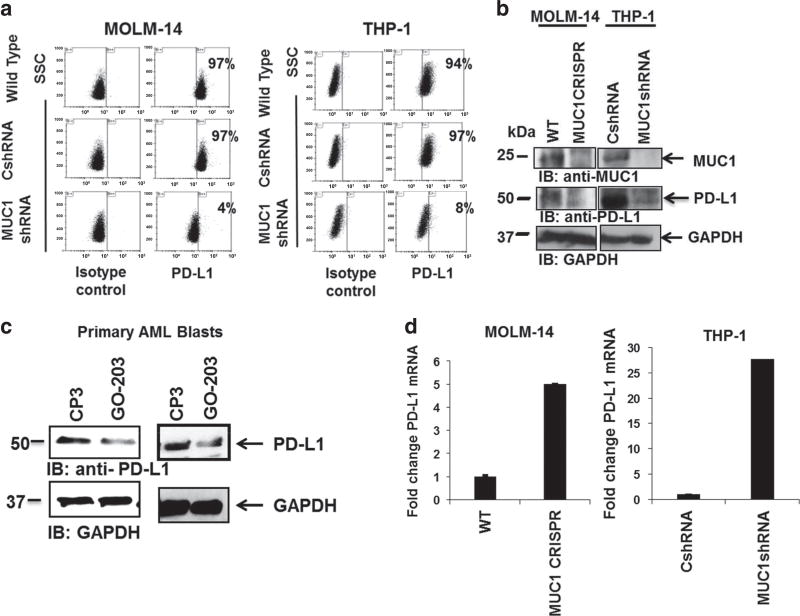
We examined the role of MUC1 in modulating expression of PD-L1, a critical mediator of T-cell exhaustion and anergy. Downregulation of MUC1 expression in MOLM-14 and THP-1 cells, via lentiviral transduction with MUC1-specific shRNA, resulted in the marked suppression of PD-L1 expression as determined by flow cytometric analysis, (Figure 1a, n = 3) and immunoblotting (Figure 1b, n = 3). To examine the effect of MUC1 inhibition on patient-derived AML cells, tumor cells were obtained from peripheral blood or BM samples from patients at diagnosis. The cells were then treated ex vivo, using a sub-lethal dose of a cellpenetrating peptide (GO-203), which blocks MUC1-C homodimerization necessary for downstream signaling. Consistent with silencing of MUC1 gene, MUC1 inhibition in patient-derived AML cells led to a significant decrease in PD-L1 expression, as demonstrated by immunoblotting (Figure 1c, n = 3). Of note, treatment of patient-derived AML cells with low dose GO-203 did not significantly affect cell viability or the expression of AML surface markers (Supplementary Figures 1A and B).
Figure 1.
MUC1 inhibition leads to decrease in PD-L1 expression. MUC1 was silenced in MOLM-14 and THP-1 AML cells using lentiviral shRNA hairpin against MUC1. As a control, MOLM-14 cells were infected with control shRNA. The cells were then evaluated for PD-L1 expression using (a) flow cytometry and (b) western blot analysis. (c) Tumor cells were obtained from patients with AML at diagnosis. The cells were then treated with 2.5 µm MUC1 inhibitor, GO-203 daily for 3 days. As a control the cells were treated with control peptide CP-3. Subsequently, the cells underwent western blot analysis for PD-L1 expression. Representative blots from patients AML1 and AML2 are shown (n =3). (d) A representative experiment demonstrating PD-L1 mRNA levels, in MUC1-silenced MOLM-14 and THP-1 cells evaluated using qPCR. Each condition was performed in triplicate (P<0.05; n=2).
To evaluate PD-L1 mRNA levels, MUC1 gene was silenced in MOLM-14 cells using CRISPR/Cas9 technology, and using MUC1-specific shRNA in THP-1 cells. Interestingly, increased levels of PDL1 mRNA was observed in MUC1-silenced AML cells as determined by qPCR analysis (Figure 1d), suggesting that MUC1 regulation of PD-L1 expression was accomplished via a post-transcriptional mechanism.
MiR-34a and miR-200c regulate PD-L1 expression in AML cells
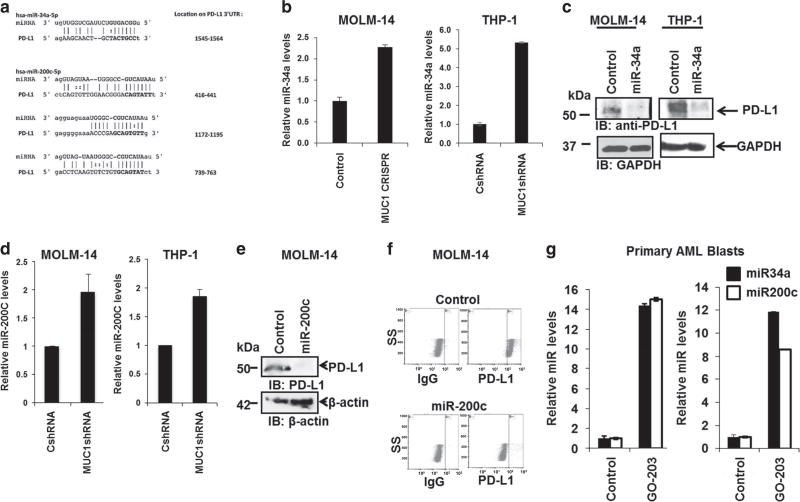
We hypothesized that MUC1 regulation of PD-L1 may be mediated by noncoding RNAs, which are known to epigenetically regulate cellular function via the binding and degradation of mRNAs with homologous sequences, providing a critical brake for protein translation. Consistent with this hypothesis, the PD-L1 3′-UTR contains multiple binding motifs for miR-200c29 and one for miR-34a4 (Figure 2a). Remarkably, MUC1 silencing in both MOLM-14 and THP-1 cells led to a significant increase in miR-34a (Figure 2b) and miR-200c (Figure 2d), as determined by qPCR analysis. Accordingly, ectopic miR-34a expression in MOLM-14 and THP-1 human AML cells via lentiviral transduction of miR-34a-mimic, resulted in the near abrogation of PD-L1 expression (Figure 2c). Similar results were seen with ectopic overexpression of miR-200c which led to significant downregulation of PD-L1 expression in MOLM-14 cells, as demonstrated by western blot (Figure 2e), and flow cytometric analysis (Figure 2f). To confirm the direct effect of miR-34a and miR-200c on the PD-L1 expression luciferase reporter assay was performed, showing repression of PD-L1 expression as demonstrated in Supplementary Figure 2a.
Figure 2.
miR-34a and miR-200c regulate PD-L1 expression in AML cells. (a) The seed sequences of miR-34a and miR-200c on the 3′-UTR PD-L1, transcript variant 1, NM_014143.3, were identified by RegRNA: a regulatory RNA motifs and element web server. MUC1 was silenced in MOLM-14 cells using the CRISPR/Cas9 technology and in THP-1 cells using transduction with MUC1-specific shRNA. (b) Relative levels of miR-34a were detected in MOLM-14 and THP-1 cells using qPCR. miR-34a was overexpressed in MOLM-14 and THP-1 cells using lentiviral transduction. (c) PD-L1 levels were evaluated using western blot analysis in miR-34a overexpressed or control MOLM-14 and THP-1 AML cells. (d) Relative levels of miR-200c were detected in MOLM-14 and THP-1 cells using qPCR. MiR-200c was overexpressed in MOLM-14 cells using lentiviral transduction. PD-L1 levels were evaluated in control and miR-200c overexpressed cells using (e) western blotting and (f) flow cytometry. Tumor cells were obtained from PB or BM aspirated of patients with AML at diagnosis. The cells were then treated with 2.5 µm MUC1 inhibitor, GO-203 daily for 3 days. As a control the cells were treated with control peptide CP-3. (g) Subsequently, relative levels of miR-34a and miR200c were detected in these cells compared to controls using qPCR. Three patients were evaluated, the bar graphs represent patients AML2 and AML3 (P<0.05).
