TO THE EDITOR
The mouse hair follicle (HF) is an excellent model system for studying stem cell lineages and their behaviors. During homeostasis, HF undergoes cyclic rounds of active growth (anagen), apoptosis-driven regression (catagen), and rest (telogen) (Cotsarelis, 2006). The mouse anagen follicle is a complex structure comprising multiple sheaths of epithelial cell layers encapsulating the hair shaft: the outer root sheath (ORS), the companion layer, the inner root sheath, and the hair shaft (Muller-Rover et al., 2001) (Figure 1b). The HF stem cells are localized in an upper permanent region (bulge) in the ORS. They are the source of the hair germ, as early progenitor cells that make the transit-amplifying progenitor cells called the matrix, which in turn produce terminally differentiated post-mitotic cells in the inner layers within the mature HFs during anagen. In catagen, the lower hair bulb regresses, and the hair germ and bulge region of the HF remain in telogen. The telogen bulge is composed of two cell layers: (i) a keratin (K) 14+ ORS housing the stem cells (SCs), and (ii) a K6+ inner layer (IL) of terminally differentiated cells. The telogen bulge K6+ IL cells (TBIL) can act as epithelial niche cells that maintain HF stem cell quiescence by sending growth inhibitory signals to the adjacent SCs (Hsu et al., 2011). These cells have also been shown to be epithelial niche to support tumor growth. However, because there are no existing mouse Cre lines to exclusively label the TBIL niche cells, specific isolation or gene targeting of these cells to understand the crosstalk between the SCs and their underlining epithelial niche cells has been challenging. This crosstalk can be relevant to understanding how the SC environment can be manipulated to maintain the hair differentiation ability and long-term potential of epithelial SCs for regenerative therapies. Recently we reported the activity of previously generated Slc1a3-CreER transgene (Nathans, 2010) in the mouse interfollicular epidermis in a subpopulation of more actively dividing cells (Sada et al., 2016). Here we show that Slc1a3-CreER is also induced in the HF during anagen, and report the resulting labeling patterns at different hair cycle stages of TBIL, to further examine its role as maintenance niche and its crosstalk with the HF stem cells.
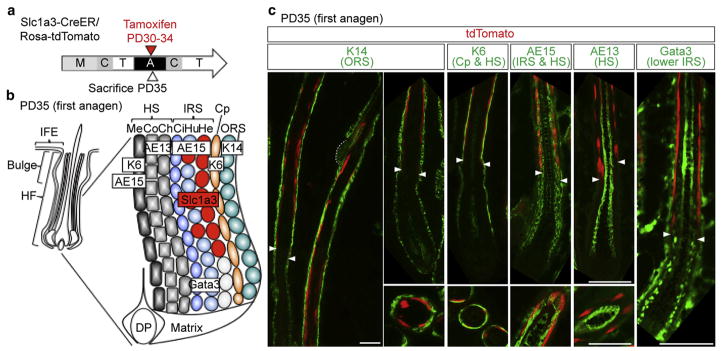
Figure 1. Slc1a3-CreER specifically marks the inner root sheath in the anagen follicle.
(a) Scheme for lineage tracing of Slc1a3-CreER marked cells. (b) Structure of the anagen hair follicle. Appropriate markers for each layer are shown in boxed text. (c) Immunostaining with lineage markers indicated in (b). Upper panels show sagittal sections and lower panels show cross sections of the hair follicle. The dashed line indicates the bulge.
Arrowheads represent the boundary between the upper and lower IRS. Scale bars = 50 μm. A, anagen; C, catagen; Ch, cuticle of hair shaft; Ci, cuticle of IRS; Co, cortex of hair shaft; Cp, companion layer; DP, dermal papilla; He, Henle’s layer; HF, hair follicle; HS, hair shaft; Hu, Huxley’s layer; IFE, inter-follicular epidermis; IRS, inner root sheath; M, morphogenesis; Me, medulla; ORS, outer root sheath; PD, postnatal day; T, telogen.
Slc1a3-CreER/Rosa-tdTomato double transgenic mice were injected tamoxifen 1×/day at postnatal day (PD) 30–34 (anagen) to obtain maximum labeling efficiency (Figure 1a). We observed tdTomato marking in the interfollicular epidermis, which we characterize elsewhere (Sada et al., 2016), and within the inner lineages of the HF at PD35. Immunostaining of skin sections with known HF differentiation markers (Figure 1b) revealed colocalization of tdTomato+ cells with cells in AE15+ cells in Henle’s and Huxley’s layers of the inner root sheath, in the upper region of the HF not marked by Gata3, another inner root sheath marker (Kaufman et al., 2003), which is confined to the lower follicle (Figure 1c, Supplementary Figures S1 and S2a, d online). Unlike previously reported CreER lines (Hsu et al., 2011; Takeda et al., 2013), Slc1a3-CreER did not label any other epithelial compartments within anagen HFs, including bulge, matrix, the ORS, the companion layer, or the hair shaft (Figure 1c, Supplementary Figures S1 and S2a, d). Thus, we identified Slc1a3-CreER as a marking tool confined to a subset of upper inner root sheath cells within the anagen HF.