Furthermore, to examine the effect of MUC1 inhibition on miR-34a and miR-200c in patient-derived AML cells, tumor cells were isolated from patients with AML at diagnosis and treated with sub-lethal doses of MUC1 inhibitor GO-203. Indeed, MUC1 inhibition led to a significant increase in miR-34a and miR-200c levels as demonstrated by qPCR and shown in Figure 2g (n = 3). In summary, MUC1 negatively regulates expression of miR-34a and miR-200c attenuating their interference with PD-L1 translation, resulting in increased PD-L1 expression by AML cells. In contrast, silencing of MUC1 results in a corresponding increase in miR-34a and miR-200c, leading to decreased expression of PD-L1. MUC1 inhibition leads to increase in DICER expression via c-Jun We then sought to determine the mechanism by which MUC1 signaling negatively regulates expression of miR-34a and miR-−200c, as these microRNAs represent a diverse species of noncoding RNAs that do not share a common promoter region.
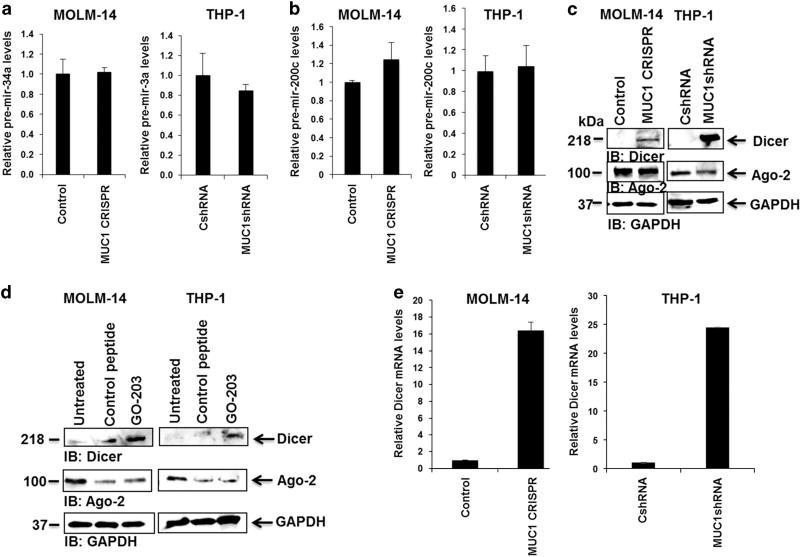
We first examined the effect of MUC1 silencing on microRNA-processing. MUC1-silenced MOLM-14 and THP-1 cells were analyzed for precursor miR-34a and miR-200c, to determine if MUC1 signaling effects miR-34a and miR-200c maturation to their functional conformation. The results demonstrated that silencing of MUC1 did not result in alteration of levels of the precursor miRNAs pre-mir-34a (Figure 3a) or pre-mir-200c (Figure 3b), suggesting the regulatory effect of MUC1 on these microRNAs occurs at the post-transcriptional stage.
Figure 3.
MUC1 inhibition leads to increase in DICER expression. MUC1 was silenced in MOLM-14 cells using CRISPR/Cas9 technology and in THP-1 cells using transduction with MUC1-specific shRNA. Relative levels of (a) precursor mir-34a and (b) precursor mir-200c were detected using qPCR in MOLM-14 and THP-1 cells with silenced MUC1 levels compared to control cells (n=2). (c) DICER and Ago-2 levels were assessed using western blot analysis in control and MUC1-silenced MOLM-14 and THP-1 cells (n=3). (d) MOLM-14 and THP-1 cells were treated with 2 µm GO-203 or control peptide daily for 3 days. DICER and Ago-2 levels were detected using western blot analysis (n =2). (e) DICER mRNA was detected using qPCR in MOLM-14 and THP-1 cells with silenced MUC1 levels compared to appropriate control cells (n=2).
We subsequently investigated whether MUC1 signaling impacted the expression of DICER, an RNAase III that forms a critical component of the RNA-induced silencing complex, required for the generation of functionally competent microRNAs. We demonstrated the novel finding that the MUC1 oncoprotein regulates DICER expression impacting activation of noncoding RNAs in a class wide effect. MUC1 silencing resulted in increased expression of DICER protein in the MOLM-14 and THP-1 AML cell lines (Figure 3c). Similarly, expression of DICER was increased following exposure of AML cells to the MUC1 inhibitor, GO-203. Of note, there was no change in the expression of the Argonaut 2 (Ago 2) protein, the 'catalytic engine' of the RNA-induced silencing complex (Figure 3d). Interestingly, MUC1 silencing of both MOLM-14 and THP-1 cells lines resulted in the dramatic increase in DICER mRNA levels by qPCR analysis (Figure 3e), consistent with regulation at the transcriptional level.
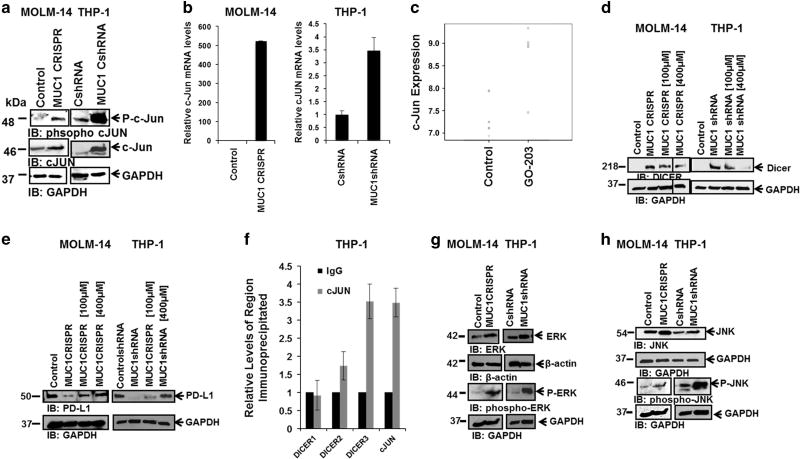
c-Jun, a member of the AP-1 transcription factor family, is known to have a binding site in the DICER1 gene promoter37 and has been shown to mediate DICER expression in Breast Cancer38 and T-cell leukemia.37 Immunoblots of MUC1-silenced AML cells demonstrated an increase in total and phosphorylated forms of c-Jun (Figure 4a, n = 2). Consistent with this finding, MUC1-silenced AML cell lines demonstrated increased levels of c-Jun mRNA by qPCR analysis (Figure 4b, n = 2). Furthermore, c-Jun was one of only 10 genes that demonstrated significant differential expression in a microarray analysis of gene expression of three paired GO-203 or control peptide-treated AML patient-derived samples, with MUC1 inhibition resulting in a statistically significant increase in c-Jun mRNA levels (false discovery rate-adjusted P = 0.003; Figure 4c). An increase in c-Jun mRNA levels following MUC1 inhibition using GO-203 in patient-derived AML cells was validated using qPCR as demonstrated in Supplementary Figure 2B (n = 3).
Figure 4.
Increase in DICER expression is mediated via c-Jun. MUC1 was silenced in MOLM-14 cells using CRISPR/Cas9 technology and in THP-1 cells using transduction with MUC1-specific shRNA. (a) The cells underwent western blot analysis for phospho c-Jun and c-Jun expression in MUC1-silenced and control AML cells. (b) Relative c-Jun mRNA levels were evaluated using qPCR analysis in MOLM-14 and THP-1 MUC1-silenced and control cells (n=2). (c) AML cells were obtained from BM aspirates from patient AML6 at diagnosis. The cells were then treated daily for 3 days with 2.5 µm of GO-203 or control peptide. Affymetrix analysis (Santa Clara, CA, USA) was performed for mRNA expression and the relative expression of c-Jun is presented for three patients. (d) MUC1-silenced MOLM-14 and THP-1 cells were treated with 100 and 400 µm c-Jun inhibitor for 30min. DICER levels were evaluated using western blot analysis (n=2). (e) Control and MUC1-silenced MOLM-14 and THP-1 cells were treated with 100 and 400 µm c-Jun inhibitor for 30min. PD-L1 levels were evaluated using western blot analysis (n=2). (f) c-Jun or IgG was immunoprecipitated from the nuclear lysate of 1 × 106 million THP-1 cells and ChIP was used to demonstrate DICER promoter enrichment at DICER site #3. The positive control is the c-Jun promoter. All values have been normalized to the enrichment detected using IgG and are expressed as the average of three experiments. All conditions were performed in triplicates. (g) Control and MUC1-silenced MOLM-14 and THP-1 cells were analyzed for ERK and phospho-ERK expression using western blot analysis (n=2). (h) Control and MUC1-silenced MOLM-14 and THP-1 cells were analyzed for JNK and phospho-JNK expression using western blot analysis (n=2). ChIP, chromatin immunoprecipitation.
Interestingly, MUC1-silenced MOLM-14 and THP-1 cells treated for 30 min with 100 and 400 um of a peptide inhibitor of c-Jun activity, demonstrated a dose dependent decrease in DICER protein expression, confirming the central role of c-Jun in mediating MUC1 regulation of DICER. (Figures 4d, n = 3).
To examine the downstream effect of c-Jun signaling on PD-L1 expression, MOLM-14 and THP-1 AML cells were treated with c-Jun inhibitor, as described above. Importantly, c-Jun inhibition of MUC1-silenced MOLM-14 and THP-1 cells led to an upregulation of PD-L1 expression, putatively via the effect of c-Jun on DICER expression and regulation of miRNAs (Figure 4e, n = 2).
We next sought to determine whether c-Jun directly regulates DICER expression. Chromatin immunoprecipitation using c-Jun was performed in THP-1 AML cells. The results demonstrated enrichment of DICER DNA at the third predicted site in the DICER promoter, indicating that c-Jun directly promotes DICER transcription in AML (Figure 4f, n = 3).
To further investigate the mechanism by which MUC1 affects DICER expression, we evaluated potential mediators of MUC1 regulation of c-Jun. MUC1-silenced MOLM-14 and THP-1 cells were assessed for their expression of ERK and JNK; described mediators of c-Jun transcription39–41 and phosphorylation,42,43 respectively. MUC1 silencing resulted in increased levels of total and phosphorylated ERK and JNK as demonstrated by immunoblot analysis (Figures 4g and h; n = 2).
MUC1 inhibition leads to increase in microRNAs in AML cells
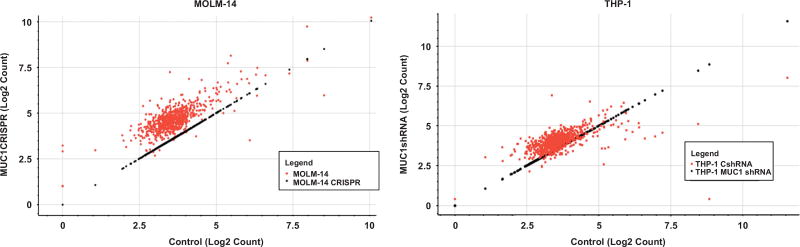
The finding that MUC1 regulates expression of DICER suggests that MUC1 exerts a class effect on the production of functionally mature miRNAs. To investigate this hypothesis, we performed an array to identify other microRNAs that were similarly impacted by silencing of MUC1. MicroRNA array of MUC1-silenced MOLM-14 and THP-1 cells demonstrated a profound global upregulation of a substantial proportion of microRNAs (Figure 5), including miR-34a and miR-200c. Of the panel of 801 miRNAs arrayed, MUC1-silenced MOLM-14 cells showed a statistically significant increase in 340 out of 801 (42.4% at false discovery rate of 0.05). Concordantly, MUC1-silenced THP-1 cells showed an increase in 154 out of 801 (19.2%) microRNAs.
Figure 5.
MUC1 silencing leads to increase in microRNAs in AML cells. MUC1 was silenced in MOLM-14 cells using CRISPR/Cas9 technology and in THP-1 cells using transduction with MUC1-specific shRNA. RNA was isolated from the MUC1-silenced and control MOLM-14 and THP-1 AML cells and underwent NanoString analysis for microRNA expression in triplicates. Scatter plot of log2 count of microRNAs in MUC1-silenced vs. control cells are demonstrated.
MUC1 inhibition exerts an immunoregulatory effect in AML cells in vivo and in vitro
Having demonstrated that MUC1 regulates PD-L1 expression in AML cells, we subsequently examined whether targeting MUC1 similarly affects PD-L1 expression in vivo, leading to increased immunogenicity in an immune competent mouse model.
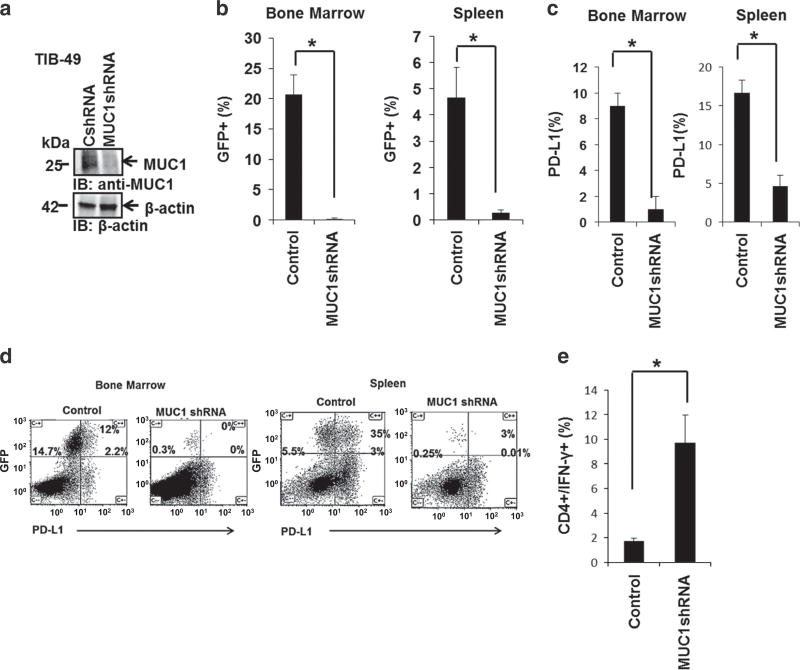
C57BL/6 mice were inoculated with 100 × 103 GFP-transduced control or MUC1-silenced syngeneic TIB-49 AML cells by retroorbital injections (Figure 6a). Animals were analyzed 14 days following inoculation, due to symptomatic disease or death in mice challenged with control shRNA TIB-49 AML cells. Leukemia engraftment, as manifested by the infiltration of GFP+ cells, was noted in the BM and spleen of these animals. Inoculation of animals with MUC1-silenced TIB-49 cells led to significantly decreased engraftment of GFP+ AML cells in BM and spleen of analyzed mice, as demonstrated in two independent experiments (Figures 6b, n=3, P<0.05). MUC1 silencing was confirmed to persist in AML cells isolated from BM of analyzed animals 14 days post inoculation using qPCR (Supplementary Figure 2C).