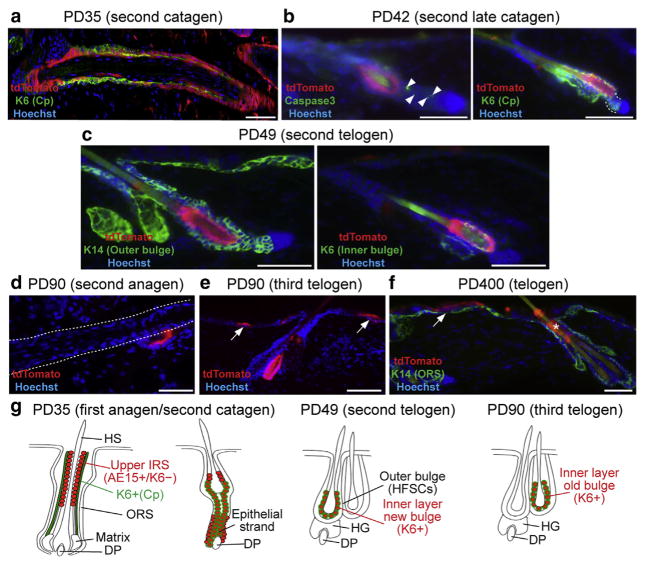
At PD35 some regions of back skin displayed HFs with catagen morphology, which showed a distinct pattern of the Slc1a3-CreER marked cells (Figure 2a, Supplementary Figure S3a online). In the upper part of the catagen follicles, tdTomato+ cells were inner to K6+ companion layer cells, similar to anagen. In contrast, in the lower part of the HF, tdTomato+ cells were colocalized with K6 as well as K14 immunostaining (Supplementary Figures S2a and S3a). By PD42, HFs were in late catagen (Figure 2b) when multiple Caspase3+ cells, indicative of apoptosis, were detected in the lower portion of the HF as expected (Mesa et al., 2015); tdTomato+ cells were not present in the epithelial strand and were visible in the middle portion of the follicle surrounding the new shaft and being positive for K6 (Figure 2b).
Figure 2. Slc1a3-CreER marked cells are a source for the telogen bulge K6+ inner layer.
(a–f) Images of immunostained lineage traced skin tamoxifen injected as shown in Figure 1 at ages and stages indicated. Confocal images of Z-stacks are shown in Supplementary Figure S3a–d. Arrowheads in (b) represent cell death. The dashed line in (b) represents the lower part of the follicle, where tdTomato+ cells were not colocalized with K6. The dashed line in (d) bounds the anagen follicle. Note localization of tdTomato+ cells in the old bulge in this image. The asterisk in (f) shows remnants of tdTomato+ cells shed through the infundibulum. Arrows represent tdTomato+ clones in the interfollicular epidermis. Scale bars = 50 μm. (g) Summary cartoon with tdTomato in red and K6 in green. Abbreviations are the same as in the Figure 1 legend.
This K6 expression remained in tdTomato+ cells at PD49, the subsequent telogen, when importantly these cells filled the majority (approximately 90%) of the TBIL (Figure 2c, Supplementary Figures S2b, c, e and S3b, c, d). No tdTomato signal was detected in tamoxifen negative controls in the short and long term after tamoxifen induction (Sada et al., 2016; Supplementary Figure S3e). During the next hair cycle, the tdTomato+ TBIL did not contribute to differentiated hair lineages, as expected of terminally differentiated cells (Hsu et al., 2011; Takeda et al., 2013) (Figure 2d), and remained as the IL in the old bulge by the third telogen (Figure 2e). tdTomato+ cells were still present in the interfollicular epidermis as reported (Sada et al., 2016) but in the HF were lost after long-term chase, with some remnants detected in the infundibulum (Figure 2f). These data demonstrate that the cells marked by the Slc1a3-CreER induction at PD30–34 give rise to the majority of the K6+ TBIL (Figure 2g). The cellular source of the TBIL may include directly differentiating SCs in the ORS (Hsu et al., 2011), or cells located within a restricted region at the base of the bulb (Takeda et al., 2013). Our study suggests that Slc1a3-CreER can be a useful tool to specifically label, isolate, and gene target different HF compartments during the hair cycle. In particular, the TBIL niche cells can be efficiently and highly specifically labeled, which will facilitate a more in-depth study of their cellular and molecular function in physiological and pathological conditions such as tumors.
MATERIALS AND METHODS
Mice
All mouse experiments were carried out according to Cornell University Institutional Animal Care and Use Committee guidelines. For lineage tracing, Slc1a3-CreER (Nathans, 2010) (The Jackson Laboratory, #012586) mice were crossed with Rosa-tdTomato (Madisen et al., 2010) (The Jackson Laboratory, #007905) reporter mice. Mice were maintained in the C57BL/6 background. Double transgenic mice were injected with tamoxifen (100 μg/g body weight) once a day for 5 consecutive days at PD30–34. Mice are killed at indicated time points.