Figure 6.
MUC1 silencing exerts an immunoregulatory effect in AML cells in vivo. (a) MUC1 was silenced in TIB-49 mouse AML cells using MUC1-specific shRNA. C57BL/6 mice were inoculated retro-orbitally with 100 × 103 GFP syngeneic TIB-49 AML cells transduced with control shRNA (CshRNA) or MUC1-silenced TIB-49 cells (MUC1shRNA). On day 14 following the inoculation, bone marrow and spleen cells were harvested and analyzed for (b) GFP expression using flow cytometric analysis; indicating AML engraftment. Subsequently, GFP+ cells were gated and analyzed for PD-L1 expression as demonstrated in (c) summary and (d) representative FACS plots (n =3, P<0.05). Subsequently, bone marrow cells were stimulated ex vivo with TIB-49 tumor lysate. (e) Following 6 days of stimulation, CD4+ T cells underwent flow cytometric analysis for intracellular IFN-γ expression (P<0.05).
TIB-49 cells did not show PD-L1 expression in vitro (data not shown), in keeping with previous reports, which describe induction of PD-L1 expression in vivo.6 To assess the effect of MUC1 silencing on PD-L1 expression in vivo, BM and spleen cells were isolated from mice, 14 days after inoculation. Gated GFP+ tumor cells were analyzed for PD-L1 expression using flow cytometric analysis. MUC1 silencing was shown to result in a statistically significant decrease in PD-L1 levels as demonstrated in Figures 6c and d (P<0.05).
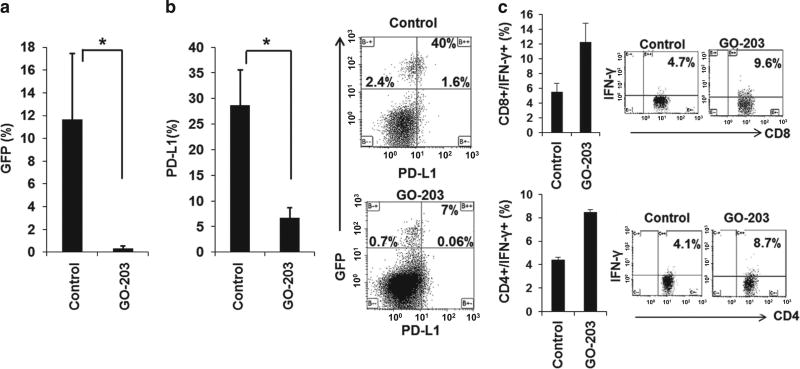
Engraftment of MUC1-silenced AML cells was associated with an expansion of AML-specific T cells as manifested by a statistically significant increase in BM derived CD4+ cells which expressed IFN-γ upon exposure to autologous tumor lysate (Figure 6e). Similarly, animals challenged with TIB-49 cells and treated with the MUC1 inhibitor GO-203 daily for 14 days showed a significantly decreased AML involvement (Figure 7a), decreased PD-L1 expression (Figure 7b) and a twofold expansion in AML-specific T cells upon ex vivo exposure to leukemia derived antigens, in the BM of analyzed animals (Figure 7c).
Figure 7.
MUC1 inhibition using peptide inhibitor, GO-203, leads to increased immunogenicity in immunocompetent murine model. C57BL/6 mice were inoculated with 100 × 103 GFP-transduced syngeneic TIB-49 AML cells using retro-orbital injections. Twenty-four hours after inoculation the mice were treated daily subcutaneously with 14 mg/kg GO-203. On day 14 following the inoculation, bone marrow and spleen cells were harvested. (a) The cells were analyzed for GFP expression using flow cytometric analysis; indicating AML engraftment. (b) GFP+ cells were gated and analyzed for PD-L1 expression, demonstrated as a mean PD-L1 expression and representative FACS plot (n= 3, P<0.05). Subsequently, bone marrow cell and spleen cells were stimulated ex vivo with TIB-49 tumor lysate. (c) Following 6 days of stimulation CD4+ and CD8+ T cells underwent flow cytometric analysis for intracellular IFN-γ expression (P<0.05).
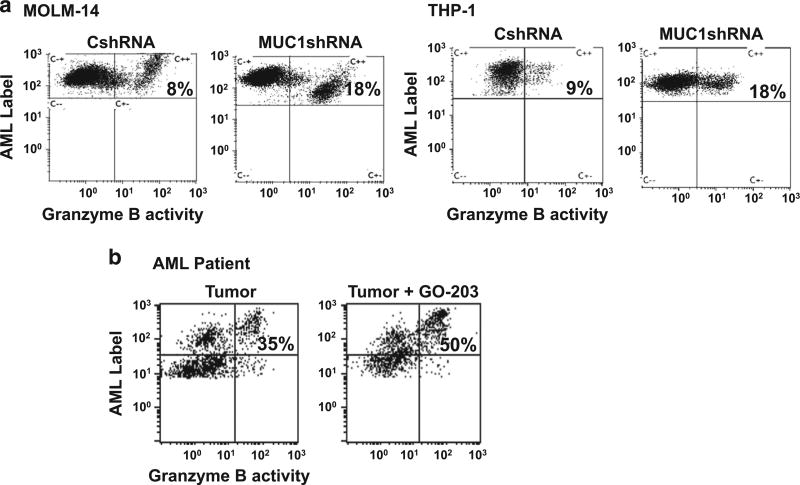
We subsequently examined the impact of targeting MUC1 on the immunogenicity of human AML cells. MUC1 was silenced in MOLM-14 and THP-1 cells using a lentiviral shRNA and compared to MOLM-14 and THP-1 cells infected with control shRNA. Silencing of MUC1 resulted in enhanced susceptibility to immune-mediated killing by alloreactive lymphocytes as determined by the cleavage of the tumor derived Granzyme B substrate in a standard cytotoxic T lymphocyte (CTL) assay (Figure 8a). We have developed a leukemia-specific vaccine in which patient-derived AML cells are fused with autologous DCs such that a broad array of tumor antigens are presented in the context of DC mediated costimulation.44 To investigate the capacity of patient-derived T cells to target autologous tumor, PBMCs were stimulated with DC/AML fusion vaccine. Consistent with the immunoregulatory effect of MUC1 and the decrease in PD-L1 levels following exposure to GO-203 in patient-derived AML blasts (Figure 1c; n = 2), treatment of patient-derived AML cells with the MUC1 inhibitor GO-203, lead to enhanced susceptibility to immune-mediated killing, by DC/AML fusion vaccine stimulated autologous T cells (Figure 8b).