Immunostaining of skin sections
Mouse dorsal skin was directly embedded in OCT compound (Tissue Tek; Sakura, Tokyo, Japan). The frozen sections (10 μm) were fixed with 4% paraformaldehyde for 10 minutes at room temperature. After blocking in normal serum, the sections were incubated overnight with primary antibodies at 4 °C. The following day, the sections were washed and incubated for 1 hour with secondary antibodies at room temperature. After washing, these sections were counterstained with Hoechst and mounted.
Primary antibodies were used at the following dilutions: rabbit anti-K14 (1:1,000, Covance), anti-K6 (1:500, Abcam), mouse AE15 (1:10, a gift from Dr Tung-Tien Sun, NYU School of Medicine, NY), mouse anti-Gata3 (1:100, Santa Cruz, HG3-31), mouse AE13 (1:50, Immunoquest), and rabbit anti-Caspase3 (1:300, R&D Systems). All secondary antibodies (FITC conjugated, Jackson ImmunoResearch) were used at a 1:500 dilution. For mouse primary antibodies, the MOM kit (Vector Laboratories) was used for blocking. Preparations were examined using a fluorescent microscope (Nikon) with ×10 and ×20 objectives and digitally imaged by a CCD 12-bit digital camera (Retiga EXi; QImaging) and IP-Lab software (MVI). The images were processed with Photoshop (Adobe) to adjust the contrast and brightness. For confocal imaging, 14-μm frozen skin sections were used. Immunostaining was performed as described above, images were captured using an LSM710 confocal microscope (Carl Zeiss) with ×40 and ×60 objectives and Z-stacks of 0.45 to 1 μm were collected. The images were analyzed and quantified using Zen black software (Carl Zeiss).
Data reproducibility
All experiments were independently performed in three different mice and the representative data are shown. For each experiment, more than 50 follicles were analyzed from both upper and lower part of back skin.
Supplementary Material
Acknowledgments
We thank Dr T. T. Sun for the AE15 antibody and the Cornell Center for Animal Resources Education facility for mouse care. We also thank Flora Eun for her help in immunostaining. Funding for this work was received from NYS-TEM Grant C024354, NIH Grant R21AR063278 to TT; a long-term fellowship from Human Frontier Science Program (HFSP) and a postdoctoral fellowship for research abroad from Japan Society for the Promotion of Science (JSPS) to AS; and a NYSTEM postdoctoral fellowship to PJ.
Abbreviations
- HF
hair follicle
- K
keratin
- IL
inner layer
- ORS
outer root sheath
- PD
postnatal day
- SC
stem cell
- TBIL
telogen bulge inner layer
Footnotes
Supplementary material is linked to the online version of the paper at www.jidonline.org, and at http://dx.doi.org/10.1016/j.jid.2017.02.974.
CONFLICT OF INTEREST
The authors state no conflict of interest.
References
- Cotsarelis G. Epithelial stem cells: a folliculocentric view. J Invest Dermatol. 2006;126:1459–68. doi: 10.1038/sj.jid.5700376. [DOI] [PubMed] [Google Scholar]
- Hsu YC, Pasolli HA, Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell. 2011;144:92–105. doi: 10.1016/j.cell.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman CK, Zhou P, Pasolli HA, Rendl M, Bolotin D, Lim KC, et al. GATA-3: an unexpected regulator of cell lineage determination in skin. Genes Dev. 2003;17:2108–22. doi: 10.1101/gad.1115203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–40. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa KR, Rompolas P, Zito G, Myung P, Sun TY, Brown S, et al. Niche-induced cell death and epithelial phagocytosis regulate hair follicle stem cell pool. Nature. 2015;522:94–7. doi: 10.1038/nature14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Rover S, Handjiski B, van der Veen C, Eichmuller S, Foitzik K, McKay IA, et al. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J Invest Dermatol. 2001;117:3–15. doi: 10.1046/j.0022-202x.2001.01377.x. [DOI] [PubMed] [Google Scholar]
- Nathans J. Generation of an inducible Slc1a3-cre/ ERT transgenic allele. MGI Direct Data Submission: [MGI Ref ID J:157151] 2010 [Google Scholar]
- Sada A, Jacob F, Leung E, Wang S, White BS, Shalloway D, et al. Defining the cellular lineage hierarchy in the interfollicular epidermis of adult skin. Nat Cell Biol. 2016;18:619–31. doi: 10.1038/ncb3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N, Jain R, Leboeuf MR, Padmanabhan A, Wang Q, Li L, et al. Hopx expression defines a subset of multipotent hair follicle stem cells and a progenitor population primed to give rise to K6+ niche cells. Development. 2013;140:1655–64. doi: 10.1242/dev.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.