Figure 8.
MUC1 silencing and inhibition leads to increased susceptibility to immune-mediated killing in human AML cells. (a) MUC1 was silenced in MOLM-14 and THP-1 cells using transduction with MUC1-specific shRNA. MOLM-14 and THP-1 cells were then labeled with APC fluorochrome and co-cultured with allogeneic PBMCs in a ratio of 1:5. Tumor lysis was detected after the addition of FITC-granzyme B substrate as a percent of APC+FITC+ cells detected by flow cytometric analysis. Representative example of allogeneic cytotoxic T lymphocyte assay depicting allogeneic PBMC response against MUC1-silenced and control AML cells (n=2). (b) Tumor cells were obtained from bone marrow aspirate from patient with AML at diagnosis. Peripheral blood was obtained from the same patient at remission, DC were generated from the PBMCs and fused with the autologous tumor cells. The PBMCs were then stimulated with the fusion cells for 6 days in a 1:5 ratio and then co-cultured with autologous tumor cells that were either pre-treated with 2.5 µm GO-203 for 3 days or control tumor cells. Tumor lysis was detected after the addition of FITC-granzyme B substrate as a percent of APC+FITC+ cells detected by flow cytometric analysis.
DISCUSSION
The BM microenvironment in patients with AML is characterized by an immunosuppressive milieu that promotes tumor tolerance, immune escape and disease growth.1,2 PD-L1 expression by tumor cells is a critical contributor to the immunosuppressive environment.6 Ligation of PD-1 on T cells in the tumor bed induces an exhausted T-cell phenotype resulting in the loss of activation, expansion and functional capacity of tumor reactive lymphocytes.45 PD-L1 is strongly expressed by AML cells3,5,6 and is also expressed by other immunosuppressive elements in the BM microenvironment.46–48 Defining the mechanisms by which PD-L1 expression is regulated is vital to better understand the evolution of immune dysregulation in AML and as a basis for the design of novel therapies to restore immune function.
The MUC1 oncogene is aberrantly expressed on solid tumors49 and in hematological malignancies11,50 including AML,25,51 and supports critical aspects of malignant transformation including resistance to apoptosis,52 cell proliferation,26 tissue invasion and metastatic potential.49 However, its role in immune regulation and tumor-mediated tolerance has not been well defined.
In the present study, we report on the novel finding that the MUC1 oncogene exerts a potent immunomodulatory effect through its regulation of PD-L1 expression on AML cells. Suppression of MUC1 expression via MUC1-specific shRNA or CRISPR-mediated gene deletion, results in the near abrogation of PD-L1 expression. In an immunocompetent murine model, ex vivo silencing of MUC1 in TIB-49 AML cells before tumor challenge, results in durable absence of PD-L1 in the engrafted leukemia cells. Of note, prior studies have shown that PD-L1 expression on AML cells is dynamic, likely regulated by factors in the microenvironment.6 The present findings suggest that the MUC1 oncogene has a critical role in regulating the immunosuppressive nature of the tumor microenvironment, via its effect on PD-L1. Of note, we have demonstrated that MUC1 expression by tumor cells may be upregulated through interactions between tumor cells and accessory cells in the microenvironment.53
Interestingly, silencing of MUC1 is paradoxically associated with an increase in PD-L1 mRNA expression, suggesting that MUC1 regulation of PD-L1 is likely mediated by a post-transcriptional mechanism. Noncoding RNAs have been identified as critical mediators of cellular function and may act as effectors of epigenetic regulation in malignant cells.29,54,55 MicroRNAs bind to the 3′-UTR sequence of candidate mRNAs leading to prevention of protein translation.28 It was previously shown that miR-200c is complementary to the 3′-UTR sequence of PD-L1 mRNA in a lung cancer model.29 In the present investigation, we demonstrate that that MUC1 negatively regulates miR-34a and miR-200c in AML and that the overexpression of these miRNAs in AML cells results in the near abrogation of PD-L1 expression.
The role of MUC1 in modulating multiple microRNA species raised the possibility of a common processing pathway being targeted by this oncogene. In the present study, we demonstrated that silencing MUC1 is associated with an increase in levels of DICER, an protein critical for mature microRNA formation.56 The generation of functionally active microRNAs is dependent on carefully scripted sequence of events beginning with the transcription of genomic DNA containing microRNA sequences, that are further processed into precursor-microRNAs (pre-micro-RNAs) by a nuclear protein complex.56–58 Pre-microRNAs are then transported to the cytoplasm, loaded onto a protein complex called the RNA-induced silencing complex composed of Dicer, Argonaute-2 (Ago-2), the Tar RNA-binding protein, and cleaved to their mature form by DICER.58 Mature microRNAs are then capable of binding to target mRNAs preventing their translation to protein.28 In the present study, we demonstrate that suppression of MUC1 is associated with increased levels of DICER, consistent with an increase in production of the functionally mature miR-34a and miR-200c. It has been previously shown that the transcription factor, c-Jun, acts as a primary regulator of DICER expression.37 We have replicated this finding in an AML model, demonstrating direct binding of c-Jun within the DICER promoter using chromatin Immunoprecipitation. Furthermore, in the present study, we demonstrate that silencing or inhibition of MUC1 results in increased levels of phosphorylated c-Jun consistent with an associated increase in DICER expression. Conversely, the introduction of a c-Jun inhibitor reversed the effect of MUC1 silencing on DICER expression in a dose dependent fashion.
To assess the mechanism by which MUC1 regulates c-Jun expression, MUC1-silenced MOLM-14 and THP-1 cells were assessed for ERK and p-ERK expression. We have previously demonstrated that MUC1 signaling regulates ERK expression and activation.59 Furthermore, ERK was reported to regulate c-Jun expression in multiple cancer models.39–41 Interestingly, MUC1 silencing in AML cells resulted in increase in total and phosphorylated ERK expression, suggesting that MUC1 regulation of c-Jun expression occurs, in part, via ERK mediated signaling. Moreover, given the marked increase in phospho c-Jun expression following MUC1 silencing, we sought to examine whether MUC1 signaling also effects c-Jun activation. Indeed, analysis of JNK expression, which has been widely shown to effect c-Jun activation,42,43 was demonstrated to be significantly increased following MUC1 silencing in both MOLM-14 and THP-1 cells. These results are consistent with reported JNK regulation by MUC1 signaling.60,61 The results demonstrate that MUC1 signaling effects c-Jun activation as well as expression.
This is the first report of an oncogene regulating DICER as a means of governing oncogenic potential. We demonstrate that MUC1 regulation of DICER is associated with a downstream effect on miR-34a and miR-200c and resultant PD-L1 expression. While genetic deletion of DICER is lethal in utero56 investigators have examined the selective deletion in subsets of cells and have demonstrated that loss of DICER may be associated with loss of function and blunting of differentiation in immune cells.62 Conflicting data regarding the role of DICER in malignancy has been observed, with some models demonstrating loss of DICER associated with disease progression.63,64 Similarly, microRNAs have been shown to mediate diverse effects in tumor models with upregulation of certain microRNA families associated with malignant transformation.65 However, given the role of microRNAs in disrupting protein translation, the loss of microRNAs due to the oncogenic regulation may result in the increased expression of critical oncoproteins. In the present study, as predicted by its effect on DICER, we demonstrate that MUC1-silencing results in a broad class effect on the generation of functionally active microRNAs. In fact, in MOLM-14 cells, MUC1 silencing was associated with an increase in 98% of microRNA species. Interestingly, MUC1 silencing did not lead to change in cell viability as determined by annexin/PI analysis in MOLM-14 and THP-1 cells.66 However, tumor cell proliferation was shown to be reduced in MOLM-14 CRISPR cells as determined by Ki67 analysis (Supplementary Figure 2D).
Consistent with its role in modulating PD-L1 expression, we demonstrate that MUC1 exerts a potent negative immunoregulatory effect in AML. In an immunocompetent AML model, animals engrafted with MUC1-silenced AML cells developed greater evidence of leukemia-specific immunity, compared to control cells, as manifested by increased levels of leukemia-specific T cells detected in the BM, determined by the percent of cells expressing IFN-γ following exposure to tumor lysate. While the higher levels of engraftment seen in animals receiving control tumor may be associated a greater degree of immune suppression, the emergence of leukemia reactive T cells also likely reflects the greater immunogenicity of AML cells in the setting of MUC1 inhibition. Consistent with this hypothesis, killing of human AML cells by alloreactive lymphocytes is enhanced in MUC1-silenced AML targets. Similarly, exposure to a MUC1 inhibitor renders AML cells more susceptible to lysis by T cells stimulated by leukemia vaccine generated by primary AML cells fused with autologous DCs.
The observation that MUC1 modulation of miR-200c and miR-34a levels regulates PD-L1 expression in AML lends itself to several areas of clinical translation. Blockade of the PD-L1/PD-1 pathway has emerged as a major area of cancer therapeutics with profound and durable responses seen in a subset of patients with advanced melanoma,67 renal cell carcinoma,7 and non-small cell lung cancer.9 The presumptive mechanism of this clinical effect is the breaking of tolerance of tumor reactive lymphocytes and the generation of tumor-specific immunity. A recent report of patients with advanced Hodgkin’s disease demonstrates that blockade of this pathway results in durable disease response in 87% of patients.10 PD-L1 expression by tumor cells is of likely prognostic importance and potentially predictive of response to antibodies that provide checkpoint blockade. Expression is likely to be dynamic, arising out of the interactions with stromal elements in the BM microenvironment. As such, staining characteristics of ex vivo cells may not be reliable and in vivo biomarker that correlates with PD-L1 expression is of potential great clinical significance.
We have developed a MUC1 inhibitor consisting of a cell-penetrating peptide that blocks MUC1 signaling by preventing dimerization necessary for translocation of the MUC1-C component from the plasma membrane to the nucleus.18 Exposure to the MUC1 inhibitor results in downregulation of PD-L1 expression in primary AML cells and potentially renders them more susceptible to T-cell-mediated recognition and lysis. Investigators have begun to explore the use of microRNAs as therapeutic agents.68 Strategies to enhance miR-34a or miR-200c expression or by the introduction of microRNA mimetics might similarly result in an enhanced immunologic milieu.
We have developed a tumor vaccine in which patient-derived AML cells are fused with autologous DCs such that a broad array of tumor antigens are presented in the context of DC mediated costimulation.44,45 Vaccination with DC/AML fusions results in the expansion of leukemia-specific T cells and protection from relapse.44 Vaccine efficacy is likely blunted by the presence of PD-L1 expression on the hybridoma cells as well as its presence as a tolerizing influence in the BM microenvironment. In an immune competent AML murine model, combining vaccination with GO-203 treatment, was shown to enhance the vaccine potency (unpublished data). We are currently exploring the use of the MUC1 inhibitor and microRNA mimetics to create an enhanced platform for the development of tumor-specific immunity using a leukemia cancer vaccine.
In summary, we have identified a novel mechanism by which the MUC1 oncoprotein upregulates PD-L1 expression by AML cells through its effect on microRNA species. We have identified a novel mechanism of tumorigenesis through the downregulation of DICER and the loss of microRNA species that regulate proteins with pro-oncogenic function such as PD-L1.
Supplementary Material
Acknowledgments
Research was supported by NIDDK P30DK046200. AJ, EA and FJS were supported by a grant from the Ludwig Center at Harvard. This study was funded, in part, by a Chief Academic Officer (CAO) Pilot Grant (2015) awarded to FS and DA.
Footnotes
CONFLICT OF INTEREST
DK holds equity in Genus Oncology and is a consultant to the company. The remaining authors declare no conflict of interest.
References
- 1.Teague RM, Kline J. Immune evasion in acute myeloid leukemia: current concepts and future directions. J Immunother Cancer. 2013;1 doi: 10.1186/2051-1426-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Dieu R, Taussig DC, Ramsay AG, Mitter R, Miraki-Moud F, Fatah R, et al. Peripheral blood T cells in acute myeloid leukemia (AML) patients at diagnosis have abnormal phenotype and genotype and form defective immune synapses with AML blasts. Blood. 2009;114:3909–3916. doi: 10.1182/blood-2009-02-206946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coles SJ, Gilmour MN, Reid R, Knapper S, Burnett AK, Man S, et al. The immunosuppressive ligands PD-L1 and CD200 are linked in AML T-cell immunosuppression: identification of a new immunotherapeutic synapse. Leukemia. 2015;29:1952–1954. doi: 10.1038/leu.2015.62. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Li J, Dong K, Lin F, Long M, Ouyang Y, et al. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal. 2015;27:443–452. doi: 10.1016/j.cellsig.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Berthon C, Driss V, Liu J, Kuranda K, Leleu X, Jouy N, et al. In acute myeloid leukemia, B7-H1 (PD-L1) protection of blasts from cytotoxic T cells is induced by TLR ligands and interferon-gamma and can be reversed using MEK inhibitors. Cancer Immunol Immunother. 2010;59:1839–1849. doi: 10.1007/s00262-010-0909-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L, Gajewski TF, Kline J. PD-1/PD-L1 interactions inhibit antitumor immune responses in a murine acute myeloid leukemia model. Blood. 2009;114:1545–1552. doi: 10.1182/blood-2009-03-206672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDermott DF, Sosman JA, Sznol M, Massard C, Gordon MS, Hamid O, et al. Atezolizumab, an anti-programmed death-ligand 1 antibody, in metastatic renal cell carcinoma: long-term safety, clinical activity, and immune correlates from a phase ia study. J Clin Oncol. 2016;34:833–842. doi: 10.1200/JCO.2015.63.7421. [DOI] [PubMed] [Google Scholar]
- 8.Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinumbased chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fehrenbacher L, Spira A, Ballinger M, Kowanetz M, Vansteenkiste J, Mazieres J, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet. 2016;387:1837–1846. doi: 10.1016/S0140-6736(16)00587-0. [DOI] [PubMed] [Google Scholar]
- 10.Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372:311–319. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi T, Makiguchi Y, Hinoda Y, Kakiuchi H, Nakagawa N, Imai K, et al. Expression of MUC1 on myeloma cells and induction of HLA-unrestricted CTL against MUC1 from a multiple myeloma patient. J Immunol. 1994;153:2102–2109. [PubMed] [Google Scholar]
- 12.Burton J, Mishina D, Cardillo T, Lew K, Rubin A, Goldenberg DM, et al. Epithelial mucin-1 (MUC1) expression and MA5 anti-MUC1 monoclonal antibody targeting in multiple myeloma. Clin Cancer Res. 1999;5:3065s–3072s. [PubMed] [Google Scholar]
- 13.Treon SP, Mollick JA, Urashima M, Teoh G, Chauhan D, Ogata A, et al. Muc-1 core protein is expressed on multiple myeloma cells and is induced by dexamethasone. Blood. 1999;93:1287–1298. [PubMed] [Google Scholar]
- 14.Paydaş S, Sahin B, Gönlüşen G, Hazar B, Zorludemir S. MUC1 expression in plasmacytoma. Leuk Res. 2001;25:221–225. doi: 10.1016/s0145-2126(00)00111-9. [DOI] [PubMed] [Google Scholar]
- 15.Cloosen S, Gratama J, van Leeuwen EBM, Senden-Gijsbers BLMG, Oving EBH, von Mensdorff-Pouilly S, et al. Cancer specific Mucin-1 glycoforms are expressed on multiple myeloma. Br J Haematol. 2006;135:513–516. doi: 10.1111/j.1365-2141.2006.06331.x. [DOI] [PubMed] [Google Scholar]
- 16.Baldus SE, Palmen C, Thiele J. MUC1 (EMA) expressing plasma cells in bone marrow infiltrated by plasma cell myeloma. Histol Histopathol. 2007;22:889–893. doi: 10.14670/HH-22.889. [DOI] [PubMed] [Google Scholar]
- 17.Tan Z, Huang Q, Zang J, Teng S-F, Chen T-R, Wei H, et al. HIF-1α activates hypoxiainduced BCL-9 expression in human colorectal cancer cells. Oncotarget. 2017;8:25885–25896. doi: 10.18632/oncotarget.8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouillez A, Rajabi H, Pitroda S, Jin C, Alam M, Kharbanda A, et al. Inhibition of MUC1-C suppresses MYC expression and attenuates malignant growth in KRAS mutant lung adenocarcinomas. Cancer Res. 2016;76:1538–1548. doi: 10.1158/0008-5472.CAN-15-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmad R, Raina D, Trivedi V, Ren J, Rajabi H, Kharbanda S, et al. MUC1 oncoprotein activates the IkappaB kinase beta complex and constitutive NF-kappaB signalling. Nat Cell Biol. 2007;9:1419–1427. doi: 10.1038/ncb1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmad R, Raina D, Joshi MD, Kawano T, Ren J, Kharbanda S, et al. MUC1-C oncoprotein functions as a direct activator of the nuclear factor-kappaB p65 transcription factor. Cancer Res. 2009;69:7013–7021. doi: 10.1158/0008-5472.CAN-09-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi H, Jin C, Rajabi H, Pitroda S, Alam M, Ahmad R, et al. MUC1-C activates the TAK1 inflammatory pathway in colon cancer. Oncogene. 2015;34:5187–5197. doi: 10.1038/onc.2014.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khodarev N, Ahmad R, Rajabi H, Pitroda S, Kufe T, McClary C, et al. Cooperativity of the MUC1 oncoprotein and STAT1 pathway in poor prognosis human breast cancer. Oncogene. 2010;29:920–929. doi: 10.1038/onc.2009.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmad R, Rajabi H, Kosugi M, Joshi MD, Alam M, Vasir B, et al. MUC1-C oncoprotein promotes STAT3 activation in an autoinductive regulatory loop. Sci Signal. 2011;4:ra9. doi: 10.1126/scisignal.2001426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin L, Ahmad R, Kosugi M, Kufe T, Vasir B, Avigan D, et al. Survival of human multiple myeloma cells is dependent on MUC1 C-terminal transmembrane subunit oncoprotein function. Mol Pharmacol. 2010;78:166–174. doi: 10.1124/mol.110.065011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stroopinsky D, Rosenblatt J, Ito K, Mills H, Yin L, Rajabi H, et al. MUC1 is a potential target for the treatment of acute myeloid leukemia stem cells. Cancer Res. 2013;73:5569–5579. doi: 10.1158/0008-5472.CAN-13-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin L, Wu Z, Avigan D, Rosenblatt J, Stone R, Kharbanda S, et al. MUC1-C oncoprotein suppresses reactive oxygen species-induced terminal differentiation of acute myelogenous leukemia cells. Blood. 2011;117:4863–4870. doi: 10.1182/blood-2010-10-296632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin L, Kosugi M, Kufe D. Inhibition of the MUC1-C oncoprotein induces multiple myeloma cell death by down-regulating TIGAR expression and depleting NADPH. Blood. 2012;119:810–816. doi: 10.1182/blood-2011-07-369686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn Y-H, Byers LA, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenblatt J, Wu Z, Vasir B, Zarwan C, Stone R, Mills H, et al. Generation of tumorspecific T lymphocytes using dendritic cell/tumor fusions and anti-CD3/CD28. J Immunother. 2010;33:155–166. doi: 10.1097/CJI.0b013e3181bed253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panchamoorthy G, Rehan H, Kharbanda A, Ahmad R, Kufe D. A monoclonal antibody against the oncogenic mucin 1 cytoplasmic domain. Hybrid. 2011;30:531–535. doi: 10.1089/hyb.2011.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang S, Tsai LT, Zhou Y, Evertts A, Xu S, Griffin MJ, et al. Identification of nuclear hormone receptor pathways causing insulin resistance by transcriptional and epigenomic analysis. Nat Cell Biol. 2015;17:44–56. doi: 10.1038/ncb3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.UCSC. UCSC Genome Browser. Available at: http://genome.ucsc.edu/
- 34.Halazonetis TD, Georgopoulos K, Greenberg ME, Leder P. c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell. 1988;55:917–924. doi: 10.1016/0092-8674(88)90147-x. [DOI] [PubMed] [Google Scholar]
- 35.Kimchi ET, Posner MC, Park JO, Darga TE, Kocherginsky M, Karrison T, et al. Progression of Barrett’s metaplasia to adenocarcinoma is associated with the suppression of the transcriptional programs of epidermal differentiation. Cancer Res. 2005;65:3146–3154. doi: 10.1158/0008-5472.CAN-04-2490. [DOI] [PubMed] [Google Scholar]
- 36.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 37.Gazon H, Belrose G, Terol M, Meniane J-C, Mesnard J-M, Césaire R, et al. Impaired expression of DICER and some microRNAs in HBZ expressing cells from acute adult T-cell leukemia patients. Oncotarget. 2016;7:30258–30275. doi: 10.18632/oncotarget.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selever J, Gu G, Lewis MT, Beyer A, Herynk MH, Covington KR, et al. Dicer-mediated upregulation of BCRP confers tamoxifen resistance in human breast cancer cells. Clin Cancer Res. 2011;17:6510–6521. doi: 10.1158/1078-0432.CCR-11-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leppä S, Saffrich R, Ansorge W, Bohmann D. Differential regulation of c-Jun by ERK and JNK during PC12 cell differentiation. EMBO J. 1998;17:4404–4413. doi: 10.1093/emboj/17.15.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng Z, Sui G, Rosa PM, Zhao W. Radiation-induced c-Jun activation depends on MEK1-ERK1/2 signaling pathway in microglial cells. PLoS One. 2012;7:e36739. doi: 10.1371/journal.pone.0036739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang W, Han D, Wan P, Pan P, Cao Y, Liu Y, et al. ERK/c-Jun recruits Tet1 to induce Zta expression and Epstein-Barr virus reactivation through DNA demethylation. Sci Rep. 2016;6:34543. doi: 10.1038/srep34543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herdegen T, Claret FX, Kallunki T, Martin-Villalba A, Winter C, Hunter T, et al. Lasting N-terminal phosphorylation of c-Jun and activation of c-Jun N-terminal kinases after neuronal injury. J Neurosci. 1998;18:5124–5135. doi: 10.1523/JNEUROSCI.18-14-05124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yarza R, Vela S, Solas M, Ramirez MJ. c-Jun N-terminal kinase (JNK) Signaling as a therapeutic target for Alzheimer’s disease. Front Pharmacol. 2016;6:321. doi: 10.3389/fphar.2015.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenblatt J, Stone R, Uhl L, Neuberg D, Vasir B, Somaiya P, et al. ASH Annual Meeting. New Orleans Convention Center; New Orleans, LI, USA: American Society of Hematology; 2013. Clinical trial evaluating DC/AML fusion cell vaccination in AML patients who achieve a chemotherapy-induced remission, Vol 122; p. 3928. [Google Scholar]
- 45.Rosenblatt J, Glotzbecker B, Mills H, Vasir B, Tzachanis D, Levine JD, et al. PD-1 blockade by CT-011, anti-PD-1 antibody, enhances ex vivo T-cell responses to autologous dendritic cell/myeloma fusion vaccine. J Immunother. 2011;34:409–418. doi: 10.1097/CJI.0b013e31821ca6ce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noman MZ, Desantis G, Janji B, Hasmim M, Karray S, Dessen P, et al. PD-L1 is a novel direct target of HIF-1α, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J Exp Med. 2014;211:781–790. doi: 10.1084/jem.20131916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Christiansson L, Söderlund S, Svensson E, Mustjoki S, Bengtsson M, Simonsson B, et al. Increased level of myeloid-derived suppressor cells, programmed death receptor ligand 1/programmed death receptor 1, and soluble CD25 in sokal high risk chronic myeloid leukemia. PLoS One. 2013;8:e55818. doi: 10.1371/journal.pone.0055818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Napolitano M, D’Alterio C, Cardone E, Trotta AM, Pecori B, Rega D, et al. Peripheral myeloid-derived suppressor and T regulatory PD-1 positive cells predict response to neoadjuvant short-course radiotherapy in rectal cancer patients. Oncotarget. 2015;6:8261–8270. doi: 10.18632/oncotarget.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kufe DW. Functional targeting of the MUC1 oncogene in human cancers. Cancer Biol Ther. 2009;8:1197–1203. doi: 10.4161/cbt.8.13.8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tagde A, Rajabi H, Bouillez A, Alam M, Gali R, Bailey S, et al. MUC1-C drives MYC in multiple myeloma. Blood. 2016;127:2587–2597. doi: 10.1182/blood-2015-07-659151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu S, Yin L, Stroopinsky D, Rajabi H, Puissant A, Stegmaier K, et al. MUC1-C oncoprotein promotes FLT3 receptor activation in acute myeloid leukemia cells. Blood. 2014;123:734–742. doi: 10.1182/blood-2013-04-493858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.David JM, Hamilton DH, Palena C. MUC1 upregulation promotes immune resistance in tumor cells undergoing brachyury-mediated epithelial-mesenchymal transition. Oncoimmunology. 2016;5:e1117738. doi: 10.1080/2162402X.2015.1117738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bar-Natan M, Stroopinsky D, Luptakova K, Coll MD, Apel A, Rajabi H, et al. Bone marrow stroma protects myeloma cells from cytotoxic damage via induction of the oncoprotein MUC1. Br J Haematol. 2017;176:929–938. doi: 10.1111/bjh.14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu Z, Li Y, Fan H, Liu Z, Pestell RG. miRNAs regulate stem cell self-renewal and differentiation. Front Genet. 2012;3:191. doi: 10.3389/fgene.2012.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim S, Song JH, Kim S, Qu P, Martin BK, Sehareen WS, et al. Loss of oncogenic miR-155 in tumor cells promotes tumor growth by enhancing C/EBP-β-mediated MDSC infiltration. Oncotarget. 2016;7:11094–11112. doi: 10.18632/oncotarget.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krill KT, Gurdziel K, Heaton JH, Simon DP, Hammer GD. Dicer deficiency reveals microRNAs predicted to control gene expression in the developing adrenal cortex. Mol Endocrinol. 2013;27:754–768. doi: 10.1210/me.2012-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Connerty P, Ahadi A, Hutvagner G. RNA binding proteins in the miRNA pathway. Int J Mol Sci. 2015;17:E31. doi: 10.3390/ijms17010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alam M, Ahmad R, Rajabi H, Kharbanda A, Kufe D. MUC1-C oncoprotein activates ERK→C/EBPβ signaling and induction of aldehyde dehydrogenase 1A1 in breast cancer cells. J Biol Chem. 2013;288:30892–30903. doi: 10.1074/jbc.M113.477158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J, Liu G, Li Q, Wang F, Xie F, Zhai R, et al. Mucin1 promotes the migration and invasion of hepatocellular carcinoma cells via JNK-mediated phosphorylation of Smad2 at the C-terminal and linker regions. Oncotarget. 2015;6:19264–19278. doi: 10.18632/oncotarget.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Q, Liu G, Yuan H, Wang J, Guo Y, Chen T, et al. Mucin1 shifts Smad3 signaling from the tumor-suppressive pSmad3C/p21<sup>WAF1<sup> pathway to the oncogenic pSmad3L/c-Myc pathway by activating JNK in human hepatocellular carcinoma cells. Oncotarget. 2015;6:4253–4265. doi: 10.18632/oncotarget.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khoshnaw SM, Rakha EA, Abdel-Fatah TM, Nolan CC, Hodi Z, Macmillan DR, et al. Loss of Dicer expression is associated with breast cancer progression and recurrence. Breast Cancer Res Treat. 2012;135:403–413. doi: 10.1007/s10549-012-2169-3. [DOI] [PubMed] [Google Scholar]
- 64.Swahari V, Nakamura A, Deshmukh M. The paradox of dicer in cancer. Mol Cell Oncol. 2016;3:e1155006. doi: 10.1080/23723556.2016.1155006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 66.Pyzer AR, Stroopinsky D, Rajabi H, Washington A, Tagde A, Coll M, et al. MUC1 mediated induction of myeloid-derived suppressor cells in patients with acute myeloid leukemia. Blood. 2017;129:1791–1801. doi: 10.1182/blood-2016-07-730614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Freeman-Keller M, Weber JS. Anti-programmed death receptor 1 immunotherapy in melanoma: rationale, evidence and clinical potential. Ther Adv Med Oncol. 2015;7:12–21. doi: 10.1177/1758834014551747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li XJ, Ren ZJ, Tang JH. MicroRNA-34a: a potential therapeutic target in human cancer. Cell Death Dis. 2014;5:e1327. doi: 10.1038/cddis.2014.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